Abstract
Arcobacter (A.) butzleri is an emerging pathogenic microorganism, whose taxonomy has been recently suggested to be emended to the Aliarcobacter (Al.) butzleri comb. nov. Despite extensive taxonomic analysis, only few fragmented studies have investigated the occurrence and the prevalence of virulence and antibiotic resistance determinants of this species in strains isolated from shellfish. Herein we report for the first time the whole genome sequencing and genomic characterization of two A. butzleri strains isolated from shellfish, with particular reference to the antibiotic, heavy metals and virulence determinants. This study supported the taxonomic assignment of these strains to the Al. butzleri species, and allowed us to identify antibiotic and metal resistance along with virulence determinants, also additional to those previously reported for the only two A. butzleri strains from different environments genomically characterized. Moreover, both strains showed resistance to β-lactams, vanocomycin, tetracycline and erythromycin and susceptibility to aminoglycosides and ciprofloxacin. Beside enlarging the availability of genomic data to perform comparative studies aimed at correlating phenotypic differences associated with ecological niche and geographic distribution with the genetic diversity of A. butzleri spp., this study reports the endowment of antibiotic and heavy metal resistance and virulence determinants of these shellfish-isolated strains. This leads to hypothesize a relatively high virulence of A. butzleri isolated from shellfish and prompt the need for a wider genomic analysis and for in vitro and in vivo studies of more strains isolated from this and other ecological niches, to unravel the mechanism of pathogenicity of this species, and the potential risk associated to their consumption.
Keywords: Aliarcobacter butzleri, Arcobacter butzleri, antibiotic and heavy metal resistance, virulence genes, genomics, food safety, emerging foodborne pathogen, shellfish
Introduction
The current validated taxonomy places the Arcobacter genus within the Campylobacteraceae family (belonging to the class Epsilonproteobacteria of the phylum Proteobacteria) together with Campylobacter and Sulfurospirillum genera. Recently, based on a wide comparative genomic analysis, Waite et al. (2017) proposed the re-classification of the class Epsilonproteobacteria with the Arcobacter genus ascribed to the Arcobacteraceae fam. nov. (type genus: Arcobacter, order: Campylobacterales, class: Campylobacteria, class. nov., phylum: Campylobacteraeota phyl. nov.) (Waite et al., 2017, 2018).
Arcobacter spp. have a wide diversity of hosts and habitats, with water being one of the main routes of transmission (Collado and Figueras, 2011). Arcobacter spp. have been indeed detected in environmental water sources, including rivers, lakes, sewage and plankton (Kim et al., 2010; Levican et al., 2013; Park et al., 2016), marine, domestic (Merga et al., 2014), and drinking water (Jalava et al., 2014; Talay et al., 2016), water distribution pipes (Phillips, 2001), groundwater (Fong et al., 2007), and recreational water (Lee et al., 2012). Four species (namely, A. butzleri, A. cryaerophilus, A. thereius, and A. skirrowii) have also been isolated in humans and animals (Banting and Figueras Salvat, 2017) and are able to cause human bacteraemia, endocarditis, peritonitis, gastroenteritis, and diarrhea (Jiang et al., 2010; Figueras et al., 2014; Arguello et al., 2015; Ferreira et al., 2016).
To date this genus includes 26 species (Ramees et al., 2017; Pérez-Cataluña et al., 2018a), which inhabit various ecological niches (Collado and Figueras, 2011; Ferreira et al., 2016). Recently, Pérez-Cataluña et al. (2018a) used a polyphasic approach to revisit the taxonomy of the genus. By setting specific cut-off values for each method (identity of 16S rRNA, genomic indexes such as ANI, AAI, and DDH, multilocus sequence analysis, etc.), they delimited genomic and phylogenetic groups and defined six different genera including Aliarcobacter gen. nov. This genus comprises seven species including also Aliarcobacter (Al.) butzleri comb. nov., whose type strain was confirmed to be LMG 10828T (ATCC 49616T; RM4018; Miller et al., 2007), and the species description as the one given by Vandamme et al. (1992) and Pérez-Cataluña et al. (2018a,b).
However, it must be considered that both the species description of A. butzleri and Al. butzleri comb. nov., were obtained by analyzing an exiguous number of A. butzleri strains (12 by Vandamme et al., 1992 and only two by Pérez-Cataluña et al., 2018a, respectively). Thus, it might be expected that the likely increasing availability of A. butzleri genomic sequences could lead this species to a further redefinition.
Recent outcomes and progress related to the pathogenicity of A. butzleri have led to the allocation of this species in the list of microbes considered a serious hazard to human health by the International Commission on Microbiological Specifications for Foods (ICMSF, 2002). A. butzleri is considered an emerging food-borne enteropathogen and, in recent years, has been associated with enteritis, severe diarrhea, bacteraemia, and septicaemia in humans and enteritis, mastitis, abortion, and stillbirth in cows, sheep, and pigs (Engberg et al., 2000; Lau et al., 2002; Collado and Figueras, 2011; Collado et al., 2014; Ferreira et al., 2014a,b, 2016; Figueras et al., 2014; Van den Abeele et al., 2014; Heimesaat et al., 2015; Gölz et al., 2016; Webb et al., 2016; Franz et al., 2018; Fusco et al., 2018; Flynn et al., 2019). Enteritis due to the ingestion of food contaminated with Arcobacter spp. can be self-limiting. Nevertheless, the severity and protraction of the symptoms might require an antibiotic treatment, which might be affected by the (multiple) antibiotic resistance of the strain, thus complicating the treat of the relevant infections. A. butzleri is the most prevalent species of this genus in meat products (chicken, pork, beef, lamb), milk, cheese, and shellfish (Figueras et al., 2011a,b; Patyal et al., 2011; Shah et al., 2012; Hausdorf et al., 2013; Lee and Choi, 2013; Rahimi, 2014; Ramees et al., 2014; Lehmann et al., 2015; Ferreira et al., 2016). The presence and persistence in these niches, also endowed by the ability to form biofilms (Ferreira et al., 2013; Girbau et al., 2017), favor its spread and transmission to shellfish and farm animal, and increase the risk associated with food consumption. Although few fragmented studies have been carried out to assess the occurrence of this species in shellfish, A. butzleri has been found as the most common species in bivalve molluscs (mussels, clams, oysters, etc.) (Nieva-Echevarria et al., 2013; Levican et al., 2014; Mottola et al., 2016; Leoni et al., 2017). This is most likely due to capture by the filter feeding process of bivalves and to a fecal contamination of the relevant environment, so that Escherichia coli has been proposed by Leoni et al. (2017) as an index organism for A. butzleri contamination in bivalve molluscs, leading to suggest that these shellfishes could be a reservoir of A. butzleri (Leoni et al., 2017). For this reason, consumption of contaminated shellfish, especially if raw or undercooked, which is still a widespread practise (Schauer Weissfeld, 2014), may be source of A. butzleri infections in humans. The ability to survive in different environments, the endowment of antibiotic resistance genes and virulence potential found by genomic approaches (Miller et al., 2007; Pérez-Cataluña et al., 2018a), as well as the genetic plasticity conferred by the presence of mobile elements, which allow the transfer of genes (Douidah et al., 2014), are important determinants for the evolution and the fitness of this and the other species of this genus.
As far as we know, extensive genome-based characterization of the species has been carried out only on two A. butzleri strains, namely, RM4018 (Culture collection n. LMG 10828T), isolated from human feces, and ED-1 isolated from microbial fuel cells (Miller et al., 2007; Pérez-Cataluña et al., 2018a). Considering that contaminated shellfish may be source of A. butzleri infection and given that the prevalence and expression of putative virulence and antibiotic resistance genes within this species may vary with the source of the strain (Douidah et al., 2012; Ferreira et al., 2014b; Girbau et al., 2015; Zacharow et al., 2015a), herein we report the antibiotic susceptibility and genomic-based characterization of two A. butzleri strains isolated from shellfish, with particular reference to the genetic determinants of the above mentioned traits of pathogenicity.
Materials and Methods
Strains and Culture Condition
Arcobacter butzleri (Ab) strains 6V and 55 were isolated on 2016 from clams (Tapes philippinarum) (Mottola et al., 2016) and mussels (Mytilus galloprovincialis) obtained from local fish market in the Apulia region (Italy). These strains were previously identified and typed by MLST (Mottola et al., 2016; Mottola, 2017). Allelic profiles and sequences are available on the Arcobacter MLST database 1 under the ID numbers 717 (Ab 6V) and 839 (Ab 55).
Pure cultures were isolated and maintained in the microbial collection of the Institute of Sciences of Food Production, CNR, Bari 2. Bacterial strains were maintained at -80°C as pure stock cultures in Brain Heart Infusion (BHI) broth (Oxoid S.p.A., Rodano, Milan, Italy) supplemented with glycerol (30% vol/vol). Cultures were streaked on Agar blood plates (Oxoid, Milan, Italy) and grown at 37°C for 48 h. Working cultures were obtained growing a single colony in 20 mL of BHI broth with 0.6% yeast extract (BHI-YE), at 37°C for 48 h.
Genome Sequencing and Assembly
DNA isolation was performed by using the Wizard® Genomic DNA Purification Kit (Promega), as previously described by Ercolini et al. (2005). The integrity, purity, and quantity of DNA were assessed as previously described by Fusco et al. (2011), by agarose gel electrophoresis, by NanoDrop-2000 (Thermo Fisher Scientific, Wilmington, DE, United States), and by Qubit 3.0 fluorometer (Life Technologies). DNA was then subjected to whole genome shotgun sequencing using the Ion S5TM library preparation workflow (Thermo Fisher Scientific, Waltman, MA, United States); 400 bp mate-paired reads were generated on the Ion S5TM System (Thermo Fisher Scientific). Duplicate reads were removed by FilterDuplicates (v5.0.0.0) Ionplugin. De novo assembly was performed by AssemblerSpades (v.5.0) IonpluginTM.
Bioinformatic Methods
Genes were predicted and annotated using PROKKA pipeline implemented in the Galaxy platform (Galaxy Tool Version 1.0.0; Afgan et al., 2016). The predicted proteins were submitted to the PFAM annotator tool within the Galaxy platform in order to predict the pfam domains. Protein ID used in the manuscript indicated those obtained by NCBI (National Center for Biotechnology Information) Prokaryotic Genome Annotation Pipeline (Tatusova et al., 2016).
Predicted proteins were assigned to Clusters of Orthologous Groups (COG) functional categories by Web CD-Search Tool (Marchler-Bauer et al., 2017) using an Expected value threshold of 0.01. COG ID were then manually mapped into functional categories3.
All the proteins sequences used in this study were retrieved from GenBank (NCBI). The homology-based relationship of Ab 55 and Ab 6V predicted proteins toward selected proteins was determined by BLASTP algorithm on the NCBI site4. Gene models were manually determined, and clustering and orientation were subsequently deduced for the closely linked genes.
Antibiotic resistance genes were predicted by BLASTP search against the Antibiotic Resistance genes Database (ARDB; Liu and Pop, 2009) and beta lactamase database (Naas et al., 2017). Genes associated with antibiotic resistance were also retrieved by keywords terms search within UniProtID entry list obtained by functional annotation.
Functional annotation, subsystem prediction, and metabolic reconstruction comparison were also performed using the RAST server (Aziz et al., 2008). Genes involved in the mechanism of resistance to heavy metals were retrieved by Poole (2017) and used as queries for BLASTP search against Ab 55 and Ab 6V proteomes.
Genetic divergence was calculated by the ANI/AAI calculator (Goris et al., 2007; Rodriguez-R and Konstantinidis, 2016) which estimates the average nucleotide/aminoacid identity (ANI/AAI) using both best hits (one-way ANI) and reciprocal best hits (two-way ANI) between genomic datasets. The Genome-to-Genome Distance Calculator (GGDC) (Meier-Kolthoff et al., 2013, 2014) web service was used to report DDH for the accurate delineation of prokaryotic subspecies and to calculate differences in G+C genomic content (available at ggdc.dsmz.de). Formula 2 alone was used for analysis, providing an estimation of DDH independent from genome lengths, as recommended by the authors of GGDC for use with any incomplete genomes (Auch et al., 2010; Meier-Kolthoff et al., 2013).
Antimicrobial Susceptibility Testing
The antimicrobial susceptibility tests for A. butzleri isolates were performed by disk diffusion and broth microdilution methods. The disk diffusion test was performed as described by Rathlavath et al. (2017) with modifications. Briefly, A. butzleri isolates were grown in 20 ml of BHI broth (Oxoid, United Kingdom) amended with 0.6% yeast extract (YE) (Biolife srl, Italy) under static condition for 48 h at 37°C and then subcultured at 1% in BHI-YE broth and incubated at 37°C for 48 h. Microbial cells were recovered after centrifugation (16,000 rcf × 6 min), washed in sterile 0.9% NaCl solution, adjusting optical density (600 nm) to 0.5. One hundred microliters of this cell suspension were then plated on 4 mm thick cation adjusted Muller Hinton agar (MHIIA, Liofilchem, Italy).
