Abstract Abstract
Two new species, Castanediellabrevis and C.monoseptata, are described, illustrated and compared with other Castanediella taxa. Evidence for the new species is provided by morphological comparison and sequence data analyses. Castanediellabrevis can be distinguished from other Castanediella species by the short hyaline conidiophores and fusiform, aseptate hyaline conidia, while C.monoseptata differs from other Castanediella species by its unbranched conidiophores and fusiform, curved, 0–1-sepatate, hyaline conidia. Phylogenetic analysis of combined ITS and LSU sequence data was carried out to determine the phylogenetic placement of the species. A synopsis of hitherto described Castanediella species is provided. In addition, Castanediella is also compared with morphologically similar-looking genera such as Idriella, Idriellopsis, Microdochium, Neoidriella, Paraidriella and Selenodriella.
Keywords: new taxa, Castanediellaceae , hyphomycetes, phylogeny, Sordariomycetes
Introduction
Hernández-Restrepo et al. (2017) introduced the family Castanediellaceae for the genus Castanediella within Xylariales and it was consolidated in recent study by Wijayawardene et al. (2018). The asexual morphs in Castanediellaceae are hyphomycetous and characterized by macronematous, mononematous or sporodochial, branched, brown to pale brown conidiophores, with monoblastic or polyblastic, sympodial, discrete, cylindrical to lageniform, hyaline to subhyaline conidiogenous cells, that produce unicellular or transversely septate, cylindrical, fusiform or lunate, hyaline conidia (Hernández-Restrepo et al. 2017).
The genus Castanediella was established by Crous et al. (2015) to accommodate C.acaciae, C.cagnizarii and C.ramosa within Xylariales genera incertae sedis. The genus contains twelve species (Costa et al. 2018; Wanasinghe et al. 2018), each characterized by branched, hyaline to pale brown conidiophores, holoblastic, sympodial conidiogenous cells and falcate, cylindrical or fusiform, 0–3-sepate, hyaline conidia (Crous et al. 2015; Costa et al. 2018).
During a survey of hyphomycetes in Thailand, two hyaline-spored hyphomycetes were collected. They were shown to belong to the genus Castanediella based on morphology and phylogeny analyses of ITS and LSU sequence data. The new species C.brevis and C.monoseptata are introduced.
Materials and methods
Collection and isolation of fungi
Dead leaves from a variety of plants in two forests (Lampang province and Chiang Mai province) were collected in 2016 in Thailand. Samples were taken to the laboratory in Zip-lock plastic bags for examination. The specimens were incubated in sterile moist chambers and examined using a Motic SMZ 168 series microscope. Fungi were removed with a needle and placed in a drop of distilled water on a slide for morphological study. Photomicrographs of fungal structures were captured with a Canon 600D digital camera attached to a Nikon ECLIPSE Ni compound microscope. All measurements were made by the Tarosoft (R) Image FrameWork program. Photo-plates were made with Adobe Photoshop CS3 (Adobe Systems, USA). Isolation of the fungi on to potato dextrose agar (PDA) was performed by the single spore isolation method (Chomnunti et al. 2014). Dried material was deposited in the Herbarium of Mae Fah Luang University (MFLU), Chiang Rai, Thailand and herbarium of Kunming Institute of Botany, Chinese Academy of Sciences (HKAS), Kunming, China. Cultures were deposited at Mae Fah Luang University Culture Collection (MFLUCC), Chiang Rai, Thailand and Kunming Institute of Botany, Chinese Academy of Sciences (KUMCC), Kunming, China. FacesofFungi and Index Fungorum numbers were registered (Jayasiri et al. 2015; Index Fungorum 2018).
DNA extraction, PCR amplification and sequencing
Genomic DNA was extracted from fungal mycelium grown on PDA or malt extract agar (MEA) at room temperature using the Fungal gDNA Kit (BioMIGA, USA) according to the manufacturer’s instructions. The internal transcribed spacer region of ribosomal DNA (ITS) and large subunit nuclear ribosomal DNA (LSU) genes were amplified via polymerase chain reaction (PCR) using the following primers: ITS5 and ITS4 (White et al. 1990) for ITS, and LR0R and LR5 (Vilgalys and Hester 1990) for LSU. The PCR products were sequenced with the same primers. The PCR amplification was performed in a 25 μL reaction volume containing 12.5 μL of 2 × Power Taq PCR MasterMix (a premix and ready to use solution, including 0.1 Units/μl Taq DNA Polymerase, 500 μM dNTP Mixture each [dATP, dCTP, dGTP, dTTP], 20 mM Tris-HCl pH 8.3, 100 Mm KCl, 3 mM MgCl2, stabilizer and enhancer), 1 μL of each primer (10 μM), 1 μL genomic DNA extract and 9.5 μL deionised water. The PCR thermal cycle program of ITS and LSU were followed as: initially 94 °C for 3 min., followed by 35 cycles of denaturation at 94 °C for 30 s, annealing at 55 °C for 50 s, elongation at 72 °C for 1 min., and final extension at 72 °C for 10 min.
