Abstract
Background
Two-hybrid screening for proteins that interact with the core domain of human topoisomerase I identified two novel proteins, BTBD1 and BTBD2, which share 80% amino acid identities.
Results
The interactions were confirmed by co-precipitation assays demonstrating the physical interaction of BTBD1 and BTBD2 with 100 kDa topoisomerase I from HeLa cells. Deletion mapping using two-hybrid and GST-pulldown assays demonstrated that less than the C-terminal half of BTBD1 is sufficient for binding topoisomerase I. The topoisomerase I sequences sufficient to bind BTBD2 were mapped to residues 215 to 329. BTBD2 with an epitope tag localized to cytoplasmic bodies. Using truncated versions that direct BTBD2 and TOP1 to the same cellular compartment, either the nucleus or the cytoplasm, co-localization was demonstrated in co-transfected Hela cells. The supercoil relaxation and DNA cleavage activities of topoisomerase I in vitro were affected little or none by co-incubation with BTBD2. Northern analysis revealed only a single sized mRNA for each BTBD1 and BTBD2 in all human tissues tested. Characterization of BTBD2 mRNA revealed a 255 nucleotide 90% GC-rich region predicted to encode the N-terminus. BTBD1 and BTBD2 are widely if not ubiquitously expressed in human tissues, and have two paralogs as well as putative orthologs in C. elegans and D. melanogaster.
Conclusions
BTBD1 and BTBD2 belong to a small family of uncharacterized proteins that appear to be specific to animals. Epitope-tagged BTBD2 localized to cytoplasmic bodies. The characterization of BTBD1 and BTBD2 and their interaction with TOP1 is underway.
Background
DNA topoisomerase I (TOP1) is a ubiquitously expressed protein that relaxes DNA supercoils generated during many transactions of DNA including replication, transcription, repair, and chromatin condensation and remodeling. TOP1 has been shown to interact with many proteins including components of the holo TFIIIC complex [1], TFIID [2], p53 [3], SV-40 large T antigen [4], nucleolin [5], the RING finger protein topors [6], the RNA splicing factors PSF/p54nrb[7] and SF2/ASF [8], the cell cycle regulatory protein ARF [9], RNA pol II complex [10,11], and HTLV type 1 tax oncoprotein [12]. TOP1 also interacts with a variety of unidentified proteins involved in its phosphorylation, ubiquitination and sumoylation [13-17].
We used the core domain of TOP1 lacking the N-terminal highly charged region, the coiled-coil linker domain and the C-terminal active site domain as bait in two hybrid screens for TOP1 interacting proteins. We obtained multiple independent cDNA clones coding for BTBD1 and BTBD2, two uncharacterized, BTB domain-containing, kelch-like proteins. The interactions of HeLa cell TOP1 with the BTBD proteins was confirmed using GST-pulldown and was further suggested by their intracellular co-localization.
Results
Identification of TOP1-interacting proteins using two-hybrid assays
The core domain of human TOP1 from residue 198 to 651 was used as the bait in two-hybrid screens [18-20]. This region of TOP1 lacks the N-terminal ~25% of the protein that is highly charged and the C-terminal domain containing the active site and the coiled-coil linker. The resulting polypeptide comprises 60% of the TOP1 sequence. A screen of a HeLa cell library yielded 56 colonies that tested positive in the two hybrid assays (growth on plates lacking histidine or uracil, lack of growth on plates containing 5-fluoroorotic acid; and induction of lacZ resulting in blue color when assayed with X-gal). Nucleotide sequence analysis of 34 of these clones revealed two cDNA contigs of 15 and 19 that predict proteins whose sequences are 80% identical. The proteins were named BTBD1 and BTBD2 according to a recent analysis of their genomic sequences [21]. The clones of each contig differed in the amount of 5' sequence present: 11/15 and 15/19 of BTBD1 and BTBD2, respectively, had different 5' ends. In the two-hybrid growth assays and β-galactosidase assays, each of the clones tested as positive as the control for strongly interacting proteins (the interaction of c-Fos with c-Jun, ProQuest, Life Technologies). Another two-hybrid screen using a different two-hybrid system (Matchmaker, Clontech) also yielded 2 clones identical to the contig of 15 clones (BTBD1).
Characterization of BTBD1 and BTBD2 mRNA
The longest BTBD1 cDNA clone obtained from two-hybrid screening (3196 bp) contains an ORF that encodes a 482 aa protein starting with an ATG that is preceded by an in-frame stop codon. Sequences in GenBank confirmed our sequence. One was obtained by the oligo-capping method (FLJ20724), and another assembled from partial cDNA and RACE fragments [21].
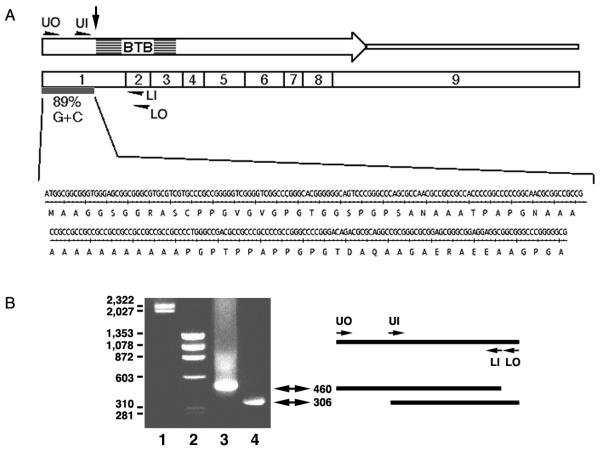
The longest BTBD2 cDNA obtained from two-hybrid screening (2413 bp) contained an ORF starting at the 5' end. To obtain the missing 5' cDNA, a nested PCR strategy was used to amplify the sequence from four independent cDNA libraries, but no extended sequences were obtained. 5'-RACE (Life Technologies) also failed to produce longer clones. This difficulty to obtain longer cDNA clones was seen by others [21]. The GC richness at the 5' end (85 nucleotides are 79% GC) may form secondary structures that block reverse transcriptase. Analysis of the BTBD2 gene immediately upstream of the 5' end of our cDNA predicts an ORF that begins with an ATG and encodes 85 amino acids in-frame with the ORF of our longest BTBD2 clone; however, the 255 nucleotides encoding these in-frame amino acids sequence are 89% GC rich. To determine if this sequence might be part of the BTBD2 mRNA, RT-PCR followed by nested PCR was used. Two antisense primers complementary to exon 2 were used to prime reverse transcription. Sense primers for PCR amplification were located within predicted exon 1 (Fig. 1A). Amplified products of 460 and 306 bp that are predicted from the mRNA template resulted (Fig. 1B). If genomic DNA had been the template, products greater than 18 kb would have been obtained. Furthermore, sequencing of the 460 bp-amplification product confirmed it to be the predicted mRNA (Fig. 1A). The cDNA starting at the predicted start codon was deposited in GenBank (AF355797).
