Abstract
Mitochondria are cellular machines essential for energy production. The biogenesis of mitochondria is a highly complex and it depends on the coordination of the nuclear and mitochondrial genome. Mitochondrial DNA (mtDNA) mutations and deletions are suspected to be associated with carcinogenesis. The most described mtDNA deletion in various human cancers is called the 4977-bp common deletion (mDNA4977) and it has been explored since two decades. In spite of that, its implication in carcinogenesis still unknown and its predictive and prognostic impact remains controversial. This review article provides an overview of some of the cellular and molecular mechanisms underlying mDNA4977 formation and a detailed summary about mDNA4977 reported in various types of cancers. The current knowledges of mDNA4977 as a prognostic and predictive marker are also discussed.
Key words: Mitochondrial DNA (mtDNA) deletion, 4977-bp mtDNA, cancer
Introduction
Cancer can be defined as an ailment characterized by progressive genomic changes, and it remains the second leading cause of death globally after cardiovascular diseases.1 According to an estimate by GLOBOCAN, in 2015 there were almost 17.5 million cancer cases worldwide and approximately 8.7 million people died due to cancer.2 It is predicted that the global cancer burden will increase by 2030, with ~26 million new cases and 17 million deaths.3
Cancer results from uncontrolled proliferation of abnormal cells due to instability of the genome.4 Genomic instability is a fundamental indicator of human cancer that can lead to an excessive accumulation in genetic alterations, including from single DNA sequence mutations to entire chromosome abnormalities.5,6
Although extensive efforts have been made by researchers to strengthen our understanding on the etiology of cancer but there is still considerable uncertainty over the precise molecular mechanism underlying cancer tumorigenesis. Because of the complexity of genomic changes within cancer cells, some researchers have shifted their focus to another genome. In addition to the nuclear genome (nDNA), it is worth to consider there is another genome that needs to be investigated.
Human mitochondrial genome (mtDNA), our other genome serves as a separate genome, which is present from several hundreds to thousands of copies per cell and their replication occur independently of the nDNA.7 Decades ago, Otto Warburg was the first scientist that proposed the relevance of mitochondria with cancer.8 Warburg’s view suggested that mitochondrial alterations in function may enhance tumor growth or promote cancer progression. 9 Since then, diverse molecular aberrations in mtDNA include point mutations, deletions, insertions, microsatellite instability, polymorphisms, and changes in mtDNA content have been identified and characterized in human cancers.
Because mtDNA is a primary target for oxidative stress, it seems that mtDNA is susceptible to such damage. In addition to DNA damage, oxidative damage to mtDNA can also result in mtDNA deletions by causing double-strand breaks in the DNA.10 Indeed, large scale deletions in mtDNA were among the first mtDNA alterations detected to cause human diseases.11-13 So far, as stated by Mitomap (http://www.mitomap.org), a mitochondrial genome database, over 150 deletions in mtDNA associated with various diseases have been reported. Among mtDNA deletions, one of the most vital that causes a huge destruction of almost onethird length of the mitochondrial genome is the 4977-bp mtDNA deletion (mDNA4977). The mDNA4977 is previously reported to be involved in myopathies, Alzheimer disease, Kearns-Sayre syndrome (KSS), chronic progressive external ophthalmoplegia (CPEO) and photoaging of the skin.11-19 Furthermore, this deletion has also been found to increase with aging in many post-mitotic human tissues.10,20
In this review, we summarize the current progress in understanding the cellular and molecular mechanisms underlying the formation of mDNA4977 and discuss its potential role in tumorigenesis. We then review recent discoveries about the reported mDNA4977 in various human neoplasias.
Mitochondrial DNA (mtDNA)
Mitochondria widely recognized as the energy powerhouses of cells are extraordinary organelles which contain their own DNA and independent biogenesis system. The structure, organization and function of mtDNA should be considered in order to understand the impact of mtDNA in cancer.
The human mtDNA is a 16,569 base-pair circular molecule that encodes for the 13 most essential genes for mitochondrial oxidative phosphorylation (OXPHOS), the primary cellular energy production system, together with the 2 ribosomal RNAs and 22 transfer RNAs for mtDNA gene expression.21,22 The entire sequence of the human mtDNA can be obtained at www.mitomap.org. It should be noted that, the only region in mtDNA where the mostly non-coding is the control region - also called the displacement- loop (D-loop) that consists of 1123 base-pair sequences. D-loop is the major control site for mtDNA expression and replication.
Although heredity of nuclear gene follows a Mendelian inheritance pattern, the transmission of mtDNA is thought to be strictly maternally inherited, due to elimination paternal mtDNA from the fertilized oocyte.23
Mitochondrial function
Mitochondria regulate several essential cellular processes including redox and calcium homeostasis control, reactive oxygen species (ROS) generation and apoptosis,24 but their most recognized duty is in the production of cellular ATP through the oxidative phosphorylation system. Mitochondrial ATP generation needs a series of multisubunit protein complexes (Complex I to Complex V), which are embedded in the inner mitochondrial membrane. Four complexes (Complex I to Complex IV) that make up the mitochondrial electron transport chain are involved in transferring electrons between the complexes to molecular oxygen as the final electron acceptor. Passage of electrons through the complexes creates a proton gradient across the inner mitochondrial membrane, which can be harnessed by Complex V (ATP synthase) to synthesize ATP from ADP and inorganic phosphate.
Defects in mitochondria are believed to be partly responsible for cancer progression and development. There was evidence from Dr. Otto Warburg in 1956 who was the first to suggest the involvement of mitochondria in the progression of cancer where he discovered a metabolic shift of cancer cells in energy production from OXPHOS to aerobic glycolysis.8,9
Alterations in mtDNA
Alterations in the mitochondrial genome have been implicated as playing a role in diverse forms of human disease and aging. It is believed that various mtDNA alterations increase with advancing age in human tissues, such as point mutations and deletions in mtDNA have been identified to accumulate in the aged human brain, skeletal muscles, heart and colon tissues.15,25 Also, mtDNA alterations have been reported as a frequent event in many human cancer studies in the types of point mutations, multiple insertions and deletions and microsatellite instability (MSI).26,27
Among the mtDNA alterations, large scale deletions in mtDNA are one of the essential mtDNA mutations that related to human diseases.28 One of the best-described large scale mtDNA deletion is the specific 4977-bp deletion in mitochondrial genome, which has been accumulated in various disorders, including mitochondrial diseases and many different types of cancer.29 The mDNA4977 has been broadly studied and has been observed to exist with increasing age in normal human brain, heart and skeletal muscle. 20 In addition, the mDNA4977 has also been exhibited to occur more frequently with increasing sun exposure in human skin, as compared with sun-protected skin.30-32
mtDNA4977 common deletion
mtDNA4977 was initially spotted in the muscle of a patient with neuromuscular diseases - Kearns-Sayre/progressive external ophthalmoplegia plus syndrome in 1989,13 and later frequently discovered to accumulate in various human tissues with aging.33-35 This genetic alteration also known as common deletion eliminates between nucleotides 8470 and 13447 of the human mitochondrial genome. mtDNA4977 removes all 5 tRNA genes (tRNAGly, tRNAArg, tRNAHis, tRNASer and tRNALeu) and 7 genes encoding 4 Complex I subunits (ND3, ND4, ND4L, partial ND5), 1 Complex IV subunit (COX III), 2 Complex V subunits (ATP6 and partial ATP8), that are crucial for supporting normal mitochondrial OXPHOS function (Figure 1).
Figure 1.
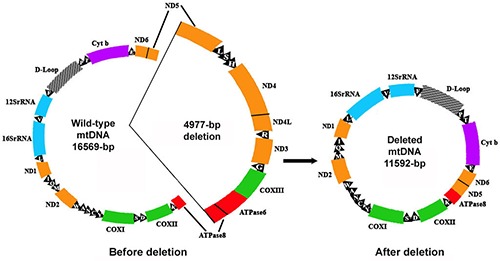
The 4977-bp human mtDNA deletion. Also known as common deletion that eliminates all 5 tRNA genes (L: tRNALeu, S: tRNASer, H: tRNAHis, R: tRNAArg, G: tRNAGly) and 7 genes encoding 4 Complex I subunits (ND3, ND4, ND4L, partial ND5), 1 Complex IV subunit (COX III), 2 Complex V subunits (ATPase6 & partial ATPase8).
mtDNA4977 has been categorized in class I deletions as these deletions almost take place within two 13-bp perfect direct repeats (ACCTCCCTCACCA) flanking the 5′- and 3′-end breakpoints, located at nucleotide positions 8470-8482 (within the ATPase 8 gene) and 13447-13459 (within the ND5 gene).36,37 One of these repeats is removed with the deleted fragment while the other is retained in mtDNA4977. Commonly, the nucleotide sequence from position 8470 to 13446 will be deleted.
