Abstract
Dietary fiber is an important food source for members of the human gut microbiome. Members of the dominant Bacteroidetes phylum capture diverse polysaccharides via the action of multiple cell surface proteins encoded within Polysaccharide Utilization Loci (PUL). The independent activities of PUL-encoded glycoside hydrolases (GH) and surface glycan-binding proteins (SGBPs) for the harvest of various glycans have been studied in detail, but how these proteins work together to coordinate uptake is poorly understood. Here, we combine genetic and biochemical approaches to discern the interplay between the BoGH9 endoglucanase and the xyloglucan-binding proteins SGBP-A and SGBP-B from the Bacteroides ovatus Xyloglucan Utilization Locus (XyGUL). The expression of BoGH9, a weakly active xyloglucanase in isolation, is required in a strain that expresses a non-binding version of SGBP-A (SGBP-A*). The crystal structure of the BoGH9 enzyme suggests the molecular basis for its robust activity on mixed-linkage β-glucan compared to xyloglucan. Yet, catalytically inactive site-directed mutants of BoGH9 fail to complement the deletion of the active BoGH9 in a SGBP-A* strain. We also find that SGBP-B is needed in an SGBP-A* background to support growth on xyloglucan, but that the nonbinding SGBP-B* protein acts in a dominant negative manner to inhibit growth on xyloglucan. We postulate a model whereby the SGBP-A, SGBP-B and BoGH9 work together at the cell surface, likely within a discrete complex, and that xyloglucan binding by SGBP-B and BoGH9 may facilitate the orientation of the xyloglucan for transfer across the outer membrane.
Keywords: Bacteroidetes, xyloglucan utilization locus, polysaccharide utilization system, Sus-like system, lipoprotein
Introduction
The Bacteroidetes phylum is a ubiquitous and dominant inhabitant of the human gut 1 Bacteroides thetaiotaomicron and Bacteroides ovatus, two representative species, devote ~20% of their genomes to encoding proteins with functions involved in carbohydrate catabolism 2. Members of the Bacteroidetes encode a diverse array of glycoside hydrolases compared to other members of the gut microbiota and these impart their broad capacity for glycan degradation 3; 4. These glycoside hydrolase (GH) genes are organized within discrete gene clusters termed Polysaccharide Utilization Loci (PUL) that also express gene products for glycan capture and import. Individual PUL are transcriptionally stimulated in the presence of their cognate glycan 5; 6. A noteworthy feature of each PUL is the presence of two genes whose products are homologous to proteins within the starch utilization system (Sus) of B. thetaiotaomicron: SusC, a putative TonB dependent transporter (TBDT) and SusD, a cell surface glycan-binding protein (SGBP). Additional SGBPs are also often encoded within PUL and contribute to glycan capture at the cell surface, but the precise roles of these proteins are variable and substrate-dependent 7; 8; 9.
B. ovatus dedicates one of its PUL to the uptake of xyloglucans (XyGs) 10, a family of prominent plant cell wall polysaccharides in commonly consumed vegetables such as tomato, lettuce and peppers 11. XyGs contain a β(1,4)-linked glucan backbone that is decorated with α(1,6) xylosyl substitutions. Depending on the source, these xylose chains can be extended with additional monosaccharides such as galactose, fucose, and arabinose 11; 12; 13. XyG is an important glycan to the human gut microbiota, as underscored by the observation that ~92% of human microbiomes contain PULs homologous to the B. ovatus Xyloglucan Utilization Locus (XyGUL) 10.
The B. ovatus XyGUL contains twelve genes responsible for the capture, import, and complete degradation of XyG (Fig 1). Most of the individual GH and SGBP components of this PUL have been structurally and functionally characterized, making this an important model system for understanding polysaccharide recognition and degradation by the Bacteroidetes 8; 10; 14 Within the XyGUL, five genes encode outer membrane proteins that initiate XyG utilization at the cell surface: a SusD-like protein (SGBP-A), an additional SGBP (SGBP-B) that mediates substrate binding, a GH9 (BoGH9), a GH5 endo-xyloglucanase (BoGH5A) that performs initial polysaccharide backbone cleavage, and a membrane-spanning SusC-like TBDT that is responsible for import of product oligosaccharides to the periplasm. Additional structural and biochemical analysis of the six periplasmic GHs has contributed to a holistic understanding of XyG disassembly into monosaccharides for cytosolic uptake and metabolism.10; 14.
Figure 1: Overview of the Xyloglucan Utilization Locus (XyGUL) in B. ovatus.
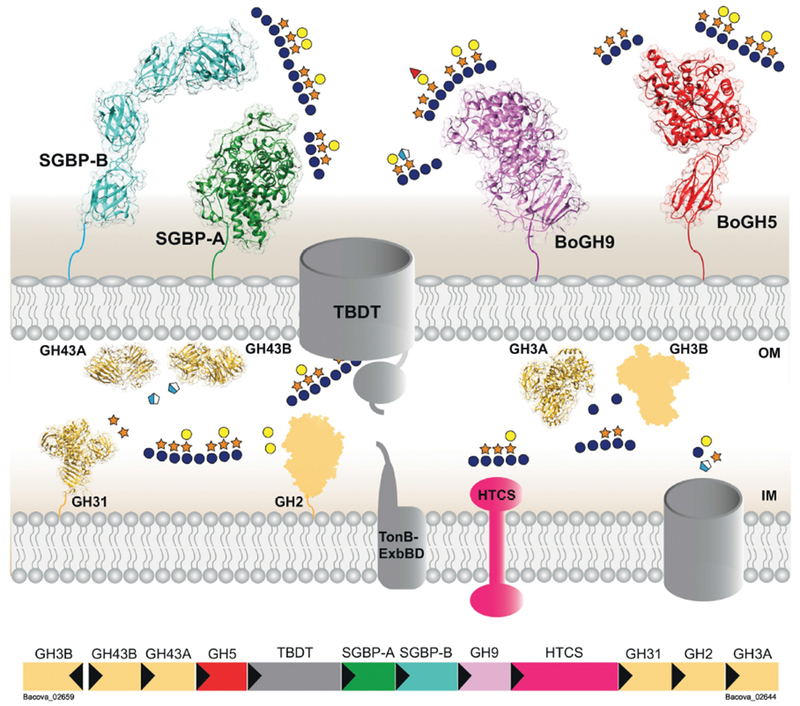
Xyloglucan is bound by the XyG-binding proteins SGBP-A and SGBP-B, and the endo-glucanases BoGH5A and BoGH9 cleave along the polysaccharide backbone to generate XyG oligosaccharides that can be imported into the cell by the TonB-dependent transporter (TBDT). Two α-arabinofuranosidases (GH43A/B), one β-galactosidase (GH2), one α-xylosidase (GH31), and two β-glucosidases (GH3A/B) disassemble these oligosaccharides into monosaccharides that enter the cytoplasm through an unknown transporter. A hybrid two-component system (HTCS)-like regulator senses an unknown XyG-derived substrate that leads to the activation of XyGUL transcription.
An outstanding question regarding the XyGUL is the function of the cell surface BoGH9. Notably, the B. ovatus XyGUL is the only XyGUL identified thus far that includes a GH9 member, and our previous study indicated that the very weak endo-(xylo)glucanase activity measured for BoGH9 was insufficient to support growth in the absence of the vanguard BoGH5A endo-xyloglucanase10. These observations led us to hypothesize that BoGH9 may have evolved toward a non-catalytic, substrate-binding function, possibly working in conjunction with the SGBPs 15 Using a combination of structural enzymology of BoGH9 and combinatorial reverse genetics with the SGBP components of the XyGUL, we demonstrate here that synergy amongst these cell surface proteins facilitates XyG capture and import, likely arising from interactions within a discrete multiprotein complex. More generally, our findings further understanding of how collaborative protein-protein and protein-carbohydrate interactions affect the utilization of common dietary glycans by a key member of gut microbiota.
Results
We recently determined the crystal structures of the XyGUL SGBP-A, SGBP-B, and BoGH5A and demonstrated that in-frame deletions of SGBP-A or BoGH5A from the B. ovatus genome abolish XyG utilization 8; 10. However, complementation of the ΔSGBP-A strain with a site-directed mutant allele encoding for SGBP-A that cannot bind XyG (SGBP-A*) restores growth on XyG. Moreover, B. ovatus expressing SGBP-A* but lacking SGBP-B displays wild-type exponential growth on XyG suggesting that glycan binding by the SGBPs is not their sole function. These findings parallel our work on the starch utilization system of B. thetaiotaomicron in which we demonstrated that a binding-deficient allele of the SGBP-A homolog SusD (SusD*) supports growth on maltooligosaccharides when co-expressed with the starch-binding deficient SGBP allele, SusE*. Together these data support that glycan-binding by SGBPs may be of secondary importance to glycan utilization while the presence of these proteins for the formation of a complex around the TBDT is primary 7; 16. Indeed, the crystal structures of two TBDT-SusD-like complexes, BT2261-2264 and BT1762-1763, demonstrate that the SusD/SGBP-A homologs close over the top of, and likely guide their substrate-binding site into, their cognate TBDT 17. That purification of BT2261-2264 included the SGBPs BT2261-2262 supports the idea that these glycan-capturing complexes may be influenced by multiple SGBP interactions beyond the cognate TBDT-SusD/SGBP-A pair.
