Abstract
Inactivation of the p53 gene is a key driver of tumorigenesis in various cancer cohorts and types. The quest for a successful p53-based therapy that holds the promise of treating more than half of the cancer population has culminated in extensive knowledge about the role and function of p53 and led to new proposed innovative strategies against p53-defective cancers. We will discuss some of these latest studies with a focus on metabolic regulation and DNA damage response and also highlight novel functions of p53 in these pathways that may provide a contemporary rationale for targeting p53 loss in tumors.
Keywords: therapeutics, metabolism, DNA damage, replication stress, p53, synthetic lethality
p53—a molecular pathway switch
While mutation of p53 abolishes the barrier against tumorigenesis, by eliminating important cell cycle checkpoints and attenuating apoptotic responses, at the same time, impairing p53 function in cells switches cell fate and pathway choices that makes them highly susceptible to certain metabolic perturbations or challenges arising from DNA transactions and damage. The networks of metabolic and DNA damage response (DDR) pathways have built in redundancies that provide the versatility for cancer cells to circumvent obstacles and escape death, even when cellular and DNA integrity is being compromised. Eliciting changes in these networks may present new opportunities for targeting the inherent vulnerability in p53-defective cancer cells and may call for nonconventional interventions for cancer therapy (Gurpinar and Vousden, 2015; Cheok and Lane, 2017).
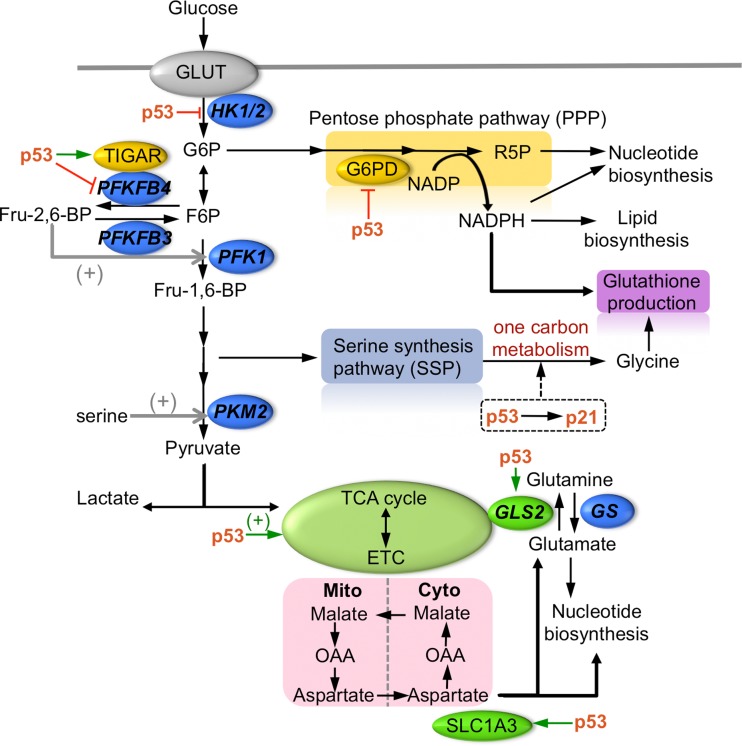
p53 is shown to restrict the pentose phosphate pathway (PPP) through multiple mechanisms (Mathupala et al., 1997; Jiang et al., 2011; Hitosugi et al., 2012; Ros et al., 2017), directing metabolites away from cellular anabolic metabolism and generally promotes conservation of energy especially under conditions of nutrient restriction. In mutant p53 cells, the switch to PPP signals an increase in biosynthesis pathways that generate a demand for NADPH and nucleotides and represents a metabolic adaption that is in line with an increase in uncontrolled cell proliferation and growth (Maddocks et al., 2013). Conversely, p53 may also promote PPP and decrease glycolysis under certain conditions that induce DNA damage and metabolic stress, indirectly through the action of TIGAR (Bensaad et al., 2006), presumably to enhance nucleic acids synthesis for DNA metabolism and decrease intracellular ROS. Therefore, the action of p53 at the bifurcation point provides a critical molecular switch determining glucose flux between glycolysis and PPP. Indeed, the ability of p53 to respond to nutrient deficiencies underlies its role as a central mediator of cellular stress response. Wild-type p53 containing cancer cells are able to cope with serine and glutamine starvation, in part by maintaining the cellular antioxidant capacity, whereas p53-deficient cancer cells are highly susceptible to serine and glutamine withdrawal (Maddocks et al., 2013; Tajan et al., 2018).
Recently, p53 has been shown to regulate critical enzymes in lipid metabolism pathways as part of its homeostatic response to changes in phospholipid biogenesis (Kumar et al., 2018). This metabolic switch impacts upon cell fate outcome by counteracting cellular events that lead to a fatty acid-induced apoptosis thus circumventing cell death. Altogether, these studies serve as some examples of how the prosurvival functions of p53 may be exploited to design innovative strategies that target p53-defective cancers.
Engineering metabolic roadblocks
Interestingly, counteracting cancer cell metabolism by engineering nutrient deficiencies has been shown to reduce tumor growth and sufficiently improve overall survival in several oncogenic mouse models tested including Apcmin/+ (intestinal), Eμ-Myc (lymophoma), and Pdx1-cre;KRasG12D/+;Trp53+/− (pancreatic) (Maddocks et al., 2017). The studies revealed that serine and glycine metabolism is critical to some types of cancers (Mattaini et al., 2016), and toggling this branch of metabolic pathway can be a strategy to selectively eliminate cancers. It seems that p53 regulate the response to serine starvation, and, consequently, mutation or loss of p53 results in a dramatic reduction in proliferation and growth that is evoked by a deficiency in G1 cell cycle arrest and antioxidant defense (Maddocks et al., 2013). Under serine starvation, cancer cells upregulate the serine biosynthetic pathway (SSP) and incite the one-carbon metabolism to deal with the need for nucleotides, ATP and methylation events (Labuschagne et al., 2014; Yang and Vousden, 2016). De novo synthesized serine is a major donor of one-carbon units to the folate cycle leading to the biosynthesis of adenosine, guanosine, and thymidylate to support the nucleotide demand in proliferative cancer cells (Davis et al., 2004; Locasale, 2013). Besides, the folate pathway contributes to the regeneration of cofactors NADH, NADPH, and ATP in the mitochondria. The finding that a multi-enzyme complex, purinosome, which promotes efficiency in purine synthesis by clustering biosynthetic enzymes in the purine synthesis pathway (An et al., 2008), is found to be localized near the mitochondria (French et al., 2016), further supports the idea that the mitochondria folate pathway is closely associated with the de novo purine synthesis pathway. More precisely, p53 promotes metabolic remodeling in response to serine starvation, in part by channeling the conversion of the limited pool of serine into reduced glutathione (GSH), rather than nucleotide synthesis, thus maintaining the antioxidant capacity in wild-type p53 cells. Conversely, cells deficient in p53 undergo a sustained change in metabolic processes that favor an increase in TCA cycle flux and higher levels of oxygen consumption which was not compensated with an increase in GSH generation, eventually resulting in increased intracellular ROS and apoptosis (Maddocks et al., 2013). Evidently, pyruvate addition alone partially elevated cell death in p53-deficient cells, as did N-acetylcysteine or GSH, but the combined supplementation restored fully the proliferation defects under the condition of serine/glycine starvation, suggesting that both impaired glycolysis and ROS generation accounted for cell death in p53-deficient cells. The dependence of cancer cells, and especially p53-defective cancer cells, on serine, provides a unique trait that can be potentially exploited either through the withdrawal of serine in diets or the inhibition of de novo serine synthesis pathway.
