Abstract
Activation of the tumour suppressor p53 upon cellular stress can induce a number of different cellular processes. The diverse actions of these processes are critical for the protective function of p53 in preventing the development of cancer. However, it is still not fully understood which process(es) activated by p53 is/are critical for tumour suppression and how this might differ depending on the type of cells undergoing neoplastic transformation and the nature of the drivers of oncogenesis. Moreover, it is not clear why upon activation of p53 some cells undergo cell cycle arrest and senescence whereas others die by apoptosis. Here we discuss some of the cellular processes that are crucial for p53-mediated tumour suppression and the factors that could impact cell fate upon p53 activation. Finally, we describe therapies aimed either at activating wild-type p53 or at changing the behaviour of mutant p53 to unleash tumour growth suppressive processes for therapeutic benefit in malignant disease.
Keywords: p53, tumour suppression, cell death, cell cycle arrest/senescence
Introduction
The tumour suppressor and transcription factor p53 has been of considerable interest since its discovery in 1979 (Kress et al., 1979; Lane and Crawford, 1979; Linzer and Levine, 1979). p53 is the most frequently mutated gene in human cancer with some cancers, such as serous ovarian cancer having an incidence of nearly 100%, whereas others, such as thyroid cancers, have only a very low incidence of p53 mutations. Cancers that do not carry mutations in p53 frequently have defects in regulators of p53, such as overexpression of its negative regulators MDM2 or MDMX (Burgess et al., 2016). The importance of p53 as a tumour suppressor is also highlighted in the Li-Fraumeni familial cancer predisposition syndrome that is caused by germline mutations in one allele of p53 resulting in a ~50% risk of cancer by the age of 35 and a ~90% lifetime risk (Sagne et al., 2014).
The p53 mRNA is expressed at readily detectable levels in most (possibly all) cells but in the absence of stress p53 protein levels are very low because it is targeted for proteasomal degradation by the E3 ubiquitin ligase, MDM2 (Oren et al., 1981). The p53 protein can be activated in response to many stresses, including activation of oncogenes or DNA lesions, and this usually involves signalling pathways that converge upon inactivation of its negative regulator, MDM2, and/or post-translational modifications (PTMs) in p53 that render it resistant to MDM2 driven degradation. Upon stimulation, p53 activates transcription of a large number of target genes—the exact number of direct targets is still debated but is reported to be around 500, with indirect target genes reaching into the thousands (Allen et al., 2014; Fischer, 2017). Both the direct and indirect targets of p53 are involved in the control of a broad range of cellular processes, including apoptotic cell death, cell cycle arrest, cellular senescence, and DNA repair. There is also evidence of emerging functions of p53 target genes in the regulation of cellular metabolism, ferroptotic cell death, and autophagy (Figure 1). Although there has been an explosion of research on p53 since its discovery, several major questions are yet to be answered: what causes the different outcomes—cell cycle arrest/senescence vs. apoptotic cell death—for a cell once p53 is activated; which of the cellular processes controlled by p53 are critical for its tumour suppressive function and how might they differ depending on cell type and oncogenic context; how can wild-type p53 and possibly mutant p53 be targeted for cancer therapy? Here we discuss these questions in context of current understanding of p53 function and its regulation.
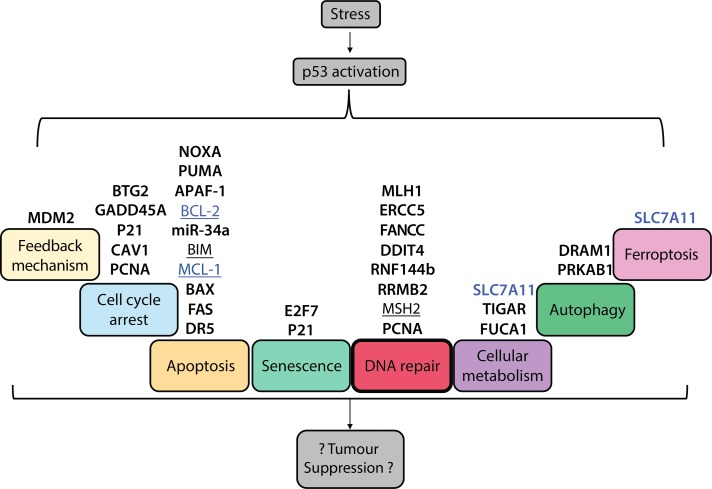
Figure 1.
Cellular processes that are impacted by p53 activation, listing some important genes that regulate these pathways. Diverse stresses activate p53 via a number of upstream signalling pathways that are not depicted here. Upon activation, p53 impacts several cellular processes through direct (bold) and/or indirect (underlined) regulation of target genes. Genes that are induced by p53 are shown in black, whereas genes that are repressed by p53 are in blue. The processes activated by p53 that are postulated to contribute to its ability to suppress tumour development include apoptosis, cell cycle arrest, cellular senescence, DNA repair, regulation of cellular metabolism, autophagy, and ferroptotic cell death.
The tumour suppressor functions of p53
Although it is now firmly established that transcriptional activation is essential for p53-mediated suppression of tumour development in diverse settings (Brady et al., 2011; Jiang et al., 2011), it remains unclear which of the cellular processes that it regulates are critical for this.
p53-induced apoptosis and other cell death pathways
Induction of apoptosis is thought to be one of the most important tumour suppressor functions exerted by p53. Indeed, some have argued that it is the only process essential for p53-mediated tumour suppression. p53 can transcriptionally activate several genes that function in either the BCL-2-regulated (also called intrinsic, stress induced, mitochondrial) or the death receptor induced (also called extrinsic) apoptotic pathways (Figure 2). Within the BCL-2-regulated pathway, p53 can directly induce the expression of the genes encoding the pro-apoptotic BH3-only proteins PUMA and NOXA, the apoptosis effector, BAX, and the caspase-9 activator, APAF-1 (Oda et al., 2000; Moroni et al., 2001; Nakano and Vousden, 2001; Robles et al., 2001; Yu et al., 2001; Chipuk et al., 2004). The BCL-2-regulated apoptotic pathway is induced when the levels of pro-apoptotic BH3-only proteins, such as PUMA and NOXA, are increased (via transcriptional or post-transcriptional processes). The BH3-only proteins can then activate the cell death effectors BAX and BAK, either directly or indirectly by neutralizing the pro-survival BCL-2 proteins (e.g. BCL-2, MCL-1) that restrain BAX and BAK in healthy cells (Czabotar et al., 2014). Activated BAX/BAK cause mitochondrial outer membrane permeabilization (MOMP) which unleashes APAF-1-assisted activation of the initiator caspase, caspase-9, and then the effector caspases that cause cell demolition (Kalkavan and Green, 2018). Thus, p53 can transcriptionally increase the expression of constituents functioning at various levels of the BCL-2-regulated apoptotic pathway. Experiments using knock-out mice have shown that the transcriptional induction of Puma and Noxa accounts, in at least certain cell types, for all p53-mediated apoptosis (Jeffers et al., 2003; Shibue et al., 2003; Villunger et al., 2003; Erlacher et al., 2005; Michalak et al., 2008). Conversely, BAX and APAF-1 can exert their functions in apoptosis even without their upregulation by p53, for example when cells are treated with glucocorticoids, anti-cancer drugs that trigger apoptosis in a p53-independent manner (Strasser et al., 1994). Thus, the p53-mediated upregulation of BAX and APAF-1 presumably serves to augment the efficiency of cell killing rather than strictly determining cell survival vs. death.
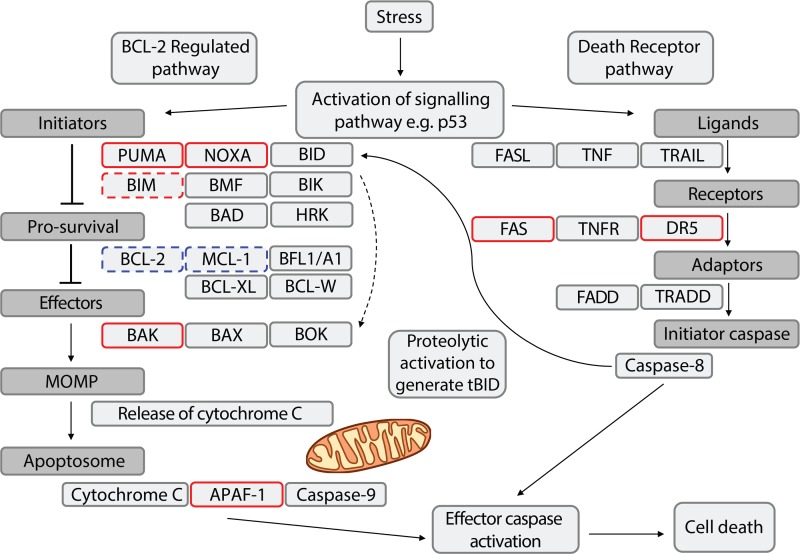
Figure 2.
p53 regulation of the apoptotic cell death pathways. Cell death regulators encoded by direct p53 target genes and transcriptionally induced are outlined in a solid red box; those encoded by indirect p53 target genes and induced are outlined in a dashed red box; those encoded by indirect p53 target genes and repressed are outlined in a dashed blue box; other cell death regulators (not controlled by p53) are outlined in grey.
