Abstract
Worldwide, colorectal cancer (CRC) is one of the leading causes of cancer-related deaths. Recent advances in high-throughput technologies have shown that the gut microbiota may have a major influence on human health, including CRC. Nonetheless, how the gut microbiota interacts with tumor cells in CRC patients is largely unknown. Studies have shown that the microbiota fills in a variety of niche metabolic pathways that the host does not possess. For example, the microbiota produces butyrate, which provides the colon's epithelial cells with about 70% of their energy needs. The typically fast proliferation of tumor cells in CRC patients drastically alters the tumor's nutrient microenvironment. Those alterations correspond to the microbiota composition and functional changes. In tumor cells, a central mediator of metabolic changes is the aberrant expression of microRNAs (miRNAs). In this study, we explored recent insights into metabolic interactions between the microbiota and tumor cells in CRC pathobiology, focusing on the role of miRNAs. These observations support our view that miRNAs may also serve as mediators of the metabolites' effects.
Keywords: host–microbiota interactions, microbiota, metabolism, metabolites, high-throughput technologies, colorectal cancer, microRNAs
Introduction
In the United States, colorectal cancer (CRC) is the third most commonly diagnosed type of cancer and the second most frequent cause of cancer-related deaths (Siegel et al., 2018). In 2018, an estimated 140,250 people will be diagnosed with CRC, and 50,630 will die from it. More than a third of CRC patients will not be alive 5 years after their diagnosis. In recent years, our understanding of the microorganisms living in the intestines (collectively called the microbiota) has grown; we now know that the microbiota plays an important role in many diseases, including CRC (Burns et al., 2015; Nakatsu et al., 2015). An average human's intestine contains more than 1014 microorganisms, including commensal bacteria, pathogenic bacteria, viruses, and fungi.
The gut microbiota actively metabolizes undigested food and substances shed from the intestinal cells, thereby generating energy and sending vital nutrients back to the host (Louis et al., 2014). Without the microbiota, the colon's epithelial cells will undergo autophagy and will fail to maintain their structure (Donohoe et al., 2011). In the normal colon, epithelial cells primarily use butyrate as energy. Tumor cells, however, require a large amount of glucose as their energy source to sustain growth, creating a large amount of lactate as the end product in the tumor microenvironment. In addition, to support the formation of new cell membranes, the tumor has increased needs for lipid biogenesis. So the change in the energy source preferred by proliferating tumor cells profoundly alters the nutrient composition of the tumor microenvironment.
In recent years, researchers have found a consistent connection between a dysfunctional gut microbiota (dysbiosis) and CRC (Shen et al., 2010; Wang et al., 2012; Burns et al., 2015; Nakatsu et al., 2015). Yet the directionality and the mediators between CRC and dysbiosis remain unclear (Yuan et al., 2018a). Given the drastic changes in the nutrient composition of the tumor microenvironment and the role of the microbiota in metabolism, there is undoubtedly a metabolic interaction between the tumor and its microbiota. In this study, we explored recent insights into metabolic interactions between the microbiota and tumor cells in CRC patients, focusing on the impact of microRNAs (miRNAs). We hypothesized that miRNAs are the mediators of the metabolites' effects (Fig. 1A).