It is becoming clear that multiple metabolic stress pathways activate p53, placing it as a central responder and regulator of cellular bioenergetics. A main part of p53’s activity is likely driven through its transcriptional function, by coordinating the expression of a plethora of genes to help bring about homeostatic regulation. Glutamine depletion induces p53 transcriptional activity, resulting in the upregulation of SLC1A3, an aspartate/glutamate transporter, that is needed to maintain nucleotides biosynthesis under conditions when glutamine is depleted, and when electron transport chain and TCA activity are likewise restricted (Tajan et al., 2018). SLC1A3 imports extracellular aspartate as an alternative source for purine and pyrimidine synthesis; therefore, although deletion of SLC1A3 did not impact upon normal cell survival under fully fed conditions, it results in a clear reduction in cell viability when glutamine is withdrawn, mimicking the effects of p53 loss. Consistent with this, high level of expression of SLC1A3 in some cancer cell types, regardless of p53 status, is thought to correlate with resistance to glutamine withdrawal, adding to the complex regulation of glutamine sensitivity and highlights the fact that other factors in addition to p53 may play a role in this (Xiang et al., 2015; Altman et al., 2016). This further warrants investigation on other factors or biomarkers of sensitivity to glutamine deprivation if SLC1A3 inhibitors or other inhibitors of aspartate metabolism are to be developed as potential therapeutics.
In addition to depriving cells of key amino acids as a way to engineer metabolic roadblocks in p53-deficient cells, direct manipulation of enzymatic activity in the glycolytic pathway has been shown to deplete p53-deficient cells (Ros et al., 2017). PFKFB4 is one of the four phosphofructokinase/fructose bisphosphatase (PFK/FBPase) proteins that differ in their tissue-specific expression and their relative kinase and phosphatase activity (Ros and Schulze, 2013). PFKFB4 controls the levels of a critical intermediate fructose-2,6-bisphosphate (Fru-2,6-BP), which in turn controls the activity of phosphofructokinase 1 (PFK1) that catalyzes the first rate limiting step in glycolysis (Figure 1). Depletion of PFKFB4 therefore increases the levels of Fru-2,6-BP which enhance glycolytic flux, and direct glucose-6-phosphate (G6P) away from PPP (Figure 1), with an overall effect on reducing the flux through PPP, and decreasing nucleotide biosynthesis and NADPH regeneration (Ros et al., 2017). Consequently, the interference with anabolic metabolism led to increased apoptosis in p53-deficient cells. A recurrent theme in how wild-type p53 modifies response to metabolic stress is the regulation of nucleotide biosynthesis, co-factor NADPH generation, and antioxidant capacity through GSH synthesis or ROS scavenging activity (Ambs et al., 1998; Yoon et al., 2004; Budanov and Karin, 2008; Bensaad et al., 2009; Cano et al., 2009; Li et al., 2012), suggesting that redox regulation and nucleic acid synthesis is somehow tightly coupled. Notably, under acute conditions of cellular stress, p53 can induce the expression of Tp53-induced glycolysis regulatory phosphatase (TIGAR) (Bensaad et al., 2006), which promotes the conversion of Fru-2,6-BP to fructose-6-phosphate (F6P), hence attenuating PFK1 glycolytic activity and resulting in an increased flux through PPP. The transient and reversible metabolic switch mediated by p53, firstly, allows cells to deal with cellular stress conditions that may promote ROS, secondly, allows cells to return to baseline conditions that disfavor high sustained PPP activity. Consistent with this, p53 imposes other metabolic checks by directly inhibiting the activity of glucose-6-phosphate dehydrogenase (G6PD) to restrain PPP activity (Jiang et al., 2011), upregulating the expression of glutaminase-2 (GLS2), that hydrolyze glutamine to glutamate, to stimulate glutaminolysis (Hu et al., 2010; Suzuki et al., 2010), and transcriptionally repressing pyruvate dehydrogenase kinase-2 (PDK2) to promote oxidative phosphorylation (Contractor and Harris, 2012). The latter leads to the speculation that loss of p53 may lead to unrestrained PDK2 activity and that the dependence on PDK2 may be therapeutically exploited (Sutendra and Michelakis, 2013). The outcomes of most of these events are aimed at enhancing mitochondrial respiration, ATP/NADPH generation, boosting GSH production, and reducing intracellular ROS. Additionally, wild-type p53 also increases antioxidant factors, such as stress inducible sestrin proteins or the p53-inducible nuclear protein 1 (TP53INP1) (Velasco-Miguel et al., 1999; Okamura et al., 2001; Budanov et al., 2002; Tomasini et al., 2002; Peeters et al., 2003).
Figure 1.
p53-dependent regulation of carbohydrate and amino acid metabolism (+). p53 regulates carbohydrate metabolism at multiple nodes in the network, contributing to changes in PPP and glycolytic pathways and the availability of NADPH, cellular glutathione, and nucleotides. Reduction of amino acids, serine and glutamine, can lead to metabolic reprogramming in part through p53 activity to promote cell survival.
The ability of p53 to help support survival and metabolic adaptation under conditions of mild and transient metabolic stress in part contributes to a widened therapeutic window for targeting p53-deficient cells. Finding ways to trigger these conditions that activate p53 prosurvival activity while subjecting p53-deficient cells to metabolic catastrophe may be a useful strategy for a broad spectrum of p53 loss-of-function mutations including both nonsense and missense p53 mutations. Yet other examples of mutant p53 being directly involved in metabolic pathways may suggest alternative methods to target mutant p53 gain-of-function. For instance, the dependency of mutant p53 on the nucleoside salvage pathway enzyme deoxycytidine kinase (dCK) to drive its oncogenic activities by maintaining a balance in the dNTP pools suggests that inhibiting dCK may achieve synthetic lethality with cells containing mutant p53 gain-of-function (Kollareddy et al., 2015).
Toggling lipid metabolism
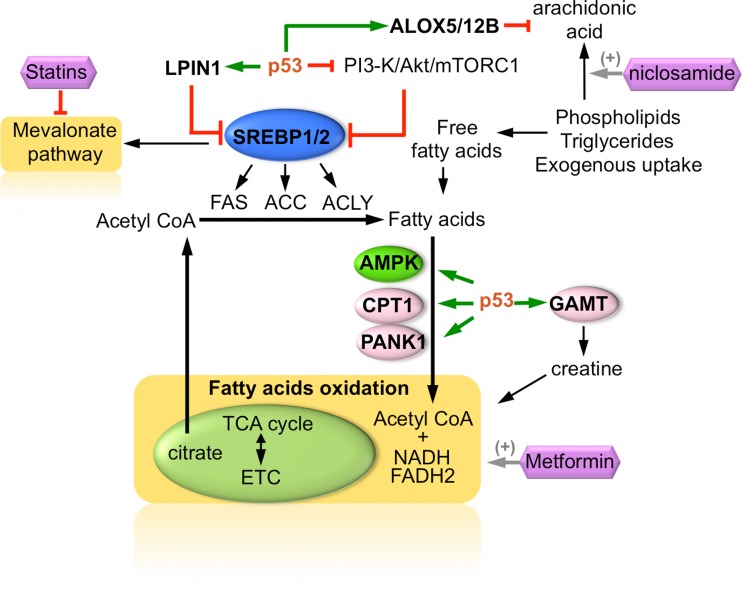
The reprogramming of cellular lipid metabolism by mutant p53 presents another attractive therapeutic target. Mutant p53 proteins were shown to alter the mevalonate pathway which disrupts the 3D architecture of mammary cells leading to a more disorganized morphology. Central to this is the interaction of mutant p53 with the sterol regulatory element-binding proteins (SREBP) transcription factors, which controls cholesterol and fatty acid synthesis (Freed-Pastor et al., 2012). Depletion of p53 reverted the mutant invasive phenotype, leading to the restoration of normal mammary architecture, as did statins, which inhibit the rate-limiting enzyme in the mevalonate pathway, HMG-CoA reductase (Freed-Pastor et al., 2012). Interestingly, statins also impaired the anchorage-independent growth of mutant p53 containing breast cancer cells in 2D culture, and reduced the growth of p53-mutant xenograft tumors, suggesting that the statins class of drugs may provide an alternative solution for targeting mutant p53 tumors (Figure 2).
