Abstract
DNA double-strand breaks (DSBs) are induced by a variety of genotoxic agents, including ionizing radiation and chemotherapy drugs for treating cancers. The elimination of DSBs proceeds via distinctive error-free and error-prone pathways. Repair by homologous recombination (HR) is largely error-free and mediated by RAD51/BRCA2 gene products. Classical non-homologous end joining (C-NHEJ) requires the Ku heterodimer and can efficiently rejoin breaks, with occasional loss or gain of DNA information. Recently, evidence has unveiled another DNA end-joining mechanism that is independent of recombination factors and Ku proteins, termed alternative non-homologous end joining (A-NHEJ). While A-NHEJ-mediated repair does not require homology, in a subtype of A-NHEJ, DSB breaks are sealed by microhomology (MH)-mediated base-pairing of DNA single strands, followed by nucleolytic trimming of DNA flaps, DNA gap filling, and DNA ligation, yielding products that are always associated with DNA deletion. This highly error-prone DSB repair pathway is termed microhomology-mediated end joining (MMEJ). Dissecting the mechanisms of MMEJ is of great interest because of its potential to destabilize the genome through gene deletions and chromosomal rearrangements in cells deficient in canonical repair pathways, including HR and C-NHEJ. In addition, evidence now suggests that MMEJ plays a physiological role in normal cells.
Keywords: DNA double strand break, Microhomology, End joining, Chromosome rearrangement, Mutagenesis
1. Introduction
Cellular DNA double-strand breaks (DSBs) arise from endogenous damage sources, such as replication errors, reactive oxygen species, and enzymatic processes, as well as exogenous damage sources, such as ionizing radiation and chemotherapeutic agents [1]. DSBs are arguably the most toxic DNA lesions, because if not repaired, a single DSB can kill a cell. Moreover, if a DSB is repaired improperly, it can induce gross chromosomal rearrangements and mutagenesis, which are the most common drivers underlying oncogenesis and a host of genetic diseases [2–5]. Cells have developed multiple mechanisms to quickly and accurately fix broken DNA – thus restoring chromosomal integrity – including homologous recombination, and classical and alternative non-homologous end joining (C-NHEJ and A-NHEJ, respectively).
In homologous recombination (HR), the break repair is directed by an intact homologous sequence, usually from sister chromatids, following DNA replication [6–9]. Four types of HR have been described: gene conversion, synthesis-dependent strand annealing (SDSA), break-induced replication (BIR), and single-strand annealing (SSA). All but SSA are dependent on the RAD51 gene, and they all initiate repair with the resection of DSB ends by a 5′-to-3′ exonuclease to produce long 3′-ended, single-stranded DNA (ssDNA) tails [10]. In gene conversion, SDSA, and BIR, the ssDNA invades the homologous donor sequence to prime repair DNA synthesis. In SSA, the complementary strands of the homologous regions flanking a DSB anneal, producing an intermediate with two non-homologous 3′-ended tails that must be removed for new DNA synthesis and ligation to occur [11,12].
Alternatively, classical non-homologous end joining (C-NHEJ) repairs DSBs by juxtaposing and ligating DNA ends, using very little (1–4 nucleotides [nt]) or no complementary base pairing [1,13–15]. This can occur throughout the cell cycle; however, it is most active in G1 [16–18]. In C-NHEJ in mammalian cells, the Ku70/Ku80 heterodimer first binds to DSB ends and recruits other NHEJ factors, including DNA-dependent protein kinase catalytic subunit (DNA-PKcs), X-ray repair cross complementing 4 (XRCC4), DNA ligase 4 (LIG4), and XRCC4-like factor (XLF), to catalyze DNA ligation across the DNA breaks [19–22]. If the DNA ends are not compatible for ligation, then DNA-end processing, such as trimming, filling-in, or blocking-end removal, ensues by one of many processing enzymes, such as Artemis, polynucleotide kinase 3′-phosphatase (PNKP), aprataxin (APTX) and PNKP-like factor (APLF), DNA polymerase mu (Polp), DNA polymerase lambda (PolA), or Werner syndrome RecQ-like helicase (WRN), prior to ligation [23–27].
Finally, alternative NHEJ (A-NHEJ), which includes microhomology (MH)-mediated end joining (MMEJ), repairs DNA DSBs by annealing 2–20-bp stretches of overlapping bases flanking the DSB [28–31]. Not all A-NHEJ produces repair junctions with MH; therefore, MMEJ likely only corresponds to a subset of A-NHEJ. Genetically, MMEJ does not require Ku70/Ku80, RAD51, BRCA2, or LIG4, but both processes require the MRE11-RAD50-NBS1 complex [MRN complex; Mre11-Rad50-Xrs2 (MRX) complex in yeast], DNA polymerase theta (Polθ) or the B and X family polymerases in yeast (Polδ and Pol4), CtBP-interacting protein (CtIP, Sae2 in yeast), poly (ADP-ribose) polymerase 1 (PARP1), ataxia telangiectasia mutated (ATM; Tel1 in yeast), and flap endonuclease 1 (FEN1) [32–40]. In this review, we will focus on the role of MMEJ as a genome destabilizer, and we will show that, despite the high mutation burden associated with MMEJ, it provides valuable physiological functions. We will also describe the pathological role of MMEJ as a back-up repair pathway, and we will discuss the emerging possibility that MMEJ is an attractive anti-cancer drug target.
2. Biochemical steps of MMEJ
Before we evaluate the current model of MMEJ, and the associated molecular and biochemical steps, we would like to highlight several recent, outstanding reviews on MMEJ. We encourage readers to refer to these reviews for additional, in-depth information about the components and attributes of MMEJ [28–31]. Throughout this review, we will primarily use the terminology for mammalian MMEJ factors, but we will list their yeast counterparts in parentheses. If the genes or proteins only exist in yeast, or in organisms other than mammals, we will designate their origin using small letters in front of their name (e.g. ySrs2; Srs2 protein in yeast).
