Abstract
Enteroendocrine cells (EECs) produce hormones such as glucagon-like peptide 1 and peptide YY that regulate food absorption, insulin secretion, and appetite. Based on the success of glucagon-like peptide 1–based therapies for type 2 diabetes and obesity, EECs are themselves the focus of drug discovery programs to enhance gut hormone secretion. The aim of this study was to identify the transcriptome and peptidome of human EECs and to provide a cross-species comparison between humans and mice. By RNA sequencing of human EECs purified by flow cytometry after cell fixation and staining, we present a first transcriptomic analysis of human EEC populations and demonstrate a strong correlation with murine counterparts. RNA sequencing was deep enough to enable identification of low-abundance transcripts such as G-protein–coupled receptors and ion channels, revealing expression in human EECs of G-protein–coupled receptors previously found to play roles in postprandial nutrient detection. With liquid chromatography–tandem mass spectrometry, we profiled the gradients of peptide hormones along the human and mouse gut, including their sequences and posttranslational modifications. The transcriptomic and peptidomic profiles of human and mouse EECs and cross-species comparison will be valuable tools for drug discovery programs and for understanding human metabolism and the endocrine impacts of bariatric surgery.
Introduction
Enteroendocrine cells (EECs) are specialized hormone-secreting cells in the intestinal epithelium that monitor the quality and quantity of ingested foods. They produce at least 20 different hormones, mostly peptides, that act in concert to coordinate digestion, peripheral nutrient disposal, and appetite through actions at local and distant target tissues. In the field of human metabolism, glucagon-like peptide 1 (GLP-1) and peptide YY (PYY) have raised particular interest because of their central and pancreatic actions controlling food intake and insulin secretion. GLP-1–based drugs are widely used for the treatment of type 2 diabetes and obesity, and new gut hormone–based therapeutics are under development, aiming to mimic the unrivalled effectiveness of gastric bypass surgery on weight loss and type 2 diabetes resolution (1).
Recent years have witnessed substantial progress in our understanding of murine EEC physiology, facilitated by the generation of transgenic mice with fluorescently labeled EECs that enable cell identification and functional characterization through a range of approaches including fluorescence-activated cell sorting (FACS), transcriptomics, and live-cell imaging (2–7). Our knowledge of human EECs, however, is limited by a lack of methods to identify and characterize this scattered cell population that only comprises ∼1% of the intestinal epithelium (2). A number of G-protein–coupled receptors (GPCRs) have been identified and characterized in murine EECs that represent promising candidates for therapeutic approaches to enhance endogenous gut hormone secretion, but tools to predict the translatability of these findings from mouse to humans would be a major advance in this field (8).
The objectives of this study were to generate transcriptomic profiles of human EECs and to compare mouse and human EECs at the transcriptomic and peptidomic levels. We sequenced EECs from humans and mice at a depth sufficient for the identification of low-abundance transcripts, including GPCRs and ion channels. With liquid chromatography–tandem mass spectrometry (LC-MS/MS), we mapped the exact sequences of different gut peptides produced along the gastrointestinal (GI) tract in humans and mice.
Research Design and Methods
Ethics
This study was conducted in accordance with the principles of the Declaration of Helsinki and good clinical practice. Human ethics approvals were given by Cambridge Central and South Research Ethics Committees (ref: 09/H0308/24, 16/EE/0338, 15/EE/0152) and the INSERM ethics committee and Agence de la Biomédecine (ref: PFS16–004). Animal work was regulated under the Animals (Scientific Procedures) Act 1986 Amendment Regulations 2012 and conducted following ethics review by the University of Cambridge Animal Welfare and Ethical Review Body.
Human Tissue Transcriptome
Sample Collection
Jejunal tissue was obtained from 11 human participants (Supplementary Table 1). Samples of human jejunum discarded during surgery were collected during total gastrectomy for treatment or prophylaxis of gastric cancer or Roux-en-Y gastric bypass for obesity. All were from the point of enteroenterostomy 50 cm distal to the ligament of Treitz. Two matched samples of jejunum and terminal ileum were collected during organ procurement from transplant donors. Data were collected on age, sex, and BMI, and participants stratified as lean versus obese (BMI >30 kg/m2). Tissue samples from different regions of the human GI tract for LC-MS/MS were obtained from Addenbrooke’s Human Research Tissue Bank and the Cambridge Biorepository for Translational Medicine.
Samples were immediately placed in cold Leibovitz’s L-15 media (Thermo Scientific, Waltham, MA) and processed to the point of fixation or homogenized and stored at −70°C within 6 h.
Tissue Preparation for FACS
FACS and RNA extraction from fixed human cells followed a modified version of the MARIS protocol (9). Intestine was rinsed in cold PBS and the muscular coat was removed. Diced mucosa was digested twice in 0.1% w/v collagenase XI (Sigma-Aldrich, St. Louis, MO) in Hanks’ Buffered Saline solution (HBSS) #9394 (Sigma-Aldrich) for 30 min each, shaking vigorously every 10 min. Supernatants were triturated, passed through a 50-μm filter and centrifuged at 300g. Pellets were resuspended in PBS and fixed in 4% w/v paraformaldehyde (PFA) at 4°C for 20 min. PFA-fixed cells were washed twice in nuclease free 1% w/v BSA in PBS, and if a FACS facility was not immediately available, they were suspended in 1% w/v BSA and 4% v/v RNAsin plus RNAse inhibitor (Promega, Fitchburg, WI) in PBS at 4°C overnight.
Cells were permeabilized with either a single 30-min incubation with 0.1% v/v Triton ×100 (Sigma-Aldrich) in 1% w/v BSA in PBS prior to antibody staining or by the addition of 0.1% w/v Saponin (Sigma-Aldrich) to solutions in all steps from this point until after the first wash postsecondary antibody staining, with identical results.
Primary antibody staining was for 1 h in 4% v/v RNAsin, 1% w/v BSA, 1% v/v goat anti-GLP-1 (sc7782; Santa Cruz, Dallas, TX), 2% v/v rabbit anti-chromogranin A (CHGA) (Ab15160; Abcam, Cambridge, U.K.), and 0.25% v/v rabbit anti-secretogranin 2 (SCG2) (Ab12241; Abcam) in PBS at 4°C. Cells were then washed twice in 1% w/v BSA and 1% v/v RNAsin, and secondary antibody staining was for 30 min in 4% v/v RNAsin, 1% w/v BSA, 0.2% v/v donkey anti-goat Alexa 555, and 0.2% v/v donkey anti-rabbit Alex 647 in PBS at 4°C. Cells were washed twice then suspended in 4% v/v RNAsin and 1% w/v BSA in PBS on ice for FACS.
