Abstract
The gut microbiota of the honey bee (Apis mellifera) offers several advantages as an experimental system for addressing how gut communities affect their hosts and for exploring the processes that determine gut community composition and dynamics. A small number of bacterial species dominate the honey bee gut community. These species are restricted to bee guts and can be grown axenically and genetically manipulated. Large numbers of microbiota-free hosts can be economically reared and then inoculated with single isolates or defined communities to examine colonization patterns and effects on host phenotypes. Honey bees have been studied extensively, due to their importance as agricultural pollinators and as models for sociality. Because of this history of bee research, the physiology, development, and behavior of honey bees is relatively well understood, and established behavioral and phenotypic assays are available. To date, studies on the honey bee gut microbiota show that it affects host nutrition, weight gain, endocrine signaling, immune function, and pathogen resistance, while perturbation of the microbiota can lead to reduced host fitness. As in humans, the microbiota is concentrated in the distal part of the gut, where it contributes to digestion and fermentation of plant cell wall components. Much like the human gut microbiota, many bee gut bacteria are specific to the bee gut and can be directly transmitted between individuals through social interaction. Although simpler than the human gut microbiota, the bee gut community presents opportunities to understand the processes that govern the assembly of specialized gut communities as well as the routes through which gut communities impact host biology.
Introduction
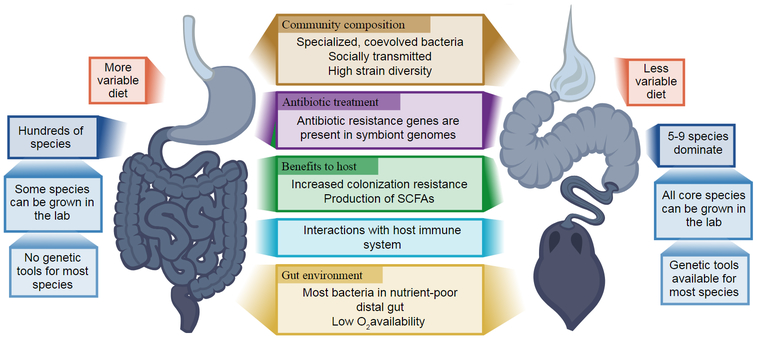
Complex microbial communities are found in virtually every site on the human body1–3, but the microbial communities associated with the gastrointestinal tract, home to the vast majority of microbes in most animals4, are of particular interest due to their diverse impacts on host health. For example, the human gut microbiota aids in food digestion, modulates the immune system, and bolsters resistance against pathogens5–7. Sequencing and sequence analysis have been used to identify correlations between microbiota composition and a variety of diseases8,9, but experimental approaches are critical in order to move beyond correlations and address cause and effect relationships. As ethical and practical considerations constrain experiments in humans, good model systems are essential for experimental study of gut microbiotas. In this review, we present the gut microbiota of the honey bee (Apis mellifera) as an experimentally tractable model system that offers numerous parallels to the human gut microbiota (Figure 1).
Figure 1.
Similarities (center) and differences (right and left sides) between the gut microbiota of humans and the gut microbiota of honey bees. SCFA, short chain fatty acids.
Over the past decade, sequencing and culture-based approaches have revealed a characteristic and relatively stable bacterial community present in the guts of all healthy adult worker honey bees. This community makes essential contributions to digestion, gut development, weight gain, and resistance to pathogens10–13 and is dominated by a limited number of bacterial lineages that live only in bee guts14–16. These species can be cultured17 and genetically manipulated18.
Here, we focus on recent publications that illustrate the utility of the bee gut microbiota as a model system for addressing general questions about microbiota functions. We provide an overview of the advantages of the honey bee as a model organism for microbiota studies, and review the composition and organization of the bee gut microbiota, recent developments in bee gut microbiota research, and parallels between the bee and human gut microbiotas. We emphasize experimental studies that use gnotobiotic hosts with defined gut communities. The diversity of the bee gut microbiota and its role in digestion of food are discussed only briefly, as these subjects have been reviewed elsewhere (e.g., 17,19,20).
Honey bees as model organisms for microbiota studies
One significant advantage of the honey bee gut microbiota as a model system is the wealth of existing knowledge of honey bee biology. For hundreds of years, humans have kept domesticated honey bees for honey and wax production and for pollination of agricultural crops. Honey bees have long been studied as models of social behavior21,22 and developmental plasticity23, and more recently as models for aging24, and behavioral disorders25. Their economic value, mainly from pollination services, is estimated in the billions of dollars annually26. Global concern over recent high seasonal mortality rates of bee hives27 has motivated research into ecological factors affecting bee health, including nutrition, toxins, pathogens, and parasites. The honey bee genome has been sequenced28, and genomic variation within the species has been surveyed29,30.
Another key advantage of honey bees as a model system for microbiota research is the availability of microbiota-free hosts, enabling investigation into how the microbiota influences host phenotypes including disease states31,32. Microbiota-free mammals can only be obtained by Caesarean section and must be maintained in specialized housing. In some insects, germ-free individuals can be generated by chemical surface sterilization of eggs33,34. However, the honey bee life cycle can be exploited to obtain large numbers of microbiota-free hosts without the use of antibiotics.
Honey bee workers undergo four distinct developmental phases: egg, larva, pupa, and adult35. The absolute abundance of gut-associated bacteria changes dramatically across these stages36. Each hive contains a single queen, able to lay 1,000–2,000 eggs per day under favorable conditions37. A fertilized egg is deposited into an individual worker cell, where it hatches into a larva that is fed by adult workers, called nurse bees38. Larval guts possess few bacterial cells, often too few to be detectable using standard PCR protocols or microscopy using fluorescent in situ hybridization36. Several bacterial species characteristic of adults have been detected from larval guts through culture-dependent or high-throughput analyses39,40, but larval gut bacteria appear to be largely acquired from food provisioned by the adult nurse bees41, and likely represent transients rather than stable colonizers. At the end of the larval stage, nurse bees construct a wax cap that seals the cell before pupation. Before the start of pupation, a septum that separated the midgut and hindgut during the larval stage is eliminated38,42. At the beginning and end of pupation42, the exoskeleton including the gut lining is shed (a process called ecdysis), eliminating any bacteria that may have been present in the larval midgut prior to pupation. Upon the completion of metamorphosis, the adult bee chews through the wax cap. Newly-emerged adult worker bees are nearly free of bacteria36,43,44 though some bacteria may be acquired as they chew45. The characteristic gut microbiota is established through social interactions with other workers during the first three days after emergence. The number of bacteria in the gut expands logarithmically until the community stabilizes at 108–109 bacterial cells around 4 days after emergence46.
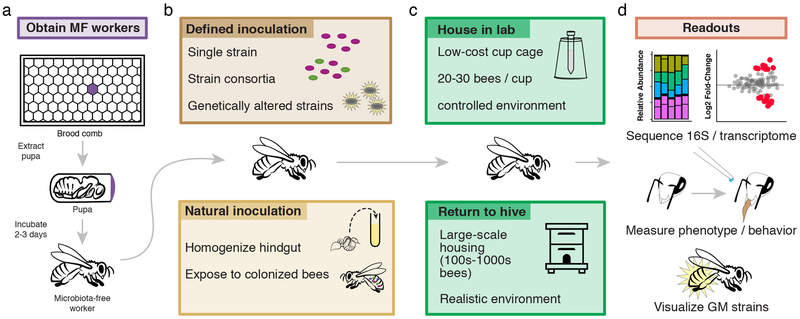
To generate gnotobiotic bees (Figure 2), hive frames with mature pupae (capped brood) are removed from the hives and transported to the laboratory, where pupae with pigmented eyes but lacking movement are aseptically removed and placed in sterile dishes. Pupae kept under sterile conditions will emerge as microbiota-free adult workers within three days46,47. Alternatively, microbiota-free workers can also be reared in the lab by rearing larvae manually48, though this approach requires more elaborate experimental infrastructure and may yield bees that are less robust. Microbiota-free workers can be inoculated orally with specific bacterial strains or with whole communities to investigate the roles of the gut microbiota in bee health, as well as the mechanisms by which microbes interact with their hosts and with one another (e.g., 46,47).
