Abstract
Lung cancer is one of the leading causes of cancer-associated death, with the etiology largely unknown. The aim of this study was to identify key driver genes with therapeutic potentials in lung adenocarcinoma (LUAD). Transcriptome microarray data from four GEO datasets (GEO: GSE7670, GSE10072, GSE68465, and GSE43458) were jointly analyzed for differentially expressed genes (DEGs). Ontologic analysis showed that most of the upregulated DEGs enriched in collagen catabolic and fibril organization processes were regulated by matrix metalloproteinases (MMPs). Matrix metalloproteinase 11 (MMP11), the highest upregulated MMP family member in LUAD-transformed cells, acted in an autocrine manner and was significantly increased in sera of LUAD patients. MMP11 depletion severely impaired LUAD cell proliferation, migration, and invasion in vitro, in line with retarded tumor growth in xenograft models. Treatment of different human LUAD cell lines with anti-MMP11 antibody significantly retarded cell growth and migration. Administration of anti-MMP11 antibody at a dose of 1 μg/g body weight significantly suppressed tumor growth in xenograft models. These findings indicate that MMP11 is a key cancer driver gene in LUAD and is an appealing target for antibody therapy.
Keywords: MMP11, lung adenocarcinoma, antibody therapy, tumor growth, migration, invasion
Introduction
Lung cancer is one of the most common malignant tumors worldwide. Surgery, radiation, and chemotherapy are three common methods for lung cancer therapy.1 However, regional therapeutic methods, including surgery and radiotherapy, have a poor effect on patients with advanced metastasis. Chemotherapy is currently a major treatment for lung cancer with metastasis, but chemotherapeutic drugs are not specific to killing tumor cells and will cause resistance in patients receiving multiple rounds of chemotherapy. With the emerging concept of “precision medicine” in recent years, more and more molecularly targeted drugs have been effectively applied,2 and the treatment of lung cancer has entered a new era. In the concept of precision medicine, key genes driving carcinogenesis could be used as therapeutic targets.3 Biological reagents or drugs specifically designed to target these genes would interfere with tumor cells and their related microenvironments, so as to eliminate or suppress the occurrence and development of cancer. A variety of therapeutic targets with different biological functions, including EGFR,4 VEGF,5 ALK,4 MET,6 KRAS,4 ROS1,7 HER2,8 and others,9 have been investigated. Accordingly, therapeutic drugs targeting specific molecules in lung cancer cells have been developed, include cell growth factor receptor inhibitors,10 angiogenesis inhibitors,11 and signal transduction inhibitors.12 Although clinical results have indicated that molecularly targeted therapy has an overall advantage, with the emergence of resistance to drugs, curing lung cancer still faces challenges,13, 14 greatly requiring development of new types of targeted therapy drugs.
Genome-wide profiling provides deep insights into tumorigenesis and has proved to be a robust way to identify pathogenic gene targets.15 Furthermore, substantial amounts of gene expression data have been created and deposited in public databases, providing a great opportunity for deciphering the molecular mechanisms of tumorigenesis.16 Two frequently used public gene expression databases, GEO (https://www.ncbi.nlm.nih.gov/geo/) and The Cancer Gene Atlas (TCGA) project (https://gdc.cancer.gov/), contain deposits of a great amount of transcriptomic data generated from microarray or high sequencing. To identify novel gene targets with therapeutic potential in lung adenocarcinoma, we investigated the gene expression pattern by integrating four independent transcriptomic microarray datasets of lung adenocarcinoma. A total of 48 common differentially expressed genes (DEGs) were identified. Collagen catabolic and fibril organization processes were the most enriched biological processes associated with 30 upregulated common DEGs. A group of four matrix metalloproteinases (MMPs) identified in this screen are functionally related to this process. After validation by TCGA data, we proposed that MMP11 was the most significantly overexpressed DEG, and it was the focus of the current study.
MMPs are a class of enzymes that degrade extracellular matrix (ECM).17 They are important tumor microenvironment regulators and play an extremely important role in tumor invasion and metastasis. MMP11, also named stromelysin-3, is a member of the stromelysin subgroup belonging to the MMP superfamily.18 MMP11 is secreted in an enzymatically active form and can be detected in patients’ serum. We proposed that MMP11 may actively participate in the regulation of the tumor microenvironment, by promoting the occurrence and development of malignant tumors. Therapeutically targeting the tumor microenvironment may have merit for improving the prognosis of cancer patients.19 In the current study, the role of MMP11 in the regulation of lung adenocarcinoma (LUAD) development was comprehensively studied by cellular functional assay in vitro and antibody therapy assay in vivo. Our results establish the potential of MMP11 as a therapeutic target for lung cancer.
Results
Bioinformatics Identifies DEGs in LUAD and Non-malignant Tissues
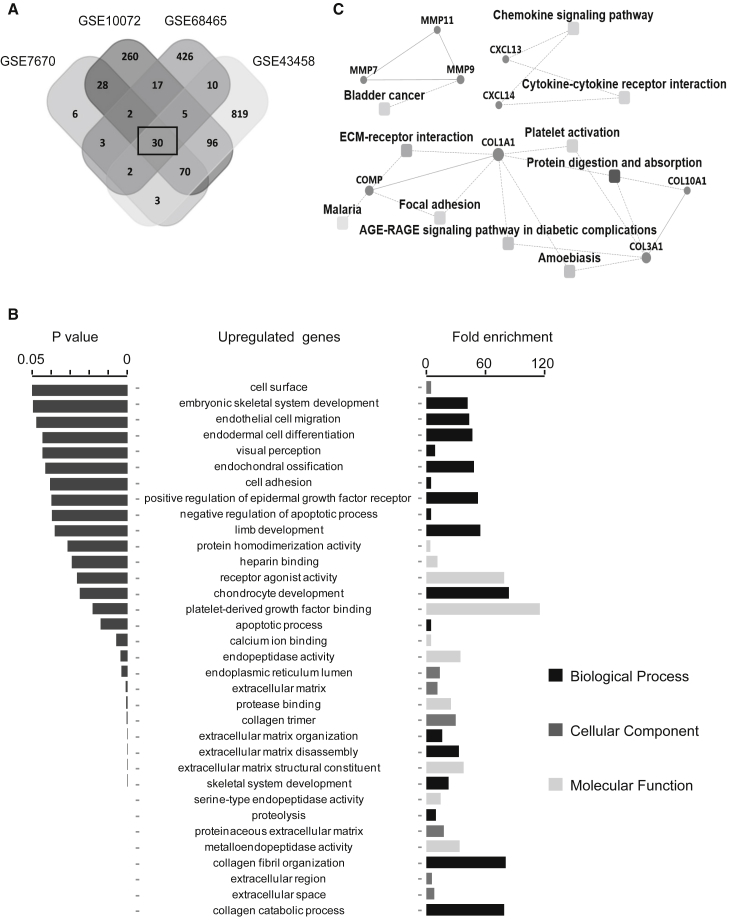
Four gene expression datasets of LUAD in the GEO database, including GSE7670, GSE10072, GSE68465, and GSE43458, were used for identification of genes differentially expressed in lung cancer. The intersecting part of the four sets of DEGs consisted of 48 genes, which were thought to be involved in development and progression of this malignancy (Table 1). Among the 48 DEGs, 30 were upregulated in tumor tissues (Figure 1A). Impressively, the gene ceacam5, encoding the most common serum biomarker CEA for diagnosis of lung cancer, was on the list of upregulated genes, providing convincing evidence of the efficacy of the DEG profile. Next, to gain insight into the biological roles of DEGs involved in cancer progression, we performed a Gene Ontology (GO) enrichment analysis and a protein-protein interaction (PPI) analysis. Most of the enriched GO terms belonged to the biological process category. Collagen catabolic and fibril organization processes were the two most relevant biological processes, and nearly half of the terms involved regulating collagen assembly or disassembly through proteolysis. In terms of the cellular component, the enriched GO terms were mainly related to the ECM. Of the 10 enriched GO molecular functions, 4 were associated with endopeptidase activity (Figure 1B). PPI combined with ontologic analysis revealed common pathways or processes associated with upregulated genes in LUAD (Figure 1C), including ECM-receptor interaction, bladder cancer, elevated chemokine-related signaling, protein digestion, and absorption ontologies. Besides the chemokine-signaling-related factors CXCL13 and CXCL14, most of the upregulated genes belong to the collagen and MMP families. We propose that the DEGs encoding MMPs or collagens found in this study may play critical roles in the development of LUAD. Large studies have demonstrated that MMPs and collagens contribute to the progression of cancer by modulating the tumor microenvironment.20, 21, 22 MMPs, as enzymes with peptidase activity taking an active role in ECM remodeling, are thought to be key players in this process.