FACS
Cell populations were sorted on a BD FACS ARIA III in the National Institute for Health Research (NIHR) Cambridge BRC Cell Phenotyping Hub or at Institut Cochin (Paris, France). Single cells positive for Alexa 647 but not Alexa 555 (i.e., CHGA/SCGA positive but GLP-1 negative) were classified as proglucagon negative (GCG−) enteroendocrine cells. Single cells positive for both Alexa 647 and Alexa 555 were classified as GCG+ enteroendocrine cells. At least 5,000 cells were collected for each positive population. Twenty thousand double-negative cells were collected as the negative (i.e., nonenteroendocrine) cell population. Cells were sorted into 2% v/v RNAsin in PBS at 4°C.
RNA Extraction
RNA was extracted using the Ambion RecoverAll Total Nucleic Acid Isolation Kit for FFPE (Ambion, Foster City, CA) with modifications to the protocol as below. The FACS-sorted cell suspension was centrifuged at 3,000g for 5 min at 4°C, and the pellet resuspended in 200 μL digestion buffer with 4 μL protease and incubated at 50°C for 3 h. The solution was then stored at −70°C for at least 12 h prior to further extraction. After thawing, RNA was extracted using the manufacturer’s protocol (including a DNase step), with the exception of performing 2× 60 μL elutions from the filter column in the final step.
The RNA solution was concentrated using a RNEasy MinElute cleanup kit (Qiagen, Hilden, Germany). RNA aliquots were diluted to 200 μL with nuclease-free water. The standard manufacturer’s protocol was followed, with the exception that 700 µL, not 500 µL, of 100% ethanol was added to the solution in step two to generate optimum binding conditions for the PFA-fragmented RNA. RNA concentration and quality was analyzed using an Agilent 2100 Bioanalyser (Agilent, Santa Clara, CA).
Sequencing
cDNA libraries were created using the Clontech SMARTer Stranded Total RNA-Seq Kit Pico Input Mammalian v1 (Takara Bio, Mountain View, CA). RNA input quantity was 5 ng, and the nonfragmentation protocol was used. The standard manufacturer’s protocol was followed, with the exception that 175 μL of AMPure beads was used for the final bead purification to ensure recovery of the small fragments of RNA arising from PFA fixation. Sixteen PCR cycles were used for amplification.
Fifty base single-end sequencing was performed using an Illumina HiSEQ 4000 at the Cancer Research UK Cambridge Institute Genomics Core.
Mouse Transcriptome
Sample Collection and Preparation for FACS
Female NeuroD1-Cre/EYFP (mixed background, 3–10 generations backcrossed with C57BL/6) and GLU-Venus mice (C57BL/6) (6,7) aged 8–10 weeks were killed by cervical dislocation (n = 3 each). Diced mucosa from the proximal 10 cm of small intestine was digested twice in 0.1% w/v collagenase XI in HBSS at 37°C for 30 min each. Cells were pelleted by centrifugation at 300g for 5 min, triturated, and passed through a 50-μm filter. Cells were stained with DAPI (1 µg/mL) for 5 min at room temperature, washed twice, and sorted in HBSS on a FACSJazz sorter at the Cambridge NIHR BRC Cell Phenotyping Hub.
FACS and RNA Extraction
All positive cells, and 20,000 negative cells, were collected separately into aliquots of 500 μL of buffer RLT+ (Qiagen), with 143 mmol/L β-mercaptoethanol. RNA was extracted using a RNeasy Micro plus kit (Qiagen) and quantified using an Agilent 2100 Bioanalyser.
Sequencing
Two nanograms of each RNA were used for cDNA amplification by SPIA amplification using the Ovation RNAseq system V2 kit (NuGEN, Redwood City, CA). One microgram of cDNA was then fragmented to ∼200 bp by sonication (Diagenode, Liege, Belgium), and adaptors for the indexing were added using the Ovation Rapid DR multiplex 1–96 kit. Samples were pooled and concentrated together with a MinElute column (Qiagen) to reach a concentration of 10 nmol/L. Single-end 50 base sequencing was performed at the Cancer Research UK Cambridge Institute Genomics Core with an Illumina Hiseq4000.
RNAseq Pipeline
Quality control and trimming of adaptors was performed using FastQC (10). Human sequenced transcripts were mapped to the human genome (GRCh37), and raw counts were generated using STAR v2.5.1. Mouse reads were aligned to the mouse genome (GRCm38) using TopHat 2.1.0, and raw counts were generated using Cufflinks 2.2.1 (11–13). Differential gene expression analysis was performed in RStudio using DESEQ2 (14). Gene annotation was pulled from the Ensembl data set held in BioMart (11). Receptor and ion channel lists were generated from the International Union of Basic and Clinical Pharmacology (IUPHAR)/British Pharmacological Society (BPS) “targets and families” list (15). Graphical output used ggplot2 and pheatmap in RStudio (16).
Comparative Transcriptomics
Mouse and human data sets were directly compared using only the 15,507 genes present in both data sets, not annotated as ribosomal, mitochondrial, or small nuclear, and described with one-to-one homology according to the Ensembl mouse-human homology data set (11). Genes with no one-to-one homology were analyzed separately. Normalized CPM (counts per million) were generated from the respective DESEQ2 models and compared for the human GCG+ population versus the murine GLU-Venus population and the human GCG− population versus the murine NeuroD1 population. Linear models were generated of the log10 CPM of the human versus murine data sets by a total least squares strategy, 99% CI calculated, and the outliers hand-searched for relevant genes.
Examining just the human samples, DESEQ2 models were generated for the following sets of jejunum samples: GCG+ versus negative, GCG− versus negative, GCG+ versus GCG−, GCG+ lean versus obese, and GCG− lean versus obese. Participant-paired DESEQ2 analyses were also performed comparing GCG+ and GCG− populations from the jejunum and ileum of the two transplant donor participants, for whom there were matched jejunum and ileum samples. An adjusted P value of ≤0.1 was defined as the cutoff for statistical significance when examining for differential gene expression between populations.
Tissue Homogenates
Small pieces (∼2 × 2 mm, 15–45 mg) of mucosa from different regions of the human or mouse GI tract were homogenized in 250 μL of 6 mol/L guanidine hydrochloride (Sigma-Aldrich) with Lyzing MatrixD (MPbio) in a FastPrep-24 for 4 × 40 s at 6 ms−1. Samples were stored at −70°C before further processing. Proteins were precipitated by adding 80% acetonitrile in water then centrifuged at 12,000g at 4°C for 5 min. The aqueous phase containing the peptides was collected, dried on a centrifugal vacuum concentrator, and stored at −70°C before analysis.