Figure 2.
Regulation of fatty acid oxidation, mevalonate pathway, and arachidonic acid metabolism by p53. SREBP transcription factors are regulated by p53 through multiple mechanisms, with an overall effect on inhibiting de novo fatty acid synthesis and the mevalonate pathway. Statins inhibit HMG-CoA synthase and are shown to inhibit growth of mutant p53 tumors, as well as restore normal mammary architecture in vitro. p53 promotes the fatty acid oxidation pathway in an AMPK-dependent manner as part of its metabolic remodeling activity in response to, for example, low glucose conditions. Metformin stimulates AMPK activity and β-oxidation of fatty acids, promoting survival in cells with wild-type p53, and impairs growth of p53-deficient cells. Niclosamide stimulates the production of arachidonic acids from phospholipids turnover. p53–ALOX5/12B axis protects cells from arachidonic acid (AA)-induced apoptosis, whereas cells lacking wild-type p53 are susceptible to AA-induced metabolic catastrophe.
The inability of p53-deficient cancer cells to execute an AMPK-induced metabolic remodeling was exploited in a study using AMPK activators. It seems that under conditions of glucose limitation, cells stimulate β-oxidation of fatty acids to cope with nutrient deprivation, and AMPK signaling pathway is needed to coordinate the way that cells utilizes resources to generate ATP when cellular bioenergetics are compromised (Jones et al., 2005). Interestingly, this switch to an AMPK-dependent β-oxidation pathway was found to be p53-regulated (Buzzai et al., 2007). AMPK activators including metformin and 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR) were found to selectively impair p53-deficient cells, resulting in an improved reduction in the growth of p53-null HCT116 tumor xenografts when compared to the parental HCT116 xenografts (Buzzai et al., 2007). Moreover, mutant p53 were shown to inhibit AMPK activation by binding to the α-subunit of AMPK thereby limiting its function (Zhou et al., 2014). The gain-of-function of mutant p53 on deregulating AMPK activity is proposed to be important for the oncogenic property of mutant p53, in mediating increased growth and invasiveness, and represent another potentially druggable protein–protein interaction for p53-based therapy.
Several other mechanisms, in addition to an AMPK/p53-dependent pathway, may account for how p53 regulates β-oxidation of fatty acids in cells. In response to nutritional stress, a ROS–ATM–p53 pathway is activated that eventually led to the induction of LPIN1 and increased fatty acid metabolism (Assaily et al., 2011). LPIN1 performs a dual function; firstly, it acts as a transcriptional cofactor in a complex with peroxisome proliferator-activated receptor α (PPARα) and PPARγ coactivator-1α (PGC-1α) to regulate fatty acid oxidation genes, and, secondly, it directly participates in the biosynthesis of triacylglycerol (Finck et al., 2006; Han et al., 2006). p53 also regulates the activity of SREBPs through multiple mechanisms, for example, by suppressing the PI3-K/Akt/mTORC1 pathway through PTEN (Feng et al., 2007) (Figure 2). In addition, disruption of p53 increases the expression of SREBP1c (Yahagi et al., 2003) and mutant p53 binds directly to SREBP2 (Freed-Pastor et al., 2012) thereby enhancing the effects of p53 loss on fatty acids and cholesterol pathway and refining the connection between p53, lipogenesis, and tumor invasiveness. Interestingly, pharmacological inhibition of SREBPs in metastatic androgen-negative prostate cancer cells bearing p53 mutations is suggested as an effective therapeutic strategy, in line with the reported oncogenic effects of mutant p53 on SREBP pathway (Li et al., 2015). Besides, p53 also promotes the transcription of other genes involved directly in fatty acid oxidation, such as carnitine palmitoyltransferase 1C (CPT1C) (Sanchez-Macedo et al., 2013), acyl-coenzyme A dehydrogenase family member 11 (ACAD11) (Jiang et al., 2015) and guanidinoacetate N-methyltransferase (GAMT) (Ide et al., 2009). Together, the mounting evidence that p53 plays a critical role in nutrient stressed condition by reprogramming cellular metabolism, suggests cellular dependencies in the absence of p53 that may be targeted through synthetic lethal strategies.
Of particular interest is the regulation of autophagy by p53; p53 induces autophagy through multiple mechanisms (Feng et al., 2005; Budanov and Karin, 2008; Lee et al., 2010), including the direct upregulation of DRAM (Crighton et al., 2007). However, because autophagy is a basic cell survival-promoting pathway that is highly regulated in cancers to sustain metabolism and homeostasis under nutrient restriction or hyper-proliferative conditions, both inhibition and activation of autophagy have found context-dependent uses in tumor inhibition (Levy et al., 2017). Nevertheless, the modulation of autophagic flux in tumors with impaired p53 functions may provide another route for therapeutic exploitation. Vakifahmetoglu-Norberg et al. (2013) provide evidence of a more direct rationale for manipulating autophagy to target p53 mutant cells. Under nutrient stress conditions when chaperone-mediated autophagy is induced, inhibition of macroautophagy led to the degradation of mutant p53, suggesting a potential way that can be pharmacologically exploited to reduce the accumulation of mutant p53 (Vakifahmetoglu-Norberg et al., 2013).
More recently, an unbiased synthetic lethal screen for FDA-approved compounds identified an anti-helminthic compound, niclosamide, in sensitizing p53-deficient cells (Kumar et al., 2018). The first-in-class inhibitor against p53 mutant tumors was found to induce mitochondrial uncoupling, leading to phospholipid turnover and arachidonic acid (AA) production as its primary mechanism in tackling p53-deficient cancer cells (Figure 2). Conversely, wild-type p53-containing cells mount a defense to the lipid metabolic catastrophe by inducing the expression of lipid oxygenation genes, ALOX5 and ALOX12B that catalyze the dioxygenation and turnover of AA, thus evading apoptosis. The arachidonate pathway lies at the intersection of many oncogenic alterations, including PTEN, IDH1 and CTNNB1 (Gatto et al., 2016), making it likely that other oncogenic events, in addition to p53 mutation, may also render tumor cells susceptible to an arachidonic acid crisis.