Figure 1.
RT-PCR identifies a highly GC-rich sequence predicted to encode the ammo terminal region of BTBD2. A. RT-PCR strategy and the sequence of the amplified region are shown: BTBD2 coding region (open arrow), 3'-UTR (narrow open bar) and exons (wide open bar, 1 – 9), sense primers (UO and UI), antisense primers (LI and LO), GC-rich region (solid bar and sequence). B. Products of the RT-PCR are shown on an ethidium bromide gel. Reverse transcription was performed with antisense primers complementary to exon 2. The schematic shows the relevant portion of the mRNA template, the RT primers (LI and LO), the sense primers (UO and UI), and the amplified products (460 and 306 bp) that correspond to the bands on the agarose gel stained with ethidium bromide (lanes 1 & 2, DNA size standards; lane 3, product of reverse transcription with LO, 1st round PCR with UO and LO, and 2nd round PCR with UO and LI; lane 4, product of reverse transcription with LO, 1st round PCR with UO and LO, and 2nd round PCR with UI and LO.
Northern blotting and in silico expression analysis
Northern blotting was used to determine the tissue distribution and sizes of the mRNA encoding BTBD1 and BTBD2 (Fig. 2). Poly (A)+RNA from human tissues (2 μg/lane) was probed with BTBD1, BTBD2 or TOP1 cDNA. Each mRNA was detected in all the tissues tested. BTBD1 mRNA was highly expressed in testes, heart and skeletal muscle. BTBD2 was more highly expressed in skeletal muscle. Only a single message size was detected for each BTBD1 (3.2 kb) and BTBD2 (2.8 kb).
Figure 2.
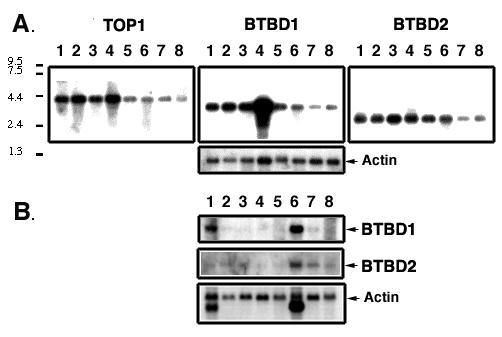
BTBD1 and BTBD2 Northern blots of human tissues. Multiple Tissue Northern (MNT™, Clontech) blots were probed with 32P-dCTP-labeled cDNA (TOP1, BTBD1 or BTBD2). Each lane contains 2 μg of polyA+RNA from each tissue. A. 1. spleen, 2. thymus, 3. prostate, 4. testes, 5. uterus, 6. small intestine, 7. colon, 8. leukocytes. B. 1. heart, 2. brain, 3. placenta, 4. lung, 5. liver, 6. skeletal muscle, 7. kidney, 8. placenta. The membranes were re-probed for actin.
BTBD1 and BTBD2 cDNA were found in libraries from the following tissues: amniotic, aorta, bone, brain, bone marrow, colon, ear, embryo, foreskin, gall bladder, germ, heart, kidney, lung, muscle, ovary, pancreas, parathyroid, placenta, skin, spleen, stomach, testis, thymus, tonsil and uterus (LocusID 53339 and 55643, respectively [22]). BTBD1 and BTBD2 was also expressed in a large number of tumor types and normal tissues present in SAGE (serial analysis of gene expression) libraries (SAGEmap, [23]).
Mapping of the regions of TOP1 and the BTBDs that interact
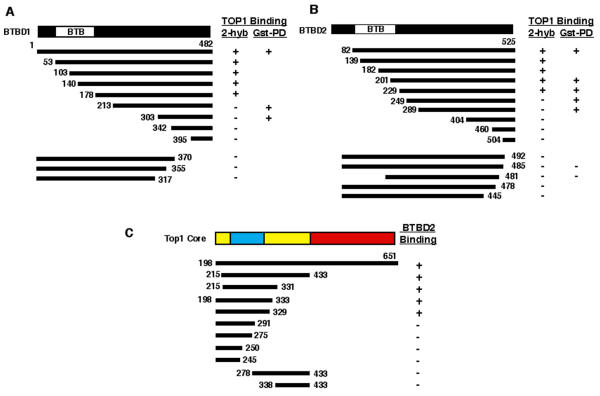
BTBD1 and BTBD2 were mapped by deletion analysis to identify amino acid sequences required for interaction with TOP1. In two-hybrid assays, N-terminal deletions of ~36 % of BTBD1 (177 aa) or 43% BTBD2 (228 aa) did not eliminate binding to TOP1 (Fig. 3A+B). In contrast, C-terminal deletions of 112 and 33 aa of BTBD1 and BTBD2, respectively, eliminated binding to TOP1 in the two-hybrid assays. Thus sequences very close to the C-terminus are required for the interaction with TOP1 by this analysis.
Figure 3.
Deletion mapping of the interacting regions of TOP1 and BTBD1 and BTBD2 using two-hybrid assays.A. Deletions of BTBD1 and their interaction with cTOP 1 (residues 198–651). B. Deletions of BTBD2 and their interaction with cTOP1. C. Deletions of cTOP1 and their interaction with BTBD2. Interactions were tested using colony formation assays on plates lacking histidine or uridine and the β-galactosidase colony color assay. The TOP1 core domain regions are shown from N- to C-terminus: subdomain I (yellow), subdomain II (blue), subdomain I (yellow) and subdomain III (red). The shortest clones obtained from two-hybrid screening were downstream of the BTB domains, beginng at residue 186 and 219 for BTBD1 and BTBD2, respectively. Results are summarized for the two-hybrid (2-hyb) and the GST-pull down assays (Gst-PD, see Fig. 4)
Two-hybrid analysis was also used to map regions of the TOP1 core domain that interact with BTBD2. The region upstream of aa 215 and downstream of aa 329 were not required for binding BTBD2 (Fig. 3C). The fragment of TOP1 from amino acid 215 to 331 was capable of binding to BTBD2. This region includes 18 aa of core subdomain I, all of core subdomain II and 10 amino acids of the second section of core subdomain I.
