Abstract
Insulin is a major autoantigen in type 1 diabetes, targeted by both CD8 and CD4 T cells. We studied an insulin-reactive T-cell receptor (TCR) α-chain transgenic NOD mouse on a TCRCα and proinsulin 2 (PI2)-deficient background, designated as A22Cα−/−PI2−/− NOD mice. These mice develop a low incidence of autoimmune diabetes. To test the role of gut microbiota on diabetes development in this model system, we treated the A22Cα−/−PI2−/− NOD mice with enrofloxacin, a broad-spectrum antibiotic. The treatment led to male mice developing accelerated diabetes. We found that enrofloxacin increased the frequency of the insulin-reactive CD8+ T cells and activated the cells in the Peyer’s patches and pancreatic lymph nodes, together with induction of immunological effects on the antigen-presenting cell populations. The composition of gut microbiota differed between the enrofloxacin-treated and untreated mice and also between the enrofloxacin-treated mice that developed diabetes compared with those that remained normoglycemic. Our results provide evidence that the composition of the gut microbiota is important for determining the expansion and activation of insulin-reactive CD8+ T cells.
Introduction
The incidence of type 1 diabetes (T1D) is increasing worldwide at a rate too rapid to be associated purely with genetic changes (1), and thus environmental factors, such as the gut microbiota, have been suggested to contribute to T1D development (2). The gut microbiota composition (3–5) and function (6) are altered in patients with T1D. In the NOD mouse model, which develops spontaneous autoimmune diabetes similar to humans, altered gut microbiota are also found in the diabetic NOD mice compared with nondiabetic NOD mice (7). Modifying the gut microbiota by fecal transfer studies (8), dietary changes (9,10), and the administration of antibiotics (dependent on type, age at which administered, and duration) (11–17) all affect diabetes development in NOD mice. Recently, we showed that islet-specific glucose-6-phosphatase catalytic subunit-related protein (IGRP)-reactive CD8+ T cells can recognize a fusobacterial peptide more strongly than their natural autoantigen (18), suggesting that islet autoimmunity can be activated by molecular mimicry. Furthermore, microbial metabolites released from the diet protect NOD mice by reducing the number of IGRP-reactive CD8+ T cells (10). Interestingly, the development of IGRP-reactive CD8+ T cells is dependent on prior insulin autoimmunity (19,20).
Proinsulin (PI) is a major autoantigen in humans (21–25) and NOD mice (26–30). PI is cleaved within the pancreatic β-cells, leading to the regulated secretion of metabolically active insulin. There are two forms of PI in mice, designated PI1 and PI2. PI2 is expressed in the thymus and pancreas, and PI2-deficient mice NOD mice developed accelerated diabetes with 100% penetrance (31). PI2 is thus considered to be important in the induction of T-cell tolerance.
G9C8 is a highly diabetogenic murine CD8+ T-cell clone that recognizes insulin B15-23 through its T-cell receptor (TCR) comprising TCRα chain (TRAV8-1/TRAJ9) and TCRβ chain (TRBV19/TRBJ2-3) gene rearrangements (32). G9C8 CD8+ T cells can be found in the islets in 4-week-old mice (29), along with other insulin-specific T cells, before islet antigen-specific T cells of other specificities are detected (33) and are required for the development of other autoantigen-specific CD8+ T cells (19,20). We previously generated a transgenic NOD mouse (line 22) with a fixed TRAV8-1/TRAJ9 TCRα chain only (29) (termed A22 for simplicity). A22 mice were bred to the PI2-deficient background to study the development and activation of insulin B15-23–reactive CD8+ T cells (34). We found that the PI2-deficient A22Cα−/− NOD mice had an increased proportion of insulin B15-23–reactive CD8+ T cells in the pancreatic draining lymph nodes (PLNs) compared with A22Cα−/− NOD mice that have normal levels of PI2. Furthermore, only male, but not female, PI2-deficient A22Cα−/− NOD mice (A22Cα−/−PI2−/− NOD) developed spontaneous diabetes.
In this study, by changing the gut microbiota, we have demonstrated that a broad-spectrum antibiotic enrofloxacin (Baytril) can alter insulin-specific CD8+ T-cell function and enable them to expand and become activated, leading to an early onset of diabetes in A22Cα−/−PI2−/− NOD mice.
Research Design and Methods
Mice
NOD/Caj mice were originally obtained from Yale University. G9Cα−/− NOD, G9Cα−/−PI2−/− NOD, and A22Cα−/−PI2−/− NOD have all been previously described and are summarized in Supplementary Table 1 (34–36). The current study used male mice from litters divided between treatment groups (Fig. 1A). Mice from several breeder pairs were mixed and housed in microisolators or scantainers with food and water ad libitum, with 12-h light and dark cycles, in the specific pathogen-free facility at Cardiff University. All procedures were performed in accordance with U.K. Home Office–approved protocols.
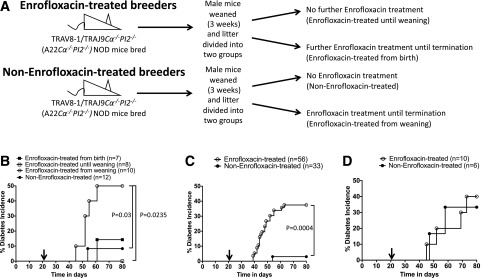
Figure 1.
Experimental design and spontaneous diabetes development. A: The experimental schema of enrofloxacin treatment, whereby A22Cα−/−PI2−/− NOD breeder mice (10 different breeder pairs were used) were treated with enrofloxacin (top) or untreated (bottom). A22Cα−/−PI2−/− NOD litters from these breeders were then randomly chosen and equally divided into enrofloxacin-treated or untreated groups to minimize any breeder effects. B: Diabetes incidence between different enrofloxacin treatment regimens of A22Cα−/−PI2−/− NOD mice outlined in A. C: Diabetes incidence of enrofloxacin-treated (from weaning) or untreated A22Cα−/−PI2−/− NOD mice. D: Diabetes incidence of enrofloxacin-treated (from weaning) or untreated G9Cα−/−PI2−/− NOD mice. Black arrows indicate the time of weaning. Statistical analysis was performed using the log-rank test.
Preparation and Administration of Enrofloxacin-Treated Water
Enrofloxacin (Bayer) was added to autoclaved, filtered water at a final concentration of 0.4 mg/mL (diluted 1:250), prepared freshly every week. Untreated mice received the same autoclaved, filtered water. Mice were treated from 3 weeks of age (at weaning) continuously until 10 weeks of age, unless otherwise stated.
Diabetes Incidence
Mice were monitored weekly for glycosuria (Bayer Diastix) from 5 weeks of age until termination. Diabetes was diagnosed after two consecutive positive glycosuria tests, confirmed by a blood glucose concentration >13.9 mmol/L (>250 mg/dL). Statistical analysis was performed using the log-rank test.