Although originally considered to be a direct transcriptional target of p53, the pro-apoptotic BH3-only protein BIM is now thought to be indirectly regulated by p53. This is supported by the finding that the increases in Bim mRNA and BIM protein levels after p53 activation occur substantially later compared to the upregulation of direct targets of p53, such as Puma and Noxa (Happo et al., 2010; Valente et al., 2016). Of note, p53 activation can indirectly cause a decrease in the pro-survival proteins BCL-2 and MCL-1, the former through transcriptional induction of miR-34 (Bommer et al., 2007), and the latter through a still largely unknown mechanism (Pietrzak and Puzianowska-Kuznicka, 2008).
The death receptor induced apoptosis pathway is initiated by the stimulation of FAS, TNF-R1 or TRAIL receptors (e.g. DR5) on the cell surface by their ligands (i.e. FAS ligand, TNF, TRAIL). This triggers the cell demolishing caspase cascade directly via FADD-mediated activation of the initiator caspase, caspase-8, and indirectly through an amplification loop that engages the BCL-2-regulated apoptotic pathway via caspase-8 mediated proteolytic activation of the BH3-only protein BID (Strasser et al., 2009). The genes encoding the death receptors FAS and DR5 are both direct transcriptional targets of p53 and their expression is increased in several cell types upon p53 activation (Friesen et al., 1996; Muller et al., 1998; Burns et al., 2001; Fei et al., 2002). Studies using a panel of transgenic and knockout mice demonstrated that only the BCL-2-regulated but not the death receptor apoptotic pathway is needed for p53 to induce cell death (Newton and Strasser, 2000). It has therefore been postulated that the p53-driven upregulation of FAS and DR5 may serve to sensitize stressed cells (e.g. ones that have sustained DNA damage) in vivo to their ligands, FAS ligand or TRAIL, respectively (Strasser et al., 2009). These ligands are expressed on cytotoxic T cells that do play a role in eliminating nascent neoplastic cells. So, the p53-mediated upregulation of FAS and DR5 may thereby contribute to preventing tumour development (Hamai et al., 2010; Liu, 2010).
Intuitively, the killing of cells undergoing neoplastic transformation appears a highly logical process for p53 to suppress tumour development—but is this actually the case? In contrast to predictions, the combined loss of PUMA and NOXA, eliminating p53-induced apoptosis, even in combination with additional loss of p21, preventing p53-induced G1/S boundary cell cycle arrest and much (albeit not all) of p53-induced cell senescence does not cause spontaneous tumour development in mice (Brady et al., 2011; Li et al., 2012; Valente et al., 2013). Loss of PUMA and even more so combined loss of PUMA and NOXA, substantially accelerate MYC-driven lymphoma development. However, this acceleration is considerably less impressive than that afforded by loss of even a single allele of p53 (Michalak et al., 2009; Valente et al., 2015). Collectively, these results indicate that p53 must suppress tumour development, at least in part, through transcriptional activation of processes in addition to the induction of apoptosis, G1/S boundary cell cycle arrest, and cell senescence. Recent genetic screens have identified a small number of p53 target genes whose loss can cooperate with loss of p53-driven cell cycle arrest and apoptosis to drive tumorigenesis. This includes several genes that function in a variety of DNA repair pathways as well as Zmat3, whose function is not known (Brady et al., 2011; Aubrey et al., 2018; Janic et al., 2018). Moreover, it was reported that induction of ferroptosis through transcriptional repression of SLC7A11, a component of the cystine/glutamate antiporter is critical for p53-mediated tumour suppression (Jiang et al., 2015). This has, however, been questioned by a recent report that p53 can actually repress ferroptosis in cancer cells under conditions of metabolic stress (Tarangelo et al., 2018).
Taking all these observations into consideration, we propose that the coordination of various DNA repair processes by p53 appears the most critical process by which it suppresses tumour development.
p53-induced cell cycle arrest and cellular senescence
The ability to induce cell cycle arrest and cellular senescence is also regarded as an important tumour suppressor function of p53. Replicative senescence (RS) was first described when it was found that most (perhaps all) primary cells can only divide for a finite number of times before they change morphology and undergo cell cycle arrest in the G0/G1 phase (Hayflick and Moorhead, 1961). This has been attributed to several factors, but mainly the shortening of telomeres with the consequent increase in DNA lesions (Kuilman et al., 2010). In at least some cells, senescence can be induced in response to certain stressors, including DNA damage from ionizing radiation or chemotherapeutic drugs, enforced expression of certain oncogenes (e.g. mutant Ras) or reactive oxygen species (ROS) (Campisi and d’Adda di Fagagna, 2007). p53 induces cellular senescence and G1/S boundary cell cycle arrest largely (albeit not exclusively) through direct transcriptional induction of p21, an inhibitor of cyclin-dependent kinases (CDKs) (Fischer, 2017). It was reported that p21 can also inhibit cell cycling indirectly by affecting the so-called DREAM complex, which represses the expression of up to 250 cell cycle-related genes (reviewed by Engeland, 2018).
Additional regulators of cell cycling that are directly transcriptionally controlled by p53 include B-cell translocation gene 2 (BTG2) and growth arrest and DNA damage inducible 45α (GADD45A) (Fischer, 2017). BTG2 activates general RNA decay and may arrest proliferation by causing a widespread reduction in proteins essential for cell cycle progression (and certain other cellular processes) (Rouault et al., 1996; Duriez et al., 2002). GADD45A is thought to mediate cell proliferation arrest by interacting with p21, PCNA, CDC2/cyclinB1 or p38 kinase (Kastan et al., 1992; Wang et al., 1999).
Although p21 expression can induce both cell cycle arrest and cellular senescence, there are differences between these two cell fates. Most importantly, cell cycle arrest is regarded as temporary and reversible, i.e. cells can re-enter the cell cycle to divide further once the stimulus for cell cycle arrest is taken away and/or damages repaired. In contrast, cellular senescence is widely regarded to be irreversible. Senescent cells can usually be identified by changes in morphology, such as doubling in volume, becoming flatter and exhibiting signs of DNA damage (Campisi, 2013). The most frequently used assays (e.g. immune-histochemistry or flow cytometry-based) for identifying senescent cells measure the levels of senescence-associated β-galactosidase (SA-β-gal), which is present at a low level in all cells but is substantially increased in senescent cells (Dimri et al., 1995). Recently developed flow cytometry-based assays test for other markers of cellular senescence, such as DEP1 and B2MG (Althubiti et al., 2014), but they have not yet become standard practice. Another major difference between reversible cell cycle arrest and cellular senescence is the induction of a senescence-associated secretory phenotype (SASP) in the latter. This involves the secretion of several cytokines (e.g. IL-6, IL-1, CSFs), chemokines (CXCL/CCL), growth factors (insulin-like), and proteases (MMPs, serine proteases) (Coppe et al., 2006; Campisi and d’Adda di Fagagna, 2007). Interestingly, a recent single cell gene expression analysis revealed that although p21 was induced in all senescent cells, the SASP was limited to a small number of cells within the senescent cell population (Wiley et al., 2017). This suggests that the SASP may drive senescence in non-SASP exhibiting neighbouring cells. The SASP associated release of cytokines and chemokines can cause inflammation and attract immune cells, which may eliminate potentially harmful cells. Cellular senescence has been identified as a critical contributor to p53-mediated tumour suppression in mutant N-Ras driven lung cancer (Feldser et al., 2010) and in liver cancer (Lujambio et al., 2013).
Of note, there is also evidence that under certain circumstances the SASP is able to promote cell motility, invasion, migration and thereby drive metastasis (Cahu et al., 2012). These functions are all regarded as hallmarks of cancer and contrary to tumour suppression (Hanahan and Weinberg, 2011). Thus, cellular senescence can be considered a double-edged sword, and in terms of cancer therapy, driving malignant cells into senescence may not always be the best strategy.
Possible reasons for differing cell fates—cell cycle arrest/cellular senescence vs. apoptosis—after p53 activation
A major question that still needs answering is what causes the different outcomes—cell cycle arrest/cellular senescence vs. apoptotic cell death—upon p53 activation. A number of theories exist to explain these differences.