Table 1.
Common DEGs Identified in Lung Adenocarcinoma
| Regulation | Gene Symbol |
|---|---|
| DEGs upregulated | COL11A1, SPINK1, MMP12, S100A2, CDH3, COL10A1, COL3A1, MMP9, AGR2, GREM1, CXCL13, TOX3, MMP7, WFDC2, COL1A1, CST1, CXCL14, TMPRSS4, COMP, SULF1, GOLM1, ABCC3, MMP11, LCN2, PPP1R14(D) FAP, ADAMDEC1, MXRA5, CEACAM5, LGSN |
| DEGs downregulated | CDH13, CTNNAL1, HL(F) SEMA5(A) NRN1, ITM2(A) GPM6(B) CX3CR1, SRPX, TSPAN7, GPC3, ADAMTS1, SOCS2, BCH(E) EDNR(B) FHL1, CPB2, CAV1 |
Figure 1.
Identification of DEGs Based on Four Datasets of Lung Adenocarcinoma
(A) Venn diagram of the overlapping parts of upregulated DEGs from four independent LUAD datasets. Thirty upregulated DEGs were common to all datasets. (B) The significantly enriched GO terms for analysis of 30 upregulated DEGs. (C) A PPI network combined with a KEGG pathway enriched in common upregulated DEGs.
MMP11 as a Serum Biomarker of LUAD
The above results indicated that four matrix metalloproteinases, including MMP11, MMP12, MMP9, and MMP7, are commonly upregulated in tumor tissues of LUAD. To further validate the results, we analyzed their expression based on transcript counts derived from the mRNA-seq data that was downloaded from TCGA. As shown in Figure 2A, the expression level of MMP11 has a 5-fold increase in tumor tissues—highly significant compared to the other three well-studied MMPs,23, 24, 25 MMP12, MMP9, and MMP7 (Figure S1). The roles of MMP11 in promoting tumorigenesis have been demonstrated in several malignancies, such as breast,26 gastric,27 and colorectal cancers,28 but there has been little research on its connection with lung cancer. Therefore, we focused on MMP11, to examine its regulation of the progression of LUAD.
Figure 2.
The Oncogenic Roles of MMP11 in Lung Adenocarcinoma
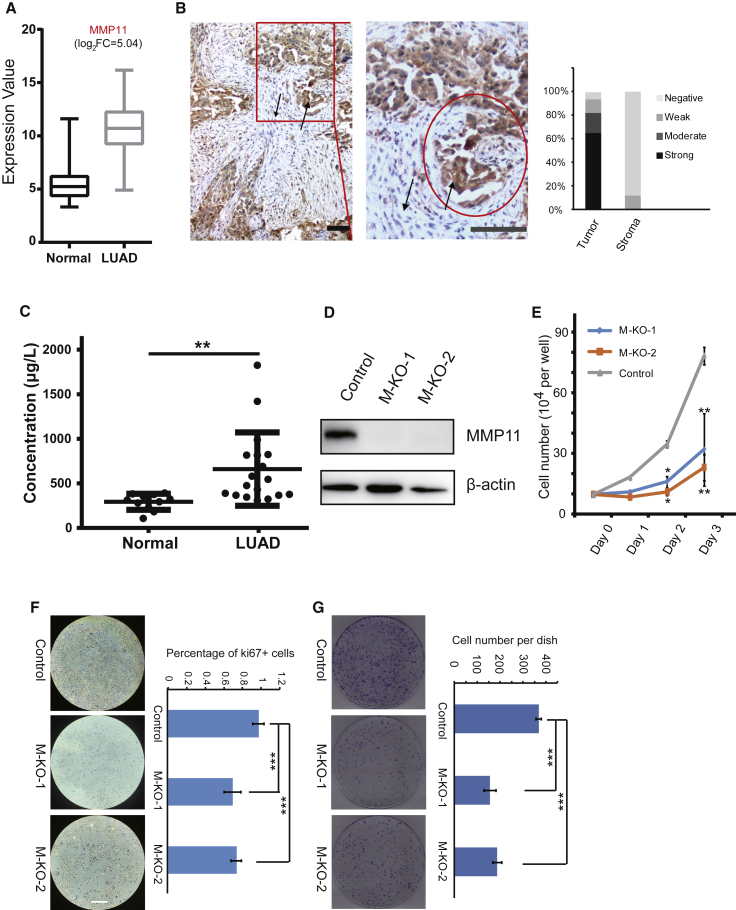
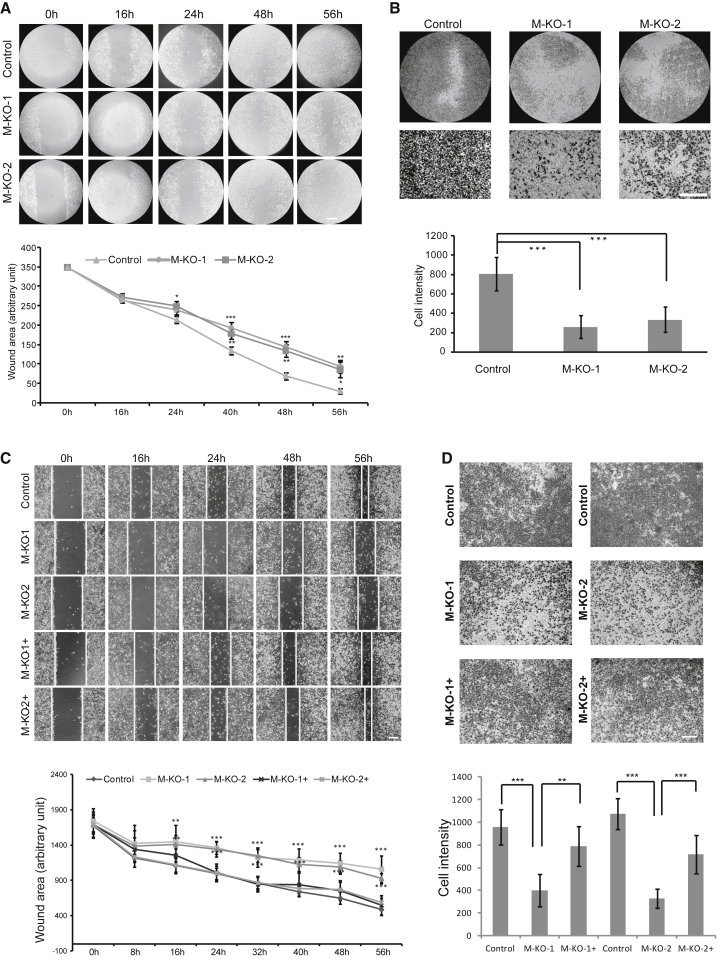
(A) Differential expression of MMP11 in LUAD datasets downloaded from the TCGA database. (B) immunohistochemical examination of MMP11 expression in 18 LUAD biopsies and fractional analysis of samples with no, weak, moderate, or strong staining for MMP11 in transformed cancer cells and stromal cells. Up arrow, cancer cells; down arrow, stromal cells. Scale bar represents 50 μm. (C) MMP11 protein levels in sera from LUAD patients were significantly upregulated. Sera included samples from 18 LUAD patients and 11 control samples from healthy individuals. (**p < 0.01). (D) Western blot of A549 cells infected with lentivirus expressing CRISPR/Cas9 control or expressing CRISPR/Cas9 targeted to MMP11. β-Actin was used as the loading control. Cells infected with lentivirus harboring empty vector or two different sequences targeted to MMP11 were the Control, M-KO-1, and M-KO-2. (E) Cellular proliferation of Control, M-KO-1, and M-KO-2 A549 cells when cultured for the indicated days. Error bars, means ± SD (*p < 0.05 and **p < 0.01 versus Control, n = 4 independent experiments). (F) Control, M-KO-1, and M-KO-2 A549 cells were immuno-reacted with Ki67 before proceeding to 3,3′-diaminobenzidine (DAB) staining. Nuclei were counterstained with hematoxylin. Representative images are shown. Scale bar, 100 μm. Quantitative plot depicting the percentage of Ki67-positive cells in at least 10 different microscopic fields. Error bars, means ± SD (***p < 0.001, n = 10). (G) Control, M-KO-1, and M-KO-2 A549 cells were subjected to a plate colony formation assay. Representative pictures for colony growth are shown (10 cm dishes), along with quantification of the number of colonies. Error bars, means ± SD (***p < 0.001, n = 3 independent experiments).