GST-pulldown assays confirm two-hybrid deletion mapping
Truncated fragments of BTBD1 and BTBD2 were tested in GST-pull down assays for their ability to bind TOP1 in nuclear extracts of HeLa cells. The GST-pulldown assay confirmed the positive interactions seen in the two-hybrid deletion mapping (Fig. 4). In addition, we found that two N-terminal truncated BTBD1 and BTBD2 were able to co-precipitate TOP1 from nuclear extracts (BTBD1, 213–482 & 303–482; BTBD2, 249–525 & 289–525) although they were negative in the two-hybrid assay. Since truncated fragments may be unstable in yeast and may not accumulate to levels sufficient to produce a positive result, we used roughly equal amounts of each truncated protein to test for binding to TOP1 in nuclear extracts. Taken together, the results of the two-hybrid and GST-pulldown assays narrow the interacting region of BTBD1 and BTBD2 to roughly the C-terminal half of each protein with critical residues within the last 50 residues.
Figure 4.
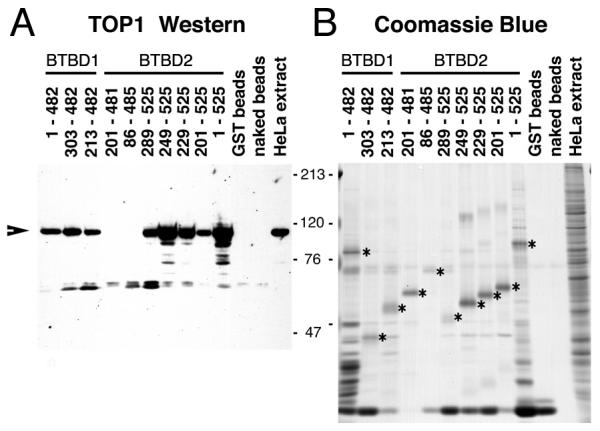
TOP1 from HeLa cells interacts with GST-BTBD1 and GST-BTBD2.A.TOP1 Western blot. HeLa cell nuclear extracts were incubated with GST or GST-BTBD2 beads. After washing, the beads were solubilized in SDS-PAGE sample buffer and subjected to SDS-PAGE and Western blotting with TOP1 antiserum. B.Coomassie Blue stained gel showing the quantity of GST and GST fusion proteins added to the binding reactions (*). METHODS: GST-BTBD1 or GST-BTBD2 was expressed and purified from E. coli extracts. ~5 μg of GST or the GST-fusion proteins were mixed with HeLa cell nuclear extracts and incubated for 2 hr at 4°C (0.25 M NaCl). The bead pellets were then washed 3 times with ice cold PBS before solubilization in SDS sample buffer. One tenth of the nuclear extract used in the binding reaction was loaded in the lanes labeled "HeLa extract". Arrow shows co-precipitated TOP1.
Effect of GST-BTBD2 on TOP1 activity
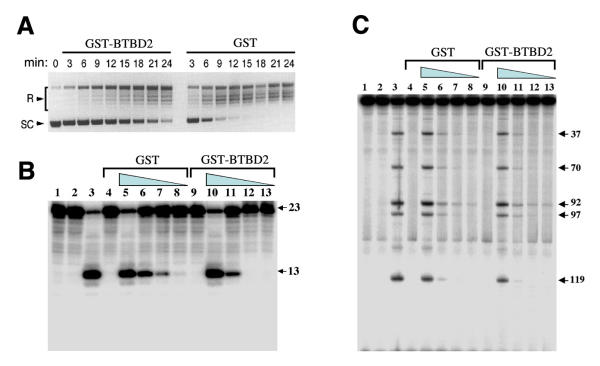
To test the effect of BTBD2 on the supercoil relaxation activity of TOP1, HeLa cell nuclear extracts containing TOP1 were incubated with GST or GST-BTBD2 (aa 86–525). GST-BTBD2 slightly inhibited the supercoil relaxation activity of TOP1, increasing the time required to completely relax the supercoiled DNA by about two-fold as compared with GST alone (Fig. 5A). The DNA cleavage assay was also used to determine if BTBD2 affected TOP1 activity. This assay measures the ability of TOP1 to cleave end-labeled DNA substrates in the presence of the anticancer drug camptothecin, which blocks the ligation step of the cleavage /religation reaction of TOP1. A 36 bp DNA containing a single strong TOP1 cleavage site [24] or a 161 bp restriction fragment [25] was used as the substrate. GST-BTBD2 addition produced a slight inhibition of TOP1 cleavage activity relative to GST alone (Fig. 5B) on the smaller substrate and had no detectable effect on cleavage activity or cleavage site selection by TOP1 on the larger substrate (5C).
Figure 5.
Effect of BTBD2 on TOP1 activity in vitro.A.Supercoil relaxation activity. HeLa cell nuclear extract containing TOP1 was mixed with GST-BTBD2 or GST alone for 15 min on ice before the addition of supercoiled DNA. The reactions were then incubated for 0–24 min at 30°C before stopping with SDS. The location of relaxed and supercoiled topoisomers are indicated (R and SC). B.DNA nicking at a strong TOP1 cleavage site. GST or GST-BTBD2 was incubated with TOP1 for 20 min at room temperature before the addition of a 36 bp 3' end-labeled DNA [24]. Cleavage reactions were continued for another 20 min before stopping with SDS. Lane 1: DNA alone; lane 2: + TOP1 (1.2 ng/reaction); lane 3: same + 10 μM camptothecin; lanes 4–8: reactions contained 0.25 μ g GST and 10 μ M camptothecin; lanes 9–13 contained 0.25 μg GST-BTBD2 + 10 μM camptothecin; lanes 4 & 9: no TOP1 added; lanes 5 & 10: 1.2 ng TOP1; lanes 6 & 11: 0.4 ng TOP1; lanes 7 & 12: 0.3 ng TOP1; lanes 8 & 13: 0.1 ng TOP1). C. As in B, but using a 161 bp substrate DNA [25]. Numbering of cleavage sites according to Pommier et al. [25] is indicated on the right side.