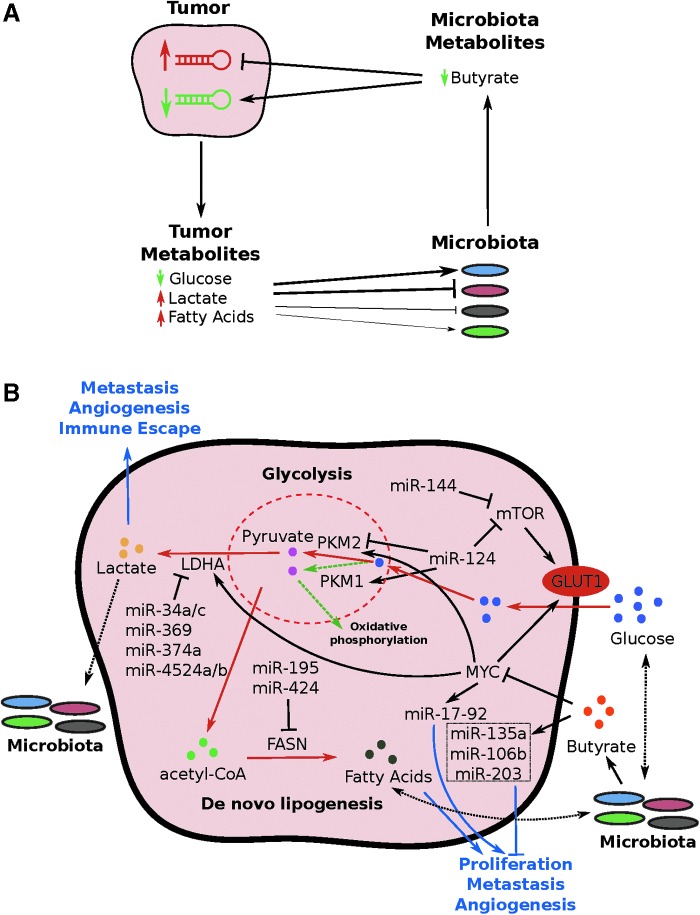
FIG. 1.
Tumor–microbiota metabolic interactions. (A) An overview of the tumor–microbiota metabolic interactions. The thickness of the line connecting tumor metabolites and microbiota shows the relative effects of tumor metabolites on microbiota composition. (B) A curated map of miRNA-mediated tumor–microbiota metabolic interactions. Red lines indicate the pathway upregulated in colorectal cancer relative to normal tissue. Blue lines indicate the overall downstream effects of the miRNAs or metabolites. Dotted green lines indicate the oxidative phosphorylation common in the normal cells. Dotted black lines indicate potential effects. miRNA, microRNA.
Tumor Nutrient Microenvironment Changes
The development of CRC entails a complex interplay between the epithelial cells, the microbiota, and the immune system in the tumor microenvironment (Tjalsma et al., 2012), and multiple signaling pathways play critical roles in both tumorigenesis and tumor progression. Tumor metabolism changes have been well studied and characterized. One of the hallmarks is an increase in glycolysis as the primary energy source, known as the Warburg effect (Warburg, 1956).
Several studies have found altered metabolite levels in both tissues and stools of CRC patients. In tissue samples (as compared with adjacent normal tissues), glucose levels were significantly lower, whereas levels of lactate and fatty acids were significantly higher (Hirayama et al., 2009; Weir et al., 2013; Brown et al., 2016). In stool samples of CRC patients, amino acid levels were higher than normal; levels of fatty acids, lower (Hirayama et al., 2009; Weir et al., 2013; Brown et al., 2016). This nutrient composition change in CRC patients correspond to the tumor's increased needs of glucose for energy and fatty acids for proliferation.
miRNAs play a critical role in regulating the metabolism of CRC patients and in sustaining the needs of tumor cells (Fig. 1B). The extracellular glucose is first transported into cells through the glucose transporter 1 (GLUT1) receptor, which is a downstream target of the mammalian target of rapamycin (mTOR) gene. In CRC patients, the mTOR gene is regulated by miR-144. Higher expression of miR-144 inhibits expression of the mTOR gene, leading to reduced glucose uptake by the tumor cells; thus, higher expression of miR-144 is associated with a good prognosis for CRC patients (Iwaya et al., 2012).
