Abstract
While it is well appreciated that loss of the p53 tumor suppressor protein promotes cancer, growing evidence indicates that increased p53 activity underlies the developmental defects in a wide range of genetic syndromes. The inherited or de novo mutations that cause these syndromes affect diverse cellular processes, such as ribosome biogenesis, DNA repair, and centriole duplication, and analysis of human patient samples and mouse models demonstrates that disrupting these cellular processes can activate the p53 pathway. Importantly, many of the developmental defects in mouse models of these syndromes can be rescued by loss of p53, indicating that inappropriate p53 activation directly contributes to their pathogenesis. A role for p53 in driving developmental defects is further supported by the observation that mouse strains with broad p53 hyperactivation, due to mutations affecting p53 pathway components, display a host of tissue-specific developmental defects, including hematopoietic, neuronal, craniofacial, cardiovascular, and pigmentation defects. Furthermore, germline activating mutations in TP53 were recently identified in two human patients exhibiting bone marrow failure and other developmental defects. Studies in mice suggest that p53 drives developmental defects by inducing apoptosis, restraining proliferation, or modulating other developmental programs in a cell type-dependent manner. Here, we review the growing body of evidence from mouse models that implicates p53 as a driver of tissue-specific developmental defects in diverse genetic syndromes.
Keywords: p53, Mdm2, development, embryo, congenital defect, syndrome, genetic disorder
Introduction
The p53 tumor suppressor protein is a transcription factor that acts as a central hub in the cellular response to stress. While normally maintained at low levels in the cell, p53 becomes stabilized and activated in response to a variety of cellular stresses, such as DNA damage and hyperproliferative signals, and induces genes involved in numerous cellular processes, including apoptosis, cell cycle arrest, DNA repair, and metabolism (Bieging et al., 2014). Given p53’s central role in regulating these diverse cellular responses, either loss of p53 or inappropriate activation of p53 can have pathological consequences. On the one hand, p53 is a potent tumor suppressor and loss of p53 allows cells to expand under adverse conditions, thereby facilitating the growth of malignant cells (Kaiser and Attardi, 2018). Loss of p53 can also perturb the development of certain tissues, with ~20% of female p53-deficient mouse embryos exhibiting neonatal lethality due to the neural tube defect exencephaly, and a subset of p53-deficient mice displaying growth retardation or subtle developmental defects affecting the palate, eyes, teeth, lungs, or kidneys (Armstrong et al., 1995; Sah et al., 1995; Kaufman et al., 1997; Baatout et al., 2002; Saifudeen et al., 2009; Rinon et al., 2011; Tateossian et al., 2015). On the other hand, inappropriate activation of p53 can trigger cellular responses such as excessive apoptosis and therefore can also profoundly disrupt tissue development and homeostasis (Van Nostrand et al., 2014; Wu and Prives, 2018). In this regard, increased p53 activity is thought to contribute to the developmental defects and premature aging phenotypes in numerous genetic syndromes and to the excessive neuronal cell death in various neurodegenerative disorders (Van Nostrand and Attardi, 2014; Lessel et al., 2017; Szybińska and Leśniak, 2017; Wu and Prives, 2018). Thus, a broad spectrum of human diseases arises as a consequence of either increased or decreased p53 activity. In this review, we will discuss the growing body of evidence implicating p53 hyperactivation as a driver of developmental defects in a wide range of genetic syndromes.
Broadly activating p53 during mouse embryogenesis promotes tissue-specific developmental defects
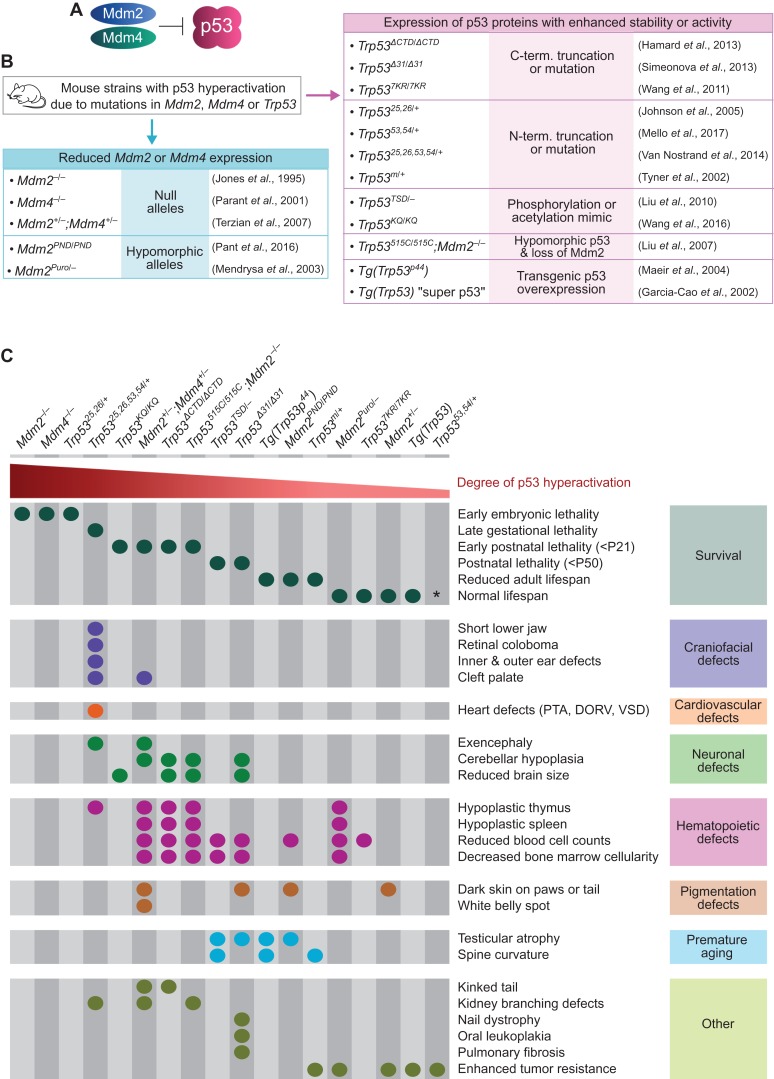
Analyses from a variety of mouse strains demonstrate that inappropriately activating p53 during embryonic and early postnatal development triggers a host of developmental defects. In these mouse strains, p53 hyperactivation has been achieved through mutations in Trp53, the gene encoding p53, or mutations in Mdm2 or Mdm4, which encode the two main negative regulators of p53 that act cooperatively to inhibit the transactivation domains of p53 and to target p53 for ubiquitin-mediated degradation (Figure 1A and B) (Perry, 2010). These mouse strains display a range of developmental phenotypes, the most striking of which occur in Mdm2- and Mdm4-null mice, which exhibit p53-dependent lethality by embryonic day 5.5 (E5.5) and E8.5, respectively (Figure 1B and C) (Jones et al., 1995; Montes de Oca Luna et al., 1995; Parant et al., 2001; Chavez-Reyes et al., 2003; Moyer et al., 2017). Mouse strains with milder levels of p53 hyperactivation—due to only partial loss of Mdm2 or Mdm4, or due to the expression of mutant p53 proteins with modestly enhanced stability or activity—survive beyond early embryogenesis and display defects only at later developmental stages (Figure 1B and C). The age of lethality of these mice varies based on the exact degree of p53 hyperactivation, with some mouse strains dying at late embryonic or early postnatal stages and displaying notable developmental defects, and other mouse strains surviving into adulthood with no overt developmental defects but instead displaying premature aging phenotypes or exhibiting enhanced tumor resistance (García-Cao et al., 2002; Tyner et al., 2002; Alt, 2003; Mendrysa et al., 2003; Maier, 2004; Johnson et al., 2005; Mendrysa, 2006; Liu et al., 2010, 2007; Terzian et al., 2007, 2011; Wang et al., 2016, 2011b; Hamard et al., 2013; Simeonova et al., 2013; Van Nostrand et al., 2014; Pant et al., 2016; Mello et al., 2017). Interestingly, the developmental defects associated with these mouse strains are highly tissue-selective and include craniofacial, cardiovascular, and neural tube defects, which manifest at embryonic stages, and hematopoietic, pigmentation, and cerebellar defects, which manifest at early postnatal stages (Figure 1C). For example, Trp5325,26,53,54/+ mice, which have elevated p53 activity due to the expression of a mutant p53 protein that binds to and stabilizes wild-type p53, die at late embryonic stages and display craniofacial, cardiovascular, and neural tube defects (Van Nostrand et al., 2014). In addition, Mdm2+/−;Mdm4+/− mice, which have elevated p53 activity due to reduced expression levels of both Mdm2 and Mdm4, die at early postnatal stages and exhibit hematopoietic defects, cerebellar hypoplasia, and skin hyperpigmentation (Terzian et al., 2007, 2011). Together, the phenotypes of the mouse strains discussed here provide strong evidence that p53 hyperactivation can trigger a host of tissue-specific developmental defects.