Phylogenetic analyses
Original sequences were checked using BioEdit version 7.0.5.3 (Hall 1999), and most reference sequences were originated from previous publications. The remaining homogenous sequences were obtained by BLAST searches (Altschul et al. 1990) from GenBank. All sequences used in this study are listed in Table 1. Alignments for each locus were done in MAFFT v7.307 online version (Katoh and Standley 2016) and manually verified in MEGA 6.06 (Tamura et al. 2013). After alignment, the concatenation of different genes was done in SequenceMatrix 1.8 (Vaidya et al. 2011). The interleaved NEXUS files for Bayesian inference analyses were formatted with AliView v1.19-beta1k (Larsson 2014). Maximum parsimony (MP), maximum likelihood (ML) and Bayesian inference (BI) were used for phylogenetic analyses.
Table 1.
GenBank accession numbers of isolates included in this study.
| Taxa | Isolatea | ITS | LSU |
|---|---|---|---|
| Castanediella acaciae | CPC 24869, CBS 139896 | NR_137985 | KR476763 |
| Castanediellabrevis | KUMCC 18-0132 | MH806361 | MH806358 |
| Castanediella cagnizarii | MUCL 41095 | KC775732 | KC775707 |
| Castanediella cagnizarii | CBS 101043 | KP859051 | KP858988 |
| Castanediella cagnizarii | CBS 542.96 | KP859054 | KP858991 |
| Castanediella camelliae | CNUFC-DLHBS5-1 | MF926620 | MF926614 |
| Castanediella camelliae | CNUFC-DLHBS5-2 | MF926621 | MF926615 |
| Castanediella communis | CPC 27631 | KY173393 | – |
| Castanediella couratarii | CBS 579.71 | NR_145250 | KP858987 |
| Castanediella eucalypti | CPC 24746, CBS 139897 | NR_137981 | KR476758 |
| Castanediella eucalypticola | CPC 26539 | NR_145254 | KX228317 |
| Castanediella eucalyptigena | CBS 143178, CPC 32055 | MG386036 | MG386089 |
| Castanediella hyalopenicillata | CPC 25873 | KX306751 | KX306780 |
| Castanediella malaysiana | CPC 24918 | NR_154810 | KX306781 |
| Castanediellamonoseptata | KUMCC 18-0133 | MH806360 | MH806357 |
| Castanediella ramosa | MUCL 39857 | KC775736 | KC775711 |
| Subsessila turbinata | MFLUCC 15-0831 | KX762288 | KX762289 |
aCBS, Centraalbureau voor Schimmelcultures, Utrecht, Netherlands; CPC, Culture collection of Pedro Crous, housed at CBS; KUMCC, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China; MFLUCC, Mae Fah Luang University Culture Collection, Chiang Rai, Thailand; MUCL, Mycothèque de l’Université Catholique de Louvian, Belgium.
The best models of evolution for each gene region were determined using Akaike information criterion (AIC) as implemented in MrModeltest v2 (Nylander 2004). The analyses’ results showed that the models GTR+I and GTR+I+G were the best ones for LSU and ITS sequence data, respectively.
MP analyses were performed in PAUP*4.0b10 (Swofford 2002) following Liu et al. (2016).
ML analyses were carried out in raxmlGUI v 1.5b1 (Silvestro and Michalak 2012) with RAxML v8.2.10 (Stamatakis 2014), using the ML + rapid bootstrap setting and the GTRGAMMAI (viz., GTR + GAMMA + I) substitution model with 1000 bootstrap replicates.
For BI analysis, Posterior probabilities (PP) (Rannala and Yang 1996; Zhaxybayeva and Gogarten 2002) were determined by Markov Chain Monte Carlo sampling (BMCMC) in MrBayes v 3.2.6 (Ronquist et al. 2012). For the combined dataset, the models were set to nst = 6 and rates = propinv for LSU and nst = 6 and rates = invgamma for ITS. Two independent analyses of two parallel runs and six simultaneous Markov chains were run for 1,000,000 generations, trees were sampled every 100th generation and the temperature value of the heated chains was set at 0.15. The first 25% sampled trees of each run were discarded as “burn-in”, and the remaining trees were used for calculating posterior probabilities (PP) in the majority rule consensus tree with the sumt command in MrBayes.
Phylogenetic trees were drawn with TreeView 1.6.6 (Page 1996).
Results
Molecular phylogeny
The aligned sequence matrix comprises LSU and ITS sequence data for 16 taxa (ingroup) and one outgroup taxon with a total of 1438 characters after alignment including the gaps, of which 120 were parsimony informative, 77 parsimony-uninformative, and 1241 characters constant. The dataset consists of thirteen species within the genus. The tree was rooted with Subsessilaturbinata (MFLUCC 15-0831). Maximum parsimony analysis resulted in two trees with TL = 391, CI = 0.657, RI = 0.642, RC = 0.422, HI = 0.343. For the Bayesian analysis, two parallel runs with six chains were run for 1,000,000 generations and trees were sampled every 100th generation, resulting in 20002 trees from two runs of which 15002 trees were used to calculate the posterior probabilities (each run resulted in 10001 trees of which 7501 trees were sampled).The MP and ML (lnL = -4041.301739) analyses based on combined LSU and ITS sequence data provided similar tree topologies, and the result of MP analysis is shown in Fig. 1.
Figure 1.
