Abstract
Recent advances in tissue engineering and 3D bioprinting have enabled construction of cell-laden scaffolds containing perfusable vascular networks. Although these methods partially address the nutrient-diffusion limitations present in engineered tissues, they are still restricted in both their viable vascular geometries and matrix material compatibility. To address this, tissue constructs are engineered via encapsulation of 3D printed, evacuable, free standing scaffolds of poly(vinyl alcohol) (PVA) in biologically derived matrices. The ease of printability and water-soluble nature of PVA grant compatibility with biologically relevant matrix materials and allow for easily repeatable generation of complex vascular patterns. This study confirms the ability of this approach to produce perfusable vascularized matrices capable of sustaining both cocultures of multiple cell types and excised tumor fragments ex vivo over multiple weeks. The study further demonstrates the ability of the approach to produce hybrid patterns allowing for coculture of vasculature and epithelial cell-lined lumens in close proximity, thereby enabling ex vivo recapitulation of gut-like systems. Taken together, the methodology is versatile, broadly applicable, and importantly, simple to use, enabling ready applicability in many research settings. It is believed that this technique has the potential to significantly accelerate progress in engineering and study of ex vivo organotypic tissue constructs.
Keywords: biologically derived matrices, bioprinting, ex vivo tissues, vascularized tissues
1. Introduction
Engineered tissue constructs have historically been promoted as a potential source of organ and tissue transplants.[1,2] More recently, there has also been increased exploration in nonclinical applications, such as elucidating mechanisms of cell–cell and cell–matrix interactions,[3] modeling disease pathologies,[4] and accelerating drug discovery and screening.[5,6] Regardless of the intended use however, the construction and applicability of engineered tissues is limited by scaling, as nutrient diffusion limitations caused by the lack of vascularization restrict tissue survival to thicknesses on the scale of a few hundred micrometers.[7]
Much effort has gone into addressing this diffusion limitation, and among the various techniques used, 3D printing has experienced significant success, due to the precise spatial control it grants during the fabrication process, along with ease of use and ability to rapidly modify geometries to enable on-demand fabrication.[2,8,9] Many variants of the technique involve deposition of cell-laden hydrogels in specific patterns[9–17] using natural materials such as alginate,[11–13,15,17] gelatin,[10,11,15,16] hyaluronic acid,[16] and decellularized extracellular matrix (ECM),[11,18] as well as synthetic polymers that include polycaprolactone (PCL),[14] poly(lactide-co-caprolactone) (PLCL),[14] and polyethylene glycol (PEG)[9,15,16] among others. The result is cellularized lattices, which may or may not be vascularized, but possess high surface area to volume ratios to increase nutrient accessibility. An alternative approach is the formation of a perfusable cellularized construct by using an evacuable fugitive ink to produce a vascular channel within a polymerized matrix.[11,19–21] Variants of this technique have been achieved using inks composed of materials such as Pluronics F127,[19] gelatin,[20] and carbohydrate glass,[21] with matrices composed of various biocompatible materials, including gelatin, fibrin, collagen, and alginate. However, their use in generating vascular geometries more complex than 2D patterns or simple 3D grids has not been well explored.[19–21] In addition, because of their material properties, most fugitive inks are compatible with only a small selection of matrix materials. For instance, Pluronics F127 solutions liquify at low temperatures,[22,23] making them difficult to use with materials such as collagen and Matrigel that require such temperatures when casting. Similarly, gelatin inks are incompatible with transglutaminase, a cross-linking enzyme commonly used to generate scaffolds from materials including collagen, gelatin, hyaluronic acid, and albumin.[24–27]
Here, we present a methodology of generating biologically derived tissue constructs containing vascular channels of complex 3D geometries via 3D printing of poly(vinyl alcohol) (PVA). PVA is known to be biocompatible,[28–30] and is usable with several deposition techniques, including selective laser sintering and fused filament printing.[31–33] In the context of cell culture, it has been used primarily as a matrix component to form porous, cell-laden hydrogels,[34,35] or to form biocompatible hydrogels via stereolithography.[28,31] More recently, PVA has also been applied as an evacuable scaffold to generate vascularized matrices, though material-compatibility has been limited thus far to gelatin.[29,30] By utilizing the water-solubility of PVA with its ability to be printed into a variety of free-standing geometries, we expanded upon its usability by encapsulating it within and evacuating it from a wide range of systematically optimized biologically derived matrices to produce complex perfused tissue structures. Notably, the methodology is both highly simple and easily reproducible, thereby making it accessible in many research settings. As such, the technique has the potential to significantly accelerate progress in tissue engineering.