Figure 1.
Broad p53 hyperactivation during mouse embryogenesis triggers tissue-specific developmental defects. (A) Mdm2 and Mdm4 are negative regulators of p53. (B) Mouse strains with increased p53 activity due to inactivating mutations in Mdm2/4 or activating/stabilizing mutations in Trp53. (C) Spectrum of phenotypes observed in mouse strains with increased p53 activity. PTA, persistent truncus arteriosus; DORV, double outlet right ventricle; VSD, ventricular septal defect; P21 and P50, postnatal day 21 and 50. *Adult Trp5353,54/+ mice appear normal and healthy, but their lifespan relative to wild-type mice has not been reported.
Inappropriate p53 activation in mice can trigger apoptosis, restrain proliferation, and modulate other developmental programs in a cell type-dependent manner
A variety of cellular mechanisms have been proposed to underlie the developmental defects in mouse strains with p53 hyperactivation. Excessive apoptosis and/or decreased proliferation have been detected in certain cell compartments in these mouse strains, such as the embryonic neuroepithelium and the neonatal bone marrow and cerebellum (Liu et al., 2007; Terzian et al., 2007; Van Nostrand et al., 2014). In addition, alterations in specific developmental programs have been reported. In particular, mice with increased p53 activity display impaired endothelial-to-mesenchymal transition of endocardial cells during heart morphogenesis (Zhang et al., 2012a). Furthermore, their keratinocytes display increased expression of the p53 target gene Kitl, which encodes a melanocyte-stimulating growth factor that promotes skin hyperpigmentation (McGowan et al., 2008; Pant et al., 2016; Chang et al., 2017). These studies suggest that inappropriate p53 activation drives developmental defects by triggering apoptosis, restraining proliferation, or modulating other developmental programs in a cell type-dependent manner during embryonic and postnatal development. The molecular basis for this cell type-specificity is not well understood. In other contexts, such as in cancer cell lines, multiple mechanisms have been proposed to explain why p53 activation elicits different responses in different cell types. These mechanisms include cell type-specific p53 post-translational modifications or expression of co-factors that influence which target genes p53 induces in a given cell type, as well as cell type-specific differences in the basal expression levels of factors required for the execution of downstream cellular responses, such as factors involved in the apoptotic pathway (Kruiswijk et al., 2015). Determining which of these mechanisms accounts for the cell type-specific responses elicited by p53 activation in embryonic and neonatal cells is an intriguing area for future research.
TP53 and MDM2 mutations can cause developmental defects and premature aging phenotypes in humans
While the analysis of various mouse strains carrying mutations in Trp53 or Mdm2 clearly indicates that inappropriate p53 activation can cause developmental defects and premature aging phenotypes in mice (Figure 1C), recent reports indicate that TP53 and MDM2 mutations can cause similar phenotypes in humans. Indeed, germline TP53 mutations were recently identified in two unrelated patients presenting with bone marrow failure, growth retardation, microcephaly, abnormal skin pigmentation, hypogonadism, and tooth anomalies (Toki et al., 2018). These patients carried heterozygous frameshift mutations that were predicted to lead to the truncation of the final 32 amino acids of the C-terminal domain of p53. In functional assays, these mutations led to an increase in the transactivation of p53 target genes, consistent with observations in mouse models that truncating the C-terminal domain of p53 leads to enhanced p53 activity (Hamard et al., 2013; Simeonova et al., 2013). Another recent report identified germline MDM2 mutations in a consanguineous family segregating premature aging phenotypes including hair graying, kidney failure, and short stature (Lessel et al., 2017). The affected family members carried a homozygous antitermination mutation leading to a 5 amino acid extension on the C-terminus of MDM2. In functional assays, this mutation compromised MDM2 activity and led to increased p53 stability and activity. Together, these studies indicate that disrupting the p53/MDM2 axis in humans can cause developmental and aging-related phenotypes. This is further supported by the finding that a common single nucleotide polymorphism in TP53 (rs1042522), which codes for either a proline (P) or arginine (R) residue at codon 72, may have subtle effects on human development and aging. Specifically, the R72 variant, which is more efficient at inducing apoptosis than the P72 variant, has been associated with a slightly shorter lifespan and an increased risk for exhibiting a low birth weight and developing neural tube defects (Dumont et al., 2003; Bojesen and Nordestgaard, 2008; Thurow et al., 2011; Arora et al., 2012). As ongoing sequencing efforts continue to identify genetic factors associated with developmental phenotypes in humans, it will be of interest to see whether additional mutations or common polymorphisms in TP53, MDM2, or MDM4 are found to be associated with isolated birth defects or to cause multi-symptom syndromes. Of note, while Li-Fraumeni syndrome is caused by mutations in TP53, these mutations are typically loss-of-function or dominant-negative, consistent with this syndrome being linked to increased cancer risk due to loss of p53 activity, rather than developmental defects or premature aging due to increased p53 activity (Valdez et al., 2017).
p53 activation is a secondary consequence of many of the mutations that cause human developmental syndromes
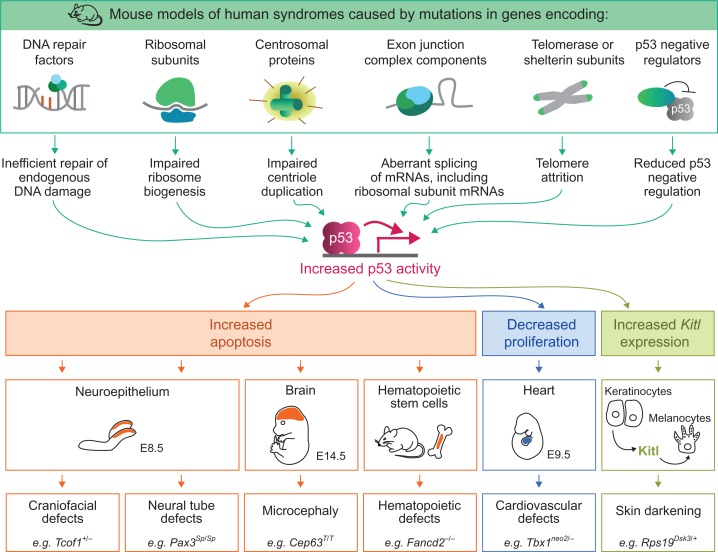
Thousands of human genetic syndromes have been identified, many of which are characterized by specific sets of developmental defects that manifest at birth or during early childhood (OMIM: https://www.omim.org). Intriguingly, a wide spectrum of these developmental syndromes has been found to be caused by mutations in genes involved in fundamental cellular functions, such as DNA repair, ribosome biogenesis, centriole duplication, RNA processing, and telomere maintenance (Yelick and Trainor, 2015; Lee et al., 2016; McMahon et al., 2016; Roake and Artandi, 2017; Nigg and Holland, 2018). A growing body of data from mouse models of these syndromes, and in some cases from human patient samples, indicate that disrupting these genes can activate p53 (Table 1 and Figure 2). For example, mutations in genes encoding DNA repair factors can lead to an accumulation of unrepaired DNA damage, which activates p53 via the DNA damage response pathway (Ceccaldi et al., 2012). Similarly, mutations that disrupt ribosome biogenesis can also activate p53. In this case, impaired ribosome biogenesis leads to the accumulation of unincorporated ribosomal subunits, which can bind to Mdm2 and prevent it from negatively regulating p53 (Bursac et al., 2014). Mutations affecting centriole duplication, RNA processing, telomere maintenance, and epigenetic regulation can also activate p53 through various mechanisms (as discussed in more detail below). Thus, while mutations in many different types of genes can cause developmental syndromes, a common theme shared by many of these mutations is that they trigger p53 activation.
Table 1.
