Abstract
Purpose
Glaucoma is characterized by optic nerve damage and retinal ganglion cell loss. The glycoprotein neuromedin B-associated (Gpnmb) gene is well-known to be involved in the glaucoma disease process. The purpose of this study is to identify a downstream gene through which Gpnmb affects the glaucoma phenotypes using a systems genetics approach.
Methods
Retinal gene expression data for the BXD recombinant inbred (RI) strains (n=75) have previously been generated in our laboratory for a glaucoma study, and these data were used for genetic and bioinformatics analysis. Expression quantitative trait locus (eQTL) mapping and genetic correlation methods were used to identify a gene downstream of Gpnmb. Gene-set enrichment analysis was used to evaluate gene function and to construct coexpression networks.
Results
The level of Gpnmb expression is associated with a highly statistically significant cis-eQTL. Stanniocalcin 1 (Stc1) has a significant trans-eQTL mapping to the Gpnmb locus. The expression of Gpnmb and Stc1 is highly correlated in the retina and other tissues, as well as with glaucoma-related phenotypes. Gene Ontology and pathway analysis showed that Stc1 and its covariates are highly associated with apoptosis, oxidative stress, and mitochondrial activity. A generated gene network indicated that Gpnmb and Stc1 are directly connected to and interact with other genes with similar biologic functions.
Conclusions
These results suggest that Stc1 may be a downstream candidate of Gpnmb, and that both genes interact with other genes in a network to develop glaucoma pathogenesis through mechanisms such as apoptosis and oxidative stress.
Introduction
Glaucoma is currently the second leading cause of blindness in the United States, and affects millions worldwide. In 2010, approximately 2.22 million people were affected by open-angle glaucoma (OAG) in the United States, and more than 70 million people worldwide had glaucoma [1-4]. Glaucoma is a genetically heterogenous group of eye diseases that results in optic nerve damage and retinal ganglion cell (RGC) loss usually associated with elevated intraocular pressure (IOP) and vision loss. Unfortunately, most cases of glaucoma are asymptomatic until after sight is lost [5]. Glaucoma damage is irreversible, and therapeutic interventions are effective only at early stages of the disease.
Pigmentary glaucoma (PG), a form of secondary open-angle glaucoma, is the second most common form of glaucoma in young adults. In this disease, pigment sloughs off the posterior iris and damages the drainage structure of the eye, which, in turn, attenuates or blocks the flow of aqueous humor [6]. In most cases, the resultant IOP increase leads to glaucoma, yet in some cases, the increase does not, indicating that additional factors, including environment, age, and genetics, may influence disease progression [7]. The exact pathogenesis of glaucoma is not fully understood. However, RGC damage, oxidative stress, ischemia and hypoxia, abnormal immunity and inflammatory reactions, and apoptosis have all been found to play a part in the progression of glaucoma [8-13].
The glycoprotein neuromedin B-associated (Gpnmb; Gene ID: 93695, OMIM: 604368) gene is a gene that is known to affect pigmentary glaucoma. Gpnmb is associated with iris pigment dispersion (IPD), which is the breakdown of the posterior iris pigment epithelium [6]. Mutations in Gpnmb and tyrosinase-related protein 1 (Tyrp1; Gene ID: 22178, OMIM: 115501) have been found to contribute to IPD and iris stromal atrophy (ISA) in DBA/2J (D2) mice [14]. D2 mice are homozygous for mutations in Gpnmb and Tyrp1, and have a higher chance of developing pigmentary glaucoma (PG) [15-17]. The eyes of D2 mice have no obvious abnormalities before 3 months of age. However, by 6 months, most D2 mice begin to develop iris depigmentation, increased IOP, and damage to the optic nerve head (ONH) [18]. Because the progression of glaucoma in D2 mice is similar to that of humans, this strain has been used as a mouse model to study pigment dispersion syndrome (PDS) and PG [18]. Using these mice, Gpnmb has been found to interact with a large network of genes to affect the presentation of glaucoma [19]. Despite these findings, the direct downstream pathway through which Gpnmb contributes to glaucoma remains unknown.
The purpose of this investigation is to identify genes and pathways through which Gpnmb works to affect the glaucoma phenotype, using a systems genetics method and BXD mouse genetic reference population (GRP), one parental strain of which is D2 mouse. After expression quantitative trait locus (eQTL) mapping and genetic correlation analysis, we identified stanniocalcin 1 (Stc1) as a downstream candidate gene of Gpnmb with high variable expression among the BXD mice. The two parental strains of BXD GRP, D2 and B6 mice, express Gpnmb and Stc1 statistically significantly differently, which provides us with an opportunity to study expression variance of Gpnmb and Stc1 among the BXD population. Using retinal gene expression data of BXD mice, we identified a genetic network involving Gpnmb and Stc1, and explored pathways through which the two genes interact to affect the glaucoma-related phenotypes.
Methods
Gene expression database access
GeneNetwork website is a public web source that houses phenotype and transcriptome data from various tissues of BXD RI mice, including the eye and the retina. The gene expression data used in this study can be accessed using the data set “Full HEI Retina Illumina V6.2 (Apr10) RankInv” that we generated through collaborative effort for a glaucoma study [20]. This data set uses data from 75 BXD strains, their parental strains B6 and D2, and both reciprocal F1 hybrids between B6 and D2. Almost all mice were young adults of 60 to 90 days of age. All animals were housed with free access to standard laboratory chow and water, maintained on a 12 h:12 h light-dark cycle, and were killed via rapid cervical dislocation as described previously [21]. This study adheres to the ARVO Statement for Use of Animals in Research. All experimental protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Tennessee Health Science Center (Memphis, TN). More detailed information on this data set, including individual mouse, tissue harvest, RNA extraction, microarray hybridization, data normalization, etc., can be found at GeneNetwork267.
Heritability calculations
The heritability of Gpnmb and Stc1 expression was calculated using broad sense heritability that compares the genetic variation between strains to the environmental variance within strains [22]. The formula used for heritability calculation is 0.5Vg / (0.5Vg + Ve), where Vg is genetic variance (variances of strain means), and Ve is environmental variance. The 0.5 factor in this ratio was applied to adjust for the twofold increase of additive genetic variance among inbred strains relative to outbred populations [23].
Expression QTL mapping
Expression QTL (eQTL) mapping was performed using the WebQTL module on our GeneNetwork website with our published methods [21,24,25]. Regression analysis was used to determine the relation between differences in an expressed trait and differences in alleles at markers across the genome. Simple interval mapping identified potential eQTLs that regulate Gpnmb and Stc1 expression levels and estimated the statistical significance at each location using known genotype data for those sites. Composite interval mapping was also performed to control for genetic variance associated with major eQTLs, and therefore, identify any secondary eQTLs that may have been otherwise masked. Each analysis produced a likelihood ratio statistic (LRS) score, providing a quantitative measure of confidence of linkage between the observed phenotype—in this case, variation in the expression levels of Gpnmb and Stc1—and known genetic markers. The genome-wide significance for each eQTL was established using a permutation test that compared the LRS of our novel site with the LRS values for 1,000–10,000 genetic permutations [26].
Downstream gene analysis
Downstream genes, or genes whose expressions are affected by differential expression of Gpnmb, were identified using multiple criteria. First, the expression of the candidate downstream gene should be significantly correlated with the expression of Gpnmb by evaluation in both retina and other tissues. The candidate gene should also have an expression level greater than 8 units in retina tissue. The candidate gene should have a significant trans-eQTL at the Gpnmb locus, located in the 10 Mb interval between 44 Mb and 54 Mb of Chromosome (Chr) 6 where the Gpnmb gene is located. The gene expression should be represented by probes that hybridize in exons or 3’-UTR region. The expression of the candidate gene should be highly correlated with eye related phenotypes. Finally, published literature must support the gene’s biological function in the retina or eye.
Gene expression correlation analysis
Genetic correlation analysis was performed by computing Pearson product-moment correlations of the strain means between the expression of Gpnmb and the expression of all other transcripts across the mouse genome to produce a set of genetically correlated genes. Genes with an expression level greater than eight and a statistically significant correlation with Gpnmb (p<0.05) were selected for further analysis. Tissue correlation is an estimate of the similarity of expression of two genes across different tissues or organs from a single individual, which eliminates any genetic or environmental effect on the gene expression difference, and helps to find a true biologic connection between any pair of genes. Tissue correlation was performed by computing Spearman rank-order correlations between the expression of Gpnmb and its downstream candidates across 20 more multiple different tissues. Literature correlation was performed by examining the correlation coefficient (r) value of genes already described using similar terminology in published papers. The Semantic Gene Organizer was used to perform literature correlation, thus further filtering for a biologic correlation between Gpnmb and other genes based on literature reports [27]. Genes with r >0.5 were considered to have a high literature correlation. Riken clones, intergenic sequences, predicted genes, and probes not associated with functional mouse genes were eliminated to create a list of genes statistically significantly correlated with Gpnmb. Later, this process was used with the identified Gpnmb downstream gene Stc1 to calculate its highly correlated genes. These computations were all performed using tools on GeneNetwork.