2. Results and Discussion
2.1. Printing 3D Vascularized Constructs Using Free-Standing PVA Scaffolds
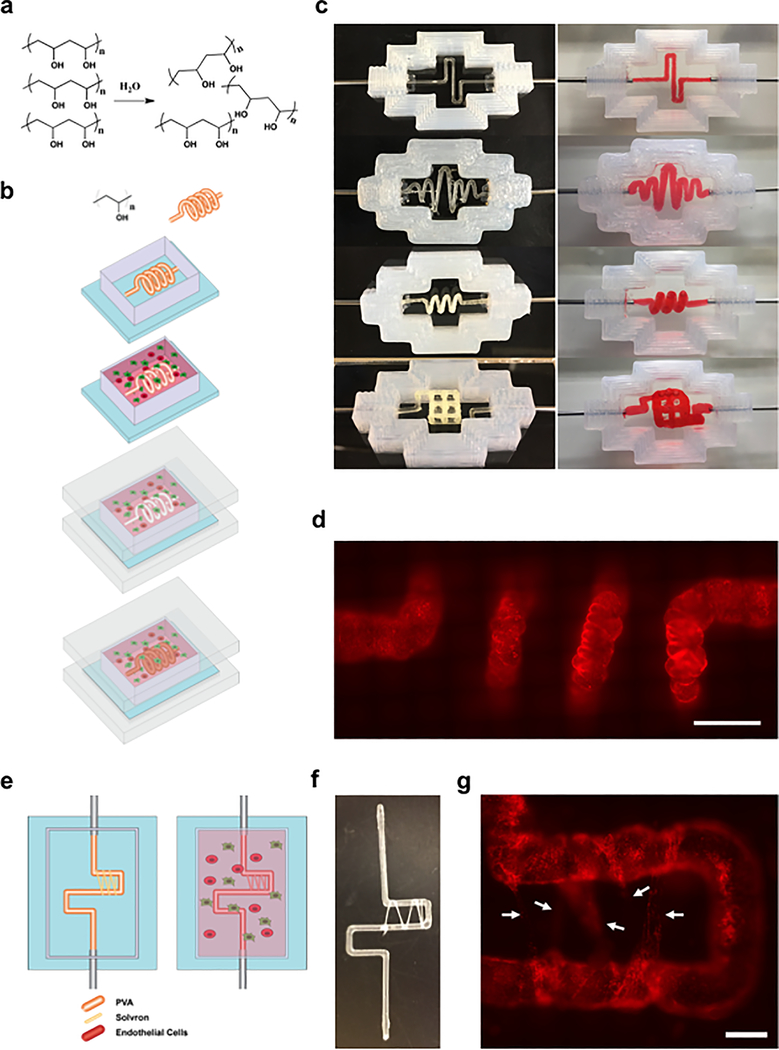
An overall schematic of our printing methodology is outlined in Figure 1a,b. In the first series of studies we assessed the viability of using PVA as a sacrificial vascular scaffold. PVA was confirmed to completely dissolve in media within a 1 h time frame (Figure S1a, Supporting Information), likely attributed to its water-soluble chemistry (Figure 1a). Next, to assess swelling properties within a hydrogel environment, 0.7 mm diameter linear PVA scaffolds were printed, embedded within a matrix of 7.5% porcine gelatin and 10 mg mL−1 fibrin, and then incubated at 37 °C for between 20 and 80 min before being evacuated with warm media. Results indicated that minimal to no swelling occurred within 20 min, and structures were able to gradually swell to twice their original diameter over 80 min (Figure S1b, Supporting Information). Finally, we confirmed the versatility of the approach via the ability to readily construct a range of viable vascular geometries embedded within a hydrogel matrix of 7.5% porcine gelatin and 10 mg mL−1 fibrin, which could in turn be evacuated successfully using warm media (Figure 1c), leaving behind the desired vascular lumens.
Figure 1.
Printing 3D vascularized constructs using PVA scaffolds: a) Chemical structure of PVA and solvation in water. b) Schematic representation of the vascularized construct manufacturing procedure. A PVA scaffold of desired geometry is printed and inserted into a silicone holder. The PVA scaffold is then encapsulated within a matrix formulation of desired composition. The matrix formulation is allowed to gelate and simultaneously, the PVA scaffold slowly dissolves. The structure is then sealed using an acrylic base and lid, and the scaffold is evacuated using warm media. Following evacuation, the resulting lumen can be seeded with endothelial or epithelial cells. c) Examples of 2D and 3D geometries that can be printed and evacuated using PVA. Images on the left show the PVA structures prior to evacuation, and images on the right show the perfused channels following evacuation. Scaffolds in this figure were encapsulated within a matrix of 7.5 wt% gelatin and 10 mg mL−1 fibrin. d) Fluorescent image of a 3D spirals-haped channel seeded with mCherry-labeled HUVECs. Scale bars: 2 mm. e) Schematic representation of dense vascular bed induction procedure. A PVA scaffold of desired geometry is printed and wrapped with PVA (Solvron) threads, before being encapsulated and dissolved. Over time, Solvron threads also dissolve, resulting in narrow channels into which endothelial cells may migrate. f) A PVA scaffold partially wrapped with Solvron thread. g) Fluorescent images of mCherry-labeled HUVECs migrating into narrow channels left by evacuated Solvron. Scale bars: 1 mm.