Cell type
Several groups have postulated that the fate of a cell upon p53 activation is controlled by the cell type, or in the case of cancer cells perhaps the cell of origin. Many haematopoietic cells, particularly lymphoid (both non-transformed as well as malignant) ones, usually undergo apoptosis upon p53 activation, whereas epithelial cells and fibroblasts generally undergo cell cycle arrest and senescence (Seluanov et al., 2001; Kojima et al., 2005; Stuhmer et al., 2005; Secchiero et al., 2006; Ventura et al., 2007; Shen and Maki, 2011). Of note, it has been reported that with respect to the induction of cellular senescence there is substantial heterogeneity at the population level (Wiley et al., 2017). Although not specifically examined, this may also be the case for apoptosis, as studies that used cell lines showed a range of cell death responses, often not reaching 100% cell killing (Tovar et al., 2006; Drakos et al., 2009, 2011; Manfe et al., 2012). An interesting question arising from this is: can a cell choose its fate if it is already pre-determined by type, and what factors can affect this decision; i.e. are cell cycle arrest/cellular senescence vs. apoptotic cell death mutually exclusive? There is evidence that this is not the case, at least in MYC-driven lymphoma cells. Upon p53 activation, for example after treatment with DNA damage inducing drugs (e.g. cyclophosphamide) or the MDM2 inhibitor nutlin-3a, these cells rapidly undergo apoptosis that relies on the pro-apoptotic BH3-only proteins PUMA, NOXA, and BIM. Interestingly, when these cells are protected from apoptosis, due to overexpression of pro-survival BCL-2 or loss of pro-apoptotic PUMA, NOXA, and BIM, they will undergo p21-dependent cell cycle arrest (Happo et al., 2010; Valente et al., 2016). This reveals that in at least this setting cell death is dominant over cell cycle arrest/cellular senescence. Conversely, some studies reported that loss of the cell cycle inhibitor p21 increases the predisposition of colon cancer-derived cell lines to undergo apoptosis upon activation of p53 (Tian et al., 2000), but this was not found in other cell types (Valente et al., 2016). Thus, it remains unclear whether induction of cell cycle arrest/cellular senescence can protect from p53-induced apoptosis.
Another important question is: can a cell that has been driven by p53 activation into senescence undergo apoptosis (Childs et al., 2014)? Pertinently, it was reported that senescent fibroblasts preferentially undergo apoptosis rather than necrosis (Seluanov et al., 2001). Moreover, senescent endothelial cells were shown to be more prone to undergoing apoptosis than their non-senescent counterparts (Hampel et al., 2004). The idea that cell cycle arrest/senescence may protect cells from undergoing apoptosis led to the discovery that senescent cells upregulate BCL-XL and BCL-W and that they can be killed, both in vitro and in vivo, when they are treated with BH3 mimetic compounds (i.e. ABT-737 or ABT-263) that inhibit both of these pro-survival proteins (Yosef et al., 2016).
If cell type is the major determinant of whether cells undergo cell cycle arrest/senescence vs. apoptotic cell death upon p53 activation, what are the factors responsible? As discussed below, this could involve different PTMs on p53, signalling pathways and factors that affect p53 function that are active in certain cells but not others or epigenetic modifications that are cell type restricted.
Stimulus for the activation of p53
It has been proposed that the nature and/or magnitude of a stimulus used to activate p53 may determine the outcome for a cell—cell cycle arrest/senescence vs. apoptotic cell death (Espinosa, 2008). For example, it was reported that early passage fibroblasts underwent senescence when treated with low doses of hydrogen peroxide but died (showing activation of caspases, a hallmark of apoptosis) when treated with high doses (Chen et al., 2000). Given that doses up to 350 μM of hydrogen peroxide were used, at least some of the cell killing might have been p53-independent and even non-apoptotic. Perplexingly, these same fibroblasts only underwent cellular senescence, but not apoptosis or other forms of cell death, at both low as well as high levels of γ-irradiation and oncogenic stimuli, such as enforced expression of mutant Ras (Krtolica et al., 2001).
p53 can be activated by diverse stimuli, including γ- or UV-irradiation, genotoxic drugs (e.g. etoposide), oxidative stress, oncogenic drivers, and the MDM2 inhibitor nutlin-3a (which activates p53 without causing DNA damage). Of note, all of these cytotoxic insults induce apoptosis in lymphoid cells but cell cycle arrest/senescence in fibroblasts (Strasser et al., 1991; Valente et al., 2016). This does not support the aforementioned concept that the nature of the stimulus determines cellular outcome after p53 activation.
Interestingly, it was reported that p53 has a higher binding affinity for the cell cycle/DNA repair gene GADD45A under reducing conditions, whereas its binding to the p21 gene was not affected. Moreover, it was shown that UV radiation reduces the binding affinity of p53 for certain of its target genes. These findings may be explained by differences in PTMs of critical amino acid residues within p53 (Buzek et al., 2002; Seo et al., 2002).
Finally, it has also been reported that the activation of distinct oncogenes can trigger distinct p53-driven cellular fates. For example, in primary fibroblasts (and certain other cell types) enforced expression of c-MYC or the viral oncogene E1A causes apoptosis whereas mutant Ras promotes cellular senescence (Debbas and White, 1993; Serrano et al., 1997; Seoane et al., 2002).
Level of the p53 protein
One school of thought proposes that the levels of p53 protein will determine which cellular response will predominate. For example, it was shown that H2O2-treated apoptotic fibroblasts exhibit ~3-fold higher levels of p53 compared to those fibroblasts undergoing senescence (Chen et al., 2000). Moreover, it was found that after exposure to a low dose stimulus, the levels of p53 fluctuated in a pulsatile manner whereas after treatment with high doses of doxorubicin cells contained stable high levels of p53 (Wu et al., 2017). Of note, only cells that had accumulated a threshold amount of p53 initiated apoptosis, the others underwent cell cycle arrest. Mapping of sites with active RNA polymerase II by using GRO-seq analysis showed that higher basal levels of target gene expression affected where on the genome p53 bound after its activation (Allen et al., 2014). This finding prompted the conclusion that higher levels of p53 mRNA at steady state correlated with lower levels of target gene induction after activation (Allen et al., 2014). In conclusion of this section, it appears fair to state that it is still not clear whether the aforementioned concepts apply to diverse cell types (or only fibroblasts) and to different p53-activating stimuli.
PTMs of the p53 protein
While MDM2-mediated ubiquitination targets p53 for proteasomal degradation and hence limits its activity, both acetylation and phosphorylation have been reported to increase the activity of p53 and impact its preference for binding to certain target genes over others. Thus, these and additional PTMs, such as neddylation, have long been thought to influence cellular outcome after p53 activation. The conformational changes in p53 have been reported to increase or decrease its binding to certain sites in promoters of genes or to prevent its binding and ubiquitination by MDM2. Unlike the common sites for mutations in p53, that are generally found in its DNA binding domain, the most common locations for PTMs reside at the N-terminus, which contains two trans-activation domains, or C-terminus, which contains the tetramerization region. Modifications on specific sites have different effects and are implemented by distinct enzymes that are themselves activated in a stimulus and/or cell type specific manner (Xu, 2003; Meek and Anderson, 2009). For example, phosphorylation of Ser15 on p53 (all residues refer to human p53) was reported to promote apoptosis, whereas phosphorylation of Ser149 will prime p53 for degradation and phosphorylation of Ser315 or Ser376 will prevent induction of apoptosis by p53 (Bech-Otschir et al., 2001).
Acetylation of Lys120 by the histone acetyl-transferase KAT5/TIP60 was shown to be critical for p53 to induce apoptosis but not for cell cycle arrest (Tang et al., 2006). This was ascribed to the observation that Lys120 acetylation is critical for p53-mediation transcriptional induction of the pro-apoptotic BH3-only protein PUMA, even though it has no impact on the expression of p21 (cell cycle arrest) or MDM2 (negative feedback regulation). Interestingly, this site in p53 is mutated in certain human cancers (Petitjean et al., 2007; Mellert et al., 2011). Proper acetylation is essential for p53-mediated tumour suppression—mice with mutations of four residues that are acetylated in p53 (so-called 4KR mutant mice) spontaneously develop tumours with a high incidence (akin to p53 knockout mice), whereas mice with mutations in only three of these residues (so-called 3KR mice) are not tumour prone (Wang et al., 2016). Thus, genes that can be controlled by 3KR mutant p53 but not by 4KR mutant p53 must be critical for tumour suppression. Of note, cells from the 3KR p53 mutant mice that are not tumour prone, are defective in the expression of PUMA, NOXA, and p21. This is consistent with the observation that mice completely deficient for these p53-activated inducers of apoptosis and G1/S boundary cell cycle arrest and cell senescence are also not tumour prone (Valente et al., 2016). One important thing to keep in mind with respect to the impact of PTMs on p53 function is that distinct cell types may differ vastly in the levels of certain p53 modifying enzymes (e.g. acetyl-transferases) and this would be predicted to dictate cellular outcomes.
