Abstract
Background
4-Hydroxyphenylacetic acid (4HPAA) is an important building block for synthesizing drugs, agrochemicals, biochemicals, etc. 4HPAA is currently produced exclusively via petrochemical processes and the process is environmentally unfriendly and unsustainable. Microbial cell factory would be an attractive approach for 4HPAA production.
Results
In the present study, we established a microbial biosynthetic system for the de novo production of 4HPAA from glucose in Escherichia coli. First, we compared different biosynthetic pathways for the production of 4HPAA. The yeast Ehrlich pathway produced the highest level of 4HPAA among these pathways that were evaluated. To increase the pathway efficiency, the yeast Ehrlich pathway enzymes were directedly evolved via error-prone PCR. Two phenylpyruvate decarboxylase ARO10 and phenylacetaldehyde dehydrogenase FeaB variants that outperformed the wild-type enzymes were obtained. These mutations increased the in vitro and in vivo catalytic efficiency for converting 4-hydroxyphenylpyruvate to 4HPAA. A tunable intergenic region (TIGR) sequence was inserted into the two evolved genes to balance their expression. Regulation of TIGR for the evolved pathway enzymes further improved the production of 4HPAA, resulting in a 1.13-fold increase in titer compared with the fusion wild-type pathway. To prevent the toxicity of a heterologous pathway to the cell, an Esa quorum-sensing (QS) circuit with both activating and repressing functions was developed for inducer-free productions of metabolites. The Esa-PesaR activation QS system was used to dynamically control the biosynthetic pathway of 4HPAA in E. coli, which achieved 17.39 ± 0.26 g/L with a molar yield of 23.2% without addition of external inducers, resulting in a 46.4% improvement of the titer compared to the statically controlled pathway.
Conclusion
We have constructed an E. coli for 4HPAA production with the highest titer to date. This study also demonstrates that the combination of directed evolution of pathway enzymes and dynamic pathway regulation using a QS circuit is a powerful strategy of metabolic engineering for the productions of metabolites.
Electronic supplementary material
The online version of this article (10.1186/s13068-019-1438-3) contains supplementary material, which is available to authorized users.
Keywords: 4-Hydroxyphenylacetic acid, Directed evolution, Dynamic pathway regulation, Quorum-sensing system, Escherichia coli
Background
Phenolic acids are aromatic acids that contain a phenol ring and at least one organic carboxylic acid group. Phenolic acids play an important role in human health and have wide applications in food, cosmetic and pharmaceutical industries. 4-Hydroxyphenylacetic acid (4HPAA) has received much attention because of its numerous applications. 4HPAA is used in the synthesis of penicillin G, atenolol, benzoprofen, and agrochemicals, etc. [1, 2]. 4HPAA is an active component of Rhodiola rosea [2] and the Chinese herbs Aster tataricus (fan hun cao). Aster tataricus is widely used in China for the treatment of pneumonia, HBV, and carcinomas [3–5]. Furthermore, 4HPAA possesses anxiolytic [6], antiplatelet [7] and hepatoprotective [8] properties. In addition, 4HPAA was considered as a potential hypopigmenting agent [9] and an inhibitor of hypertonicity and hypoxia [10].
4HPAA can be obtained by chemical synthesis from different substrates such as anisol, p-cresol, phenol, benzyl phenyl ether, or hydroxymandelic acid [11, 12]. However, the chemical routes have some drawbacks, including a requirement of elevated temperatures and pressures for the reaction, and the use of expensive solvents. Thus, biotechnological approach has been explored as an alternative. A biotransformation reaction using nitrilase has been used for 4HPAA production [2]. Koma et al. [1] engineered an Escherichia coli for the production of 4HPAA from glucose. Overexpression of the indole-3-pyruvate/phenylpyruvate decarboxylase gene ipdC from Azospirillum brasilense NBRC102289, and the phenylacetaldehyde dehydrogenase gene feaB from E. coli in a tyrosine overproducing E. coli strain resulted in the production of 6.1 mM (0.93 g/L) 4HPAA with a yield of 13.2% (mol/mol) in a shake flask culture. However, the titer of 4HPAA in engineered microorganisms is much lower than that of other aromatic compounds [13, 14]. Thus, further work is required to increase the production of 4HPAA.
Quorum-sensing (QS) systems has been considered as an auto-induction system that is regulated by cell density. The Lux and Esa QS systems are the main QS systems that have been reported to date. Recently, an Esa QS system from Pantoea stewartii has been engineered to automatically downregulate the competing pathway, significantly improving the production of myo-inositol, glucaric acid and shikimic acid [15].
In this study, we first compared different biosynthetic pathways of 4HPAA (Fig. 1). Then, the biosynthetic pathway enzymes of 4HPAA were directedly evolved and mediated using a tunable intergenic region (TIGR) sequence. Finally, the TIGR-mediated biosynthetic pathway was dynamically regulated using a quorum-sensing circuit. The resulting E. coli produced 17.39 ± 0.26 g/L 4HPAA without addition of external inducers, which is the highest value reported to date.
Fig. 1.
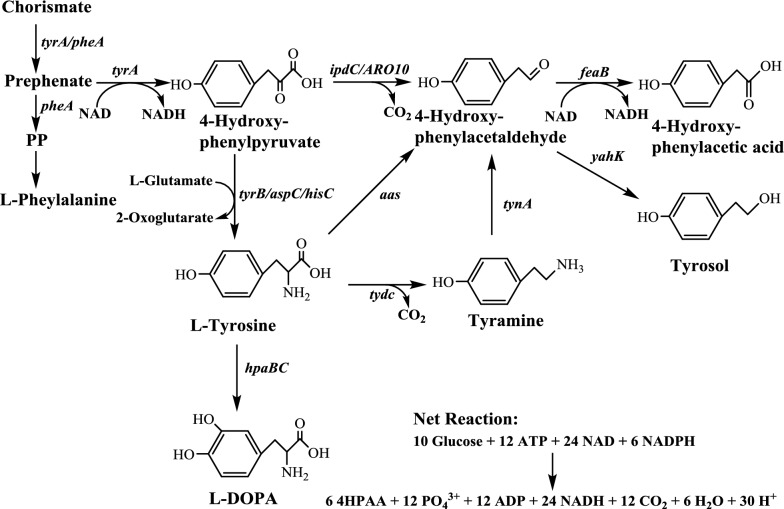
Biosynthetic pathway of 4-hydroxyphenylacetic acid (4HPAA). tyrA/pheA fused chorismate mutase/prephenate dehydrogenase gene, ipdC indole-3-pyruvate/phenylpyruvate decarboxylase gene from Azospirillum brasilense, ARO10 phenylpyruvate decarboxylase gene from Saccharomyces cerevisiae, tyrB tyrosine aminotransferase gene, aspC aspartate aminotransferase gene, hisC histidinol-phosphate aminotransferase gene, hpaBC 4-hydroxyphenylacetate 3-monooxygenase gene from E. coli W, aas aromatic aldehyde synthase gene from Petroselinum crispum, tydc tyrosine/DOPA decarboxylase gene from Pseudomonas putida KT2440, tynA tyramine oxidase gene, feaB phenylacetaldehyde dehydrogenase gene, yahK NADPH-dependent aldehyde reductase gene
Methods
Strains, plasmids and primers
The bacterial strains, plasmids, and primers used in this study are listed in Table 1. E. coli DH5α was used for gene cloning in this study. The L-DOPA overproducing strain E. coli DOPA-30N [16] was used as the host strain for the production of 4HPAA.