First, to evaluate the overexpression of MMP11 in LUAD, we performed immunohistochemistry in clinically collected biopsy samples. Although MMP11 was initially considered to be expressed by fibroblasts in tumor stroma,29 further studies indicated that it may be expressed in both tumor cells and stromal cells.18, 30, 31 Our results showed that strong staining for MMP11 was observed in LUAD-transformed cells within the tumor cell masses, with little or no staining seen in the adjacent stroma (Figure 2B), which provides evidence that LUAD tumor cells may release MMP11 protein in an autocrine manner to facilitate invasion of adjacent tissues (Figure 2B), actively exerting its biological endopeptidase activity in digesting ECM. Since MMP11 is a secreted protein, the increase of its level in the circulation may reflect oncogenic occurrence. Therefore, we examined MMP11 levels by ELISA in serum from LUAD patients compared to normal healthy individuals. There was a significant increase in MMP11 protein in sera of LUAD patients (Figure 2C), implying that MMP11 may have potential as a serum biomarker for diagnosis of LUAD.
Critical Role of MMP11 in LUAD Cell Proliferation
To characterize the oncogenic role of MMP11 in LUAD cells, we depleted MMP11 by the CRISPR/Cas9 system through a stable infection of cells with lentiviruses harboring sequence-targeting of the MMP11 gene in the A549 cell line, a widely used LUAD cell model. MMP11 knockout was efficiently achieved as almost undetectable protein of two cell clones with different sequence targeting (Figure 2D). Compared to the control cells, MMP11 depletion significantly reduced cellular proliferation rates (by >60%) (Figure 2E), and the morphology of the elongated control cells was converted to a cobblestone shape (Figure S2A), concomitant with loss of membrane ruffles and protrusions. In addition, significant reduction of labeling of the proliferation marker Ki67 was observed in MMP11-depleted cells, which was consistent with a reduced cellular proliferation rate, as indicated above (Figure 2F). Western blot showed that phosphorylated AKT was downregulated in MMP11-knockout cell lines (Figure S3), indicating that MMP11 may affect cell proliferation through the PI3K/AKT signaling cascades. A colony-formation assay was performed in both control and MMP11-knockout cells. As indicated, MMP11-depleted cells had a nearly 60% reduced number of formed cell colonies with relatively smaller size in comparison with control cells (Figure 2G). The colony formation of cancer cells is an important indicator of their malignancy, reflecting their proliferative ability.
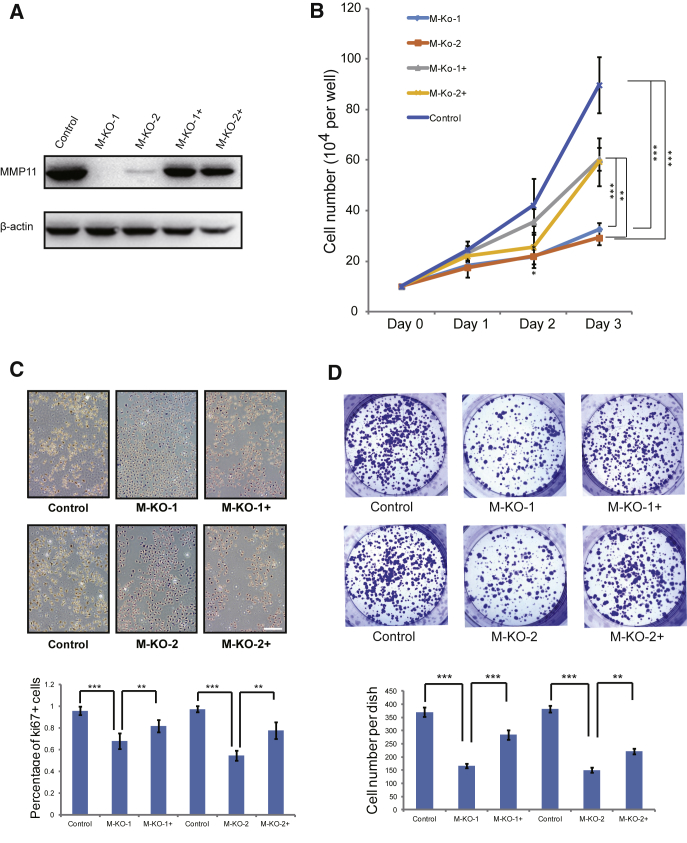
To further evaluate the oncogenic role of MMP11, another human LUAD cell line, PC9, was used for analysis. MMP11-knockout and overexpression were efficiently achieved (Figure 3A). Compared to the control cells, MMP11 depletion significantly reduced cellular proliferation rates, which were partially rescued by transiently overexpression of MMP11, especially significant on day 3 (Figure 3B). Similar cell morphological change was found in PC9 cells, compared with A549 cells (Figure S2B). In addition, significant reduction of labeling of the proliferation marker Ki67 was observed in MMP11-deficient PC9 cells, inconsistent with the results shown in A549 cells (Figure 3C). The reduction of Ki67 labeling in PC9 cells with MMP11 depletion was partially rescued by transient MMP11 overexpression (Figure 3C). A colony-formation assay was also performed in PC9 cells with MMP11 depletion. As indicated, MMP11-depleted PC9 cells had a significantly reduced number of formed cell colonies, with relative smaller size in comparison with control cells (Figure 3D), which was partially restored by overexpression of MMP11. In summary, the above results suggest that MMP11 is critical for LUAD cell proliferation.
Figure 3.
The Oncogenic Roles of MMP11 Were Validated in the LUAD Cell Line PC9
(A) Western blot in PC9 cells infected with lentivirus expressing CRISPR/Cas9 control or expressing CRISPR/Cas9 targeted to MMP11. β-Actin was used as the loading control. Cells infected with lentivirus harboring empty vector or harboring two different sequences targeting MMP11, or MMP11-depleted cells transiently transfected with MMP11 cDNA construct were named Control, M-KO-1, M-KO-2, M-KO-1+, and M-KO-2+. (B) Cellular proliferation of Control, M-KO-1, M-KO-2, M-KO-1+, and M-KO-2+ cells when cultured for the indicated days. Error bars, means ± SD (*p < 0.05, **p < 0.01 and ***p < 0.001, n = 4 independent experiment). (C) Control, M-KO-1, M-KO-2, M-KO-1+, and M-KO-2+ cells were immuno-reacted with Ki67 before proceeding to 3,3′-diaminobenzidine (DAB) staining. Nuclei were counterstained with hematoxylin. Representative images are shown. Scale bar, 100 μm. Quantitative plot depicting the percentage of Ki67-positive cells in at least 10 different microscopic fields. Error bars, means ± SD (**p < 0.01 and ***p < 0.001, n = 10). (D) Control, M-KO-1, M-KO-2, M-KO-1+, and M-KO-2+ cells were subjected to a colony-formation assay. Representative images for colony growth are shown (6-well dishes), along with quantification of the number of colonies. Error bars, means ± SD (**p < 0.01 and ***p < 0.001, n = 3 independent experiments).