Surface and Intracellular Staining
Lymphoid tissues, including spleen, PLNs, mesenteric lymph nodes (MLNs), and Peyer’s patches (PPs), were collected from 6-week-old or 10-week-old mice. Cells were homogenized and filtered. Splenic red blood cells were lysed using water, followed by 10× PBS. Then, 1 × 106 cells were incubated with TruStain FcX (BioLegend) in PBS + 0.5% BSA at 4°C for 10 min before cell staining (4°C, 30 min) using combinations of the following monoclonal antibodies (all from BioLegend unless stated): anti-B220, anti-CD4, anti-CD8α, anti-CD11b, anti-CD11c, anti-CD19, anti-CD80, anti-CD103, anti-F4/80, anti-TCRβ, anti-TCRVβ4, anti-TCRVβ5.1-5.2, anti-TCRVβ6, anti-TCRVβ8.1-8.2 (BD Biosciences), anti-TCRVβ12 (eBioscience), anti-TCRVβ11 (eBioscience), anti-TCRVβ14 (BD Biosciences), and anti-MHCII (BD Biosciences) in the presence of a viability dye (BioLegend or eBioscience).
For intracellular staining, 1–2 × 106 cells were incubated at 37°C in the presence of 50 ng/mL phorbol 12-myristate 13-acetate (PMA), 500 ng/mL ionomycin, and 2 μmol/L monensin (all from Sigma-Aldrich) for 4 h. Cells were then stained for surface molecules before fixation/permeabilization and the addition of intracellular monoclonal antibodies (all BioLegend): anti-IFN-γ, anti-IL-10, or anti-FoxP3. Cells were analyzed on a BD LSRFortessa FACS machine and subsequently analyzed with FlowJo 8.8.6 software. Statistical analysis was performed by the Student t test.
Generating Tetramers
H-2Kd-LYLVCGERG monomers or H-2Kd-AYAAAAAAV monomers (National Institutes of Health Tetramer Core Facility) were slowly defrosted on ice. Tetramers were made as described previously (37).
Tetramer Staining
Tetramer staining was conducted as previously reported (34). G9C8 transgenic T cells (35) or NY8.3 transgenic T cells (38) were used as a positive control. The minimal H-2Kd-AYAAAAAAV tetramer was used to determine nonspecific background, which was then subtracted from the H-2Kd–LYLVCGERG–Brilliant Violet 421 or H-2Kd–VYLKTNVFL–allophycocyanin tetramer staining result, respectively. Cells were analyzed as described above. Statistical analysis was performed by the Student t test.
Fecal Bacterial Extraction
Fecal samples were collected from 3-week-old and 6-week-old mice. Fecal bacteria were extracted and sequenced as previously described (8). β-Diversity was calculated to compare differences between microbial community profiles, and the data are shown as a principal coordinate analysis (PCoA). Microbial composition was analyzed by the Student t test, with β-diversity PCoA plots analyzed by analysis of similarities.
In Vitro Antigen Presentation
CD11c+ and CD11b+ cells were isolated from the spleens of 6-week-old A22Cα−/−PI2−/− NOD mice untreated or treated with enrofloxacin. CD8+ T cells were isolated by negative selection from the spleens of 6-week-old G9Cα−/− NOD mice. All cell isolations were conducted following the manufacturer’s protocols (Miltenyi). CD8+ T cells were labeled with carboxyfluorescein succinimidyl ester (CFSE) and cocultured with the antigen-presenting cells (APCs) for 48 h in a 1:1 ratio in the presence or absence of insulin B15-23 peptide. Cells were stained for surface markers and analyzed as previously described. Statistical analysis was performed by the Student t test.
In Vitro Splenocyte and Bacteria Coculture
The small intestines were harvested from 6-week-old mice and flushed with 5 mL sterile PBS. The gut contents were mixed thoroughly by vortex for 2 min, followed by centrifuging for 5 min at low speed (52g) to remove dietary residue. The supernatant was transferred to a new tube and spun. The pellet was washed twice more, and the combined supernatant was centrifuged at 469g to remove mammalian cells. Bacteria in the supernatant were pelleted by high-speed centrifugation (1,876g) for 5 min and resuspended in PBS. Bacterial concentration was measured with a spectrophotometer (Bio-Rad) and heat inactivated at 90°C for 20 min. Then 108 heat-inactivated bacteria were cocultured with splenocytes (2 × 106/mL) for 12 h before the addition of 50 ng/mL PMA and 500 ng/mL ionomycin in the presence of 2 μmol/L monensin (all Sigma-Aldrich). Cells were then stained for surface and intracellular markers 4 h later, as outlined above. Statistical analysis was performed by multiple Student t tests, corrected using the false discovery rate (FDR).
Results
Enrofloxacin Treatment Accelerates and Increases Spontaneous Diabetes Development in Male PI2-Deficient TRAV8-1/TRAJ9Cα−/− NOD Mice
Single TCRα chain transgenic NOD mice, using the TCR TRAV8–1/TRAJ9Cα−/− (designated A22Cα−/−) (34), bred with PI2-deficient mice (A22Cα−/−PI2−/−), have elevated levels of insulin B15-23–reactive CD8+ T cells and develop a low incidence of spontaneous diabetes. To test whether the gut microbiota may influence the expansion and activation of insulin B15-23–reactive CD8+ T cells, as recently reported for other autoreactive T cells (18,39,40), we administered enrofloxacin (a broad-spectrum antibiotic) to the breeder A22Cα−/−PI2−/− NOD mice throughout pregnancy or to the A22Cα−/−PI2−/− NOD pups from weaning at 3 weeks (Fig. 1A). We found that spontaneous diabetes developed in ∼50% of A22Cα−/−PI2−/− NOD male mice given enrofloxacin at the time of weaning compared with A22Cα−/−PI2−/− NOD mice never exposed to enrofloxacin (∼10%) or to A22Cα−/−PI2−/− NOD mice treated with enrofloxacin from birth (15%) or from birth to weaning only (0%) (Fig. 1B). We repeated this experiment focusing only on mice given enrofloxacin from weaning (enrofloxacin-treated) compared with untreated mice and again found a significant increase in spontaneous diabetes development in those given enrofloxacin from weaning (Fig. 1C). Whereas the A22Cα−/−PI2−/− NOD mice have the same TCRα chain as G9C8 CD8+ T cells but can pair with any endogenous TCRβ chain, the G9Cα−/−PI2−/− NOD mice express both the rearranged G9C8TCRα- and β chains. Interestingly, unlike the effect in A22Cα−/−PI2−/− NOD mice, enrofloxacin did not affect the incidence of diabetes in G9Cα−/−PI2−/− NOD mice (Fig. 1D). Thus, gut bacterial composition may alter expansion of particular TCR clonotypes, other than the clonotypic G9C8TCR, that recognize insulin B15-23, which we next investigated.
