Abstract
Objectives
Previous work showed that gag-protease-derived phenotypic susceptibility to PIs differed between HIV-1 subtype CRF02_AG/subtype G-infected patients who went on to successfully suppress viral replication versus those who experienced virological failure of lopinavir/ritonavir monotherapy as first-line treatment in a clinical trial. We analysed the relationship between PI susceptibility and outcome of second-line ART in Nigeria, where subtypes CRF02_AG/G dominate the epidemic.
Methods
Individuals who experienced second-line failure with ritonavir-boosted PI-based ART were matched (by subtype, sex, age, viral load, duration of treatment and baseline CD4 count) to those who achieved virological response (‘successes’). Successes were defined by viral load <400 copies of HIV-1 RNA/mL by week 48. Full-length Gag-protease was amplified from patient samples for in vitro phenotypic susceptibility testing, with PI susceptibility expressed as IC50 fold change (FC) relative to a subtype B reference strain.
Results
The median (IQR) lopinavir IC50 FC was 4.04 (2.49–7.89) for virological failures and 4.13 (3.14–8.17) for virological successes (P = 0.94). One patient had an FC >10 for lopinavir at baseline and experienced subsequent virological failure with ritonavir-boosted lopinavir as the PI. There was no statistically significant difference in single-round replication efficiency between the two groups (P = 0.93). There was a moderate correlation between single-round replication efficiency and FC for lopinavir (correlation coefficient 0.32).
Conclusions
We found no impact of baseline HIV-1 Gag-protease-derived phenotypic susceptibility on outcomes of PI-based second-line ART in Nigeria.
Introduction
Prevalence of virological failure for first-line antiretroviral therapy can be as high as 30%,1 with high-level resistance to NNRTI, tenofovir and cytosine analogues common in resource-limited settings and compounded by prior undisclosed ART.2,3 Second-line ART recommended by WHO comprises a ritonavir-boosted PI and two NRTIs, commonly lopinavir or atazanavir.4 PIs are the second- and last-line therapy for the majority of HIV-infected patients worldwide as access to third-line therapy is still limited.5 Virological failure with PIs as second-line therapy occurs in around 20% of individuals.6–8 In contrast to first-line therapy, with which >80% develop drug resistance mutations, only around 10%–20% develop major resistance mutations to PIs by week 48,6,7,9,10 and this proportion increases over time.5
It is known that proteins such as Gag and Env can affect susceptibility to PIs even in the absence of known major resistance mutations in the protease gene.11–15 There are limited data on changes in gag following treatment failure with PIs in the non-B subtypes that dominate low- and middle-income countries.15–20 It appears that in around 15% of patients failing boosted PI (bPI) without major protease mutations, a decrease in phenotypic susceptibility to the drug appears to occur when gag-protease is phenotyped.21–23 Therefore it is conceivable that underlying phenotypic susceptibility resulting from variation in genes such as gag and env might impact clinical responses to PI.
We previously showed that gag-protease-derived phenotypic susceptibility differed between CRF02_AG and subtype G-infected patients who went on to successfully suppress viral replication versus those who experienced virological failure (VF) of lopinavir/ritonavir monotherapy as first-line treatment in a clinical trial.12 In order to determine the relevance of this finding for real-world settings in the context of tenofovir disoproxil fumarate + lamivudine or zidovudine + lamivudine with ritonavir-boosted PI (lopinavir or atazanavir) we analysed the relationship between PI susceptibility and the outcome of PI-based second-line ART in Nigeria, where subtypes CRF02_AG and G dominate the epidemic.24
Patients and methods
Study participants
This study involved retrospectively testing samples from patients attending for HIV care at University of Abuja Teaching Hospital (UATH) who experienced second-line failure (HIV-1 RNA >1000 copies/mL after >6 months on treatment) on a lopinavir/ritonavir- or atazanavir/ritonavir-containing regimen, without any major PI mutations, who were selected as ‘cases’. They were matched to ‘controls’, who had achieved virological suppression lasting up to 12 months (HIV-1 RNA <400 copies/mL) with a similar age, sex, baseline CD4 count and duration of treatment. Baseline (pre-PI) plasma samples from these matched pairs were retrospectively retrieved.
