Abstract
The last two decades have witnessed an explosion of interest in adipocyte biology, coinciding with the upsurge of obesity and metabolic syndrome. Now we have new perspectives on the distinct developmental origins of white, brown, and beige adipocytes and their role in metabolic physiology and disease. Beyond fuel metabolism, adipocytes communicate with the immune system and other tissues by releasing diverse paracrine and endocrine factors to orchestrate adipose tissue remodeling and maintain systemic homeostasis. Significant progress has been made in delineating the regulatory networks that govern different aspects of adipocyte biology. Here we provide an overview on the emerging role of long noncoding RNAs (lncRNAs) in the regulation of adipocyte development and metabolism and discuss the implications of the RNA–protein regulatory interface in metabolic control.
Introduction
White adipocytes are responsible for energy storage, whereas brown and beige adipocytes are specialized in fuel oxidation and energy expenditure. Major progress has been made in delineating the molecular control of lineage-specific development of white, brown, and beige adipocytes (1–3), adipose tissue remodeling and inflammation (4–6), thermogenic energy expenditure (7–9), and more recently, the emerging endocrine functions of brown and beige fat (10,11). Increased adipose thermogenesis is often linked to an improved metabolic profile. Brown and beige fat thermogenesis is mediated by uncoupling protein 1 (UCP1)-dependent and UCP1-independent mechanisms; the latter includes the creatine substrate cycle (12–14) and calcium futile cycle (15). Beyond thermogenesis, brown and beige fat exert their effects on metabolic physiology through secreting endocrine factors and microRNA-containing exosomes that act on other tissues in the body (10,11,16). Neuregulin 4 (Nrg4) is a brown fat–enriched secreted factor that attenuates hepatic lipogenesis and liver injury (17–19), whereas microRNAs encapsulated in exosomes are released by brown adipocytes and may serve as important messengers for intertissue cross-talk (20,21). Adipose tissue is densely innervated by sympathetic nerve fibers (22–24). Recent work also sheds light on the mechanisms that govern adipose sympathetic innervation and plasticity (25–28).
Long noncoding RNAs (lncRNAs) are emerging as important regulators of cellular signaling and gene expression in numerous cell types. lncRNAs are long RNA transcripts (>200 bp) that do not encode proteins. Many lncRNAs contain a 5′ cap, multiple exons, and 3′ polyadenylation (29). Some of these transcripts are intergenic while others are generated from genomic regions close to or partially overlapping with protein-coding genes. Depending on the relative position with the nearby coding genes, lncRNAs can be generally categorized into intergenic, antisense, divergent, intronic, and enhancer lncRNAs (29). lncRNAs can regulate the functions of cells through a variety of mechanisms. For instance, they can function as scaffolds to bring two or more proteins into a functional ribonucleoprotein complex, as decoys to titrate a protein away from its original target, as guides to recruit chromatin modification enzymes to specific loci on chromosome, and as microRNA sponges to buffer microRNAs’ inhibitory functions on gene expression (29,30). It is noteworthy that the coding potential of lncRNAs is often assessed by computational methods based on open reading frame length, conservation, codon usage, etc. These procedures are not error proof and may misannotate some micropeptide-coding transcripts as lncRNAs (31). It is therefore important to experimentally determine the coding potential of lncRNA candidates.
Recent years have seen a rapid increase in the number of adipocyte lncRNA studies focusing on genome-wide annotation of lncRNAs, molecular and functional analyses in cultured adipocytes, and, more recently, in vivo studies to delineate their role in physiology and disease. Here, we seek to highlight the latest advances in adipose lncRNA research, discuss new insights into the emerging lncRNA–protein regulatory interface, and provide perspectives on the current challenges and future directions of this field. Readers are also referred to a few other excellent lncRNA reviews that cover additional metabolic tissues (32–35).
Annotation of Adipose Tissue lncRNAs
A key feature of lncRNAs is that they tend to be expressed in a cell type–specific manner (36), so the generic gene annotation database may not cover many lncRNAs specifically expressed in certain cell types. To fully explore the function of lncRNAs in adipose tissue, there is a need to build a comprehensive catalog of lncRNAs that are enriched and/or highly regulated in adipose tissue. Alvarez-Dominguez et al. (37) made the first effort to address this challenge. They integrated genome-wide surveys of transcription by RNA-seq and chromatin state by ChIP-seq for mouse brown adipose tissue (BAT), epididymal white adipose tissue (WAT), and inguinal WAT (iWAT), developed a computational pipeline to reconstruct adipose transcriptomes de novo, and uncovered ∼1,500 multiexonic lncRNAs in adipose tissues. Among these, 127 lncRNAs were enriched in BAT, induced during brown adipocyte differentiation and often targeted by key adipogenic transcription factors, such as PPARγ, C/EBPα, and C/EBPβ. As many as 30% of these lncRNA transcripts were mapped outside of the annotated loci; thus, these genome-wide lncRNA analyses present a unique opportunity for gene discovery.
Using a similar strategy, Zhang et al. (38) conducted RNA-seq of gluteal subcutaneous adipose tissue from 25 healthy humans and constructed a de novo noncoding transcriptome. This study revealed 1,001 putative lncRNA transcripts detectable in human adipose tissue, of which 144 lncRNAs have not been previously annotated. Interestingly, 54 of 100 lncRNAs enriched in adipose tissue harbored PPARγ and C/EBPα binding sites near their transcription start sites (38). Since sequence conservation of lncRNAs may not be prerequisite for structural and functional conservation, Zhang et al. used synteny or relative positional conservation in the genome as the evaluation criteria (39) and found that only ∼15% of mouse adipose lncRNAs had conserved orthologs. However, it is important to note that nonconserved lncRNAs may play an important role in orchestrating metabolic signaling and regulation in a species-specific manner. Zhang et al. (38) identified linc-ADAL1 as a nonconserved lncRNA that is required for human adipogenesis through its interaction with hnRNPU and IGF2BP2 at distinct subcellular locations.
Ding et al. (40) constructed a more comprehensive noncoding transcriptome by conducting deep RNA-seq for human interscapular BAT, subcutaneous WAT, and omental WAT. This study identified 3,149 lncRNAs actively transcribed in adipose tissue, of which a total of 318 lncRNAs were syntenically conserved between mouse and human. Interestingly, the expression of these lncRNAs and their nearest mRNAs also exhibited a positive correlation in both species, supporting a potential role of lncRNAs in regulating expression of the nearby coding genes in cis. One of the conserved lncRNAs was divergently transcribed from Prdm16, referred to as lnc-dPRDM16. Further in vitro and in vivo knockdown studies revealed an important role of lnc-dPRDM16 in driving expression of BAT-selective markers and genes involved in WAT browning (40). This work established a roadmap to facilitate the discovery of functionally conserved lncRNAs in adipocytes.