Human genetic syndromes with p53-dependent developmental phenotypes in mouse models.
| Human syndrome | Elevated p53 levels in human patient tissue/cells | Mouse model | ||
|---|---|---|---|---|
| Genotype | Developmental defects rescued by loss of p53a | References | ||
| Treacher Collins syndrome | N/D | Tcof1+/− | Frontonasal hypoplasia, cleft palate, microphthalmia, cranial nerve defects, neonatal lethality | Jones et al. (2008) |
| Diamond–Blackfan anemia | Yes (Dutt et al., 2011; Aspesi et al., 2017) | Rps7Zma/+ | Erythrocyte maturation delay, tail kink, white belly spot, developmental delay | Watkins-Chow et al. (2013) |
| Rps19Dsk3/+ | Lower red blood cell count, skin hyperpigmentation, reduced postnatal body weight | McGowan et al. (2008) | ||
| 5q− syndrome | Yes (Pellagatti et al. 2010; Dutt et al. 2011) | Deletion spanning Rps14 | Decreased number of hematopoietic stem cells | Barlow et al. (2010) |
| Fanconi anemia | Yes (Ceccaldi et al., 2012) | Fancd2−/− | Decreased number of hematopoietic stem cells | Ceccaldi et al. (2012) |
| Xrcc2−/− | Short tail, lethality ~E12.5 | Adam et al. (2007) | ||
| Rad51−/− | Lethality ~E6.5 | Lim and Hasty (1996) | ||
| Brca1Δ11/Δ11 | Late gestational lethality | Xu et al. (2001) | ||
| Wnt1-Cre;Brca1flox/flox | Hypoplastic craniofacial skeleton | Kitami et al. (2018) | ||
| Tyr-Cre;Brca1flox/flox | Piebald fur pigmentation | Tonks et al. (2012) | ||
| Palb2−/− | Lethality ~E8.5 | Bouwman et al. (2011) | ||
| Autosomal recessive primary microcephaly | N/D | Math1-Cre;Aspmflox/flox | Hypoplastic cerebellum | Williams et al. (2015) |
| Cit−/− | Microcephaly, ataxia | Bianchi et al. (2017) | ||
| Cenpj−/− | Lethality ~E9.5 | Bazzi and Anderson (2014) | ||
| Nestin-Cre;Cenpjflox/flox | Microcephaly | Insolera et al. (2014) | ||
| Seckel syndrome | N/D | Cep63T/T | Microcephaly | Marjanović et al. (2015) |
| Nijmegen breakage syndrome | Yes (Mlody et al., 2017) | Nestin-Cre;Nbnflox/flox | Microcephaly, cerebellar defects, ataxia, postnatal growth retardation | Frappart et al. (2005) |
| LIG4 syndrome | N/D | Lig4−/− | Late gestational lethality | Frank et al. (2000) |
| SSMED | N/D | Xrcc4−/− | Late gestational lethality | Gao et al. (2000) |
| Microlissencephaly | N/D | Nde1−/− | Microcephaly | Houlihan and Feng (2014) |
| Richieri–Costa–Pereira syndrome | N/D | Emx1-Cre; Eif4a3flox/+ | Microcephaly | Mao et al. (2016) |
| Thrombocytopenia-absent radius syndrome | N/D | Emx1-Cre; Rbm8aflox/+ | Microcephaly | Mao et al. (2016) |
| Waardenburg syndrome type 1 | N/D | Pax3Sp/Sp | Neural tube defects, heart outflow tract defects | Morgan et al. (2008), Pani (2002) |
| 22q11.2 deletion syndrome | N/D | Tbx1neo2/− | Pharyngeal arch artery defects, heart outflow tract defects | Caprio and Baldini (2014) |
| CHARGE syndrome | Yes (Van Nostrand et al., 2014) | Chd7−/− | Lethality ~E10.5 | Van Nostrand et al. (2014) |
| Dyskeratosis congenita | Yes (Pereboeva et al., 2016) | AcdAcd/Acd | Skin hyperpigmentation, hypoplastic adrenal glands, hindlimb hypoplasia, vertebral fusions | Else et al. (2009), Vlangos et al. (2009) |
| Terc−/− | Hypoplastic testes | Chin et al. (1999) | ||
aIncludes developmental defects that are fully or partially rescued by concomitant deletion of one or both alleles of Trp53.
Figure 2.
Proposed role for p53 in mouse models of human developmental syndromes. Mutations affecting a range of cellular processes trigger p53 activation. Once active, p53 is thought to drive developmental defects by inducing apoptosis, restraining proliferation, or modulating other developmental programs in specific cell compartments during embryonic or postnatal development. For each type of p53-driven developmental defect, an example of a mouse model exhibiting this defect is indicated.
Increased p53 activity contributes to the developmental defects in mouse models of numerous human developmental syndromes
The observation that many of the mutations that cause developmental syndromes can activate p53 raises the intriguing hypothesis that the phenotypes associated with these syndromes arise as a direct consequence of p53 hyperactivation. In support of this hypothesis, the developmental defects in numerous mouse models of human syndromes have been shown to arise in a p53-dependent manner (Table 1). In some of these mouse models, the developmental defects can be fully rescued by concomitant deletion of both alleles of Trp53 and partially rescued by deleting one Trp53 allele, suggesting that increased p53 activity is indeed the primary factor driving these developmental defects (Jones et al., 2008; McGowan et al., 2008). In other mouse models, complete loss of Trp53 only partially rescues the developmental defects, suggesting that both p53-dependent and p53-independent pathways contribute to the developmental phenotypes (Mao et al., 2016). Importantly, loss of p53 does not repair the primary cellular defect caused by the mutations in these mouse models. For example, in a mouse model carrying a mutation in a gene involved in ribosome biogenesis, loss of Trp53 fully rescues the associated developmental defects but does not restore the levels of ribosome biogenesis (Jones et al., 2008). This observation suggests that it is not the decrease in ribosome biogenesis per se that causes developmental defects but rather it is the increase in p53 activity that directly drives developmental defects. Collectively, these studies indicate that increased p53 activity is either fully or partially responsible for driving developmental defects in mouse models of a wide range of developmental syndromes. Here, we review the tissue-specific developmental phenotypes observed in these mouse models and their associated human syndromes, and we discuss the proposed mechanisms by which p53 becomes activated and drives these phenotypes.
Craniofacial defects in Treacher Collins syndrome
The craniofacial skeleton arises primarily from the cranial neural crest, a multipotent population that delaminates from the neuroepithelium, migrates to the facial primordia and differentiates into a variety of cell types, including those comprising skeletal and connective tissues (Bronner and Simões-Costa, 2016). Craniofacial defects are a common feature of many human developmental syndromes, including Treacher Collins syndrome, characterized by underdeveloped cheekbones and jawbones, downward slanting eyes, cleft palate, and outer ear malformations (Vincent et al., 2016). This syndrome is most commonly caused by heterozygous inactivating mutations in TCOF1, which encodes a nucleolar protein that plays a role both in ribosome biogenesis, by promoting ribosomal RNA (rRNA) transcription and pre-rRNA processing, and in DNA repair, by facilitating the recognition of DNA double-strand breaks (Sakai and Trainor, 2016). In mice, Tcof1 expression is enriched in the neuroepithelium during embryogenesis and heterozygous loss of Tcof1 leads to decreased levels of mature ribosomes and increased levels of DNA damage specifically in the neuroepithelium (Dixon et al., 2006; Jones et al., 2008; Sakai et al., 2016). Consistent with DNA damage and impaired ribosome biogenesis being known activators of p53, Tcof1+/− embryos display increased p53 immunostaining and increased apoptosis in the neuroepithelium, and generate fewer neural crest cells, most likely a direct consequence of increased neuroepithelial apoptosis (Jones et al., 2008). Strikingly, the neuroepithelial apoptosis, the decrease in neural crest formation, and the craniofacial defects in Tcof1+/− mice can all be rescued by concomitant deletion of Trp53 (Table 1) (Jones et al., 2008). Loss of p53 can also rescue the neuroepithelial apoptosis and craniofacial defects in zebrafish carrying mutations in polr1c or polr1d, which are mutated in a subset of human Treacher Collins syndrome patients and encode subunits of RNA polymerase I, which transcribes rRNA, and RNA polymerase III, which transcribes both rRNA and other non-coding RNAs (Dauwerse et al., 2011; Lau et al., 2016; Noack Watt et al., 2016). Collectively, these studies suggest that increased p53 activity, as a consequence of impaired ribosome biogenesis and perturbed DNA repair, acts as the driver of craniofacial defects in Treacher Collins syndrome.
Hematopoietic defects in Diamond–Blackfan anemia, 5q− syndrome, and Fanconi anemia
The hematopoietic syndrome Diamond–Blackfan anemia (DBA) is characterized by anemia and various other congenital defects and is caused by mutations in genes encoding ribosomal subunits, such as RPS19 and RPS7 (Arbiv et al., 2018). Consistent with impaired ribosome biogenesis being an activator of p53, increased p53 immunostaining is observed in bone marrow biopsies from patients with DBA, as well as from patients with 5q− syndrome, a related acquired hematological disorder caused by a chromosomal deletion spanning the gene encoding the RPS14 ribosomal subunit (Pellagatti et al., 2010; Dutt et al., 2011). Increased p53 immunostaining is also observed in mouse models of DBA and 5q− syndrome, and loss of Trp53 can rescue their hematopoietic defects, including the decrease in red blood cell counts in Rps19-deficient mice, the subtle delay in erythrocyte development in Rps7-deficient embryos, and the decrease in hematopoietic progenitor cells in the bone marrow of Rps14-deficient mice (Table 1) (McGowan et al., 2008; Barlow et al., 2010; Watkins-Chow et al., 2013). The hematopoietic defects in these mice are associated with increased apoptosis in hematopoietic progenitor cells, suggesting that p53 drives these defects by inducing apoptosis (McGowan et al., 2008; Barlow et al., 2010). These results suggest that inappropriate p53 activation, as a consequence of impaired ribosome biogenesis, plays a direct role in contributing to the hematopoietic defects in DBA and 5q− syndrome.