Enrofloxacin Treatment Leads to Insulin B15-23–Reactive CD8+ T-Cell Expansion, Altered TRBV Repertoire, and Activation
All T cells in A22Cα−/−PI2−/− NOD mice have the fixed TCRα chain derived originally from a CD8+ T cell, and thus, we investigated the effect of enrofloxacin treatment on shaping the CD8+ T-cell compartment. We found that in all tissues, except the PPs, there was an age-associated increase in CD8+ T cells, while CD4+ T cells were reduced (Supplementary Fig. 1A and B and Pearson et al. [34]). No significant differences in the proportion of FoxP3+ regulatory T cells (Tregs) between enrofloxacin-treated or untreated mice were observed in the PLNs (Supplementary Fig. 1C) or PPs, although there was an age-related increase in the untreated mice.
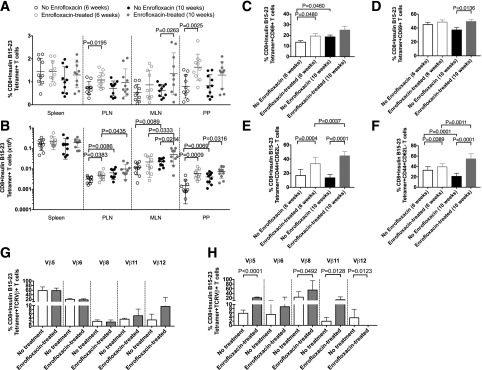
To identify the insulin-specific CD8+ T cells, we used an insulin B15-23 tetramer to investigate whether antibiotic usage affected insulin B15-23–specific CD8+ T cells in different lymphoid tissues. Interestingly, our results demonstrated a significant increase in the proportion and number of insulin B15-23 tetramer–positive CD8+ T cells in the PLNs, MLNs, and PPs of both 6-week-old and 10-week-old A22Cα−/−PI2−/− NOD mice treated with enrofloxacin compared with the untreated mice (Fig. 2A and B and Supplementary Fig. 2A). The expansion of these insulin-reactive CD8+ T cells was more pronounced in the 6-week-old mice, corresponding with the earliest development of diabetes in the enrofloxacin-treated mice. Moreover, we observed an increased percentage of activated (CD69+) tetramer-positive CD8+ T cells in the PLNs and PPs of 6-week-old and 10-week-old enrofloxacin-treated mice, respectively, compared with the untreated mice (Fig. 2C and D and Supplementary Fig. 2B). We also observed that tetramer-negative CD8+CD69+ T cells were reduced in the PPs with age in untreated mice but remained at a consistent proportion in enrofloxacin-treated mice (Supplementary Fig. 2C and D). Furthermore, we found that in both the PLNs and PPs of enrofloxacin-treated mice, the effector/memory (CD44+CD62L−) population of insulin B15-23 tetramer–positive CD8+ T cells significantly increased with age (Fig. 2E and F and Supplementary Fig. 2E). The opposite trend was observed in untreated mice. Interestingly, in tetramer-negative CD8+ T cells, we observed an increased effector/memory population only at 10 weeks of age in enrofloxacin-treated mice versus untreated mice (Supplementary Fig. 2F and G). We further analyzed TCRVβ use and found changes in multiple TCRVβ chains in both the total CD8+ T-cell repertoire and the insulin-specific CD8+ T-cell repertoire from the PLNs and PPs (Fig. 2G and H and Supplementary Figs. 3 and 4). Interestingly, no significant differences were observed in the selection of TCRVβ6 (the chain used by G9C8 T cells). These data indicate earlier activation of the insulin-reactive CD8+ T cells in the PLNs of enrofloxacin-treated mice as well as more highly activated insulin-reactive CD8+ T cells, which have an altered TCRβ repertoire, in the PPs of 10-week-old mice.
Figure 2.
Insulin B15-23 T cells are increased in number and activation with enrofloxacin treatment. A and B: Frequency and absolute number of insulin B15-23–reactive CD8+ T cells gated from live single TCRβ+CD4−CD19−CD11b−CD11c− cells. C and D: Frequency of CD69+ insulin B15-23–reactive CD8+ T-cell population from the PLNs (C) and PPs (D). Cells were gated as in A and B before subsequently gating on CD69. E and F: Frequency of insulin B15-23–reactive CD8+ T cells expressing CD44+CD62L− in the PLNs (E) and PPs (F). Cells were gated as in A and B before subsequently gating by CD44 and CD62L expression. G and H: Frequency of TCRβ chains expressed by insulin B15-23–reactive CD8+ T cells in 6-week-old A22Cα−/−PI2−/− NOD mice by enrofloxacin treatment (n = 7–10) in the PLNs (G) and PPs (H). All statistical analyses were conducted using a Student t test. Data are presented as mean ± SEM.
Enrofloxacin Alters APCs
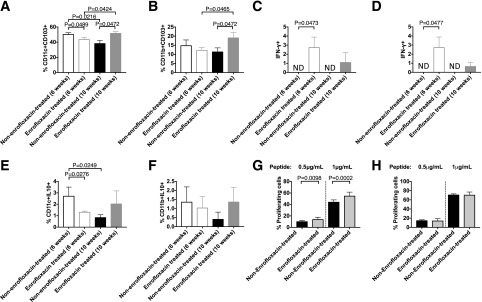
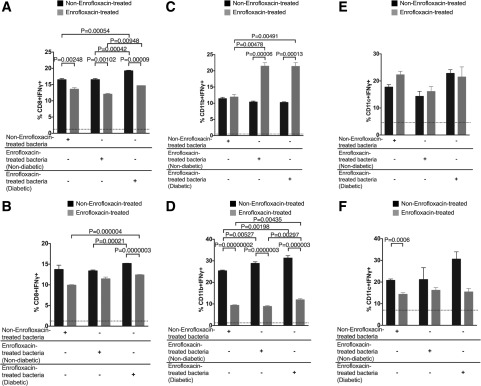
To explore the mechanism by which enrofloxacin treatment led to the increase in activated insulin B15-23–specific CD8+ T cells in A22Cα−/−PI2−/− NOD mice, we tested APCs from the PLNs and PPs. Here, we found changes in dendritic cells (DCs) and macrophages (Fig. 3A–F and Supplementary Fig. 5), as was observed in other studies using antibiotics in wild-type NOD mice (14,16). CD103 is an integrin marker of migratory DCs (41) and influenced by microbial challenge (42). We found a reduction in both CD11c+CD103+ and CD11b+CD103+ cells in the PPs of the enrofloxacin-treated mice at 6 weeks compared with untreated mice but an increase at 10 weeks, whereas these cells were reduced with age in untreated mice (Fig. 3A and B). These effects were not observed in the PLNs (data not shown). Moreover, the DCs and macrophages expressed increased interferon-γ (IFN-γ) production, particularly in 6-week-old enrofloxacin-treated mice (Fig. 3C and D). There were also fewer DCs producing interleukin-10 (IL-10), with a similar trend in the macrophages in the enrofloxacin-treated mice compared with control mice at 6 weeks (Fig. 3E and F), although not sustained by 10 weeks. Although it has been suggested that CD11b+CD11c+ cells are more inflammatory (43), we did not find any changes in CD11b+CD11c+ cells in B cells in the PPs, including IgA+ B cells (Supplementary Fig. 6).