These annotation studies have greatly expanded the lncRNA database in adipose tissue and provided a rich resource for functional analysis. It is important to note, however, that these databases are largely built with de novo assembly programs that often suffer from trade-offs between quality and size (41). Researchers should carefully validate the gene sequences and structures of their lncRNA candidates when they are using these de novo assembled annotations. It should also be noted that the functional predictions of lncRNAs in these studies were mainly based on their coexpression with mRNAs, so further experimental evidence is needed to validate their biological functions. Moreover, these studies only characterized the polyadenylated transcripts and did not offer much insight into the transcripts lacking poly (A) tails (poly A−), which represent a significant portion in the lncRNA catalog (42,43). Further annotation work should include the poly A− class of lncRNAs.
lncRNA Regulation of Brown and Beige Thermogenic Programs
Brown and beige fat development and maintenance are governed by a network of transcription factors and cofactors, including PRDM16 (44–46), EBF2 (47), PGC-1α (48), ZBTB7B (49), IRF4 (50), ZFP516 (51), and EHMT1 (52). Several lncRNAs have emerged to regulate key aspects of thermogenic adipocyte biology by interfacing with these transcriptional regulators (Table 1).
Table 1.
lncRNAs regulating brown- and beige-selective programs
| lncRNA | Role in BAT and WAT browning | Proposed mechanism of action | Ref. |
|---|---|---|---|
| Blnc1 | Positive | Interacts with EBF2 and Zbtb7b and hnRNPU to form ribonucleoprotein complexes to activate thermogenic gene expression | (49,53,54,58) |
| lncBATE1 | Positive | Interacts with hnRNPU to form a ribonucleoprotein complex | (37) |
| lncBATE10 | Positive | Decoys Celf1 from PGC-1α, thereby protecting PGC-1α mRNA from repression by Celf1 | (60) |
| UC.417 | Negative | Inhibits phosphorylation of p38MAPK | (89) |
| AK079912 | Positive | Unknown | (90) |
| H19 | Positive | Recruits PEG-inactivating H19-MBD1 complexes and acts as BAT-selective PEG gatekeeper | (63) |
| GM13133 | Negative | Unknown | (91) |
Blnc1
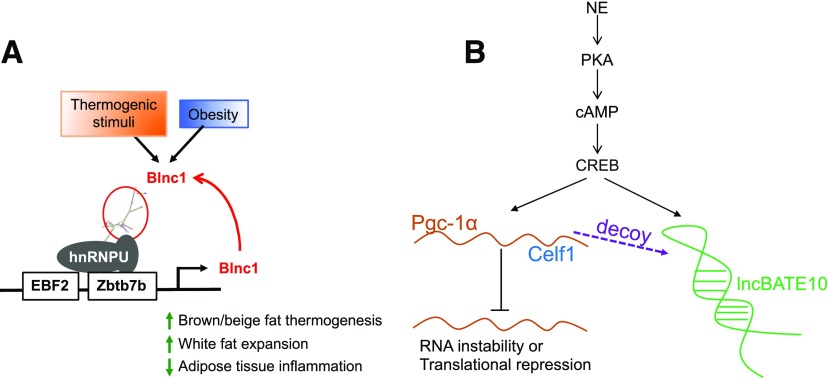
Brown fat lncRNA 1 (Blnc1) was discovered in a search for lncRNA regulators of brown and beige adipocyte differentiation. Molecular and functional studies of Blnc1 have revealed several characteristics of lncRNA regulation of adipocyte biology. A prominent feature of Blnc1 is its highly inducible expression during brown and beige adipogenesis and in response to thermogenic stimuli in vivo (53). The induction of Blnc1 during adipocyte differentiation parallels key transcriptional regulators of the thermogenic gene program, including PPARγ, PPARα, EBF2, Zbtb7b, and PGC-1α (53). RNA interference knockdown of EBF2 or Zbtb7b greatly impairs Blnc1 expression and induction of thermogenic genes in response to adrenergic stimulation. It is likely that Blnc1 serves to reinforce adipocyte differentiation toward brown and beige phenotype once the process is initiated. Blnc1 regulates brown and beige thermogenic program by interfacing with key transcription factors and cofactors through two distinct mechanisms: assembly into positive feedback regulatory loops and formation of ribonucleoprotein transcriptional complexes (Fig. 1A). In this regard, Blnc1 is localized primarily in the nucleus and physically interacts with EBF2 and Zbtb7b, transcriptional activators of thermogenic gene expression, which in turn stimulate the expression of Blnc1 itself (49,53).
Figure 1.
A: Blnc1 assembles ribonucleoprotein transcriptional complexes with hnRNPU to form a positive regulatory loop with EBF2 and Zbtb7b. B: The cAMP-CREB pathway can stimulate the transcription of lncBATE10 and Pgc-1α. lncBATE10 functions as a decoy to titrate Celf1 away from Pgc-1α mRNA. Otherwise, Celf1 will repress Celf1 mRNA and result in RNA instability or translational repression.
Molecular analyses of the Blnc1 ribonucleoprotein complexes revealed a prominent role for heterogeneous nuclear ribonucleoprotein U (hnRNPU) in interaction between lncRNA and protein factors (54). hnRNPU is a nuclear matrix protein that maintains 3-D genome architecture and associates with active chromatin (55,56). hnRNPU has been demonstrated to interact with FIRRE and lncBATE1, two lncRNAs previously implicated in the control of adipogenesis (37,57). Notably, formation of the Blnc1/hnRNPU ribonucleoprotein complex appears to be conserved between mouse and human (54). The exact mechanisms through which Blnc1 stimulates target gene transcription, however, remain to be defined at the molecular level. In a separate study, hnRNPU and several other members of the hnRNP protein family were identified as components of the Zbtb7b transcriptional complex in differentiated brown adipocytes (49). These findings illustrate a potentially central role of the hnRNP family of RNA-binding proteins in integrating lncRNA and transcriptional regulators in the control of chromatin structure and gene transcription.