Antibiotic disks, soaked with ampicillin (10 μg/disk), cefotaxime (30 μg/disk), chloramphenicol (30 μg/disk), ciprofloxacin (5 μg/disk), erythromycin (15 μg/disk), gentamicin (10 μg/disk), kanamycin (30 μg/disk), nalidixic acid (30 μg/disk), streptomycin (10 μg/disk), tetracycline (30 μg/disk), vancomycin (30 μg/disk), and penicillin G (10 units/disk) (Biolab Zrt., Hungary), were placed onto inoculated plates and incubated at 37°C under microaerophilic atmosphere (CampyGenTM Compact, Oxoid, United Kingdom), as recommended by BCCM/LMG Bacteria Collection5 for A. butzleri type strain LMG10828 (RM4018). After 48 h of incubation, inhibition zone diameters were recorded.
For those antibiotics which the tested A. butzleri strains did not provide inhibition zone at all, the minimal inhibitory concentration (MIC) was calculated by broth microdilution method as described by Riesenberg et al. (2017). After 48 h incubation at 37°C, microtitre plates were read spectrophotometrically at 600 nm using Varioskan Flash (Thermo Fisher Scientific, United States). To determine minimal bactericidal concentration (MBC), 10 μl of broth from three replicate of all wells without microbial growth was combined in a single sample spotted on MHIIA (Liofilchem, Italy) and incubated as described above up to 72 h. Since no breakpoint values are available for Arcobacter spp., classification of strains as susceptible (S), resistant (R), or intermediate (I) was defined according to zone diameter and MIC interpretive standards for Staphylococcus spp. (erythromycin, penicillin, and vancomycin) and Enterobacteriaceae (ampicillin, gentamicin, cefotaxime, ciprofloxacin, tetracycline, chloramphenicol, nalidixic acid, kanamycin, and streptomycin) (CLSI, 2015), as also reported by Elmali and Can (2017).
The A. butzleri LMG 10828T (RM4018) strain was used for comparison purposes.
Results and Discussion
General Features of A. butzleri 6V and 55 Genomes
Ab 55 and Ab 6V genomes were sequenced using a whole genome shotgun approach on an Ion S5TM platform (Thermo Fisher Scientific) generating around 671,363 and 596,333 reads with a median length of 317 and 320 bp, respectively (Table 1). Genomes were assembled using the Spades v5.0 software for a total of 32 and 46 large contigs (>500 bp) and a GC% of 26.79 and 26.85, respectively. The overall contiguity of the assembly is good, with a N50 of 211 and 129 kbp for Ab 55 and Ab 6V, respectively; the longest assembled fragment is 403 kbp in length for Ab 55 and 251 kbp for Ab 6V (performed by QUAST, available at http://quast.sourceforge.net/quast) while the total length of the assembly was around of 2.3 Mb for both genomes. These Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank under the accessions QXMK00000000 (Ab 55) and QXNB0000000 (Ab 6V). The versions described in this paper are QXMK01000000 (Ab 55) and QXNB01000000 (Ab 6V).
Table 1.
Summary of A. butzleri 55 and 6V genome sequencing and assembly results.
| A. butzleri 55 | A. butzleri 6V | |
|---|---|---|
| Total sequenced bases | 212,850,706 | 190,979,223 |
| Mean read length | 317 | 320 |
| Total length | 2,330,339 | 2,303,554 |
| Number of scaffolds | 47 | 61 |
| Largest contig | 403,569 | 251,748 |
| Number reads | 671,363 | 596,833 |
| N50 reads | 211,847 | 129,283 |
| Genome size | 2,325,213 | 2,297,763 |
| GC content | 26.79 | 26.85 |
| Predicted genes | 2395 | 2338 |
| CDS | 2344 | 2289 |
| tRNA | 46 | 44 |
| ncRNA | 2 | 2 |
| rRNA | 1, 1, 1 (5S, 16S, 23S) | 1, 1, 1 (5S, 16S, 23S) |
Genomic Analysis
The in silico MLST of the housekeeping genes retrieved from genomic sequences, confirmed in vitro results achieved by Mottola (2017), revealing that Ab 55 and Ab 6V belong to two novel sequence types, namely, ST675 and ST537, respectively, as they harbor 6/7 and 3/7 new alleles, respectively (Table 2).
Table 2.
Allelic profiles of A. butzleri isolates.
| MLST |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ID | Isolate | Species | Source | aspA | atpA | glnA | gltA | glyA | pgm | tkt | ST |
| 717 | 6V | Arcobacter butzleri | clam | 236 | 161 | 1 | 183 | 521 | 296 | 207 | 537 |
| 839 | 55 | Arcobacter butzleri | mussel | 332 | 8 | 1 | 166 | 685 | 367 | 292 | 675 |
Novel alleles and novel sequence type (ST) are indicated in bold.
Both Ab 6V and Ab 55 16S rRNA gene sequences show 100% identity with the type strain Ab RM4018 (Miller et al., 2007). ANI, AAI, and DDH analyses were performed with 22 strains within the Arcobacter group (Supplementary Table S1). Campylobacter jejuni subsp. jejuni NCTC 11168 and Helicobacter pylori 26695 were included as outgroups.
Ab 55 and Ab 6V have 98.06% nucleotide identity (Supplementary Table S2) and are comprised in the clustering including all the A. butzleri species. According to ANI, the closest relatives for both Ab 55 and Ab 6V are Ab NCTC 12481 (97.80% and 97.81 ANI, respectively) and Ab RM4018 (97.79% and 97.80 ANI). The same clustering is obtained by using AAI (Supplementary Table S3) with 97.60% between Ab 55 and Ab 6V, and 97.67 and 97.36% with Ab RM4018, respectively. DDH analysis confirmed the clustering obtained by ANI and AAI analysis, with values of 83.50 between Ab 6V and Ab 55, 81.00% between Ab 6V and Ab RM4018, and 81.30% between Ab 55 and Ab NCTC 12481 (Supplementary Table S4). As proposed by Chun et al. (2018) and, more specifically for Arcobacter spp., by On et al. (2017), these ANI and DDH values are within the range suggested to include Ab 55 and Ab 6V into the A. butzleri species. Moreover, our results support those achieved by Pérez-Cataluña et al. (2018a). Therefore, Ab 55 and Ab 6V should be placed within the Aliarcobacter gen. nov. as Al. butzleri comb. nov. (Pérez-Cataluña et al., 2018a,b), while for the definition of subspecies, a phenotypic comparison should support the in silico analyses (On et al., 2017).
Protein Functional Classification
For Ab 55, 1173 UniProtKB AC/ID identifiers retrieved by PFAM annotator tools (Galaxy Tool Version 1.0.0) were successfully mapped to 1190 UniProtKB IDs (The UniProt Consortium, 2017). In Ab 55, the retrieved list included 33 genes associated with antibiotic resistance, including beta-lactamase, multidrug efflux pump, DNA gyrase, and resistance protein, and three with antibiotic biosynthesis related to bacteriocin, six associated with drug transmembrane transporter activity, 19 with virulence, four with hemolysis, and two with quorum sensing (luxS and tqsA).
For Ab 6V, 1189 out of 1189 UniProtKB AC/ID identifiers were mapped to 1207 UniProtKB. Based on their classification, we counted three genes associated with antibiotic biosynthesis, 32 related to antibiotic resistance, including beta-lactamase, multidrug efflux pump, DNA gyrase and resistance protein, six referred to drug transmembrane transporter activity, 22 associated with virulence, three with hemolysis, and two with quorum sensing (luxS and tqsA).
One of the few differences between these strains is the presence in Ab 6V of the hemolysin tylC gene which codes for a protein with a conserved protein/cyclin M (CNNM) transmembrane domain, while, in Ab 55, we found only a precursor of hemolysin C. In both genomes, we identified the RNA methyltransferase hemolysin A (tylA), which encodes for a 16S/23S rRNA (cytidine-2′-O)-methyltransferase that is considered a virulence factor in H. pylori infection and probably acts as a pore-forming toxin (Javadi and Katzenmeier, 2016).
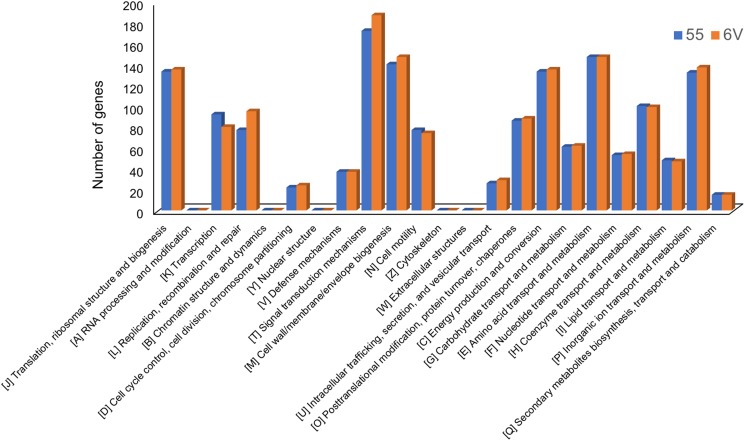
Overall, 1551 for Ab 55 and 1592 for Ab 6V predicted genes were assigned to the COG classification (Figure 1). The few differences in the distribution of genes into clusters of COG functional categories across Ab 55 and Ab 6V genomes, which emerged from Figure 1, supported the limited functional variability among these two strains, even though isolated from different samples and in different harvest seasons (Mottola et al., 2016).
FIGURE 1.
COGs functional classification of genes present in A. butzleri 55 and A. butzleri 6V genomes.
Among COG categories, the cluster signal transduction mechanisms represented the largest group for both organism (172 genes, 11.09% for Ab 55 and 187 genes, 11.75% for Ab 6V), followed by amino acid transport and metabolism cluster (147 genes, 9.48% for Ab 55 and 147 genes, 9.23% for Ab 6V), Cell wall/membrane/envelope biogenesis cluster (140 genes, 9.03% for Ab 55 and 147 genes, 9.23% for Ab 6V) and energy production and conversion cluster (133 genes, 8.58% for Ab 55 and 135 genes, 8.48% for Ab 6V). These findings suggest that our Arcobacter strains have a versatile sensory transduction system and that for energy and carbon mainly rely on amino acid catabolism rather than on sugar fermentation, which is consistent with the ecological niches they have been isolated from.
Virulence Determinants
The ability to adhere to various surfaces as well as chemotaxis, motility, and signal transduction play a pivotal role in the microbial survival and colonization of diverse ecological niches and can be involved in pathogenesis and antibiotic resistance (Richards et al., 2013; Chaban et al., 2015; Tiwari et al., 2017; Matilla and Krell, 2018).
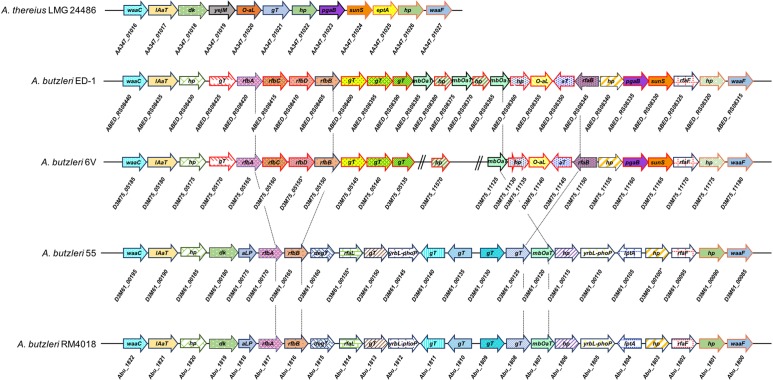
In both genomes, we identified orthologues of waaC and waaF genes, which are described as virulence determinant in A. thereius (Rovetto et al., 2017), but also in Pseudomonas aeruginosa, E. coli, Klebsiella pneumonia, and other Campylobacteraceae (Oldfield et al., 2002; DeLucia et al., 2011; Nilsson et al., 2018). waaF codes for a predicted ADP-heptose–LPS heptosyltransferase (Ab 55 D3M61_00085; Ab 6V D3M75_11180), involved in the biosynthesis of lipooligosaccharide (LOS); waaC codes for a lipopolysaccharide heptosyltransferase (Ab 55 D3M61_00195; Ab 6V D3M75_05185), which catalyzes the transfer of sugar moieties from activated donor molecules to specific acceptor molecules, forming glycosidic bonds. As in Ab RM4018 (Miller et al., 2007), which it shares the same content and organization with, in Ab 55, waaC and waaF genes are closely located suggesting the presence of a genetic cluster, whose composition is, however, different from A. thereius (Figure 2). In Ab 6V, the genetic locus comprised by waaC and waaF is similar to that of Ab ED-1 (26 genes) and contains several glycosyltransferases with no orthologues in Ab 55 and Ab RM4018. The outer genes of this locus are similar to A. thereius. However, in Ab 6V, these genes are located in three different contigs, which made only provisional, although likely, the reconstruction of the organization of the entire locus.
FIGURE 2.