After the glucose is transported into the cytosol, it undergoes glycolysis—a process regulated by the alternative splicing of pyruvate kinase (PK). Higher levels of two PK isoforms, M1 (PKM1) and M2 (PKM2), in cells will lead to increased glycolysis, instead of oxidative phosphorylation (Sun et al., 2012; Taniguchi et al., 2015a, 2015b). These studies have found that overexpressing miR-124 (another regulator of the mTOR gene) in CRC cells can lead to higher PKM1:PKM2 ratios, thus inhibiting glycolysis and control tumor cell growth. The end product of glycolysis, pyruvate, will then be metabolized into lactate by lactate dehydrogenase A (LDHA), which is commonly upregulated in CRC patients. LDHA is a rate-limiting enzyme of glycolysis. A loss in LDHA expression is thus associated with decreased adenosine triphosphate (ATP) production and cell proliferation (Wang et al., 2015). In CRC cell lines, various miRNAs—including miR-34a/c, miR-369-3p, miR-374a, and miR-4524a/b—have been shown to inhibit LDHA expression (Wang et al., 2015). The lactate produced by the tumor cells can function as signaling molecules that further affect tumor cell metastasis, angiogenesis, and immune escape (Hirschhaeuser et al., 2011).
In addition to altered glucose metabolism, CRC cells also have altered macromolecule metabolism. We postulate that the reason for the higher levels of fatty acids in CRC patients' tissue samples (relative to their stool samples) is the increased need for membrane synthesis to support cell proliferation (Hirayama et al., 2009; Weir et al., 2013; Brown et al., 2016). One of the most important genes controlling this pathway is the fatty acid synthase (FASN) gene. The enzyme encoded by the FASN gene is critical for controlling the synthesis of lipids, a process required for cell membrane formation. In breast cancer and osteosarcoma, studies have found that miR-195 and miR-424 target the FASN gene, thus inhibiting cell proliferation, invasion, and metastasis (Mao et al., 2012; Long et al., 2013; Singh et al., 2015). Both of those miRNAs are significantly upregulated in CRC tissues, according to the Cancer Genome Atlas (TCGA) data set, suggesting such miRNA-mediated lipogenesis may also happen in CRC. In addition, because FASN is potentially important to T cell immunity (Buck et al., 2015), the dynamics of the miR-424/FASN axis in tumor and immune cell function are currently being worked out. Other pathways with downstream effects on metabolism, such as PTEN and AKT/PI3K, are also modulated by miRNAs (Song et al., 2008; Schee et al., 2013; Fang et al., 2014; Wang et al., 2014; Wei et al., 2014). Based on current evidence, it is clear that aberrant miRNA expression in CRC cells profoundly alters the nutrient composition of the tumor microenvironment.
Regulation of Host miRNAs by Microbiota Metabolites
In the healthy intestinal tract, the microbiota is dominated by the Bacteroidetes and Firmicutes phyla, which together comprise about 70% of the microbiota (Burns et al., 2015). Several taxa of bacteria have been implicated in the microbiota of CRC patients. In their stool samples, at the species level, a consistently higher abundance of Bacteroides fragilis and Fusobacterium nucleatum has been found. A higher abundance of the Bacteroidetes phylum and a lower abundance of the Firmicutes phylum have been observed in CRC patients. A recent meta-analysis of various CRC microbiota data sets found, for the tissue-associated microbiota, a consistently higher abundance of F. nucleatum, Parvimonas, and Streptococcus; nine studies in that meta-analysis found a consistently lower abundance of Faecalibacterium and Ruminococcaceae (Shah et al., 2018).