The mtDNA4977 has been considered as the pathogenic mutation in human. The loss of some tRNA genes and coding regions within the deleted region disables mitochondrial protein synthesis as well as the entire mitochondrial biogenesis process. The major effect of this large deletion could cause a complete failure of ATP production and subsequently impair mitochondrial functions.38 mtDNA4977 exists in nature of heteroplasmic, with clinical signs only become apparent when the amount of deleted molecules exceeds a critical threshold.39 There has been much agreement that the generation of this common deletion because of replication error that related to the slipped mispairing between these two perfect repeats.40,41
Generation of mtDNA4977
For a long time, the sources on how deletions are generated within mtDNA genome are not well established, despite the prevalence of mtDNA deletions in human patients with mitochondrial disease and the precise molecular mechanisms underlying the formation are still under debate.
Based on the studies of deletion generation, it has been proposed that mtDNA deletions might arise either through spontaneous errors during mtDNA replication (replication slippage) or aberrancy of double-strand break repair (DBS).40-43 One possible mechanism behind this mtDNA deletion generation is replication as considered by most researchers.
Over the years, the exact mechanism behind human mtDNA replication is not completely understood and remains controversial. However, at least three major mechanisms of mtDNA replication have been proposed: i) a strand-asynchronous replication;44 ii) a strand-synchronous replication;45 and iii) a RITOLS (RNA incorporation throughout the lagging strand) model.46
The traditional strand-asynchronous (or strand-displacement) model of mammalian mtDNA replication primarily recommended by Clayton is directly based on the electron microscopic observation of replicating mtDNA intermediates.44 As stated in this model, mtDNA replication begins with the heavy strand synthesis at the origin of the heavy strand (OH) and proceeds two-thirds of the mtDNA circle prior to exposing the origin of the light strand (OL) and initiating replication of the second strand in the opposite direction. 44 The Clayton model has been challenged by Holt and colleagues mainly using data generated from the classical method of neutral two-dimensional DNA electrophoresis. The Holt model proposes that coupled mtDNA replication starts from a single origin or zone and that the leading and lagging strands are synthesized simultaneously, or synchronously.45 Later, a third model which describes long single stranded regions of mtDNA covered entirely in RNA as replication intermediate (RITOLS), has also been described.46
Most mtDNA deletions are major arc deletions with breakpoints commonly placed between OL and OH replication within the major arc. It has been previously postulated that the mechanism for mtDNA4977 formation occurs throughout an intragenomic recombination event during the slipped strand mispairing between two 13-bp perfect direct repeats.40,42,43,47 The slipped mispairing action mediates the generation of mtDNA common deletion via asynchronous replication. Asynchronous replication synthesizes a single strand to give the two repeats a chance to mispair that flank the deletion. A whole mtDNA replication initiates at the OH. Synthesis of the H-strand proceeds until the OL becomes exposed, after replication has passed two-thirds of the way around the mitochondrial genome. In the opposite direction, the L-strand can then begin to synthesize. When the upstream repeat in the parent H-strand anneals with the L-strand downstream repeat, a single-strand loop is formed. After this loop breaks followed by degradation, the Hstrand will be ligated and utilized as a template to generate a deleted mtDNA.43
A study by Birch-Machin et al. supported the above theory of the common deletion formation mechanism in human skin photoaging through forming a single-strand loop.30 The human skin may either be affected directly from prolonged exposure to ultraviolet radiation by promoting base substitutions or indirectly by introducing free radicals which consequently increasing the chances of common deletion occurrences.30
Misrepair of DSBs has been theorized as a possible cause of deletion formation including the mtDNA4977.42 The generation of mtDNA4977 has also been considered to be linked with the activity of microhomology-mediated end joining (MMEJ), which typically requires less than 12 bp of homology.48 In more recent study, Phillips et al. performed a single molecule assay of mtDNA replication based on DNA combing technique to define the underlying molecular mechanism of mtDNA4977 formation.49 According to their observations, mito-TALEN induces the breaks adjacent to the 5’ repeat and hence triggers formation of the common deletion. Finally, they clarified that mtDNA4977 was generated as a consequence of frequent fork stalling, a process which was mediated by the mitochondrial replisome, but independent of canonical DSB repair. They concluded that mtDNA4977 was not due to MMEJ or homologous recombination.49
The endogenous and exogenous source of the mtDNA4977 formation
DNA can be easily damage by endogenous (cellular metabolic pathways, ROS and errors in DNA replication) or exogenous sources (environmental factors including ionizing radiations and ultraviolet (UV) radiations from the sun). It has been believed that the increased oxidative stress in the cells results in mtDNA alterations. 50 Indeed, the hypoxic microenvironment of cancer cells triggers the production of mitochondrial ROS that can induce DNA damages and lead to genomic instability in both mitochondria and nucleus.51 As mtDNA is in close proximity to the respiratory chain, it is also permanently exposed to ROS produced as a byproduct of OXPHOS that can damage DNA and trigger accumulation of deletions.52-54 Thus, there is considerable evidence from the published literature supporting the idea. Interruption of ANT1 (adenine nucleotide translocator isoform 1) function is associated with increased ROS production, which leads to accumulation of mtDNA deletions.55 In the same manner, a partial loss of SOD2 (manganese superoxide dismutase) results in raised ROS levels and mitochondrial oxidative stress, which link to elevated mtDNA deletions.56
Researchers have discovered several important elements that are potentially linked to mtDNA4977 formation via the experimental models in vitro. In one particular pioneering study, Porteous’s group published that the bioenergetic function of cybrids containing less than 50-55% of mtDNA4977 was comparable to those cybrids with wild type mtDNA molecules.57 But, when the proportion of mtDNA4977 exceeded this threshold, resulting in decreased mitochondrial membrane potential, ATP synthesis rate and cellular ATP/ADP ratio.57A similar approach using a series of cybrids containing different proportions of mtDNA4977 associated with a chronic progressive external ophthalmoplegia (CPEO) patient has been conducted by Wei and colleagues.58 In this case, they determined that mitochondrial oxidative stress and increased mitochondrial mass and mtDNA in response to mtDNA4977 correlate with impaired respiratory function.58 In a subsequent paper from the same group, Wei’s team observed that a feed-forward, self-accelerating vicious cycle of mitochondrial ROS production could be triggered following brief, intense oxidative stress treatment in cybrids harboring mtDNA4977.59
External environmental factors are likely to influence mtDNA4977 formation. It has been considered that ROS can also be exogenously generated, mainly by ultraviolet (UV) radiations, play a role in the formation of mtDNA4977.17 Accordingly, it has been previously reported that UV exposure-induced mtDNA4977 accumulation is related to photo-aging of skin.17,30,32,60-63
The sensitivity of mtDNA to UV-generated oxidative stress has been demonstrated for skin cells in vitro and in vivo as previously published. Accumulation of mtDNA4977 is intimately associated with ROS in the dermis of both naturally and photoaged human skin in vivo.64,65 Quan et al. determined that the magnitude of the mtDNA4977 is up to 10-fold higher in photoaged skin than in naturally aged skin.65
There has been reported that singlet oxygen as generated by UVA-irradiation can cause mtDNA4977, which also detected at higher rate in photoaged skin.18 Bernerburg et al. revealed that repetitive doses of non-lethal UVA irradiation promotes the mtDNA4977 in normal human fibroblasts.18,66 In the study of Koch et al. on human keratinocytes, the level of the mtDNA4977 had a significant increased following 2 weeks of UVA irradiation.62 They also observed that the level of the mtDNA4977 decreased by 90% after prolonged culture of these irradiated cells.62 This situation could be either the replication is reduced or blocked in deleted mtDNA genomes to permit degradation of mitochondria carrying the mtDNA4977.