Genomic organization of waaC/waaF gene cluster in A. thereius LMG 24486, A. butzleri ED-1, A. butlzeri 6V, A. butzleri 55, and A. butzleri RM4018. Gene clustering is represented by the arrows superposed on the black horizontal line. Intergenic spaces are not drawn in scale. For A. thereius LMG 24486, A. butzleri ED-1, and A. butzleri RM4018, the locus tag of each gene is indicated below the respective gene arrow; for A. butlzeri 6V and A. butzleri 55, protein ID is indicated below the respective gene arrow. Red arrows in A. butlzeri ED-1 and A. butzleri 6V indicate genes with no orthologue in A. butzleri 55 and A. butlzeri RM4018. D3M75_05155 ∗ indicates pseudogene (frameshifted). lAaT: lipid A biosynthesis acylTransferase; dK: diacylglycerol kinase; yejM: inner membrane protein yejM; O-aL: O-antigen ligase; hP: hypotetical protein; gT: glycosyltransferase; pgaB: poly-beta-1,6-N-acetyl-D-glucosamine N-deacetylase; eptA: phosphoethanolamine transferase eptA; rfaF: lipopolysaccharide heptosyltransferase II; aT: acetyltransferase; sunS: glycosyltransferase sunS; aLP: alkaline phosphatase family protein; gPtT: glucose-1-phosphate thymidylyltransferase rfbA; yrbL-phoP: regulatory network protein; degT: DegT/DnrJ/EryC1/StrS family aminotransferase; rfbA: glucose-1-phosphate thymidyl transferase; rfbB: dTDP-glucose 4,6-dehydratase 1; rfbC: dTDP-4-dehydrorhamnose 3,5-epimerase; rfbD: dTDP-4-dehydrorhamnose reductase; mbOat: membrane bound O-acyl transferase; yrbL-phoP: YrbL-PhoP reg domain containing protein.
Arcobacter butzleri, as the other Arcobacter species, is motile by means of one single polar flagellum (Miller et al., 2007). The bacterial flagellar motor is comprised by a core structure (inner membrane stator complexes MotA4B2 and the C-ring), a dedicated type III secretion system (T3SS) export apparatus, the inner membrane MS-ring and the P- and L-rings. These core components are conserved across bacterial genera. Nevertheless, the architecture of flagellar motors in A. butzleri has a diverse (Rossmann and Beeby, 2018), “intermediate” (Chaban et al., 2018) motor structure. According to the analysis performed by Chaban et al. (2018), we retrieved homologous proteins of the flagellar system in Ab 55 and Ab 6V genomes (Table 3). In both strains, loci share the same genomic content and most of the flagellar genes are located within the same genomic locus. As reported by Chaban et al. (2018), the Arcobacter-type motor accessory proteins did not contain homologues of the accessory proteins FlgP, FlgQ, FlgT, while we identified the homologue FlgO, an outer membrane protein required for flagellar motility in Vibrio cholerae, highly conserved in Vibrio spp. (Zhu et al., 2017).
Table 3.
Flagellum proteins in Arcobacter butzleri genomes.
| Organism | A. butzleri RM4018 | A. butzleri 55 | A. butzleri 6V | ||
|---|---|---|---|---|---|
| Whole genome record | NC_009850.1 | QXMK00000000 | QXNB0000000 | ||
| Class | Epsilon | Epsilon | Epsilon | ||
| GenBank accession | Protein ID | ||||
| T3SS | Flagellar biosynthesis protein FlhA | FlhA | ABV68160.1 | D3M61_10075 | D3M75_09250 |
| Flagellar biosynthetic protein FlhB | FlhB | ABV68164.1 | D3M61_10055 | D3M75_09230∗ | |
| Flagellum-specific ATP synthase | FliI | ABV68162.1 | D3M61_10065 | D3M75_09240 | |
| Flagellar biosynthetic protein FliP | FliP | ABV67255.1 | D3M61_02925∗ | D3M75_03525 | |
| Flagellar biosynthetic protein FliQ | FliQ | ABV66480.1 | D3M61_08725 | D3M75_07160 | |
| Flagellar biosynthesis protein FliR | FliR | ABV68165.1 | D3M61_10050 | D3M75_09225 | |
| C-ring | Flagellar motor switch protein FliG | FliG | ABV68184.1 | D3M61_09955 | D3M75_09130 |
| Flagellar motor switch protein FliM | FliM | ABV66477.1 | D3M61_08740 | D3M75_07145 | |
| Flagellar motor switch protein FliN | FliN | ABV68182.1 | D3M61_09965 | D3M75_09140 | |
| MS-ring | Flagellar M-ring protein | FliF | ABV68185.1 | D3M61_09950 | D3M75_09125 |
| Proximal rod | Flagellar hook-basal body protein FliE | FliE | ABV68168.1 | D3M61_04895 | D3M75_09210 |
| Flagellar basal body rod protein FlgB | FlgB | ABV68186.1 | D3M61_09945 | D3M75_09120 | |
| Flagellar basal-body rod protein FlgC | FlgC | ABV68167.1 | D3M61_10040 | D3M75_09215 | |
| P-ring | Flagellar hook-associated protein | FlgI | A8ERA4.1 | D3M61_08735 | D3M75_07150 |
| L-ring | Flagellar L-ring protein precursor | FlgH | ABV66485.1 | D3M61_08700 | D3M75_07185 |
| Campy-specific | Flagellar lipoprotein | FlgP | n/a | n/a | n/a |
| Hypothetical protein | FlgQ | n/a | n/a | n/a | |
| Tetratricopeptide repeat protein | PflA | WP_012147853.1 | D3M61_10060 | D3M75_09235 | |
| Tetratricopeptide repeat protein | PflB | WP_012147861.1 | D3M61_09990 | D3M75_09165 | |
| Vibrio-specific | Flagellar assembly protein | FlgT | n/a | n/a | n/a |
| Hypothetical protein | FlgO | WP_004511148.1 | D3M61_08900 | D3M75_07635 | |
∗Pseudogene (frameshifted).
In Ab 55, the flagellar biosynthetic protein FliP (an integral membrane component) (Ferris and Minamino, 2006) was predicted as a pseudogene (frameshifted) (D3M61_02925), as the flagellar biosynthetic protein FlhB (a flagellar export component responsible for substrate specificity switching) (Minamino and Macnab, 2000) in Ab 6V (D3M75_09230). This might have occurred due to a homopolymeric region (AAAAAA) comprised in both the genomic loci which might have affected the base calling of the sequencing.
Both genomes harbor the genes cj1349 and cadF (coding for fibronectin-binding proteins CadF and Cj1349), mviN (encoding a protein essential for the peptidoglycan biosynthesis) and pldA (encoding phospholipase A), the gyrA, iroE, and irgA (iron-regulating outer membrane protein), ciaB (encoding the C. jejuni invasion antigen B) genes and the hemolysin gene tylA, while only Ab 55 harbors hecA [a member of the filamentous hemagglutinin (FHA) family], and hecB (encoding a hemolysin activation protein), as occurs in the Ab RM4018 (Miller et al., 2007).
Cia proteins (including CiaB, CiaC, and CiaD) have been suggested to be involved in promoting internalization of C. jejuni for host invasion and require a full-length flagellar filament for proper secretion (Chaban et al., 2015).
Several authors have screened A. butzleri strains for the presence of virulence genes such as ciaB, cadF, cj1349, hecA, and irgA, finding diverse prevalence of these genes in isolates from the same or different ecological niches (Collado et al., 2014; Ferreira et al., 2017; Rathlavath et al., 2017; Vicente-Martins et al., 2018). However, nucleotide sequence heterogeneity as well as PCR biases may provide false negative results thus underestimation of the actual prevalence of these genes. These drawbacks may be overcome by a genomic-based assessment, which, moreover, may allow the detection of novel (acquired) virulence genes.
Indeed, apart from the above-mentioned genes, recognized as putative virulence determinants in A. butzleri (Miller et al., 2007), we identified other genes coding for additional virulence associated protein in Ab 55 and Ab 6V genomes. Among these and present in Ab 55, Ab 6V, and Ab RM4018, we distinguished a DNA binding protein of the virulence factor B family, which contributes to the expression of virulence factors and to pathogenicity in Staphylococcus aureus (Matsumoto et al., 2010; Junecko et al., 2012), a VOC family virulence protein (glyoxalase/bleomycin resistance protein/dioxygenase superfamily domain), a conserved virulence factor B (DNA binding protein), and VirF of the AraC family of transcriptional regulators.
Only in Ab 55, with no orthologs in Ab 6V and Ab RM4018, we identified a virulence associated protein VirE (D3M61_07785), which has an 87% identity with a hypothetical protein of Ab L353 (WP_080952707.1); this was the only hit retrieved within Arcobacter genus (taxid: 28196) by BLASTP search in the NCBI database, while other results indicated identity of about 32% with a hypothetical protein of Dyella sp. 4G-K06 (WP_115495989.1) and with a primase from the Escherichia phage vB_EcoM-ep3. This sequence was also compared to metagenomic sequences comprised in the microbiome database MGNIFY (EMBL-EBI©; Mitchell et al., 2017) by BlastP analysis. Analysis showed 91% of identity with metagenomic sequence of a sample retrieved from Charlotte area wastewater treatment plants (North Carolina, United States). InterPro functional classification (Finn et al., 2017) assigned the protein the Virulence-associated E (IPR007936) family membership with a D-loop motif (Pfam:PF05272.5) and a related COG: 5545 Mobilome: prophage transposone category. The genomic locus in which the gene is located also comprises several hypothetical proteins, one Prophage CP4-57 integrase, several tRNA, one site-specific tyrosine recombinase XerC, putative DNA-invertase from lambdoid prophage Rac. The structure of this genomic locus is compatible with the presence of a genomic island (Juhas et al., 2009) and suggests the acquisition of this virulence element by a mechanism of horizontal gene transfer.
Furthermore, in Ab 6V genome, we also found one virulence sensor protein BvgS precursor and one virulence sensor histidine kinase PhoQ, which both have orthologues in Ab RM4018 (WP_012012740.1 and WP_012012731.1, respectively) but not in Ab 55.
Unique to Ab 6V, we identified two putative hemolysin activation/secretion proteins (D3M75_11460 and D3M75_03300), which have orthologues in A. butzleri ED-1, with a ShlB/FhaC/HecB family hemolysin secretion/ activation domain.
All together these findings lead to hypothesize a relatively high virulence of A. butzleri isolated from shellfish and prompt the need for a genomic analysis of more strains from this ecological niche, as well as for in vitro and in vivo studies, to unravel the mechanism of pathogenicity of this species.
Antibiotic Susceptibility and Genetic Determinants
As shown in Table 4, both genomes harbor a wide endowment of genes involved in antibiotic resistance, including transporter, efflux pump, multidrug resistance protein and methyltransferase, although with some differences. As example, Ab 55 harbors a protein predicted as a bifunctional polymyxin resistance protein ArnA (D3M61_11465), which is not present neither in Ab 6V nor in the A. butlzeri RM4018. BLASTP analysis retrieved as best hit a hypothetical protein of Campylobacter hyointestinalis (WP_111949105.1).
Table 4.
Antibiotic resistance genes in Ab 55 and Ab 6V genomes.
| Gene name | Product | UniProt KB | Present in |
|---|---|---|---|
| arnA | Bifunctional polymyxin resistance protein ArnA | O52325 | Ab 55 |
| arnB | UDP-4-amino-4-deoxy-L-arabinose-oxoglutarate (polymyxin resistance) | Q8ZNF3 | Ab 55 – Ab 6V |
| bcr | Bicyclomycin resistance protein | P28246 | Ab 55 – Ab 6V |
| bepD | Efflux pump periplasmic linker BepD precursor | Q8G2M7 | Ab 55 – Ab 6V |
| bepE | Efflux pump membrane transporter BepE | Q8G2M6 | Ab 55 – Ab 6V |
| bla | Beta-lactamase OXA-15 precursor | Q51574 | Ab 55 – Ab 6V |
| cat3 | Chloramphenicol acetyltransferase 3 | P00484 | Ab 55 – Ab 6V |
| eptA | Phosphoethanolamine transferase EptA (polymyxin resistance) | P30845 | Ab 55 – Ab 6V |
| fsr | Fosmidomycin resistance protein | P52067 | Ab 55 – Ab 6V |
| hcpC | Putative beta-lactamase HcpC precursor | O25728 | Ab 55 – Ab 6V |
| hlpA | Serine/threonine-protein kinase HipA (methicillin resistance) | P23874 | Ab 6V |
| ileS | Isoleucine-tRNA ligase (mupirocine resistance) | P00956 | Ab 55 – Ab6V |
| lmrA | Multidrug resistance ABC transporter ATP-binding and permease protein | Q9CHL8 | Ab 6V |
| - | putative metallo-hydrolase (metallo Beta-lactamase) | Q5XD24 | Ab 55 |
| macA | Macrolide export protein MacA | P75830 | Ab 55 – Ab 6V |
| macB | Macrolide export ATP-binding/permease protein MacB | P75831 | Ab 55 – Ab 6V |
| bla2 | Beta-lactamase 2 precursor | P10425 | Ab 55 – Ab 6V |
| mdtB | Multidrug resistance protein MdtB | B7NCB1 | Ab 55 – Ab 6V |
| mdtE | Multidrug resistance protein MdtE precursor | P37636 | Ab 55 |
| mexA | Multidrug resistance protein MexA | P52477 | Ab 55 – Ab 6V |
| mexB | Multidrug resistance protein MexB | P52002 | Ab 55 – Ab 6V |
| oprM | Outer membrane protein OprM | Q51487 | Ab 55 – Ab 6V |
| pbp | Beta-lactam-inducible penicillin-binding protein | P07944 | Ab 55 – Ab 6V |
| relE | mRNA interferase toxin RelE (ciprofloxacin and ampicillin) | P0C077 | Ab 55 – Ab 6V |
| rlmN | Putative dual-specificity RNA methyltransferase RlmN (ribosome target antibiotics) | Q7A600 | Ab 55 – Ab 6V |
| sttH | Streptothricin hydrolase | Q1MW86 | Ab 55 – Ab 6V |
| tetA | Tetracycline resistance protein, class C | P02981 | Ab 55 – Ab 6V |
| uppP | Undecaprenyl-diphosphatase (bacitracin resistance) | P60932 | Ab 55 – Ab 6V |
| wbpD | Group B chloramphenicol acetyltransferase | G3XD01 | Ab 55 – Ab 6V |
The UDP-4-amino-4-deoxy-L-arabinose-oxoglutarate aminotransferase arnB gene was instead identified in both Ab 55 and Ab 6V, while it is not present in A. butlzeri RM4018. The coded protein belongs to the DegT/DnrJ/EryC1/StrS aminotransferase family protein, and it is required for resistance to polymyxin and cationic antimicrobial peptides (Lee and Sousa, 2014).