Cellular localization of BTBD2 and co-localization with TOP1
To determine the localization of BTBD2 in cells, we transiently transfected HeLa cells with expression plasmids encoding either myc-tagged full length BTBD2 (m-BTBD2F) or a truncated BTBD2 having the N-terminal half of the BTB domain removed (m-BTBD2T). The full length BTBD2 localized entirely to cytoplasmic bodies (Fig. 6B, m-BTBD2F). The truncated BTBD2 localized predominantly to nuclear bodies with a minor fraction localizing to cytoplasmic bodies (Fig. 6C, m-BTBD2T).
Figure 6.
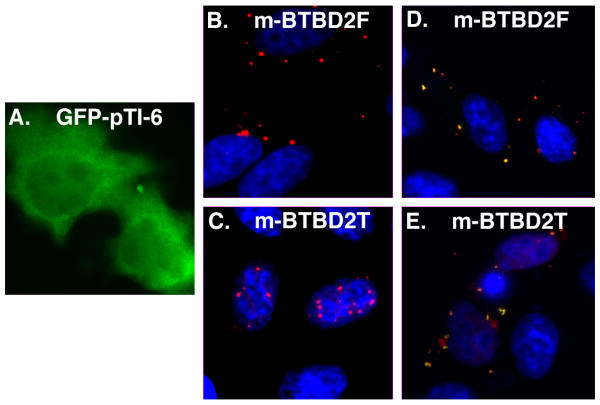
Localization of BTBD2 and co-localization with TOP1. HeLa cells were transfected with: A, GFP-tagged TOP1 lacking a nuclear localization (GFP-pTI-6); B, full length BTBD2 containing a C-terminal myc tag (m-BTBD2F); C, truncated BTBD2 having the N-terminal half the BTB domain deleted (m-BTBD2T); D, GFP-pTI-6 and m-BTBD2F; and E, GFP-pTI-6 and m-BTBD2T.
The localization of full length BTBD2 to cytoplasmic bodies was surprising because the cytoplasm is separated from the nucleus where endogenous TOP1 resides and transfected GFP-TOP1 localizes [26-30]. Because TOP1 and BTBD2 are not predominantly found in the same compartments, they may interact transiently, or they may interact only under specific conditions when they may be in the same compartment. We therefore analyzed mitotic cells, but did not observe co-localization of TOP1 and the BTBDs (not shown). We then tested for co-localization in transfected cells. Though the truncated m-BTBD2T localized to distinct nuclear bodies, its co-localization with endogenous TOP1 could not be determined against the background of endogenous TOP1 diffuse nuclear staining. However, in cells co-transfected with GFP-TOP1 and m-BTBD2T, the m-BTBD2T staining was nuclear diffuse as well as in nuclear bodies, indicating that m-BTBD2T interacts with GFP-TOP1, which localizes throughout the nucleus (not shown).
To determine if TOP1 co-localizes with BTBD2 in the cytoplasm, GFP-TOP1 lacking an NLS [30] was co-transfected with full length BTBD2 (m-BTBD2F) or truncated BTBD2 (m-BTBD2T). GFP-TOP1 lacking a NLS (GFP-pTI-6) localizes diffusely within the cytoplasm (Fig. 6A and [30]). In cells transfected with both GFP-pTI-6 and full length m-BTBD2F, the diffuse cytoplasmic GFP-pTI-6 significantly decreased, and GFP-pTI-6 co-localized with m-BTBD2F to cytoplasmic bodies (Fig. 6D). In cells transfected with GFP-pTI-6 and truncated m-BTBD2T, truncated m-BTBD2T was not in nuclear bodies as when transfected alone, rather it co-localized with GFP-pTI-6 to cytoplasmic bodies (Fig. 6E). Thus, when co-transfected with GFP-TOP1 lacking a NLS, both full length and the N-terminal deleted BTBD2 co-localized with TOP1 to cytoplasmic bodies.
Discussion
Two-hybrid screens have identified BTBD1 and BTBD2 as proteins that interact with the core domain of TOP1. BTBD1 and BTBD2 were isolated as 15 and 19 independent clones, respectively, and in two-hybrid assays they interacted as strongly as the control for strongly interacting proteins (c-Fos with c-Jun). BTBD1 and BTBD2 also bound HeLa cell TOP1 (100 kDa) in nuclear extracts by GST-pulldown assays. N-terminal truncation studies showed the C-terminal 37% to 45% of the BTBD proteins are sufficient for binding TOP1. C-terminal deletions of either protein eliminated binding to TOP1 (C-terminal truncation of 112 aa of BTBD1 or 33 aa of BTBD2 in the two-hybrid assay; or a C-terminal truncation of 40 aa of BTBD2 in GST-pulldown). Truncation of the BTB domains demonstrated that they were not required for interaction with TOP1. The region of TOP1 sufficient for binding BTBD2 was narrowed to residues 215 – 329 in the "cap" region (core subdomain II) [31]. Catalytic activities of TOP1 were affected slightly or not at all by co-incubation with GST-BTBD2 suggesting that the interaction does not regulate enzyme activity directly.
We cloned the full length BTBD1 cDNA from a HeLa cell library. This sequence is 3196 bp, and the start codon is preceded by a stop codon 6 nucleotides upstream (GenBank, FLJ20724). Our sequence is comparable to two others in GenBank, one of which was obtained by the oligo-capping method suggesting that it begins at the 5' cap [21,32].
Our repeated attempts to clone the 5' end of the BTBD2 by library screening and the RACE method were unsuccessful. We tested sequences directly upstream of the longest cDNA in the BTBD2 gene for their presence on the mRNA. RT-PCR and sequencing of the RT-PCR product demonstrated that this 255 nucleotide, 89% GC-rich sequence is present on the BTBD2 mRNA and that it shares predicted polypeptide sequence with BTBD homologues and orthologs.
