Abstract
The discovery of mutations associated with disease pathogenesis does not have to rest solely on genetic evidence.
The perennial promise of human disease genetics is the delivery of etiology-based therapies. This rests on the notion that identifying disease-causing mutations will provide a basis for determining the molecular networks that constitute the disease process—an understanding that is critical for the development of such therapies. On page 506 of this issue, Novarino et al. (1) perform what is perhaps the most complete genetic analysis of the neurological disorder hereditary spastic paraplegia (HSP), and deliver on part of this promise by creating an “HSPome,” a plausible network of proteins involved in this disease.
In many fields, the hard-won discoveries linking mutations to diseases are the foundation for investigations into molecular pathogenesis. In rare instances, mutation detection has provided immediate insight into the disease process. Usually, however, moving from gene to pathogenesis has been exceptionally difficult. Functional and mechanistic work on the molecular etiology of disease remains one of the major challenges in modern biology; this is quite understandable given the inherent limitations of traditional reductionist functional work. There have been successes, however, and these have helped form the dominant theories of pathogenesis for many diseases, including Alzheimer’s disease (2). An increasingly popular intermediate step between genetics and function is the use of pathways-based analysis. Such an approach attempts not only to produce a refined list of potential functional interactions to investigate at the bench, but also to provide a global snapshot of the landscape of a particular disease’s etiology.
Novarino et al. have executed this approach with compelling results. The underlying strategy of their study involved the identification of disease-causing mutations using the power of small, inbred families. HSPs represent a class of inherited progressive neurodegenerative disorders that manifest with stiffness and contraction in the lower limbs—a feature believed to be the result of corticospinal tract dysfunction. These diseases are extremely genetically heterogeneous. Novarino et al. examined the autosomal recessive form (AR-HSP), which was already linked to mutation of more than 20 genes (3). The authors expand this list by means of an iterative set of analyses on an initial set of 55 AR-HSP families. Using a straightforward genetic strategy of segregation, they show that disease in one-third of the families is linked to genes previously implicated in AR-HSP, but they also identify new candidate gene mutations in more than half of the remaining families. Additional mutations were identified in one-third of these new genes, providing substantive genetic support for causality. Functional analysis showed that candidate gene mutations cause a locomotor deficit in an animal model (zebrafish) of HSP, consistent with what has been described in previous HSP modeling efforts, thus further supporting pathogenicity.
Beyond identifying new genes associated with HSP, Novarino et al. constructed a pro-teome network for HSP from information available in protein interaction databases. They observed that candidate genes and known HSP genes were more highly connected within this network than expected by chance. The authors then created a network of proteins encoded by known HSP genes, their nominated candidate genes, and proximal interactors, naming this expanded network the “HSPome.” This not only provides a global view of the processes and proteins underlying HSP, but also identifies genes as new candidates to bear disease-causing mutations. Novarino et al. validated this last point by returning to genetics. They identified likely pathogenic variants in three new genes mined from the HSPome, one of which was independently implicated in HSP (4).
Novarino et al. illustrate a subtle and perhaps necessary shift in genetics work. The burden of proving mutation pathogenicity has traditionally rested on the shoulders of genetics alone, relying on association of mutations with disease, segregation of mutations within families, and independent replication of results. Given the pace and nature of genetic mutation discovery, these proofs are no longer always tenable. Instead, genetics increasingly relies on functional and bioinformatics work. This change has been contentious among those who believe that genetic evidence should be the foundation of functional work. There is admittedly potential danger in using functional work to identify candidate genes—most important is the inevitable issue of circularity where studies are limited to what investigators believe to be biologically plausible genes and proteins. This runs the risk not only of incorrectly self-affirming hypotheses, but also of ignoring new and critically important pathways because they don’t fit into a current understanding of the disease process. This is not the case in the study by Novarino et al. The authors have been careful to question the significance of their observations, and they have in some instances the reassurance of independent replication. It is exactly this type of care that the field needs to take.
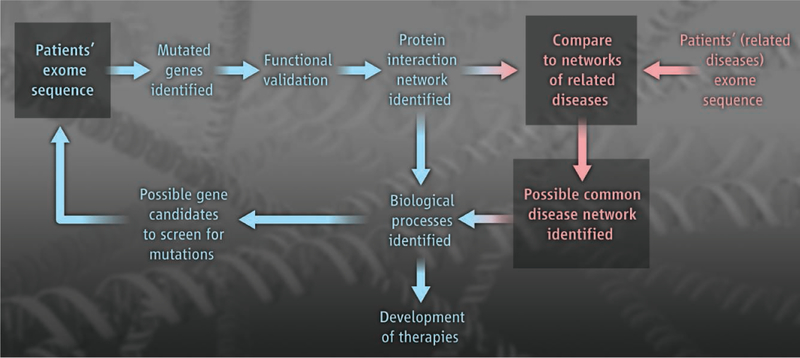
Novarino et al. also examined the relationship of the HSPome to genes implicated in other diseases, and found significant overlap with gene sets linked to Alzheimer’s disease, Parkinson’s disease, and amyotrophic lateral sclerosis. If this fascinating observation holds true, it raises a critical question: If varied neurodegenerative disorders are linked by a common pathway, what is the underlying cause of the distinct cellular vulnerability seen in these disorders? One might predict that identifying disease-linked protein networks is a key step toward understanding this phenomenon. In this regard, the type of work described by Novarino et al. shows not only the power of comprehensive genetic analysis in identifying the pathogenic networks involved in that disease but also the potential of such work to inform outside of the disease in question (see the figure).
This study clearly adds another dimension to our understanding of HSP. With this knowledge, we can turn toward fulfilling the ultimate promise of genetics: translating this understanding into etiology-based therapies.
A global view of disease pathogenesis.
The scheme illustrates an investigative approach that connects gene mutations in one disease through protein networks. This network can be used to nominate additional candidate genes and infer mechanistic overlap with other diseases. It may be possible to derive a core network that can guide the development of therapeutics.
References
- 1.Novarino G et al. , Science 343, 506 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hardy J, Selkoe DJ, Science 297, 353 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Fink JK, Acta Neuropathol. 126, 307 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oates EC et al. , Am. J. Hum. Genet 92, 965 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]