Elucidating the significance of lncRNAs in adipocyte metabolism and physiology is an important and active area of research. Adipose tissue–specific Blnc1 transgenic and knockout mouse strains have been generated to dissect its role in thermogenesis and energy balance (58). These in vivo studies revealed that Blnc1 is required for inguinal white fat browning following chronic cold acclimation. An unexpected discovery here is that Blnc1, despite its lower expression in white fat compared with brown fat, serves a critical role in healthy expansion of white adipose tissue during high-fat diet (HFD)-induced obesity. In this context, Blnc1 was found to attenuate proinflammatory signaling and obesity-associated adipose tissue inflammation in a cell-autonomous manner. Conditional inactivation of Blnc1 in the liver protects mice from nonalcoholic fatty liver disease by serving as a checkpoint for lipogenic activation (59). These in vivo studies provide compelling support for the emerging lncRNA–protein interface in the regulation of all major aspects of adipose biology.
lncBATE1 and lncBATE10
To systemically study the dynamic regulation of lncRNAs during adipocyte lineage-specific development and cell type conversion, Sun and colleagues conducted a series of transcriptome studies by RNA-seq and quantified mRNA and lncRNA changes between different fat depots (37), during WAT browning induced by chronic cold exposure, β-adrenergic agonist treatment and intense exercise, BAT activation by acute cold exposure, and BAT inactivation by thermoneutrality (60). Interrogating these data sets led to the identification of a list of BAT-enriched lncRNAs (lncBATEs). The expression of these lncBATEs was highly regulated in response to energy expenditure status in adipose tissue.
Among the most abundant lncRNAs in BAT were lncBATE1 (37) and lncBATE10 (60), both of which were enriched in BAT compared with other organs and induced during iWAT browning. Knocking down lncBATE1 in primary brown adipocyte culture slightly impaired adipogenesis but drastically reduced the BAT-selective gene expression. Overexpression of a mutant lncBATE1 resistant to siRNA knockdown could rescue the impaired expression of many BAT markers. Mechanistically, lncBATE1 physically interacts with hnRNPU; however, whether and how this interaction might affect the function of lncBATE1 warrant further investigation (37).
Compared with lncBATE1, lncBATE10 exhibits a more restricted expression pattern in BAT and more dramatic induction during browning (∼40 fold). Knockdown of lncBATE10 in primary brown and white adipocyte culture did not appear to affect adipogenesis, evidenced by adipocyte marker gene expression and lipid accumulation. In contrast, loss of lncBATE10 significantly reduced the expression of many BAT-selective genes such as Ucp1 and Pgc-1α. Adenovirus-mediated shRNA knockdown of lncBATE10 impaired iWAT browning induced by cold exposure in mice. RNA pulldown followed by mass spectrometry analysis identified CELF1 as a protein partner for lncBATE10. CELF1 binds the 3′ untranslated region of its target mRNAs, leading to mRNA degradation and translational repression (61,62). lncBATE10 served as a decoy that traps CELF1 from PGC-1α, thus protecting PGC-1α mRNA from repression by CELF1 (Fig. 1B) (60). This study not only illustrated a mechanism used by lncBATE10 to facilitate BAT program expression, but also linked lncBATE10 to an existing thermogenic signaling pathway. The lncBATE10 promoter contains several CREB-responsive elements that mediate lncBATE10 expression in response to adrenergic signaling and cAMP-CREB activation (60).
H19
Schmidt et al. (63) examined the function of a maternally imprinted lncRNA, H19, in brown fat in the context of diet-induced obesity. H19 expression was increased upon cold exposure and decreased in obesity in rodent BAT. Its expression inversely correlated with human BMI in both subcutaneous and visceral WAT. H19 knockdown by locked nucleic acid or siRNAs impaired adipogenesis, oxidative metabolism, and mitochondrial respiration in brown but not white adipocytes. In vivo, H19 transgenic mice were protected from diet-induced obesity and insulin resistance, likely due to enhanced mitochondrial biogenesis and fuel oxidation in WAT and BAT (63). Adipose tissue–specific inactivation of the H19 methylation-sensitive imprinting control regions sensitized mice to HFD-induced weight gain. Mechanistically, H19 curbs the expression of a set of paternally expressed genes such as IGF2, PEG10, and PLAGL1 by recruiting PEG-inactivating H19-MBD1 complexes (63). This study illustrates the function of a monoallelic gene in regulating the BAT gene program and metabolism, possibly by maintaining the quiescence of imprinted genes in BAT.
However, the role of H19 in brown fat development and thermogenesis was complicated by a recent report that that demonstrated a negative role of H19 in adipocyte differentiation from human bone marrow mesenchymal stem cells (BMSCs) (64). H19 was significantly downregulated during BMSC adipogenesis. Overexpression of H19 inhibited while knockdown of H19 accelerated adipogenesis. Concerning the molecular mechanism of H19 in this context, the H19-derived miR-675 targeted the 3′ untranslated region of HDAC4-6 transcripts and repressed their expression, thereby inhibiting adipogenesis. The discrepancy between these two studies may result from different biological processes examined in these studies: Schmidt et al. (63) focused on the differentiation from preadipocytes to mature adipocytes while the latter study examined the differentiation from the stem cell stage. It is also possible that although H19 was considered as a conserved gene, its biological functions are not necessarily conserved between mouse and human.
lncRNA Regulation of White Adipocyte Differentiation and Function
Adipogenesis is a fundamental aspect of adipose tissue biology. The core transcriptional circuitry that governs preadipocyte commitment and adipocyte differentiation has been elucidated in the last two decades along with the cellular origins of adipocyte progenitors (1,65). Earlier studies have revealed a long list of positive or negative regulators, particularly PPARγ and the C/EBP and KLF families of transcription factors. In the past several years, the scope of these regulators has been greatly expanded to include noncoding genes such as microRNAs (66,67) and, more recently, lncRNAs (Table 2).
Table 2.
lncRNAs regulating white adipocyte adipogenesis and function
| lncRNA | Role in adipogenesis | Proposed mechanism of action | Ref. |
|---|---|---|---|
| SRA | Positive | Coactivates PPARγ | (68–70) |
| lncRAPs | Positive | Unknown | (71) |
| FIRRE | Positive | Interacts with hnRNPU to organize nuclear structure | (57) |
| PU.1 AS | Positive | Prevents PU.1 mRNA translation through forming mRNA/AS lncRNA duplex | (92,93) |
| lnc-U90926 | Negative | Inhibits PPARγ2 promoter transactivation | (94) |
| Gm15290 | Positive | Acts as a sponge for miR-27b | (95) |
| Paral1 | Positive | Coactivates PPARγ through interaction with the paraspeckle component and hnRNP-like RNA binding protein 14 (RBM14/NCoAA) | (96) |
| TCONS_00041960 | Negative | Acts as a sponge for miR-204-5p and miR-125a-3p | (97) |
| lnc-Leptin | Positive | Mediates the distant interaction between an enhancer and leptin promoter | (73) |
| Malat1 | No | Unknown | (78) |
| GAS5 | Negative | Acts as a microRNA sponage to repress miR-21a-5p and miR-18a | (98,99) |
| AdipoQ AS lncRNA | Negative | Suppresses AdipoQ mRNA translation via formation of AdipoQ AS lncRNA/AdipoQ mRNA duplex | (100) |
| Plnc1 | Positive | Reduces the methylation level of CpG region in the PPARγ2 promoter and enhances the transcriptional activity of the promoter | (74) |
SRA1
The lncRNA steroid receptor RNA activator 1 (Sra1) is among the first described functional lncRNA in adipocytes. Knockdown of Sra1 inhibited 3T3-L1 differentiation (68), and knockout of Sra1 in mice resulted in resistance to HFD-induced obesity and glucose intolerance (69). However, according to current Ref-seq gene annotation, this gene generates three isoforms, and only one of them produces a noncoding transcript while two other isoforms encode an SRA protein (SRAP). Indeed, an earlier study has validated SRAP protein expression by Western blot (70). Because both the siRNA and the knockout approaches used in earlier studies likely impaired SRAP protein expression, the role of the noncoding Sra1 transcript in adipogenesis remains inconclusive.
