Abstract
Background
In recent years, an increasing number of studies have proved that circulating miRNAs could be used for the early diagnosis of cardiovascular diseases and even play vital roles in the evaluation of therapeutic effects or prognosis. This study was conducted to examine the correlation between serum microRNAs and hyperlipidemia to provide a theoretical basis for the early screening and intervention of atherosclerotic cardiovascular diseases (ASCVD).
Methods
The serum samples and clinical data of 122 patients with hyperlipidemia and 168 healthy subjects were collected. Related clinical information was statistically analyzed for the two groups. Expression of circulating miRNAs was detected by miRNA microarray analysis and further verified by reverse transcription-quantitative PCR (RT-qPCR).
Results
Statistical analysis of clinical information revealed a significant difference in the incidence of ASCVD between the two groups. The MiRNA microarray analysis (n = 10) showed 22 miRNAs with significantly different expression, among which 12 showed upregulation, and the others showed downregulation. Those possessing obvious differences and stable expression in the miRNA microarray, including miRNA-191-3p, miRNA-933, and miRNA-425-3p, were chosen for further investigation using RT-qPCR. The results demonstrated that several miRNAs were related to lipid metabolism disorders, especially miRNA-933. The area under the curve (AUC) of miRNA-933 in distinguishing the hyperlipidemia and ASCVD patients was 0.739 (95% CI, 0.682–0.795; P < 0.01) and 0.703 (95% CI, 0.643–0.763, P < 0.01), respectively.
Conclusions
In conclusion, miRNA-191-3p, miRNA-933, and miRNA-425-3p may be depressed in the peripheral circulation of patients with lipid metabolism disorders (mainly LDL). Circulating miRNA-933 could be a feasible predictor for ASCVD at the early stage.
Electronic supplementary material
The online version of this article (10.1186/s12944-019-1046-z) contains supplementary material, which is available to authorized users.
Introduction
Atherosclerotic cardiovascular disease (ASCVD) is defined as a group of clinical diseases mostly caused by endothelial dysfunction based on atherosclerosis (AS), including coronary heart disease, stroke, and other various cardiovascular diseases [1]. These diseases seriously threaten human life and result in high mortality and disability rates all over the world. The pathogenesis of ASCVD is extremely complex, mainly including lipid infiltration, endothelial damage reactions, and chronic inflammatory damage. The dysregulation of lipid metabolism, particularly the low-density lipoprotein (LDL), is commonly considered the foundational cause resulting in AS [1, 2]. Numerous studies have shown that LDL could enter in the intima or middle membrane through the damaged vascular endothelium, eventually forming oxidized LDL (ox-LDL), which was widely believed to be the fundamental pathological agent of atheromatous plaques [3]. A variety of factors influence the formation of atherosclerosis, including hypertension, hyperlipidemia, hyperglycemia, obesity, smoking, etc. These risk factors comprise the majority of the key targets for prevention or treatment of ASCVD, among which lipid metabolism disorders, especially the abnormal elevation of circulating LDL, is the most important point. Presently, controlling circulating LDL levels has become the most important measure to prevent ASCVD [4, 5].
MiRNAs are a class of endogenous, highly conserved, non-coding RNAs, mostly consisting of 19–24 nucleotides. MiRNAs could regulate gene expression on the posttranscriptional level by binding to the 3’UTR of mRNA, which may either inhibit the translation of protein from mRNA or promote the degradation of mRNA. These RNAs participate in nearly all physiological and pathological processes in organisms [6]. For ASCVD, various miRNAs have been reported that could affect lipid metabolism and the formation of atherosclerosis through regulating the function of vascular smooth muscle cells, endothelial cells, macrophages, and so on [7–9]. Numerous studies have proved that miRNAs could exist stably in peripheral blood circulation and have shown good physiological characteristics to tolerate different temperatures, pH, storage times, and even repeated freezing and thawing. Additional studies revealed the dynamic alterations of circulating miRNA expression could be detected under different pathological conditions, including various cardiovascular diseases, thereby indicating that detecting the different expression of miRNAs in peripheral blood circulation may help in the early diagnosis of atherosclerotic cardiovascular diseases [10, 11].
In fact, an increasing number of studies have shown that noncoding ribonucleic acids could be used for the early diagnosis of cardiovascular diseases and even play vital roles in the evaluation of therapeutic effects or prognosis [12, 13]. Wang GK et al. demonstrated that plasma miR-208a might be a novel biomarker for acute myocardial infarction [14]. Fang Y et al. found that miR-10a could regulate pro-inflammatory phenotypes in atherosclerosis-susceptible endothelium [15]. The exact involvement of circulating miRNAs in cardiovascular diseases remains to be clarified. The aim of this study was to compare circulating miRNA profiles in hyperlipidemic patients characterized by hyper-LDL-C in comparison to normolipidemic controls to provide assistance for early screening of ASCVD.
Materials and methods
Study subjects
Subjects, including healthy volunteers, outpatients, and in-patients, were recruited from October 2017 to December 2018 at Yangzhou University, Affiliated Hospital of China. According to the blood lipid level (mainly the serum LDL-C level) of patients, the subjects were divided into a hyperlipidemia group and a healthy control group.