Birch-Machin’s group study suggests that mtDNA deletions may be useful as a biomarker of cumulative UV radiation exposure in human skin with the major species have been the mtDNA4977,30,31 and a 3895-bp deletion.67-69 In addition, they also identified a higher frequency of tandem mtDNA duplications in sun-exposed human skin.32
Additionally, some exogenous chemicals or their metabolic intermediaries also have the ability to trigger either directly or indirectly oxidative DNA damage. Acrolein is a α,β-unsaturated aldehyde, and is highly toxic to mitochondria.70 The mechanisms of acrolein-induced cytotoxicity have been proposed to be related to oxidative stress and mitochondrial impairment.70-73 According to Luo et al. acrolein exposure causes dysfunction of mitochondria, indicated by reduced mitochondrial membrane potential, ATP levels and decreased oxygen consumption.73 More recently, Wang et al. revealed that acrolein promotes mitochondrial ROS over-production, resulting in the formation of the mtDNA4977 and also depletion of mtDNA content in human lung cells.74
Involvement of the mtDNA4977 in human cancers
Lee et al. detected the mtDNA4977 in oral tumors. They also observed a positive correlation between this large-scale deletion and human non-tumor oral tissues with betel quid chewing history.75 Since then, diverse association studies have been conducted in different populations, seeking a correlation between the mtDNA4977 and various types of cancer (Table 1).
Table 1.
Study of the mtDNA4977 in selected cancer sites from different populations.
| Cancer type | Country | No. of patients (N) | mtDNA4977 in cancerous tissue/blood, n (%) | mtDNA4977 in adjacent non-cancerous tissue, n (%) | mtDNA4977 in healthy normal tissue/blood, n/N (%) | Author |
|---|---|---|---|---|---|---|
| Breast | Taiwan | 60 | 3 (5%) | 28 (47%) | - | Tseng et al.76 |
| Taiwan | 60 | 3 (5%) | 29 (48.3%) | - | Tseng et al.77 | |
| China | 76 | 76 (100%) | 76 (100%) | - | Ye et al.78 | |
| Argentina | 95 | 43 (45.3%) | 70 (73.7%) | 78/199 (39.2%) | Pavicic and Richard79 | |
| Vietnam | 106 | 73 (68.8%) | 89 (84%) | - | Dimberg et al.80 | |
| China | 107(BC); | 50(46.7%)(BC) | 44(41.1%) | 10/113(8.9%) | Nie et al.85 | |
| 51(47.7%)(BC)a | ||||||
| 118(BBD) | 51(43.2%)(BBD) | |||||
| 12(10.2%)(BBD)a | ||||||
| Iraq | 26(BC) | 0(0%)(BC) | 0(0%) | - | Dhahi et al.86 | |
| 33(BBD) | 0(0%)(BBD) | |||||
| Turkey | 25 | 0(0%) | 0(0%) | - | Aral et al.88 | |
| USA | 19 | 0(0%) | 0(0%) | - | Tan et al.87 | |
| USA | 39 | 18(46.2%) | 13(33.3%) | 6/23(26.1%) | Zhu et al.84 | |
| Brazil | 17 | 4(23.5%) | 13(76.5%) | 2/17(11.8%) | Dani et al.81 | |
| Gastric | Portugal | 32 | 17(53.1%) | N/A | - | Máximo et al.90 |
| China | 108 | 86(79.6%) | 73(67.6%) | 29/56(51.8%) | Wang and Lü 91 | |
| Taiwan | 31 | 3(9.7%) | 17(55%) | - | Wu et al.83 | |
| Iran | 107 | 6(5.61%) | 18(16.8%) | - | Kamalidehghan et al.92 | |
| Brazil | 14 | 11(78.6%) | 14(100%) | 2/17(11.8%) | Dani et al.81 | |
| Colorectal | Brazil | 46 | 24(52.2%) | 38(82.6%) | 2/17(11.8%) | Dani et al.81 |
| China | 104 | 17(16.3%) | 13(12.5%) | - | Chen et al.37 | |
| Sweden | 105 | 71(67.6%) | 97(92.4%) | - | Dimberg et al.93 | |
| Vietnam | 88 | 71(80.7%) | 81(92.0%) | - | Dimberg et al.93 | |
| China | 27b | 27(100%) | - | 2/5(40%) | Li et al.94 | |
| Turkey | 25 | 0(0%) | 0(0%) | - | Aral et al.88 | |
| Hepatocellular | Japan | 28 | 7(25%) | - | 23/35(65.7%) | Fukushima et al.95 |
| China | 27 | 19(70.4%) | 12(44.4%) | - | Shao et al.97 | |
| Taiwan | 18 | 18(100%) | 18(100%) | - | Yin et al.96 | |
| China | 62 | 17(28%) | 59(95%) | 6/6(100%) | Wheelhouse et al.98 | |
| Korea | 27 | 3 (11.1%) | 23 (88.9%) | 8/8 (100%) | Gwak et al.99 | |
| China | 105 | 10(9.52%) | 0(0%) | 0/69(0%) | Guo et al.100 | |
| Esophageal | China | 19 | 17(89%) | 19/20(95%) | - | Abnet et al.101 |
| India | 39 | 2(5.1%) | 1(2.6%) | - | Upadhyay et al.102 | |
| UK | 12 | 2(16.7%) | 9/10(90%) | 1/12(8.3%) | Tan et al.103 | |
| Oral | Taiwan | 53 | 26(49.1) | 36(67.9%) | - | Lee et al.75 |
| Taiwan | 18 | 2(11.1%) | 5(27.8%) | - | Tan et al.104 | |
| Taiwan | 12 | 12(100%) | 12(100%) | - | Shieh et al.105 | |
| India | 50 | 42(84%) | - | 18/50(36%) | Pandey et al.106 | |
| Skin | Taiwan | 10(SCC) | 7(70.0%) | - | 26/53(49.1%) | Yang et al.82 |
| 7(BCC) | 5(71.4%) | |||||
| Germany | 20(SCC) | 19(95.0%) | 40/41(97.6%) | - | Kamenisch et al.107 | |
| 21(BCC) | 18(85.7%) | |||||
| Thyroid | Portugal | 5(FC) | 0(0%) | 0/5(0%)(AP) | - | Máximo et al.109 |
| 13(HCFC) | 13(100%) | 4/12(33.3%)(AP) | - | |||
| 16(PC) | 3(18.8%) | 2/16(12.5%)(AP) | - | |||
| 10(HCPC) | 10(100%) | 3/9(33.3%)(AP) | - | |||
| Lung | Turkey | 50 | 4(8%) | 3(6%) | 0/49(0%) | Aral et al.88 |
| Endometrial | China | 37 | 20(54.1%) | 22(59.5%) | 6/20(30%) | Dai et al.110 |
| Cervix | Poland | 37 | 30(81.1%) | 32(86.5%) | - | Futyma et al.111 |
| Prostate | Turkey | 21 | 4(19%) | - | 5/16(31.3%) | Kara et al.112 |
| China | 130 | 98(75.4%) | 14(10.8%) | - | Yu and Yan113 |
BC, breast carcinoma; BBD, benign breast disease; SCC, squamous cell carcinoma; BCC, basal cell carcinoma; AP, adjacent parenchyma; FC, follicular carcinoma; HCFC, hürthle cell follicular carcinoma; PC, papillary carcinoma; HCPC, hürthle cell papillary carcinoma. aBlood; bmesenteric arteries from colorectal cancer.
Breast cancer
Through a literature search of the Pubmed, Google Scholar and other databases, the majority of mtDNA4977 studies in cancer research have been focused on breast cancer.