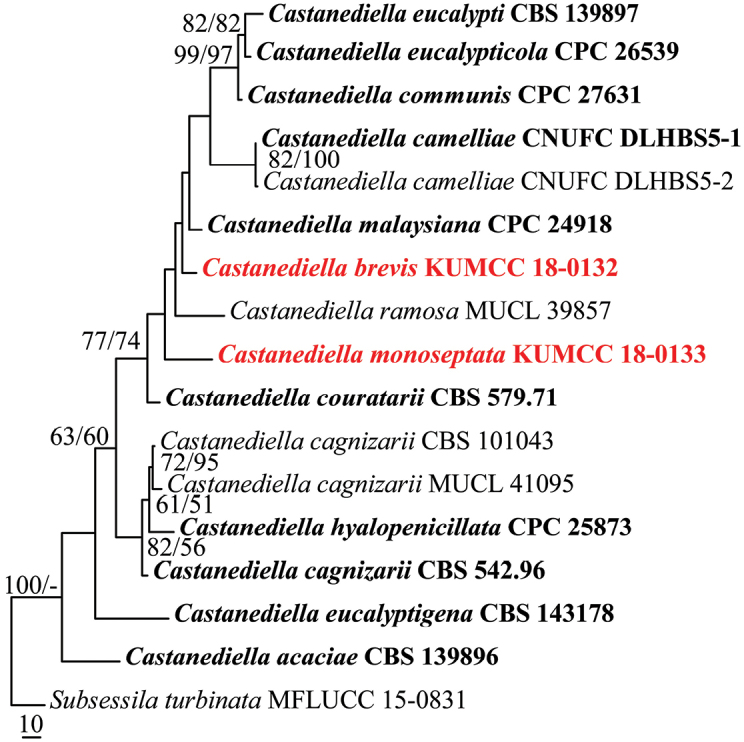
Phylogenetic tree generated from MP analysis based on combined LSU and ITS sequence data for the genus Castanediella. Bootstrap support values for maximum parsimony (MP, first set) and maximum likelihood (ML, second set) greater than 50% are indicated above or below the nodes. Ex-type strains are in bold, the new isolates are in red. The tree is rooted with Subsessilaturbinata (MFLUCC 15-0831).
The novelty of the species, Castanediellabrevis and C.monoseptata, described in this study are supported by sequence data analyses as belonging to the genus Castanediella, but with low bootstrap support values. Isolates of Castanediellabrevis and C.monoseptata formed separate clades in the phylogenetic inference, respectively. Castanediellabrevis is sister to C.malaysiana and C.ramosa, while C.monoseptata shows close phylogenetic relationship to C.couratarii and C.malaysiana. Both the new taxa can be recognized as phylogenetically distinct species and are clearly novel based on the recommendations for molecular data (Jeewon and Hyde 2016).
MP, ML and BI were used for phylogenetic analyses in this study. The tree topologies of MP and ML resulted from the combined LSU and ITS sequence data are similar, but most of the nodes are in low bootstrap support (Fig. 1). Polytomy structure was formed in the BI tree generated from the combined LSU and ITS sequence data. More sequence data, especially the protein-coding genes, e.g. TEF1-α, RPB2, β-tubulin, are required in the future study of the genus Castanediella.
Taxonomy
Castanediella brevis
s C.G. Lin & K.D. Hyde sp. nov.
MB828879
Figure 2.
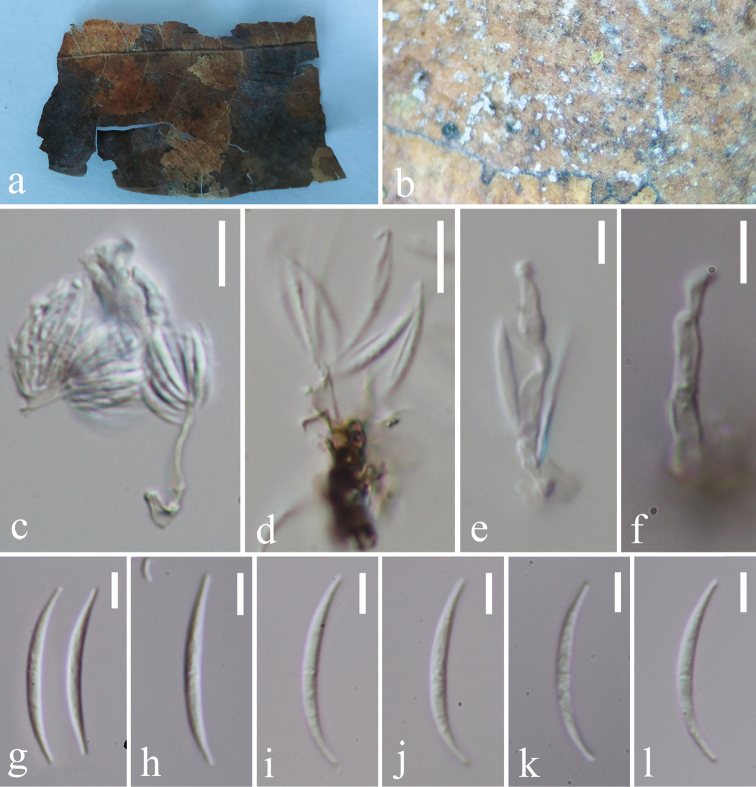
Castanediellabrevis (MFLU 18-1695, holotype) a host material b conidiophores on the host surface c–g conidiophores, conidiogenous cells with conidia h conidia. Scale bars: 10 μm (c–g), 5 μm (h).