Table 1.
Strains, plasmids and primers used in this study
| Name | Description | Source/purpose |
|---|---|---|
| Strain | ||
| E. coli DH5α | supE44 Δ(lacZYA-argF) U169 (Φ80lacZ ΔM15) hsdR17 recA endA1 gyrA96 thi-1 relA1 | Invitrogen |
| E. coli BW 25113 | lacI q rrnB T14 ΔlacZ WJ16 hsdR514 ΔaraBAD AH33 ΔrhaBAD LD78 | [17] |
| E. coli DOPA-30N | L-DOPA overproducer | [16] |
| Plasmid | ||
| pBbB2k-GFP | Expressing plasmid, BglBrick vectors, tet promoter, pBBR1 ori, kanr, Addgene plasmid #35345 | [18] |
| pBbB2k-ipdC-L-feaB | pBbB2k derivatives harboring the fusion gene cluster of ipdC from Azospirillum brasilense and feaB from E. coli with a (GSG)2 linker | This study |
| pBbB2k-ARO10-L-feaB | pBbB2k derivatives harboring the fusion gene cluster of ARO10 from Saccharomyces cerevisiae and feaB from E. coli with a (GSG)2 linker | This study |
| pBbB2k-aas-L-feaB | pBbB2k derivatives harboring the fusion gene cluster of aas from Petroselinum crispum and feaB from E. coli with a (GSG)2 linker | This study |
| pBbB2k-tydc-L-tynA-L-feaB | pBbB2k derivatives harboring the fusion gene cluster of tydc from Pseudomonas putida KT2440, tynA and feaB from E. coli with a (GSG)2 linker | This study |
| pBbB2k-ARO10*- L-feaB* | pBbB2k derivatives harboring the fusion gene cluster of the evolved ARO106D5 from S. cerevisiae and feaB2E1 from E. coli with a (GSG)2 linker | This study |
| pBbB2k-ARO10*-feaB* | pBbB2k derivatives harboring the gene cluster of the evolved ARO106D5 from S. cerevisiae and feaB2E1 from E. coli, one operon | This study |
| pBbB2k-ARO10*-TIGR-feaB* | pBbB2k derivatives harboring the TIGR-mediated gene cluster of the evolved ARO106D5 from S. cerevisiae and feaB2E1 from E. coli | This study |
| pZBK | BglBrick/ePathBrick expression vector, pBBR1 ori, P37 promoter, kanr | [19] |
| pZBK-PesaSIC-PesaRAS | Quorum-sensing plasmid, pBBR1 ori, P37 promoter, kanr | This study |
| pZBK-PesaRAS-4HPAA | pZBK-PesaSIC-PesaRAS harboring the TIGR-mediated gene cluster of the evolved ARO106D5 from S. cerevisiae and feaB2E1 from E. coli | This study |
| Primer | ||
| FeaBF | CCGGAATTCAAAAGATCTGGTATGACAGAGCCGCATGTAGCAG (EcoRI, BglII) | PCR for feaB |
| FeaBR | CGCGGATCCTTAATACCGTACACACACCGACTTAGTTTCACACCAAC (BamHI) | |
| TynAF | CCGGAATTCAAAAGATCTGGTATGGGAAGCCCCTCTCTGTATTCTG (EcoRI, BglII) | PCR for tynA |
| TynAR | CGCGGATCCACCTGAACCCTTATCTTTCTTCAGCGCCC (BamHI) | |
| ARO10F | CCGGAATTCAAAAGATCTTTTCGGAATTAAGGAGGTAATAAATATGGCACCTGTTACAATTG (EcoRI, BglII) | PCR for ARO10 |
| ARO10R | CGCGGATCCACCACTACCTTTTTTATTTCTTTTAAGTG (BamHI) | |
| AROF | GATAGAGAAAAGAATTCAAAAGATCTTTTCGGAATTAAGGAGG (EcoRI, BglII) | error-prone PCR forARO10-feaB |
| FeaR | TACTCGAGTTTGGATCCTTAATACCGTACACACACCGACTT (BamHI) | |
| epAROF | ACCTTCGATTCCGACCTCATTAAGC | error-prone PCR for ARO10 |
| epAROR | CGCGGATCCTTAATACCGAACACACACCGACTTAGTTTCACACCAAC | |
| TIGRs-F(X) | CTAGCTAGCGCCTAGCAAGATCTCCTGATCCCGGTGC (NheI) | PCR for TIGRs |
| TIGRs-R(A) | TTTTCCTTTTGCGGCCGCGGATACAGTATCTGCGGTACCCTAG (NotI) | |
| Qaro10F | CGACACCTTGTTTCCAACCG | qRT-PCR for ARO10 |
| Qaro10R | TGAAGTCACCAGGAACACCG | |
| QfeaBF | GCCAACGCTGGTGGTAAATC | qRT-PCR for feaB |
| QfeaBR | GCCTCTTCTCCATCCGCTAC | |
Construction of plasmids
The codon-optimized ipdC (GenBank accession number X99587) from A. brasilense NBRC102289, tydc (encoding tyrosine/DOPA decarboxylase, GenBank accession number AE015451) from Pseudomonas putida KT2440, and aas (encoding aromatic aldehyde synthase, GenBank accession number M96070) from Petroselinum crispum (encoding aromatic aldehyde synthase) were synthesized by GENEWIZ, Inc. (Suzhou, China). The feaB (encoding phenylacetaldehyde dehydrogenase) and tynA (encoding tyramine oxidase) genes were amplified from E. coli. The ARO10 (encoding phenylpyruvate decarboxylase) was amplified from Saccharomyces cerevisiae. The protein fusion is a common approach of substrate channeling for coordinating expression of cascade enzymes [19–21]. Thus, a linker encoding (GSG)2 was inserted into the two genes, and the TAA stop codon of the front gene was deleted. The fusion pathway was cloned into pBbB2k-GFP to obtain pBbB2k-ipdC-L-feaB, pBbB2k-ARO10-L-feaB, pBbB2k-tydc-L-tynA-L-feaB, pBbB2k-aas-L-feaB, respectively.
Generation of random mutagenesis libraries using error-prone PCR and screening
MEGAWHOP-PCR [22] was performed for the construction of libraries. The random mutagenesis libraries of the fusion ARO10-feaB gene cluster were constructed through error-prone PCR. Primer pairs AROF/FeaR were used for amplification the ARO10-feaB gene cluster using pBbB2k-ARO10-L-feaB as a template. The PCR reaction mixture (50 μL) consisted of 5 mM MgCl2, 0.3 mM MnCl2, 0.2 mM each of dATP, dGTP, dCTP and dTTP, and 2.5 U of rTaq DNA polymerase. The PCR products were then used as megaprimers to perform MEGAWHOP-PCR using pBbB2k-ARO10-L-feaB as a template. Following the MEGAWHOP-PCR, DpnI digestion (20 U) of the surplus template in the PCR reaction mixture was performed at 37 °C overnight, then DpnI was inactivated at 80 °C for 20 min. The PCR products were transformed into E. coli DH5a cells, plated on LB agar, and then pooled together after overnight growth to create a liquid library.