Interplay with other signalling pathways
Cells usually exhibit activation of several signalling pathways, not just one, and there is extensive cross-talk between different pathways (Rowland et al., 2017). It therefore appears likely that cellular outcomes after p53 activation may be substantially affected by additional signalling pathways that are active in a given cell. One could imagine that pathways that cause an increase in MCL-1 or other pro-survival BCL-2 family members or ones that suppress the expression of the pro-apoptotic BH3-only proteins could prevent apoptosis in cells that would otherwise die upon p53 activation, allowing them to instead undergo cell cycle arrest and senescence.
Of note, in malignant cells (that have wild-type p53) the outcomes of p53 activation may be impacted by mutations that activate other pathways, such as mutations that activate the MAP kinase or PI3 kinase pathways. For example, upon treatment with the MDM2 inhibitor nutlin-3a, osteosarcoma cell lines expressing wild-type p53 and carrying Mdm2 locus amplifications were found to die by apoptosis whereas those with normal Mdm2 gene copy number underwent senescence (Tovar et al., 2006). This stands in contrast to the aforementioned theory that higher levels of p53 are needed for apoptosis than for cell cycle arrest/senescence, given that cells with Mdm2 gene amplifications express abnormally high levels of MDM2 and would therefore exhibit reduced p53 levels and activation upon treatment with nutlin-3a. Accordingly, a separate study was unable to find a correlation between the levels of MDM2 and p53-induced apoptosis vs. cell senescence in haematological cancer and choriocarcinoma derived cell lines (Kitagawa et al., 2008).
p53 isoforms and p53 family members
p53 belongs to a family of transcription factors that also includes p73 and p63. At least some of the target genes of p53 can also be regulated by its two family members, but p73 and p63 can also control distinct sets of genes (Levrero et al., 2000; Danilova et al., 2008). As for p53, the levels of both p73 and p63 are increased in response to DNA damage. This occurs mainly through upregulation of E2F1, which can directly bind to the p73 promotor and increase the transcription of this gene. This is different to the regulation of p53 by E2F1, which involves an E2F1 driven increase in p14ARF, which in turn inhibits MDM2, the major negative regulator of p53 (Honda et al., 1997; Stiewe and Putzer, 2000; Zawacka-Pankau et al., 2010). Like p53, p73 and p63 can induce apoptosis and cell cycle arrest/senescence (Jost et al., 1997; Dohn et al., 2001), although this was so far shown under physiological conditions only for p63 (low dose γ-radiation of the mouse ovary) (Suh et al., 2006) but not yet for p73. Interestingly, p63 triggers apoptosis in the same way as p53, namely through transcriptional induction of Puma and Noxa (Kerr et al., 2012). p63 and p73 were reported to be critical for some p53-mediated tumour suppressive functions in certain conditions, as combined loss of p63 and p73 prevented E1A induced apoptosis that is normally driven by p53 (Flores et al., 2002). Of note, wild-type p73 was shown to bind to mutant p53 and such mixed tetramers were able to induce p21 expression and cell cycle arrest (Willis et al., 2003). This suggests that wild-type p73 can overcome the loss-of-function effect of mutations in p53 and/or impact its dominant-negative (DN) effect.
p53, p63, and p73 are all expressed as multiple isoforms due to alternative splicing and usage of multiple start sites. These isoforms are expressed in a tissue-dependent and cell stimulation affected manner. For example, p53 isoform expression varies substantially between breast cancers vs. normal breast tissue (Bourdon et al., 2005). In human-induced pluripotent stem cells, it has been shown that expression of the Δ133p53 isoform can specifically repress a subset of p53 target genes involved in cellular senescence, such as p21 and miR-34a, by displacing full-length p53 from these loci (Horikawa et al., 2017). We believe that the generation of gene-targeted mice that lack specific isoforms of p53 (as was already done for p63 and p73) is needed to unravel the complexities of how they impact cell fate, including after activation of full-length p53.
Differential impact of p53 activation on distinct cell types within a tumour
In the context of cancer, one needs to consider not only the impact of p53 activation in the malignant cells but also that in the neighbouring stromal cells (e.g. cancer-associated fibroblasts, CAFs), endothelial cells of the blood vessels that nourish the tumour and tumour infiltrating immune cells. For example, it was reported that p53-induced senescence in primary human lung fibroblasts can impact angiogenesis through the SASP associated secretion of VEGF (Coppe et al., 2006). There is also evidence that in the absence of p21 mouse embryonic fibroblasts display abnormally sustained VEGF production during hypoxia, thereby increasing angiogenesis (Farhang Ghahremani et al., 2013).
CAFs and other tumour-associated stromal cells may have a different p53 status (usually wild-type) compared to the tumour cells (often mutant p53) and due to the considerable heterogeneity detected in at least some tumours (Marusyk et al., 2012), some malignant cells may express mutant p53 but others wild-type p53. Thus, p53 activation-based anti-cancer therapies might impact distinct cell populations within a tumour in distinct ways. Tumour-associated stromal cells can themselves undergo senescence in response to stimuli that activate p53, such as treatment with certain chemotherapeutic drugs. It is conceivable that the wild-type p53-induced SASP in the stromal cells may force proliferation arrest in mutant p53 expressing malignant cells.
There is evidence that tumour cells that had undergone senescence can re-enter the cell cycle, for example after activation of the oncogene c-MYC (Xi et al., 2016). Such mutations may pre-exist before therapy in rare cancer cells or may be acquired as a consequence of genotoxic therapy. Of note, genotoxic cancer therapies preferentially impact cycling malignant cells over non-dividing cells. Thus, senescent tumour cells may be at the origin of cancer relapse after cessation of therapy, given that they will preferentially survive genotoxic drugs and due to impaired DNA repair (as a consequence of the mutations that drove their neoplastic transformation) are at risk of sustaining DNA lesions that will enhance cancer growth.
Possible implications for cancer therapy
Mutant p53 and loss of p53 in cancer
Mutations in one allele of p53 are found in ~50% of all cancers and at the time of diagnosis ~60% of those will have sustained loss of heterozygosity at the p53 locus; i.e. the malignant cells can only express mutant p53 (Liu et al., 2016; Alexandrova et al., 2017). It is important to note that the mutational landscape of p53 is rather unique for a tumour suppressor. For most other tumour suppressors, such as RB or PTEN, most if not all mutations cause a complete loss-of-function, often due to the complete absence of the protein. Conversely, for p53 most mutations are point mutations that change only one amino acid within the DNA binding domain, with so-called ‘hotspot’ mutations being more frequent than others (Freed-Pastor and Prives, 2012; Baugh et al., 2018). On the basis of the large number of mutations in p53 found in human cancers and the diversity of their functional impact, it has been postulated that single-nucleotide polymorphisms that may be fairly prevalent in the human population might have subtle but still impactful consequences for p53 function that can cause a minor but still significant increase in the risk of developing cancer (Whibley et al., 2009).
Of note, many mutant p53 proteins are considerably more stable (i.e. present at higher levels) than wild-type p53, and this is in part due to the fact that the former are unable to transcriptionally induce expression of the negative regulator, MDM2 (Midgley and Lane, 1997). Based on structure, there are two main different forms of mutant p53 proteins. In the so-called conformational mutants, the normal structure of the p53 protein is no longer maintained. These mutant p53 proteins are often degraded and the impact of these mutations is thus similar to a complete knock-out of the p53 gene although gain-of-function (GOF) effects (see below for more detailed discussion) have been reported for some conformational p53 mutant proteins (Lang et al., 2004; Olive et al., 2004). Conversely, the so-called contact mutants have a structure that is similar to that of wild-type p53 and only differ in one amino acid that contacts DNA in the regulatory sequences of target genes (Cho et al., 1994; Muller and Vousden, 2014). Both types of p53 mutants exhibit loss-of-function properties, but some of the contact mutants in p53 can also exhibit DN and/or GOF effects (Kim and Lozano, 2018). Loss-of-function usually occurs when the p53 protein is degraded, not expressed or is a conformational mutant that cannot bind to any target sequence and so all transcriptional activity is lost. The DN effect refers to the ability of some mutant p53 proteins to form mixed tetramers with wild-type p53 proteins and thereby interfere with the ability of the latter to transcriptionally activate (at least certain) target genes. This effect of course only comes into play in cells that have not yet undergone loss of heterozygosity at the p53 locus. It appears likely that this DN effect plays a crucial role during early stages of transformation in nascent neoplastic cells soon after they have acquired a mutation in one allele of p53 and still co-express wild-type and mutant p53 proteins. GOF effects refer to the ability of certain p53 mutants to exhibit de novo functions that are never exerted by wild-type p53. This is thought to involve the ability of mutant p53 to bind to other transcription factors and affect their function, thereby impacting the expression of genes that are not regulated by wild-type p53 (Kim and Lozano, 2018). The GOF effect of mutant p53 has been reported to be critical for the sustained expansion of cancers that were driven by this oncogenic lesion (Alexandrova et al., 2015). This has, however, not yet been demonstrated in an in vivo system in which mutant p53 can be inducibly removed in a tumour that arose in that particular mouse, but only in tumours grown in tissue culture or in transplant recipient mice, where anti-tumour effects of the immune system cannot be fully excluded.