The BTBD proteins contain an amino terminal BTB/POZ domain as well as a C-terminal region that is related to proteins that contain the kelch domain. Neither of these regions yields unequivocal information for functional classification. The BTB domain is a highly conserved 120 amino acid, hydrophobic-rich, protein-protein interaction domain that mediates homodimerization and heterodimerization of a large number of proteins with diverse functions [9]. The three dimensional structure of one BTB domain [33] has been characterized through structure-function studies [34]. Proteins containing BTB domains are currently categorized into three groups [35]: 1) DNA-binding proteins often containing zinc fingers in the C-terminal region, 2) actin binding proteins that contain a kelch repeat of ~50 residues that terminate in a pair of glycine residues [36] and 3) proteins that have neither an actin-binding domain nor a DNA-binding domain. The BTBD proteins belong to the last category, although they are distantly related to kelch proteins (~25% identical over ~250 aa), they lack the residues that are characteristic of kelch repeats, a double glycine sequence and a tyrosine separated from a tryptophan by precisely 6 residues [35,37].
BTBD1 and BTBD2 are closely related sharing 80% identical amino acid sequence. They are one of eleven genes in 19p13.3 → p12 with close homology to genes in 15q24 → q26 that have apparently resulted from duplications of a common ancestral gene [38]. Both BTBD1 and BTBD2 are widely if not ubiquitously expressed in human tissues. The predicted promoter regions are highly GC-rich and lack canonical TATA boxes, suggesting that they may be housekeeping genes [39,40].
Close mammalian relatives of BTBD1 and BTBD2 include the mouse orthologs that are 98% and 95% identical respectively, as well as highly similar rat and bovine partial sequences [21]. Sequence relatives in lower organisms include a C. elegans sequence with 58% identities with BTBD2 (429 residues; 2% gaps, F38H4.7), and a D. melanogaster sequence with 46% identities (437 residues, 3% gaps, CG5319). The human genome encodes two proteins with 46% (436 residues, 3% gaps, CAC22147) and 45% (497 residues, 5% gaps, AF353674) identities compared with BTBD2. These proteins comprise a small family whose members have not been functionally characterized and are evidently specific to animals because putative orthologs are not present in available fungi and plant sequence databases.
We have demonstrated an interaction between BTBD2 and TOP1 by two-hybrid, GST-pull down and intracellular co-localization analyses. Studies are underway to functionally characterize BTBD1 and BTBD2 and understand their interaction with TOP1.
Conclusions
• Fifteen and nineteen independent cDNA clones of BTBD1 and BTBD2, respectively, were isolated from two-hybrid screens and found to interact as strongly as c-Fos interacted with c-Jun in the two-hybrid assays.
• The interactions of TOP 1 with BTBD1 and BTBD2 were confirmed by deletion mapping using two-hybrid and GST-pulldown assays.
• Less than the C-terminal half of each BTBD protein lacking the BTB domain is sufficient for binding to TOP1. TOP1 from aa 215 to 329 interacted with BTBD2.
• GFP-BTBD2 localized to cytoplasmic bodies.
• BTBD2 had little effect on the supercoil relaxation or DNA cleavage activity of TOP1 in vitro.
• Co-transfected BTBD2 and TOP1 co-localized.
• The BTBD2 message contains 255 nucleotides that are 89% GC-rich and are predicted to encode the N-terminus of a 525 amino acid protein.
• BTBD1 and BTBD2 mRNA were detected in all human tissues tested. BTBD1 mRNA was highly expressed in testes, heart and skeletal muscle. BTBD2 was more highly expressed in skeletal muscle. Only a single mRNA species was present for each BTBD1 and BTBD2.
• Putative orthologs of BTBD1 and BTBD2 are found in C. elegans (58% identities, 429 residues; 2% gaps, F38H4.7) and D. melanogaster (46% identities, 437 residues, 3% gaps, CG5319).
• Two paralogs of BTBD1 and BTBD2 are present in the human genome (CAC22147 and AF353674).
Materials and Methods
Plasmid construction
To construct the bait expression plasmid (pGBD-cTOP1), the core domain of TOP1 (cTOP1, amino acids 198–651) was amplified by PCR (sense primer: 5'-CGGAATTCGAAGAGGAACAGAAGTGGAAATGG-3'; antisense primer: 5'-CGGGATCCTTAAATCTTAGTTTGCAAGTTCATCAT-3') and inserted into the vector pGBD (Clontech) cut with EcoRI and BamHI. To construct the cTOP1 expression vector (pDBleu-TOP1c), cTOP1 was excised from pGBD-cTOP1 with EcoRI and BamHI and inserted into the same sites in pDBleu (ProQuest, Life Technologies).
To construct the GST-fusion protein expression vectors, the BTBD inserts from the prey vector pPC86 were amplified using primers complementary to the BTBD coding regions with additional Sma I and EcoR I restriction sites for insertion into these sites in pGEX-2TK (Pharmacia). These primer sequences can be found below under the Two Hybrid Screens section. All constructs were confirmed by sequencing using pGEX sequencing primers (Pharmacia Cat# 27-1410-01 and 27-1411-01).
To construct the expression plasmid encoding C-terminal-myc-tagged, full length BTBD2 (m-BTBD2F), the entire coding region of BTBD2 (1–1575 bp) was amplified with primers (Upper primer: 5' ATGGCGGCGGGTGGGAGCGGCGGGCGTGCGTCGTG-3', Lower primer: 5'-GGTGTAGAAGATGACCTCGGGGATCTGGCCGTCCTCCAC-3') containing BamH I and Hind III cleavage sites and inserted into pCMV-Tag 5 vector (Stratagene) cut with BamH I and Hind III. To construct the expression plasmid encoding C-terminal-myc-tagged truncated BTBD2 having the N-terminal half the BTB domain deleted (m-BTBD2T), the coding region of BTBD2 from the second ATG to almost the end of the 3'-UTR (445–2542 bp) was amplified with primers (Upper primer: 5'-ATGTTCAACGGGGGAATGGCCACAACATC-3', Lower primer: 5'-GGGGAGGACGCAGGTTCCAGGAGT-3') containing BamH I and Hind III cleavage sites and inserted into pCMV-Tag 3 vector (Stratagene) cut with BamH I and Hind III.