Figure 3.
Direct ex vivo assessment of APC phenotype in untreated and enrofloxacin-treated A22Cα−/−PI2−/− NOD mice. CD11c+ cells or CD11b+ cells were isolated from the spleens of enrofloxacin-treated or untreated A22Cα−/−PI2−/− NOD mice and cocultured in vitro with CFSE-labeled G9CD8+ T cells for 72 h in the presence of insulin B15-23 peptide. A: Frequency of CD11c+CD103+ cells gated from live single CD19−TCRβ−MHCII+CD11b−F480− cells. B: Frequency of CD11b+CD103+ cells gated from live single CD19−TCRβ−MHCII+CD11c−F480+ cells. IFN-γ (C) and IL-10 (E) were assessed in CD11c+ cells gated from live single CD19−TCRβ−MHCII+CD11b−F480− cells. IFN-γ (D) and IL-10 (F) were assessed in CD11b+ cells gated from live single CD19−TCRβ−MHCII+CD11c−F480+ cells. All cells shown in A–F were from PPs. CFSE-labeled G9 CD8+ T cells were cocultured with CD11c+ cells (G) or CD11b+ cells (H) from enrofloxacin-treated or untreated A22Cα−/−PI2−/− NOD mice in a 1:1 ratio in the presence or absence of insulin B15-23 peptide for 48 h. Data shown were corrected for background (cells cocultured without peptide). All statistical analyses were conducted using a Student t test. All data were pooled from two independent experiments (n = 10). Data are presented as mean ± SEM. ND, not detected.
To determine whether these APC subsets were able to promote the proliferation of insulin-reactive CD8+ T cells, CFSE-labeled G9C8 CD8+ T cells were cocultured with CD11c+ cells or CD11b+ cells from donors treated with or without enrofloxacin in the presence of insulin B15-23 peptide. We found CD11c+ cells but not CD11b+ cells from enrofloxacin-treated mice promoted the proliferation of insulin-reactive CD8+ T cells (Fig. 3G and H and Supplementary Fig. 7). This was not due to a direct action of enrofloxacin on the cells, as we tested the effect of enrofloxacin in cell culture and this decreased CD69 expression in T cells at lower peptide concentrations compared with the effects seen in the absence of enrofloxacin. However, no differences were found in IFN-γ secretion (Supplementary Fig. 8), suggesting that the enrofloxacin treatment affected the APCs in the enrofloxacin-treated mice and that these played an important role in modifying T-cell function. Thus, enrofloxacin treatment induced a more inflammatory profile in some APC subsets, which could contribute to increased activation of insulin-reactive CD8+ T cells, particularly at 6 weeks of age.
Enrofloxacin Modifies the Gut Microbiota
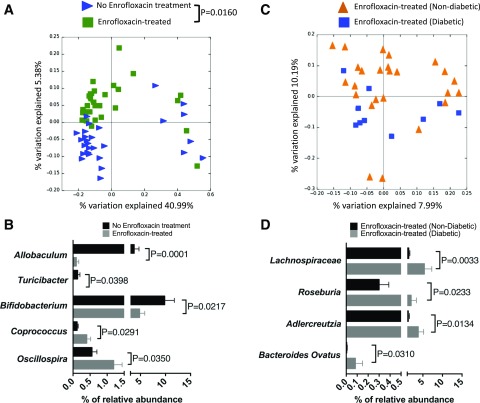
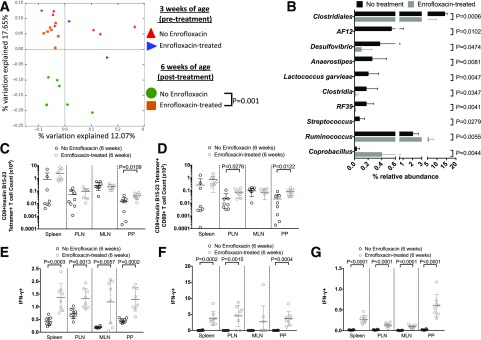
Enrofloxacin targets both gram-positive and gram-negative bacteria, and we therefore asked whether altered microbiota were driving diabetes development. We sequenced gut microbiota from the fecal pellets of 3-week-old mice (before antibiotic treatment) and 6-week-old mice (3 weeks after commencement of antibiotics) when most diabetes develops. Interestingly, α-diversity (referring to total bacterial diversity), as measured by Chao richness, was not altered in the enrofloxacin-treatment groups, regardless of diabetes development and age (Supplementary Fig. 9A). Furthermore, no significant differences in β-diversity (differences in the bacterial composition between treated and untreated mice) were seen at 3 weeks of age, confirming that there was no initial pretreatment difference in bacterial composition that could contribute to altered diabetes development (Supplementary Fig. 9B). However, we observed significant differences in 6-week-old enrofloxacin-treated compared with untreated mice (Fig. 4A). Further, enrofloxacin-treated mice had a decrease in the relative abundance of Allobaculum, Turicibacter, and Bifidobacterium but increased abundances of Coprococcus and Oscillospira (all at the genus level) compared with untreated mice (Fig. 4B). Bifidobacterium has been associated with reducing gut permeability (44), and Oscillospira is associated with increased gut permeability (45). Thus, the reduced Bifidobacterium and increased Oscillospira induced by enrofloxacin may both promote a more permeable intestinal epithelial barrier in our mice.
Figure 4.
Altered microbial composition in A22Cα−/−PI2−/− NOD mice by enrofloxacin treatment and diabetes development. A: PCoA plot of β-diversity from fecal bacteria of 6-week-old A22Cα−/−PI2−/− NOD mice that were untreated (n = 29) or treated with enrofloxacin (n = 35). B: Altered microbiota composition expressed as a percentage of relative abundance of the bacterial genus. All bacteria belong to the Firmicutes phylum except Bifidobacterium, which belongs to the Actinobacteria phylum. C: PCoA plot of β-diversity from fecal bacteria of 6-week-old A22Cα−/−PI2−/− NOD mice treated with enrofloxacin that were nondiabetic (n = 25) or diabetic (n = 10) by the end of the observation period (10 weeks old). D: Altered microbiota composition expressed as a percentage of relative abundance from the genus level except Bacteroides ovatus (species level) and Lachnospiraceae (family level). Lachnospiraceae and Roseburia (a member of Lachnospiraceae) are both members of the Firmicutes phylum; Bacteroides ovatus belongs within the Bacteroidetes phylum, whereas Adlercreutzia belongs within the Actinobacteria phylum. Analysis of similarities was used to analyze β-diversity of taxonomic families of gut microbiota in A and C. B and D were assessed for statistical significance using a Student t test.
