The enteric nervous system consists of more than a dozen types of neurons aggregated into networks of ganglia throughout the gastrointestinal tract, which regulate contractile activity, mucosal secretion, absorption, and local blood flow.1, 2 Mechanisms that contribute to remodeling of the enteric neuronal networks are of great interest. In the central nervous system, it has been suggested that microglia contribute to the fate, connectivity, and identity of neurons during development.3 Muscularis propria macrophages (MPM) within the enteric nervous system may have similar functions to microglia. Mice homozygous for the osteopetrosis mutation (Csf1op/op) which do not have MPM, have more neurons in the small intestine4 and a higher proportion of gastric neurons that express nitric oxide synthase (NOS1).5 Myenteric neurons serve diverse functions that can be indicated by their morphology, projections and the expression of marker proteins that define their “chemical code.” This study finds a previously unidentified role for MPM in altering the chemical code of myenteric neurons.
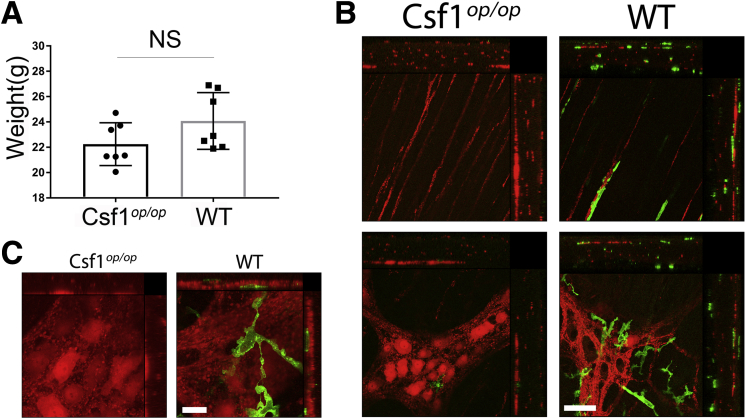
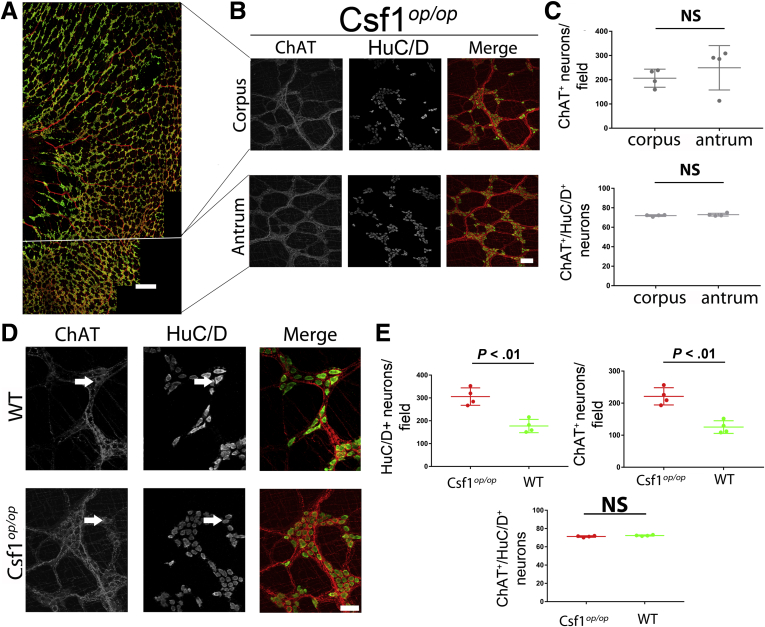
Csf1op/op mice were maintained on a specialized liquid diet to keep their weight comparable with age-matched-wild type (WT) mice (Supplementary Figure 1A). In the myenteric plexus of WT mice, populations of MPM, absent in Csf1op/op mice5 (Supplementary Figure 1B and C, Supplementary Movie 1 and 2), were associated closely with neurons, suggesting functional interactions.6 We first tested whether the number of choline acetyltransferase+ (ChAT+) neurons was affected by the absence of MPM in Csf1op/op mice (Supplementary Table 1). The density of neurons, defined by Embryonic lethal, abnormal vision, Drosophila-like protein 3/4 (HuC/D) immunoreactivity, was similar between gastric regions in both WT and Csf1op/op mice (Figure 1A–C, Supplementary Figure 2A) (Mann–Whitney test, P = NS; N = 4), yet was higher in Csf1op/op mice than in WT mice (Figure 1D and E) (P < .01, Mann–Whitney test, n = 36 fields, N = 4). Likewise, the density of ChAT+ neurons was higher in Csf1op/op mice compared with WT mice (Figure 1D and E) (P < .001, Mann–Whitney test, n = 36 fields, N = 4). However, in contrast to an increase in the percentage of NOS1+ neurons,5 the percentage of ChAT+ neurons did not differ between Csf1op/op and WT mice (Figure 1D and E) (Mann–Whitney test, n = 36 fields, N = 4). This result suggests that the presence of macrophages alters the proportion of nitrergic but not cholinergic gastric myenteric neurons.