The defects in another hematopoietic syndrome, Fanconi anemia, have also been associated with increased p53 activity. Fanconi anemia is a bone marrow failure syndrome associated with increased cancer risk and a range of congenital defects (Mamrak et al., 2017). The hematopoietic defects in Fanconi anemia patients are thought to arise in utero during the expansion of the fetal liver hematopoietic stem cell (HSC) pool and progressively worsen during childhood, culminating in overt bone marrow failure (Ceccaldi et al., 2012). Mutations in any of at least 21 different genes can cause Fanconi anemia and these genes encode proteins primarily involved in the repair of DNA interstrand crosslinks (Mamrak et al., 2017). Consistent with DNA damage being a known activator of p53, increased p53 activity is observed in blood and bone marrow cells harvested from Fanconi anemia patients as well as from Fanconi anemia fetal liver samples obtained from terminated pregnancies (Ceccaldi et al., 2012). In mouse models of Fanconi anemia, loss of Trp53 can rescue the associated hematopoietic defects, including the decrease in the number of HSCs in the bone marrow of Fancd2−/− mice and the decreased engraftment potential of bone marrow cells isolated from Fancg−/− mice (Table 1) (Ceccaldi et al., 2012). p53 is thought to drive these hematopoietic defects by triggering apoptosis or restraining proliferation, given that Fancd2−/− mice display increased apoptosis in their HSCs and that lymphoblastoid cell lines generated from Fanconi anemia patients undergo a p53-dependent cell cycle arrest in response to a brief exposure to DNA interstrand crosslinking agents (Ceccaldi et al., 2012). In addition to hematopoietic defects, loss of p53 can also fully or partially rescue other types of developmental defects in animal models of Fanconi Anemia, including the embryonic lethality, craniofacial defects, and pigmentation defects in mice deficient for Xrcc2, Rad51, Brca1, or Palb2 (Table 1), and the growth retardation, microcephaly, and eye defects in Fancd2-deficient zebrafish (Lim and Hasty, 1996; Xu et al., 2001; Liu et al., 2003; Adam et al., 2007; Bouwman et al., 2011; Tonks et al., 2012; Kitami et al., 2018). Thus, data from various animal models and human patient samples suggest that increased p53 activity, as a consequence of impaired DNA repair, contributes to the hematopoietic defects and other developmental defects in Fanconi Anemia.
Microcephaly in a variety of genetic syndromes
Microcephaly, a congenital defect in which the head circumference is reduced due to a decrease in brain size, is the defining feature of autosomal recessive primary microcephaly (MCPH for microcephaly primary hereditary). MCPH can be caused by mutations in any of at least 17 genes and overlaps phenotypically and genetically with Seckel syndrome, which is characterized by short stature and craniofacial anomalies in addition to microcephaly (Zaqout et al., 2017). Many of the genes mutated in MCPH or Seckel syndrome encode centrosomal proteins involved in centriole duplication and mitotic spindle formation, such as CENPJ, CEP63, and ASPM (Nigg and Holland, 2018). In mice, mutations in these genes trigger p53 activation in the developing brain and lead to p53-dependent microcephaly associated with excessive neuronal apoptosis (Table 1) (Insolera et al., 2014; Marjanović et al., 2015; Williams et al., 2015). It is thought that p53 is activated in these mice as a direct consequence of short delays during mitosis, which arise due to delayed mitotic spindle pole formation, although the precise pathways are not fully understood (Bazzi and Anderson, 2014). These studies suggest that mutations in genes encoding centrosomal proteins cause human microcephaly syndromes by triggering p53 activation and excessive apoptosis.
Mutations in genes involved in DNA repair have also been associated with human microcephaly syndromes. In particular, a subset of MCPH patients carry mutations in CIT, which encodes a kinase involved in both cytokinesis and DNA double-strand break repair (Li et al., 2016; Bianchi et al., 2017). In mice, loss of Cit leads to an accumulation of unrepaired DNA damage in the developing brain that triggers p53 activation and leads to apoptosis (Bianchi et al., 2017). As with other mouse models of MCPH, the reduction in brain size in Cit−/− mice can be rescued by loss of Trp53 (Table 1) (Bianchi et al., 2017). In addition to MCPH, other microcephaly-associated syndromes can be caused by mutations that affect DNA repair, including Nijmegen breakage syndrome, SSMED syndrome (short stature, microcephaly, and endocrine dysfunction), and LIG4 syndrome. These syndromes are caused by mutations in NBN, XRCC4, and LIG4, respectively, which encode the NBN protein involved in the initial processing of DNA double-strand breaks, and the XRCC4 and LIG4 proteins involved in the nonhomologous end-joining DNA repair pathway (Chrzanowska et al., 2012; de Villartay, 2015; Altmann and Gennery, 2016). In mice, loss of Trp53 can rescue the microcephaly, ataxia, and growth retardation phenotypes observed in Nbn-deficient mice, as well as the excessive neuronal apoptosis and late gestational lethality observed in Lig4- and Xrcc4-deficient mice (Table 1) (Frank et al., 2000; Gao et al., 2000; Frappart et al., 2005). Thus, a variety of mutations that affect DNA double-strand break repair pathways can trigger p53-dependent microcephaly or neuronal defects. Moreover, mice lacking Nde1, which is associated with severe microcephaly in humans, exhibit stalled DNA replication forks, leading to an increase in DNA damage, which triggers massive neuronal apoptosis and microcephaly that can be rescued by loss of Trp53 (Table 1) (Alkuraya et al., 2011; Houlihan and Feng, 2014). Together, these data suggest that p53 activation as a consequence of increased DNA damage contributes to microcephaly in multiple human syndromes.
Defects in RNA processing have also been linked to p53-dependent microcephaly. In mice, heterozygous deletion of Eif4a3 or Rbm8a in neural progenitor cells triggers p53 activation and excessive apoptosis, resulting in a microcephaly phenotype that can be partially rescued by loss of Trp53 (Table 1) (Mao et al., 2016). Eif4a3 and Rmb8a are components of the exon junction complex (EJC), which regulates multiple aspects of RNA biology, including splicing, nonsense-mediated decay, translation, and RNA localization (Hir et al., 2016). It is not clear how perturbations affecting the EJC trigger p53 activation, but one possibility is that EJC-deficiency affects the stability or splicing of ribosomal protein mRNAs, leading to impaired ribosome biogenesis, which activates p53 (Mao et al., 2016). In humans, mutations in EIF4A3 and RBM8A cause Richieri–Costa–Pereira and thrombocytopenia-absent radius syndromes, respectively, which are associated with a variety of congenital defects, including neurodevelopmental phenotypes (Albers et al., 2012; Rosenfeld et al., 2012; Favaro et al., 2014). In summary, increased p53 activity may contribute to a number of human syndromes associated with microcephaly or other neurodevelopmental phenotypes.
Cardiovascular defects in 22q11.2 deletion syndrome
The heart develops through a complex morphogenetic sequence in which the linear heart tube gives rise to the atria, ventricles, and outflow tract (Rana et al., 2013). A role for p53 in driving heart defects in human syndromes has been suggested based on the analysis of 22q11.2 deletion syndrome (22q11.2DS). This syndrome is characterized by heart defects, including heart outflow tract defects and ventricular septal defects, as well as other diverse symptoms such as cleft palate, intellectual disability, and immune system dysfunction (McDonald-McGinn et al., 2015). While the chromosomal region deleted in this syndrome includes over 40 protein-coding genes, heterozygosity for the gene encoding the TBX1 transcription factor is thought to be a major contributor to the associated phenotypes (Gao et al., 2013). Specifically, in mice, Tbx1-deficiency causes heart defects associated with a reduction in proliferation in the developing heart, and these defects can be rescued by loss of Trp53 (Table 1) (Caprio and Baldini, 2014). While the mechanisms underlying the genetic interaction between p53 and Tbx1 are not yet clear, it was proposed that they have opposing functions in the regulation of genes involved in heart development (Caprio and Baldini, 2014). Further research will be needed to elucidate the relationship between Tbx1 and p53, and to determine whether other phenotypes associated with 22q11.2DS are also p53-dependent.
Neural tube defects in Waardenburg syndrome type 1
Neural tube defects are one of the most common types of birth defects and arise when the neural tube fails to close during development, causing neural tissue to protrude from the skull (exencephaly/anencephaly) or the spinal column (spina bifida) (Zohn, 2012). While most neural tube defects occur as isolated, sporadic defects, they can also occur in the context of certain genetic syndromes, including Waardenburg syndrome type 1 (Zohn, 2012). This syndrome is caused by mutations in PAX3 and is characterized primarily by pigmentation defects and hearing loss but is also associated with spina bifida and other neural tube defects (Hart and Miriyala, 2017). In mice, Pax3-deficiency leads to increased apoptosis in the neuroepithelium and triggers the neural tube defects spina bifida and exencephaly (Pani, 2002). Pax3-deficient embryos also exhibit heart outflow tract defects and lack limb musculature (Morgan et al., 2008). Interestingly, while Pax3 is a transcription factor that regulates genes involved in multiple developmental lineages, it also has a transactivation domain-independent role as a negative regulator of p53 (Boudjadi et al., 2018). In particular, Pax3 can bind to p53 and stimulate Mdm2-mediated degradation of p53, with loss Pax3 leading to increased p53 protein levels (Wang et al., 2011a). In mice, loss of Trp53 can rescue the neural tube defects in Pax3-deficient embryos, suggesting that Pax3 plays a critical role in restraining p53 activity during neural tube closure (Table 1) (Pani, 2002). Loss of p53 also rescues the heart defects but not the limb musculature defects in Pax3-deficient mice, suggesting that p53 hyperactivation contributes to some, but not all, of the developmental phenotypes associated with Waardenburg syndrome type 1 (Table 1) (Morgan et al., 2008).