Exogenous Antibody against MMP11 Inhibits LUAD Cell Proliferation
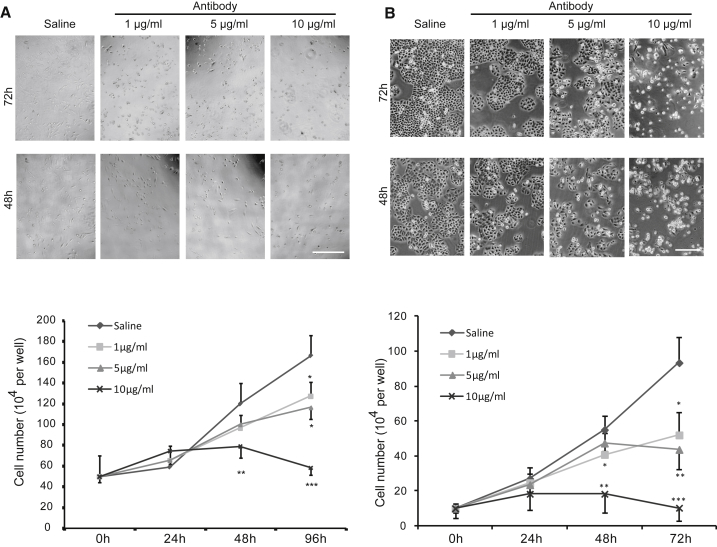
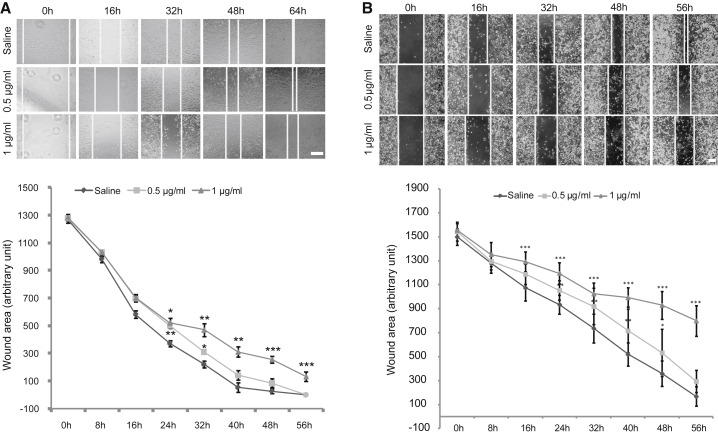
The above experiments demonstrated the inhibitory effect of endogenous depletion of MMP11 on the proliferation of LUAD cells. We considered whether inhibition of MMP11 activity by exogenously applied antibody would have the same effect on cell proliferation. Cells including A549 and PC9 were treated with different concentrations of monoclonal antibody against MMP11 or control vehicle for up to 96 h. The inhibitory effect of antibody treatment on cell proliferation was shown in an antibody concentration-dependent fashion (Figure 4). With the increase of antibody concentration and the extension of the treatment period, the cells’ status progressively deteriorated, and they started to shrink, with a morphological change from spindle to cobblestone similar to that shown in MMP11-knockout cells (Figures 4 and S2). Significant reduction of cell proliferation was observed in cells treated with antibody concentrations as low as 1 μg/mL in both A549 (Figure 4A) and PC9 cells (Figure 4B). The above results suggest that exogenous inhibition of MMP11 by antibody may be an effective therapeutic intervention for LUAD.
Figure 4.
Lung Adenocarcinoma Cells Are Responsive to MMP11 Antibody In Vitro
(A and B) Different concentrations of MMP11 antibody [0 (saline), 1, 5, or 10 μg/mL] were administrated to treat lung adenocarcinoma cell line A549 (A) or PC9 (B) for the indicated times. Representative images were obtained at 48 and 72 h. Scale bar, 100 μm. Growth inhibition was calculated by comparison of the number of cells under treatment with different doses of antibody at different time points (0, 24, 48, 72, and 96 h). As low as 1 μg/mL antibody treatment still effectively inhibited cell growth compared with the control group. Error bars, means ± SD (*p < 0.05, **p < 0.01, ***p < 0.001 versus Control (saline), n = 4 independent experiments).
MMP11 Depletion Impairs Cellular Motility and Invasion
Subsequently, we investigated whether morphological and proliferative changes in MMP11-depleted cells would affect cellular migratory and invasive behaviors. As shown in Figure 5, MMP11 depletion severely impaired the migratory ability of A549 and PC9 cells, as examined by wound-healing assay. The wound area closed significantly slower in the MMP11-depleted A549 cells compared to the control cells. Whereas the control cells almost closed the wound after culturing for 56 h, MMP11-knockout cells still left significant wound area unhealed (Figure 5A). Cell invasion is an important indicator of the metastatic ability of cancer cells, which can be evaluated by transwell invasion assay. As indicated, MMP11-depleted A549 cells exhibited significantly lower (60% reduction) infiltration rates in transwell invasion experiments than control cells (Figure 5B). To emphasize the specific role of MMP11 on LUAD cell migration and invasion, both wound-healing assay and transwell invasion assay were performed on the PC9 cell line, and similar results were obtained, indicating that MMP11 depletion severely impaired cell migration and invasion (Figures 5C and 5D). Importantly, rescue studies confirmed that transient MMP11 overexpression partially restored the migratory and invasive capacities of MMP11-deficient PC9 cells, demonstrating the specific role of MMP11 in regulation of LUAD cells’ oncogenic abilities. The above results demonstrate that MMP11 plays an essential role in LUAD cell migration and invasion.
Figure 5.
MMP11 Regulates Cancer Cell Migration and Invasion
(A) Wound-healing assay performed in control and M-KO-1 and M-KO-2 A549 cells. Representative images were obtained at the indicated time points, and results were quantified in a.u. with IPP6 software Error bars, means ± SD (*p < 0.05, **p < 0.01, ***p < 0.001 versus Control, n = 3 independent experiments). Scale bar, 50 μm. (B) Control, M-KO-1, and M-KO-2 A549 cells were subjected to transwell invasion assays. Invasive cells were imaged under a microscope, with results quantified by IPP6 software. Error bars, means ± SD (***p < 0.001, n = 5 independent experiments). Scale bar, 100 μm. (C) Wound-healing assay performed in M-KO-1, M-KO-2, M-KO-1+, and M-KO-2+ PC9 cells. Representative images are shown at indicated time points, and results were quantified in a.u. with IPP6 software Error bars, means ± SD (**p < 0.01, ***p < 0.001 versus Control, n = 3 independent experiments). Scale bar, 100 μm. (D) M-KO-1, M-KO-2, M-KO-1+, and M-KO-2+ PC9 cells were subjected to transwell invasion assays. Invasive cells were imaged under a microscope, with results quantified by IPP6 software. Error bars, means ± SD (**p < 0.01 and ***p < 0.001, n = 5 independent experiments). Scale bar, 100 μm.
Exogenous Antibody to MMP11 Inhibits Cell Migration Ability
We next investigated if exogenous treatment of A549 cells with antibody against MMP11 would affect the migratory ability of cells. Prior experiments had shown that application to the cells of a dose of antibody as low as 1 μg/mL could effectively inhibit cell proliferation. Herein, two different concentrations, 0.5 and 1 μg/mL, were selected for a wound-healing assay. As shown in Figure 6A, under both concentrations of antibody, the migratory ability of the A549 cells was decreased in a dose-dependent manner. In concurrence with the results in the A549 cell line, a similar phenomenon was found in the PC9 cell line (Figure 6B). In both cell lines, considering that the wound area did not be close, even up to 56 h, and that the cells exhibited an unhealthy cobblestone morphology, 1 μg/mL was the more preferable concentration for antibody inhibition of cell migration (Figure 6). We argue that exogenously administered MMP11 antibody may be an attractive therapy for LUAD.