Supplementary Figure 1.
(A) Weight of WT and CSf1op/op mice (P = NS; Mann–Whitney test; N = 7 mice for each group). (B and C) Major histocompatibility complex class II (MHCII) macrophages (green) and Protein gene product 9.5 (PGP 9.5) fibers in smooth muscular layers (upper panels) and myenteric plexus (lower panels). The small panels show orthogonal views generated by projecting the z-series in the x (right) and on the y plane (above). Arrows point to macrophage/fiber interactions and squares show macrophage/fiber interactions in orthogonal views. PGP 9.5 immunoreactivity was unusually bright in the cell bodies of myenteric neurons in CSf1op/op mice when compared with WT tissues. Scale bars: (B) 20 μm, (C) 10 μm.
Figure 1.
(A) Distribution of HuC/D+ and ChAT+ myenteric neurons across gastric regions.Scale bar: 200 μm. (B) Images of HuC/D+ and ChAT+ neurons in the gastric regions of Csf1op/op mice. Scale bar: 50 μm. (C) Quantification of HuC/D+ and ChAT+ neurons in the gastric regions of Csf1op/op mice (Mann–Whitney test; P = NS). (D) Images of gastric HuC/D+ and ChAT+ neurons in WT and Csf1op/op mice. Scale bar: 60 μm. Arrow indicates typical HuC/D+ and ChAT+ co-expressing neurons. (E) Quantification of HuC/D+ and ChAT+ neurons in WT and Csf1op/op mice (n = 36 fields; N = 4 mice) (Mann–Whitney test; P < .01). (C and E) Bars and whiskers indicate means ± SD and points indicate individual fields for all panels.
Supplementary Figure 2.
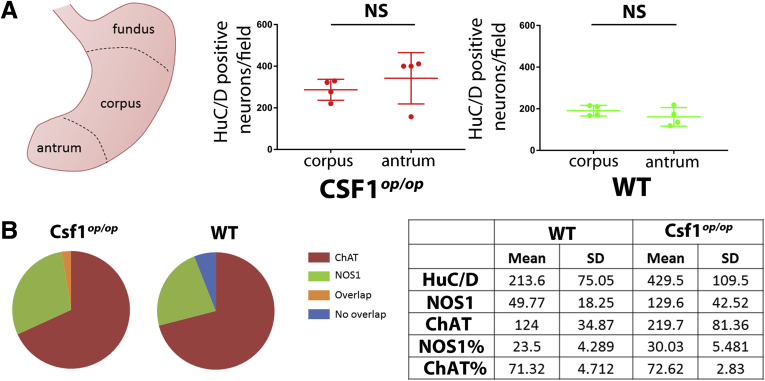
(A) Quantification of the HuC/D+ myenteric neurons in the gastric corpus and antrum of WT and Csf1op/op mice. (B) Percentage of myenteric neurons identified in Csf1op/op and WT mice. Table shows numbers per field and proportions of different types of myenteric neurons in Csf1op/op and WT mice.
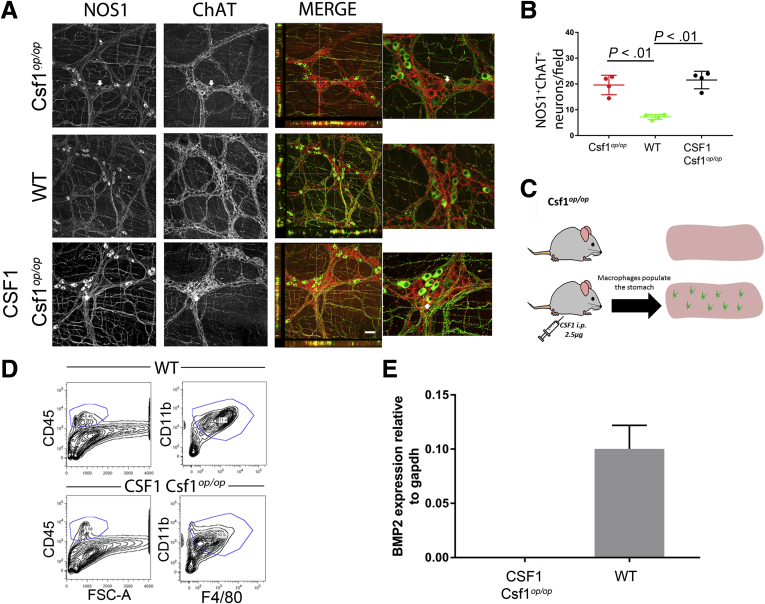
Interestingly, in Csf1op/op mice, the combined percentages of NOS1+ (30%) and ChAT+ neurons (72%) exceeded 100% (Supplementary Figure 2B), indicating partial overlap between these markers. Therefore, we investigated whether the number of NOS1+ChAT+ double-labeled neurons was changed in Csf1op/op mice. In Csf1op/op mice, Nitric Oxide Synthase 1 (NOS1+) ChAT+ neurons were more numerous than in WT mice (Figure 2A and B) (Csf1op/op: 7.8 ± 7.1 cells/field; WT, 1.7 ± 1.6 cells/field; 1-way analysis of variance; P < .001; n = 24; N = 4). This result suggests the ability of macrophages to not only modulate the neuronal number but also affect myenteric neuron differentiation. Enteric neurons are not required for bowel colonization by macrophages,7 but macrophages interact with neurons after birth, by expressing genes, such as bone morphogenetic protein 2 (BMP2), needed for macrophage-enteric neuron interaction and neuronal development.4 To test the intrinsic ability of resident macrophages to modify the neuronal chemical code by establishing functional interaction with neurons, we treated Csf1op/op with CSF1 (Colony Stimulating Factor 1) for 7 weeks to populate the stomach with macrophages (Figure 2C). In CSF1-treated Csf1op/op mice, the proportion of NOS1+ChAT+ neurons remained similar to the proportion of NOS1+ChAT+ neurons in Csf1op/op mice (Figure 2A–C) (1-way analysis of variance; n = 24; N = 4). We previously showed that repopulating macrophages in CSF1-treated Csf1op/op mice had a different phenotype from resident macrophages.5 Consistent with this observation, BMP2 was not expressed by macrophages isolated from CSF1-treated Csf1op/op mice (Antibodies and PCR primers listed in Supplementary Tables 2 and 3), whereas BMP2 was expressed by macrophages isolated from WT mice (Figure 2D and E) (Mann–Whitney test; P < .001; N = 4), as reported elsewhere.4
Figure 2.
(A)Images of NOS1+/ChAT+ neurons.Scale bar: 50 μm. Arrows show NOS1+ neurons that are also ChAT+. (B) Quantification of NOS1+ChAT+ double-labeled neurons. Points represent individual fields of view. Bars and whiskers indicate means ± SD (1-way analysis of variance; P < .01; N = 4). (C) Experimental model for CSF1 treatment. Fluorescence-activated cell sorter (FACS) strategy to isolate CD45+CD11b+F4/80+ macrophages from the gastric muscularis propria of WT (top) and CSF1-treated Csf1op/op mice (bottom). (E) BMP2 expression levels in macrophages isolated from Csf1op/op and WT mice (Mann–Whitney test; N = 3; P < .01).
During development, the chemical code of myenteric neurons changes and the overlap between NOS1 and ChAT decreases as neurons mature.8 Therefore, increased numbers of double-labeled myenteric neurons may reflect incomplete maturation of myenteric neurons in Csf1op/op mice. MPMs functionally interact with enteric neurons starting at 2 weeks of age,7 therefore the role of resident MPM in promoting myenteric neuron maturation likely happens early in life. Interestingly, MPMs that populate the gastric muscularis propria did not express BMP2, a cytokine important for establishing functional interactions between MPMs and neurons during development. Therefore, as previously suggested,4, 9 BMP2 may be required for the changes in NOS1 and ChAT expression associated with neuronal maturation.
Taken together, our results show a role for MPM in enteric neuronal maturation as indicated by the changes in chemical code in gastric myenteric neurons. The mechanisms by which MPM regulate neuronal numbers and chemical codes needs further investigation because it may be significant to the development or plasticity of the adult enteric nervous system and normal gastric function.
Acknowledgments
The authors thank Mrs Kristy Zodrow for her excellent assistance with this work; the Mayo Microscopy and Cell Analysis Core for assistance with the flow cytometry experiment; and Dr Vanda Lennon (Mayo Clinic) for supplying the HuC/D antibody used for the immunohistochemistry study.
Footnotes
Author contributions G. Cipriani was responsible for the study concept and design, acquisition, analysis, and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, and statistical analysis; M. L. Terhaar was responsible for the analysis and interpretation of the data; S. T. Eisenman was responsible for the analysis and interpretation of the data; D. R. Linden was responsible for critical revision of the manuscript for important intellectual content; A.M. Wright was responsible for the acquisition, analysis, and interpretation of the data; S. Ji was responsible for the acquisition, analysis, and interpretation of data; L. Sha was responsible for critical revision of the manuscript for important intellectual content; T. Ordog was responsible for critical revision of the manuscript for important intellectual content; J. H. Szurszewski was responsible for critical revision of the manuscript for important intellectual content; S. J. Gibbons was responsible for the study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, statistical analysis, administrative, technical, or material support, and study supervision; and G. Farrugia was responsible for the study concept and design, analysis and interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, statistical analysis, obtained funding, administrative, technical, or material support, and study supervision.
Conflicts of interest The authors disclose no conflicts.
Funding This work was supported by National Institutes of Health grants P01 DK 68055 and P30DK084567 (Mayo Clinic Center for Cell Signaling in Gastroenterology), and American Gastroenterological Association Rome Foundation award 36.
Contributor Information
Simon J. Gibbons, Email: gibbons.simon@mayo.edu.
Gianrico Farrugia, Email: Farrugia.gianrico@mayo.edu.
Supplementary Materials and Methods
Animals
These studies were approved by the Mayo Clinic Institutional Animal Care and Use Committee. Mice were humanely killed by carbon dioxide exposure followed by cervical dislocation. Mice homozygous for the Csf1op mutation and WT littermates were studied. These mice were bred in-house from a Csf1op/+ colony of hemizygous breeders with founders originating from The Jackson Laboratory (Bar Harbor, ME). Wild-type Csf1+/+ mice were identified by genotyping as previously described.1 Csf1op/op mice were maintained on a specialized wet diet (Bio-serv, Frenchtown, NJ) after weaning at 3–4 weeks of age to keep their weight comparable with age-matched WT mice (Supplementary Figure 1A). After 12 weeks of age, Csf1op/op mice were treated with CSF1 (2.5 μg intraperitoneally once daily, recombinant mouse macrophage colony stimulating factor-1 (rmM-CSF); Peprotech, Rocky Hill, NJ) (Figure 2A).
Immunolabeling
The mucosa was removed and muscularis propria was fixed with 4% paraformaldehyde in 0.1 mol/L phosphate buffer for 4 hours. Then, whole mounts were rinsed in 0.1 mol/L phosphate-buffered saline and blocked in the presence of 10% normal donkey serum in phosphate-buffered saline and 0.3% Triton X-100 (Thermo Fisher, Waltham, MA) overnight at 4°C and gastric muscularis propria was labeled with primary antibodies overnight at 4°C. After washing, the tissue was incubated with secondary antibodies (Jackson ImmunoResearch, West Grove, PA), washed, and then incubated with 4',6-diamidino-2-phenylindole dilactate (Invitrogen, Carlsbad, CA) for 30 minutes. Neurons were identified by HuC/D-immunoreactivity (ANNA1, a gift from Dr Vanda Lennon, Mayo Clinic, Rochester, MN), cholinergic neurons using a goat anti-ChAT antibody (EMD Millipore, Burlington, MA), and nitrergic neurons using a rabbit anti-NOS1 antibody (EMD Millipore). Muscularis macrophages were identified using the MHCII primary antibody (eBioscience, Waltham, MA).
Controls omitting the primary antibody and controls in double-labeling experiments that used the wrong secondary antibody were performed for all experiments. For quantification, 3 different fields were taken from the corpus and 3 from the antrum. The list of antibodies is shown in Supplementary Table 1.
Confocal Microscopy
A laser scanning confocal microscope using a 20×, numerical aperture, (NA), 0.95 XLUMPlanFl objective (Olympus, Tokyo, Japan) in Fluoview (Olympus), with the optimal confocal aperture to provide a resolution of 0.994 × 0.994 × 1.13 μm (X × Y × Z), was used. Stacks of confocal images of the entire muscularis propria were collected from 4 different mice (n = 4). For quantification of the labeling, all of the confocal image stacks were flattened into projections using the FV10-ASW Viewer (Olympus). The flattened images were renumbered in random order and the enteric neuronal number was determined while blinded to the source. All cells were counted from fields with dimensions of 636 × 636 μm.
Images used for reconstruction and orthogonal view were taken from a Zeiss LSM 780 microscope using either a 40× 1.2 NA water immersion objective at a resolution of 0.415 × 0.415 × 0.444, or a 100 × 1.4 NA oil immersion objective at a resolution of 0.133 × 0.133 × 0.373 μm per pixel. Images were analyzed using Imaris-Microscopy Image Software by Bitplane (Supplementary Figure 1A).
Isolation and Analysis of Gastric Muscularis Macrophages
Cell sorting was performed using a fluorescence activated cell sorting Aria Cell Sorter cytometer running fluorescence activated cell sorting Diva 6 software (Becton Dickinson, San Jose, CA), located in the Mayo Clinic Flow Cytometry Core Facility. Aliquots of cells were either unstained or stained with individual fluorescently labeled antibodies (Zurich, Switzerland, Supplementary Table 2) to establish instrument voltages, compensation, and appropriate gates. Each positive control tube was initially run without storing the data to ensure that the positive signals were on scale. Data were analyzed using FlowJo X software (Tree Star, Inc, Ashland, OR).
Gastric CD45+CD11b+F4/80+ cells were isolated directly into the lysing buffer provided by the RNeasy micro plus kit (Qiagen, Hilden, Germany). The extraction was performed following the instructions provided and the RNA concentration was determined by using a NanoDrop spectrophotometer. The RNA extracted was used for a real-time quantitative reverse-transcription polymerase chain reaction. The SuperScript VILO complementary DNA Synthesis Kit (Invitrogen) was used to generate complementary DNA. Quantitative reverse-transcription polymerase chain reaction was performed on complementary DNA using commercial primer sets (Supplementary Table 3) and RT2SYBR Green/ROX quantitative reverse-transcription polymerase chain reaction master mix according to the manufacturer's instructions (SABiosciences, Frederick, MD). The data were normalized to the expression of the glyceraldehyde-3-phosphate dehydrogenase by transforming the difference in threshold cycle for the gene of interest and the housekeeping gene to the second power, and expressed as the means ± SEM.
Statistics
Data are expressed as scatter plots with medians and quartiles and analyzed by the Mann–Whitney test. A P value less than .05 was considered significant. The method used for statistical analysis of 3 different groups was 1-way analysis of variance with multiple comparisons. Normality was addressed by applying D’Agostino and Pearson normality tests. Statistical analysis was performed with GraphPad Prism (GraphPad Software, La Jolla, CA).
Supplementary Table 1.
Sources of Commercial Antibodies Used in Immunohistochemistry Experiments
| Supplier | Final titer | Host | Clonality | Catalog number | Research resource initiative identifier | |
|---|---|---|---|---|---|---|
| Primary antibody | ||||||
| Embryonic lethal, abnormal vision, Drosophila-like protein 3/4 | Gift from Dr V. Lennon (Mayo Clinic) | 1:500 | Human | AB_2314657 | ||
| NOS1 | Millipore | 0.33 μg/mL | Rabbit | Polyclonal | AB5380 | AB_91824 |
| ChAT | Millipore | 1:100 | Goat | Polyclonal | AB144P | AB_2079751 |
| F4/80 direct conjugate | Thermo Fisher | 0.4 μg/mL | Rat | Polyclonal | MF 48020 | AB_10376287 |
| Major Histocompatibility Complex II | eBioscience | 1.0 μg/mL | Rat | Monoclonal | 14-5321-81 | AB_467560 |
| Protein Gene Product 9.5 | Thermo Fisher | 1:400 | Rabbit | Polyclonal | 38-1000 | AB_2533355 |
| Secondary antibody | ||||||
| Cy3 anti-goat | Jackson ImmunoResearch | 1.75 μg/mL | Donkey | Polyclonal | 705-165-147 | AB_2307351 |
| Alexa Fluor–488 anti-rat | Jackson ImmunoResearch | 2.33 μg/mL | Donkey | Polyclonal | 712-545-150 | AB_2340683 |
| Cy3 anti-rabbit | Jackson ImmunoResearch | 1.75 μg/mL | Donkey | Polyclonal | 711-165-152 | AB_2307443 |
| Cy5 anti-human | Jackson ImmunoResearch | 1.75 μg/mL | Donkey | Polyclonal | 709-175-149 | AB_2340539 |
Supplementary Table 2.
List of Antibodies Used for Sorting Experiments and List of Primers Used for Quantitative Reverse-Transcription Polymerase Chain Reaction
| Antibody | Fluorophore | Catalog number | Company |
|---|---|---|---|
| F4/80 monoclonal antibody (BM8) | Phycoerythrin--cyanine 5 | 15-4801-82 | eBioscence |
| Anti-mouse CD11b | Alexa Fluor 488 | 53-0112-82 | eBioscence |
| Anti-mouse CD45 | Alexa Fluor 450 | 48-0451-82 | eBioscence |
| Rat IgG2b K isotype control | APC | 17-4031-81 | eBioscence |
| Rat IgG2a K isotype control | PE-cyanine 7 | 25-4321-81 | eBioscence |
Supplementary Table 3.
List of Primers used for RT-PCR
| Gene symbol | Unigene title | Forward | Reverse |
|---|---|---|---|
| BMP2 | Bone morphogenetic protein 2 | GGTGATGGCTTCCTTGTACC | AGTGAGGCCCATACCAGAAG |
| Gapdh | Glyceraldehyde-3-phosphate dehydrogenase | Qiagen | Qiagen |
Supplementary Material
Macrophage (green)- and nerve fiber (red) distribution in the gastric muscularis propria.
Macrophage (green)- and nerve fiber (red) distribution in the gastric myenteric plexus.
References
- 1.Furness J.B. J Auton Nerv Syst. 2000;81:87–96. doi: 10.1016/s0165-1838(00)00127-2. [DOI] [PubMed] [Google Scholar]
- 2.Furness J.B. Nat Rev Gastroenterol Hepatol. 2012;9:286–294. doi: 10.1038/nrgastro.2012.32. [DOI] [PubMed] [Google Scholar]
- 3.Tremblay M.E. J Neurosci. 2011;31:16064–16069. doi: 10.1523/JNEUROSCI.4158-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Muller P.A. Cell. 2014;158:300–313. doi: 10.1016/j.cell.2014.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cipriani G. Gastroenterology. 2018;154:2122–2136 e12. doi: 10.1053/j.gastro.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gabanyi I. Cell. 2016;164:378–391. doi: 10.1016/j.cell.2015.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avetisyan M. Proc Natl Acad Sci U S A. 2018;115:4696–4701. doi: 10.1073/pnas.1802490115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao M.M. J Comp Neurol. 2013;521:3358–3370. doi: 10.1002/cne.23354. [DOI] [PubMed] [Google Scholar]
- 9.Anitha M. Am J Physiol Gastrointest Liver Physiol. 2010;298:G375–G383. doi: 10.1152/ajpgi.00343.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplementary References
- 1.Cipriani G. Gastroenterology. 2018;154:2122–2136 e12. doi: 10.1053/j.gastro.2018.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamaji M. PLoS One. 2015;10:e0115563. doi: 10.1371/journal.pone.0115563. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Macrophage (green)- and nerve fiber (red) distribution in the gastric muscularis propria.
Macrophage (green)- and nerve fiber (red) distribution in the gastric myenteric plexus.