Targeting the loss of p53-dependent metabolic transformation for cancer therapy is an attractive option that may offer the alternative to directly targeting mutant p53. Other cellular dependencies arising in the absence of p53 include the upregulation of specific enzymes such as phosphatidylinositol 5-phosphate 4-kinase (Emerling et al., 2013) in breast cancers harboring p53 loss or mutations, and hexokinase 2 (HK-2) in prostate cancers that bear dual deficiencies in both PTEN and p53 (Wang et al., 2014). Inhibitors to these enzymes or pathways might impair p53-mutant cancers by thwarting their metabolic adaption.
Targeting replication stress response
A number of recently published papers have shown that p53 protects the genome during S phase which broadens the tumor suppressive functions of p53 and elicits a model whereby p53 acts to prevent the occurrence of DNA damage emerging from replication processes, rather than simply respond to damage through its canonical roles in cell cycle arrest, apoptosis or repair. Exploring new mechanisms of p53’s functions in maintaining genome integrity may draw light to new ways to target loss-of-p53 by understanding the deficiencies in DNA damage/replication stress responses (RSRs) that create tumor vulnerabilities.
ATR is a key protein kinase that is activated during RSR to protect the integrity of replication forks, as well as to prevent aberrant and excessive replication origin firing, which depletes the cellular nucleotide pool and replication proteins. Extensive research shows that ATR delays cell cycle progression, stabilizes DNA replication forks (Couch et al., 2013; Zeman and Cimprich, 2014), and prevents the collapse of replication forks (Couch et al., 2013; Toledo et al., 2013). Importantly, the maintenance of genomic stability becomes dependent on a functional p53 and ATM when ATR is inhibited (Shiloh and Ziv, 2013; Bieging et al., 2014), suggesting that multiple compensatory pathways exist for genome maintenance.
A number of reports suggested that ATR inhibition may be a feasible synthetic lethal strategy to target cells which are defective in p53 and ATM. For instance, using the ATR inhibitor AZD6738 in p53- or ATM-defective chronic lymphocytic leukemia cells resulted in the accumulation of unrepaired DNA damage and consequently cell death through mitotic catastrophe (Kwok et al., 2016). The selective cytotoxicity of the ATR inhibitor, AZD6738, was observed in primary cells as well as in vivo xenograft models (Kwok et al., 2016). Deletion of ATR in p53-deficient mice has also been shown to be synthetically lethal (Ruzankina et al., 2009). Reintroducing the wild-type p53 has been shown to reduce sensitization of cells to ATR inhibition and this therefore confirms the attractiveness of exploring p53 mutation as a synthetically lethal target for ATR inhibition. In chronic lymphocytic leukemia cells deficient in p53, although they are initially resistant to most monotherapies such as chlorambucil and bendamustine even at high doses, they became sensitive when combined with the ATR inhibitor AZD6738 (Kwok et al., 2016). Therefore, the synthetic lethal interaction between ATR and p53 provides a promising therapeutic option to treat p53-deficient cancers.
Emerging knowledge suggest that p53 plays a more intricate role in replication processes than expected. Yeo et al. (2016) reported a novel function of p53 in preventing potential collisions between replication and transcription. The discovery stems from a screen of FDA-approved compounds, which revealed that topoisomerase II inhibitors are particularly toxic to p53-deficient cells. Topoisomerases are required to relieve the over- or under-winding of the DNA, that can normally occur during DNA transactions such as replication or transcription, which involves the unwinding of the DNA helix (Pommier, 2013). Positive supercoils accumulate ahead of the replication fork or transcription machinery, and a head-on collision between the two processes may result in severe topological constraints (Lin and Pasero, 2012). The exquisite sensitivity of p53-deficient cells to TOP2 poisons led to the suggestion that cells lacking p53 are predisposed to an increased frequency of replication-transcription interference (Yeo et al., 2016).
Underlying this outcome is the newfound role of p53 in regulating replication fork progression. The proposal that p53 might be involved in promoting normal replication fork progression is supported by the use of DNA fiber labeling method that provides a qualitative and quantitative assessment of the dynamics of DNA replication (Klusmann et al., 2016; Nieminuszczy et al., 2016). Deficiency in p53 impairs replication fork progression (Klusmann et al., 2016; Yeo et al., 2016). Inhibiting transcription rescued the replication defect in the p53-deficient cells and restored the replication rate to near wild-type level (Yeo et al., 2016) further demonstrating that transcription-associated replication stress is an outcome of p53 deficiency that renders cells vulnerable to TOP2 poisons. Interestingly, p53-deficient cells were found to be sensitive to the TOP2 poisons and not to other DNA-damaging drugs, suggesting that the sensitivity is not a consequence of defects in general DNA damage checkpoints. In addition, the response to TOP2 poisons was independent of p21, which debunked the possibility of a p53/p21-dependent G1 checkpoint arrest as a means of protecting against TOP2 poisons (Yeo et al., 2016).
Interestingly, inhibition of MDM2, a downstream target of p53, further reduced replication fork progression and exacerbated replication stress (Klusmann et al., 2016) even in the absence of p53, suggesting that MDM2 may promote additional fork activities that are independent of p53 (Klusmann et al., 2018). Whether the inhibition of MDM2 may further exacerbate genomic instability in a p53-deficient setting and lead to increased tumor cell death is an interesting possibility to explore. Although the inhibition of MDM2 using specific antagonists has been widely investigated as a potential therapeutic strategy against wild-type p53 tumors (Shangary and Wang, 2009; Cheok and Lane, 2012; Burgess et al., 2016; Wang et al., 2017), recent data support its alternative use in sensitizing p53-deficient cancer cells instead. Deletion of Mdm2 in T cell lymphoma or sarcomas deficient in p53 led to increased cell death and prolonged the survival of mice bearing these tumors, supporting the rationale for targeting MDM2 in p53-deficient tumors (Feeley et al., 2017). Other than MDM2, another downstream target of p53, p21, has been implicated in the maintenance of fork progression (Maya-Mendoza et al., 2018). It was suggested that fork progression exceeding 40% of the normal velocity results in DNA damage. Mechanistically, p21 cooperates with PARP in modulating fork progression by attenuating fork speed and ultimately preventing genomic instability (Maya-Mendoza et al., 2018).
p53 in fork restart pathways
p53 has also been uncovered as a key regulator in the DNA restart network. p53 recruits MRE11 nuclease that is involved in replication fork restart. On the other hand, p53 null and gain-of-function mutations were shown to exhibit defects in the restart of stalled or damaged DNA replication forks. Different p53 mutations have varying effects on replication fork stalling. For instance, p53 R172P MEFs shows a moderate increase in fork stalling when compared to p53 R172H (Roy et al., 2018). This function of p53 in replication fork restart was described to contribute to suppression of cellular sensitivity to replication stress and also showed that p53 binds directly to the ongoing and stalled DNA replication fork (Roy et al., 2018).
Hampp et al. (2016) also show that p53 participates in a novel DNA damage tolerance pathway, together with the translesion polymerase ι (POLι). It seems that p53 acts in concert with POLι to cause a deceleration of the elongating fork when it encounters a replication barrier, and this is essential for the resolution of the stalled fork by structure-specific enzymes. Together with data from Klusmann et al. (2016), the evidence suggests that not only does p53 suppress replication fork stalling but also a fraction of stalled forks is resolved to enable replication to continue (Hampp et al., 2016; Klusmann et al., 2016).
p53 has also been described to confer protection to the replicating DNA by stimulating homologous recombination (HR) during S phase in order to overcome replication fork stalling and avert fork collapse (Gatz and Wiesmuller, 2006; Ireno et al., 2014). In the absence of p53, it was demonstrated that mutagenic RAD52 and POLθ pathways hijack the stalled replication forks, resulting in increased replication associated genomic instability that was reflected in the mutational signatures of breast cancer patients (Roy et al., 2018).