Figure 6.
MMP11 Antibody Treatment Inhibits Cancer Cell Migration In Vitro
(A and B) Different concentrations of MMP11 antibody [0 (saline), 0.5, and 1 μg/mL] were applied to treat lung adenocarcinoma cell line A549 (A) or PC9 (B) for the indicated times. Representative images of a wound-healing assay are shown at different time points during treatment. Inhibition of cell migration was calculated by IPP6 software on cell images taken at various time points during treatment with different doses of antibody. A 1 μg/mL antibody treatment effectively inhibited cell migration in both A549 (A) and PC9 (B) cells compared to Control (saline). Error bars, means ± SD (*p < 0.05, **p < 0.01, ***p < 0.001 versus Control, n = 5 independent experiments). Scale bar, 100 μm.
In Vivo Tumorigenesis and Tumor Targeting
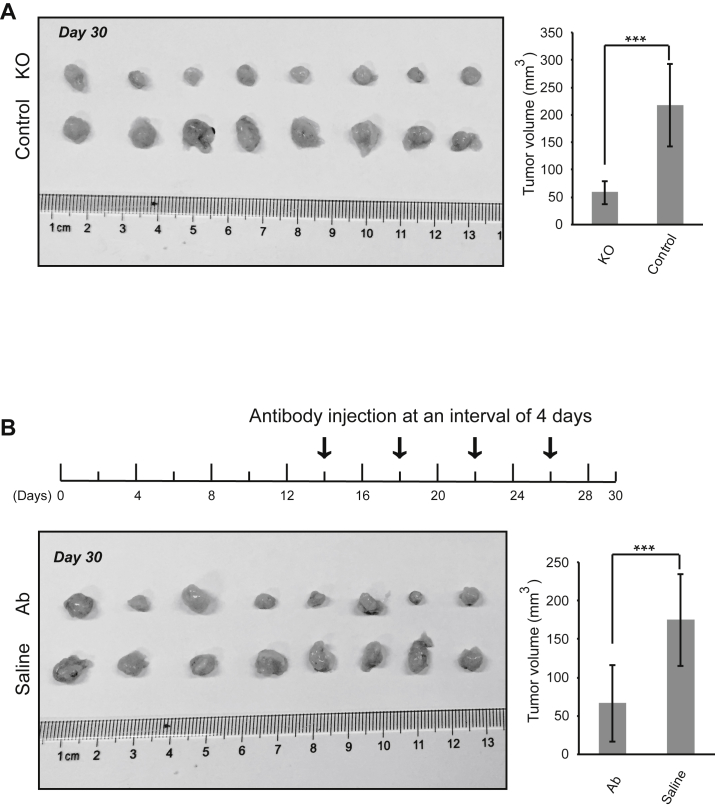
The above results demonstrated the oncogenic properties of MMP11 and the efficacy of MMP11 antibody treatment to inhibit oncogenic behaviors of LUAD tumor cells in vitro. We next sought to examine whether silencing MMP11 or antibody targeting of MMP11 affects tumor development in vivo. A xenograft model was established using the LUAD cell line A549. First, mice were injected subcutaneously with MMP11-depleted A549 cells and control normal A549 cells to develop solid tumors. Tumor xenografts were excised and analyzed at day 30. The mouse body weights were comparable over the course of treatment between the different groups of mice: those with subcutaneous injection of MMP11-depleted cells and those with control normal A549 cells (Figure S4A). Immunohistochemistry assays validated the loss of expression of MMP11 in excised tumors derived from MMP11-depleted cells (Figure S5A). Importantly, the volume of the tumor masses in mice injected with control cells was almost four times larger at day 30 than in those with the MMP11-depleted xenografts (Figure 7A), suggesting that MMP11 affects tumor development in vivo.
Figure 7.
Anti-MMP11 Antibody Retarded Tumor Formation In Vivo
(A) Nude mice were subcutaneously inoculated with Control or MMP11-deficient (KO) A549 cells, and tumor formation was monitored. Tumor sizes were calculated at day 30 (***p < 0.001, n = 8). (B) Nude mice were subcutaneously inoculated with wild-type A549 cells, and tumor formation was monitored. Beginning on day 14 (arrow), mice were injected at intervals of 4 days with 1 μg/g body weight of anti-MMP11 antibody (Ab) or vehicle Control (saline). Error bars are mean ± SD. ***p < 0.001, n = 8.
To further validate the role of MMP11 on tumorigenesis in vivo, we tested the efficacy of MMP11 antibody therapy on tumorigenic inhibition in mouse xenografts. Female BALB/c nude mice were inoculated subcutaneously with A549 cells. Beginning on day 14, anti-MMP11 antibody and sterile saline control were injected via the tail vein at intervals of 4 days. After four rounds of antibody injections up to day 30, tumor pads were excised for measurement. The mouse body weights were comparable between the groups at various time points (Figure S4B). Importantly, MMP11 antibody treatment significantly retarded LUAD tumor growth, given that tumor volumes in the antibody-treated group were two to three times smaller than those in the control group (Figure 7B). That is, antibody treatment reduced the growth of pre-established tumors by more than 60% compared with the saline control (Figure 7B), which was inconsistent with greatly reduced Ki67 expression (Figure S5B). These results suggest that MMP11 antibody therapy can effectively reduce the LUAD tumor load.
Discussion
In this study, we identified MMP11 as a tumor biomarker in LUAD, which poses the potential for its use as a therapeutic target for treatment of this malignancy. MMP11 has long been found to be associated with tumor progression in some types of cancer, such as breast, liver, and colon. To date, however, data are limited on the role of MMP11 in lung cancer. In this study, we first performed gene expression profiling on four independent datasets of LUAD and uncovered MMP11 as a potential driver gene associated with LUAD. We found that the expression of MMP11 increased in both tumor tissues and sera from LUAD patients. Functional studies indicated that MMP11 played a key role in promoting tumor cell proliferation, migration, and invasion. Most important, its suitability as a therapeutic target was assessed, using an anti-MMP11 antibody, in both in vitro and in vivo studies.
It is well known that almost all solid tumors are composed not only of tumor cells, but also of various non-tumor stromal cells, as well as cytokines, growth factor, proteases, and ECM.32 Tumor cells and stromal components are tightly linked and orchestrated with each other through extracellular elements to form the tumor microenvironment, which promotes tumorigenesis by malignantly transforming normal epithelial cells or by accelerating tumor cell growth and invasion. Generally, degradation and remodeling of the ECM is a prerequisite for tumor cell invasion of the adjacent normal parenchyma and metastasis to distant tissues.33 MMPs are a family of zinc-dependent endopeptidases with a powerful capacity for degrading ECM proteins, revealing its critical role during the process of tumorigenesis. In this study, we found that tumor cells may release MMP11 in an autocrine manner, thus actively modifying the microenvironment to adapt their malignant biological behaviors.
