Abstract
Motivation
Low-frequency DNA mutations are often confounded with technical artifacts from sample preparation and sequencing. With unique molecular identifiers (UMIs), most of the sequencing errors can be corrected. However, errors before UMI tagging, such as DNA polymerase errors during end repair and the first PCR cycle, cannot be corrected with single-strand UMIs and impose fundamental limits to UMI-based variant calling.
Results
We developed smCounter2, a UMI-based variant caller for targeted sequencing data and an upgrade from the current version of smCounter. Compared to smCounter, smCounter2 features lower detection limit that decreases from 1 to 0.5%, better overall accuracy (particularly in non-coding regions), a consistent threshold that can be applied to both deep and shallow sequencing runs, and easier use via a Docker image and code for read pre-processing. We benchmarked smCounter2 against several state-of-the-art UMI-based variant calling methods using multiple datasets and demonstrated smCounter2’s superior performance in detecting somatic variants. At the core of smCounter2 is a statistical test to determine whether the allele frequency of the putative variant is significantly above the background error rate, which was carefully modeled using an independent dataset. The improved accuracy in non-coding regions was mainly achieved using novel repetitive region filters that were specifically designed for UMI data.
Availability and implementation
The entire pipeline is available at https://github.com/qiaseq/qiaseq-dna under MIT license.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Detection of low-frequency variants is important for early cancer diagnosis and is a very active area of research. Targeted DNA sequencing generates very high coverage over a specific genomic region, therefore allowing low-frequency variants to be observed from a reasonable number of reads. However, distinguishing the observed variants from experimental artifacts is very difficult when the variants’ allele frequencies are near or below the noise level. Providing an error-correction mechanism, unique molecular identifiers (UMIs) have been implemented in several proof-of-concept studies (Jabara et al., 2011; Kennedy et al., 2014; Kukita et al., 2015; Newman et al., 2016; Peng et al., 2015; Schmitt et al., 2012) and used in translational medical research (Acuna-Hidalgo et al., 2017; Bar et al., 2017; Young et al., 2016). In these protocols, UMIs (short oligonulceotide sequences) are attached to endogenous DNA fragments by ligation or primer extension, carried along through amplification and sequencing and finally identified from the reads. Sequencing errors can be corrected by majority vote within a UMI family, because reads sharing a common UMI and random fragmentation site should be identical except for rare collision events (Liang et al., 2014) or errors within the UMI sequences. DNA polymerase errors occurring during DNA end repair and early PCR cycles (particularly the first cycle), however, cannot be corrected because all reads in the UMI would presumably carry the error. Although PCR error rates are low (, depending on the enzyme and types of substitution), they impose fundamental limits to UMI-based variant calling.
A two-step UMI-based variant calling approach that first constructs a consensus read with tools like fgbio (https://github.com/fulcrumgenomics/fgbio) and then applies one of the conventional low-frequency variant callers (Xu, 2018) to the consensus reads has been implemented in Peng et al. (2015) and Blumenstiel et al. (2017). In addition to the two-stage method, three UMI-based variant callers, DeepSNVMiner (Andrews et al., 2016), smCounter (Xu et al., 2017) and MAGERI (Shugay et al., 2017), are publicly available. DeepSNVMiner relies on heuristic thresholds to draw consensus and call variants. By default, a UMI is defined as ‘supermutant’ if 40% of its reads support a variant and two supermutants are required to confirm the variant. smCounter was released in 2016 by our group and reported above 90% sensitivity at fewer than 20 false positives per megabase for 1% variants in coding regions. smCounter’s core algorithm consists of a joint probabilistic modeling of PCR and sequencing errors. MAGERI is a collection of tools for UMI-handling, read alignment, and variant calling. The core algorithm estimates the first-cycle PCR errors as a baseline and calls variants whose allele frequencies are higher than the baseline level. MAGERI reported 93% area under curve (AUC) on variants with about 0.1% allele frequencies.
In this article, we present smCounter2, a single nucleotide variant (SNV) and short indel caller for UMI-based targeted sequencing data. smCounter2 offers significant upgrades from its predecessor (smCounter) in terms of algorithm, performance and usability. smCounter2 adopts the widely popular Beta distribution to model the background error rates and Beta-binomial distribution to model the number of non-reference UMIs. An important feature of smCounter2 is that the model parameters are dynamically adjusted for each input read set. In addition, smCounter2 uses a regression-based filter to reject artifacts in repetitive regions while retaining most of the real variants. The algorithm improvements help to push the detection limit down to 0.5% from the previously reported 1% and increase the sensitivity and specificity compared to other UMI-based methods (two-step consensus-read approach and smCounter), as shown in Section 3. For ease of use, smCounter2 has been released with a Docker container image that includes the complete read processing (using reads from a QIAGEN QIAseq DNA targeted enrichment kit as an example) and variant calling pipeline as well as all the supporting packages and dependencies.
2 Materials and methods
2.1 smCounter2 workflow
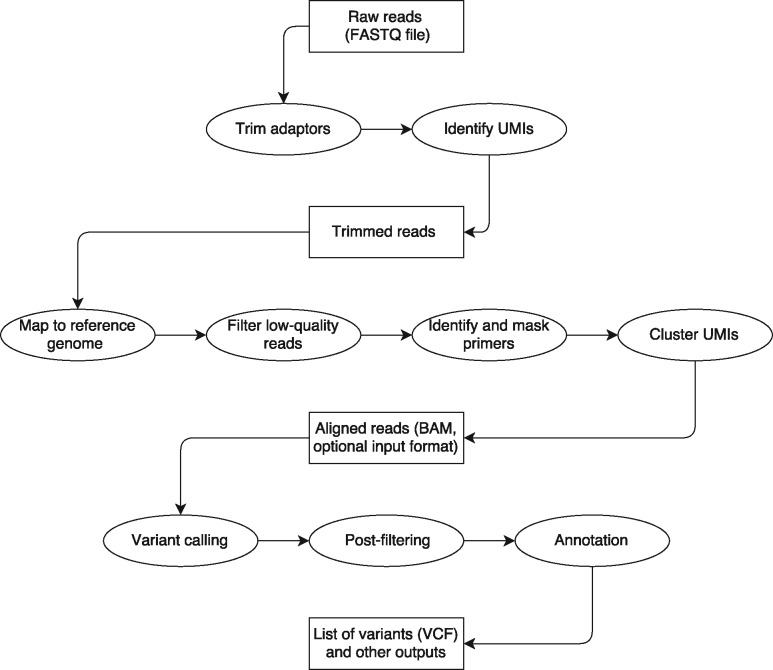
smCounter2’s workflow (Fig. 1) begins with read-processing steps that (i) remove the exogenous sequences such as PCR and sequencing adapters and UMI, (ii) identify the UMI sequence and append it to the read identifier for downstream analyses and (iii) remove short reads that lack enough endogenous sequence for mapping to the reference genome. The trimmed reads are mapped to the reference genome with BWA-MEM, followed by filtering of poorly mapped reads and soft-clipping of gene-specific primers. A UMI with much smaller read count is combined with a much larger read family if their UMIs are within edit distance of 1 and the corresponding 5' positions of aligned R2 reads are within 5 bp (i.e. at the random fragmentation site). After UMI clustering, the aligned reads (BAM format) are sent for variant calling.