The inclusion criteria for patients with hyper-LDL-C was according to the classification criteria of dyslipidemia in the population in 2016 Chinese guideline for the management of dyslipidemia in adults, defined hyper-LDL-C as serum LDL-C level ≥ 3.4 mmol/L(130 mg/dl). As there are small number of patients with isolated hyper-LDL-C in clinical, some with mixed dyslipidemia were also recruited. The exclusion criteria of the study was as follows: age > 80 years, autoimmune diseases, systemic inflammatory diseases, cancer, rheumatic heart diseases, thyroid disease, or liver and kidney dysfunction. Patients with a history of major trauma and surgery, or women who were pregnant and breastfeeding were also not included in this study. Clinical information, including personal history, heart disease, cerebrovascular disease, and genetic history information, etc., was collected from all patients. Hypertension was diagnosed as systolic pressure 140 mmHg and/or diastolic blood pressure 90 mmHg. The diagnostic criteria of diabetes were fasting plasma glucose 7.0 mmol/L or random glucose 11.1 mmol/L. The diagnosis of ASCVD was made if the patient had a history of one or more of the following: acute coronary syndromes, myocardial infarction, stable or unstable angina, coronary or other arterial revascularization, stroke, and TIA. Similar principles were used to collect other medical history information. Subjects who continuously or cumulatively smoked for more than 6 months were defined as smokers. Alcoholics were defined as individuals with an average daily consumption of alcohol equivalent to that of pure alcohol (125 g), which lasted for more than 10 years. The study was approved by the ethics committee of The First People’s Hospital of Yangzhou. All the patients were given informed consent and recruited into the ongoing prospective study.
Blood sample collection and RNA extraction
In a calm environment, fasting venous blood was collected in a test tube without anticoagulant. After centrifugation, the serum was removed and stored at − 80 °C. The miRNA expression profiles of 10 serum samples (n = 5 for each group) were detected by miRNA microarray (Agilent miRNA chip V2.4, Agilent Technologies, Santa Clara, CA, USA). Serum miRNA was isolated using a miRcute serum miRNA kit (Tiangen Biotech, Beijing, China), according to the manufacturer’s instructions. Total RNA was quantified by the NanoDrop ND-2000 (Thermo Scientific) and the RNA integrity was assessed using Agilent Bioanalyzer 2100 (Agilent Technologies). The sample labeling, microarray hybridization and washing were performed based on the manufacturer’s standard protocols. Briefly, total RNA was dephosphorylated, denaturated and then labeled with Cyanine-3-CTP. After purification the labeled RNAs were hybridized onto the microarray. After washing, the arrays were scanned with the Agilent Scanner G2505C (Agilent Technologies). Differentially expressed miRNAs were then identified through fold change (FC) as well as P value calculated using t-test. The threshold set for up- and down-regulated genes was a fold change≥2.0 and a P value≤0.05.
For RT-qPCR experiments, the purity and concentration of miRNA extracted from the serum was determined with a spectrophotometer. Only RNA samples with an A260/A280 ratio of 1.9–2.1 were used for reverse transcription of cDNA.
RT-qPCR determination
The miRcute miRNA cDNA First-Strand Synthesis kit (Tiangen Biotech) was used for reverse transcription of miRNA extracted, according to the manufacturer’s instructions. The reaction system (20 μL) contained the following: 10 μL 2 x miRNA RT reaction buffer, 2 μL miRNA RT enzyme mix, and 8 μL total RNA. The reaction conditions were as follows: 25 °C for 5 min, 42 °C for 60 min, and 95 °C for 3 min. After cDNA synthesis, DEPC water was used to dilute the product in a 1:5 ratio, and the samples were stored at − 20 °C. A miRcute-enhanced miRNA Fluorescence Quantitative Detection kit (SYBR Green), miRNA primers (miRNA-320b, miRNA-933, miRNA-191-3p, and U6), and miRcute miRNA fluorescence quantitative detection reagents were used for detection, with a fluorescence quantitative PCR (Roche, Basel, Switzerland) instrument. The 20-μL reaction system was prepared according to the manufacturer’s instructions as follows: 2 μL template cDNA (5 times dilution), 10 μL 2x miRcute miRNA premix, 0.4 μL miRNA/U6 primers, 0.4 μL reverse primer, and 7.2 μL RNase-free water. The reaction conditions were as follows: 95 °C for 15 min; 94 °C for 20 s, 64 °C for 30 s, and 72 °C for 34 s for 5 cycles, no fluorescence signal; 94 °C for 20 s; and 64 °C for 30 s for a total of 40 cycles. All samples were examined with at least two duplicates. We used U6 as an internal reference in the qPCR experiment, the relative amount of miRNA normalized to U6 was calculated with the Equation 2-ΔΔCT, in which ΔΔCT = (CT miRNA − CTU6) target − (CT miRNA − CT U6) control.
Statistical methods
The experimental data was analyzed with SPSS13.0 software (SPSS, Inc., Chicago, IL, USA). The mean ± standard deviation (x ± s) was used as the quantitative index, and differences between the two groups were compared by t tests. The count data were compared with the χ2 test. Receiver operating characteristic curves were also used to analyze relevant experimental data. P < 0.05 was considered statistically significant.
Results
We first collected and analyzed the clinical information of the patients between the hyperlipidemia and control groups. Table 1 shows the basic clinical data of the subjects of the two groups. According to the table, there were no significant differences in age, smoking history, drinking history, and incidence of diabetes between the two groups. Similarly, differences in the history of stroke, hypertension, and body mass index were also not obvious. Nevertheless, our dates revealed a significant difference in the incidence of both carotid atherosclerosis and ASCVD between the two groups. In terms of biochemical indicators, uric acid showed a significant difference between the two groups (Table 1).
Table 1.