Mass Spectrometry
Samples were extracted using a Waters HLB μElution solid‐phase extraction plate (Waters, Milford, MA) after being resuspended in 500 μL 0.1% v/v formic acid in water as described previously and analyzed after reduction/alkylation (17). Human homogenates and mouse homogenates for interspecies comparison were analyzed using nanoflow-based separation and electrospray approaches on a Thermo Fisher Ultimate 3000 Nano LC system coupled to a Q Exactive Plus Orbitrap mass spectrometer (Thermo Scientific). High-flow separation for the longitudinal mouse analysis was done as previously described (17). Downstream analysis was performed using Peaks 8.0 software (Waterloo, Ontario, Canada) against the human and the mouse Swiss-Prot databases (downloaded 26 October 2017) (18), with a fixed cysteine carbamidomethylation and variable methionine oxidation, N-terminal acetylation, and pyro-glutamate and COOH-terminal amidation modifications. Manual searches were performed for other modifications. Peptides of interest were quantified by measuring peak areas for selected m/z ranges and retention times corresponding to the peptide sequences and normalized by tissue weight.
Results
Cell and RNA Collection for Transcriptomics
Human intestinal tissue pieces were digested, PFA fixed, and stained for CHGA and SCG2 as general markers for EECs and for GLP-1 as a marker of the EEC-subpopulation known as L-cells. By flow cytometry (Supplementary Fig. 1A–C), we collected pooled cell populations that were 1) positive for CHGA, SCG2, and GLP-1 (henceforth named GCG+), 2) positive for CHGA and SCG2 but negative for GLP-1 (henceforth named GCG−), and 3) negative for all three markers (i.e., nonendocrine lineage cells, henceforth named negative). The GCG+ (L-cell) population represented ∼0.2% of all single cells examined, and the ratio of GCG+ to GCG− cells was ∼1:5.
From mice, we collected unfixed EEC populations from the upper small intestine of the mouse strain GLU-Venus (n = 3) to identify Gcg-expressing L-cells (Supplementary Fig. 1E–G) (7) and of NeuroD1-Cre/EYFP strain (6) (n = 3) to identify the total EEC population (Supplementary Fig. 1I–K). GLU-Venus positive cells represented ∼0.2% of singlets, and NeuroD1 positive cells, ∼0.6% of singlets.
RNA extracted from the purified fixed human cell populations had measured RNA integrity number (RIN) values of 2–3 (compared with >7 in the unfixed murine cells), with most RNA fragments being 25–500 bases in length (Supplementary Fig. 1D). The low RIN values of the human samples reflect the simultaneous fragmentation of ribosomal RNAs, but we predicted that the fragments were of lengths suitable for RNA sequencing using random primers and an rRNA depletion protocol. After RNA sequencing, a mean of 77% of reads from the human samples mapped to the human genome, compared with 86% of reads mapping to the murine genome in the mouse samples. The mean number of reads mapping uniquely to exons was 3.5 × 106 (range 0.8 × 106 to 8.8 × 106) in the human samples, and 3.9 × 106 (range 2.0 × 106 to 5.7 × 106) in murine samples.
Transcriptomics of Human EEC Populations
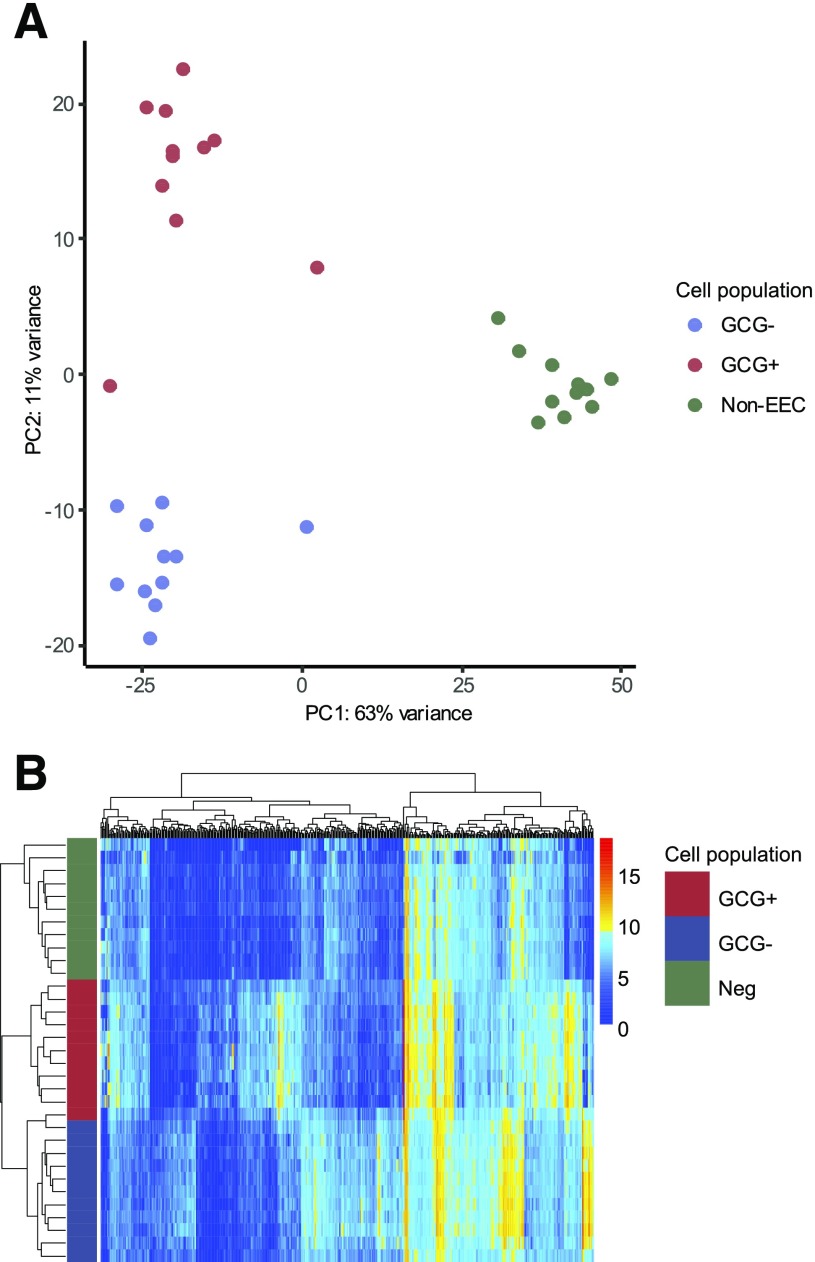
In total, we obtained individual RNA sequencing data from GCG+, GCG−, and negative cells from each of 11 human jejunum samples and 2 human ileum samples. Principle component analysis of the top 500 differentially expressed genes in the jejunal samples separated EECs (GCG+ and GCG−) from negative cells on the first component, and GCG+ cells from GCG− cells on the second component (Fig. 1A). Principle component analysis did not demonstrate clustering of samples by BMI of the donor. A heatmap of the top 500 differentially expressed genes (Fig. 1B) revealed clear transcriptomic differences between the GCG+, GCG−, and negative cells (14). A comparison between jejunal and ileal cell populations from the two matched donors is shown in Supplementary Fig. 2.