Two hybrid screens
pGBD-cTOP1 was co-transfected with the HeLa cell library (Clontech, Cat. no. HL4000AA) into strain PJ69-4A (MATa, trp 1-190 leu2-3, 112 ura3-52 his3-200 gal4 gal80 LYS2::GAL1-HIS3 GAL2-ADE2 met2::GAL7-lacZ)[41]. Strong expression of full length GALDB-TOP1 in PJ69-4A was verified by Western blotting with antibodies to both TOP1 and GALDB (not shown). Additionally, the ProQuest two-hybrid system was used because the low copy CEN plasmids may decrease the probability of obtaining weakly interacting false positives that could result from high concentrations of the two-hybrid proteins. In the ProQuest screens, pDBleu-TOP1c was co-transfected with a HeLa cell cDNA library (Life Technologies Cat. No. 11287-018) according the manufacturer protocols. Clones that tested positive in the two-hybrid assays were sequenced across the GAL4AD-prey junction into the coding regions of the prey inserts. Sequences were grouped into cDNA contigs using the SeqMan II program of DNASTAR (LASERGENE).
C-terminal deletions for mapping the interactions using two-hybrid assays were constructed using the exonuclease III/mung bean nuclease method (C-terminal deletions, Exonuclease III/Mung Bean Nuclease Deletion kit, Cat#460, NEW ENGLAND BioLabs company). N-terminal deletions, in addition to the series obtained through two-hybrid screening, were constructed by amplifying the two-hybrid cDNA clones using primers containing Sma I and EcoR I at the N- and C-terminus, respectively, and inserting them into both pPC86 and pGEX-2TK vectors. The sense primer sequences are as follows: BTBD1: U1 (395-482aa) 5'-GACCCCGGGGAATGATACCGGCTTTAGTTGTGA-3', U2 (342-482aa) 5'-GACCCCGGGCTGGGGTTACAGTGGGACGAGT-3', U3 (213-482aa) 5'-GACCCCGGGAAGTGCAGAAGGGTTTACT-3', U4 (303-482aa) 5'-GACCCCGGGAAACCTCTTTCTTCATTTTACTGT-3', The same antisense primer was used for all amplifications: 5'-CGGAATTCTTTGTGGCCATTTATGTTTTTGAC-3'. The sense primers for amplification of truncated BTBD2 fragments were: U1 (504-525aa) 5'-GACCCCGGGCTACGCGGCCGGGAACAACAATG-3', U2 (460-525aa) 5'-GACCCCGGGGGTGCTGCCCAACGTCAACTACAC-3', U3 (404-525aa) 5'-GACCCCGGGCGTGGTGGGATTTGGGCTGTATGG-3', U4 (289-525aa) 5'-GACCCCGGGCCGCTGGTCCGAGGCCGAGTGTC-3'. The same antisense primer was used for all amplifications: 5'-CGGAATTCAACCCCGTCCTGATGCTGAGAAAG-3'
Northern blots
Multiple tissue Northern blots containing 2 μg poly (A) + RNA (Human Multiple Tissue Northern Blot IV and Human Multiple Tissue Northern Blot; Catalog numbers: 7766-1 and 7760-1; CLONTECH) were hybridized with 32P-labeled human cDNA probes for TOP1(nucleotides 212–1661, GenBank accession number J03250), BTBD1 (nucleotides 84–3196, GenBank accession number AF355402) or BTBD2 (nucleotides 1229–2668, GenBank accession number AF355797) in Rapid-hyb buffers (Amersham Life Science, RPN1635). Hybridization and washing was carried out according to the manufacturer instructions.
Isolation of the BTBD2 coding region
The longest BTBD2 cDNA obtained from two-hybrid screening (2413 bp) contained an ORF starting at the 5' end. To obtain the full coding region, four independent cDNA libraries were screened using a nested PCR strategy with upper primers complementary to the vectors and two nested lower primers complementary to BTBD2 (5'-TCGTCCGAGTAGAGAAAC-3' and 5'-TCCACGTCGGGCAGCTCAATCTCC-3'). 5'-RACE (Life Technologies) was also used to try to identify longer clones using several different lower primers. Reverse transcription was also performed at 70°C using the thermophilic Tth polymerase with magnesium as the cofactor to enable it to use mRNA as a template. None of these strategies was successful.
To determine whether the mRNA contained the BTBD2 gene sequences directly upstream of the longest cDNA clone, RT-PCR was used with a sense primer complementary to the predicted start codon. The primers for reverse transcription were complementary to exon 2 (LO and LI, see below). PolyA+ RNA was reverse transcribed using Tth polymerase (Promega) at 70°C for 20 min. The resulting products were amplified using nested PCR with the Advantage-GC 2 PCR kit (CLONTECH, Cat# K-1913-Y) and two 30 cycle rounds with each cycle having two steps (denaturation 94°C, 30 sec.; annealing and extension 68°C, 3 min; UO: 5'-ATGGCGGCGGGTGGGAGCGGCGGGCGTGCGTCGTG-3'; UI: 5'-CGGGCCCCGGGACAGACG-3'; LO: 5'-CGGGCAGCTCAATCTCCGTGGATG-3'; LI: 5'-TTGTGGCCATTCCCCCGTTGAACA-3'). To amplify the 306 bp fragment, two rounds of PCR were used: the first round used the LO and UO primers, and the second round used the LO and UI. To amplify the 460 bp fragment, two rounds of PCR were also used, the LO and UO in the first round, and the LI and the UO in the second round.
GST Pull Down Assay
E. coli containing plasmids encoding GST-BTBD1 or GST-BTBD2 were grown to an O.D. of 0.3 at 25°C, and 2% EtOH was added just prior to induction with 0.1 mM IPTG for 8 hrs. E. coli extracts were prepared using a French Pressure Cell Press (American Instrument Company, Silver Spring, MD). For the GST-pull-down assay 5 μg of GST or the GST-fusion proteins were mixed with 150 μl HeLa cell nuclear extracts (5 × 106 cells/ml) and incubated for 2 hr at 4°C (0.25 M NaCl). The bead pellets were then washed 3 times with ice cold PBS before solubilization in SDS-PAGE sample buffer.