Basic clinical data of subjects in the LDL group and control group
| Terms | Hyperlipidemia group (n = 122) | Normal group (n = 168) | P-value |
|---|---|---|---|
| Age (years) | 50.06 ± 1.09 | 48.601 ± 0.84 | 0.407 |
| Sex (male/female) | 74/48 | 69/99 | 0.378 |
| History of smoking (Smoker/Nonsmoker) | 45/77 | 60/108 | 0.522 |
| History of alcoholism (Yes/No) NoNONo)Nondrinker) | 42/80 | 64/104 | 0.520 |
| Body mass index (BMI) | 24.56 ± 0.22 | 23.72 ± 0.24 | 0.064 |
| Hypertension (Yes/No) | 38/84 | 68/100 | 0.103 |
| Diabetes (Yes/ No) | 26/96 | 50/118 | 0.106 |
| Carotid plaque (Yes/ No) | 46/56 | 47/99 | 0.039 |
| Not checked | 20 | 22 | |
| History of stroke (Yes/ No) | 25/97 | 22/146 | 0.092 |
| History of coronary heart disease | 33/89 | 34/134 | 0.174 |
| ASCVD | 53/69 | 41/119 | 0.002 |
| Fatty liver | 41/81 | 34/134 | 0.101 |
| Random blood glucose | 5.81 ± 0.27 | 5.33 ± 0.06 | 0.126 |
| Creatinine | 70.6 ± 1.36 | 71.78 ± 3.17 | 0.349 |
| Urea nitrogen | 5.33 ± 0.13 | 5.60 ± 0.13 | 0.083 |
| Uric acid | 345.68 ± 7.50 | 321.4 ± 6.41 | 0.013 |
| AST | 20.4 ± 0.78 | 19.9 ± 0.42 | 0.507 |
| ALT | 24.93 ± 1.45 | 21.73 ± 1.12 | 0.111 |
| TC | 6.34 ± 0.14 | 4.37 ± 0.28 | 0.024 |
| TG | 1.77 ± 1.07 | 2.01 ± 0.59 | 0.067 |
| HDL-C | 1.16 ± 0.53 | 1.58 ± 0.31 | 0.021 |
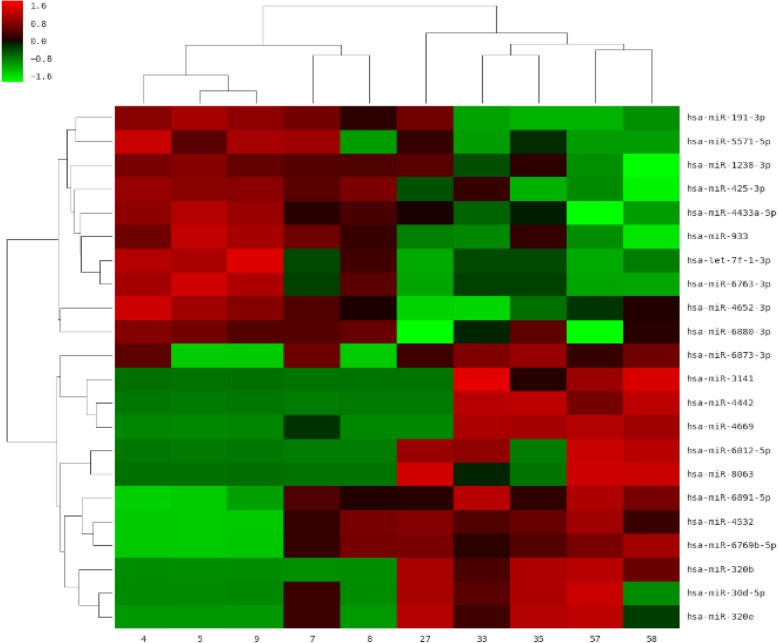
We then randomly selected 10 serum samples from subjects of the two groups, with 5 patients from the hyperlipidemia group and 5 patients from the control group, for miRNA microarray analysis. A total of 2449 miRNAs with intensity > 30 in at least one test serum were detected, as shown in the volcano plot (Fig. 1). Compared with the control group, 28 differentially expressed miRNAs were identified in the hyperlipidemia group, including 12 upregulated miRNAs and 10 downregulated miRNAs, at the criteria of FC (Fold change) ≥2 and P ≤ 0.05 (Fig. 2). Detailed differential expression of miRNAs is shown in Table 2.
Fig. 1.
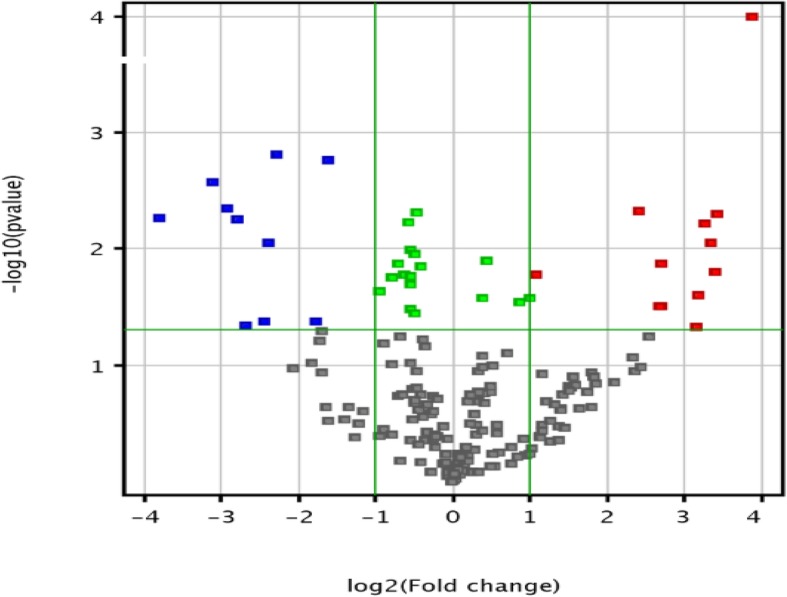
Volcano plot showing miRNAs detected by microarray. The red dots represent differential upregulation of miRNAs (fold change> 2,P > 0.05). The blue dots represent differential downregulation of miRNAs (fold change<−2,P < 0.05). The green points represent miRNAs (absolute fold change < 2,P < 0.05). The gray dots indicate the miRNAs expressed with P-value>0.05
Fig. 2.
Heat map of microarray results. The rows indicate miRNAs and columns indicate samples. The red color represents upregulation, and the green color represents downregulation. Samples 4, 5, 7, 8, and 9 were from the normal group, and samples 27, 33, 35, 57, and 58 were from the hyperlipidemia group
Table 2.