Figure 1.
Transcriptomic distinction between cell populations from human small intestine. A: Principal component (PC) analysis plot of all human samples (n = 3 cell populations from each of 11 jejunal tissue samples), differentiated by cell population. B: Pairwise analysis of key genes differentiating the cell populations from jejunum was performed using a DESEQ2 model, and the normalized results for the 500 most differentially expressed genes are presented in a heatmap (n = 3 cell populations per each of 11 participants). The y-axis is cell population identified by colored bar, and x-axis is genes.
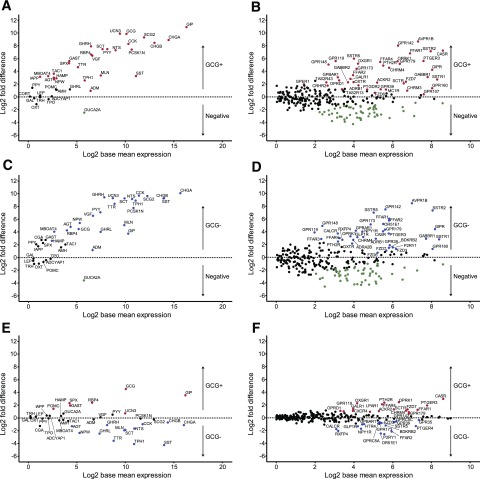
Human jejunal EEC populations expressed a wide range of hormonal transcripts (Fig. 2A, C, and E). As GLP-1 antibodies were used to purify the GCG+ cells, we were not surprised that the hormonal transcript showing the strongest differential expression between GCG+ and GCG− cells was GCG itself (Fig. 2C). Consistent with previous findings in mice (3,19–21), human GCG+ cells also expressed a range of additional hormonal transcripts including GIP (glucose-dependent insulinotropic polypeptide), CCK (cholecystokinin), NTS (neurotensin), PYY, and SCT (secretin) as well as MLN (motilin), a hormone produced by human but not mouse (3,19–22). Compared with GCG+ cells, GCG− cells had higher expression of SCT, CCK, NTS, MLN, GHRL (ghrelin), and SST (somatostatin), as well as TPH1, the enzyme responsible for serotonin biosynthesis in enterochromaffin cells. EECs also expressed the putative gut hormones UCN3 (urocortin 3), PCSK1N (ProSAAS), and NPW (neuropeptide W), as well as lower levels of RNAs encoding peptides not classically described as gut hormones, such as VGF, GHRH, and ADM.
Figure 2.
Transcripts enriched in human EECs. Enrichment vs. expression plots for human jejunum EEC populations. Enrichment is presented as the log2-fold difference between the cell populations indicated, and expression is presented as the log2 base mean normalized expression extracted from the DESEQ2 model. A and B: GCG+ vs. negative. C and D: GCG− vs. negative. E and F: GCG+ vs. GCG−. Hormones and granins are shown in A, C, and E; receptors and ion channels are shown in B, D, and F. Red, enriched (adjusted P ≤ 0.1 in DESEQ2 model) in GCG+; blue, enriched in GCG−; green, enriched in negative cells.
Transcripts of ∼50 GPCRs were either enriched in EECs compared with negative cells, or expressed at >100 CPM in one or both EEC populations (Fig. 2B, D, and F). Multiple GPCRs previously implicated in postprandial gut hormone secretion in mice were highly expressed in human EECs, including the fat-sensing receptors FFAR1, FFAR2, FFAR3, FFAR4, and GPR119, the amino acid–sensing receptors CASR and GPR142, the butyrate- and isovalerate-sensing OR51E1 (23), and the bile acid receptor, GPBAR1. At least four orphan GPCRs were differentially expressed in human EECs, GPR148, GPR160, GPR173, and GPR179, hinting to as-yet-undescribed pathways that may control gut hormone secretion.
Human EECs expressed transcripts for a range of ion channel subunits (Supplementary Fig. 3), consistent with previous reports that murine L-cells and enterochromaffin cells are electrically active (24). Transcription factor profiling of human GCG+ and GCG− cells is shown in Supplementary Fig. 3.
Comparison of Human Versus Mouse EEC Transcriptomes
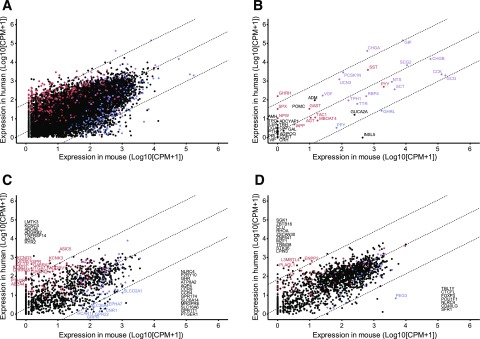
To compare the L-cell populations between humans and mice, we compared human jejunal GCG+ cells with murine upper small intestinal GLU-Venus cells (Fig. 3). Log-log plots of normalized gene expression indicated a strong correlation between the two species (R2 = 0.73) (Fig. 4). Notable genes more highly expressed in human than mouse L-cells included GIP, CHGA, ASIC5, GIPR, GPR142, SCTR, PTH2R, CHRNA5, and OPRK1, whereas genes more highly expressed in mice included Gpr174, Gpr171, Ghr, Grpr, Ptger1, Cnr1, Insl5, Gpr22, and Ghrl.
Figure 3.
Comparison between human and mouse L-cells. Human vs. mouse jejunal L-cell gene expression (log10 normalized CPM + 1; n = 11 humans, n = 3 mice). A: All genes with 1:1 homology between species, excluding mitochondrial, ribosomal, and small nuclear transcripts (n = 15,507). B: Hormones. C: GPCRs and ion channels. D: Transcription factors. Dashed lines are linear regression and 99% CI. Each dot represents normalized CPM + 1 for one gene. Red, enriched (>fourfold change) and differentially expressed (adjusted P ≤ 0.1) for human GCG+ vs. negative cell populations, but not murine GLU-Venus vs. negative cell populations in relevant DESEQ2 model. Blue, enriched and differentially expressed for murine GLU-Venus vs. negative cell populations, but not human GCG+ vs. negative cell populations. Purple, enriched and differentially expressed in both murine GLU-Venus and human GCG+ cells vs. relevant negative cell populations. Black, not enriched or differentially expressed in either human GCG+ or murine GLU-Venus cells vs. relevant negative cell populations. All genes are labeled in B, and genes outside the 99% CI are labeled in C and D, with those not differentially expressed or enriched in either population listed along the axis.