When applied to living cells, we also confirmed the presence of PVA within cell culture media had no negative impact on the growth of either human umbilical vein endothelial cells (HUVECs) or MDA-MB-231 cells (Figure S1c,e, Supporting Information). To assess viability, we used the CCK8 reagent, and toward this validated the correlation between CCK8 absorbance readouts and cell numbers in the context of our studies (Figure S1d, Supporting Information). Subsequently, we constructed vascularized constructs using 5 mg mL−1 Matrigel and 10 mg mL−1 fibrin matrices seeded with human mesenchymal stem cells (hMSCs) followed by seeding of HUVECs within the residual channels (post evacuation of PVA structures) and perfusion over multiple days. Results indicated that HUVECs adhered even with complex channel geometries (Figure 1d) and formed a monolayer around the lumen (Figure S1g, Supporting Information). Endothelial barrier functionality was confirmed via perfusion and permeability measurements of fluorescein isothiocyanate (FITC) conjugated 70 kDa dextran (Figure S1f, Supporting Information). Fixation and staining of HUVECs with mouse monoclonal anti-CD34 (Thermo Fisher) and rabbit monoclonal anti-VE-Cadherin (Cell Signaling Technologies) indicated that they expressed proper endothelial lineage markers and formed adherens junctions[36] within 10 d (Figure S1h, Supporting Information). In addition, staining of hMSCs cultured within the matrix using mouse monoclonal anti-CD105 (Thermo Fisher) and Alexa-594 phalloidin (Thermo Fisher) confirmed viability and maintenance of cell potency[37] (Figure S1i, Supporting Information).
Beyond introducing a single main vascularized channel to a matrix construct, we also assessed the viability of adding a dense and fine vascular bed. This was accomplished by wrapping water-soluble PVA-based thread (Solvron, Nitivy Co. 62T Type SS) around the main PVA scaffold (Figure 1e,f) prior to encapsulation. Because of increased fluid flow resistance exhibited by the pathways containing Solvron (100–400 μm diameter channels) in comparison to the main PVA channel, clearing of the former took place over multiple days following endothelial seeding. Results showed that seeded HUVECs migrated into the channel tracks left behind by evacuated Solvron, creating microvasculature capable of linking different portions of the primary vascular network (Figure 1g), with functionality confirmed via flow of 70 kDa FITC-dextran. Taken together, PVA based sacrificial structures enable facile construction of vascularized tissue constructs with diverse programmable 3D geometries and channel dimensions ranging from 100 to 1000 μm.
2.2. Development of an Optimally Cell-Compatible Matrix
We next sought to design a matrix environment suitable for high cell growth and promotion of diverse cellular responses while compatible with maintaining the structural stability necessary for evacuation of PVA and long-term perfusion. In this regard, previous studies have extensively made use of synthetic matrices. However, with a goal to increase similarities to the in vivo microenvironment, we primarily explored biological matrices such as collagen, fibrin,[19,20] and Matrigel.[38,39]
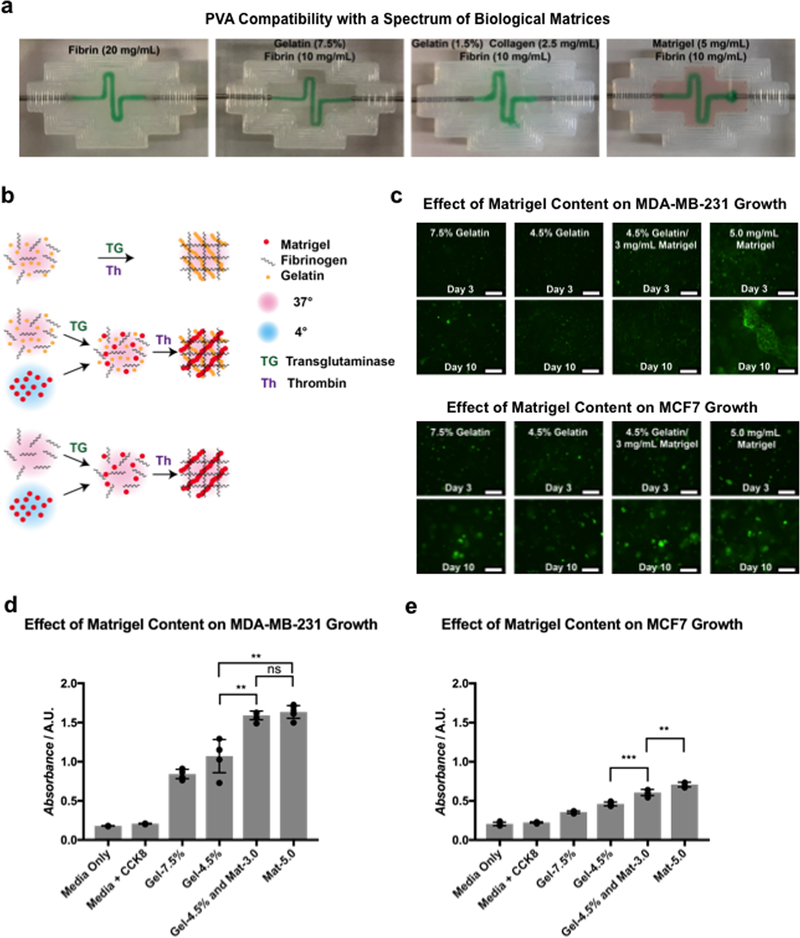
Toward this, we first examined the compatibility of our methodology with a range of matrix materials that include fibrin, gelatin, collagen, and Matrigel. Specifically, PVA scaffolds of a square wave geometry were printed, embedded in, and evacuated from matrices of varied compositions (Figure 2a), confirming viability of use. To assess the impact of the introduction of Matrigel on cell growth and viability, two breast cancer epithelial cell lines (MDA-MB-231 and MCF7) were encapsulated in matrices of varied composition and grown over 10 d. All matrices contained 10 mg mL−1 fibrin, along with gelatin, Matrigel, or a blend of the two (Figure 2b). Average hydrogel stiffness, as measured via atomic force microscopy (AFM), showed less than 1 kPa variation across all gelatin-containing conditions, while a significant reduction in stiffness was observed in the absence of gelatin (Figure S2a, Supporting Information). Growth over time was qualitatively confirmed via fluorescent microscopy (Figure 2c), and quantitative measurements of metabolic activity were obtained using the CCK8 reagent (Figure 2d,e). Results indicated that the presence of Matrigel significantly increased cell growth, both with and without gelatin, which we believe can be attributed to the various biological basement membrane components contained within it.[40] Lower absorbance from MCF7 cells can be explained by their relatively slower growth rate compared to MDA-MB-231 cells. An identical experiment was conducted with hMSCs to assess the effect on the stromal cells, with the duration increased to 20 d to account for a slower rate of growth. Both quantitative (Figure S2b, Supporting Information) and qualitative (Figure S2c, Supporting Information) results mirrored those of the breast cancer epithelial cells.