Hyperpigmentation in dyskeratosis congenita
The melanin pigment, which gives skin and hair their color, is produced by melanocytes in the epidermis. During embryogenesis, melanoblasts arise from the neural crest, migrate throughout the developing dermis of the embryo, and subsequently migrate into the epidermis and differentiate into melanocytes (Bonaventure et al., 2013). A number of mouse strains with p53 hyperactivation exhibit hyperpigmentation defects in which the skin on the paws, tails, and ears darkens during the early postnatal period due to the progressive accumulation of melanocytes in the epidermis (Figure 1C) (Terzian et al., 2011; Simeonova et al., 2013; Pant et al., 2016). Skin hyperpigmentation is also one of the phenotypes associated with the bone marrow failure and cancer-predisposition syndrome dyskeratosis congenita (Dokal, 2011). In addition to pigmentation defects, patients with dyskeratosis congenita exhibit oral leukoplakia, nail dystrophy, and pulmonary fibrosis, and interestingly these phenotypes are also observed in mouse strains with p53 hyperactivation, suggesting a role for p53 in this syndrome (Figure 1C) (Simeonova et al., 2013). In support of this notion, lymphocytes from dyskeratosis congenita patients have elevated p53 levels, and loss of Trp53 can rescue the hyperpigmentation phenotype, the skeletal defects, and the testes defects in mouse models of this syndrome (Table 1) (Chin et al., 1999; Else et al., 2009; Vlangos et al., 2009; Pereboeva et al., 2016). Given that dyskeratosis congenita is caused by mutations in genes encoding components of the telomerase or shelterin complexes essential for telomere maintenance, it is thought that telomere attrition or dysfunction triggers a DNA damage response that activates p53 in this syndrome (Roake and Artandi, 2017). Moreover, some of the genes associated with dyskeratosis congenita are involved in processing rRNAs in addition to maintaining telomeres, and studies in zebrafish models suggest that impaired ribosome biogenesis also contributes to p53 hyperactivation in this syndrome (Pereboom et al., 2011; Zhang et al., 2012b). Together, these studies suggest that p53 activation, as a consequence of telomere dysfunction and impaired ribosome biogenesis, contributes to the pathology of dyskeratosis congenita.
Interestingly, in addition to driving skin hyperpigmentation, activation of p53 can also cause decreased pigmentation. Specifically, an absence of pigment in the ventral region, which manifests as a white belly spot, is observed in some Mdm2+/−;Mdm4+/− mice and in other mouse models associated with p53 hyperactivation, including mice carrying mutations in Rps19, Rps7, Rpl24, or Rack1 (McGowan et al., 2008; Barkić et al., 2009; Volta et al., 2013; Watkins-Chow et al., 2013; Zhang et al., 2017). While it may seem paradoxical that activation of p53 can trigger both increased and decreased pigmentation, the etiology of these phenotypes is likely distinct. In particular, p53 activation is thought to promote skin darkening at postnatal stages by inducing the p53 target gene Kitl in keratinocytes, leading to increased production of the Kitl growth factor that signals to melanocytes and promotes their proliferation (McGowan et al., 2008; Pant et al., 2016; Chang et al., 2017). In contrast, p53 activation is thought to promote white belly spots by decreasing the number of melanoblasts that arise from the neuroepithelium at embryonic stages (McGowan et al., 2008; Watkins-Chow et al., 2013). Thus, hyper- and hypo-pigmentation phenotypes likely arise due to different effects of p53 activation at different developmental stages.
CHARGE syndrome
CHARGE syndrome, caused by mutations in the gene encoding the CHD7 chromatin remodeler, is an acronym for its major symptoms, including coloboma, heart defects, atresia of the choana, retarded growth and development, genital hypoplasia, and ear defects (Zentner et al., 2010). A role for p53 in CHARGE syndrome was initially proposed based on the observation that the specific set of developmental defects triggered by p53 activation in Trp5325,26,53,54/+ mice (Figure 1C) is remarkably similar to the set of defects associated with CHARGE syndrome. In particular, the specific combination of inner ear defects, outer ear defects, heart defects, and retinal coloboma observed in Trp5325,26,53,54/+ mice is characteristic of CHARGE syndrome (Zentner et al., 2010; Van Nostrand et al., 2014). Further analyses revealed that p53 activity is indeed increased in human CHARGE syndrome, with fibroblasts from CHARGE patients and thymus tissue from CHARGE fetuses displaying elevated p53 protein levels (Van Nostrand et al., 2014). Moreover, reducing the dosage of Trp53 can partially rescue the developmental delay and severe hypoplasia of E10.5 Chd7−/− mouse embryos, suggesting that increased p53 activity contributes to the phenotypes downstream of Chd7-deficiency (Van Nostrand et al., 2014). While the mechanisms by which deficiency for the Chd7 chromatin remodeler triggers p53 activation are not completely elaborated, it was found that Chd7 can bind to the Trp53 promoter, suggesting that Chd7 may repress Trp53 expression (Van Nostrand et al., 2014). Further research will be needed to elucidate these mechanisms and to further define the role for p53 in this syndrome.
Summary and future perspectives
In summary, the studies described here provide strong evidence that p53 can act as a driver of a wide spectrum of developmental defects. First, studies of mouse strains with p53 hyperactivation due to mutations in Trp53, Mdm2, or Mdm4 indicate that broadly activating p53 during embryogenesis can trigger surprisingly specific sets of developmental defects (Figure 1). Second, recent reports indicate that human patients carrying activating TP53 mutations or hypomorphic MDM2 mutations also display developmental defects. Finally, many human developmental syndromes are caused by mutations that indirectly trigger p53 activation, and data from mouse models of these syndromes suggest that this increase in p53 activity plays a direct role in contributing to their developmental defects (Figure 2 and Table 1). These studies raise a number of intriguing questions for further investigation. The observation that mouse strains with broad p53 hyperactivation display tissue-specific developmental defects suggests that only certain embryonic and neonatal cell types are sensitive to p53 activation. However, the molecular basis for the enhanced sensitivity of these specific cell types remains to be elucidated. Furthermore, the complete spectrum of cellular responses elicited by p53 activation in these cells has not yet been defined. While many of the studies discussed here indicate that inappropriate p53 activation can trigger apoptosis or restrain proliferation in certain embryonic cell types, the involvement of other p53-regulated cellular responses—such as ferroptosis, metabolic reprogramming, migration, and cellular differentiation (Kaiser and Attardi, 2018)—is not yet known. Another open question is what contributes to the phenotypic specificity of human syndromes. Although the syndromes discussed here are all associated with increased p53 activity, each syndrome is phenotypically unique, and it is not clear how p53 activation drives different phenotypes in different syndromes. It will be of interest to determine whether these phenotypic differences arise because different causative mutations trigger p53 activation in different cell compartments, at different developmental time points, or to different degrees in each of these syndromes, or whether these phenotypic differences arise due to differences in p53-independent pathways.
It will be important to determine whether the observations presented here can be leveraged for therapeutic benefit. While it is possible that direct pharmacological inhibition of p53 could prevent developmental defects in these syndromes, this approach may not be prudent given the critical role that p53 plays in tumor suppression. This approach should also be treated with caution because some phenotypes may be exacerbated by p53-inhibition. For example, in a mouse model of Fanconi Anemia, loss of Trp53 rescues the hematopoietic defects during early developmental stages but exacerbates the exhaustion of the stem cell pool in adult mice (Li et al., 2018). Thus, instead of directly inhibiting p53, strategies to rectify the upstream cellular defects and thereby prevent p53 hyperactivation may be of interest. For example, maternal antioxidant supplementation can decrease oxidative stress-induced DNA damage during embryogenesis, thereby dampening p53 activation in disorders associated with defective DNA repair (Sakai et al., 2016). Alternatively, inhibiting specific direct p53 target genes could be beneficial. For example, induction of Puma, a component of the apoptotic pathway, has been shown to be responsible for some of the phenotypes induced by p53 hyperactivation in mice, and a chemical inhibitor of Puma has recently been generated (Liu et al., 2010; Leibowitz et al., 2018). Thus, it will be of interest to determine whether specific developmental phenotypes could be rescued by Puma inhibition.