Amplification of full-length gag-protease genes
HIV-1 RNA was manually extracted from archived plasma samples using the QIAamp viral RNA extraction kit. Using previously described techniques,11,25 full-length gag-protease was amplified and cloned into a subtype B-based (p8.9NSX+) vector. Clonal sequencing of up to 10 plasmids (where possible) was performed by standard Sanger sequencing. The variant that most closely represented the consensus (obtained via next-generation sequencing as previously described6) was taken forward for phenotypic testing. Sequences were manually analysed using DNA dynamo software (http://www.bluetractorsoftware.co.uk) and MEGA v7.0 software.26 Protease sequences were analysed for PI resistance mutations using the Stanford Resistance Database (https://hivdb.stanford.edu).
PI susceptibility and infectivity assays
PI susceptibility and viral infectivity were determined using a previously described single assay. Briefly, 293T cells were co-transfected with a Gag-Pol protein expression vector (p8.9NSX+) containing cloned patient-derived gag-protease sequences, pMDG expressing vesicular stomatitis virus envelope glycoprotein (VSV-g), and pCSFLW (expressing the firefly luciferase reporter gene with HIV-1 packaging signal).
PI drug susceptibility testing was carried out as previously described.25 Transfected cells were seeded with serial dilutions of lopinavir and harvested pseudovirions were used to infect fresh 293T cells. To determine strain infectivity, transfected cells were seeded in the absence of drug. Infectivity was monitored by measuring luciferase activity 48 h after infection. Results derived from at least two independent experiments (each in duplicate) were analysed. The IC50 was calculated using GraphPad Prism 5 (GraphPad Software Inc., La Jolla, CA, USA). Susceptibility was expressed as a fold change in IC50 compared with the subtype B reference strain (p8.9NSX+). Replicative capacity of these viruses was assessed by comparing the luciferase activity of recombinant virus with that of the WT subtype B control virus in the absence of drug. Equal amounts of input plasmid DNA were used, and it has previously been shown that percentage infectivity correlates well with infectivity/ng p24 in this system.25 The PI drugs used in this study were obtained from the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH.
Ethics
Informed consent was obtained from all subjects and ethics approval for virological testing was obtained from the National Research Ethics Committee of Nigeria (NHREC/01/01/2007).
Statistical analysis
Differences in PI susceptibility were compared with the Wilcoxon rank-sum test (GraphPad Software Inc., La Jolla, CA, USA), which is robust to data that are not normally distributed.
Results
Six matched pairs of patients were included. Table 1 contains clinical and laboratory data on cases, who experienced virological failure (duration), and controls, who suppressed viral replication for 48 weeks. Of note, all pairs but one had a CD4 count <200 cells/mm3. All but one pair was treated with lopinavir-based ART (atazanavir was used in one pair). Table 2 shows NRTI and NNRTI resistance mutations detected prior to second-line initiation. All patients had lamivudine resistance [M184V/I in reverse transcriptase (RT)] and 7/12 (58.3%) had at least moderate resistance to tenofovir (3 with K65R, 3 with K70E and 1 with three thymidine analogue mutations including M41L, L210W and T215Y). All 12 individuals had high-level NNRTI resistance. Two pairs were infected with subtype G viruses and four pairs with CRF02_AG viruses (Table 2). No major mutations in protease were observed in the patients. We analysed sequences for mutations in Gag in cases and controls associated with PI susceptibility or exposure (Table 3).
Table 1.