The mtDNA4977 of 60 breast cancer and paired non-tumorous breast tissues from 60 Taiwanese patients were analyzed by Tseng et al. using regular PCR analysis.76 It was found that the mtDNA4977 was found to accumulate in non-tumorous tissues (28/60, 47%) rather than in tumor tissues (3/60, 5%). In addition, the author demonstrated that the mtDNA4977 was associated with NAD(P)H:quinone oxidoreductase 1 (NQO1) enzyme deficiency in carcinogenesis of breast cancer.77
In another study that involved a quantitative real-time PCR assay to examine the level of the mtDNA4977 in 55 primary breast cancer patients and 21 patients with benign breast disease, all of the cases were detected to be deleted in mtDNA4977.78 However, the mtDNA4977 level was lower in tumor tissues compared to adjacent normal tissues in all cases. On the other hand, Pavicic and Richard also reported that the appearance of mtDNA4977 at higher rates in non-tumoral tissue (70/95, 73.7%) than in corresponding tumoral (43/95, 45.3%) counterpart of breast cancer patients.79
In a different study, Dimberg et al. analyzed this large-scale deletion of mtDNA in 106 Vietnamese breast cancer patients by sequencing PCR products.80 They noticed that the mtDNA4977 was significantly more frequent in normal tissue in comparison with paired cancer tissue.80 This is also in accordance with the previous study of Dani et al. which demonstrated the lower proportion deletion in tumors than in adjacent non-tumoral tissues.81
In almost all of the previous studies of the mtDNA4977 in breast cancer have reported a lower frequency in cancerous tissues as compared with corresponding non-cancerous paired tissues. The lower incidence of the mtDNA4977 in cancerous tissue than in noncancerous tissue has been also reported in other different cancers. 75,82,83 The clarification of the lower incidence of the mtDNA4977 in tumors could be either the consequence of a dilution effect because of mitotic clonal expansion throughout cancer progression or an active selection mechanism that eliminates cancer cells harboring the deleted mtDNA.75
An increased accumulation of mtDNA4977 has also been reported in cancerous breast tissues. In 2004, Zhu et al. found a high mtDNA large scale deletion rate in the breast cancer tissue samples than that inthe matched normal tissues nearby to the tumor.84 In a more recent study, Nie et al. carried out a comprehensive experiment to detect the mtDNA4977 as well as mtDNA copy number alteration in breast carcinoma and benign breast disease patients.85 The authors revealed that the mtDNA4977 in the blood of breast carcinoma patients was significantly higher (51/107, 48%) than that of benign breast disease patients and healthy controls. This deletion has also been found to be correlated with a lower mtDNA content and higher MnSOD level and oxidative damage in both group patients. These outcomes conclude that the occurrence of mtDNA4977 in blood may potentially serve as a biomarker for breast cancer.
There is also a report that shows a negative finding of this mtDNA common deletion in breast cancer patients.86-88 In 2016, Dhahi et al. published the results of a study in which mtDNA4977 was not detected in any of tumor tissues of breast cancer as well as in adjacent non-tumor tissues.86 This finding is in agreement with two of the previous study by Tan et al. and Aral et al. which report the absence of mtDNA4977 in all breast tumors and their matched surrounding normal tissues.87,88
Gastric cancer
The mtDNA large scale deletions have been first described in gastric carcinomas by Maximo et al.89,90 They reported a high frequency of 4977-bp mtDNA deletion (53.1%) in 17/32 primary sporadic gastric carcinomas. Consistent with these data, Wang and Lu also detected a higher mtDNA4977 rate in gastric cancer tissues (86/108, 79.6%) than in the adjacent normal tissues (73/108, 67.6%).91
However, in contrast to this, in another study by Wu et al. only 9.7% of common mtDNA4977 was detected in gastric carcinomas compared to 55% in non-cancerous tissue samples.83 Kamalidehghan et al. also found a low frequency (6/107, 5.6%) of 4977-bp deletion in tumoral tissues of gastric cancer.92 Wu et al.’s and Kamalidehghan et al.’s findings are consistent with those for oral cancer75 and breast cancer.76
Colorectal cancer
A decreased proportion of mtDNA4977 that found in tumor tissue as compared to adjacent non-tumor tissue also has been reported in colorectal cancer.81 In 2004, Dani et al. investigated the involvement of mtDNA4977 in colorectal cancer and other types of tumors with multiplex PCR assay.81 They found that the level of the mtDNA4977 in tumor tissues of colorectal cancer (52%) was significantly less than that in adjacent non-tumor tissues (83%).81
And this observation was further supported by the finding of Chen et al. which analyzed the mtDNA4977 level in 104 colorectal cancer of Chinese patients.37 The authors revealed that the level of deletion in cancerous tissues appeared lower than those in noncancerous areas. However, they claimed that decreased this deletion level lead to a significantly increased mtDNA copy number and correlated with cancer stage.
In a separate study, Dimberg et al. also demonstrated that the mtDNA4977 was found at a significantly higher frequency in normal tissues compared to paired cancer tissues in colorectal cancer cases of Swedish and Vietnamese patients.93
However, this view is in contradiction with the study of mesenteric arteries from colorectal cancer patients. Li et al. reported that the mtDNA4977 was significantly increased in cancer-associated mesenteric arteries than healthy controls.94 The authors suggested that the presence of this large-scale deletion in blood vessels nearby cancer cells may be related to oxidative stress in the tumor microenvironment which may contribute to more mtDNA damage.94
Hepatocellular cancer
Overall, 6 published studies and findings that have examined the mtDNA4977 in patients with hepatocellular carcinoma (HCC).95-100 The large-scale deletion has been first described in human hepatic cancer samples by Fukushima et al. in 1995.95 In 2004, Yin et al. analyzed the association of HCC patients with the level of the mtDNA4977 and mtDNA content.96 They observed that this common deletion accumulated in all of the tumors and the non-tumors liver tissues of HCC patients.96 In addition, the author also found that non-cancerous liver of the HCC patients with a background of long-term alcohol intake have a significantly increased level of the mtDNA4977 and decreased mtDNA content.96 While in the same year, Shao et al. reported the first quantitative study of frequent occurrence of the mtDNA4977 in HCC patients.97 The authors suggested that this common deletion may play an essential role in HCC development and progression.
The incidence of the mtDNA4977 in HCC was much lower than that in non-cancerous hepatic tissues and this had also been reported in two previous studies by Wheelhouse et al.98 and Gwak et al.99
In a more recent research, Guo et al. used nested PCR assay to screen a common deletion of the mtDNA4977 in blood and tissues of 105 HCC patients and 69 unrelated healthy subjects.100 They detected 9.52% of HCC patients contained this deletion, while it was absent in nearby normal tissues and healthy subjects. In addition, this deletion has also been determined to be related with elevated ROS level and mtDNA copy number.100
Esophageal cancer
In 2004, Abnet et al. conducted an analysis among a high-risk population of esophageal squamous cell carcinoma in China and discovered that 89% of tumors and 95% of the adjacent normal tissues carried the mtDNA4977.101 However, in a low-risk population of esophageal cancer in northern India, Upadhyay et al. identified a low frequency of the mtDNA4977.102 In another study, Tan et al. carried out an experiment to investigate the presence of this common deletion in the progression of Barrett’s esophagus to esophageal adenocarcinoma.103 They claimed that the frequency of specimens with the mtDNA4977 increased in association with the degree of dysplasia, thus the mtDNA4977 may be a potential biomarker to predict the severity of dysplasia.103
Oral cancer
There have been very few studies conducted on the connection of this mtDNA4977 in oral carcinogenesis.75,104-106 Apart from the earliest study by Lee et al.75 that reported very impressive data on the accumulation of large-scale deletions of mtDNA in human oral tissues, similar results were also reported by Tan et al.104 in a study of Taiwanese patients with primary oral squamous cell carcinoma by the same detection approach. Another interesting study also involved Taiwanese patients carried out by Shieh et al. on paired oral cancer and precancerous lesions using the combination techniques of laser microdissection and qPCR, revealed a higher level of the mtDNA4977 in stromal non-tumor tissue compared with tumor tissue.105
In another previous study, Pandey et al. reported an interesting finding on the correlation between mtDNA4977 and polymorphism in cytochrome P450 2E1 (CYP2E1) gene in oral cancer in the North Indian population.106 Their outcomes suggested that CYP2E1gene polymorphisms coexist with mtDNA4977 can contribute to a high risk factor for oral cancer.106
Other cancers
The mtDNA4977 has also previously been investigated in several other cancers including skin cancer,82,107,108 thyroid cancer, 88,109 lung cancer,110 endometrial cancer,111 cervix cancer,112 as well as prostate cancer.113
There are a few interesting studies that show a positive association between this mtDNA common deletion and cancer cells. A 2010 study from China investigated this common deletion in prostate cancer patients.113 The study showed that there was a significantly higher prevalence of the mtDNA4977 in prostate cancer patients compared to benign prostatic hyperplasia patients. Given such a higher frequency of mtDNA4977, the author concluded that their finding is likely to be a useful biomarker for assessing the degree of malignancy in prostate cancer patients.
A 2016 study by Shen et al. examined the relationship between the levels of mtDNA4977 in blood samples from 206 melanoma patients and 219 healthy controls.108 The study revealed that the mtDNA4977 levels in melanoma cases were significantly higher than healthy controls. Furthermore, they reported elevated levels of mtDNA4977 were associated with increased risk of melanoma and suggested that mtDNA deletions may mediate the connection between sun exposure and melanoma risk.
Can mtDNA4977 be a diagnostic and prognostic biomarker for cancer?