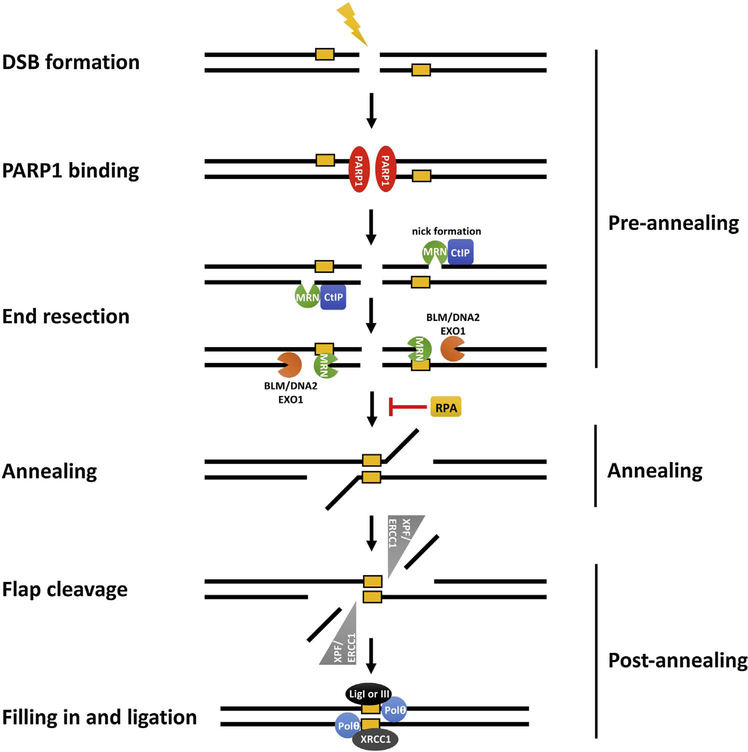
Operationally, MMEJ can be subdivided into three discrete steps: pre-annealing, annealing, and post-annealing of the flanking MHs (Fig. 1). The first step in MMEJ involves end resection to expose the flanking MHs for annealing. Resection likely operates via a common mechanism between HR and MMEJ; the MRN complex (MRX in budding yeast) along with CtIP (Sae2 in yeast), introduces a nick near the DSB lesion and degrades DNA using 3′−5′ exonuclease activity [38,41–43]. More extensive 5′−3′ resection is catalyzed by the Bloom syndrome RecQ-like helicase (BLM)/exonuclease 1 (EXO1) (Sgs1/Exo1 and Dna2 in budding yeast) [38,44–46]. Interestingly, BLM/EXO1 is not only dispensable for MMEJ, but it also suppresses MMEJ when MHs are located within 2-kb of the DSBs [38,47]. Thus, extensive resection might favor HR over MMEJ.
Fig. 1.
The basic mechanisms of MMEJ in human cells. MMEJ could be divided to three discrete steps, pre-annealing, annealing, and post-annealing of the flanking MHs. PARP1 binds to DSB ends and facilitates the recruitment of resection factors [CtIP and Mre11 complex (Mre11/Rad50/Nbs1)] to uncover MHs (shown in yellow boxes) flanking DSBs. MHs that are far from the break likely require extensive resection by BLM/EXO1 to facilitate MMEJ. Annealing of MHs, which is inhibited by single strand binding RPA complex, induces the formation of non-homologous tails/flaps. Non-homologous tails/flaps are then removed by XPF/ERCC1 nuclease before filling-in synthesis and ligation by Polθ and LigI/III, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Once exposed by resection, the flanking MHs will anneal via an unidentified mechanism. The size of the MHs, engaged in annealing, likely dictates the stability of the annealed intermediates and the repair outcome. Consistent with this idea, GC-rich MHs facilitate MMEJ in vitro [48], whereas mismatched nucleotides within MHs reduce MMEJ [49]. Finally, the ssDNA-binding, heterotrimeric replication protein A (RPA) complex, which consists of RPA70, RPA32, and RPA14, interferes with MMEJ mainly by preventing annealing of ssDNA [47,50].
The annealed intermediates are further modified by the XPF/ERCC1 (Rad1/Rad10 in yeast) structure-specific nuclease complex, which trims the non-homologous tails and produces 3′-hydroxyl termini that are eligible for extension by DNA polymerase [36,51]. The low fidelity DNA polymerase Polθ [52], which is encoded by POLQ, is capable of extending mismatched termini, ssDNA, and partial ssDNA [53]. Polθ and its fly ortholog dmMus308 have been implicated in MMEJ of eroded telomeres and DSBs induced by stressed replication forks [32,33,54,55]. In budding yeast, there is no Polθ homologue; instead, polymerase delta (Polδ) and Pol4 are important for yeast MMEJ [56,57].
Finally, the DNA ligation is catalyzed redundantly by LIG3 and LIG1, or partially by Dnl4 in yeast [36,58–60]. In mammals, MMEJ also requires X-ray repair cross complementing 1 (XRCC1) and PARP1. XRCC1/LIG3 and PARP1 have been implicated in end modification/synapsis and in recruitment of Polθ to DSB lesions to prime MMEJ [33]. Notably, the reported genetic requirements of MMEJ are surprisingly diverse and significantly divergent across species. Therefore, the ligases involved in MMEJ or the role of XRCC1 in MMEJ remain controversial [60]. Although additional studies are surely needed to resolve these discrepancies, one possible reason for these reported differences might stem from the fact that MMEJ and A-NHEJ are not necessarily part of a single pathway. Instead, MMEJ and A-NHEJ comprise several different mechanisms, each with their own genetic requirements. Adding to this complexity, another end-joining pathway, was described recently; this pathway has more resemblance to C-NHEJ and operates at G1 upon limited resection [61]. In the future, it will be necessary to reassess the precise factors involved in each of these processes and their relationship to each other.
3. Genomic consequences of MMEJ
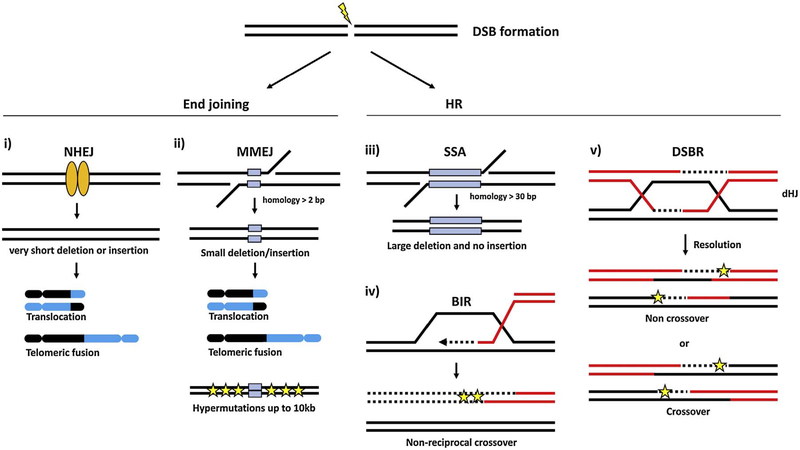
Although DSB repair pathways provide cells the capacity to avoid the more catastrophic consequences that follow repair failure, the DSB repair pathways are often mutagenic and are the primary source of gross chromosomal rearrangements and chromosomal instability upon DNA damage (Fig. 2). HR, which is regarded as the high-fidelity repair pathway, can trigger chromosomal translocations if the recombination intermediates are resolved by crossover between allelic or non-homologous chromosomes [62]. BIR and SSA can also produce sequence deletions and non-reciprocal translocations at sequences flanking the breakpoint junctions [40,63]. C-NHEJ leads to small sequence insertions or deletions (indels) at the breakpoint junction due to the end processing that occurs prior to ligation by different nucleases and polymerases [64–67]. Moreover, if more than two breaks occur simultaneously, improper joining of these DSBs by C-NHEJ can also produce chromosomal translocations and rearrangements [68].