The predicted serine/threonine-protein kinase HipA belonging to the type II toxin-antitoxin system was only retrieved in Ab 6V predicted proteome (D3M75_03575), and it is involved in the methicillin resistance. Only in Ab 55, we identified the mRNA interferase toxin RelE (D3M61_04745), which is involved in ciprofloxacin and ampicillin resistance in E. coli (Harms et al., 2017).
Table 5 shows metal resistance genes annotated in Ab 55 and Ab 6V genomes.
Table 5.
Metal resistance genes in Ab 55 and Ab 6V genomes.
| Genes | Product | Metal resistance | Present in |
|---|---|---|---|
| cadA | Cadmium, zinc, and cobalt-transporting ATPase | Cadmium, zinc, cobalt | Ab 6V – Ab 55 |
| czcB | Cobalt-zinc-cadmium resistance protein CzcB | Cadmium, zinc, cobalt | Ab 6V |
| czcD | Cadmium, cobalt and zinc/H(+)-K(+) antiporter | Cadmium, zinc, cobalt | Ab 6V – Ab 55 |
| copA_1 | Copper-exporting P-type ATPase A | Copper | Ab 6V – Ab 55 |
| copA_2 | Putative copper-importing P-type ATPase A | Copper | Ab 6V – Ab 55 |
| cusS | Sensor kinase CusS | Copper | Ab 6V – Ab 55 |
| Additional genes | |||
| arsC | Arsenate reductase | Arsenic | Ab 6V – Ab 55 |
| modA | Molybdate-binding periplasmic protein precursor | Molybdate chromate | Ab 6V – Ab 55 |
| copZ | Copper chaperone CopZ | Copper | Ab 6V – Ab 55 |
Observational and experimental studies have highlighted that exposure of bacteria to heavy metals (mainly zinc and copper), mainly due to anthropogenic environmental contamination, can induce or co-select resistance to them and to one or more antibiotics. In particular, resistance may be induced by metals (i) via co-selection resistance, when different genes coding for antibiotic and metal resistance share a close location (as in mobile genetic elements, such as integrin, plasmid, or transponson), (ii) via cross-selection, when the same genetic element encodes for both antibiotic and metal resistance, and (iii) via co-regulation, when antibiotic and metal resistance genes share the same regulatory system (Lupo et al., 2012; Seiler and Berendonk, 2012; Chenia and Jacobs, 2017; Li et al., 2017; Poole, 2017; Yu et al., 2017; Wu et al., 2018). Nevertheless, as far as we know, only Otth et al. (2005) investigated susceptibility of A. butzleri to heavy metals, finding that all the 50 tested strains were susceptible to mercury, silver, and chrome salts, whereas all were resistant to molybdenum, manganese, nickel, cobalt, lead, and iron.
Metal resistance genes share the same location in both Ab 55 and Ab 6V genomes. cadA is close to genes coding for the ferrous iron transport protein FeoA and FeoB, outer membrane efflux proteins, transcriptional regulators, and several tRNA genes, suggesting the presence of a genomic island (Juhas et al., 2009). czcB is close to gene coding for the multidrug resistance protein MstB, one permease and the gene coding for the sensor protein ZraS. One copA gene is close to the gene coding for outer membrane porin precursor and the copper sensing transcriptional repressor CsoR, while the other copA is close to the ferrous iron transport proteins FeoA and FeoB. cusS is close to the transcriptional activator protein czcR, macB, and several chaperonine, whereas arsC is located near to genes coding for plasmid stabilization system protein and genes coding for flagellum biosynthesis (fliK and flhB), and cell division protein FtsA and FtsZ. modA gene is located within a hypothetical operon including a transcriptional regulator GltR, a gene coding for a molybdenum-pterin binding protein, the regulator ModE, and the transport system permease protein ModB. copZ is close to merT gene, coding for a mercuric transport protein, a gene coding for a natural resistance-associated macrophage protein, a transcriptional regulatory protein ZraR and a sensor protein FixL.
The antimicrobial susceptibility of Arcobacter spp. isolated from various ecological niches has been investigated by several authors (Kabeya et al., 2004; Kayman et al., 2012; Ferreira et al., 2013, 2017; Scanlon et al., 2013; Collado et al., 2014; Rahimi, 2014; Yesilmen et al., 2014; Zacharow et al., 2015b; Aski et al., 2016; Van den Abeele et al., 2016; Elmali and Can, 2017; Rathlavath et al., 2017; Šilha et al., 2017; Soma et al., 2017; Vicente-Martins et al., 2018), but the lack of standardized protocols and interpretive criteria for the antimicrobial susceptibility testing (AST) of Arcobacter spp. is the major limitation for a comparable and univocal evaluation of antimicrobial resistance and susceptibility for these microorganisms (Ferreira et al., 2016). Results of the disk diffusion test are shown in Table 6, whereas in Table 7, are reported the MICs and MBCs assessed by broth microdilution method (section “Antimicrobial Susceptibility Testing”). The antimicrobial susceptibility pattern of Ab type strain LMG 10828T (ATCC 49616, RM4018), used as a reference strain for the AST in our study, was comparable to that reported by Miller et al. (2007), for 10 out of 12 tested antibiotics, i.e., gentamicin, kanamycin, streptomycin, ciprofloxacin, chloramphenicol, erythromycin, vancomycin, ampicillin, cefotaxime, and penicillin G. Slight differences were related to susceptibility and resistance toward tetracycline and nalidixic acid, reported by Miller et al. (2007); in particular, in our test, the type strain resulted intermediate resistant toward both antibiotics. As concerns MIC values, the type strain Ab LMG10828 showed results consistent with those find by Riesenberg et al. (2017). Additionally, in our study, using a wider range of concentrations for penicillin and vancomycin, we were able to assess MIC values for Ab LMG10828T toward these two antibiotics (i.e., 128 and 2048 μg/ml, respectively) that were previously reported by Riesenberg et al. (2017) as ≥64 μg/ml. According to MIC interpretive standards, A. butzleri LMG 10828T, Ab 55 and Ab 6V have been classified as resistant toward vancomycin and the three β-lactam antibiotics used in this study, i.e., cefotaxime (β-lactam cephalosporin), ampicillin, and penicillin G (β-lactam penicillins), confirming the results obtained by disk diffusion test (Tables 6, 7).
Table 6.
Disk diffusion susceptibility test for A. butzleri.
|
A. butzleri strain |
||||
|---|---|---|---|---|
| Class | Antibiotics | LMG 10828 | 55 | 6V |
| β-Lactams | Ampicillin 10 μg/disk | R∗ | R∗ | R∗ |
| Penicillin G 10 units/disk | R∗ | R∗ | R∗ | |
| Cefotaxime 30 μg/disk | R∗ | R∗ | R∗ | |
| Glycopeptides | Vancomycin 30 μg/disk | R∗ | R∗ | R∗ |
| Quinolones | Ciprofloxacin 5 μg/disk | S | S | S |
| Nalidixic acid 30 μg/disk | I | R | I | |
| Aminoglycosides | Gentamicin 10 μg/disk | S | S | S |
| Kanamycin 30 μg/disk | S | S | S | |
| Streptomycin 10 μg/disk | S | S | S | |
| Phenicol | Chloramphenicol 30 μg/disk | R | I | I |
| Tetracycline | Tetracycline 30 μg/disk | I | R | R |
| Macrolide | Erytromycin 15 μg/disk | I | R | R |
Classification as S (susceptible), I (intermediate), and R (resistant) was carried out according to zone diameter interpretive standards for Staphylococcus spp. (erytromycin and penicillin G) and Enterobacteriaceae (ampicillin, gentamicin, cefotaxime, ciprofloxacin, tetracycline, chloramphenicol, nalidixic acid, kanamycin, and streptomycin) (CLSI, 2015; Elmali and Can, 2017). To our knowledge, no vancomycin reference interpretive criteria are reported for A. butzleri. ∗No inhibition zone detected.
Table 7.
Cefotaxime, ampicillin, penicillin G, and vancomycin MIC and MBC values for A. butzleri.
| β-Lactams |
Glycopeptide |
|||||||
|---|---|---|---|---|---|---|---|---|
| A. butzleri strain | Cefotaxime μg/ml |
Ampicillin μg/ml |
Penicillin G μg/ml |
Vancomycin μg/ml |
||||
| MICa | MBCb | MICa | MBCb | MICa | MBCb | MICa | MBCb | |
| LMG 10828 | 16 | 16 | 32 | 32 | 128 | 256 | 2,048 | >2,048 |
| 55 | 128 | 128 | 256 | 256 | 1,024 | 1,024 | >2,048 | >2,048 |
| 6V | 64 | 64 | 64 | 64 | 256 | 256 | 2,048 | >2,048 |
aMinimal inhibitory concentration. bMinimal bactericidal concentration.
For A. butzleri type strain, Ab 55 and Ab 6V, the highest MIC and MBC values were observed for vancomycin, which were ≥2048 μg/ml (Table 7).
As recently reviewed by Ahmed and Baptiste (2018) in enterococci, molecular basis for vancomycin resistance phenotypes are determined by the presence of the vancomycin resistance (Van) operons (described as vanA, -B, -C, -D, -E, -G, -L, -M, and N) which may be located on the chromosome or on a plasmid. No element of the described operons was found in Ab 55 and Ab 6V genomes, as well as in Ab RM4018. However, the resistance of Gram-negative bacteria toward vancomycin may be intrinsic due to the inability of glycopeptides [molecules with high molecular weight (1450–1500 Da) and size] to pass through porins, which govern the movement of hydrophilic molecules across their outer membrane (Quintiliani and Courvalin, 1995) to reach their site of action, i.e., the cell wall (Nicolosi et al., 2010). Indeed, 100% of the A. butzleri isolates tested by Aski et al. (2016), Rathlavath et al. (2017), and Soma et al. (2017) were resistant to vancomicyn.
Both strains are resistant to all the ß-lactam antibiotics tested (ampicillin, penicillin, and cefotaxime) (Table 6). The resistance of A. buzleri isolates to β-lactams is widespread in seafood and water sources (Collado et al., 2014; Rathlavath et al., 2017; Šilha et al., 2017) as well as in other environments (Kayman et al., 2012; Ferreira et al., 2013; Rahimi, 2014; Yesilmen et al., 2014; Zacharow et al., 2015b; Aski et al., 2016; Van den Abeele et al., 2016; Elmali and Can, 2017; Vicente-Martins et al., 2018). Rathlavath et al. (2017) found that on 40 A. butzleri isolated from shellfish, 100% resulted resistant to cefotaxime and 70% to ampicillin, while among 81 A. butzleri isolated from fish, 98.7% was resistant to cefotaxime and 72.8% to ampicillin. Moreover, of 26 A. butzleri isolated from coastal water, 100% was resistant to cefotaxime and 73% to ampicillin (Rathlavath et al., 2017). Šilha et al. (2017) found that 94.4% of 18 A. butzleri isolated from water sources were resistant to ampicillin and 100% were resistant to penicillin G, while Collado et al. (2014) found that 45.2% of 62 A. butzleri isolated from bivalve molluscs were resistant to ampicillin.