Clinical data for matched patient pairs comprising virological successes and failures
| Sample pair | Age (years) |
Sex |
Baselinea CD4 count (cells/mm3) |
Second-line PI used |
Baselinea viral load (copies of HIV-1 RNA/mL) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|
| success | failure | success | failure | success | failure | success | failure | success | failure | |
| 1 | 33 | 45 | female | female | <200 | <200 | LPV | LPV | 503 951 | 140 991 |
| 2 | 43 | 36 | female | female | <200 | <200 | LPV | LPV | 39 844 | 20 178 |
| 3 | 27 | 26 | female | female | <200 | <200 | LPV | LPV | 32 284 | 271 974 |
| 4 | 47 | 39 | female | female | 200–499 | 200–499 | LPV | LPV | 228 083 | 24 693 |
| 5 | 35 | 40 | female | female | <200 | <200 | ATV | ATV | 14 487 | 274 504 |
| 6 | 34 | 33 | female | female | <200 | <200 | LPV | LPV | 39 929 | 18 056 |
LPV, lopinavir; ATV, atazanavir.
Baseline refers to pre-initiation of second-line therapy.
Table 2.
NRTI and NNRTI mutations observed at first-line failure, prior to initiation of second-line PI-based ART
| NRTI mutations | NNRTI mutations | Baselinea VL (copies of HIV-1 RNA/mL) |
HIV-1 subtype |
2L backbone |
|||||
|---|---|---|---|---|---|---|---|---|---|
| success | failure | success | failure | success | failure | ||||
| Pair 1 | success | M41L, L74LI, M184V, L210W, T215F | K101E, E138Q, G190A | 503 951 | 140 991 | CRF02_AG | CRF02_AG | TDF/FTC | TDF/FTC |
| failure | M184V | K103N | |||||||
| Pair 2 | success | E44D, D67N, T69D, K70R, M184V, T215Y | K101E, K103N | 39 844 | 20 178 | CRF02_AG | CRF02_AG | TDF/FTC | TDF/FTC |
| failure | M184V | K101E, G190A | |||||||
| Pair 3 | success | K70E, M184V | A98G, Y181C | 32 284 | 271 974 | G | G | TDF/FTC | TDF/FTC |
| failure | K70E, M184V | Y181C, G190A, H221Y | |||||||
| Pair 4 | success | K70E, Y115F, M184V | K103N | 228 083 | 24 693 | CRF02_AG | CRF02_AG | AZT/3TC | AZT/3TC |
| failure | K65R, M184V | K101E, V108I, Y181C, G190A | |||||||
| Pair 5 | success | D67N, K70R, M184V, T215F, K219E | Y188C | 14 487 | 274 504 | G | G | TDF/FTC | TDF/FTC |
| failure | K70R, M184V, K219Q | K103N, Y318F | |||||||
| Pair 6 | success | K65R, M184I | K103N, Y181C | 39 929 | 18 056 | CRF02_AG | CRF02_AG | TDF/FTC | TDF/FTC |
| failure | K65R, M184I | K103N, Y181C | |||||||
AZT, zidovudine; 3TC, lamivudine; FTC, emtricitabine; TDF, tenofovir disoproxil fumarate.
Baseline refers to pre-initiation of second-line therapy.
Table 3.