Nowadays there are still no useful or specific tumor biomarkers that can be used for detecting cancer at the early stage, as well as disease recurrence and determining a prognosis. Thus, the exploration of novel tumor biomarkers for early diagnosis, prognostic prediction and effective therapies will positively benefit cancer patients.
The most well-established and widely used biomarkers in cancer diagnosis include prostate specific antigen (PSA), carcinoembryonic antigen (CEA), serum cancer antigen (CA 125, CA 15-3 and CA 19-9), human epidermal growth factor receptor-2 (HER2/neu), alpha-fetoprotein (AFP).114 In addition, several biomarkers including isocitrate dehydrogenase 1 or 2 (IDH1/2), estrogen receptor (ER), p53, Ki-67, and O6-methylguanine-DNAmethyltransferase (MGMT) are also used in cancer as prognostic and predictive biomarkers.115 Improving the accuracy for the detection of the malignancy at an early stage remains a great challenge concerning biomarkers in diagnosing cancer.
The use of mtDNA as a potential biomarker to assess the risk and prognosis of cancer remains controversial and is still under investigation. The most well-studied of all mtDNA cancer alterations are in the displacement loop (D-loop) region of mtDNA non-coding part. This D-loop region acquires high mutation rates to determine the proliferation and progression of cell lineages.116 Previous studies have reported that D-loop mutations were associated with poor prognosis in some types of cancer.117,118 The mtDNA4977 has been reported to accumulate in many tissues during aging119 and has also been widely utilized as a biomarker of mtDNA damage.20
Up to now, the most studied mitochondrial alteration in skin was the mtDNA4977. It has been proposed that the frequency of mtDNA4977 contributes a potential biomarker of cumulative UV exposure in human skin.30,31 In 2006, Eshaghian et al. detected numerous mtDNA deletions from photoaged, tumor-free skin of multiple individuals which include the mtDNA4977.63 They concluded that mtDNA deletions may potentially be used as biomarkers of photoaging in the skin. In 2016, Powers et al. quantified the mtDNA4977 in the sun and non-sun exposed skin of five cohorts of patients and revealed abnormally high levels of mtDNA4977 in sun exposed skin samples. They claimed that mtDNA4977 could be indicative of sun exposure.120 One year later, the same group extended the study and discovered there was a possible synergistic relationship that caused the localised high levels of mtDNA deletions, presumably due to sun exposure and the prescribed drugs side effects.121
On the cancer-based research, Tan and coworkers reported a 70 specimen-study, ranging from normal to malignant esophageal tissue and proposed that this large deletion might be useful biomarker in detecting the severity of dysplasia in Barret’s esophagus.103 In a Chinese study for HCC, the authors proposed that mtDNA4977 might be served as a potential biomarker, possibly via the mtDNA copy number and oxidative stress alteration.100 In another study of three human tumor cell lines (liver, esophageal and breast), Li et al. suggested that mtDNA4977 could be one of the markers for radiation-induced damage due to the occurrences of this common deletion in all the tumor cells after γ-ray irradiation at any doses.
The mtDNA4977 has been broadly explored in various cancer types, unfortunately its predictive or prognostic role in cancer is still not well understood. Until now, not much is known about the link between mtDNA4977 and clinical outcome/prognosis in cancers. Cancer cell proliferation needs a continuous and plentiful supply of energy to fulfil their cell growth and division. This onethird length deletion of mtDNA impairs the function of the respiratory chain, leading to inhibition of cell proliferation. In previous studies, mtDNA4977 has been predominantly reported to be detected in normal tissues adjacent to cancerous tissues.75,80,82,83,93 Consistent with these findings, a recently published meta-analysis of 38 studies by Nie et al. also noticed a significantly decreased proportions of mtDNA4977 in cancerous tissue compared to adjacent non-cancerous tissue.122,123 The authors hypothesized that a greater mutational burden might be needed to initiate tumor growth at the early stages. Through clonal expansion, the cells with mutations resume proliferation which will result in a larger frequency of alterations in adjacent normal tissue than cancerous tissue. Moreover, both factors of genetic and environmental influences should also be taken into consideration.
The prognostic impact of mtDNA4977 remains debated and inconsistent. Upadhyay et al. believed that minor frequency of mtDNA4977 might have some role in tumor progression and prediction of survival outcome in esophageal cancer.102 Badie et al. observed that the mtDNA4977 was obviously not correlated with prognostic indicators among Sudanese with oral lesions.124
Recently, not on the cancer-based study, Huang et al. reported that mtDNA4977 status had no effect on the prognosis of stroke patients.125 However, they concluded that undetectable mtDNA4977 might be a marker or risk factor for ischemic stroke.125 In contrast, in more recent an Italian cohort study, about 515 patients with stable coronary artery disease, the authors demonstrated that the mtDNA4977 were highly observed and had prognostic implications in patients with major adverse cardiac events.126
It is widely accepted that deletion of mtDNA4977 participates in the pathogenesis of cancer, but there still remains some issue as to whether a mtDNA4977 contributes to cancer initiation and progression, and this remains an active area of investigation. Results in the previous studies have been inconsistent and many factors may influence the proportion of the deletion in cancer tissues. Although it is an encouraging finding across many studies, active investigations are needed to discover whether the deletion can potentially be applied as an effective biomarker for cancer. It is believed that early detection of cancer at an earlier stage may lead to improved outcomes, including can increase chances of survival.
Conclusions
This review summarizes the most studies of mtDNA4977 that have been reported in different kinds of human cancer. At the moment, significant extensive research efforts have been made to better understand the role of mtDNA4977 on the initiation, growth, and progression of cancer. The existence of different proportion of the mtDNA4977 in tumor tissue as compared with corresponding non-tumorous tissue could be classified as alterations in connection with the presence of various environmental and genetic factors. More studies and attention should be given before a clear conclusion could be achieved regarding the effect and role of this large mtDNA deletion in carcinogenesis.
Funding Statement
Funding: the authors thanks the Universiti Sains Malaysia for financial support (Research University Grant: 1001/PPSP/8012218 & 1001/PPSP/8012242).