Figure 2.
Development of optimally cell-compatible constructs based on biologically derived matrix materials: a) Images indicating successful evacuation of PVA from matrices composed of various biological materials. b) Schematic representation of the materials-testing procedure. Gelatin/fibrin matrices were formulated by mixing both components with transglutaminase, then polymerizing with thrombin at 37 °C. Matrigel/fibrin and gelatin/Matrigel/fibrin blended matrices were formulated by mixing all components except Matrigel with transglutaminase at 37 °C, then adding Matrigel and polymerizing with thrombin. Matrigel was maintained at 4 °C, while all other components were maintained at 37 °C during the procedure. c) Fluorescent images showing growth of GFP-labeled MDA-MB-231 cells and MCF-7 cells in matrices of various compositions. Scale bars: 250 μm. d,e) Absorbance measurements of MDA-MB-231 cells and MCF-7 cells grown in matrices of various compositions, obtained using a CCK8 assay (n = 4 with P values **P < 0.01 and ***P < 0.001).
In addition to designing an ideal matrix environment, we also optimized media conditions that would allow for co-culturing of multiple cell types, specifically in this case, endothelial cells within the channel and tumor cells within the stroma. Cells were grown in media formulations containing varying amounts of Dulbecco’s modified Eagle medium (DMEM), fetal bovine serum (FBS), L-glutamine, and endothelial growth medium 2 (EGM-2) (Lonza). Control media for MDA-MB-231 cells and HUVECs were DMEM supplemented with 10% FBS and 2 × 10−3 M L-glutamine, and EGM-2, respectively. Both qualitative results (Figure S2d,e, Supporting Information) and quantitative results (Figure S2f,g, Supporting Information) indicated that a 50/50 mixture of EGM-2 and DMEM supplemented with 20% FBS and 4 × 10−3 M L-glutamine maximized growth of MDA-MB-231 cells while producing no significant effect on the growth of HUVECs compared to controls in EGM-2 only.
Having established a robust system for engineering long-term culturable vascularized constructs based on biologically derived matrices we next focused on evaluating the system in the contexts of two distinct application scenarios to highlight the methodologies broad applicability. Specifically, in the first we explored the ability of the engineered vascularized tissue to sustain embedded biopsied tumor pieces long-term in a fully ex vivo setting. In the second, we expanded the system’s capabilities to engineer hybrid vascular systems supporting flow of distinct biological fluids, specifically focusing on engineering an ex vivo vascularized gut-like system.
2.3. Application of the Methodology to In Vitro Tumor Sustenance
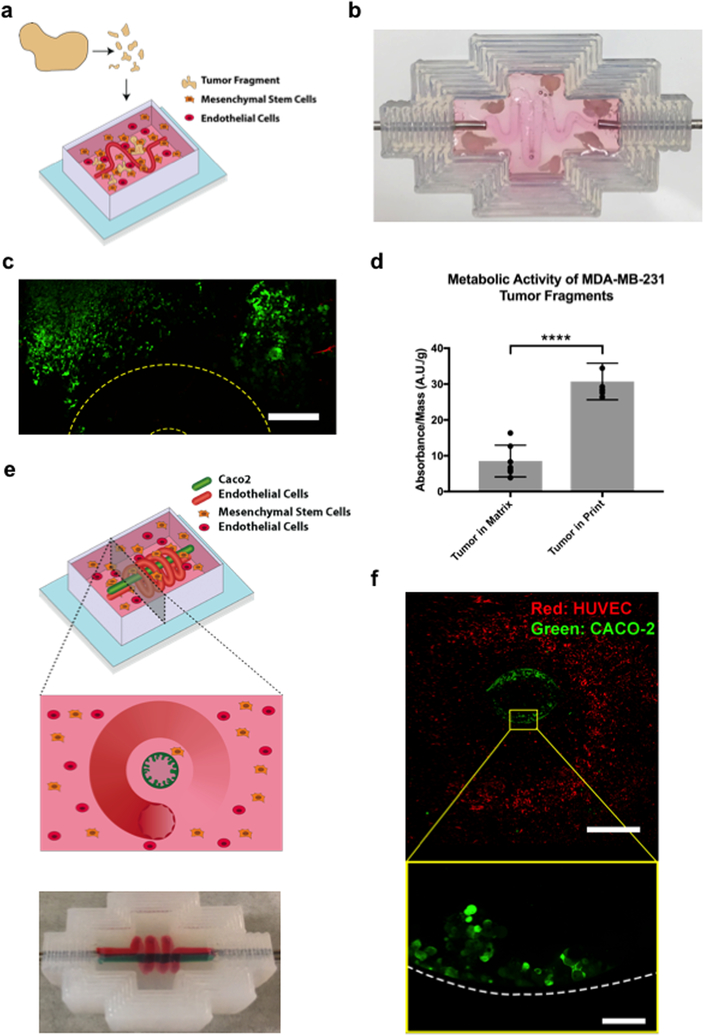
To examine the feasibility of using the vascularized matrix to sustain tumor tissue ex vivo, tumors were grown from green fluorescent protein (GFP) MDA-MB-231 cells in NOD-SCID mice, excised, fragmented, and embedded in either a vascularized tissue construct, or nonvascularized matrices of equal thickness (Figure 3a,b). Both vascularized and nonvascularized conditions used matrices of 10 mg mL−1 fibrin and 5 mg mL−1 Matrigel, and were seeded with hMSCs, which are known to have essential roles in the stromal microenvironment.[41] Vascularized tissue constructs were continuously perfused with fresh media, changed every 2 d, while nonvascularized tissue constructs had fresh media added twice a day. In both conditions, the previously identified optimum media formulation was used (1:1 mixture of EGM-2 and DMEM supplemented with 20% FBS and 4 × 10−3 M L-glutamine). After 21 d, half of the tumor fragments of all conditions were excised and cell viability was measured using the CCK8 reagent (Figure 3d). The remaining tumor fragments were embedded in optimal cutting temperature compound (OCT), cryosectioned, mounted, and imaged directly (Figure S3a–c, Supporting Information). Confocal images of perfused tumor fragments prior to extraction indicated that they were viable and living in dense tissue clusters after 21 d, and the results of CCK8 measurements confirmed significantly higher viability of fragments embedded in perfused constructs compared to nonperfused matrices. This was further reinforced by results of postsectioning GFP expression. In this context, greater expression of GFP in sectioned tissue represented greater proportions of surviving cells at the time of extraction and freezing, and images indicate that although degrees of cell death and loss of tissue integrity occur in both perfused (Figure S3c, Supporting Information) and static (Figure S3b, Supporting Information) conditions, a significantly greater proportion of cells under perfused conditions appears to have survived.