The ß-lactam resistance is generally due to the combined effects of the presence and activity of ß-lactamase genes, of the binding to targets (penicillin-binding proteins) and, in Gram-negative bacteria, of the outer-membrane permeability (Georgopapadakou, 1993). In the genomes of Ab 55 and Ab 6V, we identified three putative ß-lactamases orthologues to that of A. butlzeri RM4018 (Miller et al., 2007) [MBL fold metallo-hydrolase D3M61_00510 and D3M61_04375 (Ab 55), D3M75_05505 and D3M75_08430(Ab 6V); class D beta-lactamase D3M61_10735, D3M75_10375 (Ab 6V)] as well as penicillin binding proteins (Ab 6V D3M75_03520, D3M75_10675, D3M75_10720; Ab 55: D3M61_02930, D3M61_10940, D3M61_10985).
Furthermore, in the genomes of both strains, we retrieved the lrgAB operon, which modulates penicillin tolerance in Staphylococcus (Bayles, 2000; Groicher et al., 2000) and was previously suggested as the ß-lactam resistance enhancer in A. butzleri RM4018 (Miller et al., 2007).
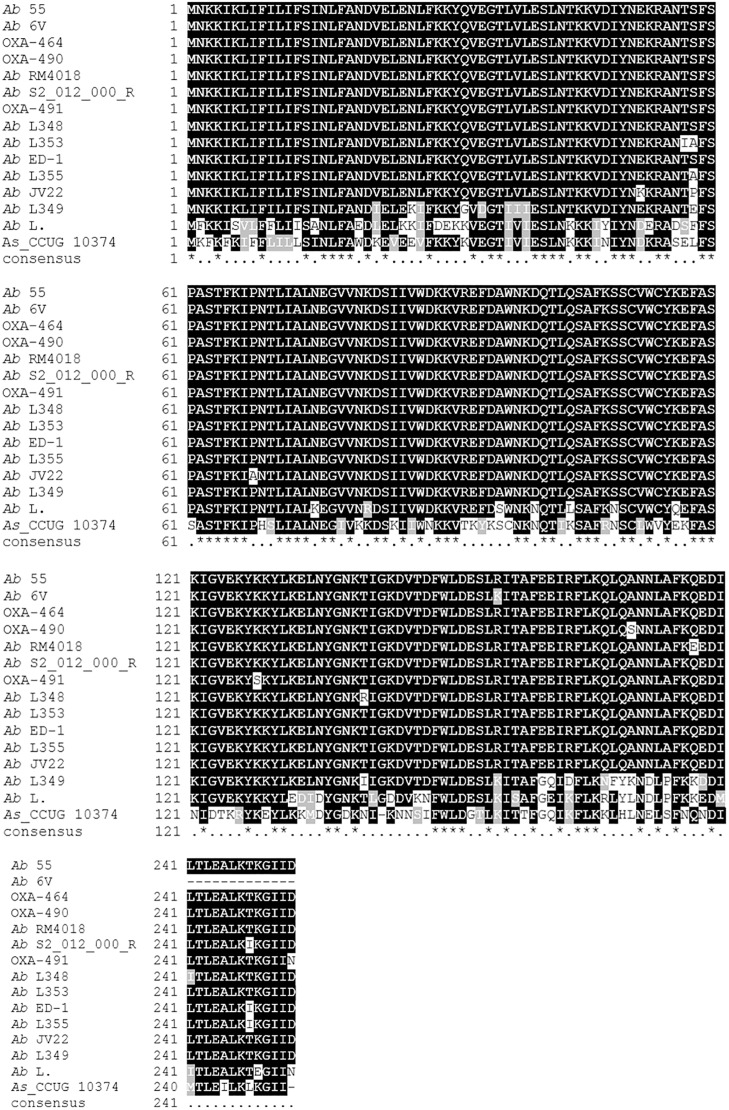
Figure 3 shows the multialignment of beta-lactamase protein in Ab 55 and Ab 6V and other Arcobacter species. In Ab 6V the predicted protein (D3M75_10375) is truncated at N-terminal due to a nucleotide mutation in the genomic locus which leads to a premature stop codon. The DNA sequence translated by EMBOSS® Sixpack shows that changing the reading frame would recover the entire protein, identical to Ab 55 D3M61_10735. The sequence of Ab 55 D3M61_10735 share 100% of identity with OXA-464, which differs with the ß-lactamase of the type strain A. butleri RM4018 for a glutamine instead of glutamic acid in position 177.
FIGURE 3.
Multialignment of OXA beta-lactamase from Arcobacter butzleri obtained by using T-Coffee web server (Di Tommaso et al., 2011). Ab 55 protein ID: D3M61_10735; Ab 6V: D3M75_10375; OXA-491, Accession: ANW35665.1; OXA-464: ANW35663.1; OXA-490: ANW35664.1. A. butzleri RM4018: WP_012013127.1; A. butzleri S2_012_000_R2_80: PZP12670.1; A. butzleri L348: WP_046997374.1; A. butzleri L353: WP_050071304.1; A. butzleri ED-1: WP_014468976.1; A. buzleri L355: WP_046997672.1; A. butzleri JV22: EFU68937.1; A. butzleri L349: WP_046993700.1; A. butzleri L.: WP_014474670.1; A. skirrowii CCUG 10374: WP_115588490.1.
Ab 55 and Ab 6V as well as the Ab type strain LMG 10828 (RM4018) were sensitive to the three aminoglycoside antibiotics used in this study namely gentamicin, kanamycin, and streptomycin (Table 6). Our results are in agreement with those obtained by Rathlavath et al. (2017) on the 147 A. butzleri isolates from seafood (40 isolates were from shellfish) and coastal water, which were all sensitive to these three antibiotics. Susceptibility to these antibiotics is also consistent to that found by other authors who reported susceptibility to streptomycin and/or gentamicin and/or kanamycin for the great majority (96.4–100%) of A. butzleri isolated from shellfish [gentamicin (Collado et al., 2014)], water sources [gentamicin and streptomycin (Šilha et al., 2017)] and other various sources [gentamicin (Kayman et al., 2012; Ferreira et al., 2013, 2017; Van den Abeele et al., 2016); gentamicin and streptomycin (Šilha et al., 2017); kanamycin and streptomycin (Kabeya et al., 2004); gentamicin, kanamycin and streptomycin (Rahimi, 2014)]. Aminoglycosides are proposed as antibiotics to be used in Arcobacter infections (Rahimi, 2014; Ferreira et al., 2016; Rathlavath et al., 2017). Nevertheless, a study reported high percentage (80%) of resistance to gentamicin and kanamycin in A. butzleri isolates from porcine samples (Scanlon et al., 2013) but only five isolates were tested. Mechanisms of bacterial resistance to this class of antibiotics are diverse including the inactivation by aminoglycoside modifying enzymes, mutation of the ribosome target, and modification of the ribosome by methyltransferase enzymes (Wilson, 2014; Garneau-Tsodikova and Labby, 2016). Resistance may also arise from mutation in the rrs encoding for 16S rRNA, even if these mutations are quite rare, as they would interfere with the vital cellular machinery.
Susceptibility to the hydrophilic fluoroquinolone ciprofloxacin or very low percentage of resistant isolates (0–3.2%) are widely reported in literature (Kayman et al., 2012; Collado et al., 2014; Rahimi, 2014; Ferreira et al., 2017; Rathlavath et al., 2017; Šilha et al., 2017; Soma et al., 2017), although some authors reported percentages ranging from 12.7 to 55.8% of A. butzleri resistant to ciprofloxacin isolated from patients with gastroenteritis, retail food products, and poultry slaughterhouse (Ferreira et al., 2013; Zacharow et al., 2015b; Van den Abeele et al., 2016; Vicente-Martins et al., 2018). The acquisition of fluoroquinolone resistance may represent a serious issue for the health care system since they are the first-choice antibiotic for treating Campylobacter infection in humans and they were suggested to be used in Arcobacter enteritis (Vandenberg et al., 2006; Collado et al., 2014; Ferreira et al., 2016).
Our strains are susceptible to ciprofloxacin. Indeed, we did not find in A. butzleri Ab 55 and Ab 6V the mutation in the quinolone resistance determining region in position 254 of the gyrA gene which causes a transition from cytosine to thymine leading to the substitution of a threonine to isoleucine (Abdelbaqi et al., 2007).
Ab LMG10828T (RM 4018) and Ab 6V were intermediate resistant to the hydrophobic quinolone nalidixic acid, whereas Ab 55 was resistant to this antibiotic. However, the absence of gyrA mutations in these strains suggests that the putative resistance could be due to the mechanisms of hydrophobic quinolones uptake as suggested by Miller et al. (2007). High percentage of resistance toward the nalidixic acid was reported for 77.5 and 83.4% of A. butzleri isolated from shellfish by Rathlavath et al. (2017) and Collado et al. (2014), respectively, for 71.6% of A. butzleri isolates from fish (Rathlavath et al., 2017) and for 57.6 and 88.9% of A. butzleri isolates from water sources in two different studies (Rathlavath et al., 2017; Šilha et al., 2017). One hundred percent of A. butzleri isolated from other various sources, namely, retail food products, porcine, slaughterhouse, and dairy plant samples, by different authors, were resistant to nalidixic acid (Scanlon et al., 2013; Elmali and Can, 2017; Ferreira et al., 2017; Vicente-Martins et al., 2018). By contrast, high percentage of susceptibility toward nalidixic acid, ranging from 50 to 77.8%, was reported by Yesilmen et al. (2014) and Kayman et al. (2012) for A. butzleri isolated from milk and dairy products and human gastroenteritis stool samples, respectively. However, no more than 10 A. butzleri isolates were tested in both studies.
Arcobacter butzleri Ab 55 and Ab 6V are intermediate resistant to chloramphenicol (Table 6). Differences about susceptibility against this antibiotic were reported among several studies (Ferreira et al., 2016); e.g., Rathlavath et al. (2017) reported high susceptibility rates ranging from 65.3 to 77.7% for 147 A. butzleri isolates from shellfish, fish, and coastal water whereas Šilha et al. (2017) reported that 66.6% of A. butzleri isolated from water sources was resistant. Also for A. butzleri isolated from various sources (poultry meat and other retail food products, poultry slaughterhouse, and animal and human stool samples) variable resistance percentages were reported ranging from 2.3 to 87.7% (Ferreira et al., 2013; Rahimi, 2014; Aski et al., 2016; Šilha et al., 2017; Soma et al., 2017; Vicente-Martins et al., 2018). Otth et al. (2004) suggested that it may depend on local differences in the usage of this antibiotic.
Chloramphenicol resistance commonly consists of its enzymatic inactivation mainly by acetyltransferases or occasionally by phosphotransferases, but additional mechanisms involve the presence of efflux pump which act as extrusion transporters, mutation or modification of the target site, and decreased outer membrane permeability. In the genomes of Ab 55 and Ab 6V, we retrieved the cat3 gene encoding for a type A chloramphenicol O-acetyltransferase (Ab 55 D3M61_09345; Ab 6V D3M75_06440), which catalyzes the acetyl-CoA-dependent acetylation of chloramphenicol and they resulted as intermediate resistant.
Our A. butzleri strains Ab 55 and Ab 6V are resistant to tetracycline (Table 6). As for chloramphenicol, tetracycline susceptibility results vary among studies, even if tetracycline is proposed for treating Arcobacter infections by different authors (Ferreira et al., 2016; Rathlavath et al., 2017). Rathlavath et al. (2017) and Šilha et al. (2017) reported that 100 and 77.8% of A. butzleri isolated from shellfish, fish, coastal water, and water sources, respectively, were susceptible to tetracycline and high susceptibility rates, ranging from 78.2 to 100% were reported also for A. butzleri isolated from poultry meat, human and animal stool samples (Rahimi, 2014; Aski et al., 2016; Šilha et al., 2017). Conversely Vicente-Martins et al. (2018) reported that 95.4% of 65 A. butzleri strains isolated from retail food products was resistant to tetracycline and also Yesilmen et al. (2014) reported that 100% of A. butzleri isolated from milk and cheese was resistant toward this antibiotic, even if only 10 isolates were tested.
Tetracycline resistance may be due to several mechanisms: efflux, modification, protection from the ribosome binding, modification of 16S rRNA at the tetracycline binding site. These mechanisms are mediated by different proteins, among which Tet(O) and Tet(M) are the most important. These proteins are paralogues of the translational GTPase EF-G which removes tetracycline from its inhibitory site on the ribosome through a GTP-dependent hydrolysis. Both are part of a larger group of proteins called ribosomal protection proteins (RPPs), which also includes Tet(Q), Tet(S), Tet(T), Tet(W), and OtrA (Chopra and Roberts, 2001). In the genomes of our strains, we retrieved proteins predicted as tetracycline resistance protein of class C (MFS transporter-multidrug efflux pump) with 27% identity with the orthologues in E. coli and elongation factors with the same domain found in the C terminus of RPPs Tet(M) and Tet(O), with 65% identity with EF-G of E. coli.