Figure 2.
Design of gnotobiotic honey bee studies. Microbiota-free (MF) hosts emerge in the lab (a) where they can be inoculated with isolated gut symbiont strains, genetically modified symbionts, or natural communities (b). These bees may be maintained in laboratory conditions or marked and returned to the hive environment (c). Destructive sampling of bees and sequencing allows for analyses of community composition and function. Alternatively, phenotypic (e.g., behavioral) assays, can reveal effects of defined gut communities on bees, while genetically modified (GM) fluorescent strains facilitate in situ imaging of bacteria (d).
Honey bees are also amenable to experiments on the effects of perturbation on an established gut microbiota. Honey bee colonies are inexpensive to maintain49 and, with 30,000–80,000 adult workers per colony37, readily provide large numbers of individuals for experiments with large sample sizes (See Box 1 for the availability and cost of beekeeping). In studies of microbiome perturbation, adult workers are collected and marked, treated in the lab with antibiotics or chemicals, and then returned to their hive of origin12. This approach can be used to study the resilience of the bee gut microbiota to perturbation and to understand the impacts of xenobiotics, including chemicals applied to crops that may be transported back to the hive by forager bees50,51, on bee behavior and health. Furthermore, isolates of bacterial pathogens, originating from sick honey bees, can be used to assess the role of the bee gut microbiota in colonization resistance12. In summary, a honey bee hive can provide hundreds to thousands of bees for experiments, including conventional and microbiota-free bees.
Box 1. The cost and feasibility of beekeeping.
Honey bee colonies can be established in almost every habitat inhabited by humans, wherever flowering plants are available for foraging, which makes bees broadly accessible as model organisms. The costs of standard hive equipment and bees are low, approximately $200-$300 per hive in the United States, and new hives can be started from healthy hives. Beekeeping methods are readily learned from local beekeepers, beekeeping clubs, or from various “how-to” manuals. Once established, most honey bee hives last for years and are largely self-sustaining.
The honey bee gut microbiome
A primary advantage of the honey bee as a model system for microbiota research is that healthy workers have a simple and specific gut microbiota that is present in honey bees collected worldwide52,53. This community is dominated by five to nine taxa, each corresponding to a species or a cluster of closely related species (Table 1), which together account for >98% of bacterial 16S rRNA gene sequences in the gut of a typical adult worker14–16. The dominance of these core taxa is consistent across bees from different hives and even different continents15,52,54–56. Once established in the adult gut, the composition of the microbiota changes little, despite seasonal changes and shifts in diet, behavior, and gene expression that occur as workers transition from nurse bees into foragers36,57. Compositional surveys using 16S rRNA genes reveal variation in the abundance of core taxa. Typically, a cluster of Lactobacillus strains collectively referred to as the Firm-5 phylotype is most abundant, followed by Lactobacillus Firm-4, Bifidobacterium spp., Gilliamella apicola, and Snodgrassella alvi (Table 1). Frischella perrara, Bartonella apis, Apibacter adventoris, and Parasaccharibacter apium are sometimes present at variable levels56,58. The occurrence of P. apium in honey bees is sporadic14,59, but it is routinely isolated both from bee guts and from other environments, including queen bee guts, honey and beebread60. While Lactobacillus Firm-4 and Firm-5 are true gut symbionts and rarely detected outside of bee guts, other Lactobacillus species can be found within the hive and on hive materials (e.g., Lactobacillus kunkeei). These Lactobacillus spp. belong to distantly related phylogenetic clusters, and genetic markers and methods should be designed to differentiate core species from environmental species when analyzing the Lactobacillus phylotypes61–64. Multiple strains from all major taxa of bee gut bacteria have been isolated (Table 1) and cultured. The relative ease of culturing these species facilitates genome sequencing and analysis to identify strain-level gene repertoires, as well as the development of genetic tools, further enabling insight into the evolution and functional roles of members of the gut microbiota11,65.
Table 1.
Bacteria occurring in honey bee guts and in the hive environment.
| Taxa | Phylum | Primary location | References |
|---|---|---|---|
| Ubiquitous gut-restricted taxa | |||
| Lactobacillus sp. Firm-5 | Firmicutes | Hindgut (ileum, rectum) | 61,132 |
| L. apis | |||
| L. helsingborgensis | |||
| L. kimbladii | |||
| L. kullabergensis | |||
| L. melliventris | |||
| Lactobacillus sp. Firm-4 | Firmicutes | Hindgut (rectum) | 61 |
| L. mellifer | |||
| L. mellis | |||
| Bifidobacterium sp. | Actinobacteria | Hindgut (rectum) | 133-135 |
| B. asteroides | |||
| B. coryneforme | |||
| B. indicum | |||
| Snodgrassella alvi | Proteobacteria | Hindgut (ileum wall) | 136 |
| Gilliamella apicola | Proteobacteria | Hindgut (ileum lumen) | 136 |
| Frischella perrara | Proteobacteria | Hindgut (pylorus, ileum) | 137 |
| Bartonella apis | Proteobacteria | Hindgut, variably present | 138 |
| Commensalibacter sp. | Proteobacteria | Hindgut, variably present | 14,139 |
| Other common taxa | |||
| Apibacter adventoris | Bacteroidetes | Adult gut | 103,140 |
| Parasaccharibacter apium | Proteobacteria | Larval gut, adult crop, queen gut, hive | 57,141 |
| Lactobacillus kunkeeii | Firmicutes | Larval and adult gut, hive, nectar | 142 |
| Fructobacillus fructosus | Firmicutes | Larval and adult gut, hive | 143 |
| Saccharibacter spp. | Proteobacteria | Bee stomach, honey, pollen | 144 |
| Opportunistic pathogens | |||
| Serratia marcescens | Proteobacteria | Adult gut | 12,145 |
| Hafnia alvei | Proteobacteria | Adult gut | 146 |
Recently developed genetic tools for manipulating bee gut microbes facilitate investigation of the molecular basis of host-microbe and microbe-microbe interactions in this system. Leonard et al. published genetic methods for heterologous gene expression and targeted disruption of multiple genes in Gram-negative bee gut microbes, including gut symbionts S. alvi, G. apicola, B. apis, and P. apium, and the opportunistic pathogen Serratia marcescens18. These methods comprise a toolkit for combinatorial assembly of broad-host-range replicative plasmids that can be transferred by conjugation into recipient bacteria, multiple functional antibiotic cassettes, inducible synthetic promoters, and CRISPR-interference. Furthermore, strains engineered to produce fluorescent proteins readily colonize microbiota-free bees in mono- and co- inoculation and can be directly imaged in the digestive tract of the bee. This methodology makes it possible to study the localization of bacteria within the bee gut without the time-consuming sample preparation and expensive probes required for fluorescence in situ hybridization. No genetic tools have been reported for the Gram-positive members of the bee gut microbiota, Bifidobacteria and Lactobacillus. However, Rangberg et al. introduced a recombinant plasmid into the hive-associated bacterium L. kunkeei and showed that engineered strains had no negative effects on bee health66,67. This was done to validate the potential of L. kunkeei for “paratransgenesis,” that is, the ability of engineered gut bacteria to improve host health and pathogen resistance. As interest in microbiome engineering for human health expands68, the bee gut microbiome provides a useful model to study the stability and function of engineered bacteria in the gut context.