The list of syndromes associated with increased p53 activity continues to grow. For example, studies in zebrafish models suggest that inappropriate p53 activation also contributes to the developmental defects in syndromes caused by mutations affecting components of the spliceosome and proteins involved in sister chromatid cohesion (Percival et al., 2015; Lei et al., 2017). In addition, emerging evidence indicates that the developmental defects caused by maternal viral infections, environmental exposures, or metabolic disorders, such as Zika virus infection, fetal alcohol spectrum disorders, and maternal diabetes, respectively, are also associated with p53 hyperactivation (Slomnicki et al., 2017; Su et al., 2017; Yuan et al., 2017). Further unraveling the role of p53 in these and other developmental conditions may ultimately lead to novel strategies for their prevention.
Acknowledgements
We thank Anthony Boutelle and Nitin Raj for critical reading of this manuscript. We apologize to the investigators whose work could not be cited owing to space constraints.
Funding
This work was supported by a March of Dimes Foundation grant #6-FY15-189 (L.D.A.), R35 grant CA197591 (L.D.A.), and Jane Coffin Childs Fund Postdoctoral Fellowship (M.E.B.).
Conflict of interest
none declared.
References
- Adam J., Deans B., and Thacker J. (2007). A role for Xrcc2 in the early stages of mouse development. DNA Repair 6, 224–234. [DOI] [PubMed] [Google Scholar]
- Albers C.A., Paul D.S., Schulze H., et al. (2012). Compound inheritance of a low-frequency regulatory SNP and a rare null mutation in exon-junction complex subunit RBM8A causes TAR syndrome. Nat. Genet. 44, 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkuraya F.S., Cai X., Emery C., et al. (2011). Human mutations in NDE1 cause extreme microcephaly with lissencephaly. Am. J. Hum. Genet. 88, 536–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alt J.R. (2003). Mdm2 haplo-insufficiency profoundly inhibits Myc-induced lymphomagenesis. EMBO J. 22, 1442–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann T., and Gennery A.R. (2016). DNA ligase IV syndrome; a review. Orphanet J. Rare Dis. 11, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbiv O.A., Cuvelier G., Klaassen R.J., et al. (2018). Molecular analysis and genotype-phenotype correlation of Diamond–Blackfan anemia. Clin. Genet. 93, 320–328. [DOI] [PubMed] [Google Scholar]
- Armstrong J.F., Kaufman M.H., Harrison D.J., et al. (1995). High-frequency developmental abnormalities in p53-deficient mice. Curr. Biol. 5, 931–936. [DOI] [PubMed] [Google Scholar]
- Arora J., Saraswathy K.N., and Deb R. (2012). Effect of maternal Tp53 gene G412C polymorphism on neural tube defects: a study from North India. Indian J. Hum. Genet. 18, 177–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aspesi A., Monteleone V., Betti M., et al. (2017). Lymphoblastoid cell lines from Diamond Blackfan anaemia patients exhibit a full ribosomal stress phenotype that is rescued by gene therapy. Sci. Rep. 7, 12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baatout S., Jacquet P., Michaux A., et al. (2002). Developmental abnormalities induced by X-irradiation in p53 deficient mice. In Vivo 16, 215–221. [PubMed] [Google Scholar]
- Barkić M., Crnomarković S., Grabusić K., et al. (2009). The p53 tumor suppressor causes congenital malformations in Rpl24-deficient mice and promotes their survival. Mol. Cell. Biol. 29, 2489–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow J.L., Drynan L.F., Hewett D.R., et al. (2010). A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q− syndrome. Nat. Med. 16, 59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzi H., and Anderson K.V. (2014). Acentriolar mitosis activates a p53-dependent apoptosis pathway in the mouse embryo. Proc. Natl Acad. Sci. USA 111, E1491–E1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi F.T., Tocco C., Pallavicini G., et al. (2017). Citron kinase deficiency leads to chromosomal instability and TP53-sensitive microcephaly. Cell Rep. 18, 1674–1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieging K.T., Mello S.S., and Attardi L.D. (2014). Unravelling mechanisms of p53-mediated tumour suppression. Nat. Rev. Cancer 14, 359–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojesen S.E., and Nordestgaard B.G. (2008). The common germline Arg72Pro polymorphism of p53 and increased longevity in humans. Cell Cycle 7, 158–163. [DOI] [PubMed] [Google Scholar]
- Bonaventure J., Domingues M.J., and Larue L. (2013). Cellular and molecular mechanisms controlling the migration of melanocytes and melanoma cells. Pigment Cell Melanoma Res. 26, 316–325. [DOI] [PubMed] [Google Scholar]
- Boudjadi S., Chatterjee B., Sun W., et al. (2018). The expression and function of PAX3 in development and disease. Gene 666, 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwman P., Drost R., Klijn C., et al. (2011). Loss of p53 partially rescues embryonic development of Palb2 knockout mice but does not foster haploinsufficiency of Palb2 in tumour suppression. J. Pathol. 224, 10–21. [DOI] [PubMed] [Google Scholar]
- Bronner M.E., and Simões-Costa M. (2016). The neural crest migrating into the twenty-first century Curr. Top. Dev. Biol. 116, 115–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bursac S., Brdovcak M.C., Donati G., et al. (2014). Activation of the tumor suppressor p53 upon impairment of ribosome biogenesis. Biochim. Biophys. Acta 1842, 817–830. [DOI] [PubMed] [Google Scholar]
- Caprio C., and Baldini A. (2014). p53 suppression partially rescues the mutant phenotype in mouse models of DiGeorge syndrome. Proc. Natl Acad. Sci. USA 111, 13385–13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi R., Parmar K., Mouly E., et al. (2012). Bone marrow failure in fanconi anemia is triggered by an exacerbated p53/p21 DNA damage response that impairs hematopoietic stem and progenitor cells. Cell Stem Cell 11, 36–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.-H., Kuo C.-J., Ito T., et al. (2017). CK1α ablation in keratinocytes induces p53-dependent, sunburn-protective skin hyperpigmentation. Proc. Natl Acad. Sci. USA 114, E8035–E8044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavez-Reyes A., Parant J.M., Amelse L.L., et al. (2003). Switching mechanisms of cell death in mdm2- and mdm4-null mice by deletion of p53 downstream targets. Cancer Res. 63, 8664–8669. [PubMed] [Google Scholar]
- Chin L., Artandi S.E., Shen Q., et al. (1999). p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell 97, 527–538. [DOI] [PubMed] [Google Scholar]
- Chrzanowska K.H., Gregorek H., Dembowska-Bagińska B., et al. (2012). Nijmegen breakage syndrome (NBS). Orphanet J. Rare Dis. 7, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauwerse J.G., Dixon J., Seland S., et al. (2011). Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat. Genet. 43, 20–22. [DOI] [PubMed] [Google Scholar]
- de Villartay J.-P. (2015). When natural mutants do not fit our expectations: the intriguing case of patients with XRCC4 mutations revealed by whole-exome sequencing. EMBO Mol. Med. 7, 862–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon J., Jones N.C., Sandell L.L., et al. (2006). Tcof1/Treacle is required for neural crest cell formation and proliferation deficiencies that cause craniofacial abnormalities. Proc. Natl Acad. Sci. USA 103, 13403–13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dokal I. (2011). Dyskeratosis congenita. Hematology 2011, 480–486. [DOI] [PubMed] [Google Scholar]
- Dumont P., Leu J.I.-J., Della Pietra A.C., et al. (2003). The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat. Genet. 33, 357–365. [DOI] [PubMed] [Google Scholar]
- Dutt S., Narla A., Lin K., et al. (2011). Haploinsufficiency for ribosomal protein genes causes selective activation of p53 in human erythroid progenitor cells. Blood 117, 2567–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Else T., Trovato A., Kim A.C., et al. (2009). Genetic p53 Deficiency partially rescues the adrenocortical dysplasia phenotype at the expense of increased tumorigenesis. Cancer Cell 15, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favaro F.P., Alvizi L., Zechi-Ceide R.M., et al. (2014). A noncoding expansion in EIF4A3 causes Richieri-Costa-Pereira Syndrome, a craniofacial disorder associated with limb defects. Am. J. Hum. Genet. 94, 120–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank K.M., Sharpless N.E., Gao Y., et al. (2000). DNA ligase IV deficiency in mice leads to defective neurogenesis and embryonic lethality via the p53 pathway. Mol. Cell 5, 993–1002. [DOI] [PubMed] [Google Scholar]
- Frappart P.-O., Tong W.-M., Demuth I., et al. (2005). An essential function for NBS1 in the prevention of ataxia and cerebellar defects. Nat. Med. 11, 538–544. [DOI] [PubMed] [Google Scholar]
- Gao Y., Ferguson D.O., Xie W., et al. (2000). Interplay of p53 and DNA-repair protein XRCC4 in tumorigenesis, genomic stability and development. Nature 404, 897–900. [DOI] [PubMed] [Google Scholar]