Structure of the XyGUL GH9
We previously determined the crystal structures of the BoGH5A and the two SGBPs of the XyGUL 8; 10. Therefore, to resolve a gap in our understanding of the structural biology of the XyGUL, and as a prelude to further biochemical and reverse genetic studies, we obtained a tertiary structure of recombinant BoGH9 by X-ray crystallography. The 1.4Å structure of BoGH9 (Rw = 16.8%, Rf = 19.7%; Table 1) is composed of two domains: an N-terminal Ig-like fold (residues 27-115) followed by the catalytic domain that has a canonical (α/α)6 fold typical of this GH family (Fig 2A). The final model was composed of residues 27-583, with only a surface loop (residues 535-545) that could not be resolved in the density.
Table 1:
X-ray Data Collection and Refinement Statistics for BoGH9
| PDB | 6DHT |
| Wavelength (nm) | 0.979 |
| Resolution range (Å) | 44.74 - 1.42 (1.47 - 1.42) |
| Space group | C2 |
| Unit cell (Å) | a = 130.65 b = 50.41, c = 95.98, β = 109° |
| Total reflections | 731375 (48647) |
| Unique reflections | 109840 (9432) |
| Multiplicity | 6.7 (5.2) |
| Completeness (%) | 98.30 (84.59) |
| Mean I/sigma(I) | 21.0 (2.1) |
| Wilson B-factor | 16.0 |
| Rmerqe | 0.081 (0.59) |
| Rmeas | 0.088 |
| CC1/2 | 0.996 (0.841) |
| CC* | 0.999 (0.956) |
| Reflections used for Rfree | 1898 |
| Rwork | 0.167 (0.28) |
| Rfree | 0.197 (0.31) |
| Number of non-hydrogen atoms | 4957 |
| macromolecules | 4344 |
| ligands | 87 |
| water | 526 |
| Protein residues | 547 |
| RMS(bonds) | 0.009 |
| RMS(angles) | 1.28 |
| Ramachandran favored (%) | 96 |
| Ramachandran outliers (%) | 0.18 |
| Clashscore | 3.22 |
| Average B-factor | |
| all atoms | 23.6 |
| macromolecules | 22.3 |
| ligands | 33.8 |
| solvent | 33.4 |
Parentheses indicate statistics for the highest resolution shell.
Figure 2: Crystal structure of BoGH9 with imidazole.
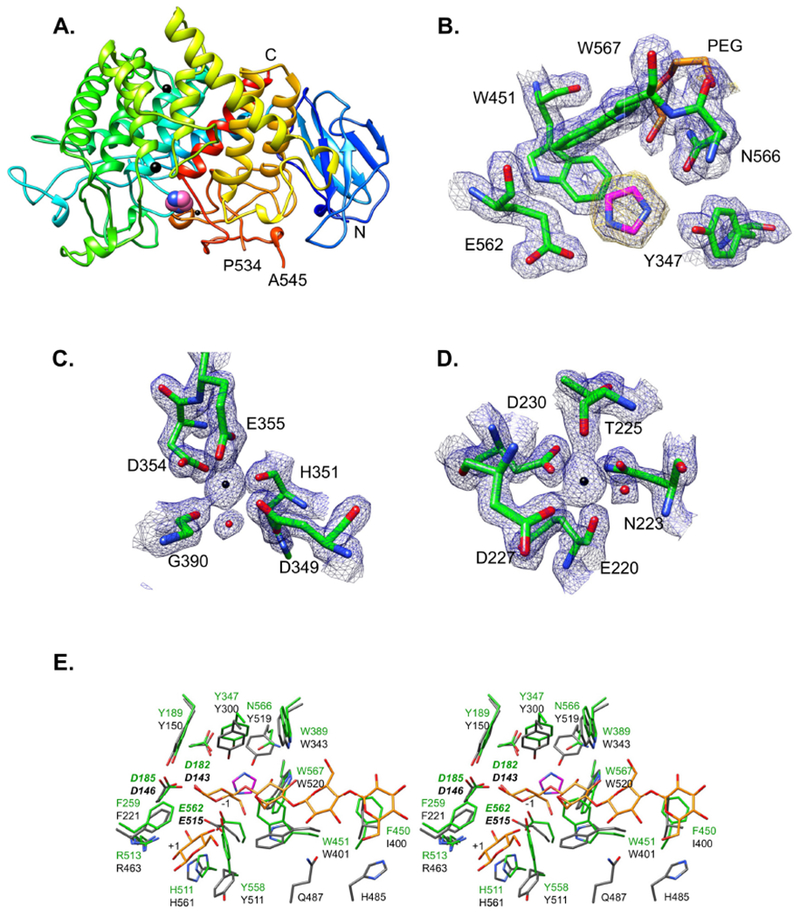
Ribbon cartoon of the BoGH9 backbone, colored blue to red from the N- to C-terminus. Two calcium ions are displayed as large black spheres, and the magnesium is displayed as a smaller black sphere. An imidazole that was trapped in the active site cleft is displayed as pink and blue spheres. The N- and C-termini are labeled, as is the ends of a disordered loop (residues 535-544) that could not be resolved. (B) Close-up view of the imidazole in the active site. Protein is displayed with |2Fo-Fc| density contoured at σ=1.3, and omit density for the imidazole is contoured at 3.0 σ. A PEG molecule (yellow) from the crystallization liquor is also displayed. (C,D) Coordination spheres of the calcium ions with |2Fo-Fc| density contoured at 1.5 σ. (E) Divergent overlay of the BoGH9 active site (green) with the AaCel9A (3H3K) active site (grey sticks) that has cellotetraose and glucose (yellow sticks). Imidazole in the BoGH9 structure is displayed as magenta and blue sticks.
The closest structural homologs of BoGH9 from a DALI search18 are Clostridium thermocellum CelD (CtCelD, PDB id 4CJ1, Z=46.5)19, Alicyclobacillus acidocaldarius cellulase Cel9A (AaCel9A PDB id 3GZK, Z=46.2)20, and a metagenomic cellulase (PDB id 3X17, Z=45.5). Both CtCelD and AaCel9A are well-characterized cellulases 21; 22 and can be superimposed onto the BoGH9 structure with RMSD values of ~1.0 Å for 378 and 366 Cα atom pairs respectively. The combination of an Ig-like N-terminus proceeded by a catalytic domain places BoGH9 within the Type C class of GH9 23, akin to AaCel9A.
Within the active site cleft of BoGH9, an imidazole and PEG molecule from the crystallization liquor were captured (Fig 2B). In addition, density was observed in other parts of the protein that represented two calcium ions and one magnesium ion, both of which were included in the crystallization liquor (as MgCl2 and CaCl2, respectively). The calcium ions are coordinated primarily by amino acid side chains, and display the expected octahedral geometry and atomic distances (~2.4Å) for this metal (Fig 2C,D). Conversely, the magnesium ion is coordinated near the surface of the enzyme by two aspartates and three water molecules, with coordination distances of ~2.1Å. Its presence here may be serendipitous, due to the inclusion of magnesium in the crystallization. Indeed, the coordinating residues of all three ions are conserved in the structure of the C. thermocellum CelD (CtCelD), and capture calcium atoms 19. These divalent cations are thought to lend structural stability to the enzyme, and none are involved directly in catalysis 24.
Comparison of the BoGH9 structure with that of AaCel9A complexed with cellotetraose and glucose (PDB 3H3K, Z-score 50.8 between catalytic domains, Supplemental Fig 1) reveals structural homology within the active site (Fig 2E). Notably, the binding of the imidazole (a known inhibitor of some β-glucosidases 25; 26, here present in high concentration, ~50mM, in the crystallization buffer), between the −1/−2 subsites results in several changes in the orientation of active site residues in BoGH9 compared to AaCel9A. The N atoms of imidazole are coordinated by Y347 and the catalytic acid E562. W451 and Y558 swing towards the imidazole to create a more hydrophobic environment, although neither provide a direct stacking interaction. The equivalent residues in AaCel9A, W401 and Y511, provide a hydrophobic platform to position glucose residues at the −2 and +1 subsites 20. Of the 19 residues that create the active site cleft in AaCel9A, only four are not conserved in BoGH9. Two of these residues are G390 and N391 in BoGH9 (A344 and D345 in AaCel9A), which are proximal to the putative −3 and −4 subsites but do not directly bind substrate in AaCel9A. F450 of BoGH9 is homologous to 1400 in AaCel9A and Y456 of CtCelD within the −4 subsite; the substitution to a Phe in BoGH9 may better support substrate interaction. Likewise, in both the BoGH9 structure and CtCelD, an Asn (as N566 in BoGH9) is occupied by Y519 in AaCel9A near the −2 subsite. Overall, BoGH9 demonstrates a high degree of structural similarity with these representative GH9 members, most notably including a narrow active-site cleft that reflects its poor tolerance of the branched β-glucan XyG (vide infra).
Regarding the disordered loop in BoGH9 extending from residues 535-545, neither the CtCelD nor a homologous metagenomic cellulase (PDB 3X17) display an extended surface loop in this region. However, the AaCel9A has a loop composed of H485-G497 that shapes the −3/−4 subsites of the active site. In AaCe9lA, H485 and Q487 provide hydrogen-bonding coordination with the hydroxyls of adjacent glucose residues. In the equivalent loop of BoGH9, G535 replaces H485 but Q537 has the same position in the sequence as Q487 of AaCel9A. The remainder of this loop sequence is not conserved between BoGH9 and AaCel9A, and the significance for activity is unknown. However, the flexibility of this loop in BoGH9, inferred from its lack of density, suggests that its unlikely to block access to the active site and restrict activity or substrate selection.