Next, the gut microbiota composition in enrofloxacin-treated A22Cα−/−PI2−/− NOD mice, which did not develop diabetes, was compared with the composition in those that later developed disease. Although there were no significant differences in β-diversity in 6-week-old enrofloxacin-treated mice, with or without diabetes, some of the samples from the diabetic mice clustered closely, whereas the others were scattered widely (Fig. 4C). However, there were significant differences at the genus, family, and species level between these two groups (Fig. 4D). These included increased relative abundances of Adlercreutzia (genus level), Roseburia (genus level), Lachnospiraceae (family level), and Bacteroidetes ovatus (species level) in those mice that later developed diabetes compared with those that were protected (Fig. 4D). Our results are in line with the findings by Krych et al. (7), who demonstrated an increased relative abundance of Firmicutes (inclusive of Coprococcus, Oscillospira, Lachnospiraceae, and Roseburia) correlated to diabetes development in wild-type NOD mice.
Enrofloxacin-Modified Gut Microbiota Promote Stronger Proinflammatory Immune Responses
To assess the functional effects of the microbiota induced by enrofloxacin treatment and diabetes development, we performed crisscross in vitro cultures of splenocytes from 6- and 10-week-old mice with heat-killed microbiota from enrofloxacin-treated diabetic and nondiabetic mice as well as from untreated nondiabetic mice (all microbiota donors were 6 weeks old) as controls. We also cultured splenocytes from each of the mouse groups, without gut bacteria, and no differences were found between any of the groups.
Gut bacteria from the diabetic enrofloxacin-treated donors induced the highest proportion of IFN-γ–producing CD8+ T cells in the splenocytes of 6- and 10-week-old untreated mice compared with the gut bacteria from nondiabetic enrofloxacin-treated and untreated mice (Fig. 5A and B and Supplementary Fig. 10). Furthermore, gut bacteria from enrofloxacin-treated donors, with or without diabetes, induced a greater frequency of IFN-γ–producing splenocyte CD11b+ cells from enrofloxacin-treated mice compared with splenocyte CD11b+ cells from untreated mice (Fig. 5C and Supplementary Fig. 11). It is interesting that the profile was reversed by 10 weeks of age, with the highest frequency of IFN-γ–producing splenocyte CD11b+ cells of untreated mice after culture with bacteria (Fig. 5D). We found no significant differences in the induction of IFN-γ–producing splenocyte CD11c+ cells of 6- or 10-week-old mice by the different sources of bacteria (Fig. 5E and F and Supplementary Fig. 12). Splenocyte CD11b+ cells from 6-week-old treated and 10-week-old untreated mice showed the strongest inflammatory response to the gut bacteria, regardless of the source (Fig. 5C and D). We found no obvious changes in CD11b+CD11c+ cells, B cells, or CD4+ T cells producing transforming growth factor-β, IL-10, IL-17a, IL-12/23, and tumor necrosis factor-α (data not shown).
Figure 5.
Effect of culture of A22Cα−/−PI2−/− NOD mouse splenocytes with small-intestinal bacteria from untreated or enrofloxacin-treated mice. Intracellular IFN-γ was investigated in CD8+ T cells (gated from TCRβ+CD19−CD4) (A and B), CD11b+ cells (gated from TCRβ−CD19−IAg7+CD11c−F480+) (C and D), and CD11c+ cells (gated from TCRβ−CD19−IAg7+CD11b−F480−) (E and F). Splenocyte donors were studied at both 6 (A, C, E) and 10 weeks of age (B, D, F). Average baseline levels of splenocyte IFN-γ–producing cells, cultured without bacteria, were similar in all treatment groups and are shown by the dotted line. The labeling beneath the graphs represents the origin of the bacteria: bacteria from non-enrofloxacin-treated mice labeled as non-enrofloxacin-treated bacteria, bacteria from nondiabetic enrofloxacin-treated mice labeled as enrofloxacin-treated bacteria (nondiabetic), and bacteria from diabetic enrofloxacin-treated mice labeled as enrofloxacin-treated bacteria (diabetic). All splenocyte cultures with bacteria displayed significant differences from those without bacteria. All data were pooled from three independent experiments (n = 8/group; from three to four different breeders) and were assessed for statistical significance using multiple Student t tests corrected using FDR. Data are plotted as mean ± SEM.
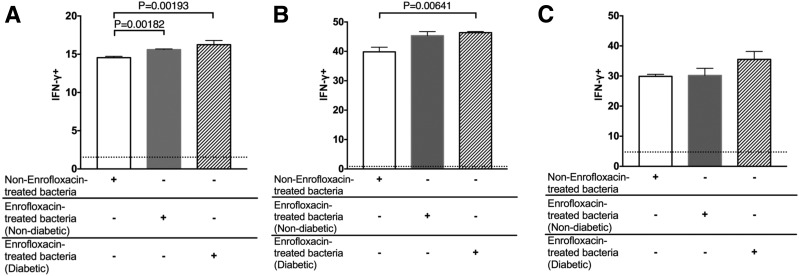
Finally, we tested the inflammatory response of splenocytes from G9Cα−/− NOD mice (expressing the G9C8TCR) to gut microbiota from enrofloxacin-treated and nontreated mice. Our results confirmed that the microbiota from enrofloxacin-treated mice were more proinflammatory, as significantly higher numbers of IFN-γ–producing G9C8 CD8+ T cells were found in response to the gut bacteria from enrofloxacin-treated mice compared with the G9C8 CD8+ T cells cultured with the microbiota from untreated mice (Fig. 6A and Supplementary Fig. 13A). We also observed enhanced IFN-γ–producing splenocyte CD11b+ cells in response to microbiota from diabetic enrofloxacin-treated mice compared with bacteria from untreated mice (Fig. 6B and Supplementary Fig. 13B), with a similar trend in splenocyte CD11c+ cells (Fig. 6C and Supplementary Fig. 13C). Together our data demonstrate that enrofloxacin treatment of A22Cα−/−PI2−/− NOD mice promotes more IFN-γ–inducing gut microbiota.
Figure 6.
Effect of culture of G9Cα−/− NOD mice splenocytes with small-intestinal bacteria from untreated or enrofloxacin-treated A22Cα−/−PI2−/− NOD mice. Intracellular IFN-γ was measured in CD8+ T cells (gated from TCRβ+CD19−CD4−) (A), CD11b+ cells (gated from TCRβ−CD19−IAg7+CD11c−F480+) (B), and CD11c+ cells (gated from TCRβ−CD19−IAg7+CD11b−F480−) (C). Average baseline levels of splenocyte IFN-γ–producing cells, cultured without bacteria, which were similar in all treatment groups, are shown by the dotted line. The labeling beneath the graphs represents the origin of the bacteria: bacteria from non-enrofloxacin-treated mice are labeled as non-enrofloxacin-treated bacteria, bacteria from nondiabetic enrofloxacin-treated mice are labeled as enrofloxacin-treated bacteria (nondiabetic), and bacteria from diabetic enrofloxacin-treated mice are labeled as enrofloxacin-treated bacteria (diabetic). All splenocyte cultures with bacteria displayed significant differences from those without bacteria. All data were pooled from two independent experiments (n = 6/group). All data were assessed for statistical significance using multiple Student t tests corrected using FDR. Data are presented as mean ± SEM.