Phenotype correlation analysis
The GeneNetwork website contains extensive phenotypic data sets ranging from behavioral to morphological to pharmacological. We queried the BXD published phenotypes database in GeneNetwork for eye-related traits, and focused the analysis on the traits that were statistically significantly correlated with Gpnmb and Stc1 expression in the retina (p<0.05). Then, we conducted 20,000 permutation tests as a more random and accurate measure of significance, and all traits that were no longer statistically significantly correlated after the permutation tests were eliminated.
Gene set enrichment analysis
To investigate Stc1 function and gene pathways through which Stc1 might play a role in retinal-related diseases, the top 500 genes with statistically significant genetic correlations (p<0.01) and literature correlations (r >0.58) with Stc1 were uploaded to the Webgestalt website for Gene Ontology (GO) and pathway analysis [28]. The p values generated from the hypergeometric test were automatically adjusted to account for multiple comparisons using the Benjamini and Hochberg correction [29]. The categories with an adjusted p value (adj P) of less than 0.05 indicated that the set of submitted genes were statistically significantly over-represented in those categories.
Gene network construction
The gene network was constructed and visualized using the Cytoscape utility through the Gene-set Cohesion Analysis Tool (GCAT). Nodes in the network represent genes, and edges between two nodes represent the cosine score of latent semantic indexing (LSI) that determines whether the functional coherence of the gene sets is larger than 0.6. The significance of the functional cohesion is evaluated by the observed number of gene relationships above a cosine threshold of 0.6 in the LSI model. The literature p value (LP) is calculated using Fisher’s exact test by comparing the cohesion of the given gene set to a random one [27].
Quantitative reverse transcription PCR
The expression levels of Gpnmb and Stc1 gene were verified with quantitative reverse transcription PCR (qRT-PCR). Retina tissue from D2 mice were harvested at 2–3 months (before the onset of glaucoma) and 6–7 months (after the onset of glaucoma) of age (≥3 samples per group). Total RNA was extracted with the RNeasy Mini Kit (Qiagen, Germantown, MD) following the manufacturer’s instructions. RNA concentration and purity were evaluated with NanoDrop (Thermo Scientific, Waltham, MA). The qRT-PCR reactions (20 μl total volume) were performed using the iTaq Universal SYBR Green One-Step Kit (Bio-Rad, Hercules, CA), including 4 μl RNA (20 ng/μl), 1.125 μl primers (5 μM), 7.5 μl 2X SYBR Green RT–PCR Reaction Mix, 0.3 μl iScript Reverse Transcriptase, and 2.075 μl Nuclease-free H2O. The RT step involved incubation at 50 °C for 10 min. The PCR cycling conditions included an initial denaturation of 95 °C for 5 min followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. The qRT-PCR reaction was performed on the CFX Connect Real-Time System (Bio-Rad). The sequences of the PCR primers were as follows: Gpnmb, forward 5′-GCT GGT CTT CGG ATG AAA ATG A-3′, reverse 5′-CCA CAA AGG TGA TAT TGG AAC CC-3′; Stc1, forward 5′-ACG AGG CGG AAC AAA ATG ATT-3′, reverse 5′-TGC ACT TTA AGC TCT CTT TGA CA-3′; Gapdh forward 5′-GGA GCC AAA AGG GTC ATC AT-3′, reverse 5′-GTG ATG GCA TGG ACT GTG GT-3′. The relative expression of each gene was normalized to Gapdh by using the ΔΔ2Ct method, and data were presented as mean ± standard deviation (SD).
Results
Gpnmb levels in the retinas of B6, D2, and BXD mice, and heritability
Gpnmb has highly variable expression among the BXD strains (Figure 1). The average expression across all BXD strains was 10.871±0.240 (mean ± standard error [SE]), with the parental B6 strain having the highest expression at 15.074±0.381 and the parental D2 strain having one of the lowest expressions at 7.560±0.473, indicating a statistically significant difference between the parental strains (p<0.01). The expression levels of most BXD strains are between B6 and D2 mice. The heritability of Gpnmb expression is 0.589, which indicates that genetic variability affects Gpnmb expression, and allows us to identify a locus that controls Gpnmb expression among BXD mice.
Figure 1.
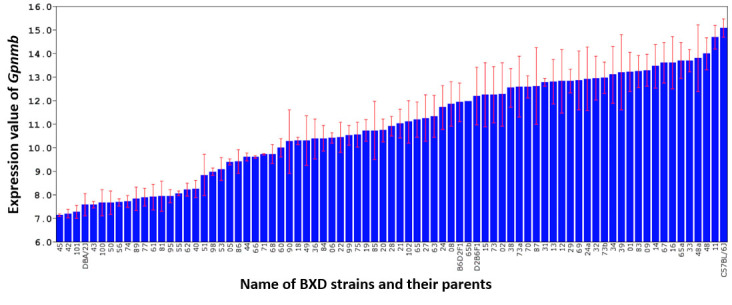
Expression level of Gpnmb in the retina of the B6 and D2 parental strains, F1 hybrids, and 75 BXD strains. The expression values for each sample were calculated using rank-invariant normalization through the BeadStudio software, and then renormalized using modified Z-scores. The x-axis denotes the strain while the y-axis denotes the expression of the strain mean on a log2 scale. Each bar represents the mean expression values ± standard error of the mean (SEM).
Gpnmb eQTL mapping and sequence variants
Simple interval mapping for Gpnmb revealed a statistically significant eQTL with an LRS of 43.0 on chromosome 6 at 49.66 megabases (Mb), which is close to where the Gpnmb gene itself is located chromosome 6 at 49.06 Mb (Figure 2). Composite interval mapping revealed no secondary locus that could modulate Gpnmb expression. This indicates that Gpnmb is cis-regulated, meaning that most of the variation in Gpnmb expression is caused by sequence variants in or near Gpnmb. Using our open access sequence data resources at GeneNetwork, we identified 37 single nucleotide polymorphisms (SNPs) in Gpnmb between the BXD parental strains (Table 1). Four SNPs are located in the coding region, including one nonsynonymous SNP, and the rest are located in the intron area. One or several of these SNPs are responsible for Gpnmb expression differences in the BXD mice.
Figure 2.
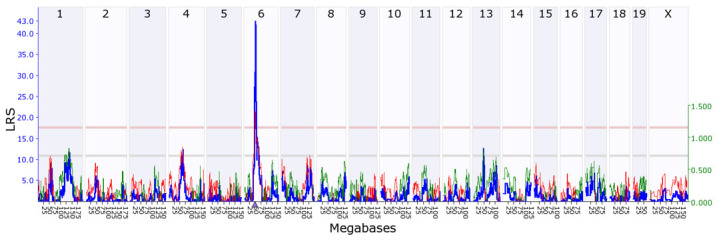
Genetic mapping of Gpnmb. A statistically significant expression quantitative trait locus (eQTL) was found at chromosome 6, which is within 1 Mb of the location of the Gpnmb gene itself (triangle), thus indicating that it is a cis-eQTL. The upper x-axis shows the chromosome, the lower x-axis shows the location in megabases, the left y-axis provides the likelihood ratio statistic (LRS) score in blue, and the right y-axis provides the additive effect. The red and green lines show the effect of the D or B allele on trait values, respectively. The pink (upper) horizontal line across the plot indicates the threshold for genome-wide statistical significance (p<0.05), and the gray (lower) horizontal line indicates a suggestive threshold (p<0.63).