TOP1 assays
Supercoil relaxation assay
HeLa nuclear extract (5 ng) in reaction buffer (100 mM Tris.HCl, pH 7.5, 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, 15 μg/ml BSA) was incubated with 5 μg of purified GST or GST-BTBD2 on ice for 15 min before the addition of 0.25 μg plasmid and incubation at 30°C for 0–24 min. Reactions were stopped by adding SDS (1% final) and proteinase K (0.5 mg/reaction).
DNA cleavage assays
Purified TOP1 was pre-incubated with GST or GST-BTBD2 for 20 min before addition of the DNA substrate, which was either an end-labeled 36 bp fragment containing a single strong TOP1 cleavage site [24], or a 161 bp PvuII-HindIII fragment of pBluescript (Stratagene) [25]). End labeling of the DNA substrates was as previously described [24,25]. Cleavage reactions were continued for 20 min before stopping with 0.5% SDS.
Abbreviations
aa, amino acid; BTB: Broad-Complex, Tramtrack and Bric-a-brac; GST, glutathione-S-transferase; NLS, nuclear localization signal; ORF, open reading frame; RT-PCR, reverse transcription – polymerase chain reaction; TOP1, DNA topoisomerase I.
Acknowledgments
Acknowledgements
We thank Dr's Yin-Yuan Mo and Bill Beck for providing the GFP-TOP1 expression plasmids and Dr. Todd Martensen for critical comments on the manuscript.
Contributor Information
Lixin Xu, Email: lixu@usuhs.mil.
Lihong Yang, Email: yanglih@mail.nih.gov.
Keiko Hashimoto, Email: khashimoto@usuhs.mil.
Melvin Anderson, Email: mander11@gl.umbc.edu.
Glenda Kohlhagen, Email: kohlhagg@mail.nih.gov.
Yves Pommier, Email: pommiery@pop.nci.nih.gov.
Peter D'Arpa, Email: pdarpa@usuhs.mil.
References
- Wang Z, Roeder RG. DNA topoisomerase I and PC4 can interact with human TFIIIC to promote both accurate termination and transcription reinitiation by RNA polymerase III. Mol Cell. 1998;1:749–757. doi: 10.1016/s1097-2765(00)80074-x. [DOI] [PubMed] [Google Scholar]
- Merino A, Madden K, Lane WS, Champoux J, Reinberg D. DNA topoisomerase I is involved in both repression and activation of transcription. Nature. 1993;365:227–232. doi: 10.1038/365227a0. [DOI] [PubMed] [Google Scholar]
- Gobert C, Skladanowski A, Larsen AK. The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53. Proc Natl Acad Sci USA. 1999;96:10355–10360. doi: 10.1073/pnas.96.18.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haluska P, Jr, Saleem A, Edwards TK, Rubin EH. Interaction between the N-terminus of human topoisomerase I and SV40 large T antigen. Nucleic Acids Res. 1998;26:1841–1847. doi: 10.1093/nar/26.7.1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards TK, Saleem A, Shaman JA, Dennis T, Gerigk C, Oliveros E, Gartenberg MR, Rubin EH. Role for Nucleolin/Nsr1 in the Cellular Localization of Topoisomerase I. J Biol Chem. 2000;275:36181–36188. doi: 10.1074/jbc.M006628200. [DOI] [PubMed] [Google Scholar]
- Haluska P, Jr, Saleem A, Rasheed Z, Ahmed F, Su EW, Liu LF, Rubin EH. Interaction between human topoisomerase I and a novel RING finger/arginine-serine protein. Nucleic Acids Res. 1999;27:2538–2544. doi: 10.1093/nar/27.12.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straub T, Grue P, Uhse A, Lisby M, Knudsen BR, Tange TO, Westergaard O, Boege F. The RNA-splicing factor PSF/p54 controls DNA-topoisomerase I activity by a direct interaction. J Biol Chem. 1998;273:26261–26264. doi: 10.1074/jbc.273.41.26261. [DOI] [PubMed] [Google Scholar]
- Labourier E, Rossi F, Gallouzi IE, Allemand E, Divita G, Tazi J. Interaction between the N-terminal domain of human DNA topoisomerase I and the arginine-serine domain of its substrate determines phosphorylation of SF2/ASF splicing factor. Nucleic Acids Res. 1998;26:2955–2962. doi: 10.1093/nar/26.12.2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayan L, Riou JF, Seite P, Migeon J, Cantereau A, Larsen CJ. Human ARF protein interacts with Topoisomerase I and stimulates its activity. Oncogene. 2001;19:836–848. doi: 10.1038/sj.onc.1204170. [DOI] [PubMed] [Google Scholar]
- Shaiu WL, Hsieh TS. Targeting to transcriptionally active loci by the hydrophilic N-terminal domain of Drosophila DNA topoisomerase I. Mol Cell Biol. 1998;18:4358–4367. doi: 10.1128/mcb.18.7.4358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaiu WL, Hu T, Hsieh TS. The hydrophilic, protease-sensitive terminal domains of eucaryotic DNA topoisomerases have essential intracellular functions. Pac Symp Biocomput. 1999:578–589. doi: 10.1142/9789814447300_0057. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Uchida-Toita M, Andoh T, Yoshida M. HTLV-1 tax oncoprotein binds to DNA topoisomerase I and inhibits its catalytic activity. Virology. 2000;270:291–298. doi: 10.1006/viro.2000.0266. [DOI] [PubMed] [Google Scholar]
- Desai SD, Li TK, Rodriguez-Bauman A, Rubin EH, Liu LF. Ubiquitin/26S proteasome-mediated degradation of topoisomerase I as a resistance mechanism to camptothecin in tumor cells. Cancer Res. 2001;61:5926–5932. [PubMed] [Google Scholar]
- Mao Y, Sun M, Desai SD, Liu LF. A possible repair response to topoisomerase-mediated DNA damage. Proc Natl Acad Sci USA. 2000;97:4046–4051. doi: 10.1073/pnas.080536597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staron K, Samuels DS. Phosphorylation of serine residues in the N-terminal domains of eukaryotic type I topoisomerases. Mol Biol Rep. 1998;25:157–161. doi: 10.1023/A:1006827925817. [DOI] [PubMed] [Google Scholar]