As mentioned previously, the microbiota is a powerhouse of metabolite production. It fills in many niche metabolic pathways that are not present in the human host. The gut microbiota produces about 70% of the energy required by the intestinal epithelial cells in the form of butyrate. Butyrate belongs to the short-chain fatty acids (SCFAs) that are produced by the gut microbiota through the fermentation of complex carbohydrates. In addition to being the major fuel source for normal intestinal epithelial cells, butyrate also functions as a histone deacetylase inhibitor (HDACi). This function is especially important in CRC patients, in part because the tumor cells switch from using butyrate to glucose as the major source of energy: the Warburg effect (Warburg, 1956). In CRC cells, high butyrate concentrations reduce MYC expression, which in turn reduces the levels of the miR-17-92 cluster miRNAs (Hu et al., 2015). The overexpression of miR-17-92a cluster in CRC cells has been shown to lead to cell proliferation, metastasis, and angiogenesis (Dews et al., 2010; Zhang et al., 2014; Ke et al., 2015). These suggest MYC/miR-17-92a cluster mediate butyrate's antitumor function in CRC cells. Butyrate also exerts antiproliferation effects on CRC in vitro, through directly regulating the miR-203, miR-106b, and miR-135a expression (Schlörmann et al., 2015; Han et al., 2016). Other members of the SCFA family, acetate and propionate, also act as HDACi. Because acetate and propionate can pass through the epithelial cells, they can exert their effect in T cells in the tumor microenvironment, by regulating the mTOR pathway (Park et al., 2015). Those effects suggest that butyrate and other SCFA members might help slow CRC progression through modulating tumor miRNA expression and the function of tumor-infiltrating T cell. Unfortunately, in the CRC microbiota, the fecal SCFA levels and the butyrate-producing bacteria levels are all lower than in the normal microbiota (Weir et al., 2013; Yuan et al., 2018a).
Conclusions and Perspectives
Our current knowledge supports the belief that gut microbes can alter tumor cells in CRC patients through the metabolites being produced. These metabolites can affect miRNA expression in the tumor cells, leading to alterations in many important signaling pathways. According to recent evidence, metabolites mediate interactions between the host's microbiota and tumor cells. Our view that miRNAs are the mediators of the metabolites' effects is supported by both experimental and computational findings, as reviewed earlier (Fig. 1B).
Currently, no tissue-level data set on the microbiota's metabolites exists. So, our laboratory recently used a nonhuman primate model to analyze global interactions between the microbiota and metabolites in healthy intestines (Yuan et al., 2018b). Doing so has brought us one step closer to finally understanding the metabolic interactions between the microbiota and the host. Of note, miRNAs are part of a complex highly dynamic web of interactions. Other recent studies suggest that miRNAs can directly affect the growth of bacteria and that tumor cells' miRNAs can affect the stromal and immune cells in the tumor microenvironment (Zhuang et al., 2012; Kohlhapp et al., 2015; Liu et al., 2016; Teng et al., 2018). A recent effort to develop a mouse model with humanized microbiota may further propel the field of tumor–microbiota metabolic interactions (Staley et al., 2017). Future research will require incorporating humanized microbiota animal models with a dual approach that leverages high-throughput genomics and metabolomics technologies.
Acknowledgments
We thank Dr. Mary Knatterud for assisting in article preparation. Because of space restrictions, we cannot cite many other significant contributions made by numerous researchers and laboratories in this potentially important and rapidly progressing field. S.S. is supported by research grants funded by the NIH R03CA219129 and C.Y. by the MnDrive—University of Minnesota Informatics Institute graduate fellowship.
Authors' Contributions
C.Y. and S.S. conceived the idea and wrote the article.
Disclosure Statement
The authors have no competing financial interests.