The mutant plasmid was recovered from the library and transferred into the L-DOPA overproducer E. coli DOPA-30N. Then, the resulting transformants were plated on LB agar with 50 mg/mL kanamycin and 120 ng/mL anhydrotetracycline. After overnight growth, individual colonies were inoculated into a 48-well deep-well microplate (4.6 mL) containing 1 mL of the fermentation medium and then incubated at 37 °C and 1000 rpm for 72 h on a MBR-420FL shaker (TAITEC Corporation, Saitama-ken, Japan). Because L-DOPA can be easily oxidized to dopachrome and then polymerized non-enzymatically to form melanin, the culture broth of the host strain E. coli DOPA-30N became black [16]. Because 4HPAA shares the same precursor (tyrosine) with L-DOPA, light color cultures were selected for further shake flask analysis of 4HPAA production.
Creating TIGR libraries and screening
To replace the linker between ARO10* and feaB* in pBbB2k-ARO10*-L-feaB* with a two-restriction enzyme sequence, the ARO10* and feaB* genes were amplified from pBbB2k-ARO10*-L-feaB*. The two genes were then cloned into pBbB2k-GFP to obtain pBbB2k-ARO10*-feaB*.
TIGRs were synthesized using PCR to assemble the oligonucleotides into chimeric DNA sequences as described by Pfleger et al. [23] and Li et al. [19]. The assembled products were purified using a nucleotide removal column, amplified using the end-specific primers TIGRs-F(X)/TIGRs-R(A), and then cloned into the NheI/NotI sites of pBbB2k-ARO10*-feaB* to obtain the plasmid libraries pBbB2k-ARO10*-TIGRs-feaB*. The plasmid libraries were transferred into component DOPA-30N to generate the mutant library.
The TIGR library was plated on LB agar plates containing 50 μg/mL kanamycin and 120 ng/mL anhydrotetracycline. The plates were then incubated at 30 °C overnight. Single colonies were inoculated into a 48-well deep-well microplate (4.6 mL) containing 1 mL of fermentation medium, and then incubated at 30 °C and 1000 rpm for 72 h on a MBR-420FL shaker (TAITEC, Japan). Lighter color cultures were selected for further shake flask analysis of 4HPAA production.
Construction of a quorum-sensing circuit
The quorum-sensing fragment containing the signal molecule acyl-homoserine lactone (AHL) synthase gene esaI, the transcriptional regulator esaRI70V, and PeasS and PesaR promoters was synthesized by GENEWIZ, Inc (Suzhou, China) and then cloned into the MluI/SalI of pZBK [19] to obtain the plasmid pZBK-PeasSIC-PeasRAS (Additional file 1: Figure S1A). The TIGR-mediated gene cluster of the evolved ARO10 and feaB was cloned into the multiple cloning site 2 (MCS2) of pZBK-PeasSIC-PeasRAS to obtain pZBK-PesaRAS-4HPAA.
Production of 4HPAA
A single colony was inoculated into 5 mL of LB medium in a falcon tube then cultured overnight at 30 °C. The overnight seed culture was then inoculated into 50 mL of fermentation medium with a starting OD600 of 0.1. The fermentation medium contained (per liter) tryptone (10 g), yeast extract (5 g), NaCl (10 g), glucose (40 g), KH2PO4 (0.6 g), Na2HPO4·7H2O (2.56 g), and 10 mL of trace element solution. The trace element solution contained (per liter) FeSO4·7H2O (10 g), ZnSO4·7H2O (2.2 g), MnSO4·4H2O (0.58 g) CuSO4·5H2O (1 g), (NH4)6Mo7O24·4H2O (0.1 g), and Na2B4O7·10H2O (0.2 g). The pH of the medium was adjusted to 7.0. The main cultures were incubated at 30 °C and 200 rpm. When the OD600 reached 2.5, the cultures were induced with 120 ng/mL anhydrotetracycline. After induction, the cultures were incubated at 30 °C and 200 rpm for 72 h.
Fed-batch fermentation was carried out in a 2 L bioreactor (MiniBox 2L*2 Parallel Bioreactor System, T&J Bio-engineering (Shanghai) Co. LTD, Shanghai, China) containing 1.2 L of fermentation medium with an initial OD600 of approximately 0.1. Fermentation was carried out at 30 °C with an airflow of 1.2 L/min and a starting agitation rate of 400 rpm. For E. coli containing the statically controlled pathway, 120 ng/mL anhydrotetracycline was added as an inducer after 6 h. The pH was controlled at 7.0 by automatic addition of NH4OH. Dissolved oxygen was maintained above 25% by adjusting the agitation rate. The feed solution (pH 7.0,) contained 500 g/L glucose and 30 g/L MgSO4·7H2O. The feed was introduced continuously into the fermenter using a pH-stat feeding strategy. Once the glucose was exhausted, the pH rose rapidly. When the pH was greater than 7.0 by 0.1 of a pH unit, the feed was automatically added to the fermenter. Samples were periodically withdrawn, and the following parameters were measured: OD600, residual glucose concentrations, and 4HPAA concentrations. Fermentation experiments were performed in duplicate.
Catalytic efficiency assays
For in vitro activity assay, E. coli DOPA-30N harboring pBbB2k-ARO10-L-feaB or pBbB2k-ARO10*-L-feaB* were pre-inoculated in 5 mL of LB medium overnight and then inoculated into 50 mL the fermentation medium containing kanamycin. The cultures were incubated at 30 °C and 200 rpm until OD600 reached 2.5 and then induced with 120 ng/mL anhydrotetracycline. After 44 h, the culture broth was disrupted directly using an Ultra-high Pressure Cell Disrupter (JN-3000Plus, Guangzhou Juneng Nano & Bio Technology Co. LTD, China), and then centrifuged to remove cell debris. The supernatant was selected as a crude enzyme solution for catalytic efficiency assay. The total protein concentration in the crude extract was determined by a Bradford assay using a Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, CA, USA). The catalytic efficiency assay was conducted as follows: 4-hydroxyphenylpyruvic acid of 10 mM was added to the crude enzyme. The reaction was conducted at 37 °C for 6 h. Then, the reaction mixture was immediately placed on ice broth to stop the reaction. The product (4HPAA) concentration in the reaction solution was determined immediately by HPLC.
For in vivo activity assay, E. coli BW25113 was transformed with pBbB2k-ARO10-L-feaB and pBbB2k-ARO10*-L-feaB*, respectively. Single colonies were pre-inoculated into 5 mL of LB medium containing kanamycin and cultured overnight at 30 °C and 200 rpm. A 500 μL overnight cultures were inoculated into 50 mL the fermentation medium containing kanamycin. The cultures were incubated at 30 °C and 200 rpm until OD600 reached 2.5 and then induced with 120 ng/mL anhydrotetracycline at 37 °C and 200 rpm. Three hours after induction, 4-hydroxyphenylpyruvic acid was added to a final concentration of 1.0 g/L and incubated at 30 °C and 200 rpm for 6 h. Growth and the concentration of 4HPAA were analyzed.