Variation in Gag cleavage and non-cleavage sites and protease
| HIV-1 subtype | Treatment outcome | Gag cleavage sites |
Gag non-cleavage sites | |||||
|---|---|---|---|---|---|---|---|---|
| MA/CA 128–137 VSQNY/ PIVQN | CA/p2 359–368 KARVL/ AEAMS | p2/NC 373–382 SATIM/ MQRGN | NC/p1 428–437 ERQAN/ FLGKI | p1/p6 444–453 RPGNF/ LQSRP | ||||
| Pair 1 | CRF02_AG | success | -----/----- | -----/----- | -+N--/----- | -----/----- | -----/P---- | E12K, R76K, R79F, H219Q, T242N, R409K, T487S |
| failure | -----/----- | -----/----- | T +N--/----- | -----/----- | -----/P---- | R76K, R79F, T81A, G248A, V370A, R409K, E468K, T487S | ||
| Pair 2 | G | success | -----/----- | -----/----- | A-A--/----- | -----/----L | -----/--N-- | E12K, R76K, R79F, G123E, H219Q, G248A, V370A, R409K, T487S |
| failure | -----/----- | -----/----- | ---+-/---S- | -----/----- | -----/--N-L | E12K, G62R, R76K, H219Q, V370A, R409K, T487S | ||
| Pair 3 | G | success | -----/----- | -----/----- | --A--/--KS- | -----/----- | -----/--N-L | E12K, G62R, R76K, H219Q, T242N, G248A, V370A, R409K, T487S |
| failure | -----/----- | -----/----- | -+NV-/--K-- | -----/----- | -----/P---- | E12K, R76K, R79F, H219Q, T242N, V370A, T371del, R409K, T456S, T487S | ||
| Pair 4 | CRF02_AG | success | +----/----- | -----/----- | T+NV-/--K-- | -----/----- | -----/P---- | R76K, R409K, T487S |
| failure | +----/----- | -----/----- | -+NV-/-K-- | -----/----- | -----/P---- | E12K, R76K, R79F, G248A, V370A, R409K, T487S | ||
| Pair 5 | G | success | -----/----- | -----/----- | A-A--/----- | -----/---R- | -----/----- | E12K, R76K, R79F, G123E, V370A, R409K, T487S |
| failure | +---F/----- | -----/----- | QPN--/I---- | -----/----V | -----/P---- | E12K, R76K, R79F, S165N, H219Q, V370A, R409K, T487S | ||
| Pair 6 | CRF02_AG | success | S----/----- | ---I-/----- | ANI--/----- | -----/----- | -----/P---- | E12K, R76K, R79F, V370A, R409K, T487S |
| failure | +----/----- | -----/----- | T+NV-/--K-- | -----/----- | -----/P---- | E12K, G248A, V370A, R409K, S451T, T487S | ||
The consensus clonal sequence at each of the Gag cleavage sites is shown for the six pairs using HXB2 numbering. Deletions are represented by +.
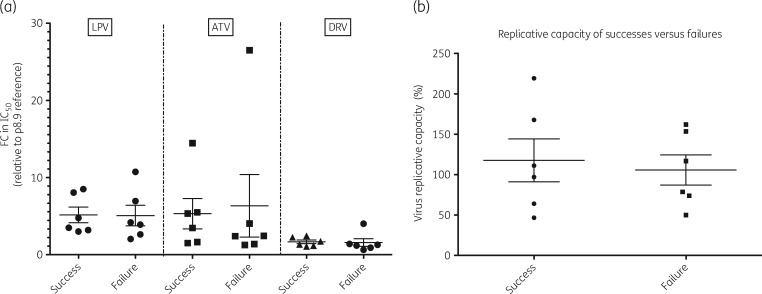
The median (IQR) lopinavir fold change (FC) was 4.04 (2.49–7.89) for virological failures and 4.13 (3.14–8.17) for virological successes (P = 0.94), as described in Figure 1(a). The median (IQR) atazanavir FC was 2.43 (1.35–9.66) for virological failures and 4.39 (1.60–7.73) for successes (P = 0.47). The median (IQR) darunavir FC was 1.234 (0.84–2.05) for virological failures and 1.529 (1.14–2.319) for successes (P = 0.47).
Figure 1.
(a) PI susceptibility relative to a subtype B reference strain, expressed as FC in IC50, and (b) single-round replication efficiency (relative to a subtype B reference strain) of patient-derived gag-protease-containing pseudoviruses derived from patients prior to initiation of second-line boosted PI treatment who either did (Success) or did not (Failure) suppress viral replication after 48 weeks. Each data point is the mean of at least two independent experiments and hairs represent mean and SD. LPV, lopinavir; ATV, atazanavir; DRV, darunavir.