Fig. 2.
Genomic instability caused by DNA DSB repair mechanisms. C-NHEJ (i) generates small sequence deletions/insertions at breakpoint junctions. MMEJ (ii) induces deletions and insertions at the breakpoint junctions and hypermutagenesis at the flanking DNA sequence up to several kilobases from the break. Both C-NHEJ and MMEJ could trigger chromosomal translocations and telomeric fusions. SSA (iii) also induces large deletions but not insertions at repair junctions. In BIR (iv) and gene conversion (v), low fidelity DNA synthesis produces mutagenesis at the flanking DNA sequence and could induce chromosomal translocations if recombination involves non-allelic template and/or crossover formation. Blue boxes represent homologies and the stars represent mutations. The dotted lines are newly synthesized DNA. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
However, among the DSB repair pathways, MMEJ appears to play a major role in DSB-induced mutagenesis. MMEJ is always mutagenic because one of the two MH regions, and the inter-MH region, will be deleted from the repair product. Analysis of ~7000 deletion breakpoints in the C. elegans revealed that the majority of ethyl methane sulfonate (EMS) or photoactivated trimethylpsolaren (UV/TMP)-induced indels are products of Polθ-mediated end joining [69]. These breakpoint junctions often contain short, sometimes templated, or non-templated, insertions that are reminiscent of the breakpoint junctions of complex copy number variants, which often contain MHs and are associated with a high frequency of missense and indel mutations in the flanking DNA due to error-prone repair synthesis [69]. The terminal transferase activity and frequent template switching of Polθ, followed by the iterative synthesis of ssDNA or partial ssDNA, might account for frequent sequence insertions at the breakpoint junctions [70]. Mammalian Pol0 also has RAD51-binding domains that can block RAD51-mediated recombination and thereby promote end joining pathways including MMEJ [71].
MMEJ has also been implicated in the formation of chromosomal translocations and gross chromosomal rearrangements in mouse, which could explain why most breakpoints from chromosomal translocations in human somatic cells and cancers feature MHs at the junctions [72,73]. In yeast, it was initially shown that, if multiple breaks form simultaneously, MMEJ mediates promiscuous end joining, leading to chromosomal translocations and rearrangements [49]. To further test the role of MMEJ in the formation of chromosomal translocation, two DSBs were induced on different chromosomes, using CRISPR-CAS9 or I-SceI nucleases, in murine and human cells deficient in C-NHEJ, and the frequencies of reciprocal translocation induction were measured [74,75]. They found that, in mouse embryonic stem cells, inactivation of the NHEJ factor XRCC4 elevates chromosomal rearrangements approximately 5-fold over those in controls, and over 60% of the XRCC4-depletion-driven translocations contain 1–4-bp MHs [74,75]. Moreover, in murine cells, the formation of chromosomal aberrations is dependent on the MMEJ repair factors CtIP and LIG3 [60,76]. However, in human cells, the formation of chromosomal translocations is not dependent on MMEJ factors but is dependent on C-NHEJ factors, including Ligase IV [74]. Recently, a unique variant of NHEJ, termed resection-dependent NHEJ (rdNHEJ), has been described [61]; this variant operates with delayed kinetics and repair of DNA ends that are resected by CtIP or Artemis in the G1 phase of the cell cycle. These repair events are distinct from MMEJ because they do not depend on PARP1 and LIG3 but do depend on DNA-PKcs, and Ligase IV. Thus, it has been suggested that rdNHEJ, rather than A-NHEJ, induces chromosomal translocation formation in human cells [61]. Alternatively, the human cell lines used for this study could be deficient in MRN/ATM function [77] and this may have influenced the contribution of A-NHEJ to the rearrangements [78].
In addition to chromosomal structural changes, MMEJ-mediated DSB repair elevates the mutation frequency at breakpoint junctions, which is likely due to the use of low fidelity polymerases in repair synthesis and/or to the formation of mutation-prone ssDNA. Indeed, hypermutagenesis frequently occurs in the sequences flanking the breakpoint junctions, which has been suggested to be due to the error-prone nature of Polθ [55]. However, in yeast, which lacks a Polθ homologue, hypermutagenesis still occurs within sequences flanking the breakpoint junctions, and this hypermutagenesis extends 7–9 kb from the junctions [79]. These data suggest that the hypermutagenesis is not solely due to Polθ; instead, it is an inherent feature of MMEJ. It is possible that the hypermutagenesis is due to the slow kinetics of MMEJ [79], inevitably leading to the formation of extensive ssDNA, which is more prone to irreversible sequence changes upon exogenous and endogenous DNA damage [80]. Interestingly, the junctions from chromosomal rearrangements and copy number variants (CNVs) are often associated with increased mutagenesis within several kilobases flanking the breakpoints, and most of these junctions harbor MHs at the breakpoints, likely resulting from MMEJ or micro-BIR [81–85].
4. Pathological functions of MMEJ and its potential as an anticancer drug target
Because of the high mutation burden and chromosome structural changes that accompany MMEJ, one would assume that MMEJ is tightly regulated, and that it would not be the first option for repairing DSBs. Indeed, accumulating evidence suggests that, when C-NHEJ or HR are deficient, MMEJ is a surprisingly robust and efficient alternative repair option.
Multiple lines of evidence suggest that MMEJ is efficient when C-NHEJ is deficient. For instance, immunoglobulin class-switch DNA recombination (CSR) [86], initiated by activation-induced cytidine deaminase (AID)-introduced DSBs in the switch (S) regions, typically involves Ku70/Ku86-mediated NHEJ [87,88]. However, in Ku-deficient B cells, MMEJ still promotes CSR, albeit at a reduced level compared with that of NHEJ, introducing MHs at the S-S junctions [86–90].
Moreover, uncapped telomere fusion can be catalyzed in a C-NHEJ- or MMEJ-dependent manner. Eukaryotic telomeres are protected by the Shelterin complex, which contains TRF1, TRF2, POT1, RAP1, TIN2, and TPP1 [91]. In murine cells, TRF2 deficiency induces checkpoint sensor 53BP1-dependent, C-NHEJ-mediated telomere fusion [92]. However, in 53BP1−/−or Lig4−/− murine cells with a TPP1-POT1 deficiency, telomere fusions are still induced under naturally shortened telomere environments, presumably via an A-NHEJ pathway [92]. Importantly, in human cells, sequencing analysis data showed that shortened telomere fusion is due to MMEJ, and not C-NHEJ [93]. Confusingly, however, subsequent work demonstrated that both LIG3 and LIG4 play roles in telomere fusions in human cells [94].