Assay
Growth was monitored by measuring the optical density at 600 nm. 4HPAA in the supernatants was analyzed using a Shimadzu HPLC system (LC-20A, Shimadzu, Japan) equipped with an Inertsil ODS-SP column (5 μm, 4.6 × 150 mm, GL Sciences Inc., Tokyo, Japan). The mobile phase was 0.2% TFA in methanol, and the flow rate was 0.5 mL/min at 30 °C. The methanol concentration was increased from 14 to 45% for 20 min, and then decreased to 14%. This concentration was then maintained for 10 min. A photodiode array detector (SPD-M20A) operating at 222 nm was used, and a standard curve was constructed from serial dilutions of a standard stock solution. Glucose concentrations were determined using glucose oxidase and a glucose assay kit (Shanghai Rongsheng Biotech Corporation, Shanghai, China).
Statistical analysis
All experiments were conducted in triplicate, and the data were averaged and presented as the mean ± standard deviation. One-way analysis of variance followed by Tukey’s test was used to determine significant differences using the OriginPro (version 7.5) package. Statistical significance was defined as p < 0.05.
Results and discussion
Screening biosynthetic pathways
Three biosynthetic pathways (Fig. 1) have been developed for the production of tyrosol; namely, the Ehrlich pathway via the ipdC [1] and the yeast ARO10 [24, 25], the aas (encoding aromatic aldehyde synthase) pathway [26] and the tydc-tyo (encoding tyrosine decarboxylase and tyramine oxidase, respectively) pathway [27]. Because 4HPAA shares the same precursor, 4-hydroxyphenylacetaldehyde (4HPAAL), with tyrosol, we first constructed these pathways and evaluated their 4HPAA productions (Fig. 1). These pathways were then introduced into the L-DOPA overproducing E. coli strain DOPA-30N for shake flask analysis of 4HPAA. No L-DOPA was detected in the four engineered strains, indicating that, the rate of 4HPAA production was higher than that for L-DOPA (data not shown). Thus, E. coli DOPA-30N was used as the parent strain for the production of 4HPAA in this study. The introduction of all pathways resulted in the production of 4HPAA (Table 2), indicating that these pathways can be used for 4HPAA production. Moreover, the yeast Ehrlich pathway produced the highest titer of 4HPAA (Table 2). The recombinant strain harboring the yeast Ehrlich pathway produced 3.08 ± 0.05 g/L 4HPAA with a molar yield of 13.4%. The same result was obtained for tyrosol. The yeast Ehrlich pathway also exhibited the highest biosynthetic efficiency for tyrosol production among the four pathways [1, 24–27]. Therefore, the yeast Ehrlich pathway was selected as the biosynthetic pathway for the production of 4HPAA in this study.
Table 2.
4-Hydroxyphenylacetic acid production in E. coli DOPA-30N harboring different biosynthetic pathways
| Host strain | Plasmid | OD600 | Titer (g/L) | Yield (%, mol/mol) |
|---|---|---|---|---|
| E. coli DOPA-30N | pBbB2k-ipdC-L-feaB | 16.28 ± 0.45 | 2.32 ± 0.05 | 11.8 ± 0.2 |
| pBbB2k-ARO10-L-feaB | 17.36 ± 0.19 | 3.08 ± 0.05 | 13.4 ± 0.2 | |
| pBbB2k-aas-L-feaB | 17.13 ± 1.44 | 2.11 ± 0.06 | 9.3 ± 0.2 | |
| pBbB2k-tydC-L-tynA-L-feaB | 15.44 ± 0.42 | 2.41 ± 0.04 | 12.1 ± 0.2 | |
| pBbB2k-ARO10*-L-feaB* | 16.68 ± 0.35 | 5.64 ± 0.06 | 22.9 ± 0.3 | |
| pBbB2k-ARO10*-TIGR-feaB* | 17.12 ± 0.32 | 6.58 ± 0.16 | 27.5 ± 0.3 |
Data represent the means of three replicates and standard deviations
Although only ipdC Ehrlich pathway was reported for the production of 4HPAA in literatures, our results demonstrated that three other pathways based on the reported tyrosol biosynthetic pathways can be used for the production of 4HPAA. Moreover, the 4HPAA titer that was obtained via our yeast Ehrlich pathway was 1.33-fold as high as that obtained via the ipdC Ehrlich pathway.
Directed evolution of the biosynthetic pathway
Independent of the enzyme structure and catalytic mechanism, directed evolution of enzyme is widely used to alter the coding sequence and improve the catalytic activities of enzymes. Thus, to increase the efficiency of the biosynthetic pathway, the fusion gene cluster of ARO10-feaB was directedly co-evolved by error-prone PCR. L-DOPA can be easily oxidized to dopachrome and then polymerized non-enzymatically to form the black pigment melanin [28]. Because the preparation of 4HPAA utilizes the same precursors (4-hydroxyphenylpyruvate/tyrosine) as L-DOPA (Fig. 1), 4HPAA biosynthesis competes with L-DOPA. The higher the activity of the fusion gene cluster, the lower the production of L-DOPA, hence, the culture broth becomes lighter. A total of 792 colonies were used for the assay of 4HPAA production in deep-well microplate cultures. Twenty-three colonies with lighter colors were selected for further shake flask analysis. As shown in Fig. 2a, strains no. 12 and 23 produced the highest level of 4HPP (4.66 ± 0.14 and 4.65 ± 0.10 g/L, respectively). These mutant plasmids, denoted as 2E1 and 4F3, were isolated and sent for sequencing. The sequencing results are presented in Additional file 1: Table S1. In plasmid 2E1, one base mutant (C1869T) in ARO10 and three base mutants (T72G, T1195C, and A1491T) were observed in feaB. In plasmid 4F3, two base mutants (A1682G and A1842G) in ARO10 and three base mutants (C165G, A609T, and T1296) were observed in feaB. None of the base mutants in ARO10 produced an amino acid change. One amino acid mutant of feaB was observed in plasmid 2E1 (I24M) and 4F3 (N55K).
Fig. 2.