References
- 1.GBD 2015. Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980-2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016;388:1459-544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Global Burden of Disease Cancer Collaboration, Fitzmaurice C, Allen C, et al. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived With Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015: A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol 2017;3:524-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thun MJ, DeLancey JO, Center MM, et al. The global burden of cancer: priorities for prevention. Carcinogenesis 2010;31:100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability— an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010;11:220-8. [DOI] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [DOI] [PubMed] [Google Scholar]
- 6.Pikor L, Thu K, Vucic E, Lam W. The detection and implication of genome instability in cancer. Cancer Metastasis Rev 2013;32:341-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lightowlers RN, Chinnery PF, Turnbull DM, Howell N. Mammalian mitochondrial genetics: heredity, heteroplasmy and disease. Trends Genet 1997;13:450-5. [DOI] [PubMed] [Google Scholar]
- 8.Warburg O. On the origin of cancer cells. Science 1956;123;309-14. [DOI] [PubMed] [Google Scholar]
- 9.Warburg O. On respiratory impairment in cancer cells. Science 1956;124:269-70. [PubMed] [Google Scholar]
- 10.Lee HC, Wei YH. Oxidative stress, mitochondrial DNA mutation, and apoptosis in aging. ExpBiol Med (Maywood) 2007;232:592-606. [PubMed] [Google Scholar]
- 11.Holt IJ, Harding AE, Morgan-Hughes JA. Deletions of muscle mitochondrial DNA in patients with mitochondrial myopathies. Nature 1988;331:717-9. [DOI] [PubMed] [Google Scholar]
- 12.Zeviani M, Moraes CT, DiMauro S, et al. Deletions of mitochondrial DNA in Kearns-Sayre syndrome. Neurology 1988;38:1339-46. [DOI] [PubMed] [Google Scholar]
- 13.Shoffner JM, Lott MT, Voljavec AS, Soueidan SA, Costigan DA, Wallace DC. Spontaneous Kearns-Sayre/chronic external ophthalmoplegia plus syndrome associated with a mitochondrial DNA deletion: a slip-replication model and metabolic therapy. Proc Natl Acad Sci U S A 1989;86:7952-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holt IJ, Harding AE, Cooper JM, et al. Mitochondrial myopathies: clinical and biochemical features of 30 patients with major deletions of muscle mitochondrial DNA. Ann Neurol 1989;26:699-708. [DOI] [PubMed] [Google Scholar]
- 15.Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res 1990;18:6927-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corral-Debrinski M, Horton T, Lott MT, et al. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics 1994;23: 471-6. [DOI] [PubMed] [Google Scholar]
- 17.Berneburg M, Gattermann N, Stege H, et al. Chronically ultraviolet- exposed human skin shows a higher mutation frequency of mitochondrial DNA as compared to unexposed skin and the hematopoietic system. Photochem Photobiol 1997; 66:271-5. [DOI] [PubMed] [Google Scholar]
- 18.Berneburg M, Grether-Beck S, Ku¨rten V, et al. Singlet oxygen mediates the UVA-induced generation of the photoagingassociated mitochondrial common deletion. J Biol Chem 1999;274:15345-9. [DOI] [PubMed] [Google Scholar]
- 19.Wallace DC, Shoffner JM, Trounce I, et al. Mitochondrial DNA mutations in human degenerative diseases and aging. Biochim Biophys Acta 1995;1271:141-51. [DOI] [PubMed] [Google Scholar]
- 20.Meissner C, Bruse P, Mohamed SA, et al. The 4977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol 2008;43:645-52. [DOI] [PubMed] [Google Scholar]
- 21.Anderson S, Bankier AT, Barrell BG, et al. Sequence and organization of the human mitochondrial genome. Nature 1981;290:457-65. [DOI] [PubMed] [Google Scholar]
- 22.Andrews RM, Kubacka I, Chinnery PF, et al. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet 1999;23:147. [DOI] [PubMed] [Google Scholar]
- 23.Pyle A, Hudson G, Wilson IJ, et al. Extreme-depth re-sequencing of mitochondrial DNA finds no evidence of paternal transmission in humans. PLOS Genet 2015;11:e1005040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wallace DC. Mitochondria and cancer. Nat Rev Cancer 2012;12:685-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Greaves LC, Nooteboom M, Elson JL, et al. Clonal expansion of early to mid-life mitochondrial DNA point mutations drives mitochondrial dysfunction during human ageing. PLoS Genet 2014;10:e1004620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mohamed Yusoff AA. Role of mitochondrial DNA mutations in brain tumors: A mini-review. J Cancer Res Ther 2015;11:535-44. [DOI] [PubMed] [Google Scholar]
- 27.Hertweck KL, Dasgupta S. The Landscape of mtDNA Modifications in Cancer: A Tale of Two Cities. Front Oncol 2017;7:262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Damas J, Samuels DC, Carneiro J, et al. Mitochondrial DNA rearrangements in health and disease—a comprehensive study. Hum Mutat 2014;35:1-14. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y, Ma Y, Bu D, et al. Deletion of a 4977-bp Fragment in the Mitochondrial Genome is Associated with Mitochondrial Disease Severity. PLoS One 2015;10:e0128624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Birch-Machin MA, Tindall M, Turner R, et al. Mitochondrial DNA deletions in human skin reflect photo- rather than chronologic aging. J Invest Dermatol 1998;110:149-52. [DOI] [PubMed] [Google Scholar]
- 31.Birch-Machin MA. Mitochondria and skin disease. Clin Exp Dermatol 2000;25:1416. [DOI] [PubMed] [Google Scholar]
- 32.Krishnan KJ, Birch-Machin MA. The incidence of both tandem duplications and the common deletion in mtDNA from three distinct categories of sun-exposed human skin and in prolonged culture of fibroblasts. J Invest Dermatol 2006;126:408-15. [DOI] [PubMed] [Google Scholar]
- 33.Yen TC, Su JH, King KL, Wei YH. Ageing-associated 5 kb deletion in human liver mitochondrial DNA. Biochem Biophys Res Commun 1991;178:124-31. [DOI] [PubMed] [Google Scholar]
- 34.Lee HC, Pang CY, Hsu HS, Wei YH. Differential accumulations of 4,977 bp deletion in mitochondrial DNA of various tissues in human ageing. Biochim Biophys Acta 1994;1226:37-43. [DOI] [PubMed] [Google Scholar]
- 35.Yang JH, Lee HC, Lin KJ, Wei YH. A specific 4977-bp deletion of mitochondrial DNA in human ageing skin. Arch Dermatol Res 1994;286:386-90. [DOI] [PubMed] [Google Scholar]
- 36.Lee HC, Wei YH. Mitochondrial DNA instability and metabolic shift in human cancers. Int J Mol Sci 2009;10:674-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen T, He J, Shen L, et al. The mitochondrial DNA 4,977-bp deletion and its implication in copy number alteration in colorectal cancer. BMC Med Genet 2011;12:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei YH, Lee CF, Lee HC, et al. Increases of mitochondrial mass and mitochondrial genome in association with enhanced oxidative stress in human cells harboring 4,977 BP-deleted mitochondrial DNA. Ann N Y Acad Sci 2001;928:97-112. [DOI] [PubMed] [Google Scholar]
- 39.Wallace DC, Chalkia D. Mitochondrial DNA genetics and the heteroplasmy conundrum in evolution and disease. Cold Spring Harb Perspect Biol 2013;5:a021220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schon EA, Rizzuto R, Moraes CT, et al. A direct repeat is a hotspot for large-scale deletion of human mitochondrial DNA. Science 1989;244:346-9. [DOI] [PubMed] [Google Scholar]
- 41.Samuels DC, Schon EA, Chinnery PF. Two direct repeats cause most human mtDNA deletions. Trends Genet 2004;20:393-8. [DOI] [PubMed] [Google Scholar]
- 42.Krishnan KJ, Reeve AK, Samuels DC, et al. What causes mitochondrial DNA deletions in human cells? Nat Genet 2008;40:275-9. [DOI] [PubMed] [Google Scholar]
- 43.Chen T, He J, Huang Y, Zhao W. The generation of mitochondrial DNA large-scale deletions in human cells. J Hum Genet 2011;56:689-94. [DOI] [PubMed] [Google Scholar]
- 44.Clayton DA. Replication of animal mitochondrial DNA. Cell 1982;28:693-705. [DOI] [PubMed] [Google Scholar]
- 45.Holt IJ, Lorimer HE, Jacobs HT. Coupled leading- and laggingstrand synthesis of mammalian mitochondrial DNA. Cell 2000;100:515-24. [DOI] [PubMed] [Google Scholar]
- 46.Yasukawa T, Reyes A, Cluett TJ, et al. Replication of vertebrate mitochondrial DNA entails transient ribonucleotide incorporation throughout the lagging strand. EMBO J 2006;25:5358-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Degoul F, Nelson I, Amselem S, et al. Different mechanisms inferred from sequences of human mitochondrial DNA deletions in ocular myopathies. Nucleic Acids Res 1991;19:493-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sfeir A, Symington LS. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem Sci. 2015;40:701-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Phillips AF, Millet AR, Tigano M, et al. Single-molecule analysis of mtDNA replication uncovers the basis of the common deletion. Mol Cell 2017;65:527-538.e6. [DOI] [PubMed] [Google Scholar]