Fig. 1.
smCounter2 workflow. Rectangular boxes represent the data files and elliptical boxes represent steps of the pipeline. Users can choose to run the whole pipeline from FASTQ to VCF or run the variant calling part only from BAM to VCF
Like many variant callers, smCounter2 walks through the region of interest and processes each position independently. At each position, the covering reads go through several quality filters and the remaining high-quality reads are grouped by putative input molecule (as determined by both the clustered UMI sequence and the random fragmentation site). A consensus base call (including indels) is drawn within a UMI if of its reads agree. The core variant calling algorithm is built on the estimation of background error rates, i.e. the baseline noise level for the data. A potential variant is identified only if the signal is well above that level (Sections 2.2 and 2.3). The potential variants are subject to post-filters, including both traditional filters such as strand bias and novel model-based, UMI-specific repetitive region filters (Section 2.4). Finally, the variants are annotated with SnpEff (Cingolani et al., 2012a) and SnpSift (Cingolani et al., 2012b) and output in VCF format.
For better flexibility, users can choose to run the variant calling part only. smCounter2 accepts both raw UMI-tagged BAM file and consensused BAM file (e.g. generated by fgbio) as input. In addition, smCounter2 can be used to verify a list of pre-called variants if a VCF file is provided.
2.2 Estimation of background error rates
Estimating the background error rates is one of the commonly used strategies in somatic variant calling. EBCall (Shiraishi et al., 2013) and shearwater (Gerstung et al., 2014) assume that each site has a distinctive error rate (predominantly sequencing errors) that follows a Beta distribution. LoLoPicker (Carrot-Zhang and Majewski, 2017) estimates site-specific sequencing error rates as fixed values. For UMI-tagged data, background errors can come from base mis-incorporation by DNA polymerase during end repair and the first-cycle PCR reaction, oxidation damage to DNA bases during sonication shearing and probe hybridization (Newman et al., 2016; Park et al., 2017), UMI mis-assignment, misalignment, and polymerase slippage (often in repetitive sequences), etc. iDES (Newman et al., 2016) characterizes the site-specific background error rates in duplex-sequencing data using Normal or Weibull distributions. The limitation of these algorithms is the requirement of many control samples for the site-specific error modeling. As an alternative, MAGERI (Shugay et al., 2017) assumes a universal Beta distribution for all sites, which may result in lower accuracy compared to site-specific error modeling, but as a trade-off requires only one control sample, if the UMI coverage is high enough to observe the background errors and enough sites are covered to reveal the full distribution of error rates.
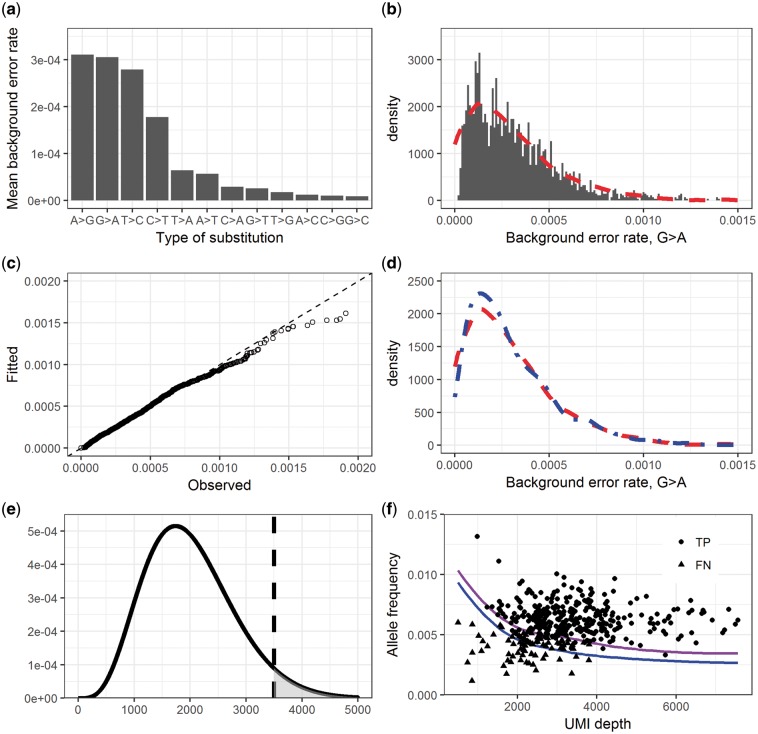
smCounter2 takes similar experimental and modeling approaches as MAGERI with important modifications. To obtain high-depth data for error profiling, we sequenced 300 ng of NA12878 DNA within a 17 kbp region using a custom QIAseq DNA panel. After excluding the known SNPs [Genome in a Bottle Consortium (Zook et al., 2014)], we calculated the error rates by base substitution at each site assuming any non-reference UMIs are background errors. The calculation process is explained in Supplementary Material, Section 2. We observed notable variation across different base substitutions and that transitions were more error-prone than transversions (Fig. 2a). We used the Beta distribution to fit the observed error rates [R fitdistrplus (Delignette-Muller and Dutang, 2015), Fig. 2b]. The quantile plot indicates good fit in general and under-estimation of the tail, possibly due to outliers (Fig. 2c). We prepared two versions of error models, one excluding singletons (UMIs with only one read pair) and the other including singletons, to accommodate deep and shallow sequencing depths. For read sets with mean read pair per UMI (rpu) , smCounter2 drops singletons to reduce errors and uses the error model without singletons. For read sets with rpu < 3, smCounter2 keeps some or all singletons (Supplementary Material, Section 4) to avoid losing too many UMIs, and uses the error model with singletons.
Fig. 2.