- Desai SD, Liu LF, Vazquez-Abad D, D'Arpa P. Ubiquitin-dependent destruction of topoisomerase I is stimulated by the antitumor drug camptothecin. J Biol Chem. 1997;272:24159–24164. doi: 10.1074/jbc.272.39.24159. [DOI] [PubMed] [Google Scholar]
- Staron K, Kowalska-Loth B, Zabek J, Czerwinski RM, Nieznanski K, Szumiel I. Topoisomerase I is differently phosphorylated in two sublines of L5178Y mouse lymphoma cells. Biochim Biophys Acta. 1995;1260:35–42. doi: 10.1016/0167-4781(94)00175-3. [DOI] [PubMed] [Google Scholar]
- Stewart L, Ireton GC, Champoux JJ. The domain organization of human topoisomerase I. J Biol Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- Stewart L, Ireton GC, Parker LH, Madden KR, Champoux JJ. Biochemical and biophysical analyses of recombinant forms of human topoisomerase I. J Biol Chem. 1996;271:7593–7601. doi: 10.1074/jbc.271.13.7593. [DOI] [PubMed] [Google Scholar]
- Stewart L, Ireton GC, Champoux JJ. Reconstitution of human topoisomerase I by fragment complementation. J Mol Biol. 1997;269:355–372. doi: 10.1006/jmbi.1997.1056. [DOI] [PubMed] [Google Scholar]
- Carim-Todd L, Sumoy L, Andreu N, Estivill X, Escarceller M. Identification and characterization of BTBD1, a novel BTB domain containing gene on human chromosome 15q24. Gene. 2001;262:275–281. doi: 10.1016/S0378-1119(00)00513-8. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lash AE, Tolstoshev CM, Wagner L, Schuler GD, Strausberg RL, Riggins GJ, Altschul SF. SAGEmap: a public gene expression resource. Genome Res. 2000;10:1051–1060. doi: 10.1101/gr.10.7.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pourquier P, Takebayashi Y, Urasaki Y, Gioffre C, Kohlhagen G, Pommier Y. Induction of topoisomerase I cleavage complexes by 1-beta-D-arabinofuranosylcytosine (ara-C) in vitro and in ara-C-treated cells. Proc Natl Acad Sci USA. 2000;97:1885–1890. doi: 10.1073/pnas.97.4.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pommier Y, Kohlhagen G, Wu C, Simmons DT. Mammalian DNA topoisomerase I activity and poisoning by camptothecin are inhibited by simian virus 40 large T antigen. Biochemistry. 1998;37:3818–3823. doi: 10.1021/bi972067d. [DOI] [PubMed] [Google Scholar]
- Muller MT, Pfund WP, Mehta VB, Trask DK. Eukaryotic type I topoisomerase is enriched in the nucleolus and catalytically active on ribosomal DNA. Embo J. 1985;4:1237–1243. doi: 10.1002/j.1460-2075.1985.tb03766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danks MK, Garrett KE, Marion RC, Whipple DO. Subcellular redistribution of DNA topoisomerase I in anaplastic astrocytoma cells treated with topotecan. Cancer Res. 1996;56:1664–1673. [PubMed] [Google Scholar]
- Wadkins RM, Danks MK, Horowitz L, Baker SD. Characterization of topotecan-mediated redistribution of DNA topoisomerase I by digital imaging microscopy. Exp Cell Res. 1998;241:332–339. doi: 10.1006/excr.1998.4033. [DOI] [PubMed] [Google Scholar]
- Zini N, Santi S, Ognibene A, Bavelloni A, Neri LM, Valmori A, Mariani E, Negri C, Astaldi-Ricotti GC, Maraldi NM. Discrete localization of different DNA topoisomerases in HeLa and K562 cell nuclei and subnuclear fractions. Exp Cell Res. 1994;210:336–348. doi: 10.1006/excr.1994.1046. [DOI] [PubMed] [Google Scholar]
- Mo YY, Wang P, Beck WT. Functional expression of human DNA topoisomerase I and its subcellular localization in HeLa cells. Exp Cell Res. 2000;256:480–490. doi: 10.1006/excr.2000.4864. [DOI] [PubMed] [Google Scholar]
- Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- Yudate HT, et al. HUNT: launch of a full-length cDNA database from the Helix Research Institute. Nucleic Acids Res. 2001;29:185–188. doi: 10.1093/nar/29.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad KF, Engel CK, Prive GG. Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci USA. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Peng H, Schultz DC, Lopez-Guisa JM, Rauscher FJ, 3rd, Marmorstein R. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 1999;59:5275–5282. [PubMed] [Google Scholar]
- Adams J, Kelso R, Cooley L. The kelch repeat superfamily of proteins: propellers of cell function. Trends Cell Biol. 2000;10:17–24. doi: 10.1016/S0962-8924(99)01673-6. [DOI] [PubMed] [Google Scholar]
- Lai F, Orelli BJ, Till BG, Godley LA, Fernald AA, Pamintuan L, Le Beau MM. Molecular characterization of KLHL3, a human homologue of the Drosophila kelch gene. Genomics. 2000;66:65–75. doi: 10.1006/geno.2000.6181. [DOI] [PubMed] [Google Scholar]
- Bomont P, Cavalier L, Blondeau F, Hamida CB, Belal S, Tazir M, Demir E, Topaloglu H, Korinthenberg R, Tuysuz B, Landrieu P, Hentati F, Koenig M. The gene encoding gigaxonin, a new member of the cytoskeletal BTB/kelch repeat family, is mutated in giant axonal neuropathy. Nat Genet. 2000;26:370–374. doi: 10.1038/81701. [DOI] [PubMed] [Google Scholar]
- Carim-Todd L, Escarceller M, Estivill X, Sumoy L. Cloning of the novel gene TM6SF1 reveals conservation of clusters of paralogous genes between human chromosomes 15q24 → q26 and 19p13.3 → p12. Cytogenet Cell Genet. 2000;90:255–260. doi: 10.1159/000056784. [DOI] [PubMed] [Google Scholar]
- Dynan WS. Understanding the molecular mechanism by which methylation influences gene expression. Trends Genet. 1989;5:35–36. doi: 10.1016/0168-9525(89)90016-4. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Miura K, Fujino Y, Iwao H, Ogita S, Yamanaka S. Evolution, structure, and expression of GNPI/Oscillin orthologous genes. Genomics. 2000;68:179–186. doi: 10.1006/geno.2000.6287. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]