Holotype.
THAILAND. Lampang: Amphoe Mueang Pan, Tambon Chae Son, on decaying leaves, 24 September 2016, Chuangen Lin, LCG 10-1 (MFLU 18-1695, holotype; HKAS 102198, isotype), ex-type living cultures KUMCC 18-0132.
GenBank number.
Etymology.
In reference to the short conidiophores.
Saprobic on plant host. Asexual morph: Colonies on substrate effuse, white. Mycelium partly superficial, composed of septate, branched, smooth, hyaline to subhyaline hyphae. Conidiophores macronematous, mononematous, solitary, erect, unbranched, straight or flexuous, short, 0–1-septate, hyaline, subcylindrical, ampulliform, smooth, often reduced to conidiogenous cells. Conidiogenous cells holoblastic, polyblastic, sympodial, integrated, terminal, subcylindrical, ampulliform, hyaline, denticulate, with 2–4 tiny protuberant denticles, 3–14 × 1.5–5.5 μm. Conidia solitary, dry, acropleurogenous, smooth, fusiform, curved, aseptate, hyaline, 12.5–21.7 × 1.2–3 μm (av. 16.95 × 2.2 μm, n = 60). Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA effuse, greyish white to dark from above and below, reaching a diam. of 5–7 cm in 30 days at 25 °C.
Notes.
Based on a megablast search of the NCBI nucleotide database using the ITS sequence of the ex-type culture, the highest similarities found were with Castanediellamalaysiana (GenBank NR_154810; identities = 526/537(98%), gaps = 1/537(0%)) and C.couratarii (GenBank KX960789; identities = 521/538(97%), gaps = 3/538(0%)). Castanediellabrevis differs from these two species by its conidiophore morphology. Castanediellacouratarii has pale brown conidiophores and longer conidiogenous cells (10.5–37 × 2–3.5 μm) whereas C.malysiana has pale brown and longer conidiophores (76–157 × 2.5–3 μm).
Among the species that produce more or less falcate and aseptate conidia, Castanediellacommunis, C.eucalypti, C.eucalypticola and C.eucalyptigena are most similar to C.brevis. However, Castanediellabrevis differs from these species by its short, unbranched and 0–1-septate conidiophores.
Castanediella monoseptata
C.G. Lin & K.D. Hyde sp. nov.
MB828881
Figure 3.
Castanediellamonoseptata (MFLU 18-1696, holotype) a host material b conidiophores on the host surface c–f conidiophores, conidiogenous cells with conidia g–l conidia. Scale bars: 10 μm (c, d), 5 μm (e–l).
Holotype.
THAILAND. Chiang Mai: on decaying leaves, 24 August 2016, Chuangen Lin, MRC 3-1 (MFLU 18-1696, holotype; HKAS 102199, isotype), ex-type living cultures KUMCC 18-0133.
GenBank number.
Etymology.
In reference to the 0–1-septate conidia
Saprobic on plant host. Asexual morph: Colonies on substrate effuse, white. Mycelium partly superficial, composed of septate, branched, hyaline to subhyaline, smooth hyphae. Conidiophores macronematous, mononematous, solitary, erect, unbranched, straight or flexuous, septate, hyaline, subcylindrical, smooth, 8–29 × 2–4 μm. Conidiogenous cells polyblastic, integrated, sympodial, subcylindrical, hyaline, with several scars. Conidia solitary, dry, acropleurogenous, smooth, fusiform, curved, 0–1-sepatate, hyaline, 15.4–25.8 × 1.5–2.3 μm (av. 23.03 × 1.98 μm, n = 45). Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 24 h. Colonies on PDA effuse, grayish white to dark from above and below, reaching a diam. of 5–7 cm in 30 days at 25 °C.
Notes.
A megablast search of the NCBI nucleotide database using the ITS sequence of the ex-type culture showed the highest similarities with uncultured Sordariales fungi (GenBank GQ268569; identities = 518/539(96%), gaps = 3/539(0%)) and Castanediellacouratarii (GenBank KX960789; identities = 516/540(96%), gaps = 4/540(0%)).
Five Castanediella species, C.cagnizarii, C.diversispora, C.hyalopenicillata, C.malaysiana and C.ramosa, were reported to produce 1-septate conidia. Castanediellamonoseptata can be distinguished from these species by its unbranched conidiophores and falcate and 15.4–25.8 × 1.5–2.3 μm conidia. Castanediellamonoseptata is phylogenetically closely related to C.couratarii and C.ramosa, but differs from both species by its conidial morphology. Castanediellacouratarii has shorter conidia (9.5–19 × 2–3 μm) are aseptate and C.ramosa has larger conidia (26–44 × 2–3 μm) that are 0–3-septate.
Discussion
In this study, two new Castanediella species, C.brevis and C.monoseptata, were identified from decaying leaves in Thailand and a synopsis of hitherto described Castanediella species is provided (Table 2).
Table 2.