- Gao S., Li X., and Amendt B.A. (2013). Understanding the role of Tbx1 as a candidate gene for 22q11.2 deletion syndrome. Curr. Allergy Asthma Rep. 13, 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cao I., García-Cao M., Martín-Caballero J., et al. (2002). ‘Super p53’ mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 21, 6225–6235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamard P.-J., Barthelery N., Hogstad B., et al. (2013). The C terminus of p53 regulates gene expression by multiple mechanisms in a target- and tissue-specific manner in vivo. Genes Dev. 27, 1868–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart J., and Miriyala K. (2017). Neural tube defects in Waardenburg syndrome: a case report and review of the literature. Am. J. Med. Genet. A 173, 2472–2477. [DOI] [PubMed] [Google Scholar]
- Hir H.L., Saulière J., and Wang Z. (2016). The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 17, 41–54. [DOI] [PubMed] [Google Scholar]
- Houlihan S.L., and Feng Y. (2014). The scaffold protein Nde1 safeguards the brain genome during S phase of early neural progenitor differentiation. eLife 3, e03297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insolera R., Bazzi H., Shao W., et al. (2014). Cortical neurogenesis in the absence of centrioles. Nat. Neurosci. 17, 1528–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson T.M., Hammond E.M., Giaccia A., et al. (2005). The p53QS transactivation-deficient mutant shows stress-specific apoptotic activity and induces embryonic lethality. Nat. Genet. 37, 145–152. [DOI] [PubMed] [Google Scholar]
- Jones N.C., Lynn M.L., Gaudenz K., et al. (2008). Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nat. Med. 14, 125–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S.N., Roe A.E., Donehower L.A., et al. (1995). Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature 378, 206–208. [DOI] [PubMed] [Google Scholar]
- Kaiser A.M., and Attardi L.D. (2018). Deconstructing networks of p53-mediated tumor suppression in vivo. Cell Death Differ. 25, 93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman M.H., Kaufman D.B., Brune R.M., et al. (1997). Analysis of fused maxillary incisor dentition in p53-deficient exencephalic mice. J. Anat. 191, 57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitami K., Kitami M., Kaku M., et al. (2018). BRCA1 and BRCA2 tumor suppressors in neural crest cells are essential for craniofacial bone development. PLoS Genet. 14, e1007340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruiswijk F., Labuschagne C.F., and Vousden K.H. (2015). p53 in survival, death and metabolic health: a lifeguard with a licence to kill. Nat. Rev. Mol. Cell Biol. 16, 393–405. [DOI] [PubMed] [Google Scholar]
- Lau M.C.C., Kwong E.M.L., Lai K.P., et al. (2016). Pathogenesis of POLR1C-dependent Type 3 Treacher Collins syndrome revealed by a zebrafish model. Biochim. Biophys. Acta 1862, 1147–1158. [DOI] [PubMed] [Google Scholar]
- Lee Y., Choi I., Kim J., et al. (2016). DNA damage to human genetic disorders with neurodevelopmental defects. J. Genet. Med. 13, 1–13. [Google Scholar]
- Lei L., Yan S.-Y., Yang R., et al. (2017). Spliceosomal protein eftud2 mutation leads to p53-dependent apoptosis in zebrafish neural progenitors. Nucleic Acids Res. 45, 3422–3436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibowitz B.J., Yang L., Wei L., et al. (2018). Targeting p53-dependent stem cell loss for intestinal chemoprotection. Sci. Transl. Med. 10, eaam7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessel D., Wu D., Trujillo C., et al. (2017). Dysfunction of the MDM2/p53 axis is linked to premature aging. J. Clin. Invest. 127, 3598–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Bielas S.L., Zaki M.S., et al. (2016). Biallelic mutations in citron kinase link mitotic cytokinesis to human primary microcephaly. Am. J. Hum. Genet. 99, 501–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Wilson A.F., Du W., et al. (2018). Cell-cycle-specific function of p53 in Fanconi anemia hematopoietic stem and progenitor cell proliferation. Stem Cell Rep. 10, 339–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim D.S., and Hasty P. (1996). A mutation in mouse rad51 results in an early embryonic lethal that is suppressed by a mutation in p53. Mol. Cell. Biol. 16, 7133–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T.X., Howlett N.G., Deng M., et al. (2003). Knockdown of zebrafish Fancd2 causes developmental abnormalities via p53-dependent apoptosis. Dev. Cell 5, 903–914. [DOI] [PubMed] [Google Scholar]
- Liu D., Ou L., Clemenson G.D., et al. (2010). Puma is required for p53-induced depletion of adult stem cells. Nat. Cell Biol. 12, 993–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Terzian T., Xiong S., et al. (2007). The p53-Mdm2 network in progenitor cell expansion during mouse postnatal development. J. Pathol. 213, 360–368. [DOI] [PubMed] [Google Scholar]
- Maier B. (2004). Modulation of mammalian life span by the short isoform of p53. Genes Dev. 18, 306–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamrak N.E., Shimamura A., and Howlett N.G. (2017). Recent discoveries in the molecular pathogenesis of the inherited bone marrow failure syndrome Fanconi anemia. Blood Rev. 31, 93–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao H., McMahon J.J., Tsai Y.-H., et al. (2016). Haploinsufficiency for core exon junction complex components disrupts embryonic neurogenesis and causes p53-mediated microcephaly. PLoS Genet. 12, e1006282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marjanović M., Sánchez-Huertas C., Terré B., et al. (2015). CEP63 deficiency promotes p53-dependent microcephaly and reveals a role for the centrosome in meiotic recombination. Nat. Commun. 6, 7676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald-McGinn D.M., Sullivan K.E., Marino B., et al. (2015). 22q11.2 deletion syndrome. Nat. Rev. Dis. Primer 1, 15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan K.A., Li J.Z., Park C.Y., et al. (2008). Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nat. Genet. 40, 963–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon J.J., Miller E.E., and Silver D.L. (2016). The exon junction complex in neural development and neurodevelopmental disease. Int. J. Dev. Neurosci. 55, 117–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello S.S., Valente L.J., Raj N., et al. (2017). A p53 super-tumor suppressor reveals a tumor suppressive p53–Ptpn14–Yap axis in pancreatic cancer. Cancer Cell 32, 460–473.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa S.M. (2006). Tumor suppression and normal aging in mice with constitutively high p53 activity. Genes Dev. 20, 16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrysa S.M., McElwee M.K., Michalowski J., et al. (2003). mdm2 is critical for inhibition of p53 during lymphopoiesis and the response to ionizing irradiation. Mol. Cell. Biol. 23, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlody B., Wruck W., Martins S., et al. (2017). Nijmegen Breakage Syndrome fibroblasts and iPSCs: cellular models for uncovering disease-associated signaling pathways and establishing a screening platform for anti-oxidants. Sci. Rep. 7, 7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montes de Oca Luna R., Wagner D.S., and Lozano G. (1995). Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature 378, 203–206. [DOI] [PubMed] [Google Scholar]
- Morgan S.C., Lee H.-Y., Relaix F., et al. (2008). Cardiac outflow tract septation failure in Pax3-deficient embryos is due to p53-dependent regulation of migrating cardiac neural crest. Mech. Dev. 125, 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer S.M., Larsson C.A., and Lozano, G. (2017). Mdm proteins: critical regulators of embryogenesis and homoeostasis. J. Mol. Cell Biol. 9, 16–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg E.A., and Holland A.J. (2018). Once and only once: mechanisms of centriole duplication and their deregulation in disease. Nat. Rev. Mol. Cell Biol. 19, 297–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noack Watt K.E., Achilleos A., Neben C.L., et al. (2016). The roles of RNA polymerase I and III subunits Polr1c and Polr1d in craniofacial development and in zebrafish models of Treacher Collins syndrome. PLoS Genet. 12, e1006187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani L., Horal M., and Loeken M.R. (2002). Rescue of neural tube defects in Pax-3-deficient embryos by p53 loss of function: implications for Pax-3- dependent development and tumorigenesis. Genes Dev. 16, 676–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pant V., Xiong S., Chau G., et al. (2016). Distinct downstream targets manifest p53-dependent pathologies in mice. Oncogene 35, 5713–5721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parant J., Chavez-Reyes A., Little N.A., et al. (2001). Rescue of embryonic lethality in Mdm4-null mice by loss of Trp53 suggests a nonoverlapping pathway with MDM2 to regulate p53. Nat. Genet. 29, 92–95. [DOI] [PubMed] [Google Scholar]
- Pellagatti A., Marafioti T., Paterson J.C., et al. (2010). Induction of p53 and up-regulation of the p53 pathway in the human 5q− syndrome. Blood 115, 2721–2723. [DOI] [PubMed] [Google Scholar]