Experimental evidence for roles of bee gut bacteria
The field of microbiota research is young, but the honey bee model system has already been used to investigate several important questions relating to the function and evolution of host-associated gut communities (Figure 3). Here we present an overview of significant findings in this system.
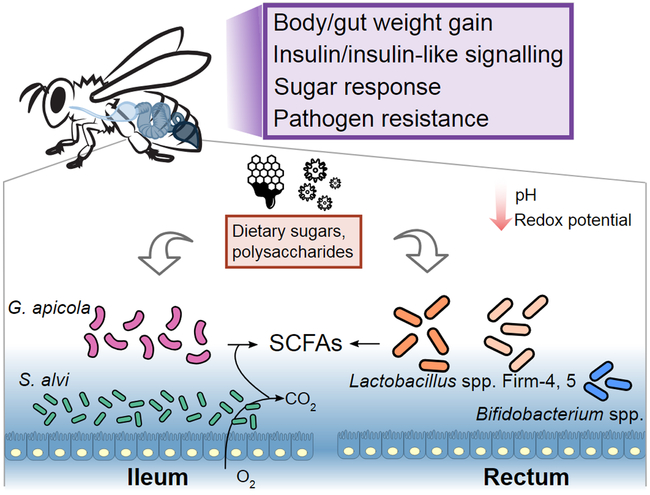
Figure 3.
Summary of the effects of the honey bee gut microbiota on host and the gut microbial metabolism. The dominant members in the ileum and rectum ferment sugars and polysaccharides from the host diet (honey and pollen) to SCFAs. Oxygen consumption by the species S. alvi, which forms a layer attached to the inner gut wall, maintains an anoxic gut environment. The presence of gut bacteria can also reduce the gut pH and redox potential. Moreover, gut bacteria have various effects on the weight gains, insulin signaling, behaviors, and pathogen resistance of the hosts.
Effects on endocrine signaling and behavior
A study comparing microbiota-free to conventional honey bee workers has shown that the gut microbiota is required for normal weight gain69 and that the midgut and ileum of conventional bees are heavier than those of microbiota-free bees (Figure 3). This effect on weight gain is associated with shifts in endocrine signaling and gene expression70,71, including changes in insulin/insulin-like signaling69 (also observed in Drosophila72) and increased levels of vitellogenin, a nutritional status regulator in honey bees73. Kešnerová et al. discovered that Bifidobacterium asteroides stimulates the production of host-derived prostaglandins and juvenile hormone derivatives known to impact bee development74. Interestingly, Schwarz et al.45 demonstrated downregulation of vitellogenin expression in gnotobiotic workers mono-inoculated with S. alvi and subsequently infected with a trypanosomatid parasite (Lotmaria passim) under hive conditions. As vitellogenin also regulates development of social behaviors in honey bees73, these observations suggest a role for the gut microbiota in influencing bee social behavior.
To date, few experiments have addressed the possible links between the honey bee gut microbiota and behavior. Gut microbes may affect host behavior by altering levels of biogenic amines, such as octopamine, dopamine and serotonin. Levels of these amines in worker brains vary seasonally, and are higher in the summer when foraging activity is highest75. Levels are significantly lower in brains of newly-emerged, microbiota-free bees relative to brains of older, conventional adults75. Conventional and microbiota-free bees do behave differently: conventional bees respond to sucrose more readily and thus feed more, which is consistent with observed shifts in insulin signaling69. These results provide compelling evidence that the gut microbiota can alter host behavior and hormonal signaling.
Intersection of host and microbiota metabolism
Gut microbes associated with humans and many other plant-eating animals utilize recalcitrant dietary glycans and complex carbohydrates as substrates for growth76,77. Pollen is a key component of the bee diet, and the only source of amino acids, fat, vitamins and minerals78. Most of these nutrients are absorbed by the host midgut, leaving only the compounds that are most difficult to digest–including pollen cell wall components such as cellulose, hemicellulose, and pectin from pollen cell walls79–to be broken down by the microbial community in the hindgut. Recent studies have illuminated the role of the bee gut microbiota in breaking down such components of the host diet and, in some cases, have identified metabolic activities associated with specific members of this community. For example, metagenomic and genomic analyses attributed the genes enabling pectin degradation to G. apicola10,11, a result that was confirmed when metabolomics analysis documented increased galacturonic acid in honey bee ileums colonized by conventional microbiota (mainly G. apicola and S. alvi) compared to ileums of microbiota-free bees69. Guts of conventional bees also contained large amounts of acetate and other short-chain fatty acids (SCFAs) relative to microbiota-free individuals69, indicating that these SCFAs are largely produced via microbial fermentation (Figure 3). In the bee gut, anoxia is maintained by S. alvi, which associates with the gut wall and uses acetate as the electron donor for consumption of O2 in vitro 69 (Figure 3). This is significant, as alteration of the gut microenvironment can strongly affect metabolic activities. Among bee gut bacteria known to produce SCFAs, Lactobacillus Firm-5 is the main producer of fermentation products succinate and pimelate, while B. asteroides is the main producer of valerate74. Using gnotobiotic bees inoculated with defined communities, Kešnerová et al.74 found that Bifidobacterium and Lactobacillus Firm-4 and Firm-5 can digest other pollen components, including flavonoids and compounds in the outer pollen wall and coat, such as ω-hydroxy acids and phenolamides. Lactobacillus Firm-5 and G. apicola release ω-hydroxy acids from the outer pollen wall, likely facilitating utilization of these compounds by Lactobacillus Firm-4 and B. asteroides74.
Though not all microbe-microbe interactions can be inferred from the abundance of particular compounds in mono-colonized models, many examples support the cross-feeding of essential metabolites between bee gut bacteria. Kešnerová et al.74 showed several complementary pathways suggestive of such cross-feeding. These involve the production and use of pyruvate, which accumulates in guts of bees mono-colonized with G. apicola, but which decreases in concentration in bee guts colonized by S. alvi, a species which lacks a functional glycolysis pathway47. The observation that the growth of S. alvi in culture is improved by the addition of supernatant from G. apicola cultures further supports the premise that these species engage in cross-feeding interactions74. All members of the bee gut community salvage nucleosides from the gut environment, except for S. alvi and B. apis, which encode a functional nucleoside biosynthetic pathway17,47,74 and may produce nucleosides utilized by other bacteria. S. alvi and B. apis also convert carboxylic acids and keto acids–malate, fumarate, citrate, and α-ketoglutarate–through the TCA cycle17,47,80.
Colonization determinants of specialized gut bacteria
Comparison of host and microbe phylogenies suggests that the assemblages of S. alvi and G. apicola strains found in social bees reflect a long evolutionary history of co-diversification with hosts and limited host switching52,81. Native strains easily outcompete non-native strains47 during colonization of microbiota-free hosts, although some strains can also colonize non-native hosts closely related to their native host species52. The gut communities of different, co-occurring Apis species remain distinct52, though some evidence supports transmission of pathogens and parasites between different bee species82. A study of S. alvi strain distributions in field-collected bees using fine-scale markers showed that no movement occurs between Apis and Bombus83, which is consistent with laboratory experiments on host specificity.