Synopsis of Castanediella species.
| Taxa | Conidiophores | Conidiogenous cells | Conidia | |||
|---|---|---|---|---|---|---|
| Shape | Size (μm) | Septa | Colour | |||
| C. acaciae | Subcylindrical, medium brown, 40–80 × 2–3 μm. | Polyblastic, ampulliform, pale brown, 10–15 × 2–3 μm. | Falcate with subobtuse ends | (8–)10–11(–12) × 1.5(–2) | 0 | Hyaline |
| C. brevis | Subcylindrical, ampulliform, hyaline, often reduced to conidiogenous cells | Polyblastic, cylindrical, hyaline, 3–14 × 1.5–5.5 μm | Fusiform, curved | 12.5–21.7 × 1.2–3.0 | 0 | Hyaline |
| C. cagnizarii | Cylindrical, brown at the base, subhyaline towards the apex, up to 45 μm long. | Polyblastic, sympodial, subhyaline, 5–22 × 3–4 μm. | Cylindrical to fusiform, curved at the ends | Two sizes, 10–15 × 2 or 20–26 × 2 | Hyaline | |
| C. camelliae | Conidiophores reduced to conidiogenous cell. | Cylindrical, ampulliform, globose to subglobose, or irregularly-shaped, 5.5–20.5 × 2–4.5 μm. | Straight to slightly curved, sometimes swollen in the middle part | 18.5–51.5 × 1.6–2.5 | Septum indistinct | Hyaline |
| C. communis | Subcylindrical, medium brown, 20–60 × 3–4 μm. | Polyblastic, subcylindrical to ampulliform, pale brown, 10–35 × 2–4 μm. | Falcate with subobtuse ends | (13–)17–20(–22) × (2–)2.5(–3) | 0 | Hyaline |
| C. couratarii | Pale brown | Lageniform to cylindrical, hyaline to pale brown, 10.5–37 × 2–3.5 μm | Lunate | 9.5–19 × 2–3 | 0 | Hyaline |
| C. diversispora | Pale brown to brown | Polyblastic, sympodial, pale brown to brown, 4–9 × 2–3.5 μm. | Type i) cylindrical, slightly uncinate at the ends, straight | Type i) 11.5–16 × 2 | Type i) 1-septate | Hyaline |
| Type ii) cylindrical to slightly subacerose, slightly uncinate at the apex, abruptly attenuated at the base, straight | Type ii) 19.5–25 × 1.5–2 | Type ii) 1-septate | ||||
| Type iii) long filiform, obtuse or rounded at the apex attenuated at the base, straight or curved | Type iii) 28.5–47 × 1 | Type iii) 1–3-septate | ||||
| C. eucalypti | Subcylindrical, medium brown, 10–30 × 3–4 μm. | Polyblastic, subcylindrical to ampulliform, pale brown, 8–25 × 2.5–4 μm. | Falcate, slightly curved, widest in middle with subobtuse ends | (15–)18–21(–23) × 2–3 | 0 | Hyaline |
| C. eucalypticola | Subcylindrical, medium brown, 5–30 × 3–5 μm. | Polyblastic, subcylindrical to ampulliform or lanceolate, pale brown, 5–20 × 3–3.5 μm. | Falcate, straight to curved, widest in the middle, apex subobtusely rounded, base truncate, 0.5 μm diam | (15–)20–26(–30) × (2.5–)3 | 0 | Hyaline |
| C. eucalyptigena | Subcylindrical, hyaline, frequently reduced to conidiogenous loci on hyphae, up to 15 μm tall, 3–5 μm diam. | Polyblastic, hyaline, ampulliform or subcylindrical, 2–10 × 2–5 μm | Falcate, tapering to acute ends that are subobtusely rounded | (13–)18–24(–30) × 2(–2.5) | 0 | Hyaline |
| C. hyalopenicillata | Cylindrical, penicillate, mono-, bi-, and terverticillate, hyaline, 24–69 × 1.5–3 μm. | Mono- and polyblastic, short cylindrical, ampulliform, hyaline, 6.5–14 × 2–4 μm | Fusiform, base pointed, apex obtuse | 14–24 × 2–3 | 0–1 | Hyaline |
| C. malaysiana | Cylindrical, biverticillate, pale brown, 76–157 × 2.5–3 μm. | Polyblastic, cylindrical, subcylindrical, hyaline, 19–28 × 2.5–3.5 μm. | Fusiform, curved, apex acuminate, and base acuminate or slightly flattened | 18–30 × 2–3 | 0–1 | Hyaline |
| C. monoseptata | Subcylindrical, unbranched, hyaline, 8–29 × 2–4 μm | Polyblastic, cylindrical, hyaline | Fusiform, curved | 15.4–25.8 × 1.5–2.3 | 0–1 | Hyaline |
| C. ramosa | Cylindrical, penicillate, brown at the base, subhyaline at the apex, up to 70 μm long | Polyblastic, subhyaline, 10–20 x 2.5–3.5 μm | Falcate | 26–44 × 2.2–3 | (0–) 1 (–3) | Hyaline |
Presently, the genus Castanediella contains 14 species, and is shown to be diverse in its habitats. Most of Castanediella species have been collected from plant leaves. Castanediellaacaciae, C.camelliae, C.communis, C.eucalypti, and C.eucalypticola were isolated from disease symptoms on different host plant leaves (Crous et al. 2015, 2016a, b; Wanasinghe et al. 2018) whereas C.cagnizarii is the only species found on decaying leaves submerged in a stream (Castañeda Ruiz et al. 2005). Some Castanediella species were reported from decaying leaves, such as C.brevis, C.cagnizarii, C.diversispora, C.hyalopenicillata and C.monoseptata (Castañeda Ruiz et al. 2005; Hernández-Restrepo et al. 2016b; Costa et al. 2018). Castanediellacouratarii was reported from dead wood (Hernández-Restrepo et al. 2016a).