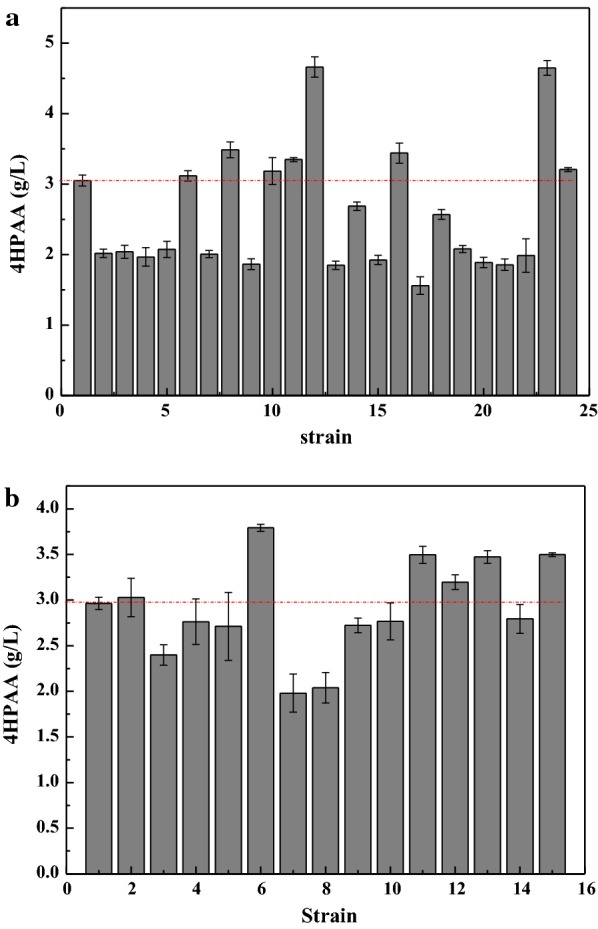
4HPAA production by E. coli DOPA-30N harboring mutant gene clusters from error-prone PCR. a The mutant gene clusters caused by the co-evolution of the ARO10-L-feaB. b The mutant gene clusters caused by the single-evolution of ARO10. Cells were grown at 30 °C and 200 rpm for 72 h. See “Methods” for other details. E. coli DOPA-30N (pBbB2k-ARO10-L-feaB) (strain no. 1) was set the control. The data represent the means of three replicates, and error bars represent standard deviations
Because no amino acid mutant in ARO10 was observed after the co-evolution of the ARO10-feaB, the ARO10 in pBbB2k-ARO10-L-feaB was single-evolved using error-prone PCR. Of 900 colonies used for the assay in deep-well microplate cultures, twelve colonies with lighter colors were selected for further shake flask analysis. As shown in Fig. 2b, strains no. 6, 11, 13, and 15 produced higher levels of 4HPAA than the wild-type strain. The mutant plasmids, denoted as 6D5, 9F5, 10A3, and 10G5, were isolated and sent for sequencing. DNA sequencing revealed some base mutations and amino acid mutations in ARO10 (Additional file 1: Table S1). Two amino acid mutations (F138L and D218G) were observed in plasmid 6D5, and one mutation (V451I) was observed in plasmid 10A3.
To investigate the effects of these mutated genes on the production of 4HPAA, we combined these mutated genes to create a biosynthetic pathway and introduced the pathway into E. coli DOPA-30N. Figure 3 shows that mutations of two genes indeed increased the production of 4HPAA. The engineered strain harboring both mutated genes produced higher levels of 4HPAA than the strain that contained only one mutated gene. The combination of the mutated ARO10 from plasmid 6D5 with the mutated feaB from plasmid 2E1 resulted in the highest titer of 4HPAA (5.64 ± 0.06 g/L). This combination resulted in approximately a 90% increase in the production of 4HPAA compared to the wild-type genes.
Fig. 3.
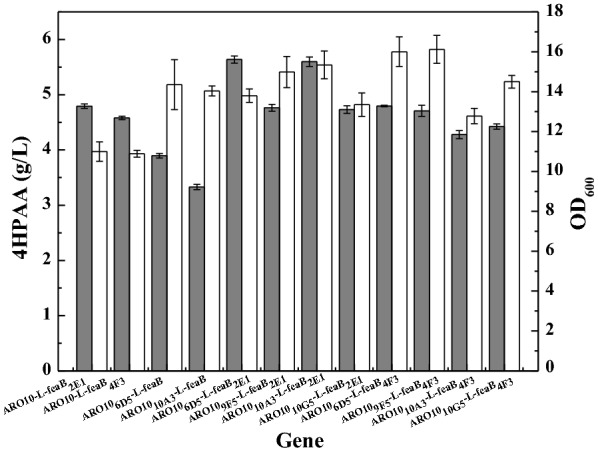
4HPAA production by E. coli DOPA-30N harboring the fusion gene cluster of different combinations of the evolved S. cerevisiae ARO10 and E. coli feaB. Growth (white bar); 4HPAA concentration (gray bar). Cells were grown at 30 °C and 200 rpm for 72 h. See “Methods” for other details. The data represent the means of three replicates, and error bars represent standard deviations
To evaluate the applicability of the evolved pathway enzymes in microbial production of 4HPAA, we first performed in vitro crude extract enzyme assays to measure their catalytic activities. 4-Hydroxyphenylpyruvic acid of 10 mM was added into the cell crude extracts of E. coli DOPA-30N harboring the wild-type or mutant plasmid, respectively. The catalytic efficiencies were measured by testing the 4HPAA formation rate. As shown in Fig. 4, directed evolution resulted in an increase in the in vitro catalytic efficiency of the pathway to 45.55 ± 2.43 mg/h/g protein from 23.67 ± 2.66 mg/h/g protein (Fig. 4), indicating that the evolved pathway was more efficient at converting 4-hydroxyphenylpyruvate to 4HPAA than the wild-type pathway in vitro.
Fig. 4.
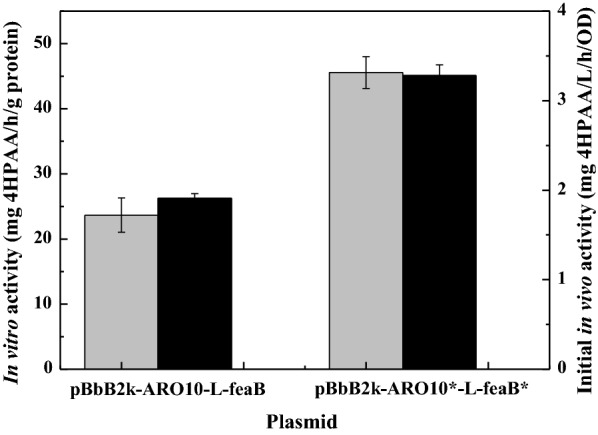
In vitro (gray bar) and initial in vivo (black bar) catalytic efficiency of the pathway. The data represent the means of three replicates, and error bars represent standard deviations
Then, the initial in vivo efficiency of converting 4-hydroxyphenylpyruvic acid to 4HPAA was analyzed through whole-cell bioconversion experiments. The results were compared in Fig. 4. The in vivo efficiency of the evolved pathway was 3.28 ± 0.12 mg/L/h/OD, while the in vivo efficiency of the parent pathway was 1.91 ± 0.05 mg/L/h/OD, indicating a 72% increment in activity level. This result is consistent with the in vitro crude enzyme assays. This result indicates that the evolved pathway was more efficient at converting 4-hydroxyphenylpyruvate to 4HPAA than the wild-type pathway both in vitro and in vivo.
Directed evolution is the most commonly used approach used to significantly improve the performance of pathway enzymes. Applying direction evolution, we isolated mutant variants with improved activity than the wild-type enzymes. The best combination of the ARO10 and FeaB mutants resulted in a 90% increase in the production of 4HPAA. The increased 4HPAA titer may be as a result of increases in the activity of pathway enzymes.