Underlying model of smCounter2. (a) Background error rates for each type of base change, averaged across the panel of M0466. (b) Modeling of the background error rates using the Beta distribution. The histogram shows the frequency of observed G>A error rates in M0466. The dashed curve is the density of the fitted Beta distribution. (c) Quantile plot to check the goodness-of-fit of the G>A error rate modeling. The observed and fitted quantiles form a 45° line in most places, indicating perfect fit. The tail skews towards ‘observed’, indicating under-estimation of the extremely high error rates. This may simply be explained by outliers, or suggests that a distribution (or mixed distributions) with heavier tail is needed. (d) A real example of parameter adjustment. The dashed curve is the originally fitted Beta distribution. The dotdashed curve with higher peak is the adjusted error model with the mean of the input data (N13532) and the original variance. (e) Illustration of the variant calling P-value. The density curve is a hypothesized Beta-binomial distribution. The vertical line indicates the observed non-reference UMI counts. The area of the shaded region is the P-value. (f) Detection limit prediction and confirmation. The top and bottom curves are the predicted site-wise detection limit for Ti and Tv/indels, respectively. The dots are the true variants in N13532 (outliers with extremely low UMI depth or high allele frequency excluded). For the dots, the y-axis represents the observed allele frequencies. Round dots are the variants detected and triangle dots are the ones not detected, concentrated in the low enrichment regions
As a distinctive feature of smCounter2, the Beta distribution parameters are adjusted for each dataset to account for the run-to-run variation. Because the true variants are unknown in the application dataset, we conservatively assumed that all non-reference alleles with VAF below 0.01 are background errors. The low DNA input in most applications impose another challenge in that few of the applications generate enough site-wise UMI coverage for any meaningful update of the error rate distribution. Fortunately, sufficient UMIs can usually be obtained by aggregating the target sites to accurately estimate the mean. Therefore, we only adjust the mean of the Beta distribution to equate the panel-wise mean and leave the dispersion unchanged (Fig. 2d). In specific, the adjusted Beta parameters are
| (1) |
| (2) |
where is the mean error rate of the current data and is the variance of the error rate from our control sample. The adjusted distribution has a mean of and variance of .
Background errors are sensitive to enrichment chemistry and DNA polymerase. The error pattern we observed in QIAseq DNA panels agrees with that in other PCR enrichment studies (Potapov and Ong, 2017; Shagin et al., 2017) but differs from hybridization capture studies (Newman et al., 2016; Park et al., 2017) where A > C and G > T errors are dominant. Also, certain high-fidelity DNA polymerases have been shown to generate tens- or hundreds-fold lower error rates (Potapov and Ong, 2017). Therefore, we did not attempt to build a universal error model by pooling data from multiple experiments with different polymerases as MAGERI did, but instead suggest users who run hybridization capture protocols or use non-QIAseq enrichment chemistry to build their own error profile. This can be done using a script provided in the Github repository.
Limited by sequencing resources, we were unable to obtain adequate site-wise UMI depth to model base substitutions with low error rates, including all transversions and some transitions. This deficit had several impacts on our modeling procedure. First, we had to assume that all transitions followed the distribution of G > A (second highest) and all transversions followed the distribution of C > T (higher than all transversions). This conservative configuration ensured that the error rates were not under-estimated, but also prevented us from reaching the theoretical detection limit. Second, we were unable to model the indel error rates because (i) indel polymerase errors occur more frequently in repetitive regions, and our panel did not include enough such regions, (ii) there are countless types of indels and we cannot model the errors by each type and (iii) indel polymerase error rates are on average lower than base substitution and we lacked the UMI depth to observe enough of them. Again, we conservatively assumed that indel error rates followed the distribution of G > A. Third, because the error rates are very low, zero non-reference UMIs were observed at some sites, especially in low enrichment regions. Depending on the percentage of such sites, we either imputed the zeros with small values or used a zero-inflated Beta distribution (a mixture of Beta distribution and a spike of zeros) instead of Beta.
2.3 Statistical model for variant calling and detection limit prediction
We treated variant calling as a hypothesis testing problem, where the null hypothesis (H0) is that all non-reference UMIs are from background errors and the alternative hypothesis (Ha) is that the non-reference UMIs are from the real variant. We assume that there are n UMIs covering a site and k of them have the same non-reference allele. Under H0, k follows a Binomial distribution Bin(n, p) where p is the background error rate. If p follows the Beta distribution with the adjusted parameter , the marginal distribution of k given is Beta-binomial. If a zero-inflated Beta distribution is used, k has a non-standard marginal distribution. To compute the P-value, we first simulated random samples of according to the distribution being used. Then for each pi we computed based on the Binomial distribution. The P-value represents the probability of observing non-reference UMIs at a wild-type site (Fig. 2e) and is approximated by
| (4) |
| (3) |
To avoid extremely small fractions, smCounter2 reports as the variant quality score.
The choice of variant calling threshold depends on the tolerance of false positive rate because if the model fits perfectly, the specificity would equal to 1 minus the P-value threshold. By default, smCounter2 aims for false positives per megabase, which is equivalent to a threshold of or . We will show in Section 3 and Supplementary Material that this threshold works well for datasets with deep and shallow UMI coverage and for variants with a range of VAFs (0.5, 1, 5% and germlines). The only exception is that, if 0.5–1% indels are of interest, we recommend lowering the Q-threshold to 2.5 to account for the overestimation of indel error rates.
Under this framework, the site-specific detection limit (sDL, the minimum allele frequency to exceed the P-value threshold) is a decreasing function of the UMI depth. It also depends on the type of variant because transitions have higher background error rates than transversions and indels. We estimate that the sDL of transitions is higher than transversions and indels on by about 0.001, or 0.1% in allele frequency. We denote as the P-value given UMI depth n, non-reference UMI count k and the type of variant . can be computed by Equation (3). The sDL is denoted as arg min k{P(n, k, t) < threshold}∕n and can be computed numerically. Importantly, the predicted sDL is the observed allele frequency that often deviates from the true allele frequency in the sample due to random enrichment bias. If we loosely define the overall detection limit as the minimum true allele frequency that the variant caller can detect with good sensitivity and specificity, the overall detection limit is usually higher than sDL. Based on our calculation, the theoretical detection limit of a QIAseq DNA panel is around 0.5% when UMI depth is between 2000 and 4000. This detection limit was confirmed experimentally by sequencing a sample with known 0.5% variants (Fig. 2f).