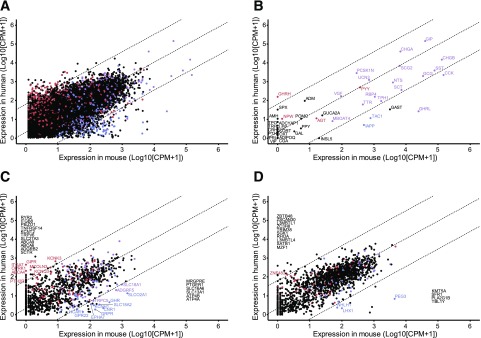
Figure 4.
Comparison between human GCG− and mouse NeuroD1 cells. Human vs. mouse jejunal EEC (GCG−) gene expression (log10 normalized CPM + 1; n = 11 humans, n = 3 mice). A: All genes with 1:1 homology between species, excluding mitochondrial, ribosomal, and small nuclear transcripts (n = 15,507). B: Hormones. C: GPCRs and ion channels. D: Transcription factors. Dashed lines are linear regression and 99% CI. Each dot represents normalized CPM + 1 for one gene. Red, enriched (>fourfold change) and differentially expressed (adjusted P ≤ 0.1) for human GCG− vs. negative cell populations, but not murine NeuroD1 vs. negative cell populations in relevant DESEQ2 model. Blue, enriched and differentially expressed for murine NeuroD1 vs. negative cell populations, but not human GCG− vs. negative cell populations. Purple, enriched and differentially expressed in both murine NeuroD1 and human GCG− cells vs. relevant negative cell populations. Black, not enriched or differentially expressed in either human GCG− or murine NeuroD1 cells vs. relevant negative cell populations. All genes are labeled in B, and genes outside the 99% CI are labeled in C and D, with those not differentially expressed or enriched in either population listed along the axis.
A similar comparison was made between human jejunal GCG− cells, representing the wider EEC population (although depleted of L-cells) and murine NeuroD1-positive cells, which similarly revealed a strong interspecies correlation (R2 = 0.74) (Fig. 4). Data on transcription factors, GPCRs, ion channels, and hormones that were EEC-enriched in either species but excluded from the comparative analysis because they did not have annotated 1:1 orthologs between mouse and human are given in Supplementary Table 2 and notably highlight MLN and GPR148 as EEC-enriched transcripts in humans that are not found in mice.
Peptidomic Analysis by LC-MS/MS
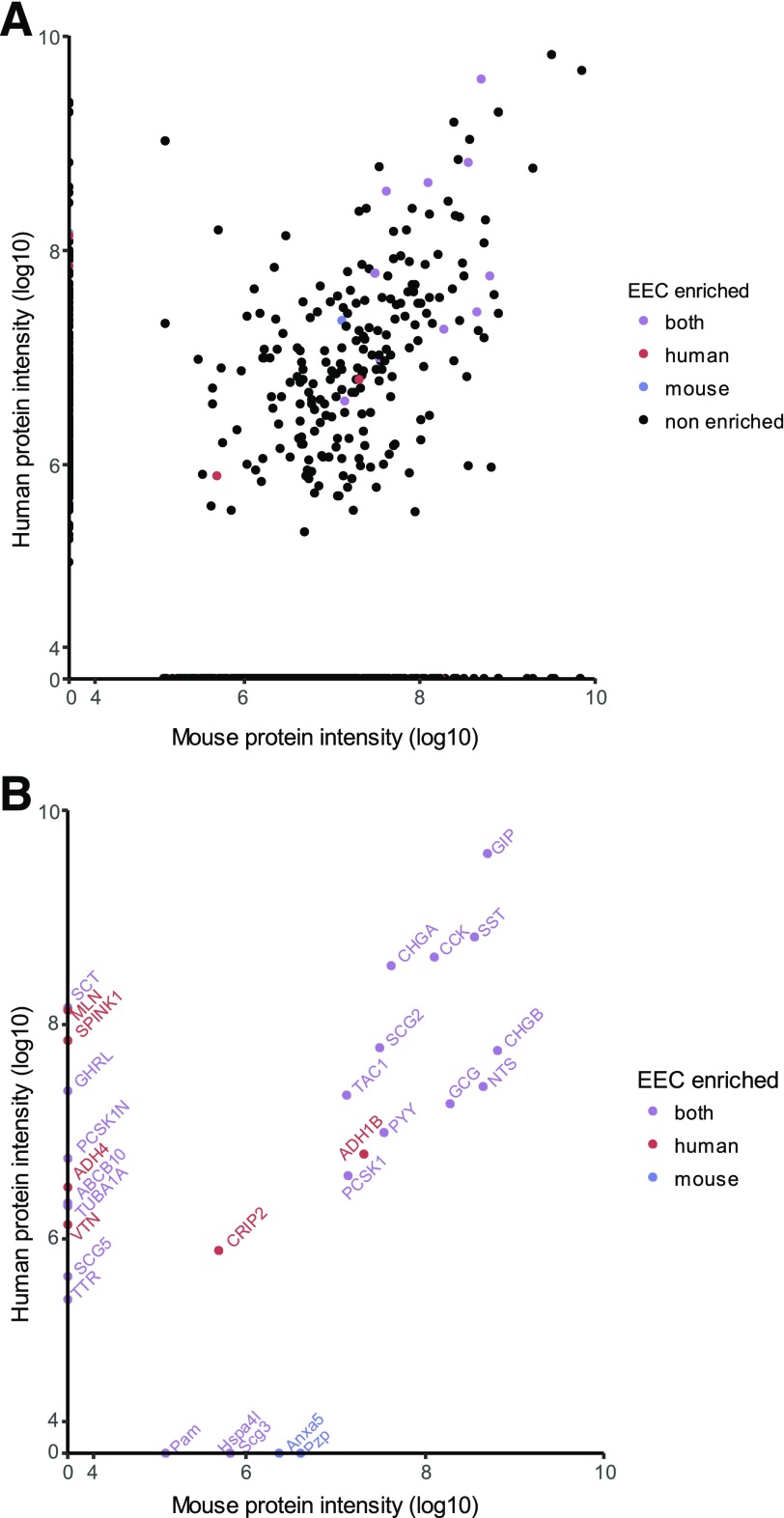
Peptide extraction and LC-MS/MS protocols were optimized for maximum peptide retrieval and identification from fresh intestinal mucosal samples (Supplementary Fig. 4) and enabled reliable detection and sequencing of peptides up to 65 amino acids in length. In the first instance, we compared human jejunum with mouse mid–small intestine and assigned peptides to their parental proteins. This method identified known EEC prohormones, granins, and enteric neuronal signaling peptides, as well as peptides derived from a variety of housekeeping proteins that likely reflected the occurrence of some tissue damage/degradation prior to homogenization. Of the 463 and 705 different proteins matched in humans and mice, respectively, 234 were common between the two species and showed good correlation (R2 = 0.54) (Fig. 5A). To identify candidate EEC-derived peptides, we restricted the analysis to peptides originating from genes that in the transcriptomic analysis showed a greater than fourfold higher expression in at least one EEC sample compared with the corresponding negative cells (Fig. 5B). Known gut hormone genes and members of the chromogranin family were mostly found to be common between mouse and human, but motilin and ghrelin were detected in human but not mouse jejunum. To search for novel candidate peptide hormones, we also examined the transcriptomic data for unannotated transcripts that had a base mean value >100 and were greater than 16-fold more highly expressed in EECs than control cells. This analysis identified MIR7–3HG (25), C1orf127, and C6orf141, but we were unable to detect corresponding peptides by LC-MS/MS.