- Percival S.M., Thomas H.R., Amsterdam A., et al. (2015). Variations in dysfunction of sister chromatid cohesion in esco2 mutant zebrafish reflect the phenotypic diversity of Roberts syndrome. Dis. Model. Mech. 8, 941–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereboeva L., Hubbard M., Goldman F.D., et al. (2016). Robust DNA damage response and elevated reactive oxygen species in TINF2-mutated dyskeratosis congenita cells. PLoS One 11, e0148793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereboom T.C., van Weele L.J., Bondt A., et al. (2011). A zebrafish model of dyskeratosis congenita reveals hematopoietic stem cell formation failure resulting from ribosomal protein-mediated p53 stabilization. Blood 118, 5458–5465. [DOI] [PubMed] [Google Scholar]
- Perry M.E. (2010). The regulation of the p53-mediated stress response by MDM2 and MDM4. Cold Spring Harb. Perspect. Biol. 2, a000968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rana M.S., Christoffels V.M., and Moorman A.F.M. (2013). A molecular and genetic outline of cardiac morphogenesis. Acta Physiol. 207, 588–615. [DOI] [PubMed] [Google Scholar]
- Rinon A., Molchadsky A., Nathan E., et al. (2011). p53 coordinates cranial neural crest cell growth and epithelial-mesenchymal transition/delamination processes. Dev. Camb. Engl. 138, 1827–1838. [DOI] [PubMed] [Google Scholar]
- Roake C.M., and Artandi S.E. (2017). Control of cellular aging, tissue function, and cancer by p53 downstream of telomeres. Cold Spring Harb. Perspect. Med. 7, a026088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld J.A., Traylor R.N., Schaefer G.B., et al. 1q21.1 Study Group 2012). Proximal microdeletions and microduplications of 1q21.1 contribute to variable abnormal phenotypes. Eur. J. Hum. Genet. 20, 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah V.P., Attardi L.D., Mulligan G.J., et al. (1995). A subset of p53-deficient embryos exhibit exencephaly. Nat. Genet. 10, 175–180. [DOI] [PubMed] [Google Scholar]
- Saifudeen Z., Dipp S., Stefkova J., et al. (2009). p53 Regulates metanephric development. J. Am. Soc. Nephrol. 20, 2328–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D., Dixon J., Achilleos A., et al. (2016). Prevention of Treacher Collins syndrome craniofacial anomalies in mouse models via maternal antioxidant supplementation. Nat. Commun. 7, 10328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D., and Trainor P.A. (2016). Face off against ROS: Tcof1/Treacle safeguards neuroepithelial cells and progenitor neural crest cells from oxidative stress during craniofacial development. Dev. Growth Differ. 58, 577–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simeonova I., Jaber S., Draskovic I., et al. (2013). Mutant mice lacking the p53 C-terminal domain model telomere syndromes. Cell Rep. 3, 2046–2058. [DOI] [PubMed] [Google Scholar]
- Slomnicki L.P., Chung D.-H., Parker A., et al. (2017). Ribosomal stress and Tp53-mediated neuronal apoptosis in response to capsid protein of the Zika virus. Sci. Rep. 7, 16652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su D., Guan L., Gao Q., et al. (2017). ROCK1/p53/NOXA signaling mediates cardiomyocyte apoptosis in response to high glucose in vitro and vivo. Biochim. Biophys. Acta 1863, 936–946. [DOI] [PubMed] [Google Scholar]
- Szybińska A., and Leśniak W. (2017). P53 dysfunction in neurodegenerative diseases—the cause or effect of pathological changes? Aging Dis. 8, 506–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tateossian H., Morse S., Simon M.M., et al. (2015). Interactions between the otitis media gene, Fbxo11, and p53 in the mouse embryonic lung. Dis. Model Mech. 8, 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T., Dumble M., Arbab F., et al. (2011). Rpl27a mutation in the sooty foot ataxia mouse phenocopies high p53 mouse models. J. Pathol. 224, 540–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzian T., Wang Y., Van Pelt C.S., et al. (2007). Haploinsufficiency of Mdm2 and Mdm4 in tumorigenesis and development. Mol. Cell. Biol. 27, 5479–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurow H.S., Haack R., Hartwig F.P., et al. (2011). TP53 gene polymorphism: importance to cancer, ethnicity and birth weight in a Brazilian cohort. J. Biosci. 36, 823–831. [DOI] [PubMed] [Google Scholar]
- Toki T., Yoshida K., Wang R., et al. (2018). De novo mutations activating germline TP53 in an inherited bone-marrow-failure syndrome. Am. J. Hum. Genet. 103, 440–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonks I.D., Walker G.J., Mould A.W., et al. (2012). Brca1 is involved in establishing murine pigmentation in a p53 and developmentally specific manner: letter to the editor. Pigment Cell Melanoma Res. 25, 530–532. [DOI] [PubMed] [Google Scholar]
- Tyner S.D., Venkatachalam S., Choi J., et al. (2002). p53 mutant mice that display early ageing-associated phenotypes. Nature 415, 45–53. [DOI] [PubMed] [Google Scholar]
- Valdez J.M., Nichols K.E., and Kesserwan C. (2017). Li-Fraumeni syndrome: a paradigm for the understanding of hereditary cancer predisposition. Br. J. Haematol. 176, 539–552. [DOI] [PubMed] [Google Scholar]
- Van Nostrand J.L., and Attardi L.D. (2014). Guilty as CHARGED: p53’s expanding role in disease. Cell Cycle 13, 3798–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand J.L., Brady C.A., Jung H., et al. (2014). Inappropriate p53 activation during development induces features of CHARGE syndrome. Nature 514, 228–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M., Geneviève D., Ostertag A., et al. (2016). Treacher Collins syndrome: a clinical and molecular study based on a large series of patients. Genet. Med. 18, 49–56. [DOI] [PubMed] [Google Scholar]
- Vlangos C.N., O’Connor B.C., Morley M.J., et al. (2009). Caudal regression in adrenocortical dysplasia (acd) mice is caused by telomere dysfunction with subsequent p53-dependent apoptosis. Dev. Biol. 334, 418–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volta V., Beugnet A., Gallo S., et al. (2013). RACK1 depletion in a mouse model causes lethality, pigmentation deficits and reduction in protein synthesis efficiency. Cell. Mol. Life Sci. 70, 1439–1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Kon N., Lasso G., et al. (2016). Acetylation-regulated interaction between p53 and SET reveals a widespread regulatory mode. Nature 538, 118–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.V., Leblanc M., Fox N., et al. (2011. b). Fine-tuning p53 activity through C-terminal modification significantly contributes to HSC homeostasis and mouse radiosensitivity. Genes Dev. 25, 1426–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.D., Morgan S.C., and Loeken M.R. (2011. a). Pax3 stimulates p53 ubiquitination and degradation independent of transcription. PLoS One 6, e29379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins-Chow D.E., Cooke J., Pidsley R., et al. (2013). Mutation of the Diamond-Blackfan anemia gene Rps7 in mouse results in morphological and neuroanatomical phenotypes. PLoS Genet. 9, e1003094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.E., Garcia I., Crowther A.J., et al. (2015). Aspm sustains postnatal cerebellar neurogenesis and medulloblastoma growth in mice. Development 142, 3921–3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu D., and Prives C. (2018). Relevance of the p53–MDM2 axis to aging. Cell Death Differ. 25, 169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Qiao W., Linke S.P., et al. (2001). Genetic interactions between tumor suppressors Brca1 and p53 in apoptosis, cell cycle and tumorigenesis. Nat. Genet. 28, 266–271. [DOI] [PubMed] [Google Scholar]
- Yelick P.C., and Trainor P.A. (2015). Ribosomopathies: global process, tissue specific defects. Rare Dis. 3, e1025185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan F., Chen X., Liu J., et al. (2017). Up-regulation of Siah1 by ethanol triggers apoptosis in neural crest cells through p38 MAPK-mediated activation of p53 signaling pathway. Arch. Toxicol. 91, 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaqout S., Morris-Rosendahl D., and Kaindl A. (2017). Autosomal recessive primary microcephaly (MCPH): an update . Neuropediatrics 48, 135–142. [DOI] [PubMed] [Google Scholar]
- Zentner G.E., Layman W.S., Martin D.M., et al. (2010). Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am. J. Med. Genet. A 152A, 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., He X., Chen L., et al. (2012. a). Synergistic regulation of p53 by Mdm2 and Mdm4 is critical in cardiac endocardial cushion morphogenesis during heart development. J. Pathol. 228, 416–428. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Morimoto K., Danilova N., et al. (2012. b). Zebrafish models for dyskeratosis congenita reveal critical roles of p53 activation contributing to hematopoietic defects through RNA processing. PLoS One 7, e30188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Xie Y., Zhou Y., et al. (2017). p53 pathway is involved in cell competition during mouse embryogenesis. Proc. Natl Acad. Sci. USA 114, 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zohn I.E. (2012). Mouse as a model for multifactorial inheritance of neural tube defects. Birth Defects Res. C Embryo Today 96, 193–205. [DOI] [PubMed] [Google Scholar]