Figure 3.
Applications of the methodology. In vitro tumor sustenance: a) Schematic showing the process by which MDA-MB-231 tumors grown in mice were excised, fragmented, and encapsulated within a vascularized construct, before being perfused over multiple weeks. b) Image showing tumor fragments encapsulated within a vascularized construct with a sinusoidal geometry. c) Fluorescent confocal images of GFP-labeled MDA-MB-231 tumor fragments after 24 d of perfusion within a vascularized construct. The location of the vascular channel is outlined in yellow. Because of the thickness and positioning of the tumor fragments, capturing the vascular channel and tumor fragments within the same plane was not possible. Scale bars: 300 μm. d) Absorbance measurements of MDA-MB-231 tumor fragments sustained over 24 d while encapsulated in either a static matrix or a perfused vascularized construct. Measurements were obtained with a CCK8 assay. To account for the variability introduced by tumor fragments of different sizes, all measurements were normalized with respect to mass (n = 6 with P-value ****P < 0.0001). Hybrid vascularized systems: e) Schematic diagram of a gut-organoid constructed by creating multiple lumens in a single matrix, designed to imitate endothelial and epithelial cocultures. The outer lumen is a 3D spiral, and is seeded with endothelial cells. The inner lumen is a channel and is seeded with gut epithelial cells. f) Image of a multichannel construct generated by evacuating more than one PVA scaffold within a single matrix. g) Fluorescent confocal images of GFP-labeled Caco-2 cells and mCherry-labeled HUVECs seeded, respectively, within an inner linear channel and an outer spiral channel of a vascularized construct. The top image shows a cross-sectional view of the construct. The bottom image shows a close-up of the Caco-2 cells, indicating formation of finger-like protrusions. Scale bars: 1 mm (top) and 100 μm (bottom).
2.4. Application of the Methodology to Generating Hybrid Vascularized Systems
Following confirmation of the system’s ability to maintain tissue viability ex vivo, we next applied it to generate a hybrid vascularized organ system in vitro. Here a modified lumenal geometry was designed consisting of a linear channel surrounded by a spiral. The central channel was seeded with Caco-2 intestinal epithelial cells, while the outer spiral was seeded with HUVECs, generating a system mimicking a vascularized gut (Figure 3e,f). Prior to the hybrid system, we examined the ability of the perfused construct to sustain an in vitro gut model by seeding evacuated channels with GFP Caco-2 cells, and then subjecting them to either static culture or perfusion over 12 d. Confocal fluorescent microscopy revealed the arrangement of a confluent Caco-2 layer on the channel interiors in both cases. However, perfusion resulted in the formation of 3D protrusion-like arrangements, while static culture did not (Figure S3d, Supporting Information). Immunostaining was performed using rabbit monoclonal anti-Na+/K+ ATPase (Thermo Fisher) and Alexa-594 phalloidin (Thermo Fisher), confirming the presence of F-actin-coated borders, as well as Na+/K+ ATPase transporters within the aforementioned projections (Figure S3e, Supporting Information). When extended to the hybrid system, this behavior was maintained, with Caco-2 cells still expressing villus-like morphology, and endothelial cells forming a channel around them (Figure 3g), with a minimum separation distance of less than 200 um. Together, these results indicate the viability of supporting cocultures of epithelial and endothelial cells, each with a unique microenvironment, in extremely close proximity.