Figure 5.
Comparison between human and mouse jejunum peptidome. Tissue samples were analyzed by nano-LC-MS/MS and compared between human jejunum (n = 4) and mouse mid–small intestine (n = 4). Peptides were assigned to their parental proteins by Peaks software, and protein intensity was calculated by Peaks v8.0 for all proteins detected corresponding to genes with 1:1 homology between human and mouse for jejunum mucosal homogenates. A: All proteins. B: Proteins from genes enriched in at least one of the species from the human and the mouse transcriptome data sets. Enrichment was defined as adjusted P ≤ 0.1, fold difference >4, and base mean expression >50 from DESEQ2 model. Colors indicate in which species the mRNA for the genes was enriched.
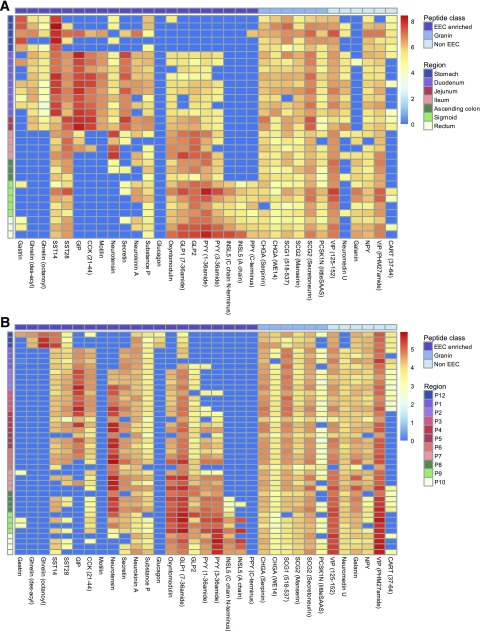
We performed a longitudinal LC-MS/MS analysis of known bioactive peptides along the mouse and human GI tracts. Most EEC peptides were identifiable in their known bioactive forms, but as our method was not optimized for very small peptides like CCK-8, we used CCK21-44 as a surrogate for CCK production. Peptides have been depicted in separate heatmaps for mouse and human (Fig. 6), separated according to their origin from EEC prohormones, granins, and non-EECs (likely reflecting enteric neural peptides). We observed longitudinal peptide profiles that broadly mirror historical immunostaining patterns (26) but additionally provide details of the exact peptide sequences and their posttranslational modifications.
Figure 6.
Longitudinal profiles of gut peptides along the human and mouse gut. Heatmap of gut peptide quantification normalized by tissue weight (log10 scale) along human (A) and mouse (B) GI tract. Sequential samples were taken at 5-cm intervals from the stomach to the rectum in mice (n = 4 for each location), and human biopsies were analyzed from the stomach (n = 5), duodenum (n = 9), jejunum (n = 2), ileum (n = 4), proximal colon (n = 3), sigmoid colon (n = 5), and rectum (n = 3). Blue indicates not detected in the sample. Rows: samples ordered from proximal to distal and color-coded by their region of origin. Mouse regions: P12, stomach lesser curvature; P1–P7, small intestine from proximal to distal, sampling every 5 cm; P8–P10, large intestine (proximal, mid, and distal). Columns: peptide (using the human name if different between human and mouse), classified by origin (purple, classic EEC hormone peptides; medium blue, peptides from granins; light blue, enteric neuron bioactive peptides).
Discussion
By RNA sequencing of fixed, FACS-purified cells from the human gut, we report here the transcriptome of human L-cells and the wider EEC population and a between-species comparison showing a strong similarity with matching cells from the mouse. LC-MS/MS–based peptidomic analysis of the human and mouse gut revealed longitudinal gradients of a range of EEC peptides, including details of their exact sequences and posttranslational modifications.
To purify human EECs for transcriptomics, cells were first fixed, permeabilized, and antibody stained, and the RNA extraction and sequencing kits were only just technically capable of dealing with PFA-modified RNA. Nevertheless, the RNAseq-derived transcriptomes of human L-cells and GCG− EECs mapped robustly onto the corresponding data from freshly purified matching murine EEC populations and the read depth was sufficient for the detection of low-abundance mRNAs, such as those encoding receptors and ion channels. Although we analyzed samples from 11 humans with varying BMI, we were unable to detect any effect of BMI on the EEC transcriptome. However, the study was not designed or powered to address the impact of BMI or diet on EECs, and further studies will be needed to examine this question.
GPCRs play key sensory roles in EECs and a number are under investigation as candidate drug targets for enhancing gut hormone secretion as a therapy for type 2 diabetes and obesity. The GPCR repertoire of human EECs largely mirrored their well-studied murine counterparts (8,27–29), including expression of receptors for amino acids (CASR, GPR142), triglyceride digestion products (FFAR1, FFAR4, GPR119), and bile acids (GPBAR1), as well as for hormones such as somatostatin (SSTR1, SSTR2, SSTR5), GIP (GIPR), and arginine vasopressin (AVPR1B). Interestingly, GPR142 was highly expressed in human EECs, supporting current studies looking to exploit its ability to stimulate both insulin and incretin hormone secretion (28).