MMEJ provides a back-up DNA repair mechanism upon HR deficiency, particularly in cancer cells [50,71]. A recent study showed that BRCA2-deficient ovarian cancer cells express high levels of POLQ, which may contribute to elevated MMEJ in these cells [71]. Therefore, inhibition of Polθ sensitizes these cells to genotoxic chemicals and PARP1 inhibitor treatment, indicating Polθ inhibition as a novel cancer therapeutic strategy. Cancer cells with BRCA1 or BRCA2 mutations are sensitive to PARP1 inhibitor treatment [95–97]; however, the treated cells eventually develop PARP1 resistance as a result of a secondary BRCA mutation, depletion of 53BP1, downregulation of PARP1, or upregulation of ABC transporters (see review in [98]). To overcome PARP1 inhibitor resistance, several studies have investigated the effects of co-treatment with PARP1 inhibitors and histone deacetylase (HDAC) inhibitors [99–103]. Evidence suggests that HDAC inhibitors decrease the expression of HR factors [103–105]. Interestingly, the deacetylase and mono-ADP-transferase SIRT6 deacetylates the MMEJ factor CtIP, which stimulates CtIP activity. SIRT6 could thus increase MMEJ activity and be a potential therapeutic target in HR-deficient cancer cells with high MMEJ activity.
There are also reports of a concomitant upregulation of MMEJ, and downregulation of C-NHEJ activity in certain cancer cells. For instance, the reciprocal translocation t(9;22)(q34;q11) results in expression of the oncogenic BCR/ABL fusion gene that causes chronic myeloid leukemia (CML). Interestingly, levels of the MMEJ factors WRN and LIG3 are increased, whereas the C-NHEJ factors Artemis and LIG4 are decreased, in CML cells [106]. Moreover, siRNA-mediated inactivation of WRN and LIG3 decreases end-joining efficiency and leads to persistent DSBs in CML cells [106]. In human bladder cancer, the levels of C-NHEJ factors are also reduced, suggesting that these cancer cells may also rely on MMEJ to repair their DSBs [107]. This increased use of the MMEJ pathway in specific cancer cells has important implications for cancer therapy.
5. Physiological role of MMEJ
Although ample evidence supports the role of MMEJ as the back-up pathway for cells deficient in C-NHEJ and HR, the physiological roles of MMEJ in cells proficient in C-NHEJ and HR are not well understood and remain debatable. Unlike DSBs induced by site-specific nucleases in experimental settings, radiation generates DSBs with complex end termini such as 3′-phosphate (P) and 3′-phosphoglycolate [108]. Prior to ligation, the 3′-Ps at DNA ends must be removed by PNKP [109], and blocking 3′-P removal at DSBs increases MMEJ approximately 3-fold in cancer cells [110]. This result suggests the intriguing possibility that intracellular DSBs, induced by radiation or physiological agents, are more dependent on MMEJ for repair. Furthermore, recent evidence suggests that mammalian mitochondrial extract relies on CtIP, FEN1, LIG3, MRE11, and PARP1, but not LIG4, to repair DSBs [111]. Notably, more than 85% of over 100 types of mitochondrial (mt) DNA deletions are flanked by short repeated sequences, suggesting that error-prone MMEJ in mtDNA may be associated with human mitochondrial disorders [112–114]. However, it should be noted that the reliance of DSB repair in mitochondria on MMEJ can – at least in part – be attributed to the fact that LIG3 is the sole ligase expressed in that organelle. Overall, the emerging theme is that if DSB breaks are not repaired quickly, then they are more likely to be the substrate of MMEJ for repair.
Alternatively, it may be that MMEJ is favored for DSB repair over other DNA repair pathways under specific conditions. Indeed, several regulatory mechanisms that favor MMEJ have been reported recently. For instance, S-phase phosphorylation of CtIP triggers MMEJ [38,115]. Moreover, deacetylation of CtIP by SIRT6 could activate CtIP resection activity and increase MMEJ [116]. In cells in G1, 53BP1 also promotes MMEJ [117], and interestingly, knockdown of 53BP1 decreases distal MMEJ (> 1000 bp distance from DSB) but not proximal MMEJ [117]. Lastly, evidence suggests that Polθ-dependent MMEJ operates primarily in S phase. One study used a reporter system with MHs flanking I-SceI-induced DSB breaks to illustrate the predominance of MMEJ in S/G2 [38]. It is possible that NHEJ and HR are both suppressed in the S phase, rendering MMEJ the sole option to repair DSBs arising during replication collapse and stress. Furthermore, because MMEJ is inhibited by extensive resection, MMEJ may be optimal under limited resection conditions; however, the physiological conditions that favor partial resection are unknown.
6. Concluding remarks
MMEJ is an emerging drug target due to its role as the back-up DSB repair pathway in C-NHEJ- or HR-deficient cancers [71,118–120]. However, accumulating evidence suggests that MMEJ is not simply a back-up for HR and C-NHEJ. MMEJ likely has bona-fide, physiological roles in DSB repair in many eukaryotic cells. Moreover, we would like to emphasize that, although MMEJ is highly destabilizing and capable of triggering chromosomal translocations and rearrangements, inactivation of the MMEJ factor Polθ results in vast chromosomal deletions and instability [34,54,121], indicating that MMEJ also provides necessary protective functions to the genome. All these results warrant more systematic and extensive analyses of the biochemical and genetic components of MMEJ in multiple models. The outcome of such studies will further elucidate the intricate network and regulation of DSB repair in human cells, thereby facilitating the development of novel therapeutic strategies to eliminate cancer cells with little or no side effects.
Acknowledgements
We are grateful to the members of Lee lab for helpful discussions. We also thank Kihoon Lee, Jaehoon Ji, Supriya Sinha and Diana Villarreal for their contributions to MMEJ studies in our laboratory.
Funding
This work was supported by NIH research grant GM71011, William and Ella Owens Medical Research Foundation and CTSA pilot grant to S.E.L and a grant from ThriveWell Cancer Foundation to E.Y.S.
Footnotes
Conflict of interest statement
The authors declare that there are no conflicts of interest.