Enrofloxacin Treatment of NOD Mice Leads to Insulin B15-23–Reactive CD8+ T-Cell Expansion and Activation Within the PPs
To assess the effect of enrofloxacin treatment on insulin B15-23–reactive CD8+ T cells in a non-TCR-restricted mouse, we treated male NOD mice with/without enrofloxacin from weaning for 3 weeks. We found enrofloxacin treatment significantly altered the gut microbiota composition after 3 weeks of treatment (Fig. 7A and B). However, the microbiota changes induced by enrofloxacin treatment were different in the NOD mice compared with the A22Cα−/−PI2−/− NOD mice (Fig. 4), likely due to the TCR restriction contributing to altered microbiota composition (Supplementary Fig. 14). We demonstrated that enrofloxacin treatment expanded the number of insulin B15-23–reactive CD8+ T cells within the PPs (Fig. 7C and Supplementary Fig. 15A and B), with a trend to an increase in proportion. In addition, we also observed an increase in the median fluorescence intensity of the tetramer staining in the PLNs of enrofloxacin-treated mice compared with untreated mice (Supplementary Fig. 15C). The number and proportion of CD69-expressing insulin-reactive CD8+ T cells was also higher in the PLNs and PPs of enrofloxacin-treated NOD mice (Fig. 7D and Supplementary Fig. 15D and E). However, these enrofloxacin-induced changes were restricted to the insulin B15-23–reactive CD8+ T cells, as we found no significant differences in the proportion, number, or activation of the IGRP-reactive CD8+ T cells (Supplementary Fig. 16). We also observed increased IFN-γ secretion by CD8+ T cells, CD11c+ cells, and CD11b+ cells in enrofloxacin-treated NOD mice compared with untreated mice in all tissues (Fig. 7E–G and Supplementary Fig. 17). Thus, our data suggest enrofloxacin treatment can promote the expansion and activation of insulin B15-23–reactive CD8+ T-cells and IFN-γ secretion in both TCRα-restricted and non-TCR-restricted NOD mice.
Figure 7.
Effect of enrofloxacin treatment on non-TCR-restricted polyclonal NOD mice. A: PCoA plot of β-diversity from fecal bacteria of 3- and 6-week-old NOD mice that were untreated or treated with enrofloxacin (n = 4–7 mice). B: Altered microbiota composition expressed as a percentage of relative abundance of bacterial genus. C: Absolute number of insulin B15-23–reactive CD8+ T cells gated from live single TCRβ+CD4−CD19−CD11b−CD11c− cells. D: Absolute number of CD69+ insulin B15-23–reactive CD8+ T cells gated as in C before subsequently gating on CD69. Frequency of IFN-γ–producing CD8+ T cells (gated from TCRβ+CD19−CD4−) (E), CD11c+ cells (gated from TCRβ−CD19−IAg7+CD11b−F480−) (F), and CD11b+ cells (gated from TCRβ−CD19−IAg7+CD11c−F480+) (G). All data were pooled from two independent experiments (n = 8/group). All statistical analyses were conducted using a Student t test. Data are presented as mean ± SEM.
Discussion
In this study we investigated how changing the gut microbiota by using a broad-spectrum antibiotic (enrofloxacin) influenced the TCRβ repertoire, together with expansion and activation of insulin-reactive CD8+ T cells, thereby leading to a higher incidence of diabetes. Our study has demonstrated that the altered gut microbiota induced a strong IFN-γ response from a number of cell types, particularly the CD8+ T cells.
Using a mouse model with a restricted insulin-reactive TCRα chain but polyclonal TCRβ chains (both insulin reactive and non–insulin reactive), we found alterations in the TCRβ repertoire influenced by gut microbiota that had not previously been reported in any diabetes-related microbiome studies. It is known that the Treg TCR repertoire can be strongly influenced by the gut microbiota composition, arising from tolerogenic antigen presentation and conversion of T cells into Tregs (46–48). Our current study demonstrates that the TCR repertoire can clearly be influenced by altering gut microbiota through antibiotic use, which in turn affects the development of diabetes. These changes in TCR repertoire likely arise due to expansions of CD8+ T cells recognizing the altered microbiota composition. Interestingly, enrofloxacin administration to G9C8TCRαβ mice, in which both G9TCRα and TCRβ were fixed, did not alter the incidence of diabetes. This suggests that the altered diabetes incidence we observed in this study was related to the increased expansion of cells with different TCRβ chains. It also emphasizes the importance of the effect that environmental factors, such as the microbiota, may have on different T-cell clonotypes. Not only is T-cell selection affected by the different gut microbial composition, but the antigen-specific T cells also are more activated and expand to increase diabetes incidence.
As other studies have shown, antibiotic treatment can influence the composition of gut microbiota (11–17). Although most of these studies investigated female NOD mice, one study focused on the male NOD mice that developed diabetes (17). Studies have shown that hormones can influence the gut microbiota and contribute to diabetes susceptibility in NOD mice (49,50). Interestingly, we previously showed that in our model system, female A22Cα−/−PI2−/− NOD mice did not develop T1D (34). Therefore, our current study was focused on male A22Cα−/−PI2−/− NOD mice. It is conceivable that the microbiota in our A22Cα−/−PI2−/− NOD mice may also be influenced by sex hormones and thus contribute to the dichotomy of diabetes development in the different sexes.
A recent study showed that IGRP-reactive CD8+ T cells can recognize a microbial mimic (18,40); it is possible that insulin-reactive CD8+ T cells may also do so, as the human insulin-reactive 1E6 clone can recognize a bacterial peptide from a common human pathogen (25). However, it is unknown whether the bacteria themselves can stimulate this T-cell clone in vivo. Thus, to date, no bacterial mimics for insulin have yet been identified that can alter diabetes development. Given the ability of autoreactive CD8+ T cells to recognize a vast number of peptides (51), that there will be some insulin mimics is highly likely. It is also possible that the changes in the composition of gut microbiota, especially Bifidobacteria and Oscillospira, altered gut permeability in our mice. Bifidobacterium can reduce gut permeability (44), while Oscillospira is associated with increasing gut permeability (45). Although it is not currently clear which specific bacteria played the dominant role in our study, we present evidence that the microbiota clearly affected insulin-reactive T-cell development, activation, expansion, and function, particularly in relation to IFN-γ secretion, leading to increased incidence of diabetes.