References
- Brown D.G., Rao S., Weir T.L., O'Malia J., Bazan M., Brown R.J., et al. (2016). Metabolomics and metabolic pathway networks from human colorectal cancers, adjacent mucosa, and stool. Cancer Metab 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck M.D., O'Sullivan D., and Pearce E.L. (2015). T cell metabolism drives immunity. J Exp Med 212, 1345–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M.B., Lynch J., Starr T.K., Knights D., and Blekhman R. (2015). Virulence genes are a signature of the microbiome in the colorectal tumor microenvironment. Genome Med 7, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews M., Fox J.L., Hultine S., Sundaram P., Wang W., Liu Y.Y., et al. (2010). The myc-miR-17 ∼ 92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res 70, 8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoe D.R., Garge N., Zhang X., Sun W., O'Connell T.M., Bunger M.K., et al. (2011). The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 13, 517–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Li H., Wang L., Hu J., Jin T., Wang J., et al. (2014). MicroRNA-17-5p promotes chemotherapeutic drug resistance and tumour metastasis of colorectal cancer by repressing PTEN expression. Oncotarget 5, 2974–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han R., Sun Q., Wu J., Zheng P., and Zhao G. (2016). Sodium butyrate upregulates miR-203 expression to exert anti-proliferation effect on colorectal cancer cells. Cell Physiol Biochem 39, 1919–1929 [DOI] [PubMed] [Google Scholar]
- Hirayama A., Kami K., Sugimoto M., Sugawara M., Toki N., Onozuka H., et al. (2009). Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res 69, 4918–4925 [DOI] [PubMed] [Google Scholar]
- Hirschhaeuser F., Sattler U.G.A., and Mueller-Klieser W. (2011). Lactate: a metabolic key player in cancer. Cancer Res 71, 6921–6925 [DOI] [PubMed] [Google Scholar]
- Hu S., Liu L., Chang E.B., Wang J.-Y., and Raufman J.-P. (2015). Butyrate inhibits pro-proliferative miR-92a by diminishing c-Myc-induced miR-17-92a cluster transcription in human colon cancer cells. Mol Cancer 14, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaya T., Yokobori T., Nishida N., Kogo R., Sudo T., Tanaka F., et al. (2012). Downregulation of miR-144 is associated with colorectal cancer progression via activation of mTOR signaling pathway. Carcinogenesis 33, 2391–2397 [DOI] [PubMed] [Google Scholar]
- Ke T.-W., Wei P.-L., Yeh K.-T., Chen W.T.-L., and Cheng Y.-W. (2015). MiR-92a promotes cell metastasis of colorectal cancer through PTEN-mediated PI3K/AKT pathway. Ann Surg Oncol 22, 2649–2655 [DOI] [PubMed] [Google Scholar]
- Kohlhapp F.J., Mitra A.K., Lengyel E., and Peter M.E. (2015). MicroRNAs as mediators and communicators between cancer cells and the tumor microenvironment. Oncogene 34, 5857–5868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., da Cunha A.P., Rezende R.M., Cialic R., Wei Z., Bry L., et al. (2016). The host shapes the gut microbiota via fecal microrna. Cell Host Microbe 19, 32–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long X.H., Mao J.H., Peng A.F., Zhou Y., Huang S.H., and Liu Z.L. (2013). Tumor suppressive microRNA-424 inhibits osteosarcoma cell migration and invasion via targeting fatty acid synthase. Exp Ther Med 5, 1048–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P., Hold G.L., and Flint H.J. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12, 661–672 [DOI] [PubMed] [Google Scholar]
- Mao J.H., Zhou R.P., Peng A.F., Liu Z.L., Huang S.H., Long X.H., et al. (2012). microRNA-195 suppresses osteosarcoma cell invasion and migration in vitro by targeting FASN. Oncol Lett 4, 1125–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu G., Li X., Zhou H., Sheng J., Wong S.H., Wu W.K.K., et al. (2015). Gut mucosal microbiome across stages of colorectal carcinogenesis. Nat Commun 6, 8727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Kim M., Kang S.G., Jannasch A.H., Cooper B., Patterson J., et al. (2015). Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol 8, 80–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schee K., Lorenz S., Worren M.M., Günther C.-C., Holden M., Hovig E., et al. (2013). Deep sequencing the microRNA transcriptome in colorectal cancer. PLoS One 8, e66165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlörmann W., Naumann S., Renner C., and Glei M. (2015). Influence of miRNA-106b and miRNA-135a on butyrate-regulated expression of p21 and Cyclin D2 in human colon adenoma cells. Genes Nutr 10, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah M.S., DeSantis T., Yamal J.