BoGH9 has substantial calcium-dependent activity on mixed-linkage β-glucans
Our previous kinetic analysis of recombinant BoGH9 revealed that the enzyme was catalytically feeble toward XyG, as well as other β-glucans. In light of the BoGH9 crystal structure, and the previous report that CtCelD of GH9 requires calcium for activity22, we revisited the enzymology of BoGH9. As observed previously,10 recombinant BoGH9 has very weak and equally poor activity on the representative β-glucans, mixed-linkage β(1-3)/β(1-4)-glucan (MLG), hydroxyethylcellulose (HEC), carboxymethylcellulose (CMC), and XyG in the absence of the addition of Ca2+ to the assay (Table 2, see Supplemental Fig 2 for carbohydrate structures). BoGH9 displays 400-fold lower specific activity towards XyG (0.042 ± 0.012 μmol/min/mg, Table 2) than the vanguard endo-xyloglucanase of the XyGUL, BoGH5A (9.3 ± 0.6 μmol/min/mg). In the presence of 1 mM Ca2+, the specific activity of BoGH9 toward XyG remains essentially unchanged (2-fold increase) while, strikingly, the specific activity toward MLG increases 8.5-fold (Table 2). The specific activities of BoGH9 toward the artificial substrates HEC and CMC are likewise increased, 5.5- and 3.8-fold respectively, in the presence of Ca2+.
Table 2.
Activity of BoGH9 against different polysaccharide substratesa
| Enzyme | Substrate | Specific activity μmol (min.mg)−1 | Specific activity (with Ca2+ 1mM) μmol (min.mg)−1 |
|---|---|---|---|
| BoGH9 | MLG | 0.042 ± 0.012 | 0.36 ± 0.05 |
| XyG | 0.023 ± 0.003 | 0.05 ± 0.01 | |
| HEC | 0.027 ± 0.004 | 0.15 ± 0.05 | |
| CMC | 0.021 ± 0.001 | 0.08 ± 0.01 |
Assays conducted in HEPES-NaOH pH 7.0. Recombinant enzymes were incubated at 37 ºC with the different tested substrates.
Product analysis revealed that, similar to BoGH5A, BoGH9 cleaves the XyG backbone endo-hydrolytically to ultimately yield XyGO1, the natural distribution of variably galactosylated Glc4-backbone-based hepta- to nonasaccharides from tamarind seed XyG (Supplemental Fig 3). BoGH9 hydrolzes MLG to the mixed-linkage tetrasaccharides G3G4G4G, G4G3G4G, and G4G4G3G, which is distinct from the exclusive G4G4G3G product specificity of GH16 MLGases 27. Notably, G3G4G4G was ultimately hydrolyzed to G3G4G in the limit digest. All β(1-4) linked cello-oligosaccharides were not produced, indicating that BoGH9 does not cleave the β(1-3)-linkages in MLG (Supplemental Fig 3).
Examination of complexes in which MLG and XyG oligosaccharide fragments were manually docked into the BoGH9 apo structure indicated the structural basis for the observed substrate specificity of the enzyme. Indeed, a BoGH9:XyGO complex could not be definitively modeled, due to significant steric clashes of highly branched oligosaccharides (e.g. from PDB ID 3ZMR) with sidechains of BoGH9 in the active-site cleft (Supplemental Figure 4A). Modeling of the MLG oligosaccharide G4G3G (e.g. from PDB ID 1ZM1) into the BoGH9 active-site was more favorable, despite the differences in the position of the −2/−3 subsites imposed by the β1,3 linkage (Supplemental Figure 4B). Compared to the conformation of the enzyme in our structure with imidazole, productive BoGH9 and substrate binding likely requires a significant rearrangement of the active site.
BoGH9 is required for growth on XyG in an SGBP-A* strain
The poor activity of BoGH9 towards XyG, even in the presence of calcium, is concordant with the inability of this enzyme to confer growth of B. ovatus on the polysaccharide in the absence of BoGH5A 10. We therefore hypothesized that BoGH9 could play a role in XyG capture rather than degradation, perhaps working in harness with the SGBPs at the cell surface. To test this, we created a set of in-frame deletions and/or point mutant alleles of the BoGH9 and SGBP-B genes in a genetically manipulatable B. ovatus Δtdk strain, hereafter referred to as wild-type 10. In particular, we designed a series of variants to completely ablate substrate binding by removal of key aromatic sidechains in the SGBPs, referred to as SGBP-A* and SGBP-B*, as well as inactive, catalytic residue variants of BoGH9 (Table 3). Mutant strains were passaged overnight in minimal media (MM) containing glucose, then back-diluted 1:200 into parallel cultures containing glucose, xylose, XyGOs, or XyG prior to monitoring growth in liquid culture. The XyGUL is not required for growth on glucose10 or xylose and none of our mutants demonstrated a kinetic growth defect or apparent difference in lag on these monosaccharides (Supplemental Fig 5).
Table 3.
Mutant XyGUL alleles used in this study
| Mutant allele | Mutations (by protein residue number) | Effect | Reference |
|---|---|---|---|
| SGBP-A* | W82A, W283A, W306A | No XyG binding | 8 |
| SGBP-B* | Y363A, W364A | No XyG binding | 8 |
| BoGH9D185N | D185N | Catalytic base mutant | This study |
| BoGH9D185A | D185A | Catalytic base mutant | This study |
| BoGH9E562Q | E562Q | Catalytic acid mutant | This study |
| BoGH9E562A | E562A | Catalytic acid mutant | This study |
While a single in-frame deletion of the gene encoding BoGH9 does not affect growth on XyG or XyGO2 (the natural distribution of Glc8-backbone-based oligosaccharides from tamarind XyG, Supplemental Fig 2)10, a strain containing a combined SGBP-A*/ΔBoGH9 mutation could no longer grow on any of the XyG substrates tested (Fig 3B-E). However, this phenotype could be rescued by complementation of the native BoGH9 gene within the operon (Supplemental Fig 6). A strain lacking all potential XyG-binding sites (SGBP-A*/ΔSGBP-B/ΔBoGH9) was likewise unable to grow on any XyG or XyGO substrate tested. It should be noted here that we have used SGBP-A* variants instead of the corresponding ΔSGBP-A variants, because deletion of the protein ablates growth on XyG, likely due to an intimate structural interaction with the TBDT of the XyGUL 17; 28. Interestingly, the ΔBoGH9 mutation alone was sufficient to produce a kinetic growth defect on XyGO1, similar to the SGBP-A* mutation (Fig 3C and 3E), whereas on XyG and XyGO2 there was no significant difference from wild-type for either mutant strain (Fig 3B and 3D). These phenotypes suggest that both SGBP-A and BoGH9 play similarly important roles in XyG/XyGO capture, and that when both of these proteins are simultaneously absent, saccharide uptake is too inefficient to support growth.
Figure 3: BoGH9 capture of XyG compensates for the loss of XyG binding by SGBP-A.
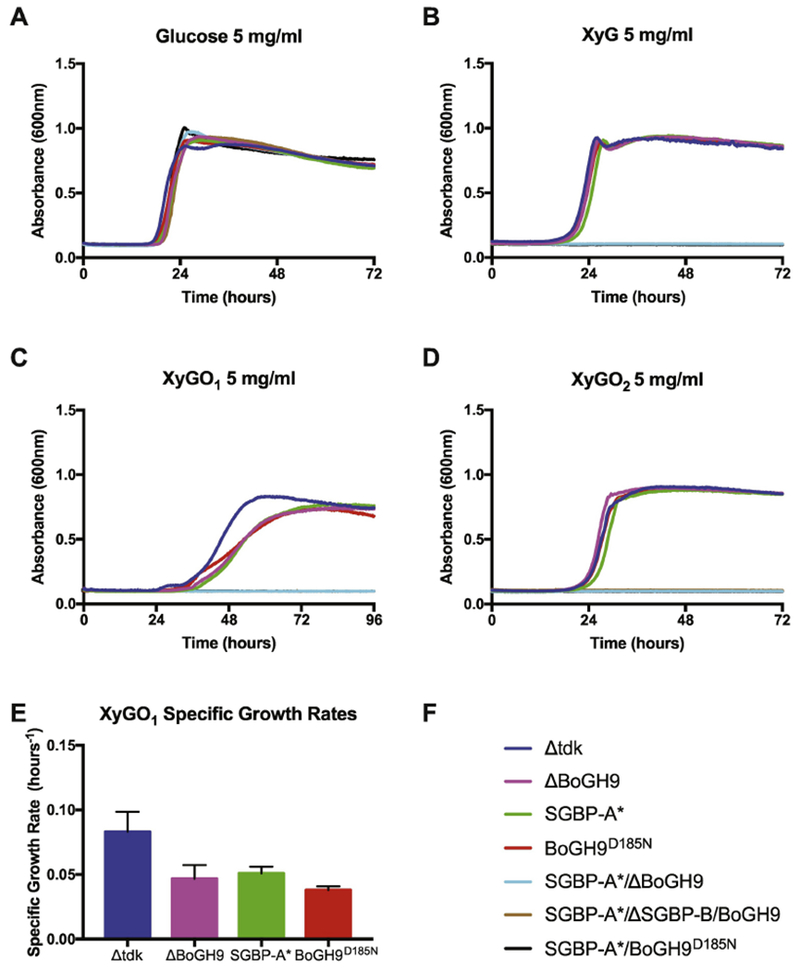
Growth of BoGH9 mutants on 5 mg/ml (A) Glucose, (B) XyG, (C) XyGO1, (D) XyGO2, and (E) Specific growth rate of select strains on XyGO1 at O.D.600 = 0.5 (F) Strain legend for panels A-E. In panels B – D the strains that do not display growth are SGBP-A*/ΔGH9 (blue), SGBP-A*/ΔSGBP-B/BoGH9 (brown), and SGBP-A*/GH9D185N (black).