- 50.Lin PH, Lee SH, Su CP, Wei YH. Oxidative damage to mitochondrial DNA in atrial muscle of patients with atrial fibrillation. Free Radic Biol Med 2003;35:1310-8. [DOI] [PubMed] [Google Scholar]
- 51.Nourazarian AR, Kangari P, Salmaninejad A. Roles of oxidative stress in the development and progression of breast cancer. Asian Pac J Cancer Prev 2014;15:4745-51. [DOI] [PubMed] [Google Scholar]
- 52.Shokolenko I, Venediktova N, Bochkareva A, et al. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res 2009;37:2539-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Singh AK, Pandey P, Tewari M, et al. Human mitochondrial genome flaws and risk of cancer. Mitochondrial DNA 2014;25:329-34. [DOI] [PubMed] [Google Scholar]
- 54.Blajszczak C, Bonini MG. Mitochondria targeting by environmental stressors: Implications for redox cellular signaling. Toxicology 2017;391:84-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subramaniam V, Golik P, Murdock DG, et al. MITOCHIP assessment of differential gene expression in the skeletal muscle of Ant1 knockout mice: coordinate regulation of OXPHOS, antioxidant, and apoptotic genes. Biochim Biophys Acta 2008;1777:666-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kokoszka JE, Coskun P, Esposito LA, Wallace DC. Increased mitochondrial oxidative stress in the Sod2 (+/-) mouse results in the age-related decline of mitochondrial function culminating in increased apoptosis. Proc Natl Acad Sci USA 2001;98:2278-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Porteous WK, James AM, Sheard PW, et al. Bioenergetic consequences of accumulating the common 4977-bp mitochondrial DNA deletion. Eur J Biochem 1998;257:192-201. [DOI] [PubMed] [Google Scholar]
- 58.Wei YH, Lee CF, Lee HC, et al. Increases of mitochondrial mass and mitochondrial genome in association with enhanced oxidative stress in human cells harboring 4,977 BP-deleted mitochondrial DNA. Ann N Y Acad Sci 2001;928:97-112. [DOI] [PubMed] [Google Scholar]
- 59.Jou MJ, Peng TI, Wu HY, Wei YH. Enhanced generation of mitochondrial reactive oxygen species in cybrids containing 4977-bp mitochondrial DNA deletion. Ann N Y Acad Sci 2005;1042:221-8. [DOI] [PubMed] [Google Scholar]
- 60.Yang JH, Lee HC, Lin KJ, Wei YH. A specific 4977-bp deletion of mitochondrial DNA in human ageing skin. Arch Dermatol Res 1994;286:386-90. [DOI] [PubMed] [Google Scholar]
- 61.Ray AJ, Turner R, Nikaido O, et al. The spectrum of mitochondrial DNA deletions is a ubiquitous marker of ultraviolet radiation exposure in human skin. J Invest Dermatol 2000;115:674-9. [DOI] [PubMed] [Google Scholar]
- 62.Koch H, Wittern KP, Bergemann J. In human keratinocytes the Common Deletion reflects donor variabilities rather than chronologic aging and can be induced by ultraviolet A irradiation. J Invest Dermatol 2001;117:892-7. [DOI] [PubMed] [Google Scholar]
- 63.Eshaghian A, Vleugels RA, Canter JA, et al. Mitochondrial DNA deletions serve as biomarkers of aging in the skin, but are typically absent in nonmelanoma skin cancers. J Invest Dermatol 2006;126:336-44. [DOI] [PubMed] [Google Scholar]
- 64.Fisher GJ, Quan T, Purohit T, et al. Collagen fragmentation promotes oxidative stress and elevates matrix metalloproteinase-1 in fibroblasts in aged human skin. Am J Pathol 2009;174:101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quan C, Cho MK, Perry D, Quan T. Age-associated reduction of cell spreading induces mitochondrial DNA common deletion by oxidative stress in human skin dermal fibroblasts: implication for human skin connective tissue aging. J Biomed Sci 2015;22:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berneburg M, Gremmel T, Kurten V, et al. Creatine supplementation normalizes mutagenesis of mitochondrial DNA as well as functional consequences. J Invest Dermatol 2005;125:213-20. [DOI] [PubMed] [Google Scholar]
- 67.Krishnan KJ, Harbottle A, Birch-Machin MA. The use of a 3895 bp mitochondrial DNA deletion as a marker for sunlight exposure in human skin. J Invest Dermatol 2004;123:1020-4. [DOI] [PubMed] [Google Scholar]
- 68.Harbottle A, Birch-Machin MA. Real-time PCR analysis of a 3895 bp mitochondrial DNA deletion in nonmelanoma skin cancer and its use as a quantitative marker for sunlight exposure in human skin. Br J Cancer 2006;94:1887-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harbottle A, Maki J, Reguly B, et al. Real-time polymerase chain reaction analysis of a 3895-bp mitochondrial DNA deletion in epithelial swabs and its use as a quantitative marker for sunlight exposure in human skin. Br J Dermatol 2010;163:1291-5. [DOI] [PubMed] [Google Scholar]
- 70.Sun L, Luo C, Long J, et al. Acrolein is a mitochondrial toxin: effects on respiratory function and enzyme activities in isolated rat liver mitochondria. Mitochondrion 2006;6:136-42. [DOI] [PubMed] [Google Scholar]
- 71.Adams JD, Klaidman LK. Acrolein-induced oxygen radical formation. Free Radic Biol Med 1993;15:187-93. [DOI] [PubMed] [Google Scholar]
- 72.Picklo MJ, Montine TJ. Acrolein inhibits respiration in isolated brain mitochondria. Biochim Biophys Acta 2001;1535:145-52. [DOI] [PubMed] [Google Scholar]
- 73.Luo J, Robinson JP, Shi R. Acrolein-induced cell death in PC12 cells: role of mitochondria-mediated oxidative stress. Neurochem Int 2005;47:449-57. [DOI] [PubMed] [Google Scholar]
- 74.Wang HT, Lin JH, Yang CH, et al. Acrolein induces mtDNA damages, mitochondrial fission and mitophagy in human lung cells. Oncotarget 2017;8:70406-70421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee HC, Yin PH, Yu TN, et al. Accumulation of mitochondrial DNA deletions in human oral tissues — effects of betel quid chewing and oral cancer. Mutat Res 2001;493:67-74. [DOI] [PubMed] [Google Scholar]
- 76.Tseng LM, Yin PH, Chi CW, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in breast cancer. Genes Chromosomes Cancer 2006;45:629-38. [DOI] [PubMed] [Google Scholar]
- 77.Tseng LM, Yin PH, Tsai YF, et al. Association between mitochondrial DNA 4,977 bp deletion and NAD(P)H:quinone oxidoreductase 1 C609T polymorphism in human breast tissues. Oncol Rep 2009;21:1169-74. [DOI] [PubMed] [Google Scholar]
- 78.Ye C, Shu XO, Wen W, et al. Quantitative analysis of mitochondrial DNA 4977-bp deletion in sporadic breast cancer and benign breast diseases. Breast Cancer Res Treat 2008;108:427-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Pavicic WH, Richard SM. Correlation analysis between mtDNA 4977-bp deletion and ageing. Mutat Res 2009;670:99-102. [DOI] [PubMed] [Google Scholar]
- 80.Dimberg J, Hong TT, Nguyen LTT, et al. Common 4977 bp deletion and novel alterations in mitochondrial DNA in Vietnamese patients with breast cancer. Springerplus 2015;4:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dani MA, Dani SU, Lima SP, et al. Less Delta mtDNA4977 than normal in various types of tumors suggests that cancer cells are essentially free of this mutation. Genet Mol Res 2004;3:395-409. [PubMed] [Google Scholar]
- 82.Yang JH, Lee HC, Chung JG, Wei YH. Mitochondrial DNA mutations in light-associated skin tumors. Anticancer Res 2004;24:1753-8. [PubMed] [Google Scholar]
- 83.Wu CW, Yin PH, Hung WY, et al. Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer. Genes Chromosomes Cancer 2005;44:19-28. [DOI] [PubMed] [Google Scholar]
- 84.Zhu W, Qin W, Sauter ER. Large-scale mitochondrial DNA deletion mutations and nuclear genome instability in human breast cancer. Cancer Detect Prev 2004;28:119-26. [DOI] [PubMed] [Google Scholar]
- 85.Nie H, Chen G, He J, et al. Mitochondrial common deletion is elevated in blood of breast cancer patients mediated by oxidative stress. Mitochondrion 2016;26:104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dhahi MAR, Jaleel YA, Mahdi QA. Screening for mitochondrial DNA A4977 common deletion mutation as predisposing marker in breast tumors in Iraqi patients. Curr Res J Biol Sci 2016;8:6-9. [Google Scholar]
- 87.Tan DJ, Bai RK, Wong LJ. Comprehensive scanning of somatic mitochondrial DNA mutations in breast cancer. Cancer Res 2002;62:972-6 [PubMed] [Google Scholar]