So far, there have been many studies on the MMPs family proteins, but the research on MMP11 has not been adequate, especially in lung cancer. Human MMP11 was first identified in the stromal cells of breast carcinoma.34 It is upregulated in colon cancer,31 and its downregulation inhibits tumor proliferation and metastasis in hepatocellular carcinoma.35 In colorectal cancer, tumor metastasis can be effectively prevented by let-7-mediated inhibition of MMP11.28 Knockdown of MMP11 also inhibits cell proliferation and metastasis in gastric cancer.27 It is also highly expressed in laryngeal cancer.36 In prostate cancer, downregulation of MMP11 can reduce tumor migration.37 Downregulation of mir125a5p in osteosarcoma targets MMP11, to suppress tumor progression,38 and MMP11 may even serve as a new prognostic factor in hormone-receptor-negative, HER2-positive breast cancers.39 Different from other MMP family proteins, MMP11 has its unique characteristics and functions. It is intracellularly processed to secrete in an active form instead of as an inactive zymogen like other MMPs. Moreover, unlike other MMPs that degrade the laminin, fibronectin, and elastin of these classic substrates, MMP11 preferentially hydrolyzes serine protease inhibitor α1-antitrypsin, insulin-like growth factor binding protein-1 (IGFBP-1),40 and collagen VI.41 IGF-1-mediated cell growth signaling was generally neutralized by IGFBP-1. MMP11 cleaves IGFBP-1 to release extra free IGF-1 in the tumor microenvironment, thus hyperactivating the IGF-1-mediated PI3k/AKT signaling cascade. Our work supports this hypothesis, since MMP11-deficient cells showed impaired proliferative ability and downregulation of phosphorylated AKT protein. Our results are also consistent Deng et al.42, which shows that knockdown of MMP11 in gastric cell lines inhibits cell growth and colony formation. It is not surprising to find that MMP11 plays roles in promoting cell migration and invasion, since MMP11 functions in ECM reconstruction. Substantial evidence supports this notion in various cell models, since the first study demonstrated that the oncogene Gli1 enhances migration and invasion via upregulation of MMP11 in breast cancer cell lines.43
Our work demonstrated that MMP11 protein levels in serum was significantly increased in patients with LUAD compared to normal healthy individuals. Because of the importance of early diagnosis of malignant tumors in decreasing mortality, this finding suggests that MMP11 is a potential biomarker for clinical diagnosis in LUAD. A prior study proved the efficacy of serum MMP11 as a diagnostic marker for gastric cancer, with higher sensitivity compared to other traditional tumor markers, such as CEA, CA199, CA242, and its family member MMP9.44 In addition, as a diagnostic marker, another important application for MMP11 in LUAD may be as a therapeutic target. As an important component of the cancer microenvironment, MMP11 is released by LUAD tumor cells, which may remodel ECM to facilitate tumorigenesis and progression of cancer. Our results indicate that anti-MMP11 antibody application could efficiently inhibit growth and migration of lung cancer cells in vitro. In vivo treatment of human LUAD xenografts with anti-MMP11 antibody strongly prevented tumor growth, which was inconsistent with in vitro data. In a previous study, immunotherapy using MMP11 as antigen exerted effective anti-tumor effects in a colon adenocarcinoma mouse model.45 Combined with our current work, MMP11 holds potential as a therapeutic target in different solid tumors.
In summary, MMP11 expression was highly expressed in tissues and sera from patients with LUAD, and MMP11 antibody therapy inhibited tumor growth in both in vitro experiments and in vivo xenografts. This work demonstrates that MMP11 may serve as a potent therapeutic target through antibody therapy for LUAD. Additional studies are required to delineate in detail the exact molecular mechanism, as well as the in vivo efficacy and safety of this therapeutic strategy.
Materials and Methods
Microarray Data Mining and Gene Expression Analysis
We analyzed microarray datasets containing LUAD patient samples from the GEO (https://www.ncbi.nlm.nih.gov/geo/) and TCGA gene expression data from the Genomic Data Commons data portal (https://portal.gdc.cancer.gov/). More details are available in Supplemental Materials and Methods.
Functional Enrichment Analysis and PPI Network Construction
For functional enrichment analysis, the online tool Database for Annotation, Visualization and Integrated Discovery (DAVID; https://david.ncifcrf.gov/) was used to perform GO analysis. A web-based tool in the OmicsBean database (http://www.omicsbean.cn) was used for PPI combined with Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis.46, 47 GO terms or pathways with p < 0.05 were considered to be significantly enriched. An ontologic model PPI network of the DEGs was constructed.
Cell Lines
The human cell lines used in this study, including A549, PC9 (LUAD), and 293FT (embryonic kidney cells), were purchased from American Type Cell Culture (ATCC) and cultured in the recommended media at 37°C and 5% CO2 in a Thermo Fisher X50 incubator.
Collection of Serum and Tumor Tissues
The collection of tumor and adjacent normal tissues and plasma from LUAD patients was approved by the Institutional Review Board of the Cancer Hospital (Chinese Academy of Sciences, Hefei, China), in accordance with the Declaration of Helsinki.
Antibodies
Mouse monoclonal antibody against MMP11 was purchased from Boster Biological Technology. Mouse monoclonal antibodies against AKT, p-AKT, ERK, p-ERK, and Vimentin were purchased from Cell Signaling Technology. Rabbit antibodies against Ki67 and β-actin were obtained from Fuzhou Maixin Biotech (China) and Sangon Biotech (China), respectively.
Immunohistochemistry and Western Blot
Immunohistochemical staining and western blot were performed according to standard protocols.48, 49 Details are available in Supplemental Materials and Methods.
Plasmid Construction, Lentivirus Packaging, and Infection
The single guide RNAs were designed with the CRISPR Design webtool (https://zlab.bio/guide-design-resources/). The oligos were annealed and cloned into the lentiviral shuttle vector lenti-CRISPRv2 (Addgene). Lentivirus packaging and infection were performed as previously described.50 An MMP11-overexpressing plasmid was constructed by subcloning of MMP11 cDNA into pcDNA3.1, subsequently verified by sequencing. More details are available in Supplemental Materials and Methods.
Proliferation Assay
Cells were seeded into 6-well plates in DMEM with 10% FBS culture medium, at the density indicated Figures 2E and 3B, and maintained at 37°C and 5% CO2 in a humidified incubator. At the indicated time points, cell were counted by trypan blue exclusion.
Plate Colony Formation Assay
The cells were diluted in gradients to the appropriate concentrations. For the A549 cells, 1,000 cells were seeded and uniformly distributed in 10 cm dishes with complete medium, and for the PC9 cells, 800 cells were seeded and uniformly distributed in 6-well dishes, then placed in an incubator for 2–3 weeks until single clones formed. Cell colonies were stained with crystal violet, and complete dishes were scanned with a scanner. The number of cell colonies was analyzed by Image-Pro Plus 6.0 (IPP6) software.
Wound-Healing and Invasion Assay
The wound-healing and invasion assay was performed as previously described.48 Details are available in Supplemental Materials and Methods.
ELISA
MMP11 levels in the serum of patients were measured with an ELISA kit (Kete Biological Technology, China). The experiment was performed according to the manufacturer’s instructions. Details are available in Supplemental Materials and Methods.
Tumor Xenografts and Treatment
Six- to eight-week-old female BALB/c nude mice were purchased from the animal model center of Nanjing University. To obtain solid tumors, each mouse was inoculated with 1 × 107 cells/mL suspended in Matrigel (1:1) via subcutaneous injection into the left or right shoulder flank. Tumor development was monitored over a period of 30 days before the mice were killed for further analysis. For antibody therapy, beginning on day 14, inoculated mice were injected via the tail vein every 4 days with 1 mg/kg anti-MMP11 antibody, or a vehicle control (sterile saline). Tumor volume (mm3) is calculated by the formula V = L × B2/2, where V is tumor volume (mm3), L is tumor length (mm), and B is tumor breadth (mm). All animal care and handling were in compliance with the guidelines of the Institutional Animal Care and Use Committee at the Hefei Institutes of Physical Science, Chinese Academy of Sciences.
Statistical Analysis
Comparisons of data were performed with a paired two-tailed Student’s t test with equal variance or one-way ANOVA with Bonferroni’s post hoc test. Data are presented as means ± SD. Results were considered to be statistically significant when *p < 0.05, **p < 0.01, or ***p < 0.001, or as indicated in the figures.
Author Contributions
W.Y. and H.W. conceived and designed the work. W.Y., H.W., H.D., L.L., and H.-Q.W. developed the experimental methodologies. H.Y., P.J., D.L., X.N., Q.D., and H.-Q.W. performed the experiments. W.Y., H.Y., P.J., H.W., and H.-Q.W. analyzed and interpreted the data. W.Y., H.W., L.L., H.-Q.W., and H.D. wrote the paper. All authors approved the submission of the final version of the manuscript.