Environmental bacteria are generally unable to colonize the honey bee gut, indicating that gut-associated bacteria have adaptations which enable them to tolerate or evade the host immune system and other stressors within the gut. An analysis of S. alvi genes required to establish within the host gut utilized transposon mutagenesis and high-throughput sequencing to identify nearly 400 genes that contribute to fitness during colonization of the bee gut84. These included genes in pathways required for colonization in mammalian models85. Among these were type IV-pili and adhesion factors, which are likely to be important for localization of S. alvi to the gut wall, amino acid biosynthesis pathways, and DNA repair and stress response pathways. These findings provide insight into the challenges that S. alvi experiences during colonization of the bee gut, including nutrient scarcity in the ileum and competition for resources.
Effects on immune functioning
Gut microbes can modulate host immune function, which could indirectly affect other microbes and may affect host fitness. Colonization by a conventional microbiota or by a single S. alvi isolate results in upregulation of the antimicrobial peptides apidaecin and hymenoptaecin in gut epithelial cells86. A more dramatic immune response occurs when F. perrara, a bacterium present in many but not all honey bees, colonizes the honey bee pylorus46, where the midgut transitions into the ileum. Colonization by F. perrara triggers the development of a ‘scab’ phenotype consisting of a dark ring around part of the gut circumference87. This dark ring is produced by melanization, a honey bee immune response88. Although F. perrara interacts with the honey bee immune system, it is not yet clear whether this interaction is harmful or beneficial, for example, via immune priming89.
Perturbation of the native microbiota
Perturbation of the normal, established gut community, using antibiotics or other disruptors, provides further insight into microbiota function. Raymann et al.12 measured the effects of treatment with tetracycline, a broad-spectrum antibiotic, on the size and composition of the honey bee gut microbiota and on host fitness. Antibiotic-treated bees showed altered relative abundance and diversity of core microbial taxa90, elevated abundance of non-core taxa, decreased survival in the hive, and increased mortality when exposed to the opportunistic pathogen S. marcescens kz11. However, none of the dominant members of the microbiota were fully eliminated after antibiotic treatment. Perturbation of the gut microbiota was harmful even in the absence of opportunistic pathogens, as conventional bees exhibited increased mortality after antibiotic treatment in sterile laboratory conditions, relative to microbiota-free bees treated with antibiotic. In another study, Li et al.91 found that perturbation of the worker gut microbiota with antibiotics lowered immune responses and elevated susceptibility to a major microsporidian parasite (Nosema ceranae) that invades through the midgut epithelium.
The honey bee gut microbiota shows parallels with the human gut microbiota
Although the importance of bee health is itself a major motivation for studying the bee gut microbiota, this system also offers the advantage of having numerous parallels to the human gut microbiota.
Specificity and evolutionary adaptation to hosts:
As in the human gut microbiota, most dominant members of the honey bee gut community are found solely in the host gut environment. Gut bacteria in both honey bees and humans are likely to be specifically adapted to these habitats, as they have coevolved with their hosts over millions of years52,92. The five most abundant bacterial species associated with the guts of modern corbiculate (social) bee hosts, which include species of Apis (Asian honey bees), Bombus (bumble bees), and Meliponini (stingless bees), most likely descend from a core community present in the shared ancestor of these bees, with subsequent strain divergence and gains and losses of taxa producing the gut communities found today52.
Transmission through social interactions:
Both bee and primate gut bacteria are primarily transmitted through social interactions46,92,93. In bees, phylogenetic analyses of bacterial strains present across corbiculate bee hosts suggest that their acquisition coincided with the transition to social lifestyles52. Core taxa of social bees are not found in solitary bees or other related insects such as wasps, nor have they been isolated from other environments14. In contrast, many invertebrate gut communities have erratic compositions dominated by bacteria from environmental sources (e.g., 94–97).
Strain variation:
Although the bee gut microbiota possesses a limited number of bacterial species, each component species exhibits extensive strain variation, which is also true of the human gut microbiota98. Deep sequencing of single-copy protein coding genes revealed high levels of S. alvi and G. apicola strain diversity in bee guts52,83,90. These two species also have large pools of accessory genes, which are not present in all strains. For example, many of the accessory genes in G. apicola strains are involved in carbohydrate metabolism47,99, but only some strains encode genes for utilization of components of pollen cell walls100 or monosaccharides that are toxic to the bee host11, suggesting that strains of the same species differ in contributions to host nutrition. Different strains have distinct assortments of Type VI Secretion System (T6SS)-associated toxin and antitoxin genes101, which may influence which combinations of strains are capable of co-colonizing a single host. Additionally, some Apibacter strains encode a T6SS similar to the T6SS used in interbacterial antagonism by Bacteroidetes species in the human gut102,103.
Pathogen resistance:
Perturbation of gut microbial communities can have negative repercussions for host health. In humans, dysbiosis, or abnormal composition or function of the microbiota, is associated with numerous diseases and can result from antibiotic treatment, poor diet, and other disturbances. For example, destabilization of the gut microbiota due to antibiotic treatment increases susceptibility to Clostridium difficile infections in humans104. Similarly, perturbation of the bee gut microbiota by antibiotics or other chemicals increases susceptibility to infection by S. marcescens12.
Role in fermentation and SCFA production:
As in humans and other animals105,106, the gut microbiota of honey bees is concentrated in the distal gut where it contributes to the digestion and fermentation of complex carbohydrate polymers derived from plant cell walls. This role of the honey bee gut microbiota contrasts with the gut communities of some other insects. For example, in Drosophila, the gut community occupies the midgut and is not implicated in digesting plant cell wall components, though it is important in immune and developmental signaling107. Even herbivorous lepidopteran larvae, which consume only plant material, appear not to rely on gut microbiota for digestion or nutrition96. Oxygen availability within the gut can influence patterns of colonization and can affect the mutualistic interactions between gut microbes108,109. Most herbivorous insects’ guts are nearly anoxic110, unlike the gut of Drosophila which contains oxygen and is dominated by aerobes111. Anoxia in the bee ileum is maintained by the respiration of S. alvi, a bacterium that associates with the ileum wall, which is fueled by acetate, the most abundant SCFA in the gut (Figure 3)69.
History of antibiotic exposure:
Long-term antibiotic use may have impacted the diversity within human gut communities and has resulted in high frequencies of resistance determinants112. Likewise, antibiotic exposure has affected gut communities of honey bees, particularly in the United States and other countries where beekeepers have used antibiotics since the late 1940’s to control or prevent larval bacterial diseases known as foulbrood113–115. This practice has resulted in high frequencies of antibiotic resistance determinants in core gut bacteria isolated from bees in the United States, in contrast to gut bacteria of honey bees from countries which do not permit the use of antibiotics in beekeeping116,117. In both human and honey bee gut communities, resistance determinants have been exchanged among community members through horizontal transfer118. Furthermore, antibiotic exposure has an immediate impact on the size and diversity of honey bee gut communities12,90.
Limitations of the honey bee as a model for gut microbiota
We have described the utility of the honey bee as a model for microbiota research, but this system does have some drawbacks and important differences from humans. As with any model system, the honey bee microbiota is most valuable as a tool for understanding general principles of microbe-microbe and microbe-host interactions. Care should be taken in any attempt to directly apply findings from the bee system to the human gut microbiota, as physiology and diet differ markedly between bees and humans and the bee gut microbiota, which has comparatively low biomass and diversity, may not reflect all of the processes that occur in the human gut microbiota119.