Another translesion polymerase, DNA polymerase η, is required for continuous replication in the presence of photoproducts (Ohmori et al., 2001; Laposa et al., 2007). Xeroderma pigmentosum variant (XP-V) cells are known to lack DNA polymerase η and are therefore hypermutable following exposure to UV (Waters et al., 1993). In these cells, deficiency in p53 alters the response to UV-induced DNA damage resulting in the accumulation of increased levels of both single-stranded and double-stranded DNA breaks. In XP-V cells containing wild-type p53, p53 induced an S phase arrest to protect cells from UV damage (Laposa et al., 2003; Sengupta and Harris, 2005). On the other hand, loss of p53 in XP-V cells sensitized cells to the cytogenetic effects of UV (Cleaver et al., 1999; Laposa et al., 2007). Crosstalks between translesion polymerases and p53 raise the interesting possibility if targeting translesion polymerases may further compromise genomic instability in p53-deficient cells and attenuate cell survival.
Targeting PI3K-like protein kinases
The combined loss of p53 and ATM is synthetically lethal in DNA damaging chemotherapy, mainly as a result of a deficiency in DNA damage-induced cell checkpoints in the p53 mutant cells. In the absence of p53, suppressing ATM sensitizes tumors to DNA damaging drugs; in p53-deficient H-rasV12 MEFs, knockdown of ATM by using shRNA impaired growth and increased apoptosis when cells were treated with cisplatin or doxorubicin (Tribius et al., 2001; Jiang et al., 2009). However, interestingly, in the presence of wild-type p53, suppression of ATM conversely protects cells from the effects of genotoxic drugs, implying that p53 status is critical for determining the outcome of ATM inhibition, in promoting or compromising the effectiveness of genotoxic chemotherapy. Therefore, evaluating the status of both p53 and ATM is beneficial in predicting the clinical response to a number of genotoxic chemotherapies.
DNA-dependent protein kinase catalytic subunit (DNA-PKcs) is one of the major components of the nonhomologous end joining (NHEJ) pathway (Weterings and Chen, 2007). It is important for V(D)J recombination during the development of T and B cell lymphocyte and therefore in mammals, DNA-PKcs deficiency causes severe combined immunodeficiency (SCID) (Blunt et al., 1996; Kurimasa et al., 1999). DNA-PKcs have been shown to affect chemosensitivity in p53-deficient cells. Loss of DNA-PKcs in p53-deficient cells increases the sensitivity of cells to doxorubicin and epirubicin. This enhanced sensitization to doxorubicin was related to G2/M checkpoint activation. Also, in p53-deficient cells, the concomitant loss of DNA-PKcs enabled cells to overcome anthracycline-resistance (Fedier et al., 2003), suggesting that inhibiting DNA-PKcs may be another viable therapeutic strategy against p53-deficient cells.
Tipping the balance between HR and NHEJ
HR and NHEJ pathways contribute towards double-stranded break repair. The two pathways are temporally distinct and are regulated by separate set of proteins; HR mostly occurs in late S and G2 phases of the cell cycle, in contrast to NHEJ, which is functional throughout the cell cycle. HR is an error-free repair process and its activity requires homologous sister chromatids, homologous chromosomes, or DNA repeats (Takata et al., 1998). p53 appears to help in maintaining genomic stability by regulating HR activity. Inactivation of p53 results in increased spontaneous and induced HR, and DNA damage-induced sister chromatid exchanges. In S phase, p53 has been shown to be associated with HR factors such as BLM, Rad52, and BRCA1/2 (Zink et al., 2002; Sengupta et al., 2003). Furthermore, it has been suggested that p53 may regulate genomic instability by preventing aberrant HR in a transactivation-independent manner (Saintigny et al., 1999; Romanova et al., 2004), and in part through its interaction with RAD51 and RAD54 (Menon and Povirk, 2014). Upon DNA damage, p53 interacts with BLM helicase at replication forks independent of its G1-S and transactivation activity in order to regulate HR (Janz and Wiesmuller, 2002; Bertrand et al., 2004). On the contrary, Rieckmann et al. (2013) find the involvement of p53 during HR to be strongly dependent on the transactivation-dependent functions of p53, primarily mediated through its cell cycle checkpoint-dependent function. Other studies however show that extrachromosomal HR is not regulated by p53 (Willers et al., 2001).
While there is evidence that p53 may regulate and prevents aberrant HR, loss of p53 also results in increased HR activity in certain context. In the absence of p53, Moureau et al. (2016) observed enhanced HR in cells treated with topoisomerase inhibitors. For instance, they show that in p53-deficient cells, there is efficient HR-mediated repair of CPT-induced DSBs (Moureau et al., 2016). In view of this, it can be speculated that inhibiting HR in this condition could sensitize p53-deficient cells. Therefore, a combined treatment of topoisomerase inhibitor and an inhibitor of HR might prove therapeutically beneficial in treating p53-deficient cancers.
Multiple studies suggest that p53 promotes NHEJ in some instances. During NHEJ, p53 blocks the annealing of single strands along flanking stretches of microhomology and therefore restricts DNA end modification (Dahm-Daphi et al., 2005). p53 has also been suggested to be involved in NHEJ as evidenced in the role played by p53 in directly enhancing the DNA end joining of short complementary ends (Tang et al., 1999). Conflicting results on the role of p53 in regulating NHEJ have been reported. While some studies suggest that p53 promotes NHEJ, others show that p53 has no effect or suppressed NHEJ (Bill et al., 1997; Yang et al., 1997; Willers et al., 2001; Dahm-Daphi et al., 2005), therefore making it unclear the exact role of p53 in NHEJ.
Interestingly, p53 is shown to promote the recruitment of 53BP1 to DNA damage sites and in turn impose limits on the accumulation of BRCA1. 53BP1 is known to inhibit DNA resection and therefore stimulate NHEJ while BRCA1 promotes DNA end resection and HR (Bunting et al., 2010; Escribano-Diaz et al., 2013). In view of this, in the absence of p53, increased BRCA1 is present at DNA damage sites to facilitate DNA repair by HR. Further support for the enhanced HR in p53-deficient cells is evidenced by the elevated percentage of RAD51 positive cells, suggesting that p53 determines pathway choice between HR and NHEJ (Moureau et al., 2016). Besides, p53 is implicated in the nuclear export of BRCA1 and regulates the nuclear-cytoplasmic distribution of BRCA1 protein in response to DNA damage. It is suggested that impairing p53 function may contribute to resistance to DNA damage by allowing nuclear retention of BRCA1 and BRCA1-repair activities, leaving one to speculate if reinstating BRCA1 subcellular redistribution may be a strategy for sensitizing p53-deficient breast cancer cells.