lnc-RAP
To systemically study the expression and function of lncRNAs in adipogenesis, Sun et al. (71) profiled the transcriptome of primary brown and white adipocytes, preadipocytes, and cultured adipocytes by RNA-seq and identified 175 lncRNAs that were regulated during adipogenesis. Many of them were enriched in adipocytes and might be regulated by PPARγ and C/EBPα. As such, these lncRNAs were named Regulated in AdiPogenesis, lnc-RAPs. Using RNA interference–mediated loss-of-function screening, 10 lnc-RAPs were identified to be required for adipogenesis in primary white adipocyte culture (71). This was the first genome-wide profiling and functional study on lncRNAs in adipocytes. The exact mechanisms through which these lnc-RAPs contribute to adipogenic regulation remain to be delineated.
lnc-RAP1 has a conserved ortholog on human X chromosome and contains multiple 156-bp repeating RNA domains. Because of this structural feature, lnc-RAP1 was later renamed as functional intergenic repeating RNA element (FIRRE). Its subcellular localization was restricted to the nucleus, distributed across a 5-Mb domain near its transcription site, and unexpectedly, lay in proximity to five other transchromosomal loci, four of which harbored genes important for adipogenesis and lipid metabolism. Biochemistry analysis showed that hnRNPU binds to FIRRE and that disruption of this interaction abolishes colocalization of FIRRE-contacting transchromosomal loci (57). Therefore, FIRRE may facilitate adipogenesis by reshaping 3-D genome organization and bringing different adipogenic loci into close proximity within the nucleus to coordinate their regulations.
lnc-Leptin
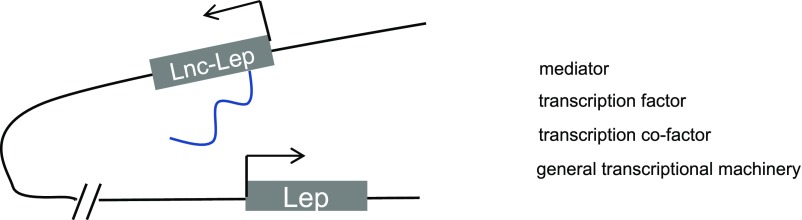
To explore the role of lncRNAs in adipose tissue during obesity, Lo et al. (72) examined the lncRNA expression changes in BAT, epididymal WAT, and iWAT with RNA-seq and identified a set of lncRNAs regulated in all three depots during obesity and in the fed versus fast state. One of these lncRNAs was located ∼20 kb upstream of Leptin, referred to as lnc-Leptin. The expression of this lncRNA was downregulated by fasting, elevated in obesity, induced by insulin, and positively correlated with Leptin expression across a variety of pathophysiological conditions. The lnc-Leptin locus does not appear to harbor H3K4 trimethylation (H3K4Me3), evidenced by ChIP-seq data, but exhibited H3K4 monomethylation (H3K4Me1) and H3K27 acetylation (H3K27Ac), a feature of active enhancers (73), suggesting that lnc-Leptin is likely an enhancer lncRNA.
Knockdown of lnc-Leptin with siRNA during primary white adipocyte differentiation inhibited adipogenesis and in the differentiated adipocytes resulted in a reduction of Leptin expression. The regulation of lnc-Leptin on Leptin was confirmed in vivo by injecting gapmer oligonucleotides targeting lnc-Leptin into iWAT. Mechanistically, chromatin conformation capture (3C) assay demonstrated that the lnc-Leptin enhancer region and the promoter of Leptin formed direct physical interaction, a common mechanism employed by enhancers to augment transcription of target genes. This enhancer–promoter interaction was diminished upon lnc-Leptin knockdown (Fig. 2). Therefore, lnc-Leptin regulates Leptin expression by mediating the formation of enhancer–promoter interaction (72).
Figure 2.
lnc-Leptin (Lnc-Lep) mediates the interaction between the enhancer and promoter of Leptin. lnc-Leptin is transcribed from an enhancer region ∼10 kb upstream Leptin promoter. In this model, the enhancer can form a long-distance loop with the promoter to promote a more active chromatin architecture for leptin transcription. lnc-Leptin is likely to play a role in the formation of the enhancer–promoter interaction.
Plnc1
A very recent study identified another adipogenic lncRNA, Plnc1, that functions by modulating Pparγ2, a key adipogenic transcriptional factor. Plnc1 is transcribed from a position ∼25 kb upstream of the Pparγ2 gene, abundantly expressed in adipose tissue, significantly induced during adipogenesis, and upregulated in obese mice. Knockdown of Plnc1 blocked adipogenesis in ST2 and BMSC cell lines, while its overexpression promoted these cells to differentiate into mature adipocytes. Mechanistically, Plnc1 increases the transcription of Pparγ2 by reducing the methylation level of CpG region in the Pparγ2 promoter (74).
Malat1
While most studies in this field focused on identifying novel noncoding regulators, it also worth reexamining the role of previously discovered lncRNAs in adipocytes. For example, Malat1, a 7-kb nuclear lncRNA, was reported to regulate cellular proliferation, was associated with cancer cell progression (75,76), and was reported to promote lipid accumulation in hepatocytes (77). Malat1 is broadly expressed in various cell types. Carter et al. (78) observed a significant reduction of Malat1 in visceral WAT from the aged men and mice. However, the significance of this lncRNA in adipose tissue remains uncertain as the Malat1 knockout strain exhibited modest effects on adipose tissue expansion and insulin resistance during aging and HFD-induced obesity (78). Despite this, reexamining previously discovered lncRNAs using mouse models could be valuable to establish their functions in adipocyte development and metabolism in vivo.
lncRNA Regulation of Human Adipocyte Biology
As discussed in the lncRNA annotation section, the majority of lncRNAs are not conserved between human and model organisms (39,40). Even for highly conserved lncRNAs, it is not necessarily the case that they will perform similar functions in different species with conserved mechanisms. While using model organisms is essential for researchers to understand the function and mechanism of lncRNAs, functional studies in human cells have the potential to reveal functional lncRNAs relevant to human biology. Recent years have seen a significant increase in the number of studies to determine the function of lncRNAs in human adipocytes (Table 3).