The observation that a GH can complement the loss of an SGBP has not been observed previously in PUL systems, to our knowledge. To test whether the enzymatic activity of BoGH9 was required to compensate for the SGBP-A* mutation, we mutated the catalytic residues inferred from the tertiary structure, D185 and E562, to generate four independent B. ovatus strains (Table 3). Recapitulating the ΔBoGH9 strain phenotype, growth of the BoGH9D185N strain on XyGO1 was noticeably slower (Fig 3C,E), while growth on XyG and XyGO2 was indistinguishable from the wild-type (Fig 3B and 3D). Analogous to the SGBP-A*/ΔBoGH9 strain, the SGBP-A*/BoGH9D185N strain was not able to grow on any of the XyG substrates tested (Figure 3B-E). Identical growth defects on XyG were obtained with the other catalytically inactive variants (Supplemental Fig 6). As observed for the ΔBoGH9 strain, none of the four active-site-directed mutant alleles interfered with growth on XyG in a wild-type background (Supplemental Fig 6). Western blot analysis using anti-BoGH9 antibodies confirmed that all BoGH9 variants were expressed at similar levels (Supplemental Fig 6D). Together, these data indicate that both the presence of BoGH9 and its enzymatic activity are required for polysaccharide utilization in the absence of a functional SGBP-A.
SGBP-B* antagonizes growth on XyG in an SGBP-A* background
The dependence of a catalytically competent BoGH9 in the SGBP-A* background was surprising, and prompted us to re-examine how the other non-catalytic XyG-binding protein, SGBP-B, contributes to growth.8 We previously reported that a ΔSGBP-B strain grows like wild-type on XyG and XyGOs, but a SGBP-A*/ΔSGBP-B strain displays a severe apparent lag (Figure 6 in reference 8, and Fig 4). Here, we extended our previous analysis by replacing the native SGBP-B with the XyG-binding deficient allele SGBP-B* (Table 3) in B. ovatus expressing wild-type SGBP-A or the binding deficient allele SGBP-A*, to determine the specific contribution of the SGBP binding site to growth. While the ΔSGBP-B and SGBP-B* strains grew normally on both XyG and XyGO2 (Fig 4B,D), a combined SGBP-A*/SGBP-B* mutant was completely unable to grow on any XyG or XyGO substrate, which we ascribe to a dominant-negative effect in which the SGBP-B* protein may interfere with the structural function of the SGBP-A* protein (vide infra).
Figure 4: XyG-binding deficient SGBP-B antagonizes growth in the absence of SGBP-A glycan binding.
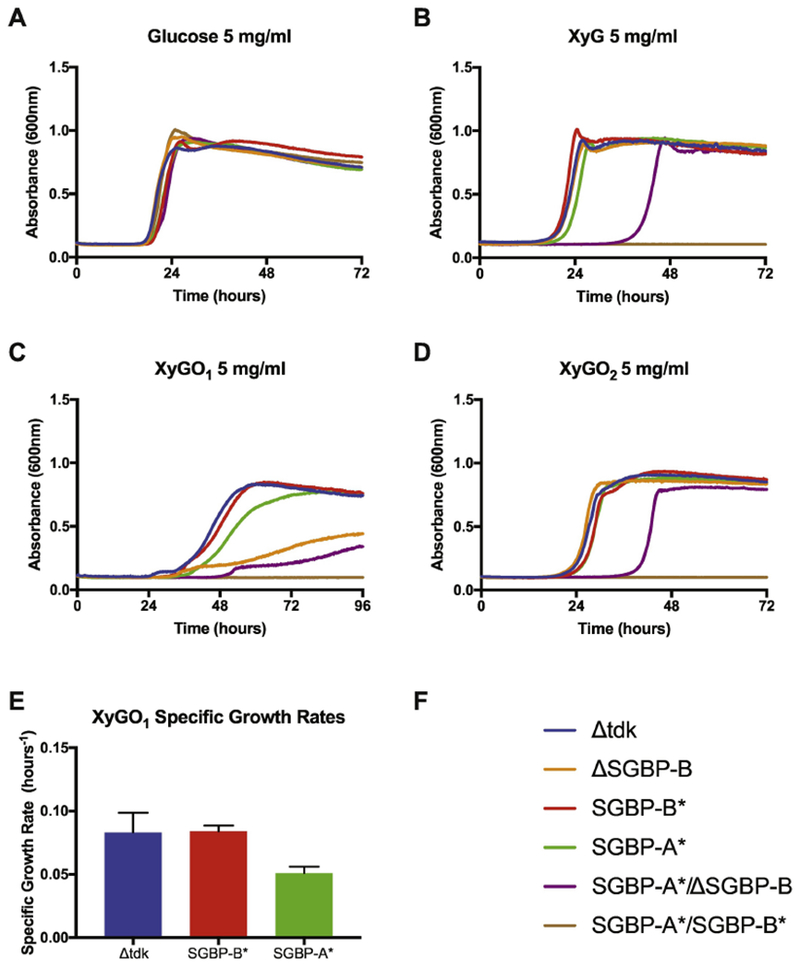
Growth of B. ovatus SGBP-B mutants on 5 mg/ml (A) Glucose, (B) XyG, (C) XyGO1 and (D) XyGO2 (E) Specific growth rate of select strains on XyGO1 at O.D.600 = 0.5 (F) Strain legend for panels A – E. In panels B – D the SGBP-A*/SGBP-B* strain that cannot grow is colored brown.
Isothermal titration calorimetry (ITC) previously indicated that SGBP-A displays negligible binding for XyGO1, whereas SGBP-B has a Kd of ~ 400 μM toward this minimal repeating motif 8. Consistent with this observation, we previously demonstrated that ΔSGBP-B strains grew less efficiently on XyGO1 than the SGBP-A* strain8. Here, the single SGBP-B* mutant grew comparably to wild-type whereas the combined mutant SGBP-A*/SGBP-B* could not grow at all on XyGO1 (Fig 4C,E). The complete loss of XyGO1 growth in the SGBP-A*/SGBP-B* strain demonstrates that at least one functional XyG-binding SGBP is required to support the capture of XyG or its oligosaccharides.
Since SGBP-A*/ΔSGBP-B but not SGBP-A*/SGBP-B* is capable of growth on XyG, albeit with an apparent lag, we reasoned that the SGBP-B* antagonizes XyG capture when combined with the XyG-binding deficient SGBP-A*. Using a pNBU2 vector modified for anhydrotetracycline (aTC)-inducible gene expression 29, we engineered our parent strain, SGBP-A*, to express either SGBP-B or SGBP-B* from the att1 site, which is located outside of the XyGUL 10. In order to ensure expression of SGBP-B or SGBP-B* during the early timepoints of the experiment, cultures were pregrown on glucose-containing medium with 2nM aTC to induce expression and then back-diluted into XyG-containing medium including 2nM aTC. Inducing the expression of SGBP-B appeared to enhance the start of exponential growth on XyG, likely by priming cells for XyG capture, whereas the expression of SGBP-B* abolished growth of the SGBP-A* similar to the SGBP-A*/SGBP-B* phenotype (Fig 5, yellow and green lines). Since the native SGBP-B gene remained intact in these experiments, these conditions should not impact the ability of cells to bind to XyG. Rather, the dominant negative effect from early induction of SGBP-B* before exposure to XyG to induce native protein expression, may be due to competition within a cell-surface protein complex. Although the XyGUL-encoded SGBP-B protein is expressed to higher levels once cells are exposed to XyG, by inducing early expression of pSGBP-B* the mutant may outcompete the native protein for a position within the uptake complex. To validate that protein expression was similar for induction of either SGBP-B or SGBP-B*, we performed a Western blot using anti-SGBP-B antibodies on whole cell lysates (Fig 5D). Since glucose does not activate expression of the native SGBP-B, we do not detect any SGBP-B in cultures without induction. Induced, glucose-grown cultures expressing aTC – inducible SGBP-B or SGBP-B* display similar levels of the protein, underscoring that the SGBP-B* protein exerts a phenotypic difference that is not due to differing levels of expression between these strains.
Figure 5. Inducible expression of SGBP-B* acts in a dominant-negative fashion to inhibit growth in an SGBP-A* strain.