One patient had an FC >10 for lopinavir at baseline and experienced subsequent virological failure on boosted lopinavir as the PI. We also measured the single-round replication efficiency of patient-derived gag-protease-containing pseudoviruses derived prior to initiation of second-line boosted PI treatment from patients who either did (success) or did not (failure) suppress viral replication after 48 weeks (Figure 1b). Mean replication efficiency relative to a subtype B reference strain was 117.7% for the successes and 105.8% for failures. There was no statistically significant difference in replication efficiency between the two groups (P = 0.93 by Mann–Whitney U-test).
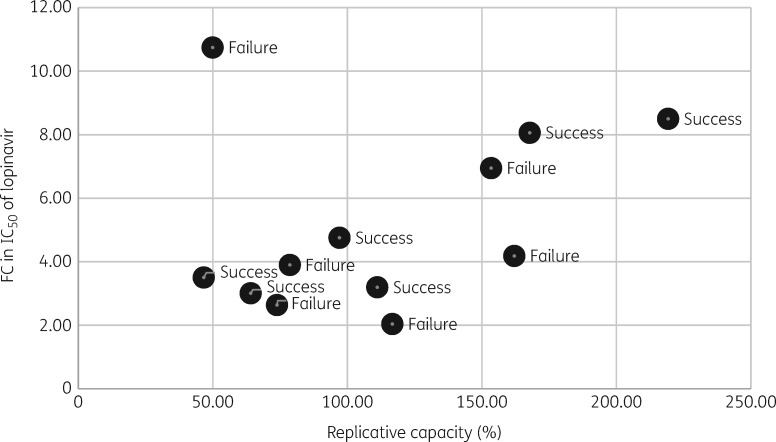
Finally, we analysed the relationship between single-round replication efficiency and FC to lopinavir in all viruses tested. There was a moderate correlation between these parameters (correlation coefficient 0.32, Figure 2). When a single outlier was excluded from analysis (FC 10.7 with replication efficiency 50.0%), the correlation coefficient increased to 0.78.
Figure 2.
Scatter plot of FC in IC50 of lopinavir relative to a subtype B reference strain, versus replicative capacity as measured over a single round of replication (also relative to a subtype B reference strain, which is represented by 100%).
Discussion
Given the contribution of the highly polymorphic Gag protein and resulting epistatic interactions to PI susceptibility, we hypothesized that patients would respond differently to these drugs, particularly in the context of extensive NRTI resistance. We previously reported an association between susceptibility to PI and outcome of first-line ritonavir-boosted lopinavir monotherapy in a clinical trial. Here we performed a similar study in patients about to start second-line combination ART, including ritonavir-boosted lopinavir or atazanavir as well as two NRTIs. We found the difference in phenotypic drug susceptibility (assessed by FC relative to a subtype B reference) was not statistically different between the virological failures (cases) and virological successes (controls) for any of the PIs tested: lopinavir, atazanavir or darunavir.
This negative result could be due to the influence of adherence, in that second-line therapy is used in patients for whom first-line therapy has failed, usually as the result of incomplete adherence. Therefore, the patient group was enriched for poor adherers, which could have overcome the effects of small differences in susceptibility.
Interestingly, we previously showed that 2/2 patients with FC >10 prior to PI monotherapy went on to virological failure.27 In this study the only patient with FC >10 for lopinavir failed treatment with this drug. Further work needs to be undertaken to explore whether a threshold FC of 10 in our assay is relevant in larger datasets.
We also showed here that replication efficiency over a single round was correlated with lopinavir susceptibility prior to initiation of the bPI. We have previously reported similar findings in replication-competent subtype C viruses that contained patient-derived gag and partial protease genes.12 These data suggest that increased replicative capacity and resistance to PI might involve an overlapping mechanism.