The genus Castanediella is morphologically similar to Idriella, Idriellopsis, Microdochium, Neoidriella, Paraidriella, Selenodriella (Seifert et al. 2011; Crous et al. 2015; Hernández-Restrepo et al. 2016a). However, these genera can be distinguished by the branching pattern of their conidiophores and conidial shape and septation (Hernández-Restrepo et al. 2016a). Castanediella differs from these genera by its branched conidiophores, ampulliform conidiogenous cells with scars instead of denticles, and filiform, 0–1-septate, straight to curved conidia (Crous et al. 2015). These similar-looking genera are phylogenetically distinct (Crous et al. 2015; Hernández-Restrepo et al. 2016a). A comparative synopsis of these genera is provided (Table 3).
Table 3.
Synopsis of Castanediella-like genera.
| Genera | Conidiophores | Conidiogenous cells | Conidia | Chlamydospores |
|---|---|---|---|---|
| Castanediella | Branched, pale brown to brown at the base and subhyaline at the apex. | Sympodial, small denticles or scars, subhyaline. | 0–1-sepate, falcate, lunate, cylindrical or fusiform, hyaline | Not observed. |
| Idriella | Brown, mostly reduced to conidiogenous cells | Denticulate, sympodial | Aseptate, lunate, curved, hyaline | Brown, uni- or pluricellular. |
| Idriellopsis | Unbranched, brown at the base, almost hyaline at the apex, mostly reduced to conidiogenous cells | Terminal, denticulate | 0–1-septate, falcate, curved, hyaline | Not observed |
| Microdochium | More or less verticillate, reduced to conidiogenous cells, hyaline | Hyaline, sympodial or percurrent, sometimes denticulate | Aseptate or multiseptate, lunate, falcate, fusiform, filiform, obovoid or subpyriform, straight or curved, hyaline | Terminal or intercalary, solitary, in chains or grouped in clusters, brown. |
| Neoidriella | Mostly unbranched, pale brown, mostly reduced to conidiogenous cells | Sympodial, denticulate, terminal. | Aseptate, cylindrical to obovoid, hyaline | Intercalary or terminal, pale brown. |
| Paraidriella | Unbranched, pale brown, mostly reduced to conidiogenous cells. | Sympodial, denticulate, terminal. | Aseptate, cylindrical to oblong, hyaline | Not observed. |
| Selenodriella | Unbranched or verticillate, brown. | Sympodial, denticulate, terminal and intercalary. | Aseptate, falcate, hyaline | Not observed |
Supplementary Material
Acknowledgements
We would like to thank Dr. Shaun Pennycook (Landcare Research Manaaki Whenua, New Zealand) for advising on the fungal names. The research is supported by the Thailand Research grants entitled The future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species (grant no: DBG6080013), Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (grant no: RDG6130001). Y. Wang would like to thank the projects of the National Natural Science Foundation of China (No. 31560489, 31500451), Talent project of Guizhou science and technology cooperation platform ([2017]5788-5), Bijie Science and Technology Project ([2016]19) and Guizhou science and technology department international cooperation base project ([2018]5806). J.K. Liu would like to thank the National Natural Science Foundation of China (NSFC 31600032) and the Science and Technology Foundation of Guizhou Province (LH [2015]7061).
Citation
Lin C-G, Bhat DJ, Liu J-K, Hyde KD, Wang Y (2019) The genus Castanediella. MycoKeys 51: 1–14. https://doi.org/10.3897/mycokeys.51.32272
Funding Statement
The research is supported by the Thailand Research grants entitled The future of specialist fungi in a changing climate: baseline data for generalist and specialist fungi associated with ants, Rhododendron species and Dracaena species (grant no: DBG6080013), Impact of climate change on fungal diversity and biogeography in the Greater Mekong Subregion (grant no: RDG6130001). Y. Wang would like to thank he projects of National Natural Science Foundation of China (No. 31560489, 31500451), Talent project of Guizhou science and technology cooperation platform ([2017]5788-5), Bijie Science and Technology Project ([2016]19) and Guizhou science and technology department international cooperation base project ([2018]5806). J.K. Liu would like to thank the National Natural Science Foundation of China (NSFC 31600032) and the Science and Technology Foundation of Guizhou Province (LH [2015]7061).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990) Basic local alignment search tool. Journal of Molecular Biology 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Castañeda Ruiz RF, Heredia GP, Arias RM, Stadler M, Minter DW. (2005) Two Hyphomycetes from submerged plant material of México. Mycotaxon 91: 333–338. [Google Scholar]