Table 1. The single nucleotide polymorphisms (SNPs) in Gpnmb.
| SNP ID | Chr | Mb | Alleles | Location | Function | B6 | D2 |
|---|---|---|---|---|---|---|---|
| rs30410930 | 6 | 49.037092 | C/T | Intron | non-coding | C | T |
| rs30802297 | 6 | 49.037483 | G/A | Intron | non-coding | G | A |
| rs30067487 | 6 | 49.037502 | T/C | Intron | non-coding | T | C |
| rs30116878 | 6 | 49.037659 | T/C | Intron | non-coding | T | C |
| rs29872743 | 6 | 49.038953 | G/T | Intron | non-coding | G | T |
| rs36282636 | 6 | 49.039072 | C/T | Intron | non-coding | C | T |
| rs30214419 | 6 | 49.039074 | A/T | Intron | non-coding | A | T |
| rs36887641 | 6 | 49.039302 | A/G | Intron | non-coding | A | G |
| rs30905960 | 6 | 49.040254 | C/T | Intron | non-coding | C | T |
| rs30068479 | 6 | 49.040837 | A/G | Intron | non-coding | A | G |
| rs29982549 | 6 | 49.041336 | A/T | Intron | non-coding | A | T |
| rs30759090 | 6 | 49.041901 | T/C | Intron | non-coding | T | C |
| rs30507850 | 6 | 49.042050 | G/A | Intron | non-coding | G | A |
| rs13478745 | 6 | 49.042801 | G/A | Exon 2 | Nonsynonymous | G | A |
| rs30847773 | 6 | 49.043742 | C/T | Intron | non-coding | C | T |
| rs30318156 | 6 | 49.044057 | T/C | Exon 3 | Synonymous | T | C |
| rs30562316 | 6 | 49.044496 | T/C | Intron | non-coding | T | C |
| rs30174757 | 6 | 49.044558 | T/C | Intron | non-coding | T | C |
| rs30029717 | 6 | 49.045223 | G/A | Intron | non-coding | G | A |
| rs30606480 | 6 | 49.045317 | A/C | Exon 4 | Synonymous | A | C |
| rs38378746 | 6 | 49.045534 | T/G | Intron | non-coding | T | G |
| rs30022295 | 6 | 49.046593 | A/C | Intron | non-coding | A | C |
| rs30898406 | 6 | 49.048592 | G/A | Intron | non-coding | G | A |
| rs30764819 | 6 | 49.050199 | A/G | Intron | non-coding | A | G |
| rs30267325 | 6 | 49.051157 | A/G | Intron | non-coding | A | G |
| rs38325869 | 6 | 49.052202 | G/A | Intron | non-coding | G | A |
| rs30219902 | 6 | 49.052349 | C/G | Intron | non-coding | C | G |
| rs30081162 | 6 | 49.053562 | A/C | Intron | non-coding | A | C |
| rs36774657 | 6 | 49.054930 | G/A | Intron | non-coding | G | A |
| rs30615648 | 6 | 49.055229 | A/G | Intron | non-coding | A | G |
| rs30667356 | 6 | 49.055666 | C/T | Exon 10 | Synonymous | C | T |
| rs30963924 | 6 | 49.055885 | A/G | Intron | non-coding | A | G |
| rs30014223 | 6 | 49.056217 | G/A | Intron | non-coding | G | A |
| rs30219354 | 6 | 49.056238 | T/C | Intron | non-coding | T | C |
| rs37661404 | 6 | 49.056774 | C/T | Intron | non-coding | C | T |
| rs30914405 | 6 | 49.056860 | T/C | Intron | non-coding | T | C |
| rs38586314 | 6 | 49.057541 | A/T | Intron | non-coding | A | T |
Stc1 is a downstream candidate target of Gpnmb
Genetic mapping showed 11 statistically significant trans-eQTLs that were located within 5 Mb of Gpnmb (chromosome 6 from 44 to 54 Mb). After filtering with genetic correlation (p<0.05) and expression level (greater than 8), four genes were found to be statistically significantly correlated with Gpnmb and highly expressed in the retina. After further filtering with tissue correlation analysis, only Stc1 was found to be statistically significantly correlated with Gpnmb (p=0.0009, Figure 3). To validate expression of Gpnmb and Stc1 in the retina, we performed qRT-PCR for D2 mice before and after the onset of glaucoma. The results showed that expression of Gpnmb in D2 mice was decreased by approximately 60% in 6- to 7-month-old D2 mice when compared with 2- to 3-month-old D2 mice (p=0.039; Figure 4). For Stc1, the expression level decreased 20% in the 6- to 7-month-old D2 mice (p=0.059). In addition, correlation analysis revealed that the expression level of Stc1 was statistically significantly correlated with Gpnmb (p=0.044). Previous literature was consulted, and Stc1 was found to be the only gene with a biologic function connected to the retina [30]. Stc1 was chosen as the best downstream candidate gene of Gpnmb for further analysis.
Figure 3.
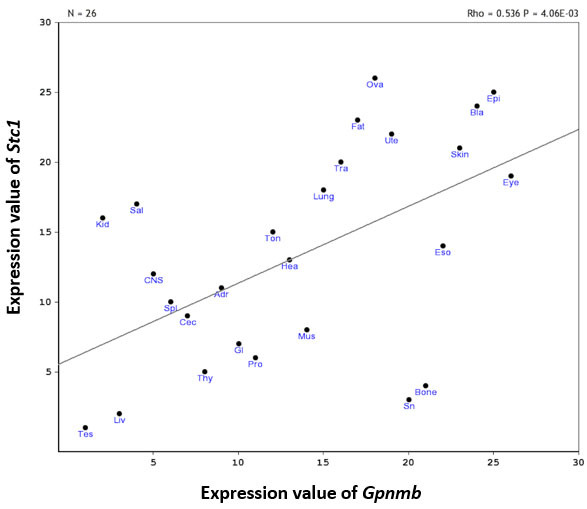
Scatterplots of Gpnmb and Stc1 expression across multiple different tissues. Each spot represents one tissue. The abbreviations for each tissue are listed in Appendix 2. There is a statistically significant positive correlation between Gpnmb and Stc1 expression (rho=0.536, p=0.00406, n=26).
Figure 4.
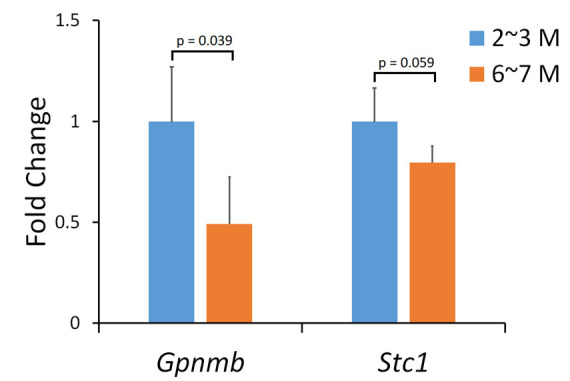
mRNA expression of Gpnmb and Stc1 in the retina of D2 mice. Each bar shows the mean relative expression value ± standard deviation (SD) using the ΔΔ cT method. The blue bar (left for each gene) shows the expression level for 2- to 3-month-old D2 mice, and the orange bar (right for each gene) shows the expression level for 6- to 7-month-old D2 mice. The y-axis denotes the fold change in the Gpnmb and Stc1 genes relative to 2-month-old D2 mice.
Stc1 expression levels in the retinas of B6, D2, and BXD mice, and heritability
Stc1 has highly variable expression among the BXD strains with the highest expression at 10.074±0.1480 in BXD 83 and the lowest expression at 8.715±0.089 in BXD51, indicating a 2.7-fold change in expression range (Figure 5). The parental B6 strain had expression of 10.074±0.1480, and the parental D2 strain had expression of 9.250±0.153, meaning that there is a statistically significant difference in expression between the two parental strains (p<0.05). Heritability of Stc1 was calculated to be 0.31, which indicates that Stc1 is affected by genetic variability, and allows us to identify a locus for Stc1.
Figure 5.
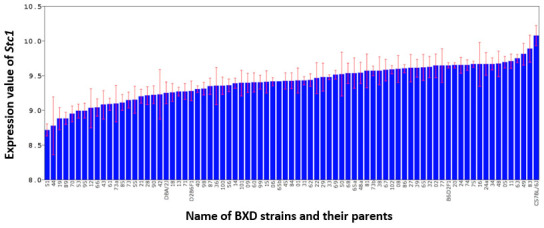
Expression level of Stc1 in the retina of the B6 and D2 parental strains, F1 hybrids, and 75 BXD strains. The expression value for each sample was calculated using the rank-invariant normalization method through BeadStudio software, and then renormalized using modified Z-scores. The x-axis denotes the strain; the y-axis denotes the expression of the strain mean on a log2 scale. Each bar represents the mean expression value ± standard error of the mean (SEM).
Stc1 eQTL mapping
Simple interval mapping for Stc1 found a statistically significant eQTL with an LRS of 19.6 (genome-wide p<0.05) on chromosome 6 at 48.92 Mb where the Gpnmb gene is located (Figure 6). The Stc1 gene itself is located on chromosome 14 at 69.04 Mb, which means that Stc1 has a trans-eQTL. Composite interval mapping showed no secondary locus that modulates Stc1 expression, suggesting that part of the expression variance in Stc1 is regulated by the DNA variants around the Gpnmb locus.