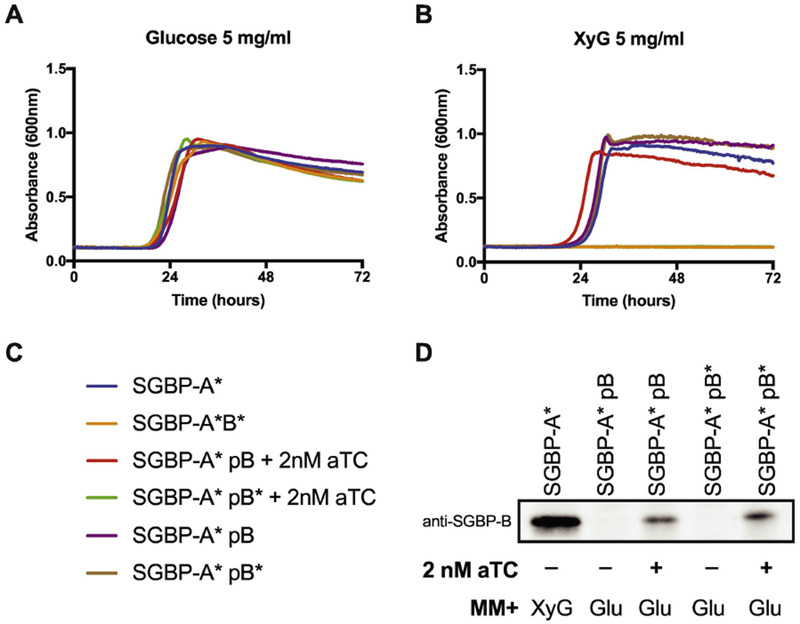
Inducible expression of SGBP-B/B* in an SGBP-A*ΔB background during growth on 5 mg/ml (A) Glucose and (B) XyG. The strains in panel B that did not grow are SGBP-A*/SGBP-B* (orange) and SGBP-A* pSGBP-B* + 2nM aTC (green). (C) Strain legend for panels A,B. (D) Western blot of SGBP-B/B* using anti-SGBP-B serum against whole cell lysates. SGBP-A* strains were cultured in minimal media containing either 5 mg/ml XyG or glucose, with or without 2 nM aTC, as noted. Cells were arrested in mid-logarithmic phase then normalized by O.D.600 before loading equal volumes in SDS-PAGE.
Discussion
The regular consumption of dietary fiber is critical for the assembly and preservation of a healthy gut microbiome. Glycans from plant cell walls transit the intestinal tract where they become depolymerized and fermented by members of the microbiota, including the dominant Bacteroidetes 3; 30; 31. This influx of carbon and energy into the gut ecosystem influences the composition of the microbiota, as well as its metabolic output 32; 33. Here, we studied the inner workings of a xyloglucan-degrading system in the model organism B. ovatus and demonstrated that the enigmatic glycoside hydrolase, BoGH9, and the cell surface glycan-binding protein, SGBP-B, impact the transport of xyloglucan in a manner that is dependent on SGBP-A XyG binding. These results imply that the relationships between the XyGUL-encoded cell surface proteins are more complex than previously appreciated, and furthermore suggest that the assembly of a multiprotein complex modulates glycan capture at the cell surface.
BoGH9 appears to also play a role in XyG capture when coexpressed with SGBP-A*. The SGBP-A*/ΔBoGH9 and SGBP-A*/BoGH9D185N strains were not able to grow on any XyG substrate tested (Fig 6A). The xylan PUL (PUL-Xyl) of B. ovatus encodes a consortium of cell surface enzymes involved in xylan debranching and degradation, but one catalytically inactive GH10 is predicted to be involved in xylan capture as opposed to hydrolysis 15. Unlike the PUL-Xyl GH10, BoGH9 requires use of its catalytic machinery in the SGBP-A* mutant, but perhaps its role in XyG capture is to locally cleave and orient XyG oligosaccharides into the TBDT in the absence of the SGBP-A binding site. Little is known about the assembly, organization, and dynamics of the proteins that form a complex around the XyGUL TBDT. A general model for glycan uptake, informed by the structures of the BT2281-2264 and BT1762-1763 complexes, predicts that SGBP-A may close down over the top of the TBDT with a XyG oligosaccharide in its binding site, thereby coordinating the substrate into the transporter 17. In an SGBP-A* strain, XyG is still utilized, but the requirement for a catalytically active BoGH9 implies that XyG fragments liberated by the cell surface BoGH5A can interact with and are perhaps hydrolyzed by, the active site of BoGH9 prior to internalization.
Figure 6: A Model for xyloglucan uptake facilitated by SGBP-B and BoGH9.
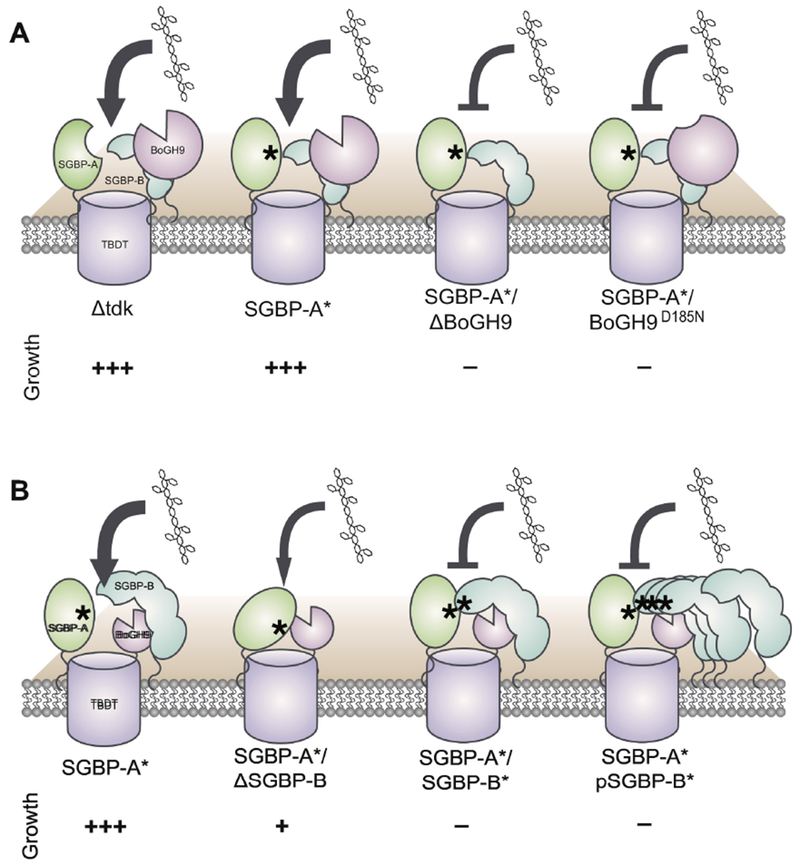
Asterisks indicate “*” versions of XyG-binding proteins that can no longer bind XyG. The relative growth of the SGBP-B and BoGH9 mutant strains on xyloglucan is displayed with more + indicating less lag time and more efficient growth. Protein sizes are not to scale and are enlarged when emphasizing proteins of interest. (A) Catalytically active BoGH9 is essential for growth on XyG in the SGBP-A* background. (B) SGBP-B is not necessary for in vitro growth on XyG, but the early induction of SGBP-B* antagonizes growth in a SGBP-A* background.
That BoGH9 is only encoded within the XyGUL of B. ovatus suggests that this enzyme has been maintained and adapted to facilitate the breakdown of XyGs at the cell surface. Acting broadly on β-glucans is typical of GH9 enzymes and strict endo-xyloglucanase activity from GH9 has only recently been characterized 34; 35. The structural determinants of the GH9 family that support the specific recognition and hydrolysis of xyloglucan compared to other β-glucans remain unknown 36. The expanded, albeit poor, activity on a range of β-glucan substrates could make BoGH9 a useful tool when B. ovatus forages polysaccharides from plant cell walls rich in cellulose and hemicelluloses. Although most PUL systems contain minimal enzyme cohorts required for their cognate substrates4; 37, the variety of enzyme activities and specificities in the XyGUL may support the liberation of xyloglucan from the plant cell wall. For example, the Gram-positive Ruminococcus cellulitycum uses a large (>1 MDa) outer membrane complex of proteins referred to as cellulosomes to bring together multiple GH9 enzymes as well as other endo- and exo- acting β-glucanases to support its catabolism of xyloglucan 35; 38; 39. Furthermore, it is possible that the activity of the BoGH9 is enhanced in its native environment at the cell surface by its proximity to the XyG-specific SGBPs, akin to how carbohydrate-binding modules can potentiate enzyme activity within contiguous polypeptides 40.
SGBP-B-like proteins encoded within PUL across the Bacteroides perform a variety of tasks during glycan capture 41. The SGBP-B-like protein from the heparin/heparin sulfate PUL of B. thetaiotaomicron appears to adapt the bacteria for growth on heparin oligosaccharides 42. SusE and SusF assist the diffusion of starch across the capsular surface for capture 43, and SusE facilitates the effective uptake of maltooligosaccharides ~7-30 glucose units long 7. SusE was found to interact in a protein complex with SusC and SusD 16, and its proximity to these proteins may impart conformational changes that influence maltooligosaccharide uptake 7. Here, the most striking phenotype for the deletion of SGBP-B in an otherwise wild-type B. ovatus is a kinetic defect in exponential growth on the smallest XyG unit, XyGO1. Conversely, a non-binding allele, SGBP-B* does not affect growth on XyGO1 in an otherwise wild-type strain suggesting that it is not just binding by SGBP-B that is important for the effective capture of this oligosaccharide. When SGBP-B is deleted in conjunction with the binding deficient SGBP-A*, an extended lag is observed on XyG and XyGO2, while a severe kinetic defect is observed on XyGO1. However, when binding deficient alleles SGBP-A* and SGBP-B* are co-expressed, cells fail to grow on any XyG substrate (Fig 6B). We believe that, like SusE during growth on starch 7, SGBP-B performs additional functions outside of binding and sequestering XyG at the cell surface and that this may be related to its interaction with the TBDT. Given the observation that the ectopic expression of SGBP-B* in the SGBP-A* mutant inhibits growth despite the presence of the wild-type SGBP-B, SGBP-B* may be outcompeting SGBP-B for an interaction near the XyGUL TBDT. Outside of the XyGUL, additional PUL-encoded surface lipoproteins like SGBP-B that assist in glycan capture may similarly need to interact with their cognate TBDT in order to impart efficiency to the capture and uptake of glycans. For example, the SGBP-B encoded by the heparin-targeting PUL (PULHep) of B. thetaiotaomicron has bee shown to interact with the PULHep SGBP-A, and this association may reflect an adaptive assembly for the capture of heparin near the TBDT 42.