Our optimized peptide extraction protocol combined with nano-LC-MS/MS analysis enabled identification of the exact peptide sequences biosynthesized in human and mouse intestinal mucosa, including posttranslational modifications, for peptides ranging from ∼8–10 to 65 amino acids in length. From the proglucagon gene, for example, we detected multiple processed and preprocessed products, including GRPP, oxyntomodulin, GLP-17-36 amide, GLP-17-37, GLP-11-37, IP131-142, IP-GLP2, and GLP-2. Intact (pancreatic-type) glucagon was detected in samples from the mouse stomach but was undetectable in the remainder of the intestine and colon from both species, conflicting with recent suggestions that the small intestine secretes intact glucagon (30) but consistent with our recent finding that postprandial glucagon concentrations were not altered following gastric bypass surgery in lean subjects despite dramatic increases in GLP-1 (31). LC-MS/MS also identified some additional peptides encoded by EEC-enriched genes, including peptides derived from PCSK1N, chromogranins, and secretogranins. Whether any of these have specific physiological roles or are simply inactive by-products of enzymatic processing of the contents of secretory vesicles requires further evaluation.
Mapping of gut hormone production along the GI tract length has previously been performed by immunostaining or extraction/immunoassays for specific peptides (26,32). Many antibody-based methods, however, do not distinguish whether a prohormone was processed or unprocessed or posttranslationally modified. Our LC-MS/MS method provides a robust mirror of previous antibody-based maps of the GI tract, while additionally assigning an exact peptide sequence to each identified peptide, clearly distinguishing, for example, oxyntomodulin from glucagon and PYY1-36 from PYY3-36. Interestingly, we identified acylated as well as nonacylated ghrelin from the human jejunum despite our previous finding that plasma acylated ghrelin levels were undetectable in humans after total gastrectomy (31). We were surprised to find high levels of PYY3-36 as well as PYY1-36 in tissue homogenates, suggesting that dipeptidyl peptidases (DPPs) are active within L-cells, although GLP-17-36 amide was much more abundant than GLP-19-36 amide. Why GLP-1 but not PYY seems protected from DPP-mediated processing in L-cells, despite both peptides being located in the same vesicular pool (33), remains unclear.
Conclusion
The methods we describe here for performing RNA sequencing of rare cell populations and LC-MS/MS–based peptidomic analysis from human surgical tissue samples have wide potential applications beyond the study of the enteroendocrine system, including islet cell biology. Although comparison of the human and mouse EEC transcriptomes revealed strong global similarities between the two species, variation at the level of individual genes could have profound implications for the use of mice as a model species in drug development programs targeting specific receptors. Our mouse/human comparative data sets provide tools for assessing the validity of using mice as a model for investigating specific signaling pathways in humans, and the human EEC GPCR-ome can be used independently as a potential source of drug targets in the human enteroendocrine system. Longitudinal peptide mapping of the GI tract by LC-MS/MS is a key step toward understanding the metabolic benefits of gastric bypass surgery, as most bariatric procedures result in a shift in the location of nutrient digestion and absorption to more distal regions of the small intestine, with consequent stimulation of the more distal EEC population and release of their distinct profiles of peptide hormones. The results presented here should guide strategies to mimic gastric bypass surgery using injectable peptide mimetics or by targeting EEC-enriched receptors to mobilize endogenous gut hormones, which are highly topical challenges for industry and academia alike.
Supplementary Material
Article Information
Acknowledgments. The authors would like to thank the surgical team in the Cambridge Oesophago-Gastric Centre in Cambridge and Sébastien Czernichow (Assistance Publique Hôpitaux de Paris, Georges Pompidou European Hospital, Paris, France) and Cyrille Ramond (Department of Endocrinology, Metabolism and Diabetes, INSERM U1016, Institut Cochin, Université Paris-Descartes, Paris, France), as well as the Cambridge Biorepository for Translational Medicine and the NIHR Cambridge Human Research Tissue Bank for assistance with the collection of donor tissue. FACS was supported by the Institut Cochin in Paris and the Cambridge NIHR BRC Cell Phenotyping Hub. RNA-sequencing was undertaken at the Cancer Research UK Cambridge Institute Genomics Core.
Funding. Metabolic Research Laboratories support was provided by the following core facilities: Disease Model Core, Genomics and Transcriptomics Core, Histology Core, Imaging Core, and NIHR BRC Core Biochemical Assay Laboratory (supported by the Medical Research Council [MRC_MC_UU_12012/5] and Wellcome Trust [100574/Z/12/Z]). G.P.R. received an Addenbrooke’s Charitable Trust/Evelyn Trust Cambridge Clinical Research Fellowship [16-69], an European Foundation for the Study of Diabetes project grant, and a Royal College of Surgeons Research Fellowship. S.G.G. is supported by a grant from the Biotechnology and Biological Sciences Research Council. Research in the laboratory of F.M.G. and F.R. is supported by the MRC (MRC_MC_UU_12012/3) and Wellcome Trust (106262/Z/14/Z and 106263/Z/14/Z) The MS instrument was funded by the MRC “Enhancing UK clinical research” grant (MR/M009041/1). P.R. was supported by a postdoctoral grant from Agence Nationale de la Recherche (Laboratoire d'Excellence Revive, Investissement d'Avenir, ANR-10-LABX-73). The laboratory of R.S. is supported by Agence Nationale de la Recherche (ANR-10-LABX-73) and the Bettencourt-Schueller Foundation. This work was partially supported by the following National Institutes of Health grants R01 DK100223-02 and R01 DK110614-01 (to A.L.).
Duality of Interest. F.M.G. is a paid consultant for and P.R. has (subsequent to his work on this project) taken up full-time employment with Kallyope (New York, NY). The laboratory of F.R. and F.M.G. hosts projects that receive funding from Medimmune. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. G.P.R., P.L., P.R., R.S., R.H.H., F.R., and F.M.G. designed the study. Tissue handling and FACS were performed by G.P.R., P.L., P.R., and L.L.G. cDNA library production and bioinformatics analysis were performed by G.P.R., P.L., P.R., M.K.L.M., and B.L., with support from G.S.H.Y. and D.C. Mass spectrometry was performed by G.P.R., P.L., R.G.K., S.G.G., and E.L.M. NeuroD1 mice were generated by A.L. and H.J.L. The manuscript was written by G.P.R., F.R., and F.M.G. All authors had editorial oversight of the final manuscript version. F.R. and F.M.G. are the guarantors of this work and, as such, had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Data Availability. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifiers PXD009788 (10.6019/PXD009788) and PXD009796 (10.6019/PXD009796) (see Vizcaíno JA, Csordas A, del-Toro N, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 2016;44:D447–D456). RNA-seq data has been deposited in the NCBI GEO repository (human, GSE114853; mouse, GSE114913).