3. Conclusions
In summary, 3D printing of PVA coupled with use of biologically derived matrices enables a robust and reproducible methodology for generating highly functional vascularized tissue constructs. Within this study, we developed an engineered tissue model that contained a vascular channel seeded with primary endothelial cells, and was capable of sustaining both cells and tissue fragments long-term. In addition, we demonstrated the viability of creating cocultures of Caco-2 gut epithelial cells with primary endothelial cells that mimicked in vivo gut-like organization patterns. As a whole, we believe that our technique offers a method to create highly complex tissue models that can be used to study a biological phenomenon. Moreover, because of the simplicity of the technique, it is highly accessible in many research settings, and as such, has the potential to significantly accelerate progress in tissue engineering.
4. Experimental Section
3D Printing of Silicone Holders and Pump Setup
Long-term perfusion of the tissue constructs was achieved using a 3-component system consisting of a media reservoir, a flow-chamber, and a peristaltic pump (Watson Marlow 205U), all connected via silicone tubing (McMaster Carr ⅛ OD Platinum 2000 Silicone). Flow chambers were constructed via extrusion-printing of silicone (Dow Corning Toray Sylgard SE1700) on glass, and contained inlet and outlet ports leading to the pump and media reservoirs.[19]
3D Printing of Free-Standing PVA Structures
All geometries of interest were designed in Autodesk Inventor and exported to the Ultimaker Cura software. Structures were then printed using the Ultimaker3 with a 0.4 mm printhead at speeds between 10 and 35 mm s−1. PVA deposited was obtained directly from Ultimaker as a 2.85 mm diameter solid-state filament with a 3860 MPa tensile modulus and a density of 1.23 g cm−3. Prior to use, PVA structures were sterilized via UV radiation.
Generating 3D Vascularized Constructs
Stock solutions of all materials were prepared prior to matrix formulation. Type A porcine skin gelatin (Sigma-Aldrich) was added to water (15 wt vol%−1), dissolved overnight at 70 °C, brought to a pH of 7.4 using NaOH solution, and passed through a 0.22 um filter (Millipore). Solutions were stored long-term at 4 °C, and warmed to 37 °C prior to use. CaCl2 (250 × 10−3 M) was prepared as a stock solution in Dulbecco’s phosphate buffered saline (dPBS) and stored at room temperature. Thrombin (Sigma-Aldrich) was prepared as a stock solution (500 U mL−1), aliquoted, stored at −20 °C, and warmed to 4 °C prior to use.[19] Matrigel (Corning) was purchased and had reported protein concentrations between 8 and 11 mg mL−1. Solutions of both bovine plasma fibrinogen (Millipore) and transglutaminase (MooGloo) were prepared immediately before use by dissolving in 37 °C dPBS at respective concentrations of 100 mg mL−1 and 50 mg mL−1.
Production of vascularized constructs required inserting a PVA-printed structure into the silicone holders with direct contact points at the inlet and outlet, then encapsulating within the formulated matrix. Matrices composed of Matrigel and fibrin were formulated using Matrigel (5 mg mL−1), fibrinogen (10 mg mL−1), transglutaminase (2 mg mL−1), CaCl2 (2.5 × 10−3 M), and thrombin (2 U mL−1), with remaining volume composed of cell-containing media. Matrices composed of Matrigel, gelatin, and fibrin were formulated from gelatin (1.5 wt% mL−1), Matrigel (4 mg mL−1), and all other components identical to those above. In either case, all components save Matrigel and thrombin were mixed at 37 °C and allowed to incubate for 30 min, after which both Matrigel and thrombin were rapidly added. The solution was mixed well, then poured into the silicone holders, and allowed to gelate at 37 °C. Gelation occurred over 1 and 2.5 h, respectively, for matrices with and without gelatin. Following gelation, PVA was evacuated via perfusion of warm media (Figure 1b), and the construct was perfused with media at ≈10–12 rpm. After several hours of perfusion, HUVECs were resuspended at a concentration of 10 × 106 cells per milliliter and injected into the vascular channel. Constructs were incubated for 30 min on either side and then left overnight without flow to allow for HUVEC adhesion. Flow was then reintroduced to remove nonadhering HUVECs from the channel.
Introduction of dense vascular beds within the constructs largely used the same procedure. However, prior to encapsulation, PVA thread (Solvron, Nitivy Co. 62T Type SS) was wrapped around the PVA scaffold (Figure 1e) and heat-sealed using a standard cauterizing pen.
AFM Measurements
Hydrogel stiffness was measured by AFM as described.[42] Nanoindentations were performed using a pyrexnitride probe with a pyramid tip (spring constant ≈0.04 N m−1, 35° half-angle opening, NanoAndMore USA Corporation, cat # PNP-TR) connected to an MFP-3D Bio Atomic Force Microscope (Oxford Instruments) mounted on a Ti-U fluorescent inverted microscope (Nikon Instruments). After calibration using a glass slide, samples were loaded on the AFM, submersed in phosphate buffered saline (PBS), and indented at a velocity of 2 μm s−1 with a trigger force of 2 nN. To ensure reproducibility, three force maps of ≈20 force measurements were performed over a 90 μm × 90 μm region per gel. In addition, measurements were made for three separate gels per condition. Elastic modulus was calculated based on a Hertz-based fit using a built-in code written in the Igor 6.34A software.