Table 3.
lncRNAs regulating human adipocyte biology
| lncRNA | Role in human adipocyte differentiation and function | Proposed mechanism of action | Ref. |
|---|---|---|---|
| HOTAIR | Positive | Unknown | (101) |
| ADINR | Positive | Binds to PA1 and recruits MLL3/4 to regulate chromatin architecture | (79) |
| linc-NFE2L3–1 | Negative | Unknown | (102) |
| H19 | Negative | Targets miR-675 and HDACs 4–6 | (64) |
| MIR31HG | Positive | Unknown | (103) |
| ASMER-1 ASMER-2 | Positive | Unknown | (80) |
| HOXA11-AS1 | Positive | Unknown | (104) |
| linc-ADAL | Positive | Interacts with hnRNPU and IGF2BP2 at distinct subcellular locations | (35) |
| RP11–20G13.3 | Positive | Unknown | (105) |
| MEG3 | Negative | Regulates miR-140-5p | (106) |
ADINR
CEBPα is a key transcriptional factor in adipogenesis, but its regulation was not fully understood. Xiao et al. (79) identified an adipogenic differentiation-induced noncoding RNA (ADINR), transcribed ∼450 bp upstream of CEBPα. Sequence analysis of the ADINR locus showed that ADINR is highly conserved in rhesus, macaque, mouse, dog, and elephant. ADINR knockdown severely impaired adipogenesis in human mesenchymal stem cells (hMSC), which could be rescued by overexpression of CEBPα. The ADINR transcript can specifically bind to PA1 and recruits MLL3/4 histone methyltransferase complexes to enhance the transcription in CEBPα locus by increasing K3K4me3 methylation but decreasing H3K27me3 modification during adipogenesis (79).
linc-ADAL
linc-ADAL was identified as a nonconserved lncRNA highly expressed in human gluteal subcutaneous adipose tissue (38). The expression of linc-ADAL is highly inducible during human adipocyte differentiation and elevated in adipose tissue from obese individuals. Using human adipocyte culture, Zhang et al. (38) demonstrated that linc-ADAL is required for activation of the adipogenic gene program and lipid accumulation in adipocytes. Mechanistically, linc-ADAL interacts with hnRNPU and IGF2BP2, two RNA-binding proteins. The latter exerts posttranscriptional regulation on a large number of cytoplasmic mRNAs, including that of PPARα. Knockdown of linc-ADAL increased PPARα protein but not mRNA expression, suggesting potential negative effects of linc-ADAL on the translation of PPARα mRNA. While cultured human adipocytes provide a readily accessible model for interrogating the function of lncRNA in adipogenesis and metabolism, an inherent challenge in studying human-specific lncRNAs is the lack of appropriate experimental systems to study their role in metabolic physiology in vivo. Future work using fat transplantation in mice and organoid culture may help solve this conundrum.
ASMER-1 and ASMER-2
The rapid expansion of transcriptome data sets from patient samples allows researchers to interrogate the relationship between lncRNA expression and metabolic traits in human adipose tissue. Gao et al. (80) profiled lncRNA expression in subcutaneous WAT from lean versus obese and insulin-resistant versus insulin-sensitive people, during adipocyte differentiation of human adipose-derived stem cells, and in stromal vascular fraction (SVF) cells versus primary adipocytes (80). These analyses led to the identification of two adipocyte-specific metabolic related lncRNAs, ASMER-1 and ASMER-2. Knockdown of either lncRNAs by antisense oligonucleotides inhibited adipogenesis, lipid mobilization, and adiponectin secretion in human adipocyte culture (80). This study serves as an example of functional lncRNA discovery by examining a large number of data sets derived from a variety of clinical conditions.
ANRIL
Several genome-wide association studies revealed that genetic variants in the locus encoding lncRNA ANRIL were associated with risk for type 2 diabetes (81–84). In a later study, Lillycrop et al. (85) aimed to determine the contribution of early-life environment to obesity risk through epigenetic processes by examining the CpG methylation in umbilical cord DNA for children with different fat mass at age 6 years. This study identified an interesting association between the level of CpG methylation in the ANRIL promoter and later childhood adiposity. This correlation persists in several other cohort studies: in birth tissues from ethnically diverse neonates, in peripheral blood from adolescents, and in adipose from adults. Moreover, promoter CpG methylation was associated with ANRIL expression in vivo and CpG mutagenesis could inhibit ANRIL promoter activity in vitro. Thus, perinatal methylation at lncRNA loci could be a marker for later adiposity, although the function of ANRIL in adipocyte metabolism and physiology has yet to be determined (85).
Conclusions and Perspectives
Remarkable progress has been made in recent years on discovering adipocyte-enriched lncRNAs, understanding their functions in adipocyte biology and revealing the mechanisms of how lncRNAs interface with protein regulators. These studies have identified a large number of lncRNAs regulated in adipocytes under different pathophysiological conditions, determined the biological function for a set of lncRNAs during adipogenesis, brown-white cell type conversion and lineage-specific development, and revealed how these lncRNA may exert their functions through diverse molecular mechanisms.
However, our understanding of lncRNAs in adipocytes is still at its infancy and several important questions remain unanswered. First, for most lncRNAs, their mechanisms of action remain largely undefined with regard to the protein partners involved and their cross-talk with key nutritional and hormonal signaling pathways. Second, most of the published studies were limited to cell culture models of mouse or human adipocytes; the roles of lncRNAs in adipose tissue metabolism and physiology need to be further explored with animal genetic models. Third, despite the fact that many single nucleotide polymorphisms (SNPs) in lncRNA loci are associated with metabolic diseases including type 2 diabetes (81–84), whether these SNPs have functional effects on lncRNA expression and whether such a SNP–lncRNA relationship can affect adipocyte biology remain currently unknown.