Conflicts of Interest
Ownership interest and patents: Hefei Institutes of Physical Science, Chinese Academy of Sciences.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81872276), the Natural Science Foundation of Anhui Province (1708085MH204), Anhui Province Key Laboratory of Medical Physics and Technology (LMPT201702), Shanxi Scholarship Council of China (2014-033), and the Natural Science Foundation of Shanxi Province (2015011132). We appreciate Mr. Jiarong Zhao and Ms. Qiuyan Sun in the Lab of Clinical Pathology of Hefei Cancer Hospital, Chinese Academy of Sciences, for help in pathologic analysis of tissue samples.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omto.2019.03.012.
Contributor Information
Hongzhi Wang, Email: wanghz@hfcas.ac.cn.
Wulin Yang, Email: yangw@cmpt.ac.cn.
Supplemental Information
References
- 1.Lemjabbar-Alaoui H., Hassan O.U., Yang Y.W., Buchanan P. Lung cancer: Biology and treatment options. Biochim. Biophys. Acta. 2015;1856:189–210. doi: 10.1016/j.bbcan.2015.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moreira A.L., Eng J. Personalized therapy for lung cancer. Chest. 2014;146:1649–1657. doi: 10.1378/chest.14-0713. [DOI] [PubMed] [Google Scholar]
- 3.Fang B., Mehran R.J., Heymach J.V., Swisher S.G. Predictive biomarkers in precision medicine and drug development against lung cancer. Chin. J. Cancer. 2015;34:295–309. doi: 10.1186/s40880-015-0028-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu S.V., Subramaniam D., Cyriac G.C., Abdul-Khalek F.J., Giaccone G. Emerging protein kinase inhibitors for non-small cell lung cancer. Expert Opin. Emerg. Drugs. 2014;19:51–65. doi: 10.1517/14728214.2014.873403. [DOI] [PubMed] [Google Scholar]
- 5.Dempke W.C., Suto T., Reck M. Targeted therapies for non-small cell lung cancer. Lung Cancer. 2010;67:257–274. doi: 10.1016/j.lungcan.2009.10.012. [DOI] [PubMed] [Google Scholar]
- 6.Liu X., Newton R.C., Scherle P.A. Development of c-MET pathway inhibitors. Expert Opin. Investig. Drugs. 2011;20:1225–1241. doi: 10.1517/13543784.2011.600687. [DOI] [PubMed] [Google Scholar]
- 7.Rolfo C., Ruiz R., Giovannetti E., Gil-Bazo I., Russo A., Passiglia F., Giallombardo M., Peeters M., Raez L. Entrectinib: a potent new TRK, ROS1, and ALK inhibitor. Expert Opin. Investig. Drugs. 2015;24:1493–1500. doi: 10.1517/13543784.2015.1096344. [DOI] [PubMed] [Google Scholar]
- 8.Mazières J., Barlesi F., Filleron T., Besse B., Monnet I., Beau-Faller M., Peters S., Dansin E., Früh M., Pless M. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2-targeted drugs: results from the European EUHER2 cohort. Ann. Oncol. 2016;27:281–286. doi: 10.1093/annonc/mdv573. [DOI] [PubMed] [Google Scholar]
- 9.Liu D., Vokes N.I., Van Allen E.M. Toward Molecularly Driven Precision Medicine in Lung Adenocarcinoma. Cancer Discov. 2017;7:555–557. doi: 10.1158/2159-8290.CD-17-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tiseo M., Loprevite M., Ardizzoni A. Epidermal growth factor receptor inhibitors: a new prospective in the treatment of lung cancer. Curr. Med. Chem. Anticancer Agents. 2004;4:139–148. doi: 10.2174/1568011043482106. [DOI] [PubMed] [Google Scholar]
- 11.Cabebe E., Wakelee H. Role of anti-angiogenesis agents in treating NSCLC: focus on bevacizumab and VEGFR tyrosine kinase inhibitors. Curr. Treat. Options Oncol. 2007;8:15–27. doi: 10.1007/s11864-007-0022-4. [DOI] [PubMed] [Google Scholar]
- 12.Yagui-Beltrán A., He B., Raz D., Kim J., Jablons D.M. Novel therapies targeting signaling pathways in lung cancer. Thorac. Surg. Clin. 2006;16:379–396, vi. doi: 10.1016/j.thorsurg.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 13.Wu D., Wang D.C., Cheng Y., Qian M., Zhang M., Shen Q., Wang X. Roles of tumor heterogeneity in the development of drug resistance: A call for precision therapy. Semin. Cancer Biol. 2017;42:13–19. doi: 10.1016/j.semcancer.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Sacco J.J., Al-Akhrass H., Wilson C.M. Challenges and Strategies in Precision Medicine for Non-Small-Cell Lung Cancer. Curr. Pharm. Des. 2016;22:4374–4385. doi: 10.2174/1381612822666160603014932. [DOI] [PubMed] [Google Scholar]
- 15.Lizardi P.M., Forloni M., Wajapeyee N. Genome-wide approaches for cancer gene discovery. Trends Biotechnol. 2011;29:558–568. doi: 10.1016/j.tibtech.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee E., Moon A. Identification of Biomarkers for Breast Cancer Using Databases. J. Cancer Prev. 2016;21:235–242. doi: 10.15430/JCP.2016.21.4.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kessenbrock K., Plaks V., Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang X., Huang S., Guo J., Zhou L., You L., Zhang T., Zhao Y. Insights into the distinct roles of MMP-11 in tumor biology and future therapeutics (Review) Int. J. Oncol. 2016;48:1783–1793. doi: 10.3892/ijo.2016.3400. [DOI] [PubMed] [Google Scholar]
- 19.Sounni N.E., Noel A. Targeting the tumor microenvironment for cancer therapy. Clin. Chem. 2013;59:85–93. doi: 10.1373/clinchem.2012.185363. [DOI] [PubMed] [Google Scholar]
- 20.Kalluri R. Basement membranes: structure, assembly and role in tumour angiogenesis. Nat. Rev. Cancer. 2003;3:422–433. doi: 10.1038/nrc1094. [DOI] [PubMed] [Google Scholar]
- 21.Velez D.O., Tsui B., Goshia T., Chute C.L., Han A., Carter H., Fraley S.I. 3D collagen architecture induces a conserved migratory and transcriptional response linked to vasculogenic mimicry. Nat. Commun. 2017;8:1651. doi: 10.1038/s41467-017-01556-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bonnans C., Chou J., Werb Z. Remodelling the extracellular matrix in development and disease. Nat. Rev. Mol. Cell Biol. 2014;15:786–801. doi: 10.1038/nrm3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv F.Z., Wang J.L., Wu Y., Chen H.F., Shen X.Y. Knockdown of MMP12 inhibits the growth and invasion of lung adenocarcinoma cells. Int. J. Immunopathol. Pharmacol. 2015;28:77–84. doi: 10.1177/0394632015572557. [DOI] [PubMed] [Google Scholar]
- 24.van Kempen L.C., Coussens L.M. MMP9 potentiates pulmonary metastasis formation. Cancer Cell. 2002;2:251–252. doi: 10.1016/s1535-6108(02)00157-5. [DOI] [PubMed] [Google Scholar]
- 25.Liu D., Nakano J., Ishikawa S., Yokomise H., Ueno M., Kadota K., Urushihara M., Huang C.L. Overexpression of matrix metalloproteinase-7 (MMP-7) correlates with tumor proliferation, and a poor prognosis in non-small cell lung cancer. Lung Cancer. 2007;58:384–391. doi: 10.1016/j.lungcan.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez-Garcia B., Eiró N., Marín L., González-Reyes S., González L.O., Lamelas M.L., Vizoso F.J. Expression and prognostic significance of fibronectin and matrix metalloproteases in breast cancer metastasis. Histopathology. 2014;64:512–522. doi: 10.1111/his.12300. [DOI] [PubMed] [Google Scholar]