Imaging
Widefield fluorescent and brightfield microscopy images were obtained using the Leica DMi8. Confocal images were obtained using the Zeiss 880 Airyscan Confocal.
Cell Culture and Media
HUVECs and hMSCs used in the study were obtained from Lonza, and were used until passages 12 and 10, respectively. HUVECs were cultured in either EGM-2 (Lonza) or EndoGRO-LS (Millipore), while hMSCs were cultured in mesenchymal stem cell growth medium (MSCGM) (Lonza) or mesenchymal stem cell expansion medium (MSCEM) (Millipore). MDA-MB-231, MCF7, and Caco-2 cells were obtained from ATCC, and were, respectively, cultured in DMEM supplemented with 10% FBS and 2 × 10−3 M L-glutamine and EMEM (ATCC) supplemented with 20% FBS.
Vascular Permeability Measurements
FITC-labeled 70 kDa dextran was flowed through channels either with or without a coating of HUVECs at a rate of 20 μL min−1, and allowed to diffuse over 3 min to obtain an initial fluorescence measure. This rate was then reduced to 5 μL min−1 for the next 30 min, with fluorescent images taken every 5 min. Permeability was calculated in accordance with Equation (1)[19,20,43]
| (1) |
Here, Ib represents the mean background fluorescence present prior to the addition of dextran, Ii and If represent the mean fluorescence at initial and final timepoints, and d represents the diameter of the channel. All image processing was performed using ImageJ.
Growing and Excising Tumors in Mice
To generate tumors, GFP-transduced MDA-MB-231 cells were injected subcutaneously into the dorsal flanks of NOD-SCID mice. 5 × 105 cells were injected in a Matrigel (5 mg mL−1) solution (200 μL),[44,45] and then allowed to grow over ≈10 weeks before being excised. All protocols conducted using mice were conducted with approval from IACUC UCSD.
Immunohistochemistry
For all immunostaining, printed constructs were extracted from holders, fixed in paraformaldehyde (4%) for 1 h, washed with three rinses of PBS for 1 h each, and blocked overnight using a solution of bovine serum albumin (BSA) (1%) and Triton-X100 (0.125%) in PBS. Constructs were then subject to a 24 h incubation with primary antibodies in blocking buffer, an overnight wash with blocking buffer, a 24 h incubation with secondary antibodies in blocking buffer, and an overnight wash with PBS.
Tissue fragments were either extracted from the matrix, or left in the matrix before being embedded in OCT and frozen in a slurry of dry ice in acetone. Blocks were stored long-term at −80 °C. Fragments were then sectioned into 14 μm slices and mounted on gelatin-coated slides for imaging.
Statistical Analysis
Statistical analysis was conducted using the GraphPad Prism 7 software, and graphical data are displayed as a mean ± standard deviation overlaid on individual data points. All data were analyzed using a two-tailed t-test with Welch’s correction, and a P-value of <0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgements
The authors thank Prof. Adam Engler for his generous help and advice on the mechanical studies of gels, and members of the Mali lab for advice and help with experiments. This work was supported by UCSD Institutional Funds, the Burroughs Wellcome Fund (1013926), the March of Dimes Foundation (5-FY15–450), the Kimmel Foundation (SKF-16–150), the ARCS/Roche Foundation Scholar Award Program in the Life Sciences, the National Science Foundation (1463689 and the Graduate Research Fellowship Program), and NIH grants (RO1GM123313, R01HG009285, RO1CA222826, R21CA217735, and R01CA206880). Imaging was accomplished with support of the UCSD Neuroscience Microscopy Shared Facility Grant (P30 NS047101). M.H. was supported in part during the project by the NHLBI as a training grant recipient (T32 HL 105373). The authors have filed a patent based on this work.
Footnotes
Conflict of Interest
The authors declare no conflict of interest. P.M. is a scientific co-founder and scientific advisory board member of Navega Therapeutics, Pretzel Therapeutics, Engine Biosciences, and Shape Therapeutics, which have no commercial interests related to this study. The terms of these arrangements have been reviewed and approved by the University of California, San Diego in accordance with its conflict of interest policies.
Supporting Information
Supporting Information is available from the Wiley Online Library or from the author.
Data Availability
The raw data required to reproduce these findings will be made available to download from http://mali.ucsd.edu. The processed data required to reproduce these findings will be made available to download from http://mali.ucsd.edu.
Contributor Information
Michael Hu, Department of Bioengineering University of California San Diego, La Jolla, CA 92093, USA.
Amir Dailamy, Department of Bioengineering University of California San Diego, La Jolla, CA 92093, USA.
Xin Yi Lei, Department of Bioengineering University of California San Diego, La Jolla, CA 92093, USA.
Udit Parekh, Department of Electrical and Computer Engineering, University of California San Diego, La Jolla, CA 92093, USA.
Daniella McDonald, Biomedical Sciences Graduate Program, University of California San Diego, La Jolla, CA 92093, USA.
Aditya Kumar, Department of Bioengineering University of California San Diego, La Jolla, CA 92093, USA.
Prashant Mali, Department of Bioengineering University of California San Diego, La Jolla, CA 92093, USA.