There is little doubt that lncRNAs are a rich source of potential new drug targets. However, given the intracellular location of lncRNAs, development of lncRNAs as drug targets for human disease may be technically challenging. Effective lncRNA targeting molecules such as siRNA and antisense oligonucleotides (ASO) were very unstable in serum and do not easily cross the cell membrane. Recent advances in the chemistry of oligonucleotide design have significantly improved the pharmacological properties of siRNA and ASO (86). These modified molecules now are more resistant to RNA degradation and can enter cells more efficiently. These technical advances have led to several U.S. Food and Drug Administration–approved RNA-based drugs including Spinraza and eteplirsen, two ASO drugs that alter mRNA splicing in neurodegenerative diseases (87,88), and the most recent siRNA drug, patisiran, that targets transthyretin (TTR) mRNA for the treatment of hereditary TTR-mediated amyloidosis. Now we are at the brink of a revolution in drug development. It is conceivable that noncoding molecular targets will emerge and that targeting noncoding genes will serve as a novel therapeutic strategy for metabolic disease in the future.
Article Information
Funding. This work was supported by the Tanoto Initiative in Diabetes Research (L.S.); National Medical Research Council’s Cooperative Basic Research Grant (NMRC/CBRG/0101/2016) and Open Fund-Individual Research Grant (NMRC/OFIRG/0062/2017); and Singapore Ministry of Education’s Tier 2 grant (MOE2017-T2-2-009) and Tier 3 grants (MOE2014-T3-1-006 to the RNA Biology Center at the Cancer Science Institute of Singapore, National University of Singapore). This work was also supported by National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases (DK102456, DK112800, and AG055379 to J.D.L.) and the American Diabetes Association (1-15-BS-118 to J.D.L.).
Duality of Interest. No potential conflicts of interest relevant to this article were reported.
References
- 1.Rosen ED, Spiegelman BM. What we talk about when we talk about fat. Cell 2014;156:20–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med 2013;19:1252–1263 [DOI] [PubMed] [Google Scholar]
- 3.Townsend KL, Tseng YH. Brown fat fuel utilization and thermogenesis. Trends Endocrinol Metab 2014;25:168–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crewe C, An YA, Scherer PE. The ominous triad of adipose tissue dysfunction: inflammation, fibrosis, and impaired angiogenesis. J Clin Invest 2017;127:74–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martinez-Santibañez G, Lumeng CN. Macrophages and the regulation of adipose tissue remodeling. Annu Rev Nutr 2014;34:57–76 [DOI] [PubMed] [Google Scholar]
- 6.Reilly SM, Saltiel AR. Adapting to obesity with adipose tissue inflammation. Nat Rev Endocrinol 2017;13:633–643 [DOI] [PubMed] [Google Scholar]
- 7.Ikeda K, Maretich P, Kajimura S. The common and distinct features of brown and beige adipocytes. Trends Endocrinol Metab 2018;29:191–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu J, Jun H, McDermott JR. Formation and activation of thermogenic fat. Trends Genet 2015;31:232–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chouchani ET, Kazak L, Spiegelman BM. New advances in adaptive thermogenesis: UCP1 and beyond. Cell Metab 2019;29:27–37 [DOI] [PubMed] [Google Scholar]
- 10.Villarroya F, Gavaldà-Navarro A, Peyrou M, Villarroya J, Giralt M. The lives and times of brown adipokines. Trends Endocrinol Metab 2017;28:855–867 [DOI] [PubMed] [Google Scholar]
- 11.Wang GX, Zhao XY, Lin JD. The brown fat secretome: metabolic functions beyond thermogenesis. Trends Endocrinol Metab 2015;26:231–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertholet AM, Kazak L, Chouchani ET, et al. Mitochondrial patch clamp of beige adipocytes reveals UCP1-positive and UCP1-negative cells both exhibiting futile creatine cycling. Cell Metab 2017;25:811–822.e4 [DOI] [PMC free article] [PubMed]
- 13.Kazak L, Chouchani ET, Jedrychowski MP, et al. A creatine-driven substrate cycle enhances energy expenditure and thermogenesis in beige fat. Cell 2015;163:643–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazak L, Chouchani ET, Lu GZ, et al. Genetic depletion of adipocyte creatine metabolism inhibits diet-induced thermogenesis and drives obesity. Cell Metab 2017;26:660–671.e3 [DOI] [PMC free article] [PubMed]
- 15.Ikeda K, Kang Q, Yoneshiro T, et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med 2017;23:1454–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen XW, Li S, Lin JD. The micro-managing fat: exosomes as a new messenger. Trends Endocrinol Metab 2017;28:541–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen Z, Wang GX, Ma SL, et al. Nrg4 promotes fuel oxidation and a healthy adipokine profile to ameliorate diet-induced metabolic disorders. Mol Metab 2017;6:863–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, Zhang P, Chen Z, et al. Hepatic neuregulin 4 signaling defines an endocrine checkpoint for steatosis-to-NASH progression. J Clin Invest 2017;127:4449–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang GX, Zhao XY, Meng ZX, et al. The brown fat-enriched secreted factor Nrg4 preserves metabolic homeostasis through attenuation of hepatic lipogenesis. Nat Med 2014;20:1436–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Y, Buyel JJ, Hanssen MJ, et al. Exosomal microRNA miR-92a concentration in serum reflects human brown fat activity. Nat Commun 2016;7:11420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomou T, Mori MA, Dreyfuss JM, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature 2017;542:450–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartness TJ, Liu Y, Shrestha YB, Ryu V. Neural innervation of white adipose tissue and the control of lipolysis. Front Neuroendocrinol 2014;35:473–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morrison SF, Madden CJ. Central nervous system regulation of brown adipose tissue. Compr Physiol 2014;4:1677–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zeng W, Pirzgalska RM, Pereira MM, et al. Sympathetic neuro-adipose connections mediate leptin-driven lipolysis. Cell 2015;163:84–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rosell M, Kaforou M, Frontini A, et al. Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am J Physiol Endocrinol Metab 2014;306:E945–E964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Sharma VP, Shen H, et al. The hepatokine Tsukushi gates energy expenditure via brown fat sympathetic innervation. Nature Metabolism 2019;1:251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chi J, Wu Z, Choi CHJ, et al. Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab 2018;27:226–236.e3 [DOI] [PubMed]
- 28.Jiang H, Ding X, Cao Y, Wang H, Zeng W. Dense intra-adipose sympathetic arborizations are essential for cold-induced beiging of mouse white adipose tissue. Cell Metab 2017;26:686–692.e3 [DOI] [PubMed]
- 29.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fatica A, Bozzoni I. Long non-coding RNAs: new players in cell differentiation and development. Nat Rev Genet 2014;15:7–21 [DOI] [PubMed] [Google Scholar]