Figure 6.
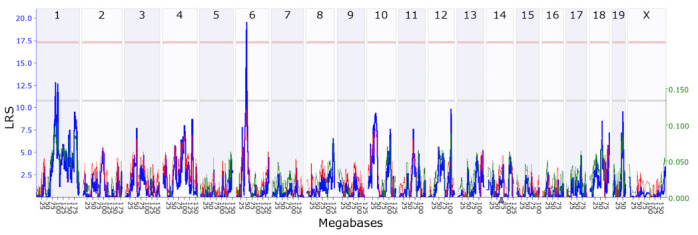
Genetic mapping of Stc1. A statistically significant expression quantitative trait locus (eQTL) was found at chromosome 6, which is different from the location of the Stc1 gene (indicated by a triangle, on chromosome 14), indicating that it is a trans-eQTL. The upper x-axis shows the chromosome, the lower x-axis shows the location in megabases, the left y-axis provides the likelihood ratio statistic (LRS) score in blue, and the right y-axis provides the additive effect. The red and green lines show the effect of the D or B allele on trait values, respectively. The pink (upper) horizontal line across the plot indicates the threshold for genome-wide statistical significance (p<0.05), and the gray (lower) horizontal line indicates a suggestive threshold (p<0.63).
Gene function enrichment
The top 500 probe sets were submitted to Webgestalt for gene function enrichment analysis. Of these probe sets, 26 could not be mapped to any Entrez gene ID or mapped to multiple gene IDs, resulting in 474 probe sets that could unambiguously map to 368 unique gene IDs. These genes were used for GO and pathway analysis.
The most statistically significant enrichment category for biologic processes was “cellular process” (n=321, adjP=2.26e-29) that includes “cell death” (n=66, adjP=1.84e-10), “programmed cell death” (n=64, adjP=1.16e-10), and “apoptotic process” (n=64, adjP=7.23e-11). The other most important and statistically significant enrichment categories included “mitochondrion” (n=67, adjP=2.01e-10) and “peroxisome” (n=8, adjP=1.86e-02) for cellular component; and “oxidoreductase activity” (n=30, adjP=0.0001), “ATP binding” (n=58, adjP=9.86e-08), “kinase activity” (n=44, adjP=1.68e-09), and “MAP kinase phosphatase activity” (n=5, adjP=3.15e-05) for molecular function. The GO results of all statistically significant enriched categories are listed in Appendix 1. Gene pathway analysis resulted in 21 statistically significant pathways (false discovery rate [FDR] p<0.05, Table 2), most of which are related to the electron transport chain, the mitochondrion, apoptosis, oxidative stress, the metabolism, the immune response, and inflammation.
Table 2. Significant enriched genes pathways.
| Pathway Name | N | FDR p value | Genes in the pathway |
|---|---|---|---|
| TCA Cycle | 7 | 0.0002 | Pdk1 Fh1 Pdk4 Idh2 Pdk2 Sdhd Dld |
| MAPK signaling pathway | 12 | 0.0031 | Dusp6 Dusp10 Stmn1 Ppp5c Dusp4 Mapk6 Akt3 Map4k4 Hspa1a Map3k12 Dusp1 Map3k5 |
| Focal Adhesion | 12 | 0.0037 | Ppp1r12a Vegfc Tnxb Rhob Pak7 Araf Mapk6 Pak4 Flt1 Akt3 Vegfb Itga9 |
| One carbon metabolism and related pathways | 6 | 0.0037 | Gclc Cth Gsr Gss Chka Gnmt |
| Glycolysis and Gluconeogenesis | 6 | 0.0037 | Tpi1 Pfkm Pfkp Slc2a3 Dld Hk1 |
| Amino Acid metabolism | 9 | 0.0041 | Fh1 Bcat1 Cth Gss Sdhd Glud1 Pdk4 Gsr Dld |
| Myometrial Relaxation and Contraction Pathways | 9 | 0.0209 | Rgs10 Ywhaz Rgs5 Camk2g Cald1 Rgs2 Prkd1 Igfbp2 Prkar1a |
| Glutathione and one carbon metabolism | 4 | 0.0214 | Gclc Cth Gsr Gss |
| Electron Transport Chain | 6 | 0.0271 | Slc25a4 Slc25a14 Atp5b Ndufs4 Atp5j Sdhd |
| Mitochondrial Gene Expression | 3 | 0.0319 | Pprc1 Tfam Nrf1 |
| Chemokine signaling pathway | 9 | 0.0319 | Nfkbib Cxcl14 Cxcl16 Prkx Akt3 Adrbk1 Cx3cl1 Nfkbia Csk |
| Calcium Regulation in the Cardiac Cell | 8 | 0.0319 | Atp1b1 Rgs10 Ywhaz Rgs5 Camk2g Rgs2 Prkd1 Prkar1a |
| Glutathione metabolism | 3 | 0.0319 | Gclc Gsr Gss |
| Apoptosis | 6 | 0.0403 | Xiap Nfkbib Bcl2l2 Dffa Nfkbia Cycs |
| EGFR1 Signaling Pathway | 10 | 0.0403 | Pebp1 Ralbp1 Araf Ceacam1 Git1 Prkd1 Dusp1 Csk Tgif1 Prkar1a |
| Insulin Signaling | 8 | 0.0408 | Mapk11 Mapk6 Pfkm Map4k4 Trib3 Gys1 Map3k12 Map3k5 |
| Regulation of Actin Cytoskeleton | 8 | 0.0408 | Ppp1r12a Pak7 Mapk6 Pak4 Limk1 Gsn Git1 Csk |
| Translation Factors | 4 | 0.0408 | Eif2b1 Eef1a2 Eif1a Eef2 |
| Oxidative Stress | 3 | 0.0408 | Gclc Gsr Txn2 |
| Integrin-mediated cell adhesion | 6 | 0.041 | Akt3 Araf Mapk6 Pak4 Csk Itga9 |
| B Cell Receptor Signaling Pathway | 9 | 0.041 | Dusp6 Plekha1 Dusp4 Pdk2 Cd81 Hnrnpk Prkd1 Nfkbia Csk |
Gene network
To generate a Gpnmb and Stc1 coexpression network, and investigate a biologic relationship through which these two genes interact in the retina, we focused on the gene group that contained Gpnmb and Stc1 within the statistically significant enrichment. We found that Gpnmb and Stc1, as well as 27 other genes, are regulated by activator protein 1 (AP-1). We then uploaded them to GCAT for functional coherence analysis and gene network construction. These genes showed statistically significant functional cohesion with a literature p value of 3.267436e-16 (Figure 7). The network graph indicated that the expression levels of Gpnmb and Stc1 are directly linked to each other. Multiple resources, such as Chilibot, GeneCard, and PubMed, were used to study the function of each member in the Gpnmb and Stc1 coexpression network. In addition to Gpnmb and Stc1, some of genes in this network (Pex5 [Gene ID: 19305, OMIM: 600414], Rgs2 [Gene ID: 19735, OMIM: 600861], and Cd68 [Gene ID: 12514, OMIM: 153634]) are already known to be related to glaucoma. Most of the other genes are highly connected to apoptosis, oxidative stress, and mitochondria.
Figure 7.
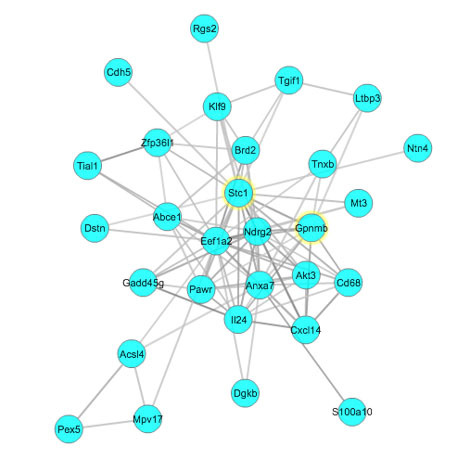
Stc1 genetic network. The Stc1 genetic network was created using the Gene-set Cohesion Analysis Tool as described in the Methods section. These genes may be functionally related. Gene symbols are located at the nodes in circles, and the lines interconnecting the nodes are based on literature correlations. The literature p value for these genes is 3.267436e-16.