Limitations
Limitations of our study include the relatively small sample size, the inclusion of more than one subtype and the possibility of viral recombination through our PCR and cloning strategy. In addition, the process of mapping next-generation sequencing reads to a consensus reference sequence to generate a patient consensus can introduce biases against variation, which may affect the identification of novel drug resistance mutations. Finally, our assay system did not incorporate the native gp160 envelope.
Despite introduction of second-generation integrase inhibitors such as dolutegravir as first-line therapy in areas where pre-treatment resistance is >10%,28,29 bPI will still be used as second-line therapy for those who fail dolutegravir-based first-line regimens. Therefore, research into determinants of responses to PI in non-B subtypes is as important as ever.
Acknowledgements
We thank Katherine Sutherland, Chris Parry, Dami Collier and Petra Mlcochova.
Funding
This work was funded by a Wellcome Trust Senior Fellowship in Clinical Science to R. K. G. (WT108082AIA) and the University College London Hospitals Biomedical Research Centre.
Transparency declarations
None to declare.
References
- 1. Boender TS, Sigaloff KC, McMahon JH. et al. Long-term virological outcomes of first-line antiretroviral therapy for HIV-1 in low- and middle-income countries: a systematic review and meta-analysis. Clin Infect Dis 2015; 61: 1453–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.TenoRes Study Group. Global epidemiology of drug resistance after failure of WHO recommended first-line regimens for adult HIV-1 infection: a multicentre retrospective cohort study. Lancet Infect Dis 2016; 16: 565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gregson J, Kaleebu P, Marconi VC. et al. Occult HIV-1 drug resistance to thymidine analogues following failure of first-line tenofovir combined with a cytosine analogue and nevirapine or efavirenz in sub Saharan Africa: a retrospective multi-centre cohort study. Lancet Infect Dis 2016; 16: 565–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO. Consolidated Guidelines on the Use of Antiretroviral Drugs for Treating and Preventing HIV Infection: Recommendations for a Public Health Approach—Second Edition http://www.who.int/hiv/pub/arv/arv-2016/en.
- 5. Rawizza HE, Chaplin B, Meloni ST. et al. Accumulation of protease mutations among patients failing second-line antiretroviral therapy and response to salvage therapy in Nigeria. PLoS One 2013; 8: e73582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Collier D, Iwuji C, Derache A. et al. Virological outcomes of second-line protease inhibitor-based treatment for human immunodeficiency virus type 1 in a high-prevalence rural South African setting: a competing-risks prospective cohort analysis. Clin Infect Dis 2017; 64: 1006–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hosseinipour MC, Gupta RK, Van Zyl G. et al. Emergence of HIV drug resistance during first- and second-line antiretroviral therapy in resource-limited settings. J Infect Dis 2013; 207 Suppl 2: S49–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ajose O, Mookerjee S, Mills EJ. et al. Treatment outcomes of patients on second-line antiretroviral therapy in resource-limited settings: a systematic review and meta-analysis. AIDS 2012; 26: 929–38. [DOI] [PubMed] [Google Scholar]
- 9. Stockdale AJ, Saunders MJ, Boyd MA. et al. Effectiveness of protease inhibitor/nucleos(t)ide reverse transcriptase inhibitor-based second-line antiretroviral therapy for the treatment of human immunodeficiency virus type 1 infection in sub-Saharan Africa: a systematic review and meta-analysis. Clin Infect Dis 2018; 66: 1846–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Doyon L, Croteau G, Thibeault D. et al. Second locus involved in human immunodeficiency virus type 1 resistance to protease inhibitors. J Virol 1996; 70: 3763–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gupta RK, Kohli A, McCormick AL. et al. Full-length HIV-1 Gag determines protease inhibitor susceptibility within in vitro assays. AIDS 2010; 24: 1651–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sutherland KA, Collier DA, Claiborne DT. et al. Wide variation in susceptibility of transmitted/founder HIV-1 subtype C isolates to protease inhibitors and association with in vitro replication efficiency. Sci Rep 2016; 6: 38153.