Concerning erythromycin, A. butzleri Ab 55 and Ab 6V are resistant to this antibioitc (Table 6). However, different percentages of erythromycin resistance were reported in several studies. In particular, A. butzleri isolated from seafood and water sources, as well as from poultry meat, animal and human stool samples, dairy plant, and cheese were found to be susceptible to this antibiotic at percentages ranging from 65 to 100% (Kayman et al., 2012; Collado et al., 2014; Aski et al., 2016; Ferreira et al., 2017; Rathlavath et al., 2017; Šilha et al., 2017). Zacharow et al. (2015b) and Soma et al. (2017) reported that 62 and 50%, respectively, of A. butzleri isolated from animals, humans, and foods of animal origin were resistant toward this antibiotic whereas Yesilmen et al. (2014) and Scanlon et al. (2013) reported that 80% of A. butzleri isolated from milk, cheese, and porcine samples, respectively, were resistant too, but no more than 10 isolates were tested.
Erythromycin resistance is due, besides the less common mutation in 23S rRNA or ribosomal proteins, to post-transcriptional methylation of an adenine residue in 23S caused by the action of erm class gene-coded methylases in Gram-positive bacteria (Kurincic et al., 2007). Ribosomal RNA small subunit methyltransferases were also present in Ab 55 (D3M61_05905) and Ab 6V (D3M75_01835) genomes with 26 and 27% identity with reference sequence of Enterococcus faecium (Accession: YP_004172630.1), respectively; this protein is also present in the genome of A. butzleri RM4018, although it results intermediate resistant in response to erythromycin. Furthermore, we identified macA and macB genes encoding for macrolide exporter proteins in both Ab 55 and Ab 6V. The single copy of 23S rRNA of both Ab 55 and Ab 6V does not present any of the identified mutation responsible for erythromycin resistance.
Conclusion
Genomic analyses herein performed allowed us to confirm the recently (Pérez-Cataluña et al., 2018a,b) suggested amendment of A. butzleri as Al. butzlerii, comb. nov.
Antimicrobial susceptibility tests defined Ab 55 and Ab 6V strains as resistant to vancomycin, tetracyclin, nalidixic acid (only Ab 55 whereas Ab 6V is intermediate resistant), erythromycin, and β-lactam antibiotics. Moreover, in our strains isolated from shellfish, we identified numerous virulence, antibiotic, and heavy metal resistance determinants, also additional to those previously found in the genome sequenced A. butzleri ED-1, isolated from fuel cell, and in the A. butzleri type strain RM 4018, isolated from human gastroenteritis (Miller et al., 2007; Pérez-Cataluña et al., 2018a), leading to hypothesize that shellfish strain may be potentially more virulent.
The findings of food-related A. butzleri support both epidemiological surveillance and food safety risk assessment and management in the shellfish industry.
Further analysis in our laboratories is ongoing to sequence and characterize other A. butleri strains isolated from shellfish and from other food matrices, in order to obtain an updated description of the species and to clarify the role of genetic endowment, as well as of the ecological niches the strain come from, in the pathogenesis of A. butzleri. The genomic sequences here presented, and the novel insights obtained in the present study appreciably contribute to achieve these goals.
Author Contributions
VF conceived the work, interpreted the data, and organized the bioinformatic work. FF performed the genomic sequencing and the bioinformatic work. DC and FB performed the antimicrobial susceptibility testing. VF and FF wrote the manuscript. All the authors contributed to the revision of the manuscript and read and approved the submitted manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00670/full#supplementary-material
References
- Abdelbaqi K., Ménard A., Prouzet-Mauleon V., Bringaud F., Lehours P., Mégraud F. (2007). Nucleotide sequence of the gyrA gene of Arcobacter species and characterization of human ciprofloxacin-resistant clinical isolates. FEMS Immunol. Med. Microbiol. 49 337–345. 10.1111/j.1574-695X.2006.00208.x [DOI] [PubMed] [Google Scholar]
- Afgan E., Baker D., van den Beek M., Blankenberg D., Bouvier D., Čech M., et al. (2016). The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 44 W3–W10. 10.1093/nar/gkw343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M. O., Baptiste K. E. (2018). Vancomycin-Resistant Enterococci: a review of antimicrobial resistance mechanisms and perspectives of human and animal health. Microbiol. Drug Resist. 24 590–606. 10.1089/mdr.2017.0147 [DOI] [PubMed] [Google Scholar]
- Arguello E., Otto C. C., Mead P., Babady N. E. (2015). Bacteremia caused by Arcobacter butzleri in an immunocompromised host. Carroll KC, ed. J. Clinic. Microbiol. 53 1448–1451. 10.1128/JCM.03450-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aski H. S., Tabatabaei M., Khoshbakht R., Raeisi M. (2016). Occurrence and antimicrobial resistance of emergent Arcobacter spp. isolated from cattle and sheep in Iran. Comp. Immunol. Microbiol. Infect. Dis. 44 37–40. 10.1016/j.cimid.2015.12.002 [DOI] [PubMed] [Google Scholar]
- Auch A. F., von Jan M., Klenk H. P., Göker M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genomic Sci. 2 117–134. 10.4056/sigs.531120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz R. K., Bartels D., Best A. A., DeJongh M., Disz T., Edwards R. A., et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 8:75. 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banting G., Figueras Salvat M. J. (2017). “Arcobacter,” in Global Water Pathogens Project, eds Rose J. B., Jiménez-Cisneros B. (East Lansing, MI: Michigan State University; ) [Google Scholar]
- Bayles K. W. (2000). The bactericidal action of penicillin: new clues to an unsolved mystery. Trends Microbiol. 8 274–278. 10.1016/S0966-842X(00)01762-5 [DOI] [PubMed] [Google Scholar]
- Chaban B., Coleman I., Beeby M. (2018). Evolution of higher torque in Campylobacter-type bacterial flagellar motors. Sci. Rep. 8:97. 10.1038/s41598-017-18115-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaban B., Hughes H. V., Beeby M. (2015). The flagellum in bacterial pathogens: for motility and a whole lot more. Semin. Cell Dev. Biol. 46 91–103. 10.1016/j.semcdb.2015.10.032 [DOI] [PubMed] [Google Scholar]
- Chenia H. Y., Jacobs A. (2017). Antimicrobial resistance, heavy metal resistance and integron content in bacteria isolated from a South African tilapia aquaculture system. Dis. Aquat. Organ 126 199–209. 10.3354/dao03173 [DOI] [PubMed] [Google Scholar]
- Chopra I., Roberts M. (2001). Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65 232–260. 10.1128/MMBR.65.2.232-260.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chun J., Oren A., Ventosa A., Christensen H., Arahal D. R., da Costa M. S., et al. (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 68 461–466. 10.1099/ijsem.0.002516 [DOI] [PubMed] [Google Scholar]
- CLSI (2015). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. CLSI document M100-S25. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Collado L., Figueras M. J. (2011). Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter. Clin. Microb. Rev. 24 174–192. 10.1128/CMR.00034-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collado L., Jara R., Vásquez N., Telsaint C. (2014). Antimicrobial resistance and virulence genes of Arcobacter isolates recovered from edible bivalve molluscs. Food Control. 46 508–512. 10.1016/j.foodcont.2014.06.013 [DOI] [Google Scholar]
- DeLucia A. M., Six D. A., Caughlan R. E., Gee P., Hunt I., Lam J. S., et al. (2011). Lipopolysaccharide (LPS) inner-core phosphates are required for complete LPS synthesis and transport to the outer membrane in Pseudomonas aeruginosa PAO1. mBio 2:e00142-11. 10.1128/mBio.00142-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P., Moretti S., Xenarios I., Orobitg M., Montanyola A., Chang J. M., et al. (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39 W13–W17. 10.1093/nar/gkr245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douidah L., De Zutter L., Van Nieuwerburgh F., Deforce D., Ingmer H.et al. (2014). Presence and analysis of plasmids in human and animal associated Arcobacter species. PLoS One 9:e85487. 10.1371/journal.pone.0085487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douidah L., de Zutter L., Baré J., De Vos P., Vandamme P., Vandenberg O., et al. (2012). Occurrence of putative virulence genes in Arcobacter species isolated from humans and animals. J. Clin. Microbiol. 50 735–741. 10.1128/JCM.05872-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmali M., Can H. Y. (2017). Occurrence and antimicrobial resistance of Arcobacter species in food and slaughterhouse samples. Food Sci. Technol. 37 280–285. 10.1590/1678-457x.19516 [DOI] [Google Scholar]
- Engberg J., On S. L., Harrington C. S., Gerner-Smidt P. (2000). Prevalence of Campylobacter, Arcobacter, Helicobacter and Sutterella spp. in human fecal samples as estimated by a reevaluation of isolation methods for Campylobacters. J. Clin. Microbiol. 38 286–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercolini D., Fusco V., Blaiotta G., Sarghini F., Coppola S. (2005). Response of Escherichia coli O157:H7, Salmonella Thyphimurium, Listeria monocytogenes and Staphylococcus aureus to the stresses occurring in model manufactures of Grana Padano cheese. J. Dairy Sci. 88 3818–3825. 10.3168/jds.S0022-0302(05)73067-8 [DOI] [PubMed] [Google Scholar]
- Ferreira S., Fraqueza M. J., Queiroz J. A., Domingues F. C., Oleastro M. (2013). Genetic diversity, antibiotic resistance and biofilm-forming ability of Arcobacter butzleri isolated from poultry and environment from a Portuguese slaughterhouse. Int. J. Food Microbiol. 162 82–88. 10.1016/j.ijfoodmicro.2013.01.003 [DOI] [PubMed] [Google Scholar]
- Ferreira S., Júlio C., Queiroz J. A., Domingues F. C., Oleastro M. (2014a). Molecular diagnosis of Arcobacter and Campylobacter in diarrhoeal samples among Portuguese patients. Diagn. Microbiol. Infect. Dis. 78 220–225. 10.1016/j.diagmicrobio.2013.11.021 [DOI] [PubMed] [Google Scholar]
- Ferreira S., Oleastro M., Domingues F. C. (2017). Occurrence, genetic diversity and antibiotic resistance of Arcobacter sp. in a dairy plant. J. Appl. Microbiol. 123 1019–1026. 10.1111/jam.13538 [DOI] [PubMed] [Google Scholar]
- Ferreira S., Queiroz J. A., Oleastro M., Domingues F. C. (2014b). Genotypic and phenotypic features of Arcobacter butzleri pathogenicity. Microb. Pathog. 76 19–25. 10.1016/j.micpath.2014.09.004 [DOI] [PubMed] [Google Scholar]
- Ferreira S., Queiroz J. A., Oleastro M., Domingues F. C. (2016). Insights in the pathogenesis and resistance of Arcobacter: a review. Crit. Rev. Microbiol. 42 364–383. 10.3109/1040841X.2014.954523 [DOI] [PubMed] [Google Scholar]
- Ferris H. U., Minamino T. (2006). Flipping the switch: bringing order to flagellar assembly. Trends Microbiol. 14 519–526. 10.1016/j.tim.2006.10.006 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Collado L., Levican A., Perez J., Solsona M. J., Yustes C. (2011a). Arcobacter molluscorum sp. nov., a new species isolated from shellfish. Syst. Appl. Microbiol. 34 105–109. 10.1016/j.syapm.2010.10.001 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Levican A., Collado L., Inza M. I., Yustes C. (2011b). Arcobacter ellisii sp. nov., isolated from mussels. Syst. Appl. Microbiol. 34 414–418. 10.1016/j.syapm.2011.04.004 [DOI] [PubMed] [Google Scholar]
- Figueras M. J., Levican A., Pujol I., Ballester F., Rabada Quilez M. J., Gomez-Bertomeu F. (2014). A severe case of persistent diarrhoea associated with Arcobacter cryaerophilus but attributed to Campylobacter sp. and a review of the clinical incidence of Arcobacter spp. New Microbes New Infect. 2 31–37. 10.1002/2052-2975.35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R. D., Attwood T. K., Babbitt P. C., Bateman A., Bork P., Bridge A. J., et al. (2017). InterPro in 2017 - beyond protein family and domain annotations. Nucleic Acids Res. 45 D190–D199. 10.1093/nar/gkw1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn K., Villarreal B. P., Barranco A., Belc N., Bjornsdottir B., Fusco V., et al. (2019). An introduction to current food safety needs. Trends Food Sci. Technol. 84 1–3. 10.1016/j.tifs.2018.09.012 [DOI] [Google Scholar]