Phenotype correlation
The results showed that Gpnmb expression is statistically significantly correlated with the optic nerve cross-sectional area, photoreceptor density, and iris pigmentation. Stc1 expression is statistically significantly correlated with optic nerve density and iris pigmentation. Among these correlated phenotypes, the iris pigmentation phenotype (graded from 0 (lowest) to 4 (highest) of iris transillumination) had a highly statistically significant negative correlation with the expression of Gpnmb and Stc1 (p<0.0005, Figure 8), indicating that Gpnmb and Stc1 play the same role in maintaining iris pigmentation.
Figure 8.
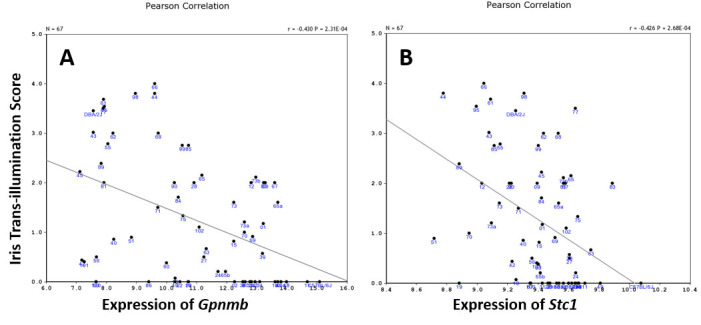
Scatterplots of correlation between iris transillumination and expression of Gpnmb and Stc1 among BXD strains. Each spot represents each BXD strain or their parental strain. There are statistically significant negative correlations between the iris transillumination score and the expression of Gpnmb (A) and Stc1 (B; p<0.0005).
Discussion
Systems genetics is an approach to understanding the flow of biologic information that underlies complex traits [31]. This approach involves analysis of sets of causal interactions among DNA variants (such as SNPs), classic traits (such as IOP), and intermediate phenotypes (such as transcripts) in populations that vary for traits of interest. The BXD recombinant inbred (RI) strains are the best mouse genetic reference population for a systems genetics study of glaucoma, because the parental strains, the D2 mouse and the B6 mouse, differ in the presentation of glaucoma. Their progeny have different phenotypes and gene expression, and provide a unique and valuable opportunity to define novel genes and molecular networks involved in glaucoma. In this study, we used the systems genetics method and BXD RI strains to analyze genes that are highly correlated with and lie downstream of Gpnmb, a gene involved with glaucoma. We found Stc1 to be highly correlated with Gpnmb, involved in the glaucoma disease process, and the best downstream candidate gene of Gpnmb. We present a genetic network for Stc1 that involves genes related to optic neuropathy, gene enrichment categories Stc1 plays a role in, and pathways through which Stc1 interacts with other genes to affect glaucoma phenotypes.
Stanniocalcin-1, encoded by Stc1, is a glycoprotein found to have antioxidant and antiapoptotic properties. Stc1 inhibits the inflammatory cascade, induces antioxidant and antiapoptotic mechanisms, and reduces reactive oxygen species [32-35]. In glaucoma, these factors have been shown to reduce IOP and provide neuroprotection in human eyes [36,37], as well as prevent the ensuing apoptosis in retinal photoreceptors [38,39]. Stc1 delays RGC apoptosis in the rat model of intraorbital optic nerve transection and decreases oxidative stress by reducing the reactive oxygen species (ROS) in the eye [40].
After gene function enrichment analysis, one of the most statistically significant biologic processes found to be associated with Stc1 was “apoptotic process,” as well as its two related pathways “chemokine signaling pathway” and “apoptosis.” Apoptosis, the programmed death of a cell, is an important process in the development of the nervous system. Apoptosis also has been implicated in various neurodegenerative diseases, such as glaucoma [41]. RGCs have been shown to die via apoptosis in experimental glaucoma, and after the axon of the nerve is severed, as defined by light microscopy [42]. Further investigations have shown that elevated IOP in the extracellular matrix of the retina is strongly correlated with RGC apoptosis in glaucoma [43]. The expression of apoptosis and inflammation-related genes, such as Il18 (Gene ID: 16173, OMIM: 600953), NF-κB (Gene ID: 18033, OMIM: 164011), Mapk1 (Gene ID: 26413, OMIM: 176948), Mmp-2 (Gene ID: 17390, OMIM: 120360), Timp-1 (Gene ID: 21857, OMIM: 305370), and apoptotic signaling components, has been found to be elevated in the DBA/2J glaucoma mouse model [44]. The present GO and pathway results support the role of Stc1 in apoptosis, and most likely in glaucoma.
One of the most statistically significant cellular components associated with Stc1 was “mitochondrion.” Neurons, which have a high-energy requirement, are dependent on mitochondria for not only their source of energy but also for calcium signaling and apoptosis. IOP elevation has also been linked to mitochondrial damage in the optic nerve head through mitochondrial fission and cristae depletion, although currently it is unknown whether IOP elevation leads to mitochondrial alterations [45]. When there is a mitochondrial malfunction, free radicals are produced in excess, which can lead to oxidative stress, common in glaucomatous tissues. This oxidative damage can damage cellular macromolecules, resulting in neuronal degeneration and RGC death [46,47]. Oxidative DNA damage has also been found to be statistically significantly increased in patients with glaucoma [48]. Further support for mitochondrial and oxidative stress involvement in Stc1 was shown in the present pathway analysis, where we found the “electron transport chain,” “mitochondrial gene expression,” and “oxidative stress” are statistically significant pathways.
Transcription factor analysis found AP-1 regulates Gpnmb and Stc1 gene expression. To study the interaction of Gpnmb and Stc1, we constructed a Gpnmb and Stc1 coexpression network based on the group of genes targeted by AP-1. The AP-1 family of transcription factors consists of homo- or heterodimers encoded by the fos and jun gene families. AP-1 regulates cell growth and differentiation, and is affected by the oxidative state of the cell [49]. AP-1 has been suggested to play a role in apoptosis, although the exact mechanism is not clear [50-52]. Considering the relationship AP-1 has with Gpnmb and Stc1 and with oxidative stress and apoptosis, AP-1 may affect the pathogenesis of glaucoma through the regulation of Gpnmb and other genes that interact with Gpnmb in this gene network.
In addition to Gpnmb, we found three genes (Pex5, Rgs2, and Cd68) in the gene network are highly correlated with glaucoma based on previous literature reports. Peroxisome biogenesis factor 5, encoded by Pex5, plays an essential role in peroxisomal protein import. The Pex5-knockout mouse model has peroxisomal metabolism defects, leading to pleomorphic mitochondria and a marked increase in ROS [53]. Zellweger syndrome spectrum is a syndrome in which there are no functional peroxisomes due to deletions or mutations in PEX (Gene ID: 5830, OMIM: 600414) genes [54]. Patients with Zellweger syndrome may exhibit characteristics such as craniofacial dysmorphia, neonatal seizures, and ocular anomalies, like cataracts, pigmentary retinopathy, and glaucoma [55,56].
Regulator of G protein signaling 2, encoded by Rgs2, acts as a GTPase activating protein. Rgs2 is widely expressed in the rat retina, particularly in the ganglion cell layer [57]. Mice with null Rgs2 expression have also been associated with lower IOP and enhanced RGC survival [58]. This is possibly because Rgs2 is a negative regulator of Gαq, which affects the constriction of smooth muscle [59-61]. Mice with null Rgs2 also had increased width in the Schlemm’s canal, which is close to a possible outflow pathway for aqueous humor in patients with open-angle glaucoma [62]. However, the exact mechanism through which Rgs2 affects IOP is not clear.
Cd68 encodes cluster of differentiation 68, a protein highly expressed in macrophages, and has been used as a marker for phagocytic amoeboid microglia in axonal injury [63,64]. Normally, Cd68 is not present in the optic nerve head. However, in glaucoma, Cd68 has been found to be present in microglia, indicating that there is phagocytic activity at the ONH and inflammatory activity in the stroma of the iris and ciliary body [65,66].
Several of the genes in the present gene network are also highly correlated with apoptosis, oxidative stress, and mitochondria based on previous literature. Il24 (Gene ID: 93672, OMIM: 604136) and Mt3 (Gene ID: 17751, OMIM: 139255) are associated with oxidative stress, Mpv17 (Gene ID: 17527, OMIM: 137960) is a mitochondrial protein, and Eef1a2 (Gene ID: 13628, OMIM: 602959) has antiapoptotic effects.