Current and potential future therapies for targeting wild-type or mutant p53
Loss or mutation in p53 does not only drive the development of diverse cancers but also renders malignant cells resistant to several anti-cancer therapeutics, including γ-radiation and chemotherapeutic drugs that elicit DNA damage (e.g. cyclophosphamide, etoposide) (Hientz et al., 2017). It is, however, noteworthy that these anti-cancer agents can also kill tumour cells in a p53-independent (Strasser et al., 1994) and this accounts, at least in part, for their ability to kill large numbers of cancer cells (e.g. lung cancer) with mutant p53 in patients, although they do not provide a cure as evidenced by the high rates of relapse. It is also noteworthy that in certain settings loss of wild-type p53 can actually enhance the efficacy of therapy (Jackson et al., 2012). A significant problem of genotoxic drugs is their ability to cause mutations that can engender relapse of cancer for which a patient is being treated or development of a new unrelated cancer that may arise many years after therapy (Swift and Golsteyn, 2014; Szikriszt et al., 2016). The latter is particularly of concern in the treatment of children, adolescents or young adults with cancer. Therefore, drugs have been developed that can activate wild-type p53 in a non-genotoxic manner by inhibiting its major negative regulator, MDM2, with nutlin-3a being the frontrunner (Vassilev et al., 2004). Nutlin-3a kills haematological cancer-derived cell lines in vitro and in mice mostly through p53-mediated induction of PUMA (and possibly also the induction of some additional pro-apoptotic BH3-only proteins, such as NOXA and BIM) (Valente et al., 2016), whereas in solid cancer-derived cell lines (e.g. osteosarcoma) nutlin-3a mostly causes proliferation arrest (Vassilev et al., 2004), presumably via induction of p21. Nutlin-3a has poor bioavailability and did not proceed to clinical trials, but since then a large number of other small molecule inhibitors of MDM2 and MDMX have been generated and several are being tested in clinical trials (Burgess et al., 2016) (Table. 1). A major limitation to MDM2 inhibitor therapies is that p53 mutation is a frequently found mechanism of resistance, although in cancers with MDM2 gene amplifications this does not appear to be a major issue (Saiki et al., 2015). Another serious limitation of MDM2 inhibitors is the predicted on-target (i.e. p53- and PUMA-driven) toxicity to vital healthy cell populations, such as those of the haematopoietic and gastro-intestinal compartments (Valente et al., 2016).
Table 1.
Clinical trials targeting the p53 pathway.
| Agent | Trial ID | Disease | Combination | Phase |
|---|---|---|---|---|
| MDM2 inhibitors | ||||
| AMG-232 | NCT03217266 | Soft tissue sarcoma | Radiation therapy | Ib |
| NCT03107780 | Glioblastoma | — | 0/I | |
| NCT03041688 | AML | Decitabine | Ib | |
| NCT03031730 | R/R multiple myeloma | Carfilzomib, lenalidomide, + dexamethasone | I | |
| Idasanutlin | NCT02545283 | R/R AML | Cytarabine | III |
| EUCTR2015-002100-83-DE | FL or DLBCL | Venetoclax + obinutuzumab or rutuximab | Ib/II | |
| NCT02633059 | Relapsed multiple myeloma | Ixazomib citrate + dexamethasone | I/II | |
| NCT03566485 | ER positive breast cancer | Atezolizumab | I/II | |
| NCT03362723 | Solid tumours | — | I | |
| DS-3032b | NCT01877382 | Advanced solid tumours or lymphomas | — | I |
| NCT02579824 | R/R multiple myeloma | — | I | |
| NCT02319369 | Heamatological malignancies | — | I | |
| RO6839921 | NCT02098967 | AML | — | I |
| ISRCTN38949950 | Prostate cancer | Abiraterone | I/II | |
| CGM097 | NCT01760525 | Advanced solid tumours | — | I |
| RO5045337 | NCT01605526 | Soft tissue sarcoma | Doxorubicin | Ib |
| NCT01143740 | Liposarcomas | — | I | |
| RO5503781 | NCT01462175 | Advanced malignancies except leukaemia | — | I |
| NCT01773408 | AML | Cytarabine | I/Ib | |
| NCT02935907 | Advanced solid tumours or lymphomas | — | I | |
| Kevetrin | NCT01664000 | Advanced solid tumours | — | I |
| BI 907828 | NCT03449381 | Advanced solid tumours | — | I |
| HDM201 | NCT02343172 | Liposarcomas | LEE011 | Ib/II |
| NCT02143635 | Advanced tumours | — | I | |
| Restoring wild-type p53 | ||||
| APR-246 | NCT00900614 | Refractory haematologic or prostate cancer | — | I |
| NCT02999893 | Oesophageal cancer | — | Ib/II | |
| NCT03391050 | Melanoma | Dabrafenib | Ib/II | |
| NCT03268382 | HGSOC | Pegylated liposomal doxorubicin | II | |
| NCT03072043 | Myeloid neoplasms | Azacitidine | Ib/II | |
| NCT02098343 | HGSOC | Carboplatin combination chemotherapy | Ib/II |
R/R, relapsed/refractory; AML, acute myeloid leukaemia; FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma; HGSOC, high grade serous ovarian cancer. Sourced from the ICTRP database and clinicaltrials.gov on July 5, 2018.
Due to the high frequency of mutations in p53 in human cancer, it has long been considered a ‘holy grail’ to develop drugs that can restore wild-type p53 tumour growth repressive functions in such malignant cells (Duffy et al., 2017). One approach involves the re-introduction of wild-type p53 protein into cancer cells using viral delivery systems. Other approaches rely on the development of small molecule compounds, such as APR-246/PRIMA-1 and related quinuclidines, that were reported to restore wild-type p53-driven tumour growth suppressive functions to 13 out of 14 mutant p53 proteins tested through direct binding. Early clinical trials of APR-246, which so far have demonstrated safety in humans, have focused on breast, ovarian, oesophageal and prostate cancers, which all have high incidence of p53 mutations (Table. 1). APR-246 was shown to induce apoptosis in certain cancer-derived cell lines in a manner dependent on mutant p53 (Bykov et al., 2005; Zandi et al., 2011; Duffy et al., 2017), but there are also reports that this compound can inhibit the growth of tumour cells in a p53-independent manner (Saha et al., 2013; Sobhani et al., 2015). Should this drug show efficacy in the aforementioned clinical trials, it will be important to clearly identify the mechanism(s) by which it stops tumour expansion.
Conclusions and perspectives
p53 is a tumour suppressor that can activate diverse known cellular responses and possibly also some still under-appreciated ones. As we have discussed above, the processes that decide between fates of a cell in which p53 has been activated, such as cell growth arrest vs. cell death, are still not resolved. The impact of cell type with different additional signalling pathways being active or silent and different p53-activating stimuli that elicit distinct PTMs on p53 are being discussed as likely determinants of cell fate. However, an overarching mechanism has not been defined and this might (at least in part) be due to the complexity arising from the ability of p53 to regulate, directly or indirectly, such a large number of target genes with such diverse functions, many of them still only poorly understood (Fischer, 2017). It is also still not fully resolved which processes activated by p53 are critical for tumour suppression. In haematological cancers, coordination of DNA repair has emerged as a highly critical process for p53-mediated tumour suppression (Janic et al., 2018), but it is possible that the relative importance of distinct p53-activated processes in tumour suppression may be different in other cell types, such as those giving rise to solid cancers. Moreover, this may be impacted by the nature of the oncogenic lesions present in the cells undergoing neoplastic transformation. Finally, the targeting of wild-type or mutant p53 for cancer therapy is still in its infancy. Here, an important question is: how can malignant cells be driven into cell death rather than cellular senescence to ensure they cannot accumulate further tumour growth promoting oncogenic lesions that may cause relapse of malignant disease? Thus, the p53 field of research is still very much alive, still has many important questions to be answered and must meet the challenge to translate its findings into effective and tolerable therapies for cancer patients.