References
- [1].Jain RK, Au P, Tam J, Duda DG, Fukumura D, Nat. Biotechnol 2005, 23, 821. [DOI] [PubMed] [Google Scholar]
- [2].Mandrycky C, Wang Z, Kim K, Kim D-H, Biotechnol. Adv 2016, 34, 422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Elson EL, Genin GM, Interface Focus 2016, 6, 20150095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Benam KH, Dauth S, Hassell B, Herland A, Jain A, Jang K-J, Karalis K, Kim HJ, MacQueen L, Mahmoodian R, Musah S, Torisawa Y, van der Meer AD, Villenave R, Yadid M, Parker KK, Ingber DE, Annu. Rev. Pathol.: Mech. Dis 2015, 10, 195. [DOI] [PubMed] [Google Scholar]
- [5].Elliott NT, Yuan F, J. Pharm. Sci 2011, 100, 59. [DOI] [PubMed] [Google Scholar]
- [6].Kimlin L, Kassis J, Virador V, Expert Opin. Drug Discovery 2013, 8, 1455. [DOI] [PubMed] [Google Scholar]
- [7].Folkman J, Nat. Med 1995, 1, 27. [DOI] [PubMed] [Google Scholar]
- [8].Murphy SV, Atala A, Nat. Biotechnol 2014, 32, 773. [DOI] [PubMed] [Google Scholar]
- [9].Wang Z, Abdulla R, Parker B, Samanipour R, Ghosh S, Kim K, Biofabrication 2015, 7, 045009. [DOI] [PubMed] [Google Scholar]
- [10].Kang H-W, Lee SJ, Ko IK, Kengla C, Yoo JJ, Atala A, Nat. Biotechnol 2016, 34, 312. [DOI] [PubMed] [Google Scholar]
- [11].Pedde RD, Mirani B, Navaei A, Styan T, Wong S, Mehrali M, Thakur A, Mohtaram NK, Bayati A, Dolatshahi-Pirouz A, Nikkhah M, Willerth SM, Akbari M, Adv. Mater 2017, 29, 1606061. [DOI] [PubMed] [Google Scholar]
- [12].Hinton TJ, Jallerat Q, Palchesko RN, Park JH, Grodzicki MS, Shue H-J, Ramadan MH, Hudson AR, Feinberg AW, Sci. Adv 2015, 1, e1500758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Park J, Lee SJ, Chung S, Lee JH, Kim WD, Lee JY, Park SA, Mater. Sci. Eng., C 2017, 71, 678. [DOI] [PubMed] [Google Scholar]
- [14].Zhang K, Fu Q, Yoo J, Chen X, Chandra P, Mo X, Song L, Atala A, Zhao W, Acta Biomater. 2017, 50, 154. [DOI] [PubMed] [Google Scholar]
- [15].Jia W, Gungor-Ozkerim PS, Zhang YS, Yue K, Zhu K, Liu W, Pi Q, Byambaa B, Dokmeci MR, Shin SR, Khademhosseini A, Biomaterials 2016, 106, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ouyang L, Highley CB, Sun W, Burdick JA, Adv. Mater 2017, 29, 1604983. [DOI] [PubMed] [Google Scholar]
- [17].Markstedt K, Mantas A, Tournier I, Martínez Ávila H, Hägg D, Gatenholm P, Biomacromolecules 2015, 16, 1489. [DOI] [PubMed] [Google Scholar]
- [18].Pati F, Jang J, Ha D-H, Won Kim S, Rhie J-W, Shim J-H, Kim D-H, Cho D-W, Nat. Commun 2014, 5, 3935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Kolesky DB, Homan KA, Skylar-Scott MA, Lewis JA, Proc. Natl. Acad. Sci. USA 2016, 113, 3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lee VK, Kim DY, Ngo H, Lee Y, Seo L, Yoo S-S, Vincent PA, Dai G, Biomaterials 2014, 35, 8092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen D-HT, Cohen DM, Toro E, Chen AA, Galie PA, Yu X, Chaturvedi R, Bhatia SN, Chen CS, Nat. Mater 2012, 11, 768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bercea M, Darie RN, Ni¸ta˘ LE, Morariu, Ind. Eng. Chem. Res 2011, 50, 4199. [Google Scholar]
- [23].Gilbert J, Washington C, Davies M, Hadgraft J, Int. J. Pharm 1987, 40, 93. [Google Scholar]
- [24].Orban JM, Wilson LB, Kofroth JA, El-Kurdi MS, Maul TM, Vorp DA, J. Biomed. Mater. Res., Part A 2004, 68A, 756. [DOI] [PubMed] [Google Scholar]
- [25].Yung CW, Wu LQ, Tullman JA, Payne GF, Bentley WE, Barbari TA, J. Biomed. Mater. Res., Part A 2007, 83A, 1039. [DOI] [PubMed] [Google Scholar]
- [26].Ranga A, Lutolf MP, Hilborn J, Ossipov DA, Biomacromolecules 2016, 17, 1553. [DOI] [PubMed] [Google Scholar]
- [27].Li P-S, Lee I. -Liang, Yu W-L, Sun J-S, Jane W-N, Shen H-H, Sci. Rep 2015, 4, 5600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Jose RR, Rodriguez MJ, Dixon TA, Omenetto F, Kaplan DL, ACS Biomater. Sci. Eng. 2016, 2, 1