Interleukin 24 (IL-24), encoded by Il24, is a proinflammatory cytokine in the IL-20 subfamily of interleukins [67]. Il24 has proapoptotic characteristics in cancer cells [68,69]. Il24 expression has been found to be expressed in the retina of DBA/2J mice [70]. Il24 also induces the production of Il6, which is upregulated in the trabecular meshwork by oxidative stress, elevated IOP, and optic nerve head injury [71-73]. Mt3 encodes metallothionein 3, which is a protein that regulates zinc levels in the central nervous system in response to the oxidative status of the cell [74,75]. Under oxidative stress, zinc levels in the cytosol and lysosomes rise, resulting in increased cell death [76]. Mpv17 codes for a mitochondrial inner membrane protein, and its absence or malfunction has been found to cause oxidative phosphorylation depletion [77]. Mpv17 has also been implicated in mitochondrial DNA (mtDNA) depletion syndromes and oxidative phosphorylation activity in yeast [78,79]. Eukaryotic translation elongation factor 1 alpha 2 (Eef1a2) has been shown to have antiapoptotic effects [80-82].
Because these genes have been associated with either glaucoma or one of the many factors that affect the glaucoma phenotype, they may be associated with the glaucoma phenotype. However, further studies are needed to clarify the relationship between Gpnmb, Stc1, and these genes.
Conclusion
In summary, the present systems genetics analysis suggested that Stc1 could be a downstream candidate gene for Gpnmb. The gene function enrichment analysis of Gpnmb and Stc1 covariant genes suggests that Gpnmb may interact with Stc1 and other genes in the network to develop glaucoma pathogenesis through mechanisms of apoptosis and oxidative stress. However, further investigation is needed to elucidate the relationship among Gpnmb, Stc1, and the genes of the proposed genetic network.
Acknowledgments
This work was supported by NIH Grant R01EY021200 (LL, PI) and Research to Prevent Blindness Stein Innovation Award (Robert Williams, PI). This study was supported in part by the by the Natural Science Foundation of Jiangsu China Grant BK2008186 and BK20161287 (HL), and Science Foundation of Nantong China Grant HS2011005 and HS2014005 (HL). We thank Dr. Robert Williams for his financial and bioinformatics support for this study. We thank Dr. David Ashbrook for his editing manuscript. Dr. Lu Lu (lulu@uthsc.edu) and Dr. Jianping Tong (tongjp2000@hotmail.com) are co-corresponding authors for this paper.
Appendix 1. The gene ontology results of all significant enriched categories.
To access the data, click or select the words “Appendix 1.”
Appendix 2. The abbreviations for each tissue in figure 3.
To access the data, click or select the words “Appendix 2.”
References
- 1.Quigley HA, Broman AT. The number of people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol. 2006;90:262–7. doi: 10.1136/bjo.2005.081224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thylefors B, Négrel AD. The global impact of glaucoma. WHO Bulletin OMS. 1994;72:323–6. [PMC free article] [PubMed] [Google Scholar]
- 3.Leske MC. The epidemiology of open-angle glaucoma: a review. Am J Epidemiol. 1983;118:166–91. doi: 10.1093/oxfordjournals.aje.a113626. [DOI] [PubMed] [Google Scholar]
- 4.Quigley HA. Number of people with glaucoma worldwide. Br J Ophthalmol. 1996;80:389–93. doi: 10.1136/bjo.80.5.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Challa P. Glaucoma Genetics. Int Ophthalmol Clin. 2008;48:73–94. doi: 10.1097/IIO.0b013e318187e71a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Howell GR, Libby RT, Marchant JK, Wilson LA, Cosma IM, Smith RS, Anderson MG, John SWM. Absence of glaucoma in DBA/2J mice homozygous for wild-type version of Gpnmb and Tyrp1. BMC Genet. 2007;8 doi: 10.1186/1471-2156-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiggs JL. Genetic Etiologies of Glaucoma. Arch Ophthal. 2007;125:30–7. doi: 10.1001/archopht.125.1.30. [DOI] [PubMed] [Google Scholar]
- 8.Yu DY, Cringle SJ, Balaratnasingam C, Morgan WH, Yu PK, Su EN. Retinal ganglion cells: Energetics, compartmentation, axonal transport, cytoskeletons and vulnerability. Prog Retin Eye Res. 2013;36:217–46. doi: 10.1016/j.preteyeres.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Chen C, Xu Y, Zhang J, Zhu J, Zhang J, Hu N, Guan H. Altered expression of nNOS/NIDD in the retina of a glaucoma model of DBA/2J mice and the intervention by nNOS inhibition. J Mol Neurosci. 2013;51:47–56. doi: 10.1007/s12031-012-9943-x. [DOI] [PubMed] [Google Scholar]
- 10.Hong S, Iizuka Y, Lee T, Kim CY, Seong GJ. Neuroprotective and neurite outgrowth effects of maltol on retinal ganglion cells under oxidative stress. Mol Vis. 2014;20:1456–62. [PMC free article] [PubMed] [Google Scholar]
- 11.Vohra R, Tsai JC, Kolko M. The role of inflammation in the pathogenesis of glaucoma. Surv Ophthalmol. 2013;58:311–20. doi: 10.1016/j.survophthal.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 12.Li H, Liang Y, Chiu K, Yuan Q, Lin B, Chang RC, Son KF. Lycium barbarum (wolfberry) reduces secondary degeneration and oxidative stress, and inhibits JNK pathway in retina after partial optic nerve transection. PLoS One. 2013;8:e68881. doi: 10.1371/journal.pone.0068881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen SD, Wang L, Zhang XL. Neuroprotection in glaucoma: present and future. Chin Med J (Engl) 2013;126:1567–77. [PubMed] [Google Scholar]
- 14.Anderson MG, Smith RS, Hawes NL, Zabaleta A, Chang B, Wiggs JL, John SW. Mutations in genes encoding melanosomal proteins cause pigmentary glaucoma in DBA/2J mice. Nat Genet. 2002;30:81–5. doi: 10.1038/ng794. [DOI] [PubMed] [Google Scholar]
- 15.Mo JS, Anderson MG, Gregory M, Smith RS, Savinova OV, Serreze DV, Ksander BR, Streilein JW, John SWM. By Altering Ocular Immune Privilege, Bone Marrow-derived Cells pathogenically Contribute to DBA/2J Pigmentary Glaucoma. J Exp Med. 2003;197:1335–44. doi: 10.1084/jem.20022041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anderson MG, Nair KS, Amonoo LA, Mehalow A, Trantow CM, Masli S, John SWM. GpnmbR150X allele must be present in bone marrow derived cells to mediate DBA/2J glaucoma. BMC Genet. 2008;9 doi: 10.1186/1471-2156-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fan W, Li X, Wang W, Mo JS, Kaplan H, Cooper NGF. Early involvement of immune/inflammatory response genes in retinal degeneration in DBA/2J mice. Ophthalmol Eye Dis. 2010;1:23–41. doi: 10.4137/oed.s2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.John SW, Smith RS, Savinova OV, Hawes NL, Chang B, Turnbull D, Davisson M, Roderick TH, Heckenlively JR. Essential iris atrophy, pigment dispersion, and glaucoma in DBA/2J mice. Invest Ophthalmol Vis Sci. 1998;39:951–62. [PubMed] [Google Scholar]
- 19.Lu H, Wang X, Pullen M, Guan J, Chen H, Sahu S, Zhang B, Chen H, Williams RW, Geisert EE, Lu L, Jablonski MM. Genetic Dissection of the Gpnmb Network in the Eye. Invest Ophthalmol Vis Sci. 2011;52:4132–42. doi: 10.1167/iovs.10-6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Freeman NE, Templeton JP, Orr WE, Lu L, Williams RW, Geisert EE. Genetic networks in the mouse retina: Growth Associated Protein 43 and Phosphatase Tensin Homolog network. Mol Vis 2011; 17:1355–1372.King R, Lu L, Williams RW, Geisert EE. Transcriptome networks in the mouse retina: An exon level BXD RI database. Mol Vis. 2015;21:1235–51. [PMC free article] [PubMed] [Google Scholar]