Fold-change and P-values of the 28 differentially expressed miRNAs
| Systematic_name | P-value | Fold-change | Regulation |
|---|---|---|---|
| hsa-miR-320b | 5.64E-06 | 14.652229 | up |
| hsa-miR-425-3p | 0.001549529 | 4.8689585 | down |
| hsa-miR-933 | 0.0017513 | 3.0571334 | down |
| hsa-miR-6763-3p | 0.002649574 | 8.668792 | down |
| hsa-let-7f-1-3p | 0.004478273 | 7.5040197 | down |
| hsa-miR-6812-5p | 0.004728028 | 5.341505 | up |
| hsa-miR-4442 | 0.00502631 | 10.697879 | up |
| hsa-miR-191-3p | 0.005391372 | 13.9024725 | down |
| hsa-miR-4652-3p | 0.005592787 | 6.952554 | down |
| hsa-miR-320e | 0.006005395 | 9.612401 | up |
| hsa-miR-4433a-5p | 0.008774483 | 5.2011013 | down |
| hsa-miR-4669 | 0.008905013 | 10.235651 | up |
| hsa-miR-3141 | 0.01339152 | 6.5122175 | up |
| hsa-miR-8063 | 0.015623125 | 10.531111 | up |
| hsa-miR-6891-5p | 0.01648203 | 2.1228352 | up |
| hsa-miR-30d-5p | 0.025248433 | 9.162879 | up |
| hsa-miR-6769b-5p | 0.031363655 | 6.3925323 | up |
| hsa-miR-4532 | 0.03136994 | 6.5774345 | up |
| hsa-miR-1238-3p | 0.042109374 | 3.418031 | down |
| hsa-miR-6880-3p | 0.042388383 | 5.4697547 | down |
| hsa-miR-5571-5p | 0.04509465 | 6.394996 | down |
| hsa-miR-6873-3p | 0.04718673 | 8.873202 | up |
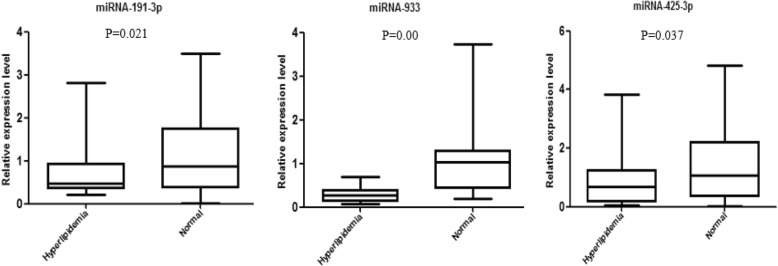
According to the P value and the FC value, three miRNAs with stable expression (miRNA-191-3p, miRNA-933, and miRNA-425-3p) as well as significant differences between the two groups were selected for further RT-qPCR validation. We randomly chose 40 other serum samples with 20 from the hyperlipidemia group and 20 from the control group for preliminary PCR validation. The results showed that the expression of all three miRNAs were downregulated, which is consistent with the results of the miRNA microarray (Fig. 3).
Fig. 3.
Expression levels of miRNA-191-3p, miRNA-933, and miRNA-425-3p were consistently downregulated in patients with hyperlipidemia compared with healthy volunteers, thereby supporting the microarray results. All of the expression levels of these three miRNAs were normalized to U6. (n = 20)
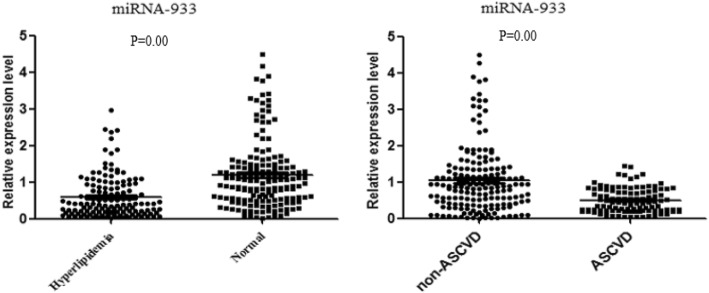
Based on the result of the preliminary PCR experiment, we selected miRNA-933 for further validation via additional experiments to detect the expression levels of miRNA-933 in serum samples from a larger sample with 290 subjects. As Fig. 4 shows, miRNA-933 was significantly downregulated in the hyperlipidemia group in comparison with the healthy volunteers (P = 0.00). Pearson correlation coefficient was also conducted to analyzed the correlation between LDL-C and miRNA-933 in the hyperlipidemia group, the result showed LDL-C and miRNA-933 were moderately correlated (R = − 0.691) (Additional file 1: Figure S1). Considering the results described above, we have sufficient reason to believe that circulating miRNA-933 is related to the disorder of serum lipid metabolism, especially hyper-LDL-C .
Fig. 4.
miRNA-933 was significantly downregulated in patients with hyperlipidemia (n = 122) compared with healthy volunteers (n = 168) (Left); The expression of miRNA-933 in ASCVD group (n = 96) was also significantly lower than that in non-ASCVD group (n = 194) (Right). Expression levels are normalized to U6. P < 0.05 was considered statistically significant
Since miRNA-933 is probably related to the disorder of serum lipid metabolism, Based on quartiles, the expression of miRNA-933 in ASCVD and non-ASCVD was further compared. The result showed that the expression of miRNA-933 in ASCVD group was significantly lower than that in non-ASCVD group (P = 0.00) (Fig. 4). After adjusting the variables P < 0.1 in univariate analysis (e.g. smoker (p = 0.079), LDL-C (P = 0.012), TC (P = 0.031), HDL (P = 0.026), and TG (P = 0.046)). Logistic regression analysis showed that the first quartile of serum miRNA-933 (OR 10.701, 95% CI, 2.834–40.48, P = 0.000), the second quartile (OR 5.255, 95% CI, 0.875–12.111; P = 0.018) were independent risk factors for ASCVD (Additional file 1: Table S1).