The unbalanced expression of multiple genes may result in the accumulation of toxic metabolic intermediates, thereby reducing product titers. Thus, Pfleger et al. [23] developed a combinatorial engineering approach for coordinating the expression of cascade enzymes by generating libraries of TIGRs. They applied the TIGR approach to balance gene expression in the MEV pathway, resulting in a sevenfold increase in the production of mevalonate. We also applied the TIGR approach to improve the production of zeaxanthin [19] and pinene [29]. Thus, we constructed a library of TIGRs to balance the expression of the evolved ARO10* and feaB*. The library of TIGRs was inserted between the two genes. A total of 1080 colonies were used for the assay of 4HPAA production in deep-well microplate cultures. Twenty colonies with lighter colors were selected for further shake flask analysis. As shown in Fig. 5a, strains nos. 4, 10, and 12 produced higher levels of 4HPAA. We again assayed the production of 4HPAA in the three strains (Fig. 5b). Strain no. 12 produced higher levels of 4HPAA, 6.58 g/L (Fig. 5b). Regulation of TIGR resulted in a 16.7% increase in the production of 4HPAA compared with the fusion evolved pathway. The TIGR in strain no. 12 was sequenced and the results are presented in Additional file 1: Table S2.
Fig. 5.
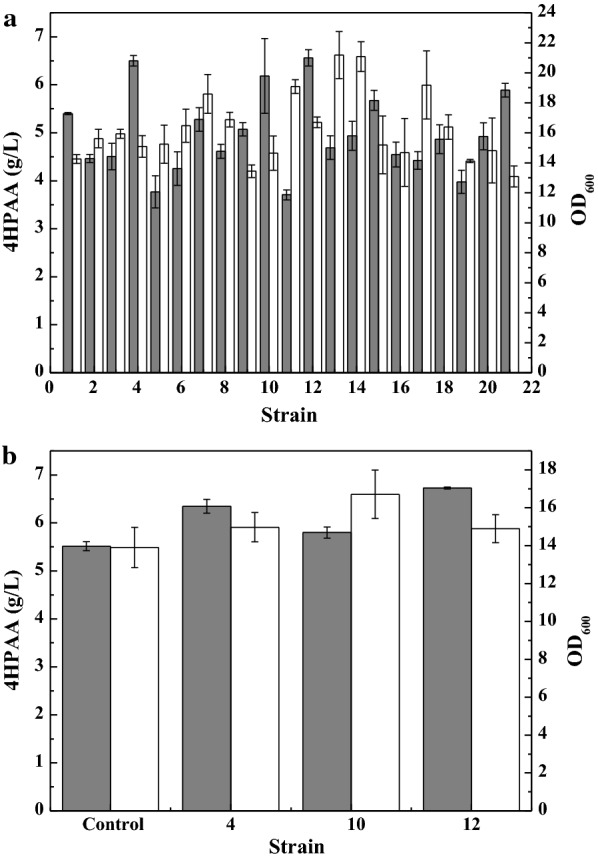
Growth (white bar) and 4HPAA production (gray bar) for E. coli DOPA-30N harboring the TIGR-mediated gene cluster of the evolved S. cerevisiae ARO10* and E. coli feaB*. a Colonies with lighter colors from the TIGR library; b strains with higher levels of 4HPAA. Cells were grown at 30 °C and 200 rpm for 72 h. See “Methods” for other details. E. coli DOPA-30N (pBbB2k-ARO10*-L-feaB*) was set the control. The data represent the means of three replicates, and error bars represent standard deviations
Dynamic regulation of the biosynthetic pathway
In the above study, the inducible expression system was used to regulate the biosynthetic pathway of 4HPAA. However, this system has some disadvantages, including leaky expression, lack of dynamic control, and the prohibitively high cost of inducers that are associated with large-scale production. QS functions to control gene expression in response to cell density, and has been applied to several biosynthetic pathways to produce glucaric acid, myo-inositol, shikimate [15], β-lactamase exoenzyme [30], isopropanol [31], and bisabolene [32] in E. coli, and para-hydroxybenzoic acid in Saccharomyces cerevisiae [33]. The Lux and Esa QS systems are the main QS systems that have been reported to date. The QS system requires two regulatory elements, a signaling molecule (also known as an autoinducer) that is constitutively produced, and a receptor protein that binds the autoinducer and acts as a transcriptional activator or repressor. The Esa QS system from P. stewartii, has been engineered for both transcriptional activation and repression by regulating EsaR binding sites on the promoter [34]. In the absence of signal molecules, 3-oxohexanoyl-homoserine lactone (AHL), the transcriptional regulator in EsaRI70V, binds the PesaS or PesaR promoter and activates or represses transcription, respectively [15, 34, 35]. Thus, we constructed an Esa QS plasmid, pZBK-PesaSIC-PesaRAS, with both activating and repressing functions (Additional file 1: Figure S1A). To characterize the function of the QS circuit, GFP and mCherry genes were successively inserted into the multiple cloning site 1 (MSC1) and MSC2 to obtain pZBK-PesaS-GFP-PesaR-mCherry (Additional file 1: Figure S1B). The relative fluorescence of GFP decreased with cell growth (Additional file 1: Figure S1C). From Additional file 1: Figure S1C, we can also see that the relative fluorescence of mCherry increased with cell growth. These results demonstrate that the PesaS-controlled gene was indeed repressed and that the PesaR-controlled gene was activated by the induction of AHL in response to cell density.
Thus, the TIGR-mediated gene cluster of the evolved ARO106D5 and feaB2E1 was cloned into the MCS2 site of the Esa QS plasmid pZBK-PesaSIC-PesaRAS to obtain pZBK–PesaRAS-4HPAA, which was then transferred into E. coli DOPA-30N. In this strain, the pathway was controlled by the PesaR promoter, indicating that the expression of the pathway was activated by the induction of AHL in response to cell density (Fig. 6a). To compare the production of 4HPAA between the recombinant strain harboring the QS-controlled and the statically controlled pathway, fed-batch cultures were conducted in 2-L bioreactors. The results are presented in Fig. 6b. The engineered strain harboring the statically controlled pathway produced 4HPAA of 11.88 ± 0.82 g/L with a molar yield of 14.1% at 84 h. The engineered strain harboring the QS-controlled pathway produced higher level of 4HPAA titer, which achieved 17.39 ± 0.26 g/L with a molar yield of 23.2% (mol/mol) at 76 h. This indicates that QS-regulation increased the production of 4HPAA by 46.4% compared to the statically controlled pathway.
Fig. 6.