2.4 Repetitive region filters based on UMI efficiency
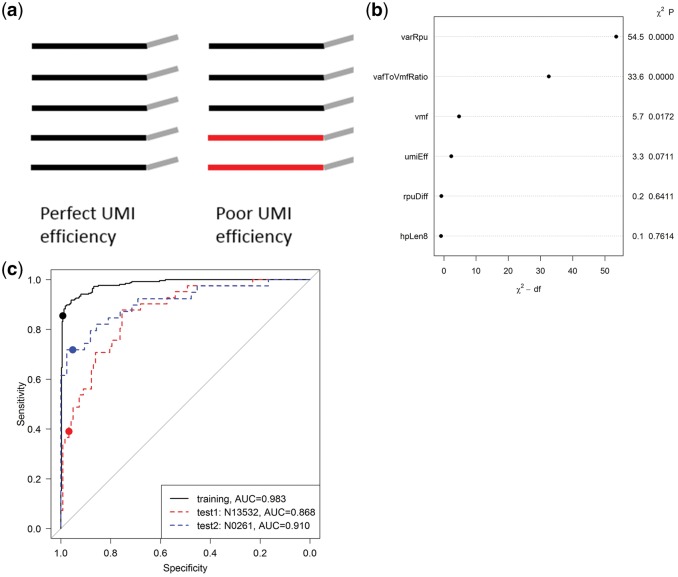
Repetitive regions such as homopolymers and microsatellites are enriched in non-coding regions where variants can have important functions from regulating gene expression to promoting diseases (Khurana et al., 2016). Unfortunately, these regions are a major source of false variant calls due to increased polymerase and mapping errors. For instance, polymerase slippage (one or more bases of the template are skipped over during base extension) occurs more frequently at homopolymers and results in false deletion calls. Reads may be incorrectly mapped to similar regions or mis-aligned if they do not span the whole repetitive sequence, both causing false variant calls. Conventional variant callers apply heuristic filters to remove false calls. For example, Strelka (Saunders et al., 2012) rejects somatic indels at homopolymers with nt or di-nucleotide repeats with nt. Recent haplotype-based variant callers such as GATK HaplotypeCaller (DePristo et al., 2011) perform local de novo assembly to avoid mapping/alignment errors in repetitive regions. However, these methods were developed for non-UMI data.smCounter2 includes a set of repetitive region filters that are specifically designed for UMI data. The filters were inspired by the observations that (i) UMIs of the false variants tend to have lower read counts and more heterogeneous reads compared to UMIs of real variants, and (ii) reads of the false variants are more likely to contradict with their UMIs’ consensus allele (usually wild-type), whereas reads of the real variants are likely to agree with their UMIs. We used the term ‘UMI efficiency’ to describe these distinctions (Fig. 3a) and quantified the UMI efficiency with four variables: (i) vafToVmfRatio, the ratio of allele frequencies based on reads and UMIs; (ii) umiEff, the proportion of reads that are concordant with their respective UMI consensus; (iii) rpuDiff, difference of read counts between variant UMIs and wild-type UMIs, adjusted by the standard deviations and (iv) varRpu, mean read fragments per variant UMI.
Fig. 3.
Training and testing of the homopolymer indel filter. (a) Illustration of UMI efficiency. The UMI on the left has perfect efficiency because all reads contributed to the consensus. The UMI on the right has low efficiency because two reads in red disagree with the majority and thus are wasted. smCounter2 requires 80% agreement to reach a consensus, so the entire UMI would be dropped and the other three reads would be wasted as well. (b) Relative importance of each predictor ranked by the explained variation minus the degree of freedom. The read pairs per variant UMI (varRpu) and the ratio between allele frequencies by read and by UMI (vafToVmfRatio) are the two variables with the most predictive power. The plot is generated with R rms package. (c) ROC curves of the logistic regression classifier. The black curve is for the training data that combined all true and false homopolymer indels in N0030, N0015, N11582 and N0164. The blue and red curves are for two test datasets N13532 and N0261, respectively. The dots represent the actual sensitivity and specificity at the cutoff, which is consistent in all three datasets
We trained and validated a logistic regression model to distinguish real homopolymers indels from artifacts. We focused our resources on this repetitive region subtype because during development, we observed that homopolymer indels were the main contributor of false positives. We combined data from several UMI-based sequencing experiments to assemble a training set with 255 GIAB high-confidence homopolymer indels with allele frequencies from 1 to 100% and 386 false positives that would otherwise be called without the filters. In addition to the UMI efficiency variables, we included sVMF (VAF based on UMI) and hpLen8 (binary variable indicating whether the repeat length ) as predictors. We found that varRpb and vafToVmfRatio were the two most important predictors in terms of explained log-likelihood (Fig. 3b). We chose the cutoff on the linear predictors to target on the highest sensitivity while maintaining 99% specificity using the R package OptimalCutpoints (López-Ratón et al., 2014). The model and cutoff were applied to two independent datasets N13532 and N0261, both containing 0.5% variants. N13532 had 41 real homopolymer indels and 122 false positives with . The predictive model achieved 39.0% sensitivity, 96.7% specificity, and 0.868 AUC. N0261 had 39 real homopolymer indels and 42 false positives with . The predictive model achieved 71.8% sensitivity, 95.2% specificity, and 0.910 AUC (Fig. 3c).
For other subtypes of variants and repetitive regions, we used heuristic thresholds as filters due to lack of training data. The model parameters and default thresholds are presented in the Supplementary Material.
3 Results
3.1 Training and validation datasets
To develop the statistical model and fine-tune the parameters, we did multiple sequencing runs using reference materials NA12878 and NA24385, both of which have high-confidence variants released by GIAB (v3.3.2 used for this study). We mixed small amounts of NA12878 DNA into NA24385 based on the amount of amplifiable DNA measured by QIAseq DNA QuantiMIZE assay to simulate low-frequency variants. The modeling of background error rates was based on M0466, a high-input, deep-sequencing run that reached over 45 000 UMI coverage per site. The selection of variant calling threshold and refinement of filter parameters were based on four datasets: N0030, N0015, N11582 and N0164. After development, we tested smCounter2 on three independent datasets: N13532, N0261 and M0253 without any modification to the algorithm and parameters. The datasets involved in this study are summarized in Table 1. A more detailed description of these datasets is provided in the Supplementary Material.
Table 1.