References
- [1].Lieber MR, The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway, Annu. Rev. Biochem 79 (2010) 181–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Bunting SF, Nussenzweig A, End-joining, translocations and cancer, Nat. Rev. Cancer 13 (2013) 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rulten SL, Caldecott KW, DNA strand break repair and neurodegeneration, DNA Repair (Amst) 12 (2013) 558–567. [DOI] [PubMed] [Google Scholar]
- [4].Phillips ER, McKinnon PJ, DNA double-strand break repair and development, Oncogene 26 (2007) 7799–7808. [DOI] [PubMed] [Google Scholar]
- [5].Bohgaki T, Bohgaki M, Hakem R, DNA double-strand break signaling and human disorders, Genome Integr. 1 (2010) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kowalczykowski SC, An overview of the molecular mechanisms of recombinational DNA repair, Cold Spring Harb. Perspect. Biol 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Paques F, Haber JE, Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae, Microbiol. Mol. Biol. Rev 63 (1999) 349–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Heyer WD, Regulation of recombination and genomic maintenance, Cold Spring Harb. Perspect. Biol 7 (2015) a016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Haber JE, Partners and pathwaysrepairing a double-strand break, Trends Genet. 16 (2000) 259–264. [DOI] [PubMed] [Google Scholar]
- [10].Symington LS, Gautier J, Double-strand break end resection and repair pathway choice, Annu. Rev. Genet 45 (2011) 247–271. [DOI] [PubMed] [Google Scholar]
- [11].Motycka TA, Bessho T, Post SM, Sung P, Tomkinson AE, Physical and functional interaction between the XPF/ERCC1 endonuclease and hRad52, J. Biol. Chem 279 (2004) 13634–13639. [DOI] [PubMed] [Google Scholar]
- [12].Symington LS, Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair, Microbiol. Mol. Biol. Rev 66 (2002) 630–670 (table of contents). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pannunzio NR, Li S, Watanabe G, Lieber MR, Non-homologous end joining often uses microhomology: implications for alternative end joining, DNA Repair (Amst) 17 (2014) 74–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Lieber MR, The mechanism of human nonhomologous DNA end joining, J. Biol. Chem 283 (2008) 1–5. [DOI] [PubMed] [Google Scholar]
- [15].Gu J, Lu H, Tsai AG, Schwarz K, Lieber MR, Single-stranded DNA ligation and XLF-stimulated incompatible DNA end ligation by the XRCC4-DNA ligase IV complex: influence of terminal DNA sequence, Nucleic Acids Res. 35 (2007) 5755–5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ira G, Pellicioli A, Balijja A, Wang X, Fiorani S, Carotenuto W, Liberi G, Bressan D, Wan L, Hollingsworth NM, Haber JE, Foiani M, DNA end resection, homologous recombination and DNA damage checkpoint activation require CDK1, Nature 431 (2004) 1011–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mao Z, Bozzella M, Seluanov A, Gorbunova V, DNA repair by nonhomologous end joining and homologous recombination during cell cycle in human cells, ABBV Cell Cycle 7 (2008) 2902–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Zhang Y, Shim EY, Davis M, Lee SE, Regulation of repair choice: cdk1 suppresses recruitment of end joining factors at DNA breaks, DNA Repair (Amst) 8 (2009) 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gottlieb TM, Jackson SP, The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen, Cell 72 (1993) 131–142. [DOI] [PubMed] [Google Scholar]
- [20].Ahnesorg P, Smith P, Jackson SP, XLF interacts with the XRCC4-DNA ligase IV complex to promote DNA nonhomologous end-joining, Cell 124 (2006) 301–313. [DOI] [PubMed] [Google Scholar]
- [21].Li Z, Otevrel T, Gao Y, Cheng HL, Seed B, Stamato TD, Taccioli GE, Alt FW, The XRCC4 gene encodes a novel protein involved in DNA double-strand break repair and V(D)J recombination, Cell 83 (1995) 1079–1089. [DOI] [PubMed] [Google Scholar]
- [22].Grawunder U, Wilm M, Wu X, Kulesza P, Wilson TE, Mann M, Lieber MR, Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells, Nature 388 (1997) 492–495. [DOI] [PubMed] [Google Scholar]
- [23].Ma Y, Pannicke U, Schwarz K, Lieber MR, Hairpin opening and overhang processing by an Artemis/DNA-dependent protein kinase complex in non-homologous end joining and V(D)J recombination, Cell 108 (2002) 781–794. [DOI] [PubMed] [Google Scholar]
- [24].Mani RS, Yu Y, Fang S, Lu M, Fanta M, Zolner AE, Tahbaz N, Ramsden DA, Litchfield DW, Lees-Miller SP, Weinfeld M, Dual modes of interaction between XRCC4 and polynucleotide kinase/phosphatase: implications for nonhomologous end joining, J. Biol. Chem 285 (2010) 37619–37629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Rulten SL, Fisher AE, Robert I, Zuma MC, Rouleau M, Ju L, Poirier G, Reina-San-Martin B, Caldecott KW, PARP-3 and APLF function together to accelerate nonhomologous end-joining, Mol. Cell 41 (2011) 33–45. [DOI] [PubMed] [Google Scholar]
- [26].Ma Y, Lu H, Tippin B, Goodman MF, Shimazaki N, Koiwai O, Hsieh CL, Schwarz K, Lieber MR, A biochemically defined system for mammalian non-homologous DNA end joining, Mol. Cell 16 (2004) 701–713. [DOI] [PubMed] [Google Scholar]
- [27].Yannone SM, Roy S, Chan DW, Murphy MB, Huang S, Campisi J, Chen DJ, Werner syndrome protein is regulated and phosphorylated by DNA-dependent protein kinase, J. Biol. Chem 276 (2001) 38242–38248. [DOI] [PubMed] [Google Scholar]
- [28].Sfeir A, Symington LS, Microhomology-mediated end joining: a back-up survival mechanism or dedicated pathway? Trends Biochem. Sci 40 (2015) 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Sinha S, Villarreal D, Shim EY, Lee SE, Risky business: microhomology-mediated end joining, Mutat. Res 788 (2016) 17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Wang H, Xu X, Microhomology-mediated end joining: new players join the team, Cell Biosci 7 (2017) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Black SJ, Kashkina E, Kent T, Pomerantz RT, DNA polymerase theta: a unique multifunctional end-joining machine, Genes (Basel) 7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chan SH, Yu AM, McVey M, Dual roles for DNA polymerase theta in alternative end-joining repair of double-strand breaks in Drosophila, PLoS Genet. 6 (2010) e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Mateos-Gomez PA, Gong F, Nair N, Miller KM, Lazzerini-Denchi E, Sfeir A, Mammalian polymerase theta promotes alternative NHEJ and suppresses recombination, Nature 518 (2015) 254–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Yousefzadeh MJ, Wyatt DW, Takata K, Mu Y, Hensley SC, Tomida J, Bylund GO, Doublie S, Johansson E, Ramsden DA, McBride KM, Wood RD, Mechanism of suppression of chromosomal instability by DNA polymerase POLQ, PLoS Genet. 