As more new and fascinating roles of p53 are being uncovered, it opens up new avenues to explore novel therapeutics in the context of synthetic lethality. Targeting tumor vulnerabilities imposed by the loss or mutation of p53 is an attractive option against a broad spectrum of p53 mutations and may be an ideal approach given the large heterogeneity of p53 mutations seen in the clinic. In addition to synthetic lethal approaches targeting loss-of-p53 function, some mutant p53-specific synthetic lethal strategies may further expand the increasing repertoire of these approaches for exploitation. Of priority is the need for proper stratification of tumor cohorts based on p53 functionality, as well as the identification and clustering of p53 mutations according to their predicted/validated responses against these new synthetic lethal approaches. With the evolving knowledge of the role of p53 in metabolism, it is hoped that strategies to utilize these p53 loss- or gain-of-function properties become more defined and soon achieves pre-clinical and clinical validation.
Acknowledgements
We sincerely apologize to all authors whose work is not cited here due to space constraints.
Funding
This work was supported by the Institute of Molecular and Cell Biology (IMCB), Agency of Science, Technology and Research (A*STAR), Singapore.
Conflict of interest
none declared.
References
- Altman B.J., Stine Z.E., and Dang C.V. (2016). From Krebs to clinic: glutamine metabolism to cancer therapy. Nat. Rev. Cancer 16, 749. [DOI] [PubMed] [Google Scholar]
- Ambs S., Ogunfusika M.O., Merriam W.G., et al. (1998). Up-regulation of inducible nitric oxide synthase expression in cancer-prone p53 knockout mice. Proc. Natl Acad. Sci. USA 95, 8823–8828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An S., Kumar R., Sheets E.D., et al. (2008). Reversible compartmentalization of de novo purine biosynthetic complexes in living cells. Science 320, 103–106. [DOI] [PubMed] [Google Scholar]
- Assaily W., Rubinger D.A., Wheaton K., et al. (2011). ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol. Cell 44, 491–501. [DOI] [PubMed] [Google Scholar]
- Bensaad K., Cheung E.C., and Vousden K.H. (2009). Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 28, 3015–3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensaad K., Tsuruta A., Selak M.A., et al. (2006). TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell 126, 107–120. [DOI] [PubMed] [Google Scholar]
- Bertrand P., Saintigny Y., and Lopez B.S. (2004). p53’s double life: transactivation-independent repression of homologous recombination. Trends Genet. 20, 235–243. [DOI] [PubMed] [Google Scholar]
- Bieging K.T., Mello S.S., and Attardi L.D. (2014). Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill C.A., Yu Y., Miselis N.R., et al. (1997). A role for p53 in DNA end rejoining by human cell extracts. Mutat. Res. 385, 21–29. [DOI] [PubMed] [Google Scholar]
- Blunt T., Gell D., Fox M., et al. (1996). Identification of a nonsense mutation in the carboxyl-terminal region of DNA-dependent protein kinase catalytic subunit in the scid mouse. Proc. Natl Acad. Sci. USA 93, 10285–10290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov A.V., and Karin M. (2008). p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134, 451–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budanov A.V., Shoshani T., Faerman A., et al. (2002). Identification of a novel stress-responsive gene Hi95 involved in regulation of cell viability. Oncogene 21, 6017–6031. [DOI] [PubMed] [Google Scholar]
- Bunting S.F., Callen E., Wong N., et al. (2010). 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A., Chia K.M., Haupt S., et al. (2016). Clinical overview of MDM2/X-targeted therapies. Front. Oncol. 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzai M., Jones R.G., Amaravadi R.K., et al. (2007). Systemic treatment with the antidiabetic drug metformin selectively impairs p53-deficient tumor cell growth. Cancer Res. 67, 6745–6752. [DOI] [PubMed] [Google Scholar]
- Cano C.E., Gommeaux J., Pietri S., et al. (2009). Tumor protein 53-induced nuclear protein 1 is a major mediator of p53 antioxidant function. Cancer Res. 69, 219–226. [DOI] [PubMed] [Google Scholar]
- Cheok C.F., and Lane D.P. (2012). Seeking synergy in p53 transcriptional activation for cancer therapy. Discov. Med. 14, 263–271. [PubMed] [Google Scholar]
- Cheok C.F., and Lane D.P. (2017). Exploiting the p53 pathway for therapy. Cold Spring Harb. Perspect. Med. 7, pii: a026310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleaver J.E., Afzal V., Feeney L., et al. (1999). Increased ultraviolet sensitivity and chromosomal instability related to P53 function in the xeroderma pigmentosum variant. Cancer Res. 59, 1102–1108. [PubMed] [Google Scholar]
- Contractor T., and Harris C.R. (2012). p53 negatively regulates transcription of the pyruvate dehydrogenase kinase Pdk2. Cancer Res. 72, 560–567. [DOI] [PubMed] [Google Scholar]
- Couch F.B., Bansbach C.E., Driscoll R., et al. (2013). ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev. 27, 1610–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crighton D., Wilkinson S., and Ryan K.M. (2007). DRAM links autophagy to p53 and programmed cell death. Autophagy 3, 72–74. [DOI] [PubMed] [Google Scholar]
- Dahm-Daphi J., Hubbe P., Horvath F., et al. (2005). Nonhomologous end-joining of site-specific but not of radiation-induced DNA double-strand breaks is reduced in the presence of wild-type p53. Oncogene 24, 1663–1672. [DOI] [PubMed] [Google Scholar]
- Davis S.R., Stacpoole P.W., Williamson J., et al. (2004). Tracer-derived total and folate-dependent homocysteine remethylation and synthesis rates in humans indicate that serine is the main one-carbon donor. Am. J. Physiol. Endocrinol. Metab. 286, E272–E279. [DOI] [PubMed] [Google Scholar]
- Emerling B.M., Hurov J.B., Poulogiannis G., et al. (2013). Depletion of a putatively druggable class of phosphatidylinositol kinases inhibits growth of p53-null tumors. Cell 155, 844–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escribano-Diaz C., Orthwein A., Fradet-Turcotte A., et al. (2013). A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883. [DOI] [PubMed] [Google Scholar]
- Fedier A., Moawad A., Haller U., et al. (2003). p53-deficient cells display increased sensitivity to anthracyclines after loss of the catalytic subunit of the DNA-dependent protein kinase. Int. J. Oncol. 23, 1431–1437. [DOI] [PubMed] [Google Scholar]
- Feeley K.P., Adams C.M., Mitra R., et al. (2017). Mdm2 is required for survival and growth of p53-deficient cancer cells. Cancer Res. 77, 3823–3833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z., Hu W., de Stanchina E., et al. (2007). The regulation of AMPK β1, TSC2, and PTEN expression by p53: stress, cell and tissue specificity, and the role of these gene products in modulating the IGF-1-AKT-mTOR pathways. Cancer Res. 67, 3043–3053. [DOI] [PubMed] [Google Scholar]
- Feng Z., Zhang H., Levine A.J., et al. (2005). The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl Acad. Sci. USA 102, 8204–8209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finck B.N., Gropler M.C., Chen Z., et al. (2006). Lipin 1 is an inducible amplifier of the hepatic PGC-1α/PPARα regulatory pathway. Cell Metab. 4, 199–210. [DOI] [PubMed] [Google Scholar]