- Fong T. T., Mansfield L. S., Wilson D. L., Schwab D. J., Molloy S. L., Rose J. B. (2007). Massive microbiological groundwater contamination associated with a waterborne outbreak in Lake Erie, South Bass Island, Ohio. Environ. Health Perspect. 115 856–864. 10.1289/ehp.9430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz C. M. A. P., den Besten H. M. W., Böhnlein C., Gareis M., Zwietering M. H., Fusco V. (2018). Microbial food safety in the 21st century: emerging challenges and foodborne pathogenic bacteria. Trends Food Sci. Technol. 81 155–158. 10.1016/j.tifs.2018.09.019 [DOI] [Google Scholar]
- Fusco V., Abriouel H., Benomar N., Kabisch J., Chieffi D., Cho G.-S., et al. (2018). “Opportunistic foodborne pathogens,” in Food Safety and Preservation: Modern Biological Approaches to Improving Consumer Health, 1st Edn, Chap. 10 eds Grumezescu A., Holban A. M. (Academic Press), 269–306. 10.1016/B978-0-12-814956-0.00010-X [DOI] [Google Scholar]
- Fusco V., Quero G. M., Morea M., Blaiotta G., Visconti A. (2011). Rapid and reliable identification of Staphylococcus aureus harbouring the enterotoxin gene cluster (egc) and quantitative detection in raw milk by real time PCR. I. J. Food Microbiol. 144 528–537. 10.1016/j.ijfoodmicro.2010.11.016 [DOI] [PubMed] [Google Scholar]
- Garneau-Tsodikova S., Labby K. J. (2016). Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. Med. Chem. Comm. 7 11–27. 10.1039/C5MD00344J [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopapadakou N. H. (1993). Penicillin-binding proteins and bacterial resistance to beta-lactams. Antimicrob. Agents Chemother. 37 2045–2053. 10.1128/AAC.37.10.2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girbau C., Guerra C., Martínez-Malaxetxebarria I., Alonso R., Fernández-Astorga A. (2015). Prevalence of ten putative virulence genes in the emerging foodborne pathogen Arcobacter isolated from food products. Food Microbiol. 52 146–149. 10.1016/j.fm.2015.07.015 [DOI] [PubMed] [Google Scholar]
- Girbau C., Martinez-Malaxetxebarria I., Muruaga G., Carmona S., Alonso R., Fernandez-Astorga A. (2017). Study of biofilm formation ability of foodborne Arcobacter butzleri under different conditions. J. Food. Prot. 80 758–762. 10.4315/0362-028X.JFP-16-505 [DOI] [PubMed] [Google Scholar]
- Gölz G., Alter T., Bereswill S., Heimesaat M. M. (2016). The immunopathogenic potential of Arcobacter butzleri - lessons from a meta-analysis of murine infection studies. PLoS One 11:e0159685. 10.1371/journal.pone.0159685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goris J., Konstantinidis K. T., Klappenbach J. A., Coenye T., Vandamme P., Tiedje J. M. (2007). DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57 81–91. 10.1099/ijs.0.64483-0 [DOI] [PubMed] [Google Scholar]
- Groicher K. H., Firek B. A., Fujimoto D. F., Bayles K. W. (2000). The Staphylococcus aureus lrgAB operon modulates murein hydrolase activity and penicillin tolerance. J. Bacteriol. 182 1794–1801. 10.1128/JB.182.7.1794-1801.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Fino C., Sørensen M. A., Semsey S., Gerdes K. (2017). Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio 8:e01964-17. 10.1128/mBio.01964-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausdorf L., Neumann M., Bergmann I., Sobiella K., Mundt K., Fröhling A., et al. (2013). Occurrence and genetic diversity of Arcobacter spp. in a spinach-processing plant and evaluation of two Arcobacter-specific quantitative PCR assay. Syst. Appl. Microbiol. 36 235–243. 10.1016/j.syapm.2013.02.003 [DOI] [PubMed] [Google Scholar]
- Heimesaat M. M., Karadas G., Alutis M., Fischer A., Kühl A. A., Breithaupt A., et al. (2015). Survey of small intestinal and systemic immune responses following murine Arcobacter butzleri infection. Gut Pathog. 7:28. 10.1186/s13099-015-0075-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- ICMSF (2002). Microorganisms in Foods. 7. Microbiological Testing in food safety management. International commission on Microbiological Specifications for Foods. New York, NY: Kluwer Academic; 10.1007/978-1-4615-0745-1 [DOI] [Google Scholar]
- Jalava K., Rintala H., Ollgren J., Maunula L., Gomez-Alvarez V., Revez J., et al. (2014). Novel microbiological and spatial statistical methods to improve strength of epidemiological evidence in a community-wide waterborne outbreak. PLoS One 9:e104713. 10.1371/journal.pone.0104713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javadi M. B., Katzenmeier G. (2016). The forgotten virulence factor: the ‘non-conventional’ hemolysin tlya and its role in Helicobacter pylori infection. Curr. Microbiol. 73 930–937. 10.1007/s00284-016-1141-6 [DOI] [PubMed] [Google Scholar]
- Jiang Z. D., DuPont H. L., Brown E. L., Nandy R. K., Ramamurthy T., Sinha A., et al. (2010). Microbial etiology of travelers’ diarrhea in Mexico, Guatemala, and India: importance of enterotoxigenic Bacteroides fragilis and Arcobacter species. J. Clin. Microbiol. 48 1417–1419. 10.1128/JCM.01709-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhas M., van der Meer J. R., Gaillard M., Harding R. M., Hood D. W., Crook D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33 376–393. 10.1111/j.1574-6976.2008.00136.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junecko J. M., Zielinska A. K., Mrak L. N., Ryan D. C., Graham J. W., Smeltzer M. S., et al. (2012). Transcribing virulence in Staphylococcus aureus. World J. Clin. Infect. Dis. 2 63–76. 10.5495/wjcid.v2.i4.63 [DOI] [Google Scholar]
- Kabeya H., Maruyama S., Morita Y., Ohsuga T., Ozawa S., Kobayashi Y., et al. (2004). Prevalence of Arcobacter species in retail meats and antimicrobial susceptibility of the isolates in Japan. Int. J. Food Microbiol. 90 303–308. 10.1016/S0168-1605(03)00322-2 [DOI] [PubMed] [Google Scholar]
- Kayman T., Abay S., Hizlisoy H., Atabay H. I., Diker K. S., Aydin F. (2012). Emerging pathogen Arcobacter spp. in acute gastroenteritis: molecular identification, antibiotic susceptibilities and genotyping of the isolated Arcobacters. J. Med. Microbiol. 61 1439–1444. 10.1099/jmm.0.044594-0 [DOI] [PubMed] [Google Scholar]
- Kim H. M., Hwang C. Y., Cho B. C. (2010). Arcobacter marinus sp. nov. Int. J. Syst. Evol. Microbiol. 60 531–536. 10.1099/ijs.0.007740-0 [DOI] [PubMed] [Google Scholar]
- Kurincic M., Botteldoorn N., Herman L., Smole Mozina S. (2007). Mechanisms of erythromycin resistance of Campylobacter spp. isolated from food, animals and humans. Int. J. Food Microbiol. 120 186–189. 10.1016/j.ijfoodmicro.2007.03.012 [DOI] [PubMed] [Google Scholar]
- Lau S. K. P., Woo P. C. Y., Teng J., Leung K. W., Yuen K. Y. (2002). Identification by 16S ribosomal RNA gene sequencing of Arcobacter bacteraemia in a patient with acute gangrenous appendicitis. Mol. Pathol. 55 182–185. 10.1136/mp.55.3.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Agidi S., Marion J. W., Lee J. (2012). Arcobacter in Lake Erie beach waters: an emerging gastrointestinal pathogen linked with human-associated fecal contamination. Appl. Environ. Microbiol. 78 5511–5519. 10.1128/AEM.08009-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Sousa M. C. (2014). Structural basis for substrate specificity in ArnB. A key enzyme in the polymyxin resistance pathway of Gram-negative bacteria. Biochemistry. 53 796–805. 10.1021/bi4015677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. H., Choi C. (2013). Survival of Arcobacter butzleri in apple and pear purees. J. Food Safety 33 333–339. 10.1111/jfs.12057 [DOI] [Google Scholar]
- Lehmann D., Alter T., Lehmann L., Uherkova S., Seidler T., Gölz G. (2015). Prevalence, virulence gene distribution and genetic diversity of Arcobacter in food samples in Germany. Berl. Munch Tierarztl. Wochenschr. 128 163–168. [PubMed] [Google Scholar]
- Leoni F., Chierichetti S., Santarelli S., Talevi G., Masini L., Bartolini C., et al. (2017). Occurrence of Arcobacter spp. and correlation with the bacterial indicator of faecal contamination Escherichia coli in bivalve molluscs from the Central Adriatic, Italy Int. J. Food Microbiol. 245 6–12. 10.1016/j.ijfoodmicro.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Levican A., Collado L., Figueras M. J. (2013). Arcobacter cloacae sp. nov. and Arcobacter suis sp. nov., two new species isolated from food and sewage. Syst. Appl. Microbiol. 36 22–27. 10.1016/j.syapm.2012.11.003 [DOI] [PubMed] [Google Scholar]
- Levican A., Collado L., Yustes C., Aguilar C., Figueras M. J. (2014). Higher water temperature and incubation under aerobic and microaerobic conditions increase the recovery and diversity of Arcobacter spp. from shellfish. Appl. Environ. Microbiol. 80 385–391. 10.1128/AEM.03014-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. G., Xia Y., Zhang T. (2017). Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 11 651–662. 10.1038/ismej.2016.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Pop M. (2009). ARDB-antibiotic resistance genes database. Nucleic Acids Res. 37 D443–D447. 10.1093/nar/gkn656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupo A., Coyne S., Berendonk T. U. (2012). Origin and evolution of antibiotic resistance: the common mechanisms of emergence and spread in water bodies. Front. Microbiol. 3:18. 10.3389/fmicb.2012.00018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A., Bo Y., Han L., He J., Lanczycki C. J., Lu S., et al. (2017). CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 45 200–203. 10.1093/nar/gkw1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matilla M. A., Krell T. (2018). The effect of bacterial chemotaxis on host infection and pathogenicity. FEMS Microbiol. Rev. 42:fux052. 10.1093/femsre/fux052 [DOI] [PubMed] [Google Scholar]
- Matsumoto Y., Xu Q., Miyazaki S., Kaito C., Farr C. L., Axelrod H. L., et al. (2010). Structure of a virulence regulatory factor CvfB reveals a novel winged helix RNA binding module. Structure 18 537–547. 10.1016/j.str.2010.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Auch A. F., Klenk H.-P., Göker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. 10.1186/1471-2105-14-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Kolthoff J. P., Göker M., Klenk H.-P. (2014). Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int. J. Syst. Evol. Microbiol. 64 352–356. 10.1099/ijs.0.056994-0 [DOI] [PubMed] [Google Scholar]
- Merga J. Y., Royden A., Pandey A. K., Williams N. J. (2014). Arcobacter spp. isolated from untreated domestic effluent. Lett. Appl. Microbiol. 59 122–126. 10.1111/lam.12256 [DOI] [PubMed] [Google Scholar]
- Miller W. G., Parker C. T., Rubenfield M., Mendz G. L., Wösten M. M., Ussery D. W., et al. (2007). The complete genome sequence and analysis of the epsilonproteobacterium Arcobacter butzleri. PLoS One 2:e1358. 10.1371/journal.pone.0001358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minamino T., Macnab R. M. (2000). FliH, a soluble component of the type III flagellar export apparatus of Salmonella, forms a complex with FliI and inhibits its ATPase activity. Mol. Microbiol. 37 1494–1503. 10.1046/j.1365-2958.2000.02106.x [DOI] [PubMed] [Google Scholar]
- Mitchell A. L., Scheremetjew M., Denise H., Potter S., Tarkowska A., Qureshi M., et al. (2017). EBI Metagenomics in 2017: enriching the analysis of microbial communities, from sequence reads to assemblies. Nucleic Acids Res. 46 D726–D735. 10.1093/nar/gkx967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottola A. (2017). Emerging Pathogen Arcobacter spp. in food: Occurrence and Genetic Diversity. Ph.D. thesis, University of Bari “Aldo Moro”, Bari. [Google Scholar]
- Mottola A., Bonerba E., Figueras M. J., Pérez-Cataluña A., Marchetti P., Serraino A., et al. (2016). Occurrence of potentially pathogenic Arcobacters in shellfish. Food Microbiol. 57 23–27. 10.1016/j.fm.2015.12.010 [DOI] [PubMed] [Google Scholar]
- Naas T., Oueslati S., Bonnin R. A., Dabos M. L., Zavala A., Dortet L., et al. (2017). Beta-Lactamase DataBase (BLDB) – Structure and Function. J. Enzyme Inhib. Med. Chem. 32 917–919. 10.1080/14756366.2017.1344235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolosi D., Scalia M., Nicolosi V. M., Pignatello R. (2010). Encapsulation in fusogenic liposomes broadens the spectrum of action of vancomycin against Gram-negative bacteria. Int. J. Antimicrob. Agent 35 553–558. 10.1016/j.ijantimicag.2010.01.015 [DOI] [PubMed] [Google Scholar]
- Nieva-Echevarria B., Martinez-Malaxetxebarria I., Girbau C., Alonso R., Fernández-Astorga A. (2013). Prevalence and genetic diversity of Arcobacter in food products in the north of Spain. J. Food Prot. 76 1447–1450. 10.4315/0362-028X.JFP-13-014 [DOI] [PubMed] [Google Scholar]