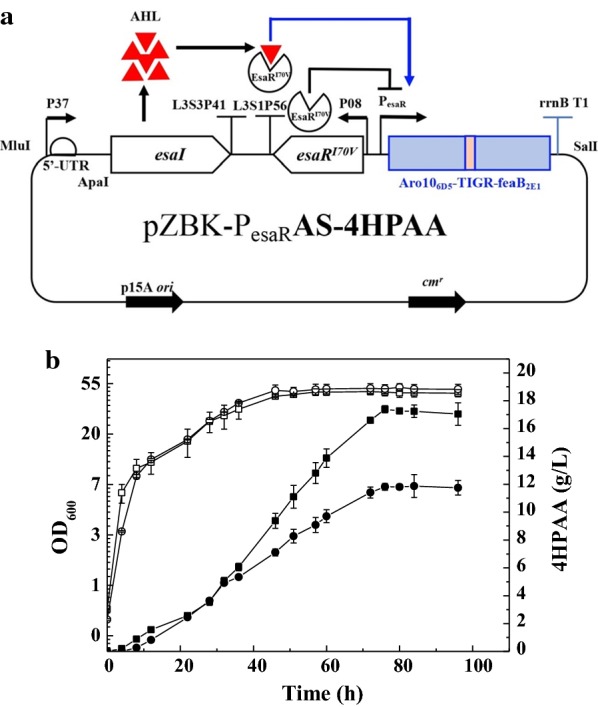
a The quorum-sensing (QS) system for 4HPAA production. In the absence of 3-oxohexanoyl-homoserine lactone (AHL, red triangle), the transcriptional regulator EsaRI70V binds the PesaR promoter and represses the transcription of the pathway enzymes. EsaI proteins synthesizes autoinducer AHL, which can diffuse in or out cells and binds to the transcriptional regulator EsaRI70V. At the threshold level of AHL, which accumulates in proportion to cell density, the transcriptional regulator EsaRI70V binds to AHL, leading to dissociation of EsaRI70Vfrom the PesaR promoter, and the EsaRI70V-AHL complex activates the transcription of the pathway enzymes. b Fed-batch cultures of E. coli DOPA-30N harboring the QS-controlled (squares) and the statically controlled (circles) pathway for 4HPAA production. Growth (open: open square, open circle); 4HPAA concentration (black: filled square, filled circle). Experiments were conducted in duplicate, and measurements are presented as the means with standard deviations
A Lux QS system from Vibrio fischeri has been used to construct a toggle switch for redirecting metabolic flux from the TCA cycle to the production of isopropanol, resulting in approximately a threefold increase in the titer [31]. This Lux QS system still requires external IPTG inducer to turn on LuxR/LuxI expression. Recently, an inducer-free Lux QS system has been used to dynamically regulate heterologous mevalonate and bisabolene biosynthetic pathways to improve the inducer-free production of bisabolene by 44% in E. coli [32]. An EsaR-PesaS QS system from P. stewartii has been constructed to improve glucaric acid in E. coli by dynamically repressing glycolysis, leading to a fivefold increase in its titer [15]. In their study, the QS system was used to repress the competing pathway, driving the flux toward the biosynthetic pathway. In our study, we constructed an Esa-PesaR activation QS system to dynamically control the biosynthetic pathway for the inducer-free production of 4HPAA, leading to a 46.4% improvement in the titer. This is the first report on an Esa activation QS system for dynamically regulating a biosynthetic pathway. The 4HPAA titer obtained in this study is 18.7-fold the maximum reported in literatures (0.93 g/L).
Conclusion
We designed three biosynthetic pathways of 4HPAA based on tyrosol synthesis. Of these pathways, the yeast Ehrlich pathway exhibited the highest efficiency for the production of 4HPAA. To increase the pathway efficiency, the yeast Ehrlich pathway enzymes were directedly evolved using error-prone PCR. After directed evolution, two ARO10 and FeaB variants that outperformed the wild-type enzymes were obtained. These mutations increased the efficiency for converting 4-hydroxyphenylpyruvate to 4HPAA both in vitro and in vivo. The TIGR regulation for the evolved pathway enzymes further improved 4HPAA production. An Esa QS circuit with both activating and repressing functions was developed. The Esa-PesaR activation QS system was used to dynamically control the biosynthetic pathway of 4HPAA, which yielded 17.39 ± 0.26 g/L 4HPAA without addition of external inducers. The titer is the highest value that has been obtained to date.
Additional file
Additional file 1: Figure S1. (A) Esa quorum-sensing (QS) plasmid. (B) Activation and repression of genes using quorum-sensing circuit. Table S1. Mutations on the evolved gene. Table S2. The sequence of the TIGR.
Authors’ contributions
YPS performed all of the experimental works. LSF and ZBY performed the fed-batch fermentation and the assay. JZL designed the study and wrote the manuscript. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Not applicable.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Funding
This work was funded by the National Natural Science Foundation of China (Grant No. 21276289 and 201808248), the Natural Science Foundation of Guangdong Province (No. 2015A030311036 and 2018A030310255), the Project of the Scientific and Technical Program of Guangdong Province (No. 2015A010107004) and the Project of the Scientific and Technical Program of Guangzhou (No. 201607010028) for their financial support.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Yu-Ping Shen, Email: shenyup@mail2.sysu.edu.cn.
Lai San Fong, Email: fongls@mail2.sysu.edu.cn.
Zhi-Bo Yan, Email: yanzhb3@mail2.sysu.edu.cn.
Jian-Zhong Liu, Phone: +86-20-84110115, Email: lssljz@mail.sysu.edu.cn.
References
- 1.Koma D, Yamanaka H, Moriyoshi K, Ohmoto T, Sakai K. Production of aromatic compounds by metabolically engineered Escherichia coli with an expanded shikimate pathway. Appl Environ Microb. 2012;78(17):6203–6216. doi: 10.1128/AEM.01148-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thakur N, Kumar V, Thakur S, Sharma N, Sheetal, Bhalla TC. Biotransformation of 4-hydroxyphenylacetonitrile to 4-hydroxyphenylacetic acid using whole cell arylacetonitrilase of Alcaligenes faecalis MTCC 12629. Process Biochem. 2018;73:117–123. [Google Scholar]
- 3.Ng TB, Liu F, Lu YH, Cheng CHK, Wang ZT. Antioxidant activity of compounds from the medicinal herb Aster tataricus. Comp Biochem Phys C. 2003;136(2):109–115. doi: 10.1016/s1532-0456(03)00170-4. [DOI] [PubMed] [Google Scholar]
- 4.Du L, Mei HF, Yin X, Xing YQ. Delayed growth of glioma by a polysaccharide from Aster tataricus involve upregulation of Bax/Bcl-2 ratio, activation of caspase-3/8/9, and downregulation of the Akt. Tumor Biol. 2014;35(3):1819–1825. doi: 10.1007/s13277-013-1243-8. [DOI] [PubMed] [Google Scholar]
- 5.Yen MH, Lee JJ, Yeh CF, Wang KC, Chiang YW, Chiang LC, Chang JS. Yakammaoto inhibited human coxsackievirus B4 (CVB4)-induced airway and renal tubular injuries by preventing viral attachment, internalization, and replication. J Ethnopharmacol. 2014;151(3):1056–1063. doi: 10.1016/j.jep.2013.11.049. [DOI] [PubMed] [Google Scholar]
- 6.Vissiennon C, Nieber K, Kelber O, Butterweck V. Route of administration determines the anxiolytic activity of the flavonols kaempferol, quercetin and myricetin—are they prodrugs? J Nutr Biochem. 2012;23(7):733–740. doi: 10.1016/j.jnutbio.2011.03.017. [DOI] [PubMed] [Google Scholar]
- 7.Kim DH, Jung EA, Sohng IS, Han JA, Kim TH, Han MJ. Intestinal bacterial metabolism of flavonoids and its relation to some biological activities. Arch Pharm Res. 1998;21:17–23. doi: 10.1007/BF03216747. [DOI] [PubMed] [Google Scholar]
- 8.Zhao HQ, Jiang ZH, Chang XM, Xue HT, Yahefu W, Zhang XY. 4-Hydroxyphenylacetic acid prevents acute APAP-induced liver injury by increasing phase ii and antioxidant enzymes in mice. Front Pharmacol. 2018;9:653. doi: 10.3389/fphar.2018.00653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen KC, Chang CS, Chien YC, Wang HW, Wu WC, Wu CS, Chiang HM. Tyrosol and its analogues inhibit alpha-melanocyte-stimulating hormone induced melanogenesis. Int J Mol Sci. 2013;14(12):23420–23440. doi: 10.3390/ijms141223420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu ZY, Xi RG, Zhang ZR, Li WP, Liu Y, Jin FG, Wang XB. 4-Hydroxyphenylacetic acid attenuated inflammation and edema via suppressing HIF-1 alpha in seawater aspiration-induced lung injury in rats. Int J Mol Sci. 2014;15(7):12861–12884. doi: 10.3390/ijms150712861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mutsukado M, Yamada S. Methods for preparing 4-hydroxyphenylacetic acids. United States Patent 4412082; 1983.