Acknowledgements
The authors thank all of their colleagues in the Molecular Genetics of Cancer Division at The Walter and Eliza Hall Institute for insightful discussions.
Funding
Work in the authors’ laboratories is supported by the National Health and Medical Research Council, Australia (Program Grant 1113133; Fellowship 1116937; Project grants 1086291 and 1143105), The Leukemia and Lymphoma Society (Specialised Center of Research, SCOR 7015-18), The Cancer Council Victoria (Venture Grant and Grants in Aid 1102104 and 1147328), The Leukaemia Foundation of Australia (Special Grant in Burkitt Lymphoma), the Victorian Cancer Agency (Mid-Career Fellowship MCRF17028), and a PhD fellowship from Melbourne University. This work was made possible through Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS.
Conflict of interest
none declared.
References
- Alexandrova E.M., Mirza S.A., Xu S., et al. (2017). p53 loss-of-heterozygosity is a necessary prerequisite for mutant p53 stabilization and gain-of-function in vivo. Cell Death Dis. 8, e2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova E.M., Yallowitz A.R., Li D., et al. (2015). Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 523, 352–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen M.A., Andrysik Z., Dengler V.L., et al. (2014). Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. eLife 3, e02200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Althubiti M., Lezina L., Carrera S., et al. (2014). Characterization of novel markers of senescence and their prognostic potential in cancer. Cell Death Dis. 5, e1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubrey B.J., Janic A., Chen Y.A., et al. (2018). Mutant TRP53 exerts a target gene selective dominant negative effect to drive tumor development. Genes Dev. 32, 1420–1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baugh E.H., Ke H., Levine A.J., et al. (2018). Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 25, 154–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir D., Kraft R., Huang X., et al. (2001). COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J. 20, 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommer G.T., Gerin I., Feng Y., et al. (2007). p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr. Biol. 17, 1298–1307. [DOI] [PubMed] [Google Scholar]
- Bourdon J.C., Fernandes K., Murray-Zmijewski F., et al. (2005). p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 19, 2122–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady C.A., Jiang D., Mello S.S., et al. (2011). Distinct p53 transcriptional programs dictate acute DNA-damage responses and tumor suppression. Cell 145, 571–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess A., Chia K.M., Haupt S., et al. (2016). Clinical overview of MDM2/X-targeted therapies. Front. Oncol. 6, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns T.F., Bernhard E.J., and El-Deiry W.S. (2001). Tissue specific expression of p53 target genes suggests a key role for KILLER/DR5 in p53-dependent apoptosis in vivo. Oncogene 20, 4601–4612. [DOI] [PubMed] [Google Scholar]
- Buzek J., Latonen L., Kurki S., et al. (2002). Redox state of tumor suppressor p53 regulates its sequence-specific DNA binding in DNA-damaged cells by cysteine 277. Nucleic Acids Res. 30, 2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bykov V.J., Zache N., Stridh H., et al. (2005). PRIMA-1(MET) synergizes with cisplatin to induce tumor cell apoptosis. Oncogene 24, 3484–3491. [DOI] [PubMed] [Google Scholar]
- Cahu J., Bustany S., and Sola B. (2012). Senescence-associated secretory phenotype favors the emergence of cancer stem-like cells. Cell Death Dis. 3, e446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. (2013). Aging, cellular senescence, and cancer. Annu. Rev. Physiol. 75, 685–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J., and d’Adda di Fagagna F. (2007). Cellular senescence: when bad things happen to good cells. Nat. Rev. Mol. Cell Biol. 8, 729–740. [DOI] [PubMed] [Google Scholar]
- Chen Q.M., Liu J., and Merrett J.B. (2000). Apoptosis or senescence-like growth arrest: influence of cell-cycle position, p53, p21 and bax in H2O2 response of normal human fibroblasts. Biochem. J. 347, 543–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs B.G., Baker D.J., Kirkland J.L., et al. (2014). Senescence and apoptosis: dueling or complementary cell fates? EMBO Rep. 15, 1139–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chipuk J.E., Kuwana T., Bouchier-Hayes L., et al. (2004). Direct activation of Bax by p53 mediates mitochondrial membrane permeabilization and apoptosis. Science 303, 1010–1014. [DOI] [PubMed] [Google Scholar]
- Cho Y., Gorina S., Jeffrey P.D., et al. (1994). Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science 265, 346–355. [DOI] [PubMed] [Google Scholar]
- Coppe J.P., Kauser K., Campisi J., et al. (2006). Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J. Biol. Chem. 281, 29568–29574. [DOI] [PubMed] [Google Scholar]
- Czabotar P.E., Lessene G., Strasser A., et al. (2014). Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 15, 49–63. [DOI] [PubMed] [Google Scholar]
- Danilova N., Sakamoto K.M., and Lin S. (2008). p53 family in development. Mech. Dev. 125, 919–931. [DOI] [PubMed] [Google Scholar]
- Debbas M., and White E. (1993). Wild-type p53 mediates apoptosis by E1A, which is inhibited by E1B. Genes Dev. 7, 546–554. [DOI] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., et al. (1995). A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl Acad. Sci. USA 92, 9363–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohn M., Zhang S., and Chen X. (2001). p63α and ΔNp63α can induce cell cycle arrest and apoptosis and differentially regulate p53 target genes. Oncogene 20, 3193–3205. [DOI] [PubMed] [Google Scholar]
- Drakos E., Atsaves V., Schlette E., et al. (2009). The therapeutic potential of p53 reactivation by nutlin-3a in ALK+ anaplastic large cell lymphoma with wild-type or mutated p53. Leukemia 23, 2290–2299. [DOI] [PubMed] [Google Scholar]
- Drakos E., Singh R.R., Rassidakis G.Z., et al. (2011). Activation of the p53 pathway by the MDM2 inhibitor nutlin-3a overcomes BCL2 overexpression in a preclinical model of diffuse large B-cell lymphoma associated with t(14;18)(q32;q21). Leukemia 25, 856–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy M.J., Synnott N.C., and Crown J. (2017). Mutant p53 as a target for cancer treatment. Eur. J. Cancer 83, 258–265. [DOI] [PubMed] [Google Scholar]
- Duriez C., Falette N., Audoynaud C., et al. (2002). The human BTG2/TIS21/PC3 gene: genomic structure, transcriptional regulation and evaluation as a candidate tumor suppressor gene. Gene 282, 207–214. [DOI] [PubMed] [Google Scholar]
- Engeland K. (2018). Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. 25, 114–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlacher M., Michalak E.M., Kelly P.N., et al. (2005). BH3-only proteins Puma and Bim are rate-limiting for γ-radiation and glucocorticoid-induced apoptosis of lymphoid cells in vivo. Blood 106, 4131–4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa J.M. (2008). Mechanisms of regulatory diversity within the p53 transcriptional network. Oncogene 27, 4013–4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farhang Ghahremani M., Goossens S., Nittner D., et al. (2013). p53 promotes VEGF expression and angiogenesis in the absence of an intact p21-Rb pathway. Cell Death Differ. 20, 888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei P., Bernhard E.J., and El-Deiry W.S. (2002). Tissue-specific induction of p53 targets in vivo. Cancer Res. 62, 7316–7327. [PubMed] [Google Scholar]
- Feldser D.M., Kostova K.K., Winslow M.M., et al. (2010). Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature 468, 572–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M. (2017). Census and evaluation of p53 target genes. Oncogene 36, 3943–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores E.R., Tsai K.Y., Crowley D., et al. (2002). p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature 416, 560–564. [DOI] [PubMed] [Google Scholar]
- Freed-Pastor W.A., and Prives C. (2012). Mutant p53: one name, many proteins. Genes Dev. 26, 1268–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen C., Herr I., Krammer P.H., et al. (1996). Involvement of the CD95 (APO-1/Fas) receptor/ligand system in drug-induced apoptosis in leukemia cells. Nat. Med. 2, 574–577. [DOI] [PubMed] [Google Scholar]
- Hamai A., Benlalam H., Meslin F., et al. (2010). Immune surveillance of human cancer: if the cytotoxic T-lymphocytes play the music, does the tumoral system call the tune? Tissue Antigens 75, 1–8. [DOI] [PubMed] [Google Scholar]