Practical considerations when planning honey bee experiments include the seasonal life cycle of the honey bee and the challenges posed by bee genetics. We have considered rate of brood production as an advantage of the honey bee system because of the potential to perform experiments with large sample sizes. However, honey bee brood production is seasonal and the queen’s oviposition rate is drastically reduced in winter, when the number of workers in the colony can drop below 10,000 bees. Thus, newly emerged, microbiota-free workers are not available from outdoor hives during winter months in most geographic regions. Methods exist for in vitro rearing of honey bees in the laboratory through artificial feeding of larvae48,120, and for maintaining whole colonies indoors, but these add to the complexity and expense of rearing, and bees produced through these approaches may have developmental abnormalities.
Seasonal shifts also subject bees to fluctuations in environmental conditions, food availability, and nutritional requirements 121, which can affect gut community composition and potentially influence experimental results122. Abundances of honey bee pathogens also vary seasonally123. Both pathogens and gut bacteria alter expression of immune genes, including antimicrobial peptides86,124, so studies of the microbiota could be confounded by the presence of a pathogen or vice versa. Another consideration is that, while the method of obtaining microbiota-free bees (discussed above) is effective for obtaining workers with little to no bacteria in their guts, it does not prevent these workers from developing viral infections. Honey bee viruses, such as Deformed Wing Virus (DWV), are common and can be transmitted to bees by parasitic Varroa destructor mites during pupation124.
Honey bees are not amenable to genetic manipulation or crossing experiments, due to their reproductive division of labor. A single queen can mate with multiple males, or can be artificially inseminated with a single male’s sperm125, so workers from the same hive are genetically similar as half- or full-sisters. Workers from different hives may have different genetic backgrounds, so experiments should control for source hive. The only tool currently available for genetic manipulation of honey bees is an injectable piggyBac-derived transposon cassette126, and its use is not widespread. Generation of transgenic honey bees using CRISPR/Cas9 is an alternative, but to date only one study has reported successful production of mutants with this approach127. Maintaining lines generated using either strategy would likely be more difficult than in other models, such as Drosophila, due to the need to propagate queens.
In the absence of stable transgenic lines, the use of RNA-interference (RNAi) to modulate expression of target genes is widespread in insect research and established in honey bees128,129. Feeding or injecting bees with double stranded RNA (dsRNA) results in gene knockdown, though, as with other insects, the magnitude and effectiveness of knockdown is variable130. Honey bees also possess a sequence-non-specific antiviral response, which requires the inclusion of appropriate controls in RNAi studies131. Regardless, RNAi approaches will allow researchers to selectively downregulate honey bee genes, such as those encoding immune effectors or immune signaling pathways, to assess their functions in structuring the gut microbial community.
Conclusions and future outlook
The studies summarized in this review demonstrate the many advantages of the honey bee model system for microbiota research. These advantages include intrinsic properties of the gut microbiota, such as its simple and specific composition, and established methods for cultivating and genetically manipulating the core gut bacterial species. Other benefits of this system stem from the biology of the host, such as the ability to experimentally colonize large numbers of gnotobiotic host animals and the ease of rearing bees economically. Years of bee research in multiple fields of study has built a knowledge base that includes genomic, ecological, behavioral, developmental, and physiological information as well as experimental protocols.
One motivation for studying the bee gut microbiota is the increasing evidence for its role in bee health: such studies may enable improvements in beekeeping that help to prevent damaging colony losses and preserve these important pollinators. Additionally, the bee gut microbiota has numerous similarities to that of humans (Figure 1), and may be useful for establishing general rules of microbiome function.
References
- 1.Ghannoum MA et al. Characterization of the oral fungal microbiome (mycobiome) in healthy individuals. PLoS Pathog 6, e1000713 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Findley K et al. Topographic diversity of fungal and bacterial communities in human skin. Nature 498, 367–370 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drell T et al. Characterization of the vaginal micro- and mycobiome in asymptomatic reproductive-age Estonian women. PLoS One 8, e54379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Donaldson GP, Lee SM & Mazmanian SK Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol 14, 20–32 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trompette A et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med 20, 159–166 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Rooks MG & Garrett WS Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol 16, 341–352 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamada N, Chen GY, Inohara N & Nunez G Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol 14, 685–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qin N et al. Alterations of the human gut microbiome in liver cirrhosis. Nature 513, 59–64 (2014). [DOI] [PubMed] [Google Scholar]
- 9.Zitvogel L et al. Cancer and the gut microbiota: an unexpected link. Sci. Transl. Med 7, 271ps271 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel P, Martinson VG & Moran NA Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 109, 11002–11007 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zheng H et al. Metabolism of toxic sugars by strains of the bee gut symbiont Gilliamella apicola. mBio 7, e01326–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Raymann K, Shaffer Z & Moran NA Antibiotic exposure perturbs the gut microbiota and elevates mortality in honeybees. PLoS Biol 15, e2001861 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee FJ, Rusch DB, Stewart FJ, Mattila HR & Newton IL Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ. Microbiol 17, 796–815 (2015). [DOI] [PubMed] [Google Scholar]
- 14.Martinson VG et al. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol 20, 619–628 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Moran NA, Hansen AK, Powell JE & Sabree ZL Distinctive gut microbiota of honey bees assessed using deep sampling from individual worker bees. PLoS One 7, e36393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sabree ZL, Hansen AK & Moran NA Independent studies using deep sequencing resolve the same set of core bacterial species dominating gut communities of honey bees. PLoS One 7, e41250 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kwong WK & Moran NA Gut microbial communities of social bees. Nat. Rev. Microbiol 14, 374–384 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leonard SP et al. Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol 7, 1279–1290 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonilla-Rosso G & Engel P Functional roles and metabolic niches in the honey bee gut microbiota. Curr. Opin. Microbiol 43, 69–76 (2018). [DOI] [PubMed] [Google Scholar]
- 20.Anderson KE & Ricigliano VA Honey bee gut dysbiosis: a novel context of disease ecology. Curr. Opin. Insect Sci 22, 125–132 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Zayed A & Robinson GE Understanding the relationship between brain gene expression and social behavior: lessons from the honey bee. Annu. Rev. Genet 46, 591–615 (2012). [DOI] [PubMed] [Google Scholar]