-M., Weir T., Ryan E.P., Cope J.L., et al. (2018). Re-purposing 16S rRNA gene sequence data from within case paired tumor biopsy and tumor-adjacent biopsy or fecal samples to identify microbial markers for colorectal cancer. PLoS One 13, e0207002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X.J., Rawls J.F., Randall T., Burcal L., Mpande C.N., Jenkins N., et al. (2010). Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 1, 138–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R.L., Miller K.D., and Jemal A. (2018). Cancer statistics, 2018. CA Cancer J. Clin 68, 7–30 [DOI] [PubMed] [Google Scholar]
- Singh R., Yadav V., Kumar S., and Saini N. (2015). MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep 5, 17454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song B., Wang Y., Kudo K., Gavin E.J., Xi Y., and Ju J. (2008). miR-192 regulates dihydrofolate reductase and cellular proliferation through the p53-microRNA circuit. Clin Cancer Res 14, 8080–8086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley C., Kaiser T., Beura L.K., Hamilton M.J., Weingarden A.R., Bobr A., et al. (2017). Stable engraftment of human microbiota into mice with a single oral gavage following antibiotic conditioning. Microbiome 5, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Zhao X., Zhou Y., and Hu Y. (2012). miR-124, miR-137 and miR-340 regulate colorectal cancer growth via inhibition of the Warburg effect. Oncol Rep 28, 1346–1352 [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Sugito N., Kumazaki M., Shinohara H., Yamada N., Matsuhashi N., et al. (2015a). Positive feedback of DDX6/c-Myc/PTB1 regulated by miR-124 contributes to maintenance of the Warburg effect in colon cancer cells. Biochim Biophys Acta 1852, 1971–1980 [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Sugito N., Kumazaki M., Shinohara H., Yamada N., Nakagawa Y., et al. (2015b). MicroRNA-124 inhibits cancer cell growth through PTB1/PKM1/PKM2 feedback cascade in colorectal cancer. Cancer Lett 363, 17–27 [DOI] [PubMed] [Google Scholar]
- Teng Y., Ren Y., Sayed M., Hu X., Lei C., Kumar A., et al. (2018). Plant-derived exosomal MicroRNAs shape the gut microbiota. Cell Host Microbe 24, 637–652.e8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjalsma H., Boleij A., Marchesi J.R., and Dutilh B.E. (2012). A bacterial driver-passenger model for colorectal cancer: beyond the usual suspects. Nat Rev Microbiol 10, 575–582 [DOI] [PubMed] [Google Scholar]
- Wang J., Wang H., Liu A., Fang C., Hao J., and Wang Z. (2015). Lactate dehydrogenase A negatively regulated by miRNAs promotes aerobic glycolysis and is increased in colorectal cancer. Oncotarget 6, 19456–19468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Cai G., Qiu Y., Fei N., Zhang M., Pang X., et al. (2012). Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J 6, 320–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang Q., Li M., Jiang S., and Wang X. (2014). MicroRNA-375 inhibits colorectal cancer growth by targeting PIK3CA. Biochem Biophys Res Commun 444, 199–204 [DOI] [PubMed] [Google Scholar]
- Warburg O. (1956). On the origin of cancer cells. Science 123, 309–314 [DOI] [PubMed] [Google Scholar]
- Wei Z., Cui L., Mei Z., Liu M., and Zhang D. (2014). miR-181a mediates metabolic shift in colon cancer cells via the PTEN/AKT pathway. FEBS Lett 588, 1773–1779 [DOI] [PubMed] [Google Scholar]
- Weir T.L., Manter D.K., Sheflin A.M., Barnett B.A., Heuberger A.L., and Ryan E.P. (2013). Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS One 8, e70803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Burns M.B., Subramanian S., and Blekhman R. (2018a). Interaction between host MicroRNAs and the gut microbiota in colorectal cancer. MSystems 3, e00205-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C., Graham M., and Subramanian S. (2018b). Microbiota-metabolites interactions in non-human primate gastrointestinal tract. BioRxiv. [Epub ahead of print]; DOI: 10.1101/454496 [DOI] [Google Scholar]
- Zhang G., Zhou H., Xiao H., Liu Z., Tian H., and Zhou T. (2014). MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig Dis Sci 59, 98–107 [DOI] [PubMed] [Google Scholar]
- Zhuang G., Wu X., Jiang Z., Kasman I., Yao J., Guan Y., et al. (2012). Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J 31, 3513–3523 [DOI] [PMC free article] [PubMed] [Google Scholar]