- 21.King R, Lu L, Williams RW, Geisert EE. Transcriptome networks in the mouse retina: An exon level BXD RI database. Mol Vis. 2015;21:1235–51. [PMC free article] [PubMed] [Google Scholar]
- 22.Hegmann JP, Possidente B. Estimating genetic correlations from inbred strains. Behav Genet. 1981;11:103–14. doi: 10.1007/BF01065621. [DOI] [PubMed] [Google Scholar]
- 23.Geisert EE, Lu L, Freeman-Anderson NE, Templeton JP, Wang X, Gu W, Jiao Y, Williams RW. Gene expression in the mouse eye: an online resource for genetics using 103 strains of mice. Mol Vis. 2009;15:1730–63. [PMC free article] [PubMed] [Google Scholar]
- 24.Chesler EJ, Lu L, Shou S, Qu Y, Gu J, Wang J, Hsu HC, Mountz JD, Baldwin NE, Langston MA, Threadgill DW, Manly KF, Williams RW. Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat Genet. 2005;37:233–42. doi: 10.1038/ng1518. [DOI] [PubMed] [Google Scholar]
- 25.Lu H, Lu L, Williams RW, Jablonski MM. Iris transillumination defect and its gene modulators do not correlate with intraocular pressure in the BXD family of mice. Mol Vis. 2016;22:224–33. [PMC free article] [PubMed] [Google Scholar]
- 26.Churchill GA, Doerge RW. Empirical Threshold Values for Quantitative Trait Mapping. Genetics. 1994;138:963–71. doi: 10.1093/genetics/138.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Homayouni R, Heinrich K, Wei L, Berry MW. Gene clustering by latent semantic indexing of MEDLINE abstracts. Bioinformatics. 2005;21:104–15. doi: 10.1093/bioinformatics/bth464. [DOI] [PubMed] [Google Scholar]
- 28.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. JR Stat Soc. 1995;57:289–300. [Google Scholar]
- 30.Kim SH, Ko JH, Yun JH, Kim JA, Kim TE, Lee HJ, Kim SH, Park KH, Oh JY. Stanniocalcin-1 protects retinal ganglion cells by inhibiting apoptosis and oxidative damage. PLoS One. 2013;8:e63749. doi: 10.1371/journal.pone.0063749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Civelek M, Lusis AJ. Systems genetics approaches to understand complex traits. Nat Rev Genet. 2014;15:34–48. doi: 10.1038/nrg3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang SE, Wu CP, Wu SY, Peng CK, Perng WC, Kang BH, Chu SJ, Huang KL. Stanniocalcin-1 ameliorates lipopolysaccharide-induced pulmonary oxidative stress, inflammation, and apoptosis in mice. Free Radic Biol Med. 2014;71:321–31. doi: 10.1016/j.freeradbiomed.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 33.Sheikh-Hamad D. Mammalian stanniocalcin-1 activates mitochondrial antioxidant pathways: new paradigms for regulation of macrophages and endothelium. Am J Physiol Renal Physiol. 2010;298:F248–54. doi: 10.1152/ajprenal.00260.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang L, Belousova T, Chen M, DiMattia G, Liu D, Sheikh-Hamad D. Overexpression of stanniocalcin-1 inhibits reactive oxygen species and renal ischemia/reperfusion injury in mice. Kidney Int. 2012;82:867–77. doi: 10.1038/ki.2012.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu L, Guo R, Hui L, Ye Y, Xiang J, Wan C, Zou M, Ma R, Sun X, Yang S, Guo D. Stanniocalcin-1 protects bovine intestinal epithelial cells from oxidative stress-induced damage. J Vet Sci. 2014;15:475–83. doi: 10.4142/jvs.2014.15.4.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roddy GW, Viker KB, Winkler NS, Bahler CK, Bradley HH, Sheikh-Hamad D, Chowdhury UR, Stamer WD, Fautsch MP. Stanniocalcin-1 Is an Ocular Hypotensive Agent and a Downstream Effector Molecule That Is Necessary for the Intraocular Pressure-Lowering Effects of Latanoprost. Invest Ophthalmol Vis Sci. 2017;58:2715–24. doi: 10.1167/iovs.16-21004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roddy GW, Bahler CK, Holman BH, Fautsch MP. The neuroprotective protein Stanniocalcin-1 (STC-1) has ocular hypotensive properties in the human anterior segment organ culture model. Invest Ophthal Vis Sci 2014; 55. [Google Scholar]
- 38.Wang Y, Huang L, Abdelrahim M, Cai Q, Truong A, Bick R, Poindexter B, Sheikh-Hamad D. Stanniocalcin-1 suppresses superoxide generation in macrophages through induction of mitochondrial UCP2. J Leukoc Biol. 2009;86:981–988. doi: 10.1189/jlb.0708454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roddy GW, Rosa RH, Oh JY, Ylostalo JH, Bartosh TJ, Choi H, Lee RH, Yasumura D, Ahern K, Nielsen G, Matthes MT, LaVail MM, Prockop DJ. Stanniocalcin-1 Rescued Photoreceptor Degeneration in Two Rat Models of Inherited Retinal Degeneration. Mol Ther. 2012;20:788–97. doi: 10.1038/mt.2011.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim SJ, Ko JH, Yun J, Kim J, Kim TE, Lee HJ, Kim SH, Park KH, Oh JO.Stanniocalcin-1 Protects Retinal Ganglion Cells by Inhibiting Apoptosis and Oxidative Damage. PLoS One 20138e63749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nickells RW. Apoptosis of Retinal Ganglion Cells in Glaucoma: An Update of the Molecular Pathways Involved in Cell Death. Surv Ophthalmol. 1999;43:S151–61. doi: 10.1016/s0039-6257(99)00029-6. [DOI] [PubMed] [Google Scholar]
- 42.Quigley HA, Nickells RW, Kerrigan LA, Pease ME, Thibault DJ, Zack DJ. Retinal Ganglion Cell Death in Experimental Glaucoma and After Axotomy Occurs by Apoptosis. Invest Opthal Vis Sci. 1995;36:774–86. [PubMed] [Google Scholar]
- 43.Guo L, Moss SE, Alexander RA, Ali RR, Fitzke FW, Cordeiro MF. Retinal Ganglion Cell Apoptosis in Glaucoma Is Related to Intraocular Pressure and IOP-Induced Effects on Extracellular Matrix. Invest Ophthalmol Vis Sci. 2005;46:175–82. doi: 10.1167/iovs.04-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhou X, Li F, Kong L, Tomita H, Li C, Cao W. Involvement of Inflammation, Degradation, and Apoptosis in a Mouse Model of Glaucoma. J Biochem. 2005;280:31240–8. doi: 10.1074/jbc.M502641200. [DOI] [PubMed] [Google Scholar]
- 45.Ju WK, Kim KY, Lindsey JD, Angert M, Duong-Polk KX, Scott RT, Kim JJ, Kukhmazov I, Ellisman MH, Perkins GA, Weinreb RN. Intraoculat pressure elevation induces mitochondrial fission and triggers OPA1 release in glaucomatous optic nerve. Invest Ophthalmol Vis Sci. 2008;49:4903–11. doi: 10.1167/iovs.07-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tezel G. Oxidative stress in glaucomatous neurodegeneration: mechanisms and consequences. Prog Retin Eye Res. 2006;25:490–513. doi: 10.1016/j.preteyeres.2006.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izzotti A, Bagnis A, Saccà SC. The role of oxidative stress in glaucoma. Mutat Res. 2006;612:105–14. doi: 10.1016/j.mrrev.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 48.Izzotti A, Saccà SC, Cartiglia C, De Flora S. Oxidative deoxyribonucleic acid damage in the eyes of glaucoma patients. Am J Med. 2003;114:638–46. doi: 10.1016/s0002-9343(03)00114-1. [DOI] [PubMed] [Google Scholar]
- 49.Mazière C, Conte M, Degonville J, Ali D, Mazière J. Cellular Enrichment with Polyunsaturated Fatty Acids Induces an Oxidative Stress and Activates the Transcription Factors AP1 and NFκB. Biochem and Piophys Res Comm. 1999;265:116–22. doi: 10.1006/bbrc.1999.1644. [DOI] [PubMed] [Google Scholar]
- 50.Karin M, Liu Z, Zandi E. AP-1 function and regulation. Curr Opin Cell Biol. 1997;9:240–6. doi: 10.1016/s0955-0674(97)80068-3. [DOI] [PubMed] [Google Scholar]