- 22.von Frisch K The dance language and orientation of bees. (Harvard University Press, 1967). [Google Scholar]
- 23.Robinson GE, Page RE Jr., Strambi C & Strambi A Hormonal and genetic control of behavioral integration in honey bee colonies. Science 246, 109–112 (1989). [DOI] [PubMed] [Google Scholar]
- 24.Page RE & Peng CY Aging and development in social insects with emphasis on the honey bee, Apis mellifera L. Exp. Gerontol 36, 695–711 (2001). [DOI] [PubMed] [Google Scholar]
- 25.Shpigler HY et al. Deep evolutionary conservation of autism-related genes. Proc. Natl. Acad. Sci. USA 114, 9653–9658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gallai N, Salles JM, Settele J & Vaissiere BE Economic valuation of the vulnerability of world agriculture confronted with pollinator decline. Ecol. Econ 68, 810–821 (2009). [Google Scholar]
- 27.Stokstad E Entomology. The case of the empty hives. Science 316, 970–972 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Consortium HGS Insights into social insects from the genome of the honeybee Apis mellifera. Nature 443, 931–949 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wallberg A et al. A worldwide survey of genome sequence variation provides insight into the evolutionary history of the honeybee Apis mellifera. Nat. Genet 46, 1081–1088 (2014). [DOI] [PubMed] [Google Scholar]
- 30.Nelson RM, Wallberg A, Simoes ZLP, Lawson DJ & Webster MT Genomewide analysis of admixture and adaptation in the Africanized honeybee. Mol. Ecol 26, 3603–3617 (2017). [DOI] [PubMed] [Google Scholar]
- 31.Goodman AL et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc. Natl. Acad. Sci. USA 108, 6252–6257 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridaura VK et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton DR & Bradley RE Sr. An integrated system for the production of gnotobiotic Anopheles quadrimaculatus. J. Invertebr. Pathol 30, 318–324 (1977). [DOI] [PubMed] [Google Scholar]
- 34.Dillon R & Charnley K Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res. Microbiol 153, 503–509 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Oertel E Metamorphosis in the honeybee. J. Morphol 50, 295–339 (1930). [Google Scholar]
- 36.Martinson VG, Moy J & Moran NA Establishment of characteristic gut bacteria during development of the honeybee worker. Appl. Environ. Microbiol 78, 2830–2840 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bodenheimer FS Studies in Animal Populations. II. Seasonal population-trends of the honey-bee. Q. Rev. Biol 12, 406–425 (1937). [Google Scholar]
- 38.Winston ML The biology of the honey bee. (Harvard University Press, 1987). [Google Scholar]
- 39.Hroncova Z et al. Variation in honey bee gut microbial diversity affected by ontogenetic stage, age and geographic location. PLoS One 10, e0118707 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vojvodic S, Rehan SM & Anderson KE Microbial gut diversity of Africanized and European honey bee larval instars. PLoS One 8, e72106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nelson JA, Lineburg B & Sturtevant AP Growth and feeding of honeybee larvae. (United States Department of Agriculture, 1924). [Google Scholar]
- 42.Jay SC The development of honeybees in their cells. J. Apic. Res 2, 117–134 (2015). [Google Scholar]
- 43.White PB The normal bacterial flora of the bee. J. Pathol. Bacteriol 24, 64–78 (1921). [Google Scholar]
- 44.Gilliam M Microbial sterility of the intestinal content of the immature honey bee, Apis mellifera. Ann. Entomol. Soc. Am 64, 315–316 (1971). [Google Scholar]
- 45.Schwarz RS, Moran NA & Evans JD Early gut colonizers shape parasite susceptibility and microbiota composition in honey bee workers. Proc. Natl. Acad. Sci. USA 113, 9345–9350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powell JE, Martinson VG, Urban-Mead K & Moran NA Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol 80, 7378–7387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kwong WK, Engel P, Koch H & Moran NA Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. USA 111, 11509–11514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schmehl DR, Tomé HVV, Mortensen AN, Martins GF & Ellis JD Protocol for the in vitro rearing of honey bee (Apis mellifera L.) workers. J. Apic. Res 55, 113–129 (2016). [Google Scholar]
- 49.Crane E Bees and beekeeping: science, practice, and world resources. (Comstock Pub. Associates, 1990). [Google Scholar]
- 50.Kasiotis KM, Anagnostopoulos C, Anastasiadou P & Machera K Pesticide residues in honeybees, honey and bee pollen by LC-MS/MS screening: reported death incidents in honeybees. Sci. Total Environ 485–486, 633–642 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Rumkee JCO, Becher MA, Thorbek P & Osborne JL Modeling Effects of Honeybee Behaviors on the Distribution of Pesticide in Nectar within a Hive and Resultant in-Hive Exposure. Environ. Sci. Technol 51, 6908–6917 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Kwong WK et al. Dynamic microbiome evolution in social bees. Sci. Adv 3, e1600513 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cox-Foster DL et al. A metagenomic survey of microbes in honey bee colony collapse disorder. Science 318, 283–287 (2007). [DOI] [PubMed] [Google Scholar]
- 54.Disayathanoowat T, Young JP, Helgason T & Chantawannakul P T-RFLP analysis of bacterial communities in the midguts of Apis mellifera and Apis cerana honey bees in Thailand. FEMS Microbiol. Ecol 79, 273–281 (2012). [DOI] [PubMed] [Google Scholar]
- 55.Ahn K, Xie X, Riddle J, Pettis J & Huang ZY Effects of Long Distance Transportation on Honey Bee Physiology. Psyche 2012, 9 (2012). [Google Scholar]
- 56.Jeyaprakash A, Hoy MA & Allsopp MH Bacterial diversity in worker adults of Apis mellifera capensis and Apis mellifera scutellata (Insecta: Hymenoptera) assessed using 16S rRNA sequences. J. Invertebr. Pathol 84, 96–103 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Corby-Harris V et al. Origin and effect of Alpha 2.2 Acetobacteraceae in honey bee larvae and description of Parasaccharibacter apium gen. nov., sp. nov. Appl. Environ. Microbiol 80, 7460–7472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mohr KI & Tebbe CC Diversity and phylotype consistency of bacteria in the guts of three bee species (Apoidea) at an oilseed rape field. Environ. Microbiol 8, 258–272 (2006). [DOI] [PubMed] [Google Scholar]
- 59.Anderson KE, Rodrigues PA, Mott BM, Maes P & Corby-Harris V Ecological succession in the honey bee gut: shift in Lactobacillus strain dominance during early adult development. Microb. Ecol 71, 1008–1019 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Anderson KE et al. Microbial ecology of the hive and pollination landscape: bacterial associates from floral nectar, the alimentary tract and stored food of honey bees (Apis mellifera). PLoS One 8, e83125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Olofsson TC, Alsterfjord M, Nilson B, Butler E & Vasquez A Lactobacillus apinorum sp. nov., Lactobacillus mellifer sp. nov., Lactobacillus mellis sp. nov., Lactobacillus melliventris sp. nov., Lactobacillus kimbladii sp. nov., Lactobacillus helsingborgensis sp. nov. and Lactobacillus kullabergensis sp. nov., isolated from the honey stomach of the honeybee Apis mellifera. Int. J. Syst. Evol. Microbiol 64, 3109–3119 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rokop ZP, Horton MA & Newton IL Interactions between cooccurring lactic acid bacteria in honey bee hives. Appl. Environ. Microbiol 81, 7261–7270 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Milani C et al. Phylotype-level profiling of lactobacilli in highly complex environments by means of an ITS-based metagenomic approach. Appl. Environ. Microbiol (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Raymann K et al. Imidacloprid decreases honey bee survival rates but does not affect the gut microbiome. Appl. Environ. Microbiol 84 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Segers FH, Kešnerová L, Kosoy M & Engel P Genomic changes associated with the evolutionary transition of an insect gut symbiont into a blood-borne pathogen. ISME J. 11, 1232–1244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rangberg A, Diep DB, Rudi K & Amdam GV Paratransgenesis: an approach to improve colony health and molecular insight in honey bees (Apis mellifera)? Integr. Comp. Biol 52, 89–99 (2012). [DOI] [PubMed] [Google Scholar]