In conclusion, this study describes roles for the B. ovatus GH9 and SGBP-B in the capture of XyG and possible modulation of the SGBP-A/TBDT complex that were previously unappreciated. The activities surrounding carbohydrate recognition by SGBPs and cell surface GHs are part of a fundamental nutrient acquisition paradigm in the gut Bacteroidetes that ultimately influences both the gut community composition and its collective metabolic activity. Further studies characterizing the interactions between these sets of proteins may facilitate the development of therapeutics to fine-tune carbohydrate catabolism by prominent gut bacteria to treat disease and promote health.
Materials and Methods
Bacterial strains and culture conditions
To generate all of the mutant XyGUL strains (Table 3), the B. ovatus ATCC-8483 Δtdk (ΔBACOVA_03071) strain was employed to facilitate allelic exchange, as previously described 44; 45, and is wild-type for the XyGUL. Mutations were generated using the counterselectable allelic exchange vector pExchange-tdk 44. The primers used in this study were synthesized by IDT DNA Technologies and are described in Supplemental Table 1. For site-directed mutagenesis of the BoGH9 gene, overlapping primers were designed to contain the mutation of interest within the pExchange vector, and then introduced into B. ovatus ATCC-8483 Δtdk (ΔBACOVA_03071) via allelic exchange as described. Final mutations were confirmed by sequencing.
B. ovatus was cultured in a Coy anaerobic chamber (5% H2/10% CO2/85% N2) from freezer stocks into tryptone-yeast extract-glucose (TYG) medium 46, then passaged into Bacteroides minimal media (MM) including 5 mg ml−1 glucose (Sigma) as the sole carbon source prior to kinetic growth experiments on 5 mg ml−1 XyGO1 (Megazyme), 5 mg ml−1 XyGO2 (Megazyme), or 0.5-5 mg ml−1 Tamarind xyloglucan (Megazyme) 6. MM-glucose-grown overnight cultures were back-diluted 1:200 into parallel cultures of glucose and the test substrate to ensure that cultures started at the same relative O.D.600 and to resolve differences in apparent lag before exponential growth. Kinetic growth experiments were performed at 37°C in 96 well plates and O.D.600 were recorded every 10-30 min. All growth experiments were performed in triplicate.
Western blotting
B. ovatus strains were grown in MM containing 5 mg ml−1 glucose or xyloglucan and were harvested at O.D.600=0.8. Cultures were normalized by O.D. and equal volumes of cells were lysed in SDS sample buffer and before being loading onto SDS-PAGE. SGBP-B and BoGH9 was detected in B. ovatus whole cell lysates by western blot using custom rabbit polyclonal primary antibodies and horseradish peroxidase conjugated goat anti-Rabbit IgG secondary antibody (Sigma) 45.
Recombinant gene cloning, expression, and protein purification
The gene encoding BoGH9 (Bacova_02649) amino acids 21 – 587 was PCR amplified from Bacteroides ovatus ATCC 8483 genomic DNA using the Q5 high fidelity polymerase (NEB). The PCR product containing appropriate pMCSG complementary sequences was ligated in an SspI linearized pMCSG53 vector providing an N-terminal His6-tag using Ligation Independent Cloning (LIC) strategy 47. Successful cloning was confirmed by colony PCR (GoTaq polymerase from Promega) and sequencing (Genewiz).
The recombinant GH9 enzyme used in this study was produced in E. coli BL21 (DE3) cells cultured in Terrific Broth (TB) containing ampicillin (50 μg.ml−1) at 37 °C (200 rpm). Cells were grown to mid-exponential growth phase (O.D.600 ≈ 0.4 to 0.6). Overexpression was induced by adding isopropyl β-D-thiogalactopyranoside (IPTG) to a final concentration of 0.5 mM and the cultures were further grown at 16 °C (200 rpm) for 18 h. The cells were harvested by centrifugation, sonicated and His6-tagged recombinant proteins were purified using a HisTrap IMAC FF nickel-nitrilotriacetic acid column (GE Healthcare) utilizing a gradient elution up to 100% elution buffer containing 20 mM Sodium Phosphate, pH 7.4, 500 mM NaCl, and 500 mM imidazole in an BioLogic FPLC system (BioRad). The purity of the recombinant protein was determined by SDS/PAGE and the identity of the expressed protein was confirmed by intact mass spectrometry 48Protein concentration was determined from the calculated molar extinction coefficient of 117370 M−1 cm−1 at 280 nm using an Epoch Microplate Spectrophotometer (BioTek).BoGH5A was purified as previously described 10.
Enzyme kinetic analysis
Polysaccharide hydrolysis was quantified using a Bicinchoninic acid (BCA) reducing-sugar assay as described previously 49 versus glucose standards (25–150 μM). To determine specific activity values of BoGH9 enzyme toward XyG, MLG, HEC, and CMC, a final concentration of 0.2164 μM of the recombinant purified enzyme was incubated with 1 mg ml−1 of polysaccharide in 200 μL reaction mixtures in 50 mM HEPES-NaOH buffer, pH 7.0. The effect of calcium ions on activity was determined by adding 1 mM CaCl2 to the buffer reaction. Reaction mixtures were incubated at 37°C for 25 minutes (MLG) or 30 minutes (XyG, HEC, CMC) prior to the BCA assay; all assays were performed in triplicate. The specific activity of BoGH5A enzyme toward XyG was determined by incubating triplicate reactions of 2.61 nM of the recombinant purified enzyme with 1 mg ml−1 of the polysaccharide in the optimum citrate buffer, pH 6.5, for 10 minutes at 37°C.
Carbohydrate analytical method
HPAEC-PAD carbohydrate analysis was performed on a Dionex ICS-5000 HPLC system operated by Chromeleon software version 7 (Dionex Corp., Sunnyvale, CA, USA). The injection volume was 10 μM and a Dionex Carbopac PA200 column (Thermo Scientific) with a guard column was used for all the samples separations. Solvent A was ultrapure water, solvent B was 1 M sodium hydroxide, and solvent C was 1 M sodium acetate prepared from anhydrous Bio Ultra-grade solid (Sigma). The following gradient was used: 0 – 5 min, 10 % B and 3.5 % solvent C; 5 – 12 min, 10% B and a linear gradient from 3.5–30% C; 12 –12.1 min, 50 % B, 50 % C; 12.1 – 13 min, exponential gradient (curve setting 9) of B and C back to initial conditions; 13 – 17 min, initial conditions 34.
To determine the mode of action of the enzyme, 3 μM of the recombinant enzyme was incubated with 0.25 mg.mL−1 polysaccharide at 37 °C in a 0.5 mL reaction mixture containing 50 mM HEPES-NaOH buffer (pH 7.0) with 1 mM CaCl2. The reaction was stopped at different time points by adding 100 μL of 95 °C water and heating to 95 °C for 10 min to a 100 μL aliquot of the reaction mixture. The reaction mixture was then diluted two times prior to product analysis by HPAEC-PAD.
BoGH9 crystallization and structure determination
The BoGH9 structure (res 27-583) presented here was determined via molecular replacement using a partially refined model, GH9-MR, obtained from crystals with translation lattice disorder and could not be completely refined. GH9-MR was expressed from the pET-YSBLLIC vector in BL21 Star (DE3) cells as described for BoGH43A by Hemsworth et al (2016) 14. After growth, cells were pelleted via centrifiguation and resuspended in Buffer A-His (50mM Tris pH 7.4, 0.5 M NaCl, 30mM imidazole and lysed via sonication. The lysate was clarified via centrifugation (38,000g for 30 min) and then applied to a 5 ml HisTrap FastFlow Ni-NTA column (GE Healthcare). A linear gradient of imidazole to a final concentration of 300 mM in Buffer A-His was used to elute the protein. Fractions containing the GH9-MR protein were pooled after inspection via SDS-PAGE, and concentrated using a Sartorius 30 kDa MWCO concentrator. The concentrated sample was applied to a 16/60 Superdex 200 column pre-equllibrated with 10 mM Tris pH 8.0, 250 mM NaCl. Peak fractions were combined after inspection via SDS-PAGE and dialyzed into 10mM Tris pH 8.0, 50 mM NaCl. Crystals of the GH9-MR were obtained via hanging drop vapor diffusion by mixing the protein (42mg/ml with XyG02) 1:1 with a crystallization solution composed of 0.1 M ammonium acetate, 0.1M Bis-Tris pH 5.5, and 10-20% PEG 10,000. Crystals were flash frozen in a cryo-protectant composed of mother liquor supplemented with 20% ethylene glycol and x-ray data were collected at Diamond Light Source. The x-ray data were indexed and integrated with XDS 50 with all subsequent scaling and processing steps performed in CCP451 . A partially built structure was obtained via molecular replacement in PHASER 52 using PDB id 1CLC as the search model. Due to translational lattice disorder the 1.7Å model (residues 30-581) was not refined beyond Rw=29.4%, Rf=34.9%, but provided an excellent starting model for the structure for the final BoGH9 structure.