- Chomnunti P, Hongsanan S, Aguirre-Hudson B, Tian Q, Persoh D, Dhami MK, Alias AS, Xu J, Liu X, Stadler M, Hyde KD. (2014) The sooty moulds. Fungal Diversity 66: 1–36. 10.1007/s13225-014-0278-5 [DOI] [Google Scholar]
- Costa PM, Barbosa MA, Da Silva GV, Sosa D, Pérez-Martinez S, Castañeda-Ruiz RF, Malosso E. (2018) Castanedielladiversispora sp. nov. from the Brazilian Atlantic Forest. Mycotaxon 133: 63–69. [Google Scholar]
- Crous PW, Wingfield MJ, Burgess TI, Hardy GESJ, Crane C, Barrett S, Cano-Lira JF, Leroux JJ, Thangavel R, Guarro J, Stchigel AM, Martín MP, Alfredo DS, Barber PA, Barreto RW, Baseia IG, Cano-Canals J, Cheewangkoon R, Ferreira RJ, Gené J, Lechat C, Moreno G, Roets F, Shivas RG, Sousa JO, Tan YP, Wiederhold NP, Abell SE, Accioly T, Albizu JL, Alves JL, Antoniolli ZI, Aplin N, Araújo J, Arzanlou M, Bezerra JDP, Bouchara JP, Carlavilla JR, Castillo A, Castroagudín VL, Ceresini PC, Claridge GF, Coelho G, Coimbra VRM, Costa LA, da Cunha KC, da silva SS, Daniel R, de beer ZW, Dueñas M, Edwards J, Enwistle P, Fiuza PO, Fournier J, García D, Gibertoni TB, Giraud S, Guevara-Suarez M, Gusmão LFP, Haituk S, Heykoop M, Hirooka Y, Hofmann TA, Houbraken J, Hughes DP, Kautmanová I, Koppel O, Koukol O, Larsson E, Latha KPD, Lee DH, Lisboa DO, Lisboa WS, López-Villalba Á, Maciel JLN, Manimohan P, Manjón JL, Marincowitz S, Marney TS, Meijer M, Miller AN, Olariaga I, Paiva LM, Piepenbring M, Poveda-Molero JC, Raj KNA, Raja HA, Rougeron A, Salcedo I, Samadi R, Santos TAB, Scarlett K, Seifert KA, Shuttleworth LA, Silva GA, Silva M, Siqueira JPZ, Souza-Motta CM, Stephenson SL. (2016a) Fungal Planet description sheets: 469–557. Persoonia - Molecular Phylogeny and Evolution of Fungi 37: 218–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, Hernández-Restrepo M, Sutton DA, Acharya K, Barber PA, Boekhout T, Dimitrov RA, Duenas M, Dutta AK, Gene J, Gouliamova DE, Groenewald M, Lombard L, Morozova OV, Sarkar J, Smith MT, Stchigel AM, Wiederhold NP, Alexandrova AV, Antelmi I, Armengol J, Barnes I, Cano-Lira JF, Castaneda Ruiz RF, Contu M, Courtecuisse PR, da Silveira AL, Decock CA, de Goes A, Edathodu J, Ercole E, Firmino AC, Fouriem A, Fournier J, Furtado EL, Geering ADW, Gershenzon J, Giraldo A, Gramaje D, Hammerbacher A, He XL, Haryadi D, Khemmuk W, Kovalenko AE, Krawczynski R, Laich F, Lechat C, Lopes UP, Madrid H, Malysheva EF, Marin-Felix Y, Martin MP, Mostert L, Nigro F, Pereira OL, Picillo B, Pinho DB, Popov ES, Pelaez CAR, Rooney-Latham S, Sandoval-Denis M, Shivas RG, Silva V, Stoilova-Disheval MM, Telleria MT, Ullah C, Unsickern SB, van der Merwe NA, Vizzini A, Wagner HG, Wong PTW, Wood AR, Groenewald JZ. (2015) Fungal Planet description sheets: 320–370. Persoonia 34: 167–266. 10.3767/003158515x688433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Richardson DM, Leroux JJ, Strasberg D, Edwards J, Roets F, Hubka V, Taylor PWJ, Heykoop M, Martín MP, Moreno G, Sutton DA, Wiederhold NP, Barnes CW, Carlavilla JR, Gené J, Giraldo A, Guarnaccia V, Guarro J, Hernández-Restrepo M, Kola, Ik M, Manjón JL, Pascoe IG, Popov ES, Sandoval-Denis M, Woudenberg JHC, Acharya K, Alexandrova AV, Alvarado P, Barbosa RN, Baseia IG, Blanchette RA, Boekhout T, Burgess TI, Cano-Lira JF, moková A, Dimitrov RA, Dyakov MY, Dueñas M, Dutta AK, Esteve-Raventós F, Fedosova AG, Fournier J, Gamboa P, Gouliamova DE, Grebenc T, Groenewald M, Hanse B, Hardy GESJ, Held BW, Jurjevi, Kaewgrajang T, Latha KPD, Lombard L, Luangsa-ard JJ, Lysková P, Mallátová N, Manimohan P, Miller AN, Mirabolfathy M, Morozova OV, Obodai M, Oliveira NT, Ordóñez ME, Otto EC, Paloi S, Peterson SW, Phosri C, Roux J, Salazar WA, Sánchez A, Sarria GA, Shin HD, Silva BDB, Silva GA, Smith MT, Souza-Motta CM, Stchigel AM, Stoilova-Disheva MM, Sulzbacher MA, Telleria MT, Toapanta C, Traba JM, Valenzuela-Lopez N, Watling R, Groenewald JZ. (2016b) Fungal Planet description sheets: 400–468. Persoonia - Molecular Phylogeny and Evolution of Fungi 36: 316–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 41: 95–98. [Google Scholar]