10 (2014) e1004654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J, CtIP promotes microhomology-mediated alternative end joining during class-switch recombination, Nat. Struct. Mol. Biol 18 (2011) 75–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Ma JL, Kim EM, Haber JE, Lee SE, Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences, Mol. Cell. Biol 23 (2003) 8820–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Rahal EA, Henricksen LA, Li Y, Williams RS, Tainer JA, Dixon K, ATM regulates Mre11-dependent DNA end-degradation and microhomology-mediated end joining, ABBV Cell Cycle 9 (2010) 2866–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Truong LN, Li Y, Shi LZ, Hwang PY, He J, Wang H, Razavian N, Berns MW, Wu X, Microhomology-mediated End Joining and Homologous Recombination share the initial end resection step to repair DNA double-strand breaks in mammalian cells, Proc. Natl. Acad. Sci. U. S. A 110 (2013) 7720–7725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Sharma S, Javadekar SM, Pandey M, Srivastava M, Kumari R, Raghavan SC, Homology and enzymatic requirements of microhomology-dependent alternative end joining, Cell. Death. Dis 6 (2015) e1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Bhargava R, Onyango DO, Stark JM, Regulation of single-strand annealing and its role in genome maintenance, Trends Genet. 32 (2016) 566–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Takeda S, Nakamura K, Taniguchi Y, Paull TT, Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination, Mol. Cell 28 (2007) 351–352. [DOI] [PubMed] [Google Scholar]
- [42].Cannavo E, Cejka P, Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks, Nature 514 (2014) 122–125. [DOI] [PubMed] [Google Scholar]
- [43].Paull TT, Making the best of the loose ends: mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection, DNA Repair (Amst) 9 (2010) 1283–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gravel S, Chapman JR, Magill C, Jackson SP, DNAhelicases Sgsl and BLM promote DNA double-strand break resection, Genes Dev. 22 (2008) 2767–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhu Z, Chung WH, Shim EY, Lee SE, Ira G, Sgsl helicase and two nucleases Dna2 and Exol resect DNA double-strand break ends, Cell 134 (2008) 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Mimitou EP, Symington LS, Sae2, Exo1 and Sgs1 collaborate in DNA double-strand break processing, Nature 455 (2008) 770–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Deng SK, Gibb B, de Almeida MJ, Greene EC, Symington LS, RPA antagonizes microhomology-mediated repair of DNA double-strand breaks, Nat. Struct. Mol. Biol 21 (2014) 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kent T, Chandramouly G, McDevitt SM, Ozdemir AY, Pomerantz RT, Mechanism of microhomology-mediated end-joining promoted by human DNA polymerase theta, Nat. Struct. Mol. Biol 22 (2015) 230–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Villarreal DD, Lee K, Deem A, Shim EY, Malkova A, Lee SE, Microhomology directs diverse DNA break repair pathways and chromosomal translocations, PLoS Genet. 8 (2012) e1003026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ahrabi S, Sarkar S, Pfister SX, Pirovano G, Higgins GS, Porter AC, Humphrey TC, A role for human homologous recombination factors in suppressing microhomology-mediated end joining, Nucleic Acids Res. 44 (2016) 5743–5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Ahmad A, Robinson AR, Duensing A, van Drunen E, Beverloo HB, Weisberg DB, Hasty P, Hoeijmakers JH, Niedernhofer LJ, ERCC1-XPF endonuclease facilitates DNA double-strand break repair, Mol. Cell. Biol 28 (2008) 5082–5092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Arana ME, Seki M, Wood RD, Rogozin IB, Kunkel TA, Low-fidelity DNA synthesis by human DNA polymerase theta, Nucleic Acids Res. 36 (2008) 3847–3856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Hogg M, Sauer-Eriksson AE, Johansson E, Promiscuous DNA synthesis by human DNA polymerase theta, Nucleic Acids Res. 40 (2012) 2611–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Roerink SF, van Schendel R, Tijsterman M, Polymerase theta-mediated end joining of replication-associated DNA breaks in C. elegans, Genome Res. 24 (2014) 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yu AM, McVey M, Synthesis-dependent microhomology-mediated end joining accounts for multiple types of repair junctions, Nucleic Acids Res. 38 (2010) 5706–5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lee K, Lee SE, Saccharomyces cerevisiae Sae2- and Tel1-dependent single-strand DNA formation at DNA break promotes microhomology-mediated end joining, Genetics 176 (2007) 2003–2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Meyer D, Fu BX, Heyer WD, DNA polymerases delta and lambda cooperate in repairing double-strand breaks by microhomology-mediated end-joining in Saccharomyces cerevisiae, Proc. Natl. Acad. Sci. U. S. A 112 (2015) E6907–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang H, Rosidi B, Perrault R, Wang M, Zhang L, Windhofer F, Iliakis G, DNA ligase III as a candidate component of backup pathways of nonhomologous end joining, Cancer Res. 65 (2005) 4020–4030. [DOI] [PubMed] [Google Scholar]
- [59].Liang L, Deng L, Nguyen SC, Zhao X, Maulion CD, Shao C, Tischfield JA, Human DNA ligases I and III, but not ligase IV, are required for microhomology-mediated end joining of DNA double-strand breaks, Nucleic Acids Res. 36 (2008) 3297–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, Gregory PD, Holmes MC, Jasin M, DNA ligase III promotes alternative nonhomologous end-joining during chromosomal translocation formation, PLoS Genet. 7 (2011) e1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Biehs R, Steinlage M, Barton O, Juhasz S, Kunzel J, Spies J, Shibata A, Jeggo PA, Lobrich M, DNA double-strand Break resection occurs during non-homologous end joining in G1 but is distinct from resection during homologous recombination, Mol. Cell 65 (2017) 671–684 ( e675). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Guirouilh-Barbat J, Lambert S, Bertrand P, Lopez BS, Is homologous recombination really an error-free process? Front. Genet 5 (2014) 175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Malkova A, Ira G, Break-induced replication: functions and molecular mechanism, Curr. Opin. Genet. Dev 23 (2013) 271–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bahmed K, Seth A, Nitiss KC, Nitiss JL, End-processing during non-homologous end-joining: a role for exonuclease 1, Nucleic Acids Res. 39 (2011) 970–978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wilson TE, Lieber MR, Efficient processing of DNA ends during yeast non-homologous end joining. Evidence for a DNA polymerase beta (Pol4)-dependent pathway, J. Biol. Chem 274 (1999) 23599–23609. [DOI] [PubMed] [Google Scholar]