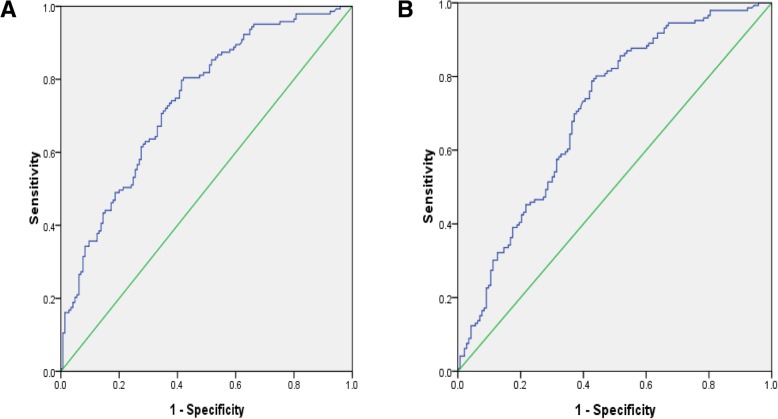
we continuously performed ROC curve analyses to investigate the usefulness of miRNA-933 as a diagnostic marker for ASCVD. The area under the curve (AUC) was 0.739 (95% CI, 0.682–0.795; P = 0.00) in distinguishing the hyperlipidemia patients (Fig. 5a), as well as 0.703 (95% CI, 0.643–0.763; P = 0.00) in distinguishing the ASCVD patients (Fig. 5b).
Fig. 5.
Graphs showing receiver operating characteristic curve analyses of serum miRNA-933 levels. a In the analysis for distinguishing hyperlipidemia patients from healthy volunteers, the area under the curve (AUC) was 0.739 (95% CI, 0.682–0.795; P = 0.00); b In distinguishing the ASCVD patients, the AUC was 0.703 (95% CI, 0.643–0.763; P = 0.00). P < 0.05 was considered statistically significant
Discussion
ASCVD, known as the main reason of death among elderly people, generally includes the following: acute coronary syndromes, history of myocardial infarction, stable or unstable angina, coronary or other arterial revascularization, stroke, TIA, or peripheral arterial disease presumed to be of atherosclerotic origin, according to the 2013 ACC/AHA guidelines [1]. The dysregulation of lipid metabolism, especially the abnormal elevation of LDL-C, is considered to be the main pathological basis of ASCVD [16]. At present, the circulating level of LDL-C is the most extensive diagnostic indicator for assessing and predicting the risk of atherosclerosis, and it has been globally acknowledged that reducing circulating LDL-C levels will effectively help in preventing ASCVD [14, 17]. Therefore, early intervention to inhibit circulating LDL-C levels has causally become more and more important.
In this study, we first statistically analyzed the clinical data of patients with abnormal elevation of LDL-C and normal controls. The results showed that the rates of ASCVD and carotid atherosclerosis were significantly higher in the hyperlipidemia group compared with the healthy control (P-values = 0.002, 0.039, respectively), thereby demonstrating the high correlation between circulating LDL-C with ASCVD. Our data also showed there were no significant differences in age, smoking history, body mass index, cerebrovascular disease, hypertension, diabetes, coronary heart disease, or other factors between the two groups.
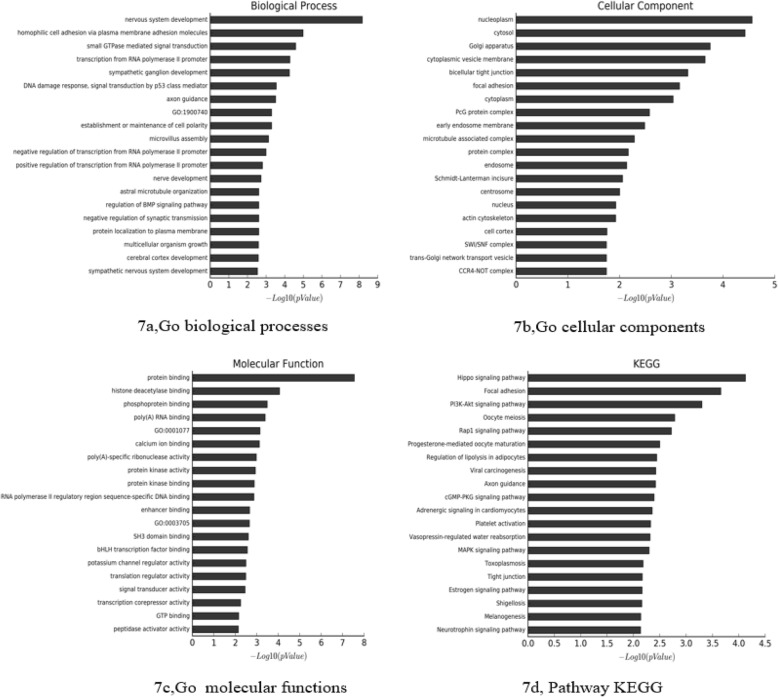
MiRNAs play a vital role in regulating life activities. Circulating miRNAs have been proved to be closely related to the development of many diseases, such as lung cancer, breast cancer, diabetes, metabolic syndrome, and heart failure, gradually becoming reliable biomarkers for various diseases [16, 18, 19]. There are many miRNAs that participate in the pathogenesis of atherosclerosis, mainly involved in lipid metabolism, inflammation, and endothelial dysfunction, thereby indicating that miRNAs are important factors in regulating the occurrence and development of atherosclerosis [20–22]. Circulating miRNAs were commonly considered to be derived from myocardial injury and atherosclerotic plaque rupture. Numerous studies have reported certain specific abnormal expression of circulating miRNAs of patients with ASCVD, suggesting that circulating miRNAs could be used as potential markers for the diagnosis of ASCVD [23, 24]. In this study, we used miRNA microarrays to analyze the differences of serum miRNA levels between the two groups (Additional file 1: Table S2 and Figure S2). We found that at least 12 miRNAs were upregulated in the LDL group, while 10 miRNAs were downregulated. Target genes of these differentially expressed miRNAs were the intersection predicted with 3 databases (Targetscan, microRNAorg, PITA). Using this method, a total of 616 target genes were found (Fig. 6). We used GO analysis to analyze all the target genes by DAVID, and we chose GOTERM_BP_FAT, GOTERM_CC_FAT, and GOTERM_MF_FAT. The analysis showed that the target genes were enriched in regulation of nervous system development, homophilic cell adhesion via plasma membrane adhesion molecules, small GTPase mediated signal transduction, mtranscription from RNA polymerase II promoter, sympathetic