- Hernández-Restrepo M, Gené J, Castañeda-Ruiz RF, Mena-Portales J, Crous PW, Guarro J. (2017) Phylogeny of saprobic microfungi from Southern Europe. Studies in Mycology 86: 53–97. 10.1016/j.simyco.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Groenewald JZ, Crous PW. (2016a) Taxonomic and phylogenetic re-evaluation of Microdochium, Monographella and Idriella. Persoonia – Molecular Phylogeny and Evolution of Fungi 36: 57–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernández-Restrepo M, Schumacher RK, Wingfield MJ, Ahmad I, Cai L, Duong TA, Edwards J, Gené J, Groenewald JZ, Jabeen S. (2016b) Fungal Systematics and Evolution: FUSE 2. Sydowia 68: 193–230. [Google Scholar]
- Jayasiri SC, Hyde KD, Ariyawansa HA, Bhat J, Buyck B, Cai L, Dai Y-C, Abd-Elsalam KA, Ertz D, Hidayat I, Jeewon R, Jones EBG, Bahkali AH, Karunarathna SC, Liu J-K, Luangsa-ard JJ, Lumbsch HT, Maharachchikumbura SSN, McKenzie EHC, Moncalvo J-M, Ghobad-Nejhad M, Nilsson H, Pang K-L, Pereira OL, Phillips AJL, Raspé O, Rollins AW, Romero AI, Etayo J, Selçuk F, Stephenson SL, Suetrong S, Taylor JE, Tsui CKM, Vizzini A, Abdel-Wahab MA, Wen T-C, Boonmee S, Dai DQ, Daranagama DA, Dissanayake AJ, Ekanayaka AH, Fryar SC, Hongsanan S, Jayawardena RS, Li W-J, Perera RH, Phookamsak R, Silva NI, Thambugala KM, Tian Q, Wijayawardene NN, Zhao R-L, Zhao Q, Kang J-C, Promputtha I. (2015) The Faces of Fungi database: fungal names linked with morphology, phylogeny and human impacts. Fungal Diversity 74: 3–18. 10.1007/s13225-015-0351-8 [DOI] [Google Scholar]
- Jeewon R, Hyde KD. (2016) Establishing species boundaries and new taxa among fungi: recommendations to resolve taxonomic ambiguities. Mycosphere 7: 1669–1677. 10.5943/mycosphere/7/11/4 [DOI] [Google Scholar]
- Katoh K, Standley DM. (2016) A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32: 1933–1942. 10.1093/bioinformatics/btw108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsson A. (2014) AliView: a fast and lightweight alignment viewer and editor for large data sets. Bioinformatics 30: 3276–3278. 10.1093/bioinformatics/btu531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JK, Yang J, Maharachchikumbura SSN, McKenzie EHC, Jones EBG, Hyde KD, Liu ZY. (2016) Novel chaetosphaeriaceous hyphomycetes from aquatic habitats. Mycological Progress 15: 1157–1167. [Google Scholar]
- Nylander J. (2004) MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University.
- Page RDM. (1996) TreeView: an application to display phylogenetic trees on personal computers. Computer Applications in the Biosciences 12: 357–358. 10.1093/bioinformatics/12.4.357 [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. (1996) Probability distribution of molecular evolutionary trees: A new method of phylogenetic inference. Journal of Molecular Evolution 43: 304–311. 10.1007/BF02338839 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology 61: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert KA, Morgan-Jones G, Gams W, Kendrick B. (2011) The genera of hyphomycetes. CBS-KNAW Fungal Biodiversity Centre, Utrecht, The Netherlands.
- Silvestro D, Michalak I. (2012) raxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution 12: 335–337. [Google Scholar]
- Stamatakis A. (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30: 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. (2002) PAUP*: Phylogenetic Analysis Using Parsimony and other methods, version 4.0 b10. Sinauer Associates.
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya G, Lohman DJ, Meier R. (2011) SequenceMatrix: concatenation software for the fast assembly of multi-gene datasets with character set and codon information. Cladistics 27: 171–180. 10.1111/j.1096-0031.2010.00329.x [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanasinghe DN, Phukhamsakda C, Hyde KD, Jeewon R, Lee HB, Gareth Jones EB, Tibpromma S, Tennakoon DS, Dissanayake AJ, Jayasiri SC, Gafforov Y, Camporesi E, Bulgakov TS, Ekanayake AH, Perera RH, Samarakoon MC, Goonasekara ID, Mapook A, Li W-J, Senanayake IC, Li J, Norphanphoun C, Doilom M, Bahkali AH, Xu J, Mortimer PE, Tibell L, Tibell S, Karunarathna SC. (2018) Fungal diversity notes 709–839: taxonomic and phylogenetic contributions to fungal taxa with an emphasis on fungi on Rosaceae. Fungal Diversity 89: 1–236. 10.1007/s13225-018-0395-7 [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ. (Eds) PCR protocols: a guide to methods and applications.Academic Press, California, 315–322.
- Wijayawardene NN, Hyde KD, Lumbsch HT, Liu JK, Maharachchikumbura SSN, Ekanayaka AH, Tian Q, Phookamsak R. (2018) Outline of Ascomycota: 2017. Fungal Diversity 88: 167–263. 10.1007/s13225-018-0394-8 [DOI] [Google Scholar]
- Zhaxybayeva O, Gogarten JP. (2002) Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3: 4–4. 10.1186/1471-2164-3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.