- 12.Vallejos JC, Christidis Y. Preparation process for hydroxyphenylacetic acids. United States Patent US005395964A; 1995.
- 13.Averesch NJH, Kromer JO. Metabolic engineering of the shikimate pathway for production of aromatics and derived compounds-present and future strain construction strategies. Front Bioeng Biotechnol. 2018;6:32. doi: 10.3389/fbioe.2018.00032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, Shen XL, Rey J, Yuan QP, Yan YJ. Recent advances in microbial production of aromatic natural products and their derivatives. Appl Microbiol Biotechnol. 2018;102(1):47–61. doi: 10.1007/s00253-017-8599-4. [DOI] [PubMed] [Google Scholar]
- 15.Gupta A, Reizman IMB, Reisch CR, Prather KLJ. Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat Biotechnol. 2017;35(3):273–279. doi: 10.1038/nbt.3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wei T, Cheng BY, Liu JZ. Genome engineering Escherichia coli for L-DOPA overproduction from glucose. Sci Rep-Uk. 2016;6:30080. doi: 10.1038/srep30080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97(12):6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee TS, Krupa RA, Zhang FZ, Hajimorad M, Holtz WJ, Prasad N, Lee SK, Keasling JD. BglBrick vectors and datasheets: a synthetic biology platform for gene expression. J Biol Eng. 2011;5(1):12. doi: 10.1186/1754-1611-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li XR, Tian GQ, Shen HJ, Liu JZ. Metabolic engineering of Escherichia coli to produce zeaxanthin. J Ind Microbiol Biotechnol. 2015;42(4):627–636. doi: 10.1007/s10295-014-1565-6. [DOI] [PubMed] [Google Scholar]
- 20.Wang C, Yoon SH, Jang HJ, Chung YR, Kim JY, Choi ES, Kim SW. Metabolic engineering of Escherichia coli for alpha-farnesene production. Metab Eng. 2011;13(6):648–655. doi: 10.1016/j.ymben.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Kirby J, Nishimoto M, Chow RWN, Baidoo EEK, Wang G, Martin J, Schackwitz W, Chan R, Fortman JL, Keasling JD. Enhancing terpene yield from sugars via novel routes to 1-deoxy-d-xylulose 5-phosphate. Appl Environ Microb. 2015;81(1):130–138. doi: 10.1128/AEM.02920-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyazaki K. MEGAWHOP cloning: a method of creating random mutagenesis libraries via megaprimer PCR of whole plasmids. Method Enzymol. 2011;498:399–406. doi: 10.1016/B978-0-12-385120-8.00017-6. [DOI] [PubMed] [Google Scholar]
- 23.Pfleger BF, Pitera DJ, Smolke CD, Keasling JD. Combinatorial engineering of intergenic regions in operons tunes expression of multiple genes. Nat Biotechnol. 2006;24(8):1027–1032. doi: 10.1038/nbt1226. [DOI] [PubMed] [Google Scholar]
- 24.Li XL, Chen ZY, Wu YF, Yan YJ, Sun XX, Yuan QP. Establishing an artificial pathway for efficient biosynthesis of hydroxytyrosol. ACS Synth Biol. 2018;7(2):647–654. doi: 10.1021/acssynbio.7b00385. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Li XB, Jiang JL, Liu ZN, Qiao B, Li FF, Cheng JS, Sun XC, Yuan YJ, Qiao JJ, et al. Convergent engineering of syntrophic Escherichia coli coculture for efficient production of glycosides. Metab Eng. 2018;47:243–253. doi: 10.1016/j.ymben.2018.03.016. [DOI] [PubMed] [Google Scholar]
- 26.Chung D, Kim SY, Ahn JH. Production of three phenylethanoids, tyrosol, hydroxytyrosol, and salidroside, using plant genes expressing in Escherichia coli. Sci Rep-Uk. 2017;7:2578. doi: 10.1038/s41598-017-02042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satoh Y, Tajima K, Munekata M, Keasling JD, Lee TS. Engineering of a tyrosol-producing pathway, utilizing simple sugar and the central metabolic tyrosine, Escherichia coli. J Agric Food Chem. 2012;60(4):979–984. doi: 10.1021/jf203256f. [DOI] [PubMed] [Google Scholar]
- 28.Claus H, Decker H. Bacterial tyrosinases. Syst Appl Microbiol. 2006;29(1):3–14. doi: 10.1016/j.syapm.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 29.Niu FX, He X, Wu YQ, Liu JZ. Enhancing production of pinene in Escherichia coli by using a combination of tolerance, evolution, and modular co-culture engineering. Front Microbiol. 2018;9:1623. doi: 10.3389/fmicb.2018.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pai A, Tanouchi Y, You LC. Optimality and robustness in quorum sensing (QS)-mediated regulation of a costly public good enzyme. Proc Natl Acad Sci USA. 2012;109(48):19810–19815. doi: 10.1073/pnas.1211072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soma Y, Hanai T. Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab Eng. 2015;30:7–15. doi: 10.1016/j.ymben.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 32.Kim EM, Woo HM, Tian T, Yilmaz S, Javidpour P, Keasling JD, Lee TS. Autonomous control of metabolic state by a quorum sensing (QS)-mediated regulator for bisabolene production in engineered E. coli. Metab Eng. 2017;44:325–336. doi: 10.1016/j.ymben.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Williams TC, Averesch NJH, Winter G, Plan MR, Vickers CE, Nielsen LK, Kromer JO. Quorum-sensing linked RNA interference for dynamic metabolic pathway control in Saccharomyces cerevisiae. Metab Eng. 2015;29:124–134. doi: 10.1016/j.ymben.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 34.Shong J, Collins CH. Engineering the esaR promoter for tunable quorum sensing-dependent gene expression. ACS Synth Biol. 2013;2(10):568–575. doi: 10.1021/sb4000433. [DOI] [PubMed] [Google Scholar]
- 35.Shong J, Huang YM, Bystroff C, Collins CH. Directed evolution of the quorum-sensing regulator EsaR for increased signal sensitivity. ACS Chem Biol. 2013;8(4):789–795. doi: 10.1021/cb3006402. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. (A) Esa quorum-sensing (QS) plasmid. (B) Activation and repression of genes using quorum-sensing circuit. Table S1. Mutations on the evolved gene. Table S2. The sequence of the TIGR.
Data Availability Statement
Not applicable.