- Hampel B., Malisan F., Niederegger H., et al. (2004). Differential regulation of apoptotic cell death in senescent human cells. Exp. Gerontol. 39, 1713–1721. [DOI] [PubMed] [Google Scholar]
- Hanahan D., and Weinberg R.A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674. [DOI] [PubMed] [Google Scholar]
- Happo L., Cragg M.S., Phipson B., et al. (2010). Maximal killing of lymphoma cells by DNA-damage inducing therapy requires not only the p53 targets Puma and Noxa but also Bim. Blood 116, 5256–5267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayflick L., and Moorhead P.S. (1961). The serial cultivation of human diploid cell strains. Exp. Cell Res. 25, 585–621. [DOI] [PubMed] [Google Scholar]
- Hientz K., Mohr A., Bhakta-Guha D., et al. (2017). The role of p53 in cancer drug resistance and targeted chemotherapy. Oncotarget 8, 8921–8946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda R., Tanaka H., and Yasuda H. (1997). Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27. [DOI] [PubMed] [Google Scholar]
- Horikawa I., Park K.Y., Isogaya K., et al. (2017). Δ133p53 represses p53-inducible senescence genes and enhances the generation of human induced pluripotent stem cells. Cell Death Differ. 24, 1017–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson J.G., Pant V., Li Q., et al. (2012). p53-mediated senescence impairs the apoptotic response to chemotherapy and clinical outcome in breast cancer. Cancer Cell 21, 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janic A., Valente L.J., Wakefield M.J., et al. (2018). DNA repair processes are critical mediators of p53-dependent tumor suppression. Nat. Med. 24, 947–953. [DOI] [PubMed] [Google Scholar]
- Jeffers J.R., Parganas E., Lee Y., et al. (2003). Puma is an essential mediator of p53-dependent and -independent apoptotic pathways. Cancer Cell 4, 321–328. [DOI] [PubMed] [Google Scholar]
- Jiang D., Brady C.A., Johnson T.M., et al. (2011). Full p53 transcriptional activation potential is dispensable for tumor suppression in diverse lineages. Proc. Natl Acad. Sci. USA 108, 17123–17128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Kon N., Li T., et al. (2015). Ferroptosis as a p53-mediated activity during tumour suppression. Nature 520, 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost C.A., Marin M.C., and Kaelin W.G. Jr. (1997). p73 is a simian [correction of human] p53-related protein that can induce apoptosis. Nature 389, 191–194. [DOI] [PubMed] [Google Scholar]
- Kalkavan H., and Green D.R. (2018). MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 25, 46–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kastan M.B., Zhan Q., El-Deiry W.S., et al. (1992). A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell 71, 587–597. [DOI] [PubMed] [Google Scholar]
- Kerr J.B., Hutt K.J., Michalak E.M., et al. (2012). DNA damage-induced primordial follicle oocyte apoptosis and loss of fertility require TAp63-mediated induction of Puma and Noxa. Mol. Cell 48, 343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.P., and Lozano G. (2018). Mutant p53 partners in crime. Cell Death Differ. 25, 161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa M., Aonuma M., Lee S.H., et al. (2008). E2F-1 transcriptional activity is a critical determinant of Mdm2 antagonist-induced apoptosis in human tumor cell lines. Oncogene 27, 5303–5314. [DOI] [PubMed] [Google Scholar]
- Kojima K., Konopleva M., Samudio I.J., et al. (2005). MDM2 antagonists induce p53-dependent apoptosis in AML: implications for leukemia therapy. Blood 106, 3150–3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kress M., May E., Cassingena R., et al. (1979). Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol. 31, 472–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A., Parrinello S., Lockett S., et al. (2001). Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl Acad. Sci. USA 98, 12072–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T., Michaloglou C., Mooi W.J., et al. (2010). The essence of senescence. Genes Dev. 24, 2463–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane D.P., and Crawford L.V. (1979). T antigen is bound to a host protein in SV40-transformed cells. Nature 278, 261–263. [DOI] [PubMed] [Google Scholar]
- Lang G.A., Iwakuma T., Suh Y.A., et al. (2004). Gain of function of a p53 hot spot mutation in a mouse model of Li-Fraumeni syndrome. Cell 119, 861–872. [DOI] [PubMed] [Google Scholar]
- Levrero M., De Laurenzi V., Costanzo A., et al. (2000). The p53/p63/p73 family of transcription factors: overlapping and distinct functions. J. Cell Sci. 113, 1661–1670. [DOI] [PubMed] [Google Scholar]
- Li T., Kon N., Jiang L., et al. (2012). Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 149, 1269–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linzer D.I., and Levine A.J. (1979). Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 17, 43–52. [DOI] [PubMed] [Google Scholar]
- Liu K. (2010). Role of apoptosis resistance in immune evasion and metastasis of colorectal cancer. World J. Gastrointest. Oncol. 2, 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Chen C., Xu Z., et al. (2016). Deletions linked to TP53 loss drive cancer through p53-independent mechanisms. Nature 531, 471–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A., Akkari L., Simon J., et al. (2013). Non-cell-autonomous tumor suppression by p53. Cell 153, 449–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfe V., Biskup E., Johansen P., et al. (2012). MDM2 inhibitor nutlin-3a induces apoptosis and senescence in cutaneous T-cell lymphoma: role of p53. J. Invest. Dermatol. 132, 1487–1496. [DOI] [PubMed] [Google Scholar]
- Marusyk A., Almendro V., and Polyak K. (2012). Intra-tumour heterogeneity: a looking glass for cancer? Nat. Rev. Cancer 12, 323–334. [DOI] [PubMed] [Google Scholar]
- Meek D.W., and Anderson C.W. (2009). Posttranslational modification of p53: cooperative integrators of function. Cold Spring Harb. Perspect. Biol. 1, a000950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellert H.S., Stanek T.J., Sykes S.M., et al. (2011). Deacetylation of the DNA-binding domain regulates p53-mediated apoptosis. J. Biol. Chem. 286, 4264–4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak E.M., Jansen E.S., Happo L., et al. (2009). Puma and to a lesser extent Noxa are suppressors of Myc-induced lymphomagenesis. Cell Death Differ. 16, 684–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalak E.M., Villunger A., Adams J.M., et al. (2008). In several cell types the tumour suppressor p53 induces apoptosis largely via Puma but Noxa can contribute. Cell Death Differ. 15, 1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midgley C.A., and Lane D.P. (1997). p53 protein stability in tumour cells is not determined by mutation but is dependent on Mdm2 binding. Oncogene 15, 1179–1189. [DOI] [PubMed] [Google Scholar]
- Moroni M.C., Hickman E.S., Denchi E.L., et al. (2001). Apaf-1 is a transcriptional target for E2F and p53. Nat. Cell Biol. 3, 552–558. [DOI] [PubMed] [Google Scholar]
- Muller M., Wilder S., Bannasch D., et al. (1998). p53 activates the CD95 (APO-1/Fas) gene in response to DNA damage by anticancer drugs. J. Exp. Med. 188, 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P.A., and Vousden K.H. (2014). Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano K., and Vousden K.H. (2001). PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 7, 683–694. [DOI] [PubMed] [Google Scholar]
- Newton K., and Strasser A. (2000). Ionizing radiation and chemotherapeutic drugs induce apoptosis in lymphocytes in the absence of fas or FADD/MORT1 signaling: implications for cancer therapy. J. Exp. Med. 191, 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda E., Ohki R., Murasawa H., et al. (2000). Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 288, 1053–1058. [DOI] [PubMed] [Google Scholar]
- Olive K.P., Tuveson D.A., Ruhe Z.C., et al. (2004). Mutant p53 gain of function in two mouse models of Li-Fraumeni syndrome. Cell 119, 847–860. [DOI] [PubMed] [Google Scholar]
- Oren M., Maltzman W., and Levine A.J. (1981). Post-translational regulation of the 54K cellular tumor antigen in normal and transformed cells. Mol. Cell. Biol. 1, 101–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitjean A., Mathe E., Kato S., et al. (2007). Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 28, 622–629. [DOI] [PubMed] [Google Scholar]
- Pietrzak M., and Puzianowska-Kuznicka M. (2008). p53-dependent repression of the human MCL-1 gene encoding an anti-apoptotic member of the BCL-2 family: the role of Sp1 and of basic transcription factor binding sites in the MCL-1 promoter. Biol. Chem. 389, 383–393. [DOI] [PubMed] [Google Scholar]