Key statistics of the datasets used for training and testing of smCounter2
| Dataset | Purpose | Sample | Target region (bp) | Mean UMI depth | Mean read pairs per UMI | VAF (%) | SNVs | Indels |
|---|---|---|---|---|---|---|---|---|
| M0466 | Training | 0.2% NA12878 | 17 859 | 45 335 | 3.2 | 0.1 | 87 | 0 |
| N0030 | Training | 2% NA12878 | 1 032 301 | 3612 | 8.6 | 1 | 363 | 56 |
| N0015 | Training | 10% NA12878 | 406 846 | 4825 | 8.5 | 5 | 4412 | 369 |
| N11582 | Training | 100% NA24385 | 1 094 204 | 479 | 2.6 | 50 or 100 | 729 | 49 |
| N0164 | Training | 1–20% NA12878 | 66 661 | 3692 | 11.5 | 0.5–10 | 237 | 177 |
| N13532 | Test | 1% NA12878 | 928 315 | 4040 | 7.6 | 0.5 | 293 | 164 |
| N0261 | Test | 1% NA12878 | 45 299 | 3384 | 13.8 | 0.5 | 5 | 269 |
| M0253 | Test | 50% HDx Tru-Q 7 | 38 370 | 4980 | 13.0 | 36 (with MNPs) | 1 |
3.2 Benchmarking 0.5% variant calling performance using mixed GIAB samples
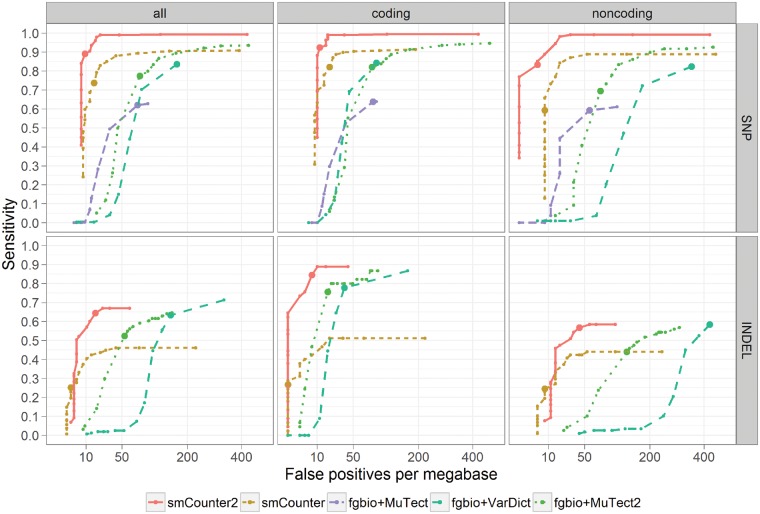
We benchmarked smCounter2 against six state-of-the-art UMI variant calling algorithms (fgbio+MuTect, fgbio+MuTect2, fgbio+VarDict, MAGERI, DeepSNVMiner and smCounter) on N13532, which contained 0.5% NA12878 variants. The first three algorithms represent the two-step approach discussed in Section 1. We first constructed consensus reads from the aligned reads (BAM file) using fgbio’s CallMolecularConsensusReads and FilterConsensusReads functions and then applied three popular low-frequency variant callers, MuTect, MuTect2 (Cibulskis et al., 2013, and VarDict (Lai et al., 2016), on the consensus reads. MAGERI, DeepSNVMiner and smCounter are representative UMI-aware variant callers. The results (Fig. 4), stratified by type of variant (SNV and indel) and genomic region (all, coding and non-coding), were measured by sensitivity and false positives per megabase (FP/Mbp, or ) at several thresholds. smCounter2 outperformed the other methods in all categories. In coding regions, smCounter2 achieved 92.4% sensitivity at 12 FP/Mbp for SNVs and 84.4% sensitivity at 7 FP/Mbp for indels (Table 2). In non-coding regions, smCounter2 was able to maintain comparable accuracy for SNVs (83.3% sensitivity at 4 FP/Mbp), but produced lower sensitivity (56.8%) and higher false positive rate (42 FP/Mbp) for indels. In the indel-enriched dataset N0261, smCounter2 produced consistent sensitivity (81.4% in coding and 61.3% in non-coding) and seemingly higher FP/Mbp (0 in coding and 114 in non-coding). However, FP/Mbp in N0261 was based on a very small target region (45 kbp) and therefore provides a less accurate specificity estimate.
Fig. 4.
Benchmarking smCounter2, smCounter, fgbio+MuTect, fgbio+VarDict and fgbio+MuTect2 on 0.5% variants in N13532. The performance is measured by false positives per megabase (x-axis) and sensitivity (y-axis), stratified by type of variant (SNV and indel) and region (coding, non-coding, and all). The ROC curves are generated by varying the threshold for each method: Q-score for smCounter2, prediction index for smCounter, likelihood ratio for MuTect and MuTect2 and minimum allele frequency for VarDict. MuTect does not detect indels so is not included in the indel comparison
Table 2.
smCounter2 performance in detecting 0.5, 1, 5 and 50–100% variants, stratified by type of variant (SNV and indel) and genomic region (coding and non-coding)
| Dataset | Region | Type | TP | FP | FN | TPR (%) | FP/Mbp | PPV (%) | HC size (bp) |
|---|---|---|---|---|---|---|---|---|---|
| N13532 | Coding | SNV | 171 | 7 | 14 | 92.4 | 12 | 96.1 | 591 154 |
| (0.5%, test) | Indel | 38 | 4 | 7 | 84.4 | 7 | 90.5 | 591 154 | |
| Non-coding | SNV | 90 | 1 | 18 | 83.3 | 4 | 98.9 | 259 162 | |
| Indel | 67 | 11 | 51 | 56.8 | 42 | 85.9 | 259 162 | ||
| N0261 | Coding | Indel | 35 | 0 | 8 | 81.4 | 0 | 100.0 | 6119 |
| (0.5%, test) | Non-coding | Indel | 138 | 4 | 87 | 61.3 | 114 | 97.2 | 35 172 |
| M0253 | All | SNV/MNV | 32 | — | 4 | 88.9 | — | — | 38 370 |
| (0.5–30%, test) | Indel | 0 | — | 1 | 0.0 | — | — | 38 370 | |
| N0030 | Coding | SNV | 214 | 5 | 4 | 98.2 | 7 | 97.7 | 694 189 |
| (1%, training) | Indel | 36 | 1 | 3 | 92.3 | 1 | 97.3 | 694 189 | |
| Non-coding | SNV | 137 | 3 | 8 | 94.5 | 13 | 97.9 | 236 687 | |
| Indel | 12 | 3 | 5 | 70.6 | 13 | 80.0 | 236 687 | ||
| N0015 | Coding | SNV | 528 | 0 | 4 | 99.2 | 0 | 100.0 | 35 718 |
| (5%, training) | Indel | 9 | 0 | 1 | 90.0 | 0 | 100.0 | 35 718 | |
| Non-coding | SNV | 3851 | 7 | 29 | 99.3 | 24 | 99.8 | 297 805 | |
| Indel | 285 | 13 | 74 | 79.4 | 44 | 95.6 | 297 805 | ||
| N11582 | Coding | SNV | 421 | 2 | 0 | 100.0 | 3 | 99.5 | 682 483 |
| (50–100%, | Indel | 4 | 0 | 0 | 100.0 | 0 | 100.0 | 682 483 | |
| training) | Non-coding | SNV | 301 | 1 | 7 | 97.7 | 4 | 99.7 | 269 761 |
| Indel | 34 | 1 | 11 | 75.6 | 4 | 97.1 | 269 761 |
Notes: The metrics were generated with the default thresholds ( for indels in N13532, N0261, M0253 and for all other cases). The allele frequency and the purpose of the dataset are displayed under the dataset name. All performance metrics are measured on GIAB high-confidence regions only, the sizes of which are presented in the last column.