Prior Presentation. A non–peer-reviewed version of this article was submitted to the bioRxiv preprint server (doi: https://doi.org/10.1101/374579) on 23 July 2018.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-0883/-/DC1.
See accompanying article, p. 904.
References
- 1.Holst JJ, Madsbad S. Mechanisms of surgical control of type 2 diabetes: GLP-1 is key factor. Surg Obes Relat Dis 2016;12:1236–1242 [DOI] [PubMed]
- 2.Gribble FM, Reimann F. Enteroendocrine cells: chemosensors in the intestinal epithelium. Annu Rev Physiol 2016;78:277–299 [DOI] [PubMed] [Google Scholar]
- 3.Glass LL, Calero-Nieto FJ, Jawaid W, et al. Single-cell RNA-sequencing reveals a distinct population of proglucagon-expressing cells specific to the mouse upper small intestine. Mol Metab 2017;6:1296–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Svendsen B, Pais R, Engelstoft MS, et al. GLP1- and GIP-producing cells rarely overlap and differ by bombesin receptor-2 expression and responsiveness. J Endocrinol 2016;228:39–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adriaenssens A, Lam BY, Billing L, et al. A transcriptome-led exploration of molecular mechanisms regulating somatostatin-producing D-cells in the gastric epithelium. Endocrinology 2015;156:3924–3936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li HJ, Kapoor A, Giel-Moloney M, Rindi G, Leiter AB. Notch signaling differentially regulates the cell fate of early endocrine precursor cells and their maturing descendants in the mouse pancreas and intestine. Dev Biol 2012;371:156–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reimann F, Habib AM, Tolhurst G, Parker HE, Rogers GJ, Gribble FM. Glucose sensing in L cells: a primary cell study. Cell Metab 2008;8:532–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reimann F, Tolhurst G, Gribble FM. G-protein-coupled receptors in intestinal chemosensation. Cell Metab 2012;15:421–431 [DOI] [PubMed] [Google Scholar]
- 9.Hrvatin S, Deng F, O’Donnell CW, Gifford DK, Melton DA. MARIS: method for analyzing RNA following intracellular sorting. PLoS One 2014;9:e89459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Babraham Bioinformatics. FastQC [Internet], 2010. Available from http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 1 February 2010
- 11.Ensembl. BioMart [Internet], 2018. Available from https://www.ensembl.org/biomart/martview/c68208e4252974c97a2be0f8947c62b7. Accessed 9 October 2018
- 12.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trapnell C, Williams BA, Pertea G, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.IUPHAR/BPS Guide to Pharmacology. Targets and families list [Internet], 2018. Available from http://www.guidetopharmacology.org/download.jsp. Accessed 28 February 2018
- 16.Wickham H. ggplot2: Elegant Graphics for Data Analysis. New York, Springer-Verlag, 2009 [Google Scholar]
- 17.Kay RG, Galvin S, Larraufie P, Reimann F, Gribble FM. Liquid chromatography/mass spectrometry based detection and semi-quantitative analysis of INSL5 in human and murine tissues. Rapid Commun Mass Spectrom 2017;31:1963–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.UniProt. SwissProt/UniProt database [Internet], 2016. Available from www.uniprot.org. Accessed 26 October 2017
- 19.Habib AM, Richards P, Cairns LS, et al. Overlap of endocrine hormone expression in the mouse intestine revealed by transcriptional profiling and flow cytometry. Endocrinology 2012;153:3054–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egerod KL, Engelstoft MS, Grunddal KV, et al. A major lineage of enteroendocrine cells coexpress CCK, secretin, GIP, GLP-1, PYY, and neurotensin but not somatostatin. Endocrinology 2012;153:5782–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grunddal KV, Ratner CF, Svendsen B, et al. neurotensin is coexpressed, coreleased, and acts together with GLP-1 and PYY in enteroendocrine control of metabolism. Endocrinology 2016;157:176–194 [DOI] [PubMed] [Google Scholar]
- 22.Sanger GJ, Wang Y, Hobson A, Broad J. Motilin: towards a new understanding of the gastrointestinal neuropharmacology and therapeutic use of motilin receptor agonists. Br J Pharmacol 2013;170:1323–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bellono NW, Bayrer JR, Leitch DB, et al. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell 2017;170:185–198.e16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers GJ, Tolhurst G, Ramzan A, et al. Electrical activity-triggered glucagon-like peptide-1 secretion from primary murine L-cells. J Physiol 2011;589:1081–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka S, Tatsumi K, Okubo K, et al. Expression profile of active genes in the human pituitary gland. J Mol Endocrinol 2002;28:33–44 [DOI] [PubMed] [Google Scholar]
- 26.Sjölund K, Sandén G, Håkanson R, Sundler F. Endocrine cells in human intestine: an immunocytochemical study. Gastroenterology 1983;85:1120–1130 [PubMed] [Google Scholar]
- 27.Diakogiannaki E, Pais R, Tolhurst G, et al. Oligopeptides stimulate glucagon-like peptide-1 secretion in mice through proton-coupled uptake and the calcium-sensing receptor. Diabetologia 2013;56:2688–2696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin HV, Efanov AM, Fang X, et al. GPR142 controls tryptophan-induced insulin and incretin hormone secretion to improve glucose metabolism. PLoS One 2016;11:e0157298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Psichas A, Larraufie PF, Goldspink DA, Gribble FM, Reimann F. Chylomicrons stimulate incretin secretion in mouse and human cells. Diabetologia 2017;60:2475–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Knop FK. EJE PRIZE 2018: a gut feeling about glucagon. Eur J Endocrinol 2018;178:R267–R280 [DOI] [PubMed] [Google Scholar]
- 31.Roberts GP, Kay RG, Howard J, Hardwick RH, Reimann F, Gribble FM. Gastrectomy with Roux-en-Y reconstruction as a lean model of bariatric surgery. Surg Obes Relat Dis 2018;14:562–568 [DOI] [PMC free article] [PubMed]
- 32.Baldissera FG, Holst JJ, Jensen SL, Krarup T. Distribution and molecular forms of peptides containing somatostatin immunodeterminants in extracts from the entire gastrointestinal tract of man and pig. Biochim Biophys Acta 1985;838:132–143 [DOI] [PubMed] [Google Scholar]
- 33.Billing LJ, Smith CA, Larraufie P, et al. Co-storage and release of insulin-like peptide-5, glucagon-like peptide-1 and peptideYY from murine and human colonic enteroendocrine cells. Mol Metab 2018;16:65–75 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.