- Nilsson I., Prathapam R., Grove K., Lapointe G., Six D. A. (2018). The sialic acid transporter NanT is necessary and sufficient for uptake of 3-deoxy-d-manno-oct-2-ulosonic acid (Kdo) and its azido analog in Escherichia coli. Mol. Microbiol. 110 204–218. 10.1111/mmi.14098 [DOI] [PubMed] [Google Scholar]
- Oldfield N. J., Moran A. P., Millar L. A., Prendergast M. M., Ketley J. M. (2002). Characterization of the Campylobacter jejuni heptosyltransferase II gene, waaF, provides genetic evidence that extracellular polysaccharide is lipid A core independent. J. Bacteriol. 184 2100–2107. 10.1128/JB.184.8.2100-2107.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- On S. L. W., Miller W. G., Houf K., Fox J. G., Vandamme P. (2017). Minimal standards for describing new species belonging to the families Campylobacteraceae and Helicobacteraceae: Campylobacter, Arcobacter, Helicobacter and Wolinella spp. Int. J. Syst. Evol. Microbiol. 67 5296–5311. 10.1099/ijsem.0.002255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otth L., Solís G., Wilson M., Fernández H. (2005). Susceptibility of Arcobacter butzleri to heavy metals. Braz. J. Microbiol. 36 286–288. 10.1590/S1517-83822005000300015 [DOI] [Google Scholar]
- Otth L., Wilson M., Cancino R., Fernández H. (2004). In vitro susceptibility of Arcobacter butzleri to six antimicrobial drugs. Arch. Med. Vet. 36 207–210. 10.4067/S0301-732X2004000200012 [DOI] [Google Scholar]
- Park S., Jung Y. T., Kim S., Yoon J. H. (2016). Arcobacter acticola sp. nov isolated from seawater on the East Sea in South Korea. J. Microbiol. 54 655–659. 10.1007/s12275-016-6268-4 [DOI] [PubMed] [Google Scholar]
- Patyal A., Rathore R. S., Mohan H. V., Dhama K., Kumar A. (2011). Prevalence of Arcobacter spp. in humans, animals and foods of animal origin including sea food from India. Transboundary Emerg. Dis. 58 402–410. 10.1111/j.1865-1682.2011.01221.x [DOI] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Salas-Massó N., Figueras M. J. (2018a). Arcobacter canalis sp. nov., isolated from a water canal contaminated with urban sewage. Int. J. Syst. Evol. Microbiol. 68 1258–1264. 10.1099/ijsem.0.002662 [DOI] [PubMed] [Google Scholar]
- Pérez-Cataluña A., Salas-Massó N., Diéguez A. L., Balboa S., Lema A., Romalde J. L., et al. (2018b). Corrigendum: Revisiting the taxonomy of the genus Arcobacter: getting order from the chaos. Front. Microbiol. 9:3123. 10.3389/fmicb.2018.03123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips C. A. (2001). Arcobacters as emerging human foodborne pathogens. Food Control. 12 1–6. 10.1016/S0956-7135(00)00024-4 [DOI] [Google Scholar]
- Poole K. (2017). At the nexus of antibiotics and metals: the impact of Cu and Zn on antibiotic activity and resistance. Trends Microbiol. 25 820–832. 10.1016/j.tim.2017.04.010 [DOI] [PubMed] [Google Scholar]
- Quintiliani R., Jr., Courvalin P. (1995). “Mechanisms of resistance to antimicrobial agents,” in Manual of Clinical Microbiology, 6th Edn, eds Murray P. R., Baron E. J., Pfaller M. A., Tenover F. C., Yolken R. H. (Washington, DC: ASM Press; ), 1319. [Google Scholar]
- Rahimi E. (2014). Prevalence and antimicrobial resistance of Arcobacter species isolated from poultry meat in Iran. Br. Poult. Sci. 55 174–180. 10.1080/00071668.2013.878783 [DOI] [PubMed] [Google Scholar]
- Ramees T. P., Dhama K., Karthik K., Rathore R. S., Kumar A., Saminathan M., et al. (2017). Arcobacter: an emerging food-borne zoonotic pathogen, its public health concerns and advances in diagnosis and control – a comprehensive review. Vet. Q. 37 136–161. 10.1080/01652176.2017.1323355 [DOI] [PubMed] [Google Scholar]
- Ramees T. P., Rathore R. S., Bagalkot P. S., Ravi Kumar G. V. P. P. S., Mohan H. V., Anoopraj R., et al. (2014). Real- time PCR detection of Arcobacter butzleri and Arcobacter cryaerophilus in chicken meat samples. J. Pure Appl. Microbiol. 8 3165–3169. 10.1089/fpd.2009.0368 [DOI] [PubMed] [Google Scholar]
- Rathlavath S., Kohli V., Singh A. S., Lekshmi M., Tripathi G., Kumar S., et al. (2017). Virulence genotypes and antimicrobial susceptibility patterns of Arcobacter butzleri isolated from seafood and its environment. Int. J. Food Microbiol. 263 32–37. 10.1016/j.ijfoodmicro.2017.10.005 [DOI] [PubMed] [Google Scholar]
- Richards V. P., Lefébure T., Pavinski Bitar P. D., Stanhope M. J. (2013). Comparative characterization of the virulence gene clusters (lipooligosaccharide [LOS] and capsular polysaccharide [CPS]) for Campylobacter coli, Campylobacter jejuni subsp. jejuni and related Campylobacter species. Infect. Genet. Evol. 14 200–213. 10.1016/j.meegid.2012.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riesenberg A., Frömke C., Stingl K., Feßler A. T., Gölz G., Glocker E. O., et al. (2017). Antimicrobial susceptibility testing of Arcobacter butzleri: development and application of a new protocol for broth microdilution. J. Antimicrob. Chemother. 72 2769–2774. 10.1093/jac/dkx211 [DOI] [PubMed] [Google Scholar]
- Rodriguez-R L. M., Konstantinidis K. T. (2016). The enveomics collection: a toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ. Preprints 4:e1900v1. [Google Scholar]
- Rossmann F. M., Beeby M. (2018). Insights into the evolution of bacterial flagellar motors from high-throughput in situ electron cryotomography and subtomogram averaging. Acta Crystallogr. D. Struct. Biol. 74 585–594. 10.1107/S2059798318007945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovetto F., Carlier A., Van den Abeele A. M., Illeghems K., Van Nieuwerburgh F., Cocolin L., et al. (2017). Characterization of the emerging zoonotic pathogen Arcobacter thereius by whole genome sequencing and comparative genomics. PLoS One 12:e0180493. 10.1371/journal.pone.0180493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah A. H., Saleha A. A, Murugaiyah M., Zunita Z., Memon A. A. (2012). Prevalence and distribution of Arcobacter spp. in raw milk and retail raw beef. J. Food Protect. 75 1474–1478. 10.4315/0362-028X.JFP-11-487 [DOI] [PubMed] [Google Scholar]
- Scanlon K. A., Cagney C., Walsh D., McNulty D., Carroll A., McNamara E. B., et al. (2013). Occurrence and characteristics of fastidious Campylobacteraceae species in porcine samples. Int. J. Food Microbiol. 163 6–13. 10.1016/j.ijfoodmicro.2013.02.004 [DOI] [PubMed] [Google Scholar]
- Schauer Weissfeld A. (2014). Infections from eating raw or undercooked seafood. Clin. Microbiol. Newsl. 36 17–21. 10.1016/j.clinmicnews.2014.01.004 [DOI] [Google Scholar]
- Seiler C., Berendonk T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. 10.3389/fmicb.2012.00399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šilha D., Pejchalová M., Šilhová L. (2017). Susceptibility to 18 drugs and multidrug resistance of Arcobacter isolates from different sources within the Czech Republic. J. Glob. Antimicrob. Resist. 9:74e77. 10.1016/j.jgar.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Soma S. M., Srinivasa R. T., Bindu K. C. H., Subramanyam K. V., Mohammad S. N. (2017). Antibiogram of Arcobacter species isolated from animals, foods of animal origin and humans in Andhra Pradesh, India. Int. J. Sci. Environ. Tech. 6 1260–1269. 10.14202/vetworld.2017.716-720 [DOI] [Google Scholar]
- Talay F., Molva C., Atabay H. I. (2016). Isolation and identification of Arcobacter species from environmental and drinking water samples. Folia Microbiol. 61 479–484. 10.1007/s12223-016-0460-0 [DOI] [PubMed] [Google Scholar]
- Tatusova T., DiCuccio M., Badretdin A., Chetvernin V., Nawrocki E. P., Zaslavsky L., et al. (2016). NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44 6614–6624. 10.1093/nar/gkw569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UniProt Consortium (2017). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45 D158–D169. 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S., Jamal S. B., Hassan S. S., Carvalho P. V. S. D., Almeida S., Barh D., et al. (2017). Two-component signal transduction systems of pathogenic bacteria as targets for antimicrobial therapy: an overview. Front. Microbiol. 8:1878. 10.3389/fmicb.2017.01878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abeele A. M., Vogelaers D., Van Hende J., Houf K. (2014). Prevalence of Arcobacter species among humans, Belgium, 2008–2013. Emerg. Infect. Dis. 20 1731–1734. 10.3201/eid2010.140433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Abeele A. M., Vogelaers D., Vanleare E., Houf K. (2016). Antimicrobial susceptibility testing of Arcobacter butzleri and Arcobacter cryaerophilus strains isolated from Belgian patients. J. Antimicrob. Chemother. 71 1241–1244. 10.1093/jac/dkv483 [DOI] [PubMed] [Google Scholar]
- Vandamme P., Vancanneyt M., Pot B., Mels L., Hoste B., Dewettinck D., et al. (1992). Polyphasic taxonomic study of the emended genus Arcobacter with Arcobacter butzleri comb. nov. and Arcobacter skirrowii sp. nov., an Aerotolerant bacterium isolated from veterinary specimens. Int. J. Syst. Bacteriol. 42 344–345. 10.1099/00207713-42-3-344 [DOI] [PubMed] [Google Scholar]
- Vandenberg O., Houf K., Douat N., Vlaes L., Retore P., Butzler J. P., et al. (2006). Antimicrobial susceptibility of clinical isolates of non-jejuni/coli Campylobacters and Arcobacters from Belgium. J. Antimicrob. Chemot. 57 908–913. 10.1093/jac/dkl080 [DOI] [PubMed] [Google Scholar]
- Vicente-Martins S., Oleastro M., Domingues F. C., Ferreira S. (2018). Arcobacter spp. at retail food from Portugal: prevalence, genotyping and antibiotics resistance. Food Control. 85 107–112. 10.1016/j.foodcont.2017.09.024 [DOI] [Google Scholar]
- Waite D. W., Vanwonterghem I., Rinke C., Parks D. H., Zhang Y., Takai K., et al. (2017). Comparative genomic analysis of the class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front. Microbiol. 8:682 10.3389/fmicb.2017.00682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite D. W., Vanwonterghem I., Rinke C., Parks D. H., Zhang Y., Takai K., et al. (2018). Erratum: Addendum: comparative genomic analysis of the Class Epsilonproteobacteria and proposed reclassification to Epsilonbacteraeota (phyl. nov.). Front. Microbiol. 18:772. 10.3389/fmicb.2018.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb A. L., Boras V. F., Kruczkiewicz P., Selinger L. B., Taboada E. N., Inglis G. D. (2016). Comparative detection and quantification of Arcobacter butzleri in stools from diarrheic and non-diarrheic human beings in southwestern Alberta, Canada. J. Clinic. Microbiol. 54 1082–1088. 10.1128/JCM.03202-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. N. (2014). Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12 35–48. 10.1038/nrmicro3155 [DOI] [PubMed] [Google Scholar]
- Wu D., Dolfing J., Xie B. (2018). Bacterial perspectives on the dissemination of antibiotic resistance genes in domestic wastewater bio-treatment systems: beneficiary to victim. Appl. Microbiol. Biotechnol. 102 597–604. 10.1007/s00253-017-8665-y [DOI] [PubMed] [Google Scholar]
- Yesilmen S., Vural A., Erkan M. E., Yildirim I. H. (2014). Prevalence and antimicrobial susceptibility of Arcobacter species in cow milk, water buffalo milk and fresh village cheese. Int. J. Food Microbiol. 188 11–14. 10.1016/j.ijfoodmicro.2014.07.006 [DOI] [PubMed] [Google Scholar]
- Yu Z., Gunn L., Wall P., Fanning S. (2017). Antimicrobial resistance and its association with tolerance to heavy metals in agriculture production. Food Microbiol. 64 23–32. 10.1016/j.fm.2016.12.009 [DOI] [PubMed] [Google Scholar]
- Zacharow I., Bystroń J., Wałecka-Zacharska E., Podkowik M., Bania J. (2015a). Genetic diversity and incidence of virulence-associated genes of Arcobacter butzleri and Arcobacter cryaerophilus isolates from pork, beef, and chicken meat in Poland. Biomed. Res. Int. 2015:ID956507. 10.1155/2015/956507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharow I., Bystroń J., Wałecka-Zacharska E., Podkowik M., Bania J. (2015b). Prevalence and antimicrobial resistance of Arcobacter butzleri and Arcobacter cryaerophilus isolates from retail meat in lower Silesia region, Poland. Pol. J. Vet. Sci. 18 63–69. [DOI] [PubMed] [Google Scholar]
- Zhu S., Nishikino T., Hu B., Kojima S., Homma M., Liu J. (2017). Molecular architecture of the sheathed polar flagellum in Vibrio alginolyticus. Proc. Natl. Acad. Sci. U.S.A. 114 10966–10971. 10.1073/pnas.1712489114 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.