- 67.Rangberg A, Mathiesen G, Amdam GV & Diep DB The paratransgenic potential of Lactobacillus kunkeei in the honey bee Apis mellifera. Benef. Microbes 6, 513–523 (2015). [DOI] [PubMed] [Google Scholar]
- 68.Sonnenburg JL Microbiome engineering. Nature 518, S10 (2015). [DOI] [PubMed] [Google Scholar]
- 69.Zheng H, Powell JE, Steele MI, Dietrich C & Moran NA Honeybee gut microbiota promotes host weight gain via bacterial metabolism and hormonal signaling. Proc. Natl. Acad. Sci. USA 114, 4775–4780 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ihle KE, Baker NA & Amdam GV Insulin-like peptide response to nutritional input in honey bee workers. J. Insect. Physiol 69, 49–55 (2014). [DOI] [PubMed] [Google Scholar]
- 71.Kannan K & Fridell YWC Functional implications of Drosophila insulin-like peptides in metabolism aging, and dietary restriction. Front. Physiol 4 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shin SC et al. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science 334, 670–674 (2011). [DOI] [PubMed] [Google Scholar]
- 73.Nelson CM, Ihle KE, Fondrk MK, Page RE & Amdam GV The gene vitellogenin has multiple coordinating effects on social organization. PLoS Biol 5, 673–677 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kešnerová L et al. Disentangling metabolic functions of bacteria in the honey bee gut. PLoS Biol 15, e2003467 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harris JW & Woodring J Effects of stress, age, season, and source colony on levels of octopamine, dopamine and serotonin in the honey bee (Apis mellifera L.) brain. J. Insect Physiol 38, 29–35 (1992). [Google Scholar]
- 76.Flint HJ, Scott KP, Louis P & Duncan SH The role of the gut microbiota in nutrition and health. Nat. Rev. Gastro. Hepat 9, 577–589 (2012). [DOI] [PubMed] [Google Scholar]
- 77.Flint HJ, Bayer EA, Rincon MT, Lamed R & White BA Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat. Rev. Microbiol 6, 121–131 (2008). [DOI] [PubMed] [Google Scholar]
- 78.Frias BED, Barbosa CD & Lourenco AP Pollen nutrition in honey bees (Apis mellifera): impact on adult health. Apidologie 47, 15–25 (2016). [Google Scholar]
- 79.Mollet JC, Leroux C, Dardelle F & Lehner A Cell wall composition, biosynthesis and remodeling during pollen tube growth. Plants (Basel) 2, 107–147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kwong WK, Zheng H & Moran NA Convergent evolution of a modified, acetate-driven TCA cycle in bacteria. Nat Microbiol 2, 17067 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Koch H, Abrol DP, Li J & Schmid-Hempel P Diversity and evolutionary patterns of bacterial gut associates of corbiculate bees. Mol Ecol 22, 2028–2044 (2013). [DOI] [PubMed] [Google Scholar]
- 82.Fürst MA, McMahon DP, Osborne JL, Paxton RJ & Brown MJ Disease associations between honeybees and bumblebees as a threat to wild pollinators. Nature 506, 364–366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Powell E, Ratnayeke N & Moran NA Strain diversity and host specificity in a specialized gut symbiont of honeybees and bumblebees. Mol. Ecol 25, 4461–4471 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Powell JE, Leonard SP, Kwong WK, Engel P & Moran NA Genome-wide screen identifies host colonization determinants in a bacterial gut symbiont. Proc. Natl. Acad. Sci. USA 113, 13887–13892 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Goodman AL et al. Identifying genetic determinants needed to establish a human gut symbiont its habitat. Cell Host Microbe 6, 279–289 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kwong WK, Mancenido AL & Moran NA Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci 4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Engel P, Bartlett KD & Moran NA The bacterium Frischella perrara causes scab formation in the gut of its honeybee host. mBio 6, e00193–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tang HP Regulation and function of the melanization reaction in Drosophila. Fly 3, 105–111 (2009). [DOI] [PubMed] [Google Scholar]
- 89.Emery O, Schmidt K & Engel P Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol 26, 2576–2590 (2017). [DOI] [PubMed] [Google Scholar]
- 90.Raymann K, Bobay LM & Moran NA Antibiotics reduce genetic diversity of core species in the honeybee gut microbiome. Mol. Ecol 8, 2057–2066 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li JH et al. New evidence showing that the destruction of gut bacteria by antibiotic treatment could increase the honey bee's vulnerability to Nosema infection. PLoS One 12, e0187505 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Moeller AH et al. Cospeciation of gut microbiota with hominids. Science 353, 380–382 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tung J et al. Social networks predict gut microbiome composition in wild baboons. eLife 4, 305224 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wong AC, Chaston JM & Douglas AE The inconstant gut microbiota of Drosophila species revealed by 16S rRNA gene analysis. ISME J 7, 1922–1932 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.van Tol S & Dimopoulos G in Advances in Insect Physiology Vol. 51 (ed Raikhel AS) 243–291 (Academic Press, 2016). [Google Scholar]
- 96.Hammer TJ, Janzen DH, Hallwachs W, Jaffe SP & Fierer N Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. USA 114, 9641–9646 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McFrederick QS et al. Flowers and wild megachilid bees share microbes. Microb. Ecol 73, 188–200 (2017). [DOI] [PubMed] [Google Scholar]
- 98.Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Engel P, Stepanauskas R & Moran NA Hidden diversity in honey bee gut symbionts detected by single-cell genomics. PLoS. Genet 10, e1004596 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Engel P & Moran NA The gut microbiota of insects - diversity in structure and function. FEMS Microbiol. Rev 37, 699–735 (2013). [DOI] [PubMed] [Google Scholar]
- 101.Steele MI, Kwong WK, Whiteley M & Moran NA Diversification of Type VI secretion system toxins reveals ancient antagonism among bee gut microbes. mBio 8, e01630–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Russell AB et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kwong WK, Steele MI & Moran NA Genome sequences of Apibacter spp., gut symbionts of Asian honey bees. Genome Biol. Evol 10, 1174–1179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Carlucci C, Petrof EO & Allen-Vercoe E Fecal microbiota-based therapeutics for recurrent Clostridium difficile Infection, ulcerative colitis and obesity. EBioMedicine 13, 37–45 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mountfort DO, Campbell J & Clements KD Hindgut fermentation in three species of marine herbivorous fish. Appl. Environ. Microbiol 68, 1374–1380 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bauer E et al. Physicochemical conditions, metabolites and community structure of the bacterial microbiota in the gut of wood-feeding cockroaches (Blaberidae: Panesthiinae). Fems Microbiol. Ecol 91, 1–14 (2015). [DOI] [PubMed] [Google Scholar]
- 107.Buchon N, Broderick NA & Lemaitre B Gut homeostasis in a microbial world: insights from Drosophila melanogaster. Nat. Rev. Microbiol 11, 615–626 (2013). [DOI] [PubMed] [Google Scholar]
- 108.Tegtmeier D, Thompson CL, Schauer C & Brune A Oxygen affects gut bacterial colonization and metabolic activities in a gnotobiotic cockroach model. Appl. Environ. Microbiol 82, 1080–1089 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Heinken A & Thiele I Anoxic conditions promote species-specific mutualism between gut microbes in silico. Appl. Environ. Microbiol 81, 4049–4061 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Johnson KS & R, V.B. Oxygen levels in the gut lumens of herbivorous insects. J. Insect Physiol. 46, 897–903 (2000). [DOI] [PubMed] [Google Scholar]