ganglion development and other biological processes in the GOTERM_BP_FAT (Fig. 7(a)), and were enriched in nucleoplasm, cytosol, golgi apparatus, cytoplasmic vesicle membrane, bicellular tight junction and other cellular components in the GOTERM_CC_FAT (Fig. 7(b)). In the GOTERM_MF_FAT, the target genes were enriched in protein binding, histone deacetylase binding, phosphoprotein binding, poly(A) RNA binding and other molecular functions (Fig. 7(c)). Finally, we performed the pathway analysis to analyze all the target genes by the KEGG. The results showed that in the classical pathway database KEGG, the target genes were enriched in hippo signaling pathway, focal adhesion, PI3K-Akt signaling pathway, oocyte meiosis, rap1 signaling pathway and other pathways (Fig. 7(d)). By analyzing Pathway analysis results, we found that among the top 20 signal pathways with the most target gene enrichment, 13 pathways contain the target gene of miRNA-933, including: CCDC97, BDNF, CAND1, DMRTA2, MAP4K4, PRKACB, MID1IP1, SLC16A2, ADCY9, FCHSD2, LRPAP1, etc. Pharmacogenomics studies have shown that the ADCY9 genotype determines the effects of CETP (cholesteryl ester transfer protein) inhibitor dalcetrapib on cardiovascular events and atherosclerosis [25], although the specific mechanism is not clear. BDNF was also reported play an important role in coronary atherosclerosis development [26]. However, whether miRNA-933 participates in the regulation of LDL-C is still not clear. Nevertheless, many studies mentioned that miRNA-933 could stably exist in the peripheral blood circulation [27, 28]. Previous studies on serum miRNA, such as acute myocardial infarction, coronary heart disease, and cerebral stroke, etc. [29–31], have proved that miRNA-933 could not only exist stably in the peripheral blood but also possessed significant differences when compared with control groups. However, few of these studies conducted further research on the correlation between circulating miRNA-933 and hyperlipidemia. Therefore we then conducted a preliminary exploration of the relationship between miRNA-933 and hyper-LDL-C and ASCVD.
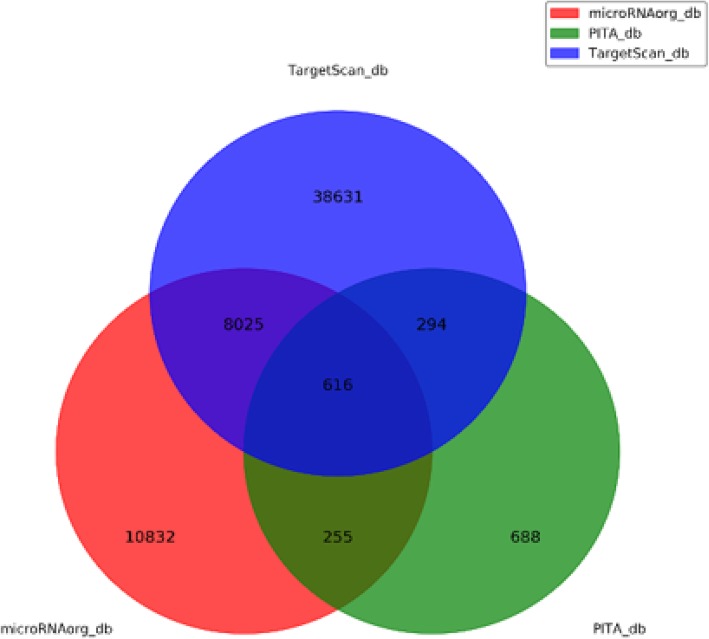
Fig. 6.
Target genes of differentially expressed miRNAs were the intersection predicted with 3 databases (Targetscan, microRNAorg, PITA)
Fig. 7.
Bioinformation analysis of the differently expressed microRNAs and their predicted targets a: GO biological process; b: GO cellular component; c: GO molecular function; d: Pathway KEGG)
Through miRNA microarrays, 22 miRNAs were found differentially expressed between the two groups. In order to verify the results, three miRNAs with relatively stable expression (i.e., miRNA-191-3p, miRNA-933, and miRNA-425-3p), as well as significant differences between the two groups, were selected for further PCR validation. The preliminary verification results of miR-191-3p, miR-933, and miR-425-3p were consistent with the miRNA microarray; all revealed a downregulation tendency. Further study of miRNA-933 in both groups verified the result, thereby suggesting that circulating miRNA-933 is probably related with a disorder of serum lipid metabolism (Additional file 1: Table S3). We then analyzed miRNA-933 expression between ASCVD and non-ASCVD group. The result showed miRNA-933 were a independent predictors associated with ASCVD by multivariate analysis. ROC curve analysis showed the area under the curve (AUC) was 0.739 (95% CI, 0.682–0.795; P = 0.00) and 0.703 (95% CI, 0.643–0.763; P = 0.00) in distinguishing the hyperlipidemia and the ASCVD patients, respectively. From the perspective of clinical application, We believe miRNA-933 could be helpful to identify patients with ASCVD at early stages. Additionally, as all the samples of this study were collected from clinical patients, we think these miRNAs may be more meaningful than the miRNAs found from animal models and therefore would be more helpful to reveal the potential mechanisms for lipid metabolism disorders. The limitation of this study include that the sample of the study was small and the results should be further confirmed in a large cohort patients.