- Freed-Pastor W.A., Mizuno H., Zhao X., et al. (2012). Mutant p53 disrupts mammary tissue architecture via the mevalonate pathway. Cell 148, 244–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French J.B., Jones S.A., Deng H., et al. (2016). Spatial colocalization and functional link of purinosomes with mitochondria. Science 351, 733–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto F., Schulze A., and Nielsen J. (2016). Systematic analysis reveals that cancer mutations converge on deregulated metabolism of arachidonate and xenobiotics. Cell Rep. 16, 878–895. [DOI] [PubMed] [Google Scholar]
- Gatz S.A., and Wiesmuller L. (2006). p53 in recombination and repair. Cell Death Differ. 13, 1003–1016. [DOI] [PubMed] [Google Scholar]
- Gurpinar E., and Vousden K.H. (2015). Hitting cancers’ weak spots: vulnerabilities imposed by p53 mutation. Trends Cell Biol. 25, 486–495. [DOI] [PubMed] [Google Scholar]
- Hampp S., Kiessling T., Buechle K., et al. (2016). DNA damage tolerance pathway involving DNA polymerase ι and the tumor suppressor p53 regulates DNA replication fork progression. Proc. Natl Acad. Sci. USA 113, E4311–E4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han G.S., Wu W.I., and Carman G.M. (2006). The Saccharomyces cerevisiae Lipin homolog is a Mg2+-dependent phosphatidate phosphatase enzyme. J. Biol. Chem. 281, 9210–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T., Zhou L., Elf S., et al. (2012). Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell 22, 585–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Zhang C., Wu R., et al. (2010). Glutaminase 2, a novel p53 target gene regulating energy metabolism and antioxidant function. Proc. Natl Acad. Sci. USA 107, 7455–7460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T., Brown-Endres L., Chu K., et al. (2009). GAMT, a p53-inducible modulator of apoptosis, is critical for the adaptive response to nutrient stress. Mol. Cell 36, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ireno I.C., Wiehe R.S., Stahl A.I., et al. (2014). Modulation of the poly (ADP-ribose) polymerase inhibitor response and DNA recombination in breast cancer cells by drugs affecting endogenous wild-type p53. Carcinogenesis 35, 2273–2282. [DOI] [PubMed] [Google Scholar]
- Janz C., and Wiesmuller L. (2002). Wild-type p53 inhibits replication-associated homologous recombination. Oncogene 21, 5929–5933. [DOI] [PubMed] [Google Scholar]
- Jiang P., Du W., Wang X., et al. (2011). p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat. Cell Biol. 13, 310–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D., LaGory E.L., Kenzelmann Broz D., et al. (2015). Analysis of p53 transactivation domain mutants reveals Acad11 as a metabolic target important for p53 pro-survival function. Cell Rep. 10, 1096–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Reinhardt H.C., Bartkova J., et al. (2009). The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 23, 1895–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.G., Plas D.R., Kubek S., et al. (2005). AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol. Cell 18, 283–293. [DOI] [PubMed] [Google Scholar]
- Klusmann I., Rodewald S., Muller L., et al. (2016). p53 activity results in DNA replication fork processivity. Cell Rep. 17, 1845–1857. [DOI] [PubMed] [Google Scholar]
- Klusmann I., Wohlberedt K., Magerhans A., et al. (2018). Chromatin modifiers Mdm2 and RNF2 prevent RNA:DNA hybrids that impair DNA replication. Proc. Natl Acad. Sci. USA 115, E11311–E11320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollareddy M., Dimitrova E., Vallabhaneni K.C., et al. (2015). Regulation of nucleotide metabolism by mutant p53 contributes to its gain-of-function activities. Nat. Commun. 6, 7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R., Coronel L., Somalanka B., et al. (2018). Mitochondrial uncoupling reveals a novel therapeutic opportunity for p53-defective cancers. Nat. Commun. 9, 3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurimasa A., Ouyang H., Dong L.J., et al. (1999). Catalytic subunit of DNA-dependent protein kinase: impact on lymphocyte development and tumorigenesis. Proc. Natl Acad. Sci. USA 96, 1403–1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok M., Davies N., Agathanggelou A., et al. (2016). ATR inhibition induces synthetic lethality and overcomes chemoresistance in TP53- or ATM-defective chronic lymphocytic leukemia cells. Blood 127, 582–595. [DOI] [PubMed] [Google Scholar]
- Labuschagne C.F., van den Broek N.J., Mackay G.M., et al. (2014). Serine, but not glycine, supports one-carbon metabolism and proliferation of cancer cells. Cell Rep. 7, 1248–1258. [DOI] [PubMed] [Google Scholar]
- Laposa R.R., Feeney L., and Cleaver J.E. (2003). Recapitulation of the cellular xeroderma pigmentosum-variant phenotypes using short interfering RNA for DNA polymerase H. Cancer Res. 63, 3909–3912. [PubMed] [Google Scholar]
- Laposa R.R., Feeney L., Crowley E., et al. (2007). p53 suppression overwhelms DNA polymerase η deficiency in determining the cellular UV DNA damage response. DNA Repair 6, 1794–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Budanov A.V., Park E.J., et al. (2010). Sestrin as a feedback inhibitor of TOR that prevents age-related pathologies. Science 327, 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy J.M.M., Towers C.G., and Thorburn A. (2017). Targeting autophagy in cancer. Nat. Rev. Cancer 17, 528–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Kon N., Jiang L., et al. (2012). Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wu J.B., Chung L.W., et al. (2015). Anti-cancer efficacy of SREBP inhibitor, alone or in combination with docetaxel, in prostate cancer harboring p53 mutations. Oncotarget 6, 41018–41032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y.L., and Pasero P. (2012). Interference between DNA replication and transcription as a cause of genomic instability. Curr. Genomics 13, 65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locasale J.W. (2013). Serine, glycine and one-carbon units: cancer metabolism in full circle. Nat. Rev. Cancer 13, 572–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddocks O.D.K., Athineos D., Cheung E.C., et al. (2017). Modulating the therapeutic response of tumours to dietary serine and glycine starvation. Nature 544, 372–376. [DOI] [PubMed] [Google Scholar]
- Maddocks O.D., Berkers C.R., Mason S.M., et al. (2013). Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature 493, 542–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathupala S.P., Heese C., and Pedersen P.L. (1997). Glucose catabolism in cancer cells. The type II hexokinase promoter contains functionally active response elements for the tumor suppressor p53. J. Biol. Chem. 272, 22776–22780. [DOI] [PubMed] [Google Scholar]
- Mattaini K.R., Sullivan M.R., and Vander Heiden M.G. (2016). The importance of serine metabolism in cancer. J. Cell Biol. 214, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maya-Mendoza A., Moudry P., Merchut-Maya J.M., et al. (2018). High speed of fork progression induces DNA replication stress and genomic instability. Nature 559, 279–284. [DOI] [PubMed] [Google Scholar]
- Menon V., and Povirk L. (2014). Involvement of p53 in the repair of DNA double strand breaks: multifaceted Roles of p53 in homologous recombination repair (HRR) and non-homologous end joining (NHEJ). Subcell. Biochem. 85, 321–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moureau S., Luessing J., Harte E.C., et al. (2016). A role for the p53 tumour suppressor in regulating the balance between homologous recombination and non-homologous end joining. Open Biol. 6, pii: 160225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieminuszczy J., Schwab R.A., and Niedzwiedz W. (2016). The DNA fibre technique—tracking helicases at work. Methods 108, 92–98. [DOI] [PubMed] [Google Scholar]