- [66].Daley JM, Laan RL, Suresh A, Wilson TE, DNA joint dependence of pol X family polymerase action in nonhomologous end joining, J. Biol. Chem 280 (2005) 29030–29037. [DOI] [PubMed] [Google Scholar]
- [67].Tseng SF, Gabriel A, Teng SC, Proofreading activity of DNA polymerase Pol2 mediates 3’-end processing during nonhomologous end joining in yeast, PLoS Genet. 4 (2008) e1000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Richardson C, Jasin M, Frequent chromosomal translocations induced by DNA double-strand breaks, Nature 405 (2000) 697–700. [DOI] [PubMed] [Google Scholar]
- [69].van Schendel R, van Heteren J, Welten R, Tijsterman M, Genomic scars generated by polymerase theta reveal the versatile mechanism of alternative end-joining, PLoS Genet. 12 (2016) e1006368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kent T, Mateos-Gomez PA, Sfeir A, Pomerantz RT, Polymerase theta is a robust terminal transferase that oscillates between three different mechanisms during end-joining, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Ceccaldi R, Liu JC, Amunugama R, Hajdu I, Primack B, Petalcorin MI, O’Connor KW, Konstantinopoulos PA, Elledge SJ, Boulton SJ, Yusufzai T, D’Andrea AD, Homologous-recombination-deficient tumours are dependent on Poltheta-mediated repair, Nature 518 (2015) 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Chiarle R, Zhang Y, Frock RL, Lewis SM, Molinie B, Ho YJ, Myers DR, Choi VW, Compagno M, Malkin DJ, Neuberg D, Monti S, Giallourakis CC, Gostissa M, Alt FW, Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells, Cell 147 (2011) 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M, Chromosomal translocations induced at specified loci in human stem cells, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 10620–10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Ghezraoui H, Piganeau M, Renouf B, Renaud JB, Sallmyr A, Ruis B, Oh S, Tomkinson AE, Hendrickson EA, Giovannangeli C, Jasin M, Brunet E, Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining, Mol. Cell 55 (2014) 829–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Simsek D, Jasin M, Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4-ligase IV during chromosomal translocation formation, Nat. Struct. Mol. Biol 17 (2010) 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Zhang Y, Jasin M, An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway, Nat. Struct. Mol. Biol 18 (2011) 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Garner KM, Eastman A, Variations in Mre11/Rad50/Nbs1 status and DNA damage-induced S-phase arrest in the cell lines of the NCI60 panel, BMC Cancer 11 (206) (2011) 201–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bhargava R, Carson CR, Lee G, Stark JM, Contribution of canonical non-homologous end joining to chromosomal rearrangements is enhanced by ATM kinase deficiency, Proc. Natl. Acad. Sci. U. S. A 114 (2017) 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Sinha S, Li F, Villarreal D, Shim JH, Yoon S, Myung K, Shim EY, Lee SE, Microhomology-mediated end joining induces hypermutagenesis at breakpoint junctions, PLoS Genet. 13 (2017) e1006714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Roberts SA, Sterling J, Thompson C, Harris S, Mav D, Shah R, Klimczak LJ, Kryukov GV, Malc E, Mieczkowski PA, Resnick MA, Gordenin DA, Clustered mutations in yeast and in human cancers can arise from damaged long single-strand DNA regions, Mol. Cell 46 (2012) 424–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hastings PJ, Ira G, Lupski JR, A microhomology-mediated break-induced replication model for the origin of human copy number variation, PLoS Genet. 5 (2009) e1000327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Ottaviani D, LeCain M, Sheer D, The role of microhomology in genomic structural variation, Trends Genet. 30 (2014) 85–94. [DOI] [PubMed] [Google Scholar]
- [83].Lawson AR, Hindley GF, Forshew T, Tatevossian RG, Jamie GA, Kelly GP, Neale GA, Ma J, Jones TA, Ellison DW, Sheer D, RAF gene fusion breakpoints in pediatric brain tumors are characterized by significant enrichment of sequence microhomology, Genome Res. 21 (2011) 505–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Francis NJ, McNicholas B, Awan A, Waldron M, Reddan D, Sadlier D, Kavanagh D, Strain L, Marchbank KJ, Harris CL, Goodship TH, A novel hybrid CFH/CFHR3 gene generated by a microhomology-mediated deletion in familial atypical hemolytic uremic syndrome, Blood 119 (2012) 591–601. [DOI] [PubMed] [Google Scholar]
- [85].Weier C, Haffner MC, Mosbruger T, Esopi DM, Hicks J, Zheng Q, Fedor H, Isaacs WB, De Marzo AM, Nelson WG, Yegnasubramanian S, Nucleotide resolution analysis of TMPRSS2 and ERG rearrangements in prostate cancer, J. Pathol 230 (2013) 174–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Yan CT, Boboila C, Souza EK, Franco S, Hickernell TR, Murphy M, Gumaste S, Geyer M, Zarrin AA, Manis JP, Rajewsky K, Alt FW, IgH class switching and translocations use a robust non-classical end-joining pathway, Nature 449 (2007) 478–482. [DOI] [PubMed] [Google Scholar]
- [87].Boboila C, Yan C, Wesemann DR, Jankovic M, Wang JH, Manis J, Nussenzweig A, Nussenzweig M, Alt FW, Alternative end-joining catalyzes class switch recombination in the absence of both Ku70 and DNA ligase 4, J. Exp. Med 207 (2010) 417–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Han L, Yu K, Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV-deficient B cells, J. Exp. Med 205 (2008) 2745–2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Alt FW, Zhang Y, Meng FL, Guo C, Schwer B, Mechanisms of programmed DNA lesions and genomic instability in the immune system, Cell 152 (2013) 417–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Boboila C, Jankovic M, Yan CT, Wang JH, Wesemann DR, Zhang T, Fazeli A, Feldman L, Nussenzweig A, Nussenzweig M, Alt FW, Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 3034–3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Diotti R, Loayza D, Shelterin complex and associated factors at human telomeres, Nucleus 2 (2011) 119–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Rai R, Zheng H, He H, Luo Y, Multani A, Carpenter PB, Chang S, The function of classical and alternative non-homologous end-joining pathways in the fusion of dysfunctional telomeres, EMBO J. 29 (2010) 2598–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Capper R, Britt-Compton B, Tankimanova M, Rowson J, Letsolo B, Man S, Haughton M, Baird DM, The nature of telomere fusion and a definition of the critical telomere length in human cells, Genes Dev. 21 (2007) 2495–2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Jones RE, Oh S, Grimstead JW, Zimbric J, Roger L, Heppel NH, Ashelford KE, Liddiard K, Hendrickson EA, Baird DM, Escape from telomere-driven crisis is DNA ligase III dependent, Cell Rep. 8 (2014) 1063–1076. [DOI] [PubMed] [Google Scholar]