- 31.Makarewich CA, Olson EN. Mining for micropeptides. Trends Cell Biol 2017;27:685–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol 2015;11:151–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao XY, Lin JD. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci 2015;40:586–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kornfeld JW, Brüning JC. Regulation of metabolism by long, non-coding RNAs. Front Genet 2014;5:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen C, Cui Q, Zhang X, et al. Long non-coding RNAs regulation in adipogenesis and lipid metabolism: emerging insights in obesity. Cell Signal 2018;51:47–58 [DOI] [PubMed] [Google Scholar]
- 36.Cabili MN, Trapnell C, Goff L, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 2011;25:1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alvarez-Dominguez JR, Bai Z, Xu D, et al. De novo reconstruction of adipose tissue transcriptomes reveals long non-coding rna regulators of brown adipocyte development. Cell Metab 2015;21:764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Xue C, Lin J, et al. Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci Transl Med 2018;10:eaar5987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet 2014;30:121–123 [DOI] [PubMed] [Google Scholar]
- 40.Ding C, Lim YC, Chia SY, et al. De novo reconstruction of human adipose transcriptome reveals conserved lncRNAs as regulators of brown adipogenesis. Nat Commun 2018;9:1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uszczynska-Ratajczak B, Lagarde J, Frankish A, Guigó R, Johnson R. Towards a complete map of the human long non-coding RNA transcriptome. Nat Rev Genet 2018;19:535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang L, Duff MO, Graveley BR, Carmichael GG, Chen LL. Genomewide characterization of non-polyadenylated RNAs. Genome Biol 2011;12:R16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wu Q, Kim YC, Lu J, et al. Poly A- transcripts expressed in HeLa cells. PLoS One 2008;3:e2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cohen P, Levy JD, Zhang Y, et al. Ablation of PRDM16 and beige adipose causes metabolic dysfunction and a subcutaneous to visceral fat switch. Cell 2014;156:304–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harms MJ, Ishibashi J, Wang W, et al. Prdm16 is required for the maintenance of brown adipocyte identity and function in adult mice. Cell Metab 2014;19:593–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seale P, Kajimura S, Yang W, et al. Transcriptional control of brown fat determination by PRDM16. Cell Metab 2007;6:38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rajakumari S, Wu J, Ishibashi J, et al. EBF2 determines and maintains brown adipocyte identity. Cell Metab 2013;17:562–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab 2005;1:361–370 [DOI] [PubMed] [Google Scholar]
- 49.Li S, Mi L, Yu L, et al. Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc Natl Acad Sci USA 2017;114:E7111–E7120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kong X, Banks A, Liu T, et al. IRF4 is a key thermogenic transcriptional partner of PGC-1α. Cell 2014;158:69–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dempersmier J, Sambeat A, Gulyaeva O, et al. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol Cell 2015;57:235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ohno H, Shinoda K, Ohyama K, Sharp LZ, Kajimura S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 2013;504:163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell 2014;55:372–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mi L, Zhao XY, Li S, Yang G, Lin JD. Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol Metab 2016;6:101–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan H, Lv P, Huo X, et al. The nuclear matrix protein HNRNPU maintains 3D genome architecture globally in mouse hepatocytes. Genome Res 2018;28:192–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geuens T, Bouhy D, Timmerman V. The hnRNP family: insights into their role in health and disease. Hum Genet 2016;135:851–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hacisuleyman E, Goff LA, Trapnell C, et al. Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 2014;21:198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao XY, Li S, DelProposto JL, et al. The long noncoding RNA Blnc1 orchestrates homeostatic adipose tissue remodeling to preserve metabolic health. Mol Metab 2018;14:60–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao XY, Xiong X, Liu T, et al. Long noncoding RNA licensing of obesity-linked hepatic lipogenesis and NAFLD pathogenesis. Nat Commun 2018;9:2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bai Z, Chai XR, Yoon MJ, et al. Dynamic transcriptome changes during adipose tissue energy expenditure reveal critical roles for long noncoding RNA regulators. PLoS Biol 2017;15:e2002176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Talwar S, Balasubramanian S, Sundaramurthy S, et al. Overexpression of RNA-binding protein CELF1 prevents apoptosis and destabilizes pro-apoptotic mRNAs in oral cancer cells. RNA Biol 2013;10:277–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vlasova IA, Bohjanen PR. Posttranscriptional regulation of gene networks by GU-rich elements and CELF proteins. RNA Biol 2008;5:201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmidt E, Dhaouadi I, Gaziano I, et al. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat Commun 2018;9:3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang Y, Zheng Y, Jin C, Li X, Jia L, Li W. Long non-coding RNA H19 inhibits adipocyte differentiation of bone marrow mesenchymal stem cells through epigenetic modulation of histone deacetylases. Sci Rep 2016;6:28897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rosen ED, MacDougald OA. Adipocyte differentiation from the inside out. Nat Rev Mol Cell Biol 2006;7:885–896 [DOI] [PubMed] [Google Scholar]
- 66.Xie H, Lim B, Lodish HF. MicroRNAs induced during adipogenesis that accelerate fat cell development are downregulated in obesity. Diabetes 2009;58:1050–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alexander R, Lodish H, Sun L. MicroRNAs in adipogenesis and as therapeutic targets for obesity. Expert Opin Ther Targets 2011;15:623–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu B, Gerin I, Miao H, et al. Multiple roles for the non-coding RNA SRA in regulation of adipogenesis and insulin sensitivity. PLoS One 2010;5:e14199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu S, Sheng L, Miao H, et al. SRA gene knockout protects against diet-induced obesity and improves glucose tolerance. J Biol Chem 2014;289:13000–13009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chooniedass-Kothari S, Emberley E, Hamedani MK, et al. The steroid receptor RNA activator is the first functional RNA encoding a protein. FEBS Lett 2004;566:43–47 [DOI] [PubMed] [Google Scholar]
- 71.Sun L, Goff LA, Trapnell C, et al. Long noncoding RNAs regulate adipogenesis. Proc Natl Acad Sci U S A 2013;110:3387–3392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lo KA, Huang S, Walet ACE, et al. adipocyte long-noncoding rna transcriptome analysis of obese mice identified Lnc-leptin, which regulates leptin. Diabetes 2018;67:1045–1056 [DOI] [PubMed] [Google Scholar]