Our investigation of other subsets of immune cells revealed relatively small changes in non-CD8+ T cells. However, we found changes in macrophages and DCs within the PPs and PLNs linked to CD103 expression, as well as altered IFN-γ and IL-10 secretion. Furthermore, upon bacterial stimulation, the macrophages and DCs exhibited enhanced IFN-γ secretion. Our data suggest a direct effect of the altered composition of microbiota on immune cells to secrete proinflammatory cytokines. It is interesting that age-related differences were observed between treatment groups; for example, enrofloxacin-treated CD11b+ cells from 6-week-old mice exposed to microbiota produced more IFN-γ compared with untreated mice. However, this trend was reversed at 10 weeks of age. These time-dependent effects may arise from changes to immune cell and microbiota interactions in vivo, and this may include effects from microbial changes associated with aging as well as changes to gut permeability. Furthermore, we have also demonstrated that CD11c+ cells from enrofloxacin-treated mice can promote insulin-specific CD8+ T-cell proliferation in vitro compared with cells from untreated donors.
In conclusion, we suggest that insulin-reactive pathogenic CD8+ T cells can be activated and expanded by altered gut bacteria as a result of antibiotic treatment. The altered gut microbiota also promote inflammatory APCs, which in turn facilitate the expansion and activation of a number of different clonotypes of insulin-reactive CD8+ T cells, leading to the development of accelerated autoimmune diabetes in our NOD transgenic mouse model of autoimmune diabetes. We also confirmed similar effects in non-TCR transgenic NOD mice, in which we observed that enrofloxacin treatment promoted the expansion and activation of insulin-reactive CD8+ T cells and also promoted IFN-γ secretion. Our study provides evidence that the gut microbiota play an important role in activating the insulin-specific autoimmune response early in life, which then later affects diabetes development.
Supplementary Material
Article Information
Acknowledgments. The authors thank the National Institutes of Health Tetramer Core Facility for provision of the H-2Kd-LYLVCGERG tetramer and minimal binding tetramers H-2Kd-AYAAAAAAV. The authors thank Karl Hager (Lab Medicine, Yale School of Medicine, Yale University) for assistance with 16S rRNA sequencing.
Funding. J.A.P. was a recipient of a Diabetes UK PhD studentship and subsequently a Fulbright-Diabetes UK postdoctoral research scholarship. The work was funded by National Institutes of Health grants (DK-092882, DK-100500, and P30-DK-45735) to L.W. and by a European Foundation for the Study of Diabetes grant (NN 2014_6) and Medical Research Council grant (G0901155) to F.S.W.
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
Author Contributions. J.A.P., D.K., J.D., J.P., J.W.-S., S.C., M.L., and L.C.d.R. performed the experiments, J.A.P., L.W., and F.S.W. designed the experiments, analyzed the results, and wrote the manuscript. L.W. and F.S.W. supervised the study. The project was conceived by F.S.W. F.S.W. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form as poster presentations at the Immunology of Diabetes Society Congress 2018, London, U.K., 25–29 October 2018, and at the Keystone Symposia on Molecular and Cellular Biology: Gut Microbiota Modulation of Host Physiology: The Search for Mechanism, Keystone, CO, 1–6 March 2015.
Footnotes
This article contains Supplementary Data online at http://diabetes.diabetesjournals.org/lookup/suppl/doi:10.2337/db18-0487/-/DC1.
References
- 1.Gillespie KM, Bain SC, Barnett AH, et al. The rising incidence of childhood type 1 diabetes and reduced contribution of high-risk HLA haplotypes. Lancet 2004;364:1699–1700 [DOI] [PubMed] [Google Scholar]
- 2.Wen L, Ley RE, Volchkov PY, et al. Innate immunity and intestinal microbiota in the development of type 1 diabetes. Nature 2008;455:1109–1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.de Goffau MC, Luopajärvi K, Knip M, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes 2013;62:1238–1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Goffau MC, Fuentes S, van den Bogert B, et al. Aberrant gut microbiota composition at the onset of type 1 diabetes in young children. Diabetologia 2014;57:1569–1577 [DOI] [PubMed] [Google Scholar]
- 5.Kostic AD, Gevers D, Siljander H, et al.; DIABIMMUNE Study Group . The dynamics of the human infant gut microbiome in development and in progression toward type 1 diabetes. Cell Host Microbe 2015;17:260–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vatanen T, Kostic AD, d’Hennezel E, et al.; DIABIMMUNE Study Group . Variation in microbiome LPS immunogenicity contributes to autoimmunity in humans. Cell 2016;165:842–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krych Ł, Nielsen DS, Hansen AK, Hansen CH. Gut microbial markers are associated with diabetes onset, regulatory imbalance, and IFN-γ level in NOD mice. Gut Microbes 2015;6:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peng J, Narasimhan S, Marchesi JR, Benson A, Wong FS, Wen L. Long term effect of gut microbiota transfer on diabetes development. J Autoimmun 2014;53:85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marietta EV, Gomez AM, Yeoman C, et al. Low incidence of spontaneous type 1 diabetes in non-obese diabetic mice raised on gluten-free diets is associated with changes in the intestinal microbiome. PLoS One 2013;8:e78687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mariño E, Richards JL, McLeod KH, et al. Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol 2017;18:552–562 [DOI] [PubMed] [Google Scholar]
- 11.Hansen CH, Krych L, Nielsen DS, et al. Early life treatment with vancomycin propagates Akkermansia muciniphila and reduces diabetes incidence in the NOD mouse. Diabetologia 2012;55:2285–2294 [DOI] [PubMed] [Google Scholar]
- 12.Tormo-Badia N, Håkansson Å, Vasudevan K, Molin G, Ahrné S, Cilio CM. Antibiotic treatment of pregnant non-obese diabetic mice leads to altered gut microbiota and intestinal immunological changes in the offspring. Scand J Immunol 2014;80:250–260 [DOI] [PubMed] [Google Scholar]
- 13.Candon S, Perez-Arroyo A, Marquet C, et al. Antibiotics in early life alter the gut microbiome and increase disease incidence in a spontaneous mouse model of autoimmune insulin-dependent diabetes [published correction appears in PLoS One 2016;11:e0147888]. PLoS One 2015;10:e0125448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu Y, Peng J, Tai N, et al. Maternal antibiotic treatment protects offspring from diabetes development in nonobese diabetic mice by generation of tolerogenic APCs. J Immunol 2015;195:4176–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nobel YR, Cox LM, Kirigin FF, et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat Commun 2015;6:7486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y, Jin P, Peng J, Zhang X, Wong FS, Wen L. Different immunological responses to early-life antibiotic exposure affecting autoimmune diabetes development in NOD mice. J Autoimmun 2016;72:47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livanos AE, Greiner TU, Vangay P, et al. Antibiotic-mediated gut microbiome perturbation accelerates development of type 1 diabetes in mice. Nat Microbiol 2016;1:16140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tai N, Peng J, Liu F, et al. Microbial antigen mimics activate diabetogenic CD8 T cells in NOD mice. J Exp Med 2016;213:2129–2146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krishnamurthy B, Dudek NL, McKenzie MD, et al. Responses against islet antigens in NOD mice are prevented by tolerance to proinsulin but not IGRP. J Clin Invest 2006;116:3258–3265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krishnamurthy B, Mariana L, Gellert SA, et al. Autoimmunity to both proinsulin and IGRP is required for diabetes in nonobese diabetic 8.3 TCR transgenic mice. J Immunol 2008;180:4458–4464 [DOI] [PubMed] [Google Scholar]