- 51.Bossy-Wetzel E, Bakiri L, Yaniv M. Induction of apoptosis by the transcription factor c-Jun. EMBO J. 1997;16:1695–709. doi: 10.1093/emboj/16.7.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jacobs-Helber SM, Wickrema A, Birrer MJ, Sawyer ST. AP1 Regulation of Proliferation and Initiation of Apoptosis in Erythropoietin-Dependent Erythroid Cells. Mol Cell Biol. 1998;18:3699–707. doi: 10.1128/mcb.18.7.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Baumgart E, Vanhorebeek I, Grabenbauer M, Borgers M, Declercq PE, Fahimi HD, Baes M. Mitochondrial Alterations Caused by Defective Peroxisomal Biogenesis in a Mouse Model for Zellweger Syndrome (PEX5 Knockout Mouse). Am J Pathol. 2001;159:1477–94. doi: 10.1016/S0002-9440(10)62534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gould SJ, Valle D. Peroxisome biogenesis disorders: genetics and cell biology. Trends Genet. 2000;16:340–5. doi: 10.1016/s0168-9525(00)02056-4. [DOI] [PubMed] [Google Scholar]
- 55.Barkovich AJ, Peck WW. MR of Zellweger Syndrome. AJNR Am J Neuroradiol. 1997;18:1163–70. [PMC free article] [PubMed] [Google Scholar]
- 56.Haddad R, Font RL, Friendly DS. Cerebro-hepato-renal Syndrome of Zellweger. Arch Ophthalmol. 1976;94:1927–30. doi: 10.1001/archopht.1976.03910040633011. [DOI] [PubMed] [Google Scholar]
- 57.Ji M, Zhao W, Dong L, Miao Y, Yang X, Sun X, Wang Z. RGS2 and RGS4 modulate melatonin-induced potentiation of glycine currents in rat retinal ganglion cells. Brain Res. 2011;1411:1–8. doi: 10.1016/j.brainres.2011.07.008. [DOI] [PubMed] [Google Scholar]
- 58.Inoue MM, Inoue T, Epstein DL, Blumer KJ, Rao PV. Intraocular Pressure (IOP) Changes in RGS-2 (Regulator of G-Protein Signaling) Null Mice. Invest Ophthalmol Vis Sci. 2008;49 [Google Scholar]
- 59.Heximer SP, Watson N, Linder ME, Blumer KJ, Hepler JR. RGS2/G0S8 is a selective inhibitor of Gqalpha function. Proc Natl Acad Sci USA. 1997;94:14389–93. doi: 10.1073/pnas.94.26.14389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wettschureck N, Offermanns S. Mammalian G proteins and their cell type specific functions. Physiol Rev. 2005;85:1159–204. doi: 10.1152/physrev.00003.2005. [DOI] [PubMed] [Google Scholar]
- 61.Somlyo AP, Somlyo AV. Ca2+ sensitivity of smooth muscle and nonmuscle myosin II: modulated by G proteins, kinases, and myosin phosphatase. Physiol Rev. 2003;83:1325–58. doi: 10.1152/physrev.00023.2003. [DOI] [PubMed] [Google Scholar]
- 62.Inoue-Mochita M, Inoue T, Epstein DL, Blumer KJ, Rao PV. RGS2-deficient mice exhibit decreased intraocular pressure and increased retinal ganglion cell survival. Mol Vis. 2009;15:495–504. [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitt AB, Brook GA, Buss A, Nacimiento W, Noth J, Kreutzberg GW. Dynamics of microglial activation in the spinal cord after cerebral infarction are revealed by expression of MHC class II antigen. Neuropathol Appl Neurobiol. 1998;24:167–76. doi: 10.1046/j.1365-2990.1998.00103.x. [DOI] [PubMed] [Google Scholar]
- 64.Arends YM, Duyckaerts C, Rozemuller JM, Eikelenboom P, Hauw JJ. Microglia, amyloid, and dementia in Alzheimer disease: a correlative study. Neurobiol Aging. 2000;21:39–47. doi: 10.1016/s0197-4580(00)00094-4. [DOI] [PubMed] [Google Scholar]
- 65.Yuan L, Neufeld AH. Activated Microglia in the Human Glaucomatous Optic Nerve Head. J Neurosci Res. 2001;64:523–32. doi: 10.1002/jnr.1104. [DOI] [PubMed] [Google Scholar]
- 66.Zenkel M, Lewczuk P, Jünemann A, Kruse FE, Naumann GO, Schlötzer-Schrehardt U. Proinflammatory Cytokines Are Involved in the Initiation of the Abnormal Matrix Process in Pseudoexfoliation Syndrome/Glaucoma. Am J Pathol. 2010;176:2868–79. doi: 10.2353/ajpath.2010.090914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rutz S, Wang X, Ouyang W. The IL-20 subfamily of cytokines—from host defence to tissue homeostasis. Nat Rev Immunol. 2014;14:783–95. doi: 10.1038/nri3766. [DOI] [PubMed] [Google Scholar]
- 68.Sauane M, Gopalkrishnan RV, Sarkar D, Su Z, Lebedeva IV, Dent P, Pestka S, Fisher PB. MDA-7/IL-24: novel cancer growth suppressing and apoptosis inducing cytokine. Cytokine Growth Factor Rev. 2003;14:35–51. doi: 10.1016/s1359-6101(02)00074-6. [DOI] [PubMed] [Google Scholar]
- 69.Gupta P, Su Z, Lebedeva IV, Sarkar D, Sauane M, Emdad L, Bachelor MA, Grant S, Curiel DT, Dent P, Fisher PB. Mda-7/IL024: Multifunctional cancer-specific apoptosis-inducing cytokine. Pharmacol Ther. 2006;111:596–628. doi: 10.1016/j.pharmthera.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wirtz MK, Keller KE. The Role of the IL-20 Subfamily in Glaucoma. Mediators Inflamm. 2016;2016 doi: 10.1155/2016/4083735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mochizuki H, Murphy CJ, Brandt JD, Kiuchi Y, Russell P. Altered stability of mRNAs associated with glaucoma progression in human trabecular meshwork cells following oxidative stress. Invest Ophthalmol Vis Sci. 2012;53:1734–41. doi: 10.1167/iovs.12-7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gonzalez P, Epstein DL, Borras T. Genes upregulated in the human trabecular meshwork in response to elevated intraocular pressure. Invest Ophthalmol Vis Sci. 2000;41:352–61. [PubMed] [Google Scholar]
- 73.Johnson EC, Doser T, Cepurna WO, Dyck JA, Jia L, Guo Y, Lambert WS, Morrison JC. Cell proliferation and interleukin-6-type cytokine signaling are implicated by gene expression responses in early optic nerve head injury in rat glaucoma. Invest Ophthalmol Vis Sci. 2011;52:504–18. doi: 10.1167/iovs.10-5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Maret W. The function of zinc metallothionein: a link between cellular zinc and redox state. J Nutr. 2000;130:1455S–8S. doi: 10.1093/jn/130.5.1455S. [DOI] [PubMed] [Google Scholar]
- 75.Maret W. Cellular zinc and redox states converge in the metallothionein/thionein pair. J Nutr. 2003;133:1460S–2S. doi: 10.1093/jn/133.5.1460S. [DOI] [PubMed] [Google Scholar]
- 76.Lee SJ, Koh J. Roles of zinc and metallothionein-3 in oxidative stress-induced lysosomal dysfunction, cell death, and autophagy in neurons and astrocytes. Mol Brain. 2010;3:30. doi: 10.1186/1756-6606-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Spinazzola A, Viscomi C, Fernandez-Vizarra E, Carrara F, D’Adamo P, Calvo S, Marsano RM, Donnini C, Weiher H, Strisciuglio P, Parini R, Sarzi E, Chan A, DiMauro S, Rötig A, Gasparini P, Ferrero I, Mootha VK, Tiranti V, Zeviani M. MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38:570–5. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- 78.Dallabona C, Marsano RM, Arzuffi P, Ghezzi D, Mancini P, Zeviani M, Ferrero I, Donnini C. Sym1, the yeast ortholog of the MPV17 human disease protein, is a stress-induced bioenergetics and morphogenetic mitochondrial modulator. Hum Mol Genet. 2010;19:1098–107. doi: 10.1093/hmg/ddp581. [DOI] [PubMed] [Google Scholar]
- 79.El-Hattab AW, Scaglia F. Mitochondrial DNA Depletion Syndromes: Review and Updates of Genetic Basis, Manifestations, and Therapeutic Options. Neurotherapeutics. 2013;10:186–98. doi: 10.1007/s13311-013-0177-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruest LB, Marcotte R, Wang E. Peptide elongation factor eEF1A–2/S1 expression in cultured differentiated myotubes and its protective effect against caspase-3-mediated apoptosis. J Biol Chem. 2002;277:5418–25. doi: 10.1074/jbc.M110685200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chang R, Wang E. Mouse translation elongation factor eEF1A–2 interacts with Prdx-I to protect cells against apoptotic death induced by oxidative stress. J Cell Biochem. 2007;100:267–78. doi: 10.1002/jcb.20969. [DOI] [PubMed] [Google Scholar]
- 82.Li Z, Qi C, Shin D, Zingone A, Newbery HJ, Kovalchuk AL, Abbott CM, Morse HC.Eef1a2 Promotes Cell Growth, Inhibits Apoptosis and Activates JAK/STAT and AKT Signaling in Mouse Plasmacytomas. PLoS One 20105e10755 [DOI] [PMC free article] [PubMed] [Google Scholar]