- 111.Cox CR & Gilmore MS Native microbial colonization of Drosophila melanogaster and its use as a model of Enterococcus faecalis pathogenesis. Infect. Immun 75, 1565–1576 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Blaser MJ Antibiotic use and its consequences for the normal microbiome. Science 352, 544–545 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Reybroeck W, Daeseleire E, De Brabander HF & Herman L Antimicrobials in beekeeping. Vet. Microbiol 158, 1–11 (2012). [DOI] [PubMed] [Google Scholar]
- 114.Evans JD & Armstrong T Inhibition of the American foulbrood bacterium, Paenibacillus larvae larvae, by bacteria isolated from honey bees. J. Apic. Res 44, 168–171 (2015). [Google Scholar]
- 115.Evans JD & Spivak M Socialized medicine: individual and communal disease barriers in honey bees. J. Invertebr. Pathol 103, 62–72 (2010). [DOI] [PubMed] [Google Scholar]
- 116.Tian B, Fadhil NH, Powell JE, Kwong WK & Moran NA Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. mBio 3, e00377–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ludvigsen J, Porcellato D, L'Abee-Lund TM, Amdam GV & Rudi K Geographically widespread honeybee-gut symbiont subgroups show locally distinct antibiotic-resistant patterns. Mol. Ecol 26, 6590–6607 (2017). [DOI] [PubMed] [Google Scholar]
- 118.Ludvigsen J, Amdam GV, Rudi K & L'Abee-Lund TM Detection and characterization of streptomycin resistance (strA-strB) in a honeybee gut symbiont (Snodgrassella alvi) and the associated risk of antibiotic resistance transfer. Microb. Ecol doi: 10.1007/s00248-018-1171-7 (2018). [DOI] [PubMed] [Google Scholar]
- 119.Thursby E & Juge N Introduction to the human gut microbiota. Biochem. J. 474, 1823–1836 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Crailsheim K et al. Standard methods for artificial rearing of Apis mellifera larvae. J. Apic. Res 52, 1–16 (2013). [Google Scholar]
- 121.Bonoan RE, O'Connor LD & Starks PT Seasonality of honey bee (Apis mellifera) micronutrient supplementation and environmental limitation. J. Insect. Physiol 107, 23–28 (2018). [DOI] [PubMed] [Google Scholar]
- 122.Ludvigsen J et al. Shifts in the midgut/pyloric microbiota composition within a honey bee apiary throughout a season. Microbes Environ 30, 235–244 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Glenny W et al. Honey bee (Apis mellifera) colony health and pathogen composition in migratory beekeeping operations involved in California almond pollination. PLoS One 12, e0182814 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zanni V, Galbraith DA, Annoscia D, Grozinger CM & Nazzi F Transcriptional signatures of parasitization and markers of colony decline in Varroa-infested honey bees (Apis mellifera). Insect Biochem. Mol. Biol 87, 1–13 (2017). [DOI] [PubMed] [Google Scholar]
- 125.Laidlaw HH Artificial insemination of the queen bee (Apis mellifera L.): morphological basis and results. J. Morphol 74, 429–465 (1944). [Google Scholar]
- 126.Schulte C, Theilenberg E, Muller-Borg M, Gempe T & Beye M Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). Proc. Natl. Acad. Sci. USA 111, 9003–9008 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kohno H, Suenami S, Takeuchi H, Sasaki T & Kubo T Production of knockout mutants by CRISPR/Cas9 in the european honeybee, Apis mellifera L. Zoolog. Sci 33, 505–512 (2016). [DOI] [PubMed] [Google Scholar]
- 128.Marco Antonio DS, Guidugli-Lazzarini KR, do Nascimento AM, Simoes ZL & Hartfelder K RNAi-mediated silencing of vitellogenin gene function turns honeybee (Apis mellifera) workers into extremely precocious foragers. Naturwissenschaften 95, 953–961 (2008). [DOI] [PubMed] [Google Scholar]
- 129.Chen YP & Evans JD Managed pollinator CAP coordinated agricultural project A national research and extension initiative to reverse pollinator decline RNAi in treating honey bee diseases. American Bee Journal 152, 1171–1173 (2012). [Google Scholar]
- 130.Yu N et al. Delivery of dsRNA for RNAi in insects: an overview and future directions. Insect Sci 20, 4–14 (2013). [DOI] [PubMed] [Google Scholar]
- 131.Flenniken ML & Andino R Non-specific dsRNA-mediated antiviral response in the honey bee. PLoS One 8, e77263 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Killer J, Dubna S, Sedlacek I & Svec P Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int. J. Syst. Evol. Microbiol 64, 152–157 (2014). [DOI] [PubMed] [Google Scholar]
- 133.Bottacini F et al. Bifidobacterium asteroides PRL2011 genome analysis reveals clues for colonization of the insect gut. PLoS One 7, e44229 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ellegaard KM et al. Extensive intra-phylotype diversity in lactobacilli and bifidobacteria from the honeybee gut. BMC Genomics 16, 1–22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Scardovi V & Trovatelli LD New species of bifid bacteria from Apis mellifica L. and Apis indica F. A contribution to the taxonomy and biochemistry of the genus Bifidobacterium. Zentralbl. Bakteriol. Parasitenkd. Infektionskr. Hyg 123, 64–88 (1969). [PubMed] [Google Scholar]
- 136.Kwong WK & Moran NA Cultivation and characterization of the gut symbionts of honey bees and bumble bees: description of Snodgrassella alvi gen. nov., sp. nov., a member of the family Neisseriaceae of the Betaproteobacteria, and Gilliamella apicola gen. nov., sp. nov., a member of Orbaceae fam. nov., Orbales ord. nov., a sister taxon to the order 'Enterobacteriales' of the Gammaproteobacteria. Int. J. Syst. Evol. Microbiol 63, 2008–2018 (2013). [DOI] [PubMed] [Google Scholar]
- 137.Engel P, Kwong WK & Moran NA Frischella perrara gen. nov., sp. nov., a gammaproteobacterium isolated from the gut of the honeybee, Apis mellifera. Int. J. Syst. Evol. Microbiol 63, 3646–3651 (2013). [DOI] [PubMed] [Google Scholar]
- 138.Kešnerová L, Moritz R & Engel P Bartonella apis sp. nov., a honey bee gut symbiont of the class Alphaproteobacteria. Int. J. Syst. Evol. Microbiol 66, 414–421 (2016). [DOI] [PubMed] [Google Scholar]
- 139.Roh SW et al. Phylogenetic characterization of two novel commensal bacteria involved with innate immune homeostasis in Drosophila melanogaster. Appl. Environ. Microbiol 74, 6171–6177 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kwong WK & Moran NA Apibacter adventoris gen. nov., sp. nov., a member of the phylum Bacteroidetes isolated from honey bees. Int. J. Syst. Evol. Microbiol 66, 1323–1329 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Corby-Harris V & Anderson KE Draft genome sequences of four Parasaccharibacter apium strains isolated from honey bees. Genome Announc. 6, e00165–18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Endo A et al. Characterization and emended description of Lactobacillus kunkeei as a fructophilic lactic acid bacterium. Int. J. Syst. Evol. Microbiol 62, 500–504 (2012). [DOI] [PubMed] [Google Scholar]
- 143.Endo A & Okada S Reclassification of the genus Leuconostoc and proposals of Fructobacillus fructosus gen. nov., comb. nov., Fructobacillus durionis comb. nov., Fructobacillus ficulneus comb. nov. and Fructobacillus pseudoficulneus comb. nov. Int. J. Syst. Evol. Microbiol 58, 2195–2205 (2008). [DOI] [PubMed] [Google Scholar]
- 144.Chouaia B et al. Acetic acid bacteria genomes reveal functional traits for adaptation to life in insect guts. Genome Biol. Evol 6, 912–920 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Burritt NL et al. Sepsis and hemocyte loss in honey bees (Apis mellifera) Infected with Serratia marcescens strain sicaria. PLoS One 11, e0167752 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Tian B & Moran NA Genome sequence of Hafnia alvei bta3_1, a bacterium with antimicrobial properties isolated from honey bee gut. Genome Announc 4, e00439–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]