- 73.Natoli G, Andrau JC. Noncoding transcription at enhancers: general principles and functional models. Annu Rev Genet 2012;46:1–19 [DOI] [PubMed] [Google Scholar]
- 74.Zhu E, Zhang J, Li Y, Yuan H, Zhou J, Wang B. Long noncoding RNA Plnc1 controls adipocyte differentiation by regulating peroxisome proliferator-activated receptor γ. FASEB J 2019;33:2396–2408 [DOI] [PubMed] [Google Scholar]
- 75.Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res 2013;73:1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet 2013;9:e1003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep 2016;6:22640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carter S, Miard S, Boivin L, Sallé-Lefort S, Picard F. Loss of Malat1 does not modify age- or diet-induced adipose tissue accretion and insulin resistance in mice. PLoS One 2018;13:e0196603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xiao T, Liu L, Li H, et al. Long noncoding RNA ADINR regulates adipogenesis by transcriptionally activating C/EBPα. Stem Cell Reports 2015;5:856–865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gao H, Kerr A, Jiao H, et al. Long non-coding RNAs associated with metabolic traits in human white adipose tissue. EBioMedicine 2018;30:248–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Saxena R, Voight BF, Lyssenko V, et al.; Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University, and Novartis Institutes of BioMedical Research . Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336 [DOI] [PubMed] [Google Scholar]
- 82.Pasmant E, Sabbagh A, Vidaud M, Bièche I. ANRIL, a long, noncoding RNA, is an unexpected major hotspot in GWAS. FASEB J 2011;25:444–448 [DOI] [PubMed] [Google Scholar]
- 83.Scott LJ, Mohlke KL, Bonnycastle LL, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science 2007;316:1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zeggini E, Weedon MN, Lindgren CM, et al.; Wellcome Trust Case Control Consortium (WTCCC) . Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science 2007;316:1336–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lillycrop K, Murray R, Cheong C, et al.; EpiGen Consortium . ANRIL promoter DNA methylation: a perinatal marker for later adiposity. EBioMedicine 2017;19:60–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Khvorova A, Watts JK. The chemical evolution of oligonucleotide therapies of clinical utility. Nat Biotechnol 2017;35:238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Finkel RS, Mercuri E, Darras BT, et al.; ENDEAR Study Group . Nusinersen versus sham control in infantile-onset spinal muscular atrophy. N Engl J Med 2017;377:1723–1732 [DOI] [PubMed] [Google Scholar]
- 88.Cirak S, Arechavala-Gomeza V, Guglieri M, et al. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment: an open-label, phase 2, dose-escalation study. Lancet 2011;378:595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Cui X, You L, Li Y, et al. A transcribed ultraconserved noncoding RNA, uc.417, serves as a negative regulator of brown adipose tissue thermogenesis. FASEB J 2016;30:4301–4312 [DOI] [PubMed] [Google Scholar]
- 90.Xiong Y, Yue F, Jia Z, et al. A novel brown adipocyte-enriched long non-coding RNA that is required for brown adipocyte differentiation and sufficient to drive thermogenic gene program in white adipocytes. Biochim Biophys Acta Mol Cell Biol Lipids 2018;1863:409–419 [DOI] [PMC free article] [PubMed]
- 91.You L, Zhou Y, Cui X, et al. GM13133 is a negative regulator in mouse white adipocytes differentiation and drives the characteristics of brown adipocytes. J Cell Physiol 2018;233:313–324 [DOI] [PubMed] [Google Scholar]
- 92.Pang WJ, Lin LG, Xiong Y, et al. Knockdown of PU.1 AS lncRNA inhibits adipogenesis through enhancing PU.1 mRNA translation. J Cell Biochem 2013;114:2500–2512 [DOI] [PubMed] [Google Scholar]
- 93.Wei N, Wang Y, Xu RX, et al. PU.1 antisense lncRNA against its mRNA translation promotes adipogenesis in porcine preadipocytes. Anim Genet 2015;46:133–140 [DOI] [PubMed] [Google Scholar]
- 94.Chen J, Liu Y, Lu S, et al. The role and possible mechanism of lncRNA U90926 in modulating 3T3-L1 preadipocyte differentiation. Int J Obes 2017;41:299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu W, Ma C, Yang B, Yin C, Zhang B, Xiao Y. LncRNA Gm15290 sponges miR-27b to promote PPARγ-induced fat deposition and contribute to body weight gain in mice. Biochem Biophys Res Commun 2017;493:1168–1175 [DOI] [PubMed] [Google Scholar]
- 96.Firmin FF, Oger F, Gheeraert C, et al. The RBM14/CoAA-interacting, long intergenic non-coding RNA Paral1 regulates adipogenesis and coactivates the nuclear receptor PPARγ. Sci Rep 2017;7:14087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shang G, Wang Y, Xu Y, et al. Long non-coding RNA TCONS_00041960 enhances osteogenesis and inhibits adipogenesis of rat bone marrow mesenchymal stem cell by targeting miR-204-5p and miR-125a-3p. J Cell Physiol 2018;233:6041–6051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu H, Li H, Jin L, et al. Long noncoding RNA GAS5 suppresses 3T3-L1 cells adipogenesis through miR-21a-5p/PTEN signal pathway. DNA Cell Biol 2018;37:767–777 [DOI] [PubMed] [Google Scholar]
- 99.Li M, Xie Z, Wang P, et al. The long noncoding RNA GAS5 negatively regulates the adipogenic differentiation of MSCs by modulating the miR-18a/CTGF axis as a ceRNA. Cell Death Dis 2018;9:554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cai R, Sun Y, Qimuge N, et al. Adiponectin AS lncRNA inhibits adipogenesis by transferring from nucleus to cytoplasm and attenuating Adiponectin mRNA translation. Biochim Biophys Acta Mol Cell Biol Lipids 2018;1863:420-432 [DOI] [PubMed]
- 101.Divoux A, Karastergiou K, Xie H, et al. Identification of a novel lncRNA in gluteal adipose tissue and evidence for its positive effect on preadipocyte differentiation. Obesity (Silver Spring) 2014;22:1781–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ballantyne RL, Zhang X, Nuñez S, et al. Genome-wide interrogation reveals hundreds of long intergenic noncoding RNAs that associate with cardiometabolic traits. Hum Mol Genet 2016;25:3125–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang Y, Jin C, Zheng Y, et al. Knockdown of lncRNA MIR31HG inhibits adipocyte differentiation of human adipose-derived stem cells via histone modification of FABP4. Sci Rep 2017;7:8080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Nuermaimaiti N, Liu J, Liang X, et al. Effect of lncRNA HOXA11-AS1 on adipocyte differentiation in human adipose-derived stem cells. Biochem Biophys Res Commun 2018;495:1878–1884 [DOI] [PubMed] [Google Scholar]
- 105.Liu Y, Ji Y, Li M, et al. Integrated analysis of long noncoding RNA and mRNA expression profile in children with obesity by microarray analysis. Sci Rep 2018;8:8750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Li Z, Jin C, Chen S, et al. Long non-coding RNA MEG3 inhibits adipogenesis and promotes osteogenesis of human adipose-derived mesenchymal stem cells via miR-140-5p. Mol Cell Biochem 2017;433:51–60 [DOI] [PubMed] [Google Scholar]