- 21.Schloot NC, Willemen S, Duinkerken G, de Vries RR, Roep BO. Cloned T cells from a recent onset IDDM patient reactive with insulin B-chain. J Autoimmun 1998;11:169–175 [DOI] [PubMed] [Google Scholar]
- 22.Semana G, Gausling R, Jackson RA, Hafler DA. T cell autoreactivity to proinsulin epitopes in diabetic patients and healthy subjects. J Autoimmun 1999;12:259–267 [DOI] [PubMed] [Google Scholar]
- 23.Skowera A, Ellis RJ, Varela-Calviño R, et al. CTLs are targeted to kill beta cells in patients with type 1 diabetes through recognition of a glucose-regulated preproinsulin epitope [published correction appears in J Clin Invest 2009;119:2844]. J Clin Invest 2008;118:3390–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bulek AM, Cole DK, Skowera A, et al. Structural basis for the killing of human beta cells by CD8(+) T cells in type 1 diabetes. Nat Immunol 2012;13:283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cole DK, Bulek AM, Dolton G, et al. Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity. J Clin Invest 2016;126:2191–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wegmann DR, Norbury-Glaser M, Daniel D. Insulin-specific T cells are a predominant component of islet infiltrates in pre-diabetic NOD mice. Eur J Immunol 1994;24:1853–1857 [DOI] [PubMed] [Google Scholar]
- 27.Wegmann DR, Gill RG, Norbury-Glaser M, Schloot N, Daniel D. Analysis of the spontaneous T cell response to insulin in NOD mice. J Autoimmun 1994;7:833–843 [DOI] [PubMed] [Google Scholar]
- 28.Zekzer D, Wong FS, Wen L, et al. Inhibition of diabetes by an insulin-reactive CD4 T-cell clone in the nonobese diabetic mouse. Diabetes 1997;46:1124–1132 [DOI] [PubMed] [Google Scholar]
- 29.Wong FS, Karttunen J, Dumont C, et al. Identification of an MHC class I-restricted autoantigen in type 1 diabetes by screening an organ-specific cDNA library. Nat Med 1999;5:1026–1031 [DOI] [PubMed] [Google Scholar]
- 30.Nakayama M, Abiru N, Moriyama H, et al. Prime role for an insulin epitope in the development of type 1 diabetes in NOD mice. Nature 2005;435:220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thébault-Baumont K, Dubois-Laforgue D, Krief P, et al. Acceleration of type 1 diabetes mellitus in proinsulin 2-deficient NOD mice. J Clin Invest 2003;111:851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong FS, Visintin I, Wen L, Flavell RA, Janeway CA Jr. CD8 T cell clones from young nonobese diabetic (NOD) islets can transfer rapid onset of diabetes in NOD mice in the absence of CD4 cells. J Exp Med 1996;183:67–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lieberman SM, Takaki T, Han B, Santamaria P, Serreze DV, DiLorenzo TP. Individual nonobese diabetic mice exhibit unique patterns of CD8+ T cell reactivity to three islet antigens, including the newly identified widely expressed dystrophia myotonica kinase. J Immunol 2004;173:6727–6734 [DOI] [PubMed] [Google Scholar]
- 34.Pearson JA, Thayer TC, McLaren JE, et al. Proinsulin expression shapes the TCR repertoire but fails to control the development of low-avidity insulin-reactive CD8+ T cells. Diabetes 2016;65:1679–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong FS, Siew LK, Scott G, et al. Activation of insulin-reactive CD8 T-cells for development of autoimmune diabetes. Diabetes 2009;58:1156–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thayer TC, Pearson JA, De Leenheer E, et al. Peripheral proinsulin expression controls low-avidity proinsulin-reactive CD8 T cells in type 1 diabetes. Diabetes 2016;65:3429–3439 [DOI] [PubMed] [Google Scholar]
- 37.Pearson JA, Wong FS. Identification of islet antigen-specific CD8 T cells using MHCI-peptide tetramer reagents in the non obese diabetic (NOD) mouse model of type 1 diabetes. Methods Mol Biol 2016;1433:119–125 [DOI] [PubMed] [Google Scholar]
- 38.Verdaguer J, Schmidt D, Amrani A, Anderson B, Averill N, Santamaria P. Spontaneous autoimmune diabetes in monoclonal T cell nonobese diabetic mice. J Exp Med 1997;186:1663–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Horai R, Zárate-Bladés CR, Dillenburg-Pilla P, et al. Microbiota-dependent activation of an autoreactive T cell receptor provokes autoimmunity in an immunologically privileged site. Immunity 2015;43:343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hebbandi Nanjundappa R, Ronchi F, Wang J, et al. A gut microbial mimic that hijacks diabetogenic autoreactivity to suppress colitis. Cell 2017;171:655–667.e17 [DOI] [PubMed] [Google Scholar]
- 41.Sung SS, Fu SM, Rose CE Jr., Gaskin F, Ju ST, Beaty SR. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J Immunol 2006;176:2161–2172 [DOI] [PubMed] [Google Scholar]
- 42.Fonseca DM, Hand TW, Han SJ, et al. Microbiota-dependent sequelae of acute infection compromise tissue-specific immunity. Cell 2015;163:354–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saxena V, Ondr JK, Magnusen AF, Munn DH, Katz JD. The countervailing actions of myeloid and plasmacytoid dendritic cells control autoimmune diabetes in the nonobese diabetic mouse. J Immunol 2007;179:5041–5053 [DOI] [PubMed] [Google Scholar]
- 44.Fukuda S, Toh H, Hase K, et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011;469:543–547 [DOI] [PubMed] [Google Scholar]
- 45.Lam YY, Ha CW, Campbell CR, et al. Increased gut permeability and microbiota change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS One 2012;7:e34233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol 2006;7:401–410 [DOI] [PubMed] [Google Scholar]
- 47.Lathrop SK, Bloom SM, Rao SM, et al. Peripheral education of the immune system by colonic commensal microbiota. Nature 2011;478:250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cebula A, Seweryn M, Rempala GA, et al. Thymus-derived regulatory T cells contribute to tolerance to commensal microbiota. Nature 2013;497:258–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yurkovetskiy L, Burrows M, Khan AA, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013;39:400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markle JG, Frank DN, Mortin-Toth S, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013;339:1084–1088 [DOI] [PubMed] [Google Scholar]
- 51.Wooldridge L, Ekeruche-Makinde J, van den Berg HA, et al. A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem 2012;287:1168–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.