- 88.Aral C, Akkiprik M, Kaya H, et al. Mitochondrial DNA common deletion is not associated with thyroid, breast and colorectal tumors in Turkish patients. Genet Mol Biol 2010;33:1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Maximo V, Soares P, Seruca R, Sobrinho-Simoes M. Comments on: mutations inmitochondrial control region DNA in gastric tumours of Japanese patients, Tamura, et al. Eur J Cancer 1999, 35, 316-319. Eur J Cancer 1999;35:1407-8. [DOI] [PubMed] [Google Scholar]
- 90.Maximo V, Soares P, Seruca R, et al. Microsatellite instability, mitochondrial DNA large deletions, and mitochondrial DNA mutations in gastric carcinoma. Genes Chromosomes Cancer 2001;32:136-43 [DOI] [PubMed] [Google Scholar]
- 91.Wang J, Lu YY. Mitochondrial DNA 4977-bp deletion correlated with reactive oxygen species production and manganese superoxide dismutase expression in gastric tumor cells. Chin Med J (Engl) 2009;122:431-6. [PubMed] [Google Scholar]
- 92.Kamalidehghan B, Houshmand M, Ismail P, et al. Delta mtDNA4977 is more common in non-tumoral cells from gastric cancer sample. Arch Med Res 2006;37:730-5. [DOI] [PubMed] [Google Scholar]
- 93.Dimberg J, Hong TT, Skarstedt M, et al. Novel and differential accumulation of mitochondrial DNA deletions in Swedish and Vietnamese patients with colorectal cancer. Anticancer Res 2014;34:147. [PubMed] [Google Scholar]
- 94.Li T, Chen GL, Lan H, et al. Prevalence of the 4977-bp and 4408-bpmitochondrial DNA deletions in mesenteric arteries from patients with colorectal cancer. Mitochondrial DNA A DNA Mapp Seq Anal 2016;27:3774-6. [DOI] [PubMed] [Google Scholar]
- 95.Fukushima S, Honda K, Awane M, et al. The frequency of 4977 base pair deletion of mitochondrial DNA in various types of liver disease and in normal liver. Hepatology 1995;21:1547-51. [PubMed] [Google Scholar]
- 96.Yin PH, Lee HC, Chau GY, et al. Alteration of the copy number and deletion of mitochondrial DNA in human hepatocellular carcinoma. Br J Cancer 2004;90:2390-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Shao JY, Gao HY, Li YH, et al. Quantitative detection of common deletion of mitochondrial DNA in hepatocellular carcinoma and hepatocellular nodular hyperplasia. World J Gastroenterol 2004;10:1560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wheelhouse NM, Lai PB, Wigmore SJ, et al. Mitochondrial Dloop mutations and deletion profiles of cancerous and noncancerous liver tissue inhepatitis B virus-infected liver. Br J Cancer 2005;92:1268-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gwak GY, Lee DH, Moon TG, et al. The correlation of hepatitis B virus pre-S mutation with mitochondrial D-loopmutations and common deletions in hepatocellular carcinoma. Hepatogastroenterology 2011;58:522-8. [PubMed] [Google Scholar]
- 100.Guo ZS, Jin CL, Yao ZJ, et al. Analysis of the mitochondrial 4977 Bp deletion in patients with hepatocellular carcinoma. Balkan J Med Genet 2017;20:81-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Abnet CC, Huppi K, Carrera A, et al. Control region muta tions and the ‘common deletion’ are frequent inthe mitochondrial DNA of patients with esophageal squamous cell carcinoma. BMC Cancer 2004;4:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Upadhyay R, Jain M, Kumar S, et al. Role of mitochondrial DNA 4977-bp deletions in esophageal cancer susceptibility and prognosis in a northern Indian population. Cancer Genet Cytogenet 2009;195:175-8. [DOI] [PubMed] [Google Scholar]
- 103.Tan BH, Skipworth RJ, Stephens NA, et al. Frequency of the mitochondrial DNA 4977bp deletion in oesophageal mucosa during the progression of Barrett’s oesophagus. Eur J Cancer 2009;45:736-40. [DOI] [PubMed] [Google Scholar]
- 104.Tan DJ, Chang J, Chen WL, et al. Novel heteroplasmic frameshift and missense somatic mitochondrial DNA mutations in oral cancer of betel quid chewers. Genes Chromosomes Cancer 2003;37:186-94. [DOI] [PubMed] [Google Scholar]
- 105.Shieh DB, Chou WP, Wei YH, et al. Mitochondrial DNA 4,977-bp deletion in paired oral cancer and precancerous lesions revealed by laser microdissection and real-time quantitative PCR. Ann N Y Acad Sci 2004;1011:154-67. [DOI] [PubMed] [Google Scholar]
- 106.Pandey R, Mehrotra D, Catapano C, et al. Association of mitochondrial deoxyribonucleic acid mutation with polymorphism in CYP2E1 gene in oral carcinogenesis. J Oral Biol Craniofac Res 2012;2:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kamenisch Y, Wenz J, Metzler G, et al. The mitochondrial DNA common deletion is present in most basal and squamous cell carcinoma samples isolated by laser capture microdissection but generally at reduced rather than increased levels. J Invest Dermatol 2007;127:486-90. [DOI] [PubMed] [Google Scholar]
- 108.Shen J, Wan J, Huff C, et al. Mitochondrial DNA 4977-base pair common deletion in blood leukocytes and melanoma risk. Pigment Cell Melanoma Res 2016;29:372-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Maximo V, Soares P, Lima J, et al. Mitochondrial DNA somatic mutations (point mutations and large deletions) and mitochondrial DNA variants in human thyroid pathology: a study with emphasis on Hurthle cell tumors. Am J Pathol 2002;160:1857-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dai JG, Xiao YB, Min JX, et al. Mitochondrial DNA 4977 BP deletion mutations in lung carcinoma. Indian J Cancer 2006;43:20-5. [DOI] [PubMed] [Google Scholar]
- 111.Futyma K, Putowski L, Cybulski M, et al. The prevalence of mtDNA4977 deletion in primary human endometrial carcinomas and matched control samples. Oncol Rep 2008;20:683-8. [PubMed] [Google Scholar]
- 112.Kara M, Tatar A, Borekci B, et al. Mitochondrial DNA 4977 bp deletion in chronic cervicitis and cervix cancers. Balkan J Med Genet 2012;15:25-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Yu JJ, Yan T. Effect of mtDNA mutation on tumor malignant degree in patients with prostate cancer. Aging Male 2010;13:159-65. [DOI] [PubMed] [Google Scholar]
- 114.Even-Desrumeaux K, Baty D, Chames P. State of the art in tumor antigen and biomarker discovery. Cancers (Basel) 2011;3:2554-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Karsy M, Neil JA, Guan J, et al. A practical review of prognostic correlations of molecular biomarkers in glioblastoma. Neurosurg Focus 2015;38:E4. [DOI] [PubMed] [Google Scholar]
- 116.Masuda S, Kadowaki T, Kumaki N, et al. Analysis of gene alterations of mitochondrial DNA D-loop regions to determine breast cancer clonality. Br J Cancer 2012;107:2016-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lievre A, Chapusot C, Bouvier AM, et al. Clinical value of mitochondrial mutations in colorectal cancer. J Clin Oncol 2005;23:3517-25. [DOI] [PubMed] [Google Scholar]
- 118.Kuo SJ, Chen M, Ma GC, et al. Number of somatic mutations in the mitochondrial D-loop region indicates poor prognosis in breast cancer, independent of TP53 mutation. Cancer Genet Cytogenet 2010;201:94-101. [DOI] [PubMed] [Google Scholar]
- 119.Schroeder P, Gremmel T, Berneburg M, Krutmann J. Partial depletion of mitochondrial DNA from human skin fibroblasts induces a gene expression profile reminiscent of photoaged skin. J Invest Dermatol 2008;128:2297-303. [DOI] [PubMed] [Google Scholar]
- 120.Powers JM, Murphy G, Ralph N, et al. Mitochondrial DNA deletion percentage in sun exposed and non sun exposed skin. J Photochem Photobiol B 2016;165:277-82. [DOI] [PubMed] [Google Scholar]
- 121.Powers JM, Murphy G, Ralph N, et al. Polypharmacy and sun exposure: Implications for mitochondrial DNA deletions in skin. J Photochem Photobiol B 2017;173:397-403. [DOI] [PubMed] [Google Scholar]
- 122.Li J, Wang Y, DU L, et al. Nested PCR for mtDNA-4977-bp deletion and comet assay for DNA damage - a combined method for radiosensitivity evaluation of tumor cells. Oncol Lett 2014;7:1083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nie H, Shu H, Vartak R, et al. Mitochondrial common deletion, a potential biomarker for cancer occurrence, is selected against in cancer background: a meta-analysis of 38 studies. PLoS One 2013;8:e67953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.BadieI HDMA, Abdelbadie A, Munsoor MM, Khalid KE. Mitochondrial DNA 4977 bp deletion among Sudanese oral lesions. ejpmr 2015;2:32-43. [Google Scholar]
- 125.Huang YH, Chen CM, Lee YS, et al. Detection of mitochondrial DNA with 4977 bp deletion in leukocytes of patients with ischemic stroke. PLoS One 2018;13:e0193175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vecoli C, Borghini A, Pulignani S, et al. Prognostic value of mitochondrial DNA(4977) deletion and mitochondrial DNA copy number in patients with stable coronary artery disease. Atherosclerosis 2018;276:91-97 [DOI] [PubMed] [Google Scholar]