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rabi SA, Laird GM, Durand CM. et al. Multi-step inhibition explains HIV-1 protease inhibitor pharmacodynamics and resistance. J Clin Invest 2013; 123: 3848–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nijhuis M, van Maarseveen NM, Lastere S. et al. A novel substrate-based HIV-1 protease inhibitor drug resistance mechanism. PLoS Med 2007; 4: e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fun A, Wensing AM, Verheyen J. et al. Human immunodeficiency virus Gag and protease: partners in resistance. Retrovirology 2012; 9: 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sutherland KA, Goodall RL, McCormick A. et al. Gag-protease sequence evolution following protease inhibitor monotherapy treatment failure in HIV-1 viruses circulating in East Africa. AIDS Res Hum Retroviruses 2015; 31: 1032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Giandhari J, Basson AE, Coovadia A. et al. Genetic changes in HIV-1 Gag-protease associated with protease inhibitor-based therapy failure in paediatric patients. AIDS Res Hum Retroviruses 2015; 31: 776–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li G, Verheyen J, Theys K. et al. HIV-1 Gag C-terminal amino acid substitutions emerging under selective pressure of protease inhibitors in patient populations infected with different HIV-1 subtypes. Retrovirology 2014; 11: 79.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jinnopat P, Isarangkura-na-ayuthaya P, Utachee P. et al. Impact of amino acid variations in Gag and protease of HIV type 1 CRF01_AE strains on drug susceptibility of virus to protease inhibitors. J Acquir Immune Defic Syndr 2009; 52: 320–8. [DOI] [PubMed] [Google Scholar]
- 20. Coetzer M, Ledingham L, Diero L. et al. Gp41 and Gag amino acids linked to HIV-1 protease inhibitor-based second-line failure in HIV-1 subtype A from Western Kenya. J Intern AIDS Soc 2017; 20: e25024.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sutherland KA, Mbisa JL, Cane PA. et al. Contribution of Gag and protease to variation in susceptibility to protease inhibitors between different strains of subtype B HIV-1. J Gen Virol 2014; 95: 190–200. [DOI] [PubMed] [Google Scholar]
- 22. Sutherland KA, Mbisa JL, Ghosn J. et al. Phenotypic characterization of virological failure following lopinavir/ritonavir monotherapy using full-length gag-protease genes. J Antimicrob Chemother 2014; 69: 3340–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Giandhari J, Basson AE, Sutherland K. et al. Contribution of Gag and protease to HIV-1 phenotypic drug resistance in pediatric patients failing protease inhibitor-based therapy. Antimicrob Agents Chemother 2016; 60: 2248–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chaplin B, Akanmu AS, Inzaule SC. et al. Association between HIV-1 subtype and drug resistance in Nigerian infants. J Antimicrob Chemother 2019; 74: 172–6. [DOI] [PubMed] [Google Scholar]
- 25. Parry CM, Kohli A, Boinett CJ. et al. Gag determinants of fitness and drug susceptibility in protease inhibitor-resistant human immunodeficiency virus type 1. J Virol 2009; 83: 9094–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tamura K, Stecher G, Peterson D. et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30: 2725–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sutherland KA, Ghosn J, Gregson J. et al. HIV-1 subtype influences susceptibility and response to monotherapy with the protease inhibitor lopinavir/ritonavir. J Antimicrob Chemother 2015; 70: 243–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.WHO. Guidelines on the Public Health Response to Pretreatment HIV Drug Resistance http://who.int/hiv/pub/guidelines/hivdr-guidelines-2017/.
- 29. Gupta RK, Gregson J, Parkin N. et al. HIV-1 drug resistance before initiation or re-initiation of first-line antiretroviral therapy in low-income and middle-income countries: a systematic review and meta-regression analysis. Lancet Infect Dis 2018; 18: 346–55. [DOI] [PMC free article] [PubMed] [Google Scholar]