Conclusion
In summary, our study screened out several miRNAs (miRNA-191-3p, miRNA-933, and miRNA-425-3p etc.) with differential expression in the peripheral circulation of patients with lipid metabolism disorders (mainly LDL-C). Circulating miRNA-933 could be a feasible predictor for ASCVD at the early stage.
Additional file
Table S1. Logistic regression to predict ASCVD. Table S2. Basic information of RNA sample quality. Table S3. Logistic regression to predict hyper-LDL-C. Figure S1. Pearson correlation to analyzed the correlation between LDL-C and miR-933. Figure S2. The Electrophoregram of miRNA assessed for microarray. (DOCX 137 kb)
Acknowledgments
Not applicable.
Funding
Supported by The National Key Research and Development Program of China (Grant No.2016YFE0126000)
Supported by The National Natural Science Foundation of China (Grant No.81570392)
Supported by The Foundation of Jiangsu Provincial Commission of Health and Family Planning (QNRC2016353)
Supported by Jiangsu Students’ Innovation and Entrepreneurship Training Program (201811117005Z)
Availability of data and materials
The authors confirm that all materials described in the manuscript are fully available to any scientist wishing to use them, without restriction.
Abbreviations
- ADCY9
Adenylate cyclase 9
- ALT
Alanine transaminase
- AS
Atherosclerosis
- ASCVD
Atherosclerotic cardiovascular disease
- AST
Aspartate aminotransferase
- BDNF
brain derived neurotrophic factor
- BMI
Body mass index
- CAND1
Cullin associated and neddylation dissociated 1
- CCDC97
Coiled-coil domain containing 97
- CVD
Cardiovascular disease
- DMRTA2
DMRT like family A2
- FCHSD2
FCH and double SH3 domains 2
- HDL-C
High density lipoprotein cholesterol
- LDL-C
Low-density lipoprotein cholesterol
- LRPAP1
LDL receptor related protein associated protein 1
- MAP4K4
Mitogen-activated protein kinase kinase kinase kinase 4
- MID1IP1
MID1 interacting protein 1
- PRKACB
Protein kinase cAMP-activated catalytic subunit beta
- SD
Standard deviation
- SLC16A2
Solute carrier family 16 member 2
- TC
Total cholesterol
- TG
Triglyceride
Authors’ contributions
XJ, ZC and YW performed all experiments and were responsible for data and drafting the article. JL and XZ designed the study and contributed to preparation of the manuscript. XW, LC, TY, XT, YL, HC, and ZD assisted in sample collection and data interpretation. JF and MC helped revise the article. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The study was approved by the ethics committee of the Affiliated Hospital of Yangzhou University. Written informed consent was obtained from each participant.
Consent for publication
All authors agree to publish this article in the Journal of Lipids in Health and Disease.
Competing interests
The authors declare that they have no competing interests.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jiang Xu, Email: 290019406@qq.com.
Zixuan Chen, Email: 852082367@qq.com.
Yingge Wang, Email: 1531961610@qq.com.
Xiaohong Wang, Email: 644303497@qq.com.
Lu Chen, Email: xj290019406@sina.com.
Tingting Yuan, Email: xxj290019406@sina.com.
Xiangming Tang, Email: xxjj290019406@qq.com.
Yaoyao Lu, Email: luyaoyao0608@sina.com.
Hongmei Chen, Email: chenhongmei0608@sina.com.
Miaolei Chen, Email: 1027385508@qq.com.
Zuowei Duan, Email: 876194075@qq.com.
Jianglin Fan, Email: jianglin@yamanashi.ac.jp.
Jingyan Liang, Phone: +86 15905270970, Email: jyliang@yzu.edu.cn.
Xinjiang Zhang, Phone: +86 18952723788, Email: lzhangxinjiang@sina.com.
References
- 1.Stone NJ, et al. ACC/AHA guideline on the treatment of blood cholesterol to reduce atherosclerotic cardiovascu2013 Lar risk in adults: a report of the American College of Cardiology/American Heart Association task force on practice guidelines. Circulation. 2014;129(25 Suppl 2):S1–45. doi: 10.1161/01.cir.0000437738.63853.7a. [DOI] [PubMed] [Google Scholar]
- 2.Williams KJ, Tabas I. The response-to-retention hypothesis of early atherogenesis. Arterioscler Thromb Vasc Biol. 1995;15(5):551–561. doi: 10.1161/01.ATV.15.5.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Camejo G. The interaction of lipids and lipoproteins with the intercellular matrix of arterial tissue: its possible role in atherogenesis. Adv Lipid Res. 1982;19:1–53. doi: 10.1016/B978-0-12-024919-0.50007-2. [DOI] [PubMed] [Google Scholar]
- 4.Ofori-Asenso R, et al. LDL-cholesterol is the only clinically relevant biomarker for atherosclerotic cardiovascular disease (ASCVD) risk. Clin Pharmacol Ther. 2018. [DOI] [PubMed]
- 5.Li WL, Hua LG, Qu P, Yan WH, Ming C, Jun YD, Yuan LD, Nan N. NLRP3 inflammasome: a novel link between lipoproteins and atherosclerosis. Arch Med Sci. 2016;12(5):950–958. doi: 10.5114/aoms.2016.61356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang J, Yang X. The function of miRNA in cardiac hypertrophy. Cell Mol Life Sci. 2012;69:3561–3570. doi: 10.1007/s00018-012-1126-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sahebkar A, Maffioli P, Banach M, Derosa G. Editorial: MicroRNA-33 inhibition: a potential adjunct to statin therapy? Curr Vasc Pharmacol. 2016;14(4):321–322. doi: 10.2174/1570161114999160513150737. [DOI] [PubMed] [Google Scholar]