We did not show DeepSNVMiner and MAGERI’s performance in Figure 4. DeepSNVMiner generated 7654 FP/Mbp to achieve 86% sensitivity for SNVs at the default setting. Similar or worse performance was achieved at other settings that we tested. Because this level of false positive rate is much higher than other methods (<200FP/Mbp at similar sensitivity), it would be hard to put the ROC curves in the same figure. For MAGERI, it is unfair to compare its performance with smCounter2 using QIAseq data. MAGERI’s error model is based only on primer extension assays from a mix of DNA polymerases including several high-fidelity enzymes (Shagin et al., 2017), while smCounter2’s error model is specific to the entire QIAseq targeted DNA panel workflow, including DNA fragmentation, end repair and PCR enrichment steps. Because the MAGERI error model does not include errors introduced at the typical DNA fragmentation and end repair process (their assays do not have those steps), MAGERI’s background error rates are lower than those in smCounter2. For example, the mean error rate of A > G and T > C used by MAGERI is per base (https://github.com/mikessh/mageri-paper/blob/master/error_model/basic_error_model.pdf) and about per base for smCounter2 (Fig. 2a). Therefore, with QIAseq data, MAGERI will produce more false positives due to under-estimation of the error rate. We included MAGERI’s ROC curve in the Supplementary Figure S2 to illustrate the point that the error models are specific to each NGS workflow and need to be empirically established for different workflows.
We applied smCounter2 on the same fgbio consensus reads that were used with MuTect/MuTect2 and VarDict. As expected, fgbio+smCounter2_consensus achieved lower sensitivity and specificity than smCounter2 on the raw reads (Supplementary Fig. S2). One reason is that many smCounter2-specific filters cannot be used in this case because the UMI efficiency metrics are not computed by fgbio and therefore lost after consensus. We had to use smCounter’s filters for the fgbio consensus reads. However, despite having the same filters and a better statistical model, fgbio+smCounter2_consensus was still outperformed by smCounter. This can possibly be changed by further fine-tuning the parameters of fgbio and smCounter2. But on the other hand, it illustrates the challenge of the two-stage approaches for UMI-based variant calling, which is harmonizing the consensus and variant calling algorithms, as pointed out in Xu et al. (2017) and Shugay et al. (2017).
We used the default setting for smCounter and adjusted the parameters of fgbio, MuTect and VarDict based on our experience of working with them. However, given the infinite parameter space, we cannot claim that the results reported here reflect their optimal performance. Several variant calling thresholds were used to investigate the sensitivity-specificity trade-off and draw the ROC curves. For fgbio+MuTect/MuTect2, we used MuTect and MuTect2’s likelihood ratio score as threshold. For fgbio+VarDict, we set VarDict’s minimum allele frequency (−f). For MAGERI, we did not use the seemingly obvious threshold ‘Q-score’ because they were not allowed to exceed 100 for computational reasons, and even a Q-score of 100 was overly sensitive and generated too many false calls. Instead, we held Q-score constant at 100 and varied the number of reads in a UMI (-defaultOverseq). The parameters and thresholds used in this study are listed in the Supplementary Material, Section 4.
3.3 Detecting variants in (possibly) shallow sequencing runs
smCounter2 achieved good sensitivity on 1, 5, 50 and 100% variants as well (Table 2, datasets N0030, N0015, N11582). The biggest advantage for smCounter2 was in non-coding regions due to the repetitive region filters. Compared to smCounter, for 1% non-coding variants, smCounter2’s sensitivity increased from 75.2 to 94.5% for SNVs and from 23.5 to 70.6% for indels (Supplementary Fig. S3). For 5% non-coding variants, smCounter2’s sensitivity increased from 95.1 to 99.3% for SNVs and from 58.2 to 79.4% for indels (Supplementary Fig. S4). For 50 and 100% non-coding variants, smCounter2’s sensitivity increased from 89.0 to 97.7% for SNVs and from 42.2 to 75.6% for indels (Supplementary Fig. S5). Both smCounter2 and smCounter outperformed fgbio+MuTect and fgbio+VarDict on 1 and 5% variants in all categories. For germline variants, however, smCounter2 had lower sensitivity for non-coding indels compared to fgbio+HaplotypeCaller (75.6% versus 88.9%). This demonstrated the advantage of a haplotype-based strategy in difficult regions. Other than for non-coding indels, the two methods achieved comparable accuracy in other categories.
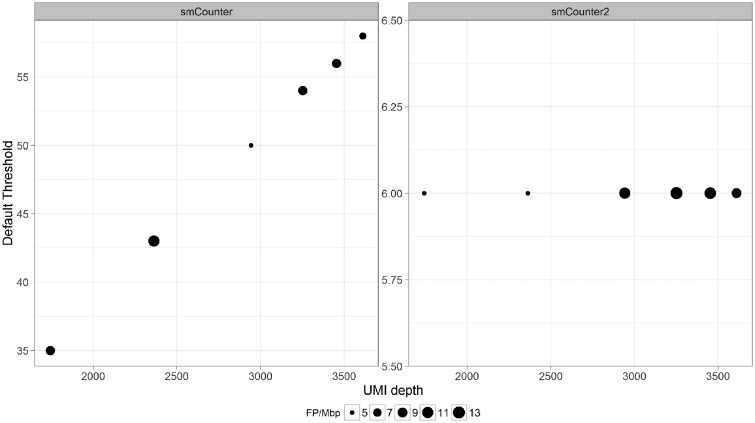
To test smCounter2’s robustness under low sequencing capacity, we in silico downsampled N0030 to 80, 60, 40, 20 and 10% of reads to mimic a range of sequencing and UMI depths. smCounter2 outperformed other methods in all sub-samples (Supplementary Figs S6–S10). The downsample series also demonstrated that smCounter2’s constant threshold can maintain consistently low false positive rates for SNV across a range of UMI depths (Fig. 5). In contrast, smCounter’s default threshold must move linearly with the UMI depth to maintain a certain level of false positive rate. Similarly, MuTect’s threshold based on the likelihood ratio needs to be adjusted for datasets with varying read depth. smCounter2’s invariant threshold allows users to apply the default setting to a wide range of sequencing and sample input conditions.