Hence, alternate crystals of BoGH9 were obtained from the Morpheus screen via hanging drop vapor diffusion at room temperature. BoGH9 protein (15.9 mg/ml) was mixed 1:1 with solution A2 (0.06 M MgCl2 & CaCl2, 0.1M imidazole-MES buffer pH 6.5, 30% mix of ethylene glycol and PEG 8000). Crystals were flash-frozen by plunging into liquid nitrogen and did not require additional cryoprotection. X-ray data were collected at the Life Sciences Collaborative Access Team (LSCAT) beamline ID-F of the Advanced Photon Source at Argonne National Laboratory. Data were processed in HKL2000 and scaled with Scalepack 53. Molecular replacement was performed in Phaser-MR 52 from the Phenix package 54 using the GH9MR structure as the search model, as described above. The final BoGH9 structure was refined with Phenix.refine 55.
Supplementary Material
Supplemental Figure 1. Superposition of the BoGH9 and AaCel9A. Superposition of the AaCel9A (PDB 3H3K) with cellotetraose and glucose and the BoGH9 structure. AaCel9A is displayed in grey ribbon with glucose residues shown in thick black and red sticks, while BoGH9 is displayed in green ribbon with imidazole displayed as blue and magenta spheres. The RMSD of the structures is 1.1Å over 366 atom pairs and was generated in Chimera 56.
Supplemental Figure 2. Glycans utilized in this study. XyGO1 from Megazyme represents a mixture of XXXG, XXLG and XLLG oligosaccharides where D-Glcp is assigned G, α-D-Xylp-(1-6)-β-D-Glcp segments are named X and β-D-Galp-(1-2)-α-D-Xylp-(1-6)-β-D-Glcp is assigned L. XyGO2, also from Megazyme, is treated with β-galactosidase.
Supplemental Figure 3. HPAEC-PAD product analysis of BoGH9. (A) mixed-linkage beta-glucan (B) xyloglucan. The asterisk indicates peaks due to enzyme buffer components.
Supplemental Figure 4. Manual docking of XXXG and G4G4G3G to BoGH9. The position of cellotetraose from an overlay of AaCel9A with cellotetraose (PDB 3H3K) onto the BoGH9 structure was used as a template to manually model the products XXXG and G4G3G into the active site of BoGH9. Modeling was performed in Coot. A. To model XXXG (from PDB 3ZMR), the side chains of F450, W451,Y558 in BoGH9 were moved into alternate conformations that mimicked the position of similarly positioned side chains in AaCel9A and to minimize negative sterics with the ligand. Pink lines indicate clashing interactions for VDWs overlap > 0.8Å between the protein and ligand as calculated in Chimera. B. To model G4G3G (from PDB 1ZM1), the side chains of W451 and Y558 were moved as described in A. Pink lines indicate clashing interactions for VDWs overlap > 0.8Å between the protein and ligand as calculated in Chimera 56.
Supplemental Figure 5. XyGUL mutants used in this study grown on 5mg/ml xylose. (A) Corresponds to growths from Fig 3. (B) Corresponds to growths from Fig 4.
Supplemental Figure 6. Catalytically active BoGH9 is required for growth in an SGBP-A* strain. Growth of BoGH9 catalytic mutants on 5 mg/ml (A) Glucose and (B) XyG. (C) Strain legend for panels A,B. (D) Western blot of BoGH9 using anti-GH9 antibodies against whole cell lysates. GH9 strains were cultured in minimal media containing 5 mg/ml and were arrested in mid-logarithmic phase then normalized by O.D.600 before loading in SDS-PAGE.
Highlights.
Xyloglucan (Xyg) is a dietary fiber utilized by the gut symbiont Bacteroides ovatus.
How XyG is captured at the cell surface by SGBP-A, -B and BoGH9 is unclear.
The BoGH9 structure reveals the molecular basis for activity.
Functional BoGH9 and SGBP-B are required for XyG uptake in an SGBPA mutant
SGBP-A, -B and BoGH9 work together, and SGBP-B and BoGH9 may orient XyG for uptake.
Acknowledgements
We thank the members of the Koropatkin and Martens labs at the University of Michigan for their support and advice during the design and execution of these studies. Research in Ann Arbor was provided by the National Institutes of Health (NIH R01 GM118475 to N.M.K.). M.H.F. was partially supported by a predoctoral fellowship from the Cellular Biotechnology Training Program (T32GM008353). Research in Vancouver was supported by the Canadian Institutes of Health Research (CIHR), the Canada Foundation for Innovation, and the British Columbia Knowledge Development Fund. Waters Corporation is gratefully acknowledged for the provision of the LC-MS system used in this study. GJD is funded by the Royal Society through the Royal Society Ken Murray Professorship. GJD/GRH were supported by the Biotechnology and Biological Sciences Research Council BBSRC (grants BB/L021633/1 and BB/I014802/1).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The coordinates for the BoGH9 structure have been deposited in the Protein Data Bank (PDB) with the accession 6DHT.
References
- 1.Ding T & Schloss PD (2014). Dynamics and associations of microbial community types across the human body. Nature 509, 357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martens EC, Lowe EC, Chiang H, Pudlo NA, Wu M, McNulty NP, Abbott DW, Henrissat B, Gilbert HJ, Bolam DN & Gordon JI (2011). Recognition and degradation of plant cell wall polysaccharides by two human gut symbionts. PLoS Biol 9, e1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.El Kaoutari A, Armougom F, Gordon JI, Raoult D & Henrissat B (2013). The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat Rev Microbiol 11, 497–504. [DOI] [PubMed] [Google Scholar]
- 4.Grondin JM, Tamura K, Dejean G, Abbott DW & Brumer H (2017). Polysaccharide Utilization Loci: Fueling microbial communities. J Bacteriol 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sonnenburg JL, Xu J, Leip DD, Chen CH, Westover BP, Weatherford J, Buhler JD & Gordon JI (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–9. [DOI] [PubMed] [Google Scholar]
- 6.Martens EC, Chiang HC & Gordon JI (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foley MH, Martens EC & Koropatkin NM (2018). SusE facilitates starch uptake independent of starch binding in B. thetaiotaomicron. Mol Microbiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tauzin AS, Kwiatkowski KJ, Orlovsky NI, Smith CJ, Creagh AL, Haynes CA, Wawrzak Z, Brumer H & Koropatkin NM (2016). Molecular dissection of xyloglucan recognition in a prominent human gut symbiont. MBio 7, e02134–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sonnenburg ED, Zheng H, Joglekar P, Higginbottom SK, Firbank SJ, Bolam DN & Sonnenburg JL (2010). Specificity of polysaccharide use in intestinal Bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsbrink J, Rogers TE, Hemsworth GR, McKee LS, Tauzin AS, Spadiut O, Klinter S, Pudlo NA, Urs K, Koropatkin NM, Creagh AL, Haynes CA, Kelly AG, Cederholm SN, Davies GJ, Martens EC & Brumer H (2014). A discrete genetic locus confers xyloglucan metabolism in select human gut Bacteroidetes. Nature 506, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffman M, Jia Z, Pena MJ, Cash M, Harper A, Blackburn AR 2nd, Darvill A & York WS. (2005). Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340, 1826–40. [DOI] [PubMed] [Google Scholar]
- 12.Hsieh YS & Harris PJ (2009). Xyloglucans of monocotyledons have diverse structures. Mol Plant 2, 943–65. [DOI] [PubMed] [Google Scholar]
- 13.Pauly M & Keegstra K (2016). Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu Rev Plant Biol 67, 235–59. [DOI] [PubMed] [Google Scholar]
- 14.Hemsworth GR, Thompson AJ, Stepper J, Sobala LF, Coyle T, Larsbrink J, Spadiut O, Goddard-Borger ED, Stubbs KA, Brumer H & Davies GJ (2016). Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut. Open Biol 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Basle A, Morland C, Day AM, Zheng H, Rogers TE, Thompson P, Hawkins AR, Yadav MP, Henrissat B, Martens EC, Dupree P, Gilbert HJ & Bolam DN (2015). Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6, 7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuson HH, Foley MH, Koropatkin NM & Biteen JS (2018). The starch utilization system assembles around stationary starch-binding proteins. Biophys J. 115, 242–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glenwright AJ, Pothula KR, Bhamidimarri SP, Chorev DS, Basle A, Firbank SJ, Zheng H, Robinson CV, Winterhalter M, Kleinekathofer U, Bolam DN & van den Berg B (2017). Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature 541, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holm L & Sander C (1995). Dali: a network tool for protein structure comparison. Trends Biochem Sci 20, 478–80. [DOI] [PubMed] [Google Scholar]
- 19.Correa A, Pacheco S, Mechaly AE, Obal G, Behar G, Mouratou B, Oppezzo P, Alzari PM & Pecorari F (2014). Potent and specific inhibition of glycosidases by small artificial binding proteins (affitins). PLoS One 9, e97438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckert K, Vigouroux A, Lo Leggio L & Morera S (2009). Crystal structures of A. acidocaldarius endoglucanase Cel9A in complex with cello-oligosaccharides: strong −1 and −2 subsites mimic cellobiohydrolase activity. J Mol Biol 394, 61–70. [DOI] [PubMed] [Google Scholar]
- 21.Eckert K, Zielinski F, Lo Leggio L & Schneider E (2002). Gene cloning, sequencing, and characterization of a family 9 endoglucanase (CelA) with an unusual pattern of activity from the thermoacidophile Alicyclobacillus acidocaldarius ATCC27009. Appl Microbiol Biotechnol 60, 428–36. [DOI] [PubMed] [Google Scholar]
- 22.Peng J, Wang W, Jiang Y, Liu M, Zhang H & Shao W (2011). Enhanced soluble expression of a thermostable cellulase from Clostridium thermocellum in Escherichia coli. Curr Microbiol 63, 523–30. [DOI] [PubMed] [Google Scholar]