- Robles A.I., Bemmels N.A., Foraker A.B., et al. (2001). APAF-1 is a transcriptional target of p53 in DNA damage-induced apoptosis. Cancer Res. 61, 6660–6664. [PubMed] [Google Scholar]
- Rouault J.P., Falette N., Guehenneux F., et al. (1996). Identification of BTG2, an antiproliferative p53-dependent component of the DNA damage cellular response pathway. Nat. Genet. 14, 482–486. [DOI] [PubMed] [Google Scholar]
- Rowland M.A., Greenbaum J.M., and Deeds E.J. (2017). Crosstalk and the evolvability of intracellular communication. Nat. Commun. 8, 16009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagne C., Marcel V., Bota M., et al. (2014). Age at cancer onset in germline TP53 mutation carriers: association with polymorphisms in predicted G-quadruplex structures. Carcinogenesis 35, 807–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha M.N., Jiang H., Yang Y., et al. (2013). PRIMA-1Met/APR-246 displays high antitumor activity in multiple myeloma by induction of p73 and Noxa. Mol. Cancer Ther. 12, 2331–2341. [DOI] [PubMed] [Google Scholar]
- Saiki A.Y., Caenepeel S., Cosgrove E., et al. (2015). Identifying the determinants of response to MDM2 inhibition. Oncotarget 6, 7701–7712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Secchiero P., Barbarotto E., Tiribelli M., et al. (2006). Functional integrity of the p53-mediated apoptotic pathway induced by the nongenotoxic agent nutlin-3 in B-cell chronic lymphocytic leukemia (B-CLL). Blood 107, 4122–4129. [DOI] [PubMed] [Google Scholar]
- Seluanov A., Gorbunova V., Falcovitz A., et al. (2001). Change of the death pathway in senescent human fibroblasts in response to DNA damage is caused by an inability to stabilize p53. Mol. Cell. Biol. 21, 1552–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo Y.R., Kelley M.R., and Smith M.L. (2002). Selenomethionine regulation of p53 by a ref1-dependent redox mechanism. Proc. Natl Acad. Sci. USA 99, 14548–14553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seoane J., Le H.V., and Massague J. (2002). Myc suppression of the p21(Cip1) Cdk inhibitor influences the outcome of the p53 response to DNA damage. Nature 419, 729–734. [DOI] [PubMed] [Google Scholar]
- Serrano M., Lin A.W., McCurrach M.E., et al. (1997). Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell 88, 593–602. [DOI] [PubMed] [Google Scholar]
- Shen H., and Maki C.G. (2011). Pharmacologic activation of p53 by small-molecule MDM2 antagonists. Curr. Pharm. Des. 17, 560–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibue T., Takeda K., Oda E., et al. (2003). Integral role of Noxa in p53-mediated apoptotic response. Genes Dev. 17, 2233–2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobhani M., Abdi J., Manujendra S.N., et al. (2015). PRIMA-1Met induces apoptosis in Waldenstrom’s Macroglobulinemia cells independent of p53. Cancer Biol. Ther. 16, 799–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiewe T., and Putzer B.M. (2000). Role of the p53-homologue p73 in E2F1-induced apoptosis. Nat. Genet. 26, 464–469. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., and Cory S. (1991). Bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell 67, 889–899. [DOI] [PubMed] [Google Scholar]
- Strasser A., Harris A.W., Jacks T., et al. (1994). DNA damage can induce apoptosis in proliferating lymphoid cells via p53-independent mechanisms inhibitable by Bcl-2. Cell 79, 329–339. [DOI] [PubMed] [Google Scholar]
- Strasser A., Jost P.J., and Nagata S. (2009). The many roles of FAS receptor signaling in the immune system. Immunity 30, 180–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuhmer T., Chatterjee M., Hildebrandt M., et al. (2005). Nongenotoxic activation of the p53 pathway as a therapeutic strategy for multiple myeloma. Blood 106, 3609–3617. [DOI] [PubMed] [Google Scholar]
- Suh E.K., Yang A., Kettenbach A., et al. (2006). p63 protects the female germ line during meiotic arrest. Nature 444, 624–628. [DOI] [PubMed] [Google Scholar]
- Swift L.H., and Golsteyn R.M. (2014). Genotoxic anti-cancer agents and their relationship to DNA damage, mitosis, and checkpoint adaptation in proliferating cancer cells. Int. J. Mol. Sci. 15, 3403–3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szikriszt B., Poti A., Pipek O., et al. (2016). A comprehensive survey of the mutagenic impact of common cancer cytotoxics. Genome Biol. 17, 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y., Luo J., Zhang W., et al. (2006). Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 24, 827–839. [DOI] [PubMed] [Google Scholar]
- Tarangelo A., Magtanong L., Bieging-Rolett K.T., et al. (2018). p53 suppresses metabolic stress-induced ferroptosis in cancer cells. Cell Rep. 22, 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H., Wittmack E.K., and Jorgensen T.J. (2000). p21WAF1/CIP1 antisense therapy radiosensitizes human colon cancer by converting growth arrest to apoptosis. Cancer Res. 60, 679–684. [PubMed] [Google Scholar]
- Tovar C., Rosinski J., Filipovic Z., et al. (2006). Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl Acad. Sci. USA 103, 1888–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valente L.J., Aubrey B.J., Herold M.J., et al. (2016). Therapeutic response to non-genotoxic activation of p53 by Nutlin3a is driven by PUMA-mediated apoptosis in lymphoma cells. Cell Rep. 14, 1858–1866. [DOI] [PubMed] [Google Scholar]
- Valente L.J., Grabow S., Vandenberg C.J., et al. (2015). Combined loss of PUMA and p21 accelerates c-MYC-driven lymphoma development considerably less than loss of one allele of p53. Oncogene 35, 3866–71. [DOI] [PubMed] [Google Scholar]
- Valente L.J., Gray D.H., Michalak E.M., et al. (2013). p53 efficiently suppresses tumor development in the complete absence of its cell-cycle inhibitory and proapoptotic effectors p21, Puma, and Noxa. Cell Rep. 3, 1339–1345. [DOI] [PubMed] [Google Scholar]
- Vassilev L.T., Vu B.T., Graves B., et al. (2004). In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 303, 844–848. [DOI] [PubMed] [Google Scholar]
- Ventura A., Kirsch D.G., McLaughlin M.E., et al. (2007). Restoration of p53 function leads to tumour regression in vivo. Nature 445, 661–665. [DOI] [PubMed] [Google Scholar]
- Villunger A., Michalak E.M., Coultas L., et al. (2003). p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science 302, 1036–1038. [DOI] [PubMed] [Google Scholar]
- Wang S.J., Li D., Ou Y., et al. (2016). Acetylation is crucial for p53-mediated ferroptosis and tumor suppression. Cell Rep. 17, 366–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.W., Zhan Q., Coursen J.D., et al. (1999). GADD45 induction of a G2/M cell cycle checkpoint. Proc. Natl Acad. Sci. USA 96, 3706–3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whibley C., Pharoah P.D., and Hollstein M. (2009). p53 polymorphisms: cancer implications. Nat. Rev. Cancer 9, 95–107. [DOI] [PubMed] [Google Scholar]
- Wiley C.D., Flynn J.M., Morrissey C., et al. (2017). Analysis of individual cells identifies cell-to-cell variability following induction of cellular senescence. Aging Cell 16, 1043–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willis A.C., Pipes T., Zhu J., et al. (2003). p73 can suppress the proliferation of cells that express mutant p53. Oncogene 22, 5481–5495. [DOI] [PubMed] [Google Scholar]
- Wu M., Ye H., Tang Z., et al. (2017). p53 dynamics orchestrates with binding affinity to target genes for cell fate decision. Cell Death Dis. 8, e3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z., Yao M., Li Y., et al. (2016). Guttiferone K impedes cell cycle re-entry of quiescent prostate cancer cells via stabilization of FBXW7 and subsequent c-MYC degradation. Cell Death Dis. 7, e2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y. (2003). Regulation of p53 responses by post-translational modifications. Cell Death Differ. 10, 400–403. [DOI] [PubMed] [Google Scholar]
- Yosef R., Pilpel N., Tokarsky-Amiel R., et al. (2016). Directed elimination of senescent cells by inhibition of BCL-W and BCL-XL. Nat. Commun. 7, 11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhang L., Hwang P.M., et al. (2001). PUMA induces the rapid apoptosis of colorectal cancer cells. Mol. Cell 7, 673–682. [DOI] [PubMed] [Google Scholar]
- Zandi R., Selivanova G., Christensen C.L., et al. (2011). PRIMA-1Met/APR-246 induces apoptosis and tumor growth delay in small cell lung cancer expressing mutant p53. Clin. Cancer Res. 17, 2830–2841. [DOI] [PubMed] [Google Scholar]
- Zawacka-Pankau J., Kostecka A., Sznarkowska A., et al. (2010). p73 tumor suppressor protein: a close relative of p53 not only in structure but also in anti-cancer approach? Cell Cycle 9, 720–728. [DOI] [PubMed] [Google Scholar]