- Ohmori H., Friedberg E.C., Fuchs R.P., et al. (2001). The Y-family of DNA polymerases. Mol. Cell 8, 7–8. [DOI] [PubMed] [Google Scholar]
- Okamura S., Arakawa H., Tanaka T., et al. (2001). p53DINP1, a p53-inducible gene, regulates p53-dependent apoptosis. Mol. Cell 8, 85–94. [DOI] [PubMed] [Google Scholar]
- Peeters H., Debeer P., Bairoch A., et al. (2003). PA26 is a candidate gene for heterotaxia in humans: identification of a novel PA26-related gene family in human and mouse. Hum. Genet. 112, 573–580. [DOI] [PubMed] [Google Scholar]
- Pommier Y. (2013). Drugging topoisomerases: lessons and challenges. ACS Chem. Biol. 8, 82–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieckmann T., Kriegs M., Nitsch L., et al. (2013). p53 modulates homologous recombination at I-SceI-induced double-strand breaks through cell-cycle regulation. Oncogene 32, 968–975. [DOI] [PubMed] [Google Scholar]
- Romanova L.Y., Willers H., Blagosklonny M.V., et al. (2004). The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene 23, 9025–9033. [DOI] [PubMed] [Google Scholar]
- Ros S., Floter J., Kaymak I., et al. (2017). 6-Phosphofructo-2-kinase/fructose-2,6-biphosphatase 4 is essential for p53-null cancer cells. Oncogene 36, 3287–3299. [DOI] [PubMed] [Google Scholar]
- Ros S., and Schulze A. (2013). Balancing glycolytic flux: the role of 6-phosphofructo-2-kinase/fructose 2,6-bisphosphatases in cancer metabolism. Cancer Metab. 1, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Tomaszowski K.H., Luzwick J.W., et al. (2018). p53 orchestrates DNA replication restart homeostasis by suppressing mutagenic RAD52 and POLθ pathways. eLife 7, pii: e31723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y., Schoppy D.W., Asare A., et al. (2009). Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat. Genet. 41, 1144–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y., Rouillard D., Chaput B., et al. (1999). Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene 18, 3553–3563. [DOI] [PubMed] [Google Scholar]
- Sanchez-Macedo N., Feng J., Faubert B., et al. (2013). Depletion of the novel p53-target gene carnitine palmitoyltransferase 1C delays tumor growth in the neurofibromatosis type I tumor model. Cell Death Differ. 20, 659–668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S., and Harris C.C. (2005). p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 6, 44–55. [DOI] [PubMed] [Google Scholar]
- Sengupta S., Linke S.P., Pedeux R., et al. (2003). BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 22, 1210–1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shangary S., and Wang S. (2009). Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu. Rev. Pharmacol. Toxicol. 49, 223–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiloh Y., and Ziv Y. (2013). The ATM protein kinase: regulating the cellular response to genotoxic stress, and more. Nat. Rev. Mol. Cell Biol. 14, 197–210. [PubMed] [Google Scholar]
- Sutendra G., and Michelakis E.D. (2013). Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front. Oncol. 3, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki S., Tanaka T., Poyurovsky M.V., et al. (2010). Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proc. Natl Acad. Sci. USA 107, 7461–7466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajan M., Hock A.K., Blagih J., et al. (2018). A Role for p53 in the Adaptation to Glutamine Starvation through the Expression of SLC1A3. Cell Metab. 28, 721–736.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata M., Sasaki M.S., Sonoda E., et al. (1998). Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 17, 5497–5508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W., Willers H., and Powell S.N. (1999). p53 directly enhances rejoining of DNA double-strand breaks with cohesive ends in γ-irradiated mouse fibroblasts. Cancer Res. 59, 2562–2565. [PubMed] [Google Scholar]
- Toledo L.I., Altmeyer M., Rask M.B., et al. (2013). ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103. [DOI] [PubMed] [Google Scholar]
- Tomasini R., Samir A.A., Pebusque M.J., et al. (2002). P53-dependent expression of the stress-induced protein (SIP). Eur. J. Cell Biol. 81, 294–301. [DOI] [PubMed] [Google Scholar]
- Tribius S., Pidel A., and Casper D. (2001). ATM protein expression correlates with radioresistance in primary glioblastoma cells in culture. Int. J. Radiat. Oncol. Biol. Phys. 50, 511–523. [DOI] [PubMed] [Google Scholar]
- Vakifahmetoglu-Norberg H., Kim M., Xia H.G., et al. (2013). Chaperone-mediated autophagy degrades mutant p53. Genes Dev. 27, 1718–1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco-Miguel S., Buckbinder L., Jean P., et al. (1999). PA26, a novel target of the p53 tumor suppressor and member of the GADD family of DNA damage and growth arrest inducible genes. Oncogene 18, 127–137. [DOI] [PubMed] [Google Scholar]
- Wang L., Xiong H., Wu F., et al. (2014). Hexokinase 2-mediated Warburg effect is required for PTEN- and p53-deficiency-driven prostate cancer growth. Cell Rep. 8, 1461–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Zhao Y., Aguilar A., et al. (2017). Targeting the MDM2-p53 protein-protein interaction for new cancer therapy: progress and challenges. Cold Spring Harb. Perspect. Med. 7, pii: a026245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters H.L., Seetharam S., Seidman M.M., et al. (1993). Ultraviolet hypermutability of a shuttle vector propagated in xeroderma pigmentosum variant cells. J. Invest. Dermatol. 101, 744–748. [DOI] [PubMed] [Google Scholar]
- Weterings E., and Chen D.J. (2007). DNA-dependent protein kinase in nonhomologous end joining: a lock with multiple keys? J. Cell Biol. 179, 183–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willers H., McCarthy E.E., Hubbe P., et al. (2001). Homologous recombination in extrachromosomal plasmid substrates is not suppressed by p53. Carcinogenesis 22, 1757–1763. [DOI] [PubMed] [Google Scholar]
- Xiang Y., Stine Z.E., Xia J., et al. (2015). Targeted inhibition of tumor-specific glutaminase diminishes cell-autonomous tumorigenesis. J. Clin. Invest. 125, 2293–2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahagi N., Shimano H., Matsuzaka T., et al. (2003). p53 activation in adipocytes of obese mice. J. Biol. Chem. 278, 25395–25400. [DOI] [PubMed] [Google Scholar]
- Yang T., Namba H., Hara T., et al. (1997). p53 induced by ionizing radiation mediates DNA end-jointing activity, but not apoptosis of thyroid cells. Oncogene 14, 1511–1519. [DOI] [PubMed] [Google Scholar]
- Yang M., and Vousden K.H. (2016). Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer 16, 650–662. [DOI] [PubMed] [Google Scholar]
- Yeo C.Q.X., Alexander I., Lin Z., et al. (2016). p53 maintains genomic stability by preventing interference between transcription and replication. Cell Rep. 15, 132–146. [DOI] [PubMed] [Google Scholar]
- Yoon K.A., Nakamura Y., and Arakawa H. (2004). Identification of ALDH4 as a p53-inducible gene and its protective role in cellular stresses. J. Hum. Genet. 49, 134–140. [DOI] [PubMed] [Google Scholar]
- Zeman M.K., and Cimprich K.A. (2014). Causes and consequences of replication stress. Nat. Cell Biol. 16, 2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou G., Wang J., Zhao M., et al. (2014). Gain-of-function mutant p53 promotes cell growth and cancer cell metabolism via inhibition of AMPK activation. Mol. Cell 54, 960–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zink D., Mayr C., Janz C., et al. (2002). Association of p53 and MSH2 with recombinative repair complexes during S phase. Oncogene 21, 4788–4800. [DOI] [PubMed] [Google Scholar]