Fig. 5.
Default thresholds of smCounter and smCounter2 at different UMI depths and associated false positive rates based on the downsample series of N0030. smCounter’s threshold moves linearly with the UMI depth and is determined using an empirical formula . smCounter2’s threshold is constant at 6. The false positive rates for SNV are well controlled (between 5 and 13 FP/Mbp, represented by the point size) using both methods
It is important to note that the results described in Sections 3.2 and 3.3 are measured over GIAB high-confidence region. smCounter2’s performance in GIAB-difficult regions is unknown, both absolutely and in comparison to other variant callers. We also note that the results in Section 3.3 are based on training datasets only. We have not tested smCounter2 on independent 1% or above variants.
3.4 Detecting complex cancer mutations using horizon Tru-Q samples
The performance data described so far were based on diluted NA12878 or pure NA24385, all of which contained germline variants. To test smCounter2 on low-frequency cancer mutations, we sequenced the Tru-Q 7 reference standard (Horizon Dx) that contained verified 1.0% (and above) onco-specific mutations. The sample was diluted 1:1 in Tru-Q 0 (wild-type, Horizon Dx) to simulate 0.5% variants. For this dataset (M0253), smCounter2 detected 32 out of 36 SNV/MNVs (88.9%) and narrowly missed the only deletion (Q = 2.49 for threshold of 2.5). Because not all variants in the Tru-Q samples are known, we cannot evaluate specificity using this dataset. The list of variants in this dataset, along with the observed VAF and smCounter2 results, can be found in Supplementary File M0253.HDx.Q7.vcf.
The Tru-Q sample contains some complex multi-allelic variants that are challenging for variant callers that are not haplotype-aware. For example, there are four variants A > C, A > T, AC > CT and AC > TT at one position (chr7: 140453136, GRCh37) and a C > T point mutation at the next position. smCounter2 detected the three SNVs but failed to recognize the two MNVs.
4 Discussion
4.1 Improvement over smCounter
In this paper, we described smCounter2, the next version of our UMI-based variant caller. Compared to the previous version of smCounter, smCounter2 features lower detection limit, higher accuracy, consistent threshold and better usability.
smCounter2 pushed the detection limit of QIAseq targeted DNA panels from smCounter’s 1% down to 0.5%. smCounter2 achieved a lower detection limit because the background error rates were accurately estimated for specific base incorporation errors. The statistical model allows smCounter2 to quantify the deviation from real variants to the background errors using P-values. Therefore, the ambiguous variants whose allele frequencies are close to the background error rates can be called by smCounter2 with reasonable confidence. Importantly, 0.5% is a not an algorithm limit, but rather a chemistry limit. We believe that smCounter2 can achieve even lower detection limits for other chemistry with lower background error rate.
smCounter2 has higher accuracy than its predecessor for both SNVs and indels, in both coding and non-coding regions, for both deep and shallow sequencing runs, and for both low-frequency () and germline variants (Fig. 4 and Supplementary Figs S2–S10). In particular, for 0.5% coding region mutations, smCounter2 achieved over 92% sensitivity for SNVs and 84% for indels in coding regions at the cost of about 10 false positives per megabase, a significant improvement compared to smCounter’s 82% sensitivity for SNVs and 27% for indels at similar false positive rate. The accuracy improvement is due to the modeling of background error rates and, particularly in non-coding regions, UMI-based repetitive region filters. The filters catch false positives in the repetitive regions that pass the P-value threshold but have low ‘UMI efficiency’, a novel concept that we have proved to be useful in distinguishing real variants from artifacts. Particularly for indels in homopolymers, smCounter2 employs a logistic regression classifier that was trained and validated with separate datasets.
smCounter2 has a more consistent variant calling threshold ( for 0.5–1% indels and for other cases) that is independent from the UMI depth, unlike smCounter or MuTect whose optimal threshold must move with the UMI or read depth. This is because smCounter evaluates potential variants by the number of non-reference UMIs, while smCounter2 evaluates potential variants by the proportion of non-reference UMIs. Moreover, because smCounter2 performs a statistical test at each site, UMI depth has already been accounted for in the P-value. A higher UMI depth will result in better power of detection without raising the threshold. The consistent threshold makes it easier to benchmark smCounter2 with independent datasets. As pointed out by Xu (2018), benchmarking studies face the challenge of tuning the variant callers for different datasets.
smCounter2 is also easier to use than smCounter. The read-processing code has been released together with the variant caller, making smCounter2 a complete pipeline from FASTQ to VCF. Some users may prefer to use their own read-processing script because read structures may differ from protocol to protocol. These users can run the variant caller only with the BAM file as input, if UMIs are properly tagged in the BAM. In addition, smCounter2 accepts UMI-consensused BAM files or pre-called variants in VCF format as input. Last but not least, smCounter2 is released as a Docker container image so that users do not need to install the dependencies manually.
4.2 Comparison with other UMI-based variant callers
smCounter2 achieved better accuracy over other UMI-based variant callers in most of our benchmarking datasets (Fig. 4 and Supplementary Figs S2–S4, S6–S10) except for non-coding germline indels where smCounter2 was outperformed by fgbio+HaplotypeCaller (Supplementary Fig. S5). Compared to the two-stage approach, smCounter2 requires less tuning and achieves better detection accuracy with low-frequency variants. In contrast to MAGERI’s strategy of pooling data from several polymerases, smCounter2’s error model is developed using a single dataset with very deep coverage. Library preparation method and DNA polymerase have a large impact on the background error rates. Therefore we believe that profiling the errors per individual polymerase and protocol is a better approach. Furthermore, smCounter2 adjusts the error model for each individual dataset, making it a Bayesian-like procedure where the final error model is determined by both the prior knowledge and the data.