- [95].Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, Martin NM, Jackson SP, Smith GC, Ashworth A, Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy, Nature 434 (2005) 917–921. [DOI] [PubMed] [Google Scholar]
- [96].Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O’Connor MJ, Ashworth A, Carmichael J, Kaye SB, Schellens JH, de Bono JS, Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers, N. Engl. J. Med 361 (2009) 123–134. [DOI] [PubMed] [Google Scholar]
- [97].Konstantinopoulos PA, Ceccaldi R, Shapiro GI, D’Andrea AD, Homologous recombination deficiency: exploiting the fundamental vulnerability of ovarian cancer, Cancer Discov. 5 (2015) 1137–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Lord CJ, Ashworth A, Mechanisms of resistance to therapies targeting BRCA-mutant cancers, Nat. Med 19 (2013) 1381–1388. [DOI] [PubMed] [Google Scholar]
- [99].Gaymes TJ, Shall S, MacPherson LJ, Twine NA, Lea NC, Farzaneh F, Mufti GJ, Inhibitors of poly ADP-ribose polymerase (PARP) induce apoptosis of myeloid leukemic cells: potential for therapy of myeloid leukemia and myelodysplastic syndromes, Haematologica 94 (2009) 638–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Zhang JX, Li DQ, He AR, Motwani M, Vasiliou V, Eswaran J, Mishra L, Kumar R, Synergistic inhibition of hepatocellular carcinoma growth by co-targeting chromatin modifying enzymes and poly (ADP-ribose) polymerases, Hepatology 55 (2012) 1840–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Chao OS, Goodman OB Jr., Synergistic loss of prostate cancer cell viability by coinhibition of HDAC and PARP, Mol. Cancer Res 12 (2014) 1755–1766. [DOI] [PubMed] [Google Scholar]
- [102].Ha K, Fiskus W, Choi DS, Bhaskara S, Cerchietti L, Devaraj SG, Shah B, Sharma S, Chang JC, Melnick AM, Hiebert S, Bhalla KN, Histone deacetylase inhibitor treatment induces ‘BRCAness’ and synergistic lethality with PARP inhibitor and cisplatin against human triple negative breast cancer cells, Oncotarget 5 (2014) 5637–5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Konstantinopoulos PA, Wilson AJ, Saskowski J, Wass E, Khabele D, Suberoylanilide hydroxamic acid (SAHA) enhances olaparib activity by targeting homologous recombination DNA repair in ovarian cancer, Gynecol. Oncol 133 (2014) 599–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Kachhap SK, Rosmus N, Collis SJ, Kortenhorst MS, Wissing MD, Hedayati M, Shabbeer S, Mendonca J, Deangelis J, Marchionni L, Lin J, Hoti N, Nortier JW, DeWeese TL, Hammers H, Carducci MA, Downregulation of homologous recombination DNA repair genes by HDAC inhibition in prostate cancer is mediated through the E2F1 transcription factor, PLoS One 5 (2010) e11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Adimoolam S, Sirisawad M, Chen J, Thiemann P, Ford JM, Buggy JJ, HDAC inhibitor PCI-24781 decreases RAD51 expression and inhibits homologous recombination, Proc. Natl. Acad. Sci. U. S. A 104 (2007) 19482–19487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Sallmyr A, Tomkinson AE, Rassool FV, Up-regulation of WRN and DNA ligase IIIalpha in chronic myeloid leukemia: consequences for the repair of DNA double-strand breaks, Blood 112 (2008) 1413–1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Bentley J, Diggle CP, Harnden P, Knowles MA, Kiltie AE, DNA double strand break repair in human bladder cancer is error prone and involves microhomology-associated end-joining, Nucleic Acids Res. 32 (2004) 5249–5259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Henner WD, Rodriguez LO, Hecht SM, Haseltine WA, gamma Ray induced deoxyribonucleic acid strand breaks. 3’ Glycolate termini, J. Biol. Chem 258 (1983) 711–713. [PubMed] [Google Scholar]
- [109].Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN, Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair, Trends Biochem. Sci 36 (2011) 262–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Dutta A, Eckelmann B, Adhikari S, Ahmed KM, Sengupta S, Pandey A, Hegde PM, Tsai MS, Tainer JA, Weinfeld M, Hegde ML, Mitra S, Microhomology-mediated end joining is activated in irradiated human cells due to phosphorylation-dependent formation of the XRCC1 repair complex, Nucleic Acids Res. 45 (2017) 2585–2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Tadi SK, Sebastian R, Dahal S, Babu RK, Choudhary B, Raghavan SC, Microhomology-mediated end joining is the principal mediator of double-strand break repair during mitochondrial DNA lesions, Mol. Biol. Cell 27 (2016) 223–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Schon EA, Rizzuto R, Moraes CT, Nakase H, Zeviani M, DiMauro S, A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA, Science 244 (1989) 346–349. [DOI] [PubMed] [Google Scholar]
- [113].Samuels DC, Schon EA, Chinnery PF, Two direct repeats cause most human mtDNA deletions, Trends Genet. 20 (2004) 393–398. [DOI] [PubMed] [Google Scholar]
- [114].Chen T, He J, Huang Y, Zhao W, The generation of mitochondrial DNA large-scale deletions in human cells, J. Hum. Genet 56 (2011) 689–694. [DOI] [PubMed] [Google Scholar]
- [115].Yun MH, Hiom K, CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle, Nature 459 (2009) 460–463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Mao Z, Hine C, Tian X, Van Meter M, Au M, Vaidya A, Seluanov A, Gorbunova V, SIRT6 promotes DNA repair under stress by activating PARP1, Science 332 (2011) 1443–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Xiong X, Du Z, Wang Y, Feng Z, Fan P, Yan C, Willers H, Zhang J, 53BP1 promotes microhomology-mediated end-joining in G1-phase cells, Nucleic Acids Res. 43 (2015) 1659–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Ledermann JA, PARP inhibitors in ovarian cancer, Ann. Oncol 27 (Suppl. 1) (2016) i40–i44. [DOI] [PubMed] [Google Scholar]
- [119].Ledermann JA, Drew Y, Kristeleit RS, Homologous recombination deficiency and ovarian cancer, Eur. J. Cancer 60 (2016) 49–58. [DOI] [PubMed] [Google Scholar]
- [120].Liu FW, Tewari KS, New targeted agents in gynecologic cancers: synthetic lethality, homologous recombination deficiency, and PARP inhibitors, Curr. Treat. Options Oncol 17 (2016) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Yousefzadeh MJ, Wood RD, DNA polymerase POLQ and cellular defense against DNA damage, DNA Repair (Amst) 12 (2013) 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]