- 23.Bayer EA, Shoham Y & Lamed R (2000). Cellulose-decomposing prokaryotes and their enzyme systems In The Prokaryotes: An Involving Electronic Resource for the Microbial Community (Dworkin M, Falkow S, Rosenberg E, Schleifer K-H & Stackebrandt E, eds.), Vol. 2 Springer Verlag, New York, NY. [Google Scholar]
- 24.Chauvaux S, Souchon H, Alzari PM, Chariot P & Beguin P (1995). Structural and functional analysis of the metal-binding sites of Clostridium thermocellum endoglucanase CelD. J Biol Chem 270, 9757–62. [DOI] [PubMed] [Google Scholar]
- 25.Li YK & Byers LD (1989). Inhibition of beta-glucosidase by imidazoles. Biochim Biophys Acta 999, 227–32. [DOI] [PubMed] [Google Scholar]
- 26.Field RA, Haines AH, Chrystal EJ & Luszniak MC (1991). Histidines, histamines and imidazoles as glycosidase inhibitors. Biochem J 274 (Pt 3), 885–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Hemsworth GR, Dejean G, Rogers TE, Pudlo NA, Urs K, Jain N, Davies GJ, Martens EC & Brumer H (2017). Molecular mechanism by which prominent human gut Bacteroidetes utilize mixed-linkage beta-glucans, major health-promoting cereal polysaccharides. Cell Rep 21, 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tauzin AS, Kwiatkowski KJ, Orlovsky NI, Smith CJ, Creagh AL, Haynes CA, Wawrzak Z, Brumer H & Koropatkin NM (2016). Molecular dissection of xyloglucan recognition in a prominent human gut symbiont MBio 7, e02134–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lim B, Zimmermann M, Barry NA & Goodman AL (2017). Engineered regulatory systems modulate gene expression of human commensals in the gut. Cell 169, 547–558 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamaker BR & Tuncil YE (2014). A perspective on the complexity of dietary fiber structures and their potential effect on the gut microbiota. J Mol Biol 426, 3838–50. [DOI] [PubMed] [Google Scholar]
- 31.Koropatkin NM, Cameron EA & Martens EC (2012). How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol 10, 323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, Biddinger SB, Dutton RJ & Turnbaugh PJ (2014). Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Natarajan N & Pluznick JL (2014). From microbe to man: the role of microbial short chain fatty acid metabolites in host cell biology. Am J Physiol Cell Physiol 307, C979–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eklof JM, Ruda MC & Brumer H (2012). Distinguishing xyloglucanase activity in endo-beta(1-->4)glucanases. Methods Enzymol 510, 97–120. [DOI] [PubMed] [Google Scholar]
- 35.Ravachol J, Borne R, Tardif C, de Philip P & Fierobe HP (2014). Characterization of all family-9 glycoside hydrolases synthesized by the cellulosome-producing bacterium Clostridium cellulolyticum. J Biol Chem 289, 7335–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Attia MA & Brumer H (2016). Recent structural insights into the enzymology of the ubiquitous plant cell wall glycan xyloglucan. Curr Opin Struct Biol 40, 43–53. [DOI] [PubMed] [Google Scholar]
- 37.Martens EC, Kelly AG, Tauzin AS & Brumer H (2014). The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol 426, 3851–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayer EA, Lamed R, White BA & Flint HJ (2008). From cellulosomes to cellulosomics. Chem Rec 8, 364–77. [DOI] [PubMed] [Google Scholar]
- 39.Ravachol J, de Philip P, Borne R, Mansuelle P, Mate MJ, Perret S & Fierobe HP (2016). Mechanisms involved in xyloglucan catabolism by the cellulosome-producing bacterium Ruminiclostridium cellulolyticum. Sci Rep 6, 22770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boraston AB, Bolam DN, Gilbert HJ & Davies GJ (2004). Carbohydrate-binding modules: fine-tuning polysaccharide recognition. Biochem J 382, 769–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Foley MH, Cockburn DW & Koropatkin NM (2016). The Sus operon: a model system for starch uptake by the human gut Bacteroidetes. Cell Mol Life Sci 73, 2603–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cartmell A, Lowe EC, Basle A, Firbank SJ, Ndeh DA, Murray H, Terrapon N, Lombard V, Henrissat B, Turnbull JE, Czjzek M, Gilbert HJ & Bolam DN (2017). How members of the human gut microbiota overcome the sulfation problem posed by glycosaminoglycans. Proc Natl Acad Sci U S A 114, 7037–7042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cameron EA, Kwiatkowski KJ, Lee BH, Hamaker BR, Koropatkin NM & Martens EC (2014). Multifunctional nutrient-binding proteins adapt human symbiotic bacteria for glycan competition in the gut by separately promoting enhanced sensing and catalysis. MBio 5, e01441–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koropatkin NM, Martens EC, Gordon JI & Smith TJ (2008). Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure 16, 1105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cameron EA, Maynard MA, Smith CJ, Smith TJ, Koropatkin NM & Martens EC (2012). Multidomain carbohydrate-binding proteins involved in Bacteroides thetaiotaomicron starch metabolism. J Biol Chem 287, 34614–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Holdeman LV, Cato ED & Moore WEC (1977). Anaerobe Laboratory Manual. (Moore WEC, Ed.), Virginia Polytechnic Institute and State University Anaerobe Laboratory, Blacksburg, Va. [Google Scholar]
- 47.Eschenfeldt WH, Lucy S, Millard CS, Joachimiak A & Mark ID (2009). A family of LIC vectors for high-throughput cloning and purification of proteins. Methods Mol Biol 498, 105–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sundqvist G, Stenvall M, Berglund H, Ottosson J & Brumer H (2007). A general, robust method for the quality control of intact proteins using LC-ESI-MS. J Chromatogr B Analyt Technol Biomed Life Sci 852, 188–94. [DOI] [PubMed] [Google Scholar]
- 49.Arnal G, Attia MA, Asohan J & Brumer H (2017). A low-volume, parallel copper-bicinchoninic acid (BCA) assay for glycoside hydrolases. Methods Mol Biol 1588, 3–14. [DOI] [PubMed] [Google Scholar]
- 50.Kabsch W (2010). Integration, scaling, space-group assignment and post-refinement. Acta Crystallogr D Biol Crystallogr 66, 133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Othman RA, Moghadasian MH & Jones PJ (2011). Cholesterol-lowering effects of oat beta-glucan. Nutr Rev 69, 299–309. [DOI] [PubMed] [Google Scholar]
- 52.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC & Read RJ (2007). Phaser crystallographic software. J Appl Crystallogr 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Otwinowski Z & Minor W (1997). [20] Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 54.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC & Zwart PH (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Afonine PV, Grosse-Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH & Adams PD (2012). Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC & Ferrin TE (2004). UCSF Chimera - A Visualization System for Exploratory Research and Analysis. J. Comput. Chem 25, 1605–1612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Superposition of the BoGH9 and AaCel9A. Superposition of the AaCel9A (PDB 3H3K) with cellotetraose and glucose and the BoGH9 structure. AaCel9A is displayed in grey ribbon with glucose residues shown in thick black and red sticks, while BoGH9 is displayed in green ribbon with imidazole displayed as blue and magenta spheres. The RMSD of the structures is 1.1Å over 366 atom pairs and was generated in Chimera 56.
Supplemental Figure 2. Glycans utilized in this study. XyGO1 from Megazyme represents a mixture of XXXG, XXLG and XLLG oligosaccharides where D-Glcp is assigned G, α-D-Xylp-(1-6)-β-D-Glcp segments are named X and β-D-Galp-(1-2)-α-D-Xylp-(1-6)-β-D-Glcp is assigned L. XyGO2, also from Megazyme, is treated with β-galactosidase.
Supplemental Figure 3. HPAEC-PAD product analysis of BoGH9. (A) mixed-linkage beta-glucan (B) xyloglucan. The asterisk indicates peaks due to enzyme buffer components.
Supplemental Figure 4. Manual docking of XXXG and G4G4G3G to BoGH9. The position of cellotetraose from an overlay of AaCel9A with cellotetraose (PDB 3H3K) onto the BoGH9 structure was used as a template to manually model the products XXXG and G4G3G into the active site of BoGH9. Modeling was performed in Coot. A. To model XXXG (from PDB 3ZMR), the side chains of F450, W451,Y558 in BoGH9 were moved into alternate conformations that mimicked the position of similarly positioned side chains in AaCel9A and to minimize negative sterics with the ligand. Pink lines indicate clashing interactions for VDWs overlap > 0.8Å between the protein and ligand as calculated in Chimera. B. To model G4G3G (from PDB 1ZM1), the side chains of W451 and Y558 were moved as described in A. Pink lines indicate clashing interactions for VDWs overlap > 0.8Å between the protein and ligand as calculated in Chimera 56.
Supplemental Figure 5. XyGUL mutants used in this study grown on 5mg/ml xylose. (A) Corresponds to growths from Fig 3. (B) Corresponds to growths from Fig 4.
Supplemental Figure 6. Catalytically active BoGH9 is required for growth in an SGBP-A* strain. Growth of BoGH9 catalytic mutants on 5 mg/ml (A) Glucose and (B) XyG. (C) Strain legend for panels A,B. (D) Western blot of BoGH9 using anti-GH9 antibodies against whole cell lysates. GH9 strains were cultured in minimal media containing 5 mg/ml and were arrested in mid-logarithmic phase then normalized by O.D.600 before loading in SDS-PAGE.