4.3 Limitations
smCounter2 has several limitations. First, the error model is specific to the QIAseq targeted panel sequencing protocol, which uses integrated DNA fragmentation plus end repair process and single primer PCR enrichment. Without further tests, we are less certain if the error model holds for other types of library preparation and enrichment protocols. We are more certain, however, that our error model would not fit the data generated by hybridization capture enrichment due to distinct base errors from hybridization chemistry. We have released the modeling code and encourage users, who want to use smCounter2 on non-QIAseq panel data, to re-estimate the background error rates if datasets with sufficient UMI depth are available. Second, limited by resources, we were not able to generate data with enough UMI depth to accurately estimate the transversion and indel error rates. This deficit prevented the variant caller from reaching the assay’s theoretical detection limit. However, as we continue to generate data, we will update the error models with more precise parameters. Third, the germline indel calling accuracy, especially in non-coding regions, is lower than the two-step approach of fgbio+HaplotypeCaller. Although smCounter2 has very efficient repetitive region filters, it still adopts a base-by-base variant calling strategy and relies on the mapping, which is error-prone in repetitive regions. Haplotype-aware variant callers such as HaplotypeCaller are more effective in repetitive and variant-dense regions because they perform local assembly and no longer rely on the local reference genome alignment information. Fourth, smCounter2 has difficulty in handling very complex variants. For example, it failed to report all minor alleles of the complex, multi-allelic variant in Section 3.4. This can potentially be solved by including haplotype-aware features. We have not tested smCounter2’s reliability in detecting variants with three or more minor alleles, partly because these variants are not observed frequently. By default, smCounter2 reports bi- and tri-allelic variants only. Fifth, the benchmarking study was based on reference standards. We have not demonstrated smCounter2’s performance using real tumor samples and therefore cannot claim clinical utility. We hope smCounter2 will be used in both translational and clinical studies and look forward to feedback from users.
Additional files and availability of data
The high-confidence heterozygous NA12878-not-NA24385 variants (GIAB v3.3.2) in N13532, N0261, N0030, N0015, high-confidence NA24385 variants in N11582 and verified Tru-Q 7 variants in M0253 are available in VCF format.
N0015 and N0030 reads have been published in Xu et al. (2017) and are available in Sequence Read Archive (SRA) under accession number SRX1742693. M0253, N13532, N0261 and N11582 are available in SRA under study number SRP153933.
Supplementary Material
Acknowledgements
We thank the reviewers for their helpful comments.
Funding
This work was supported by the R&D funding from QIAGEN Sciences.
Conflict of Interest: All authors are employees of QIAGEN Sciences. We declare that our employment with QIAGEN did not influence our interpretation of data or presentation of information.
References
- Acuna-Hidalgo R. et al. (2017) Ultra-sensitive sequencing identifies high prevalence of clonal hematopoiesis-associated mutations throughout adult life. Am. J. Hum. Genet., 101, 50–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews T.D. et al. (2016) Deepsnvminer: a sequence analysis tool to detect emergent, rare mutations in subsets of cell populations. PeerJ, 4, e2074.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar D.Z. et al. (2017) A novel somatic mutation achieves partial rescue in a child with hutchinson-gilford progeria syndrome. J. Med. Genet., 54, 212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenstiel B. et al. (2017). Understanding low allele variant detection in heterogeneous samples, required read coverage and the utility of unique molecular indices (umis). http://genomics.broadinstitute.org/data-sheets/POS_UnderstandingLowAlleleVariantDetection_AGBT_2017.pdf.
- Carrot-Zhang J., Majewski J. (2017) Lolopicker: detecting low allelic-fraction variants from low-quality cancer samples. Oncotarget, 8, 37032.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K. et al. (2013) Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol., 31, 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P. et al. (2012a) A program for annotating and predicting the effects of single nucleotide polymorphisms, snpeff: snps in the genome of drosophila melanogaster strain w1118; iso-2; iso-3. Fly, 6, 80–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P. et al. (2012b) Using drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, snpsift. Frontiers in Genetics, 3, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delignette-Muller M.L., Dutang C. (2015) fitdistrplus: an r package for fitting distributions. J. Stat. Softw., 64, 1–34. [Google Scholar]
- DePristo M.A. et al. (2011) A framework for variation discovery and genotyping using next-generation dna sequencing data. Nat. Genet., 43, 491.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstung M. et al. (2014) Subclonal variant calling with multiple samples and prior knowledge. Bioinformatics, 30, 1198–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabara C.B. et al. (2011) Accurate sampling and deep sequencing of the hiv-1 protease gene using a primer id. Proc. Natl. Acad. Sci. USA, 108, 20166–20171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy S.R. et al. (2014) Detecting ultralow-frequency mutations by duplex sequencing. Nat. Protoc., 9, 2586–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khurana E. et al. (2016) Role of non-coding sequence variants in cancer. Nat. Rev. Genet., 17, 93.. [DOI] [PubMed] [Google Scholar]
- Kukita Y. et al. (2015) High-fidelity target sequencing of individual molecules identified using barcode sequences: de novo detection and absolute quantitation of mutations in plasma cell-free dna from cancer patients. DNA Res., 22, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z. et al. (2016) Vardict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res., 44, e108–e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang R.H. et al. (2014) Theoretical and experimental assessment of degenerate primer tagging in ultra-deep applications of next-generation sequencing. Nucleic Acids Res., 42, e98.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ratón M. et al. (2014) Optimalcutpoints: an r package for selecting optimal cutpoints in diagnostic tests. J. Stat. Softw., 61, 1–36. [Google Scholar]
- Newman A.M. et al. (2016) Integrated digital error suppression for improved detection of circulating tumor dna. Nat. Biotechnol., 34, 547–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park G. et al. (2017) Characterization of background noise in capture-based targeted sequencing data. Genome Biol., 18, 136.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Q. et al. (2015) Reducing amplification artifacts in high multiplex amplicon sequencing by using molecular barcodes. BMC Genomics, 16, 589.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potapov V., Ong J.L. (2017) Examining sources of error in pcr by single-molecule sequencing. PLoS One, 12, e0169774.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders C.T. et al. (2012) Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics, 28, 1811–1817. [DOI] [PubMed] [Google Scholar]
- Schmitt M.W. et al. (2012) Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. USA, 109, 14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shagin D.A. et al. (2017) A high-throughput assay for quantitative measurement of pcr errors. Sci. Rep., 7, 2718.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiraishi Y. et al. (2013) An empirical bayesian framework for somatic mutation detection from cancer genome sequencing data. Nucleic Acids Res., 41, e89–e89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shugay M. et al. (2017) Mageri: computational pipeline for molecular-barcoded targeted resequencing. PLoS Comput. Biol., 13, e1005480.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. (2018) A review of somatic single nucleotide variant calling algorithms for next-generation sequencing data. Comput. Struct. Biotechnol. J., 16, 15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C. et al. (2017) Detecting very low allele fraction variants using targeted dna sequencing and a novel molecular barcode-aware variant caller. BMC Genomics, 18, 5.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young A.L. et al. (2016) Clonal haematopoiesis harbouring aml-associated mutations is ubiquitous in healthy adults. Nat. Commun., 7, 12484.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zook J.M. et al. (2014) Integrating human sequence data sets provides a resource of benchmark snp and indel genotype calls. Nat. Biotechnol., 32, 246.. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.