- 9.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in drosophila. Cell. 2003;113:25–36. doi: 10.1016/S0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 10.Mohajeri M, Banach M, Atkin SL, Butler AE, Ruscica M, Watts GF. Sahebkar MicroRNAs: Novel Molecular Targets and Response Modulators of Statin Therapy. Trends Pharmacol Sci. 2018;39(11):967–981. doi: 10.1016/j.tips.2018.09.005. [DOI] [PubMed] [Google Scholar]
- 11.Momtazi AA, Banach M, Pirro M, Stein EA, Sahebkar A. MicroRNAs: new therapeutic targets for familial hypercholesterolemia? Clin Rev Allergy Immunol. 2018;54(2):224–233. doi: 10.1007/s12016-017-8611-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazidi M, Penson P, Gluba-Brzozka A, Rysz J, Banach M. Relationship between long noncoding RNAs and physiological risk factors of cardiovascular disease. J Clin Lipidol. 2017;11(3):617–623. doi: 10.1016/j.jacl.2017.03.009. [DOI] [PubMed] [Google Scholar]
- 13.Navickas R, et al. Identifying circulating microRNAs as biomarkers of cardiovascular disease: a systematic review. Cardiovasc Res. 2016;111(4):322–337. doi: 10.1093/cvr/cvw174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montalbo R, et al. Prognostic value of circulating microRNAs in upper tract urinary carcinoma. Oncotarget. 2018;9(24):16691–16700. doi: 10.18632/oncotarget.24672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chanudet E, et al. Large-scale genome-wide screening of circulating microRNAs in clear cell renal cell carcinoma reveals specific signatures in late-stage disease. Int J Cancer. 2017;141(9):1730–1740. doi: 10.1002/ijc.30845. [DOI] [PubMed] [Google Scholar]
- 16.Keller A, et al. Toward the blood-borne miRNome of human diseases. Nat Methods. 2011;8(10):841–843. doi: 10.1038/nmeth.1682. [DOI] [PubMed] [Google Scholar]
- 17.Okerson T, et al. Effect of 2013 ACC/AHA blood cholesterol guidelines on statin treatment patterns and low-density lipoprotein cholesterol in atherosclerotic cardiovascular disease patients. J Am Heart Assoc. 2017;6(3). [DOI] [PMC free article] [PubMed]
- 18.Scholer N, et al. Serum microRNAs as a novel class of biomarkers: a comprehensive review of the literature. Exp Hematol. 2010;38(12):1126–1130. doi: 10.1016/j.exphem.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 20.Kandhro AH, et al. The MicroRNA interaction network of lipid diseases. Front Genet. 2017;8:116. doi: 10.3389/fgene.2017.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayner KJ, Fernandez-Hernando C, Moore KJ. MicroRNAs regulating lipid metabolism in atherogenesis. Thromb Haemost. 2012;107(4):642–647. doi: 10.1160/TH11-10-0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Romaine SP, et al. MicroRNAs in cardiovascular disease: an introduction for clinicians. Heart. 2015;101(12):921–928. doi: 10.1136/heartjnl-2013-305402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fichtlscherer S, et al. Circulating microRNAs in patients with coronary artery disease. Circ Res. 2010;107(5):677–684. doi: 10.1161/CIRCRESAHA.109.215566. [DOI] [PubMed] [Google Scholar]
- 24.Tijsen AJ, Pinto YM, Creemers EE. Circulating microRNAs as diagnostic biomarkers for cardiovascular diseases. Am J Physiol Heart Circ Physiol. 2012;303(9):H1085–H1095. doi: 10.1152/ajpheart.00191.2012. [DOI] [PubMed] [Google Scholar]
- 25.Yohann Rautureau, et al. ADCY9 (Adenylate Cyclase Type 9) Inactivation Protects From Atherosclerosis Only in the Absence of CETP (Cholesteryl Ester Transfer Protein). Circulation. Apr 19, 2018;138:1677–1692. [DOI] [PubMed]
- 26.Chaldakov GN, Fiore M, Stankulov IS, et al. NGF, BDNF, leptin, and mast cells in human coronary atherosclerosis and metabolic syndrome. Arch Physiol Biochem. 2001;109:357–360. doi: 10.1076/apab.109.4.357.4249. [DOI] [PubMed] [Google Scholar]
- 27.Tan KS, et al. Expression profile of MicroRNAs in young stroke patients. PLoS One. 2009;4(11):e7689. doi: 10.1371/journal.pone.0007689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bostjancic E, Zidar N, Glavac D. MicroRNA microarray expression profiling in human myocardial infarction. Dis Markers. 2009;27(6):255–268. doi: 10.1155/2009/641082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schwed Lustgarten DE, et al. Use of circulating tumor cell technology (CELLSEARCH) for the diagnosis of malignant pleural effusions. Ann Am Thorac Soc. 2013;10(6):582–589. doi: 10.1513/AnnalsATS.201303-068OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vijayan M, Reddy PH. Peripheral biomarkers of stroke: focus on circulatory microRNAs. Biochim Biophys Acta. 2016;1862(10):1984–1993. doi: 10.1016/j.bbadis.2016.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Samanta S, et al. MicroRNA: a new therapeutic strategy for cardiovascular diseases. Trends Cardiovasc Med. 2016;26(5):407–419. doi: 10.1016/j.tcm.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Logistic regression to predict ASCVD. Table S2. Basic information of RNA sample quality. Table S3. Logistic regression to predict hyper-LDL-C. Figure S1. Pearson correlation to analyzed the correlation between LDL-C and miR-933. Figure S2. The Electrophoregram of miRNA assessed for microarray. (DOCX 137 kb)
Data Availability Statement
The authors confirm that all materials described in the manuscript are fully available to any scientist wishing to use them, without restriction.