- 27.Kou Y.B., Zhang S.Y., Zhao B.L., Ding R., Liu H., Li S. Knockdown of MMP11 inhibits proliferation and invasion of gastric cancer cells. Int. J. Immunopathol. Pharmacol. 2013;26:361–370. doi: 10.1177/039463201302600209. [DOI] [PubMed] [Google Scholar]
- 28.Han H.B., Gu J., Zuo H.J., Chen Z.G., Zhao W., Li M., Ji D.B., Lu Y.Y., Zhang Z.Q. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J. Pathol. 2012;226:544–555. doi: 10.1002/path.3014. [DOI] [PubMed] [Google Scholar]
- 29.Basset P., Okada A., Chenard M.P., Kannan R., Stoll I., Anglard P., Bellocq J.-P., Rio M.C. Matrix metalloproteinases as stromal effectors of human carcinoma progression: Therapeutic implications. Matrix Biol. 1997;15:535–541. doi: 10.1016/s0945-053x(97)90028-7. [DOI] [PubMed] [Google Scholar]
- 30.von Marschall Z., Riecken E.O., Rosewicz S. Stromelysin 3 is overexpressed in human pancreatic carcinoma and regulated by retinoic acid in pancreatic carcinoma cell lines. Gut. 1998;43:692–698. doi: 10.1136/gut.43.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barrasa J.I., Olmo N., Santiago-Gómez A., Lecona E., Anglard P., Turnay J., Lizarbe M.A. Histone deacetylase inhibitors upregulate MMP11 gene expression through Sp1/Smad complexes in human colon adenocarcinoma cells. Biochim. Biophys. Acta. 2012;1823:570–581. doi: 10.1016/j.bbamcr.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 32.Belli C., Trapani D., Viale G., D’Amico P., Duso B.A., Della Vigna P., Orsi F., Curigliano G. Targeting the microenvironment in solid tumors. Cancer Treat. Rev. 2018;65:22–32. doi: 10.1016/j.ctrv.2018.02.004. [DOI] [PubMed] [Google Scholar]
- 33.Mittal R., Patel A.P., Debs L.H., Nguyen D., Patel K., Grati M., Mittal J., Yan D., Chapagain P., Liu X.Z. Intricate Functions of Matrix Metalloproteinases in Physiological and Pathological Conditions. J. Cell. Physiol. 2016;231:2599–2621. doi: 10.1002/jcp.25430. [DOI] [PubMed] [Google Scholar]
- 34.Basset P., Bellocq J.P., Wolf C., Stoll I., Hutin P., Limacher J.M., Podhajcer O.L., Chenard M.P., Rio M.C., Chambon P. A novel metalloproteinase gene specifically expressed in stromal cells of breast carcinomas. Nature. 1990;348:699–704. doi: 10.1038/348699a0. [DOI] [PubMed] [Google Scholar]
- 35.Bi Q., Tang S., Xia L., Du R., Fan R., Gao L., Jin J., Liang S., Chen Z., Xu G. Ectopic expression of MiR-125a inhibits the proliferation and metastasis of hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS ONE. 2012;7:e40169. doi: 10.1371/journal.pone.0040169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z., Ding S., Zhong Q., Li G., Zhang Y., Huang X.C. Significance of MMP11 and P14(ARF) expressions in clinical outcomes of patients with laryngeal cancer. Int. J. Clin. Exp. Med. 2015;8:15581–15590. [PMC free article] [PubMed] [Google Scholar]
- 37.Wan X., Pu H., Huang W., Yang S., Zhang Y., Kong Z., Yang Z., Zhao P., Li A., Li T., Li Y. Androgen-induced miR-135a acts as a tumor suppressor through downregulating RBAK and MMP11, and mediates resistance to androgen deprivation therapy. Oncotarget. 2016;7:51284–51300. doi: 10.18632/oncotarget.9992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Waresijiang N., Sun J., Abuduaini R., Jiang T., Zhou W., Yuan H. The downregulation of miR-125a-5p functions as a tumor suppressor by directly targeting MMP-11 in osteosarcoma. Mol. Med. Rep. 2016;13:4859–4864. doi: 10.3892/mmr.2016.5141. [DOI] [PubMed] [Google Scholar]
- 39.Han J., Choi Y.L., Kim H., Choi J.Y., Lee S.K., Lee J.E., Choi J.S., Park S., Choi J.S., Kim Y.D. MMP11 and CD2 as novel prognostic factors in hormone receptor-negative, HER2-positive breast cancer. Breast Cancer Res. Treat. 2017;164:41–56. doi: 10.1007/s10549-017-4234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mañes S., Mira E., Barbacid M.M., Ciprés A., Fernández-Resa P., Buesa J.M., Mérida I., Aracil M., Márquez G., Martínez-A C. Identification of insulin-like growth factor-binding protein-1 as a potential physiological substrate for human stromelysin-3. J. Biol. Chem. 1997;272:25706–25712. doi: 10.1074/jbc.272.41.25706. [DOI] [PubMed] [Google Scholar]
- 41.Motrescu E.R., Blaise S., Etique N., Messaddeq N., Chenard M.P., Stoll I., Tomasetto C., Rio M.C. Matrix metalloproteinase-11/stromelysin-3 exhibits collagenolytic function against collagen VI under normal and malignant conditions. Oncogene. 2008;27:6347–6355. doi: 10.1038/onc.2008.218. [DOI] [PubMed] [Google Scholar]
- 42.Deng H., Guo R.F., Li W.M., Zhao M., Lu Y.Y. Matrix metalloproteinase 11 depletion inhibits cell proliferation in gastric cancer cells. Biochem. Biophys. Res. Commun. 2005;326:274–281. doi: 10.1016/j.bbrc.2004.11.027. [DOI] [PubMed] [Google Scholar]
- 43.Kwon Y.J., Hurst D.R., Steg A.D., Yuan K., Vaidya K.S., Welch D.R., Frost A.R. Gli1 enhances migration and invasion via up-regulation of MMP-11 and promotes metastasis in ERα negative breast cancer cell lines. Clin. Exp. Metastasis. 2011;28:437–449. doi: 10.1007/s10585-011-9382-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang Y.-H., Deng H., Li W.-M., Zhang Q.-Y., Hu X.-T., Xiao B., Zhu H.-H., Geng P.-L., Lu Y.-Y. Identification of matrix metalloproteinase 11 as a predictive tumor marker in serum based on gene expression profiling. Clin. Cancer Res. 2008;14:74–81. doi: 10.1158/1078-0432.CCR-07-1179. [DOI] [PubMed] [Google Scholar]
- 45.Peruzzi D., Mori F., Conforti A., Lazzaro D., De Rinaldis E., Ciliberto G., La Monica N., Aurisicchio L. MMP11: a novel target antigen for cancer immunotherapy. Clin. Cancer Res. 2009;15:4104–4113. doi: 10.1158/1078-0432.CCR-08-3226. [DOI] [PubMed] [Google Scholar]
- 46.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 47.Kanehisa M., Goto S., Sato Y., Furumichi M., Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chakraborty S., Lakshmanan M., Swa H.L., Chen J., Zhang X., Ong Y.S., Loo L.S., Akıncılar S.C., Gunaratne J., Tergaonkar V. An oncogenic role of Agrin in regulating focal adhesion integrity in hepatocellular carcinoma. Nat. Commun. 2015;6:6184. doi: 10.1038/ncomms7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nilsson R., Jain M., Madhusudhan N., Sheppard N.G., Strittmatter L., Kampf C., Huang J., Asplund A., Mootha V.K. Metabolic enzyme expression highlights a key role for MTHFD2 and the mitochondrial folate pathway in cancer. Nat. Commun. 2014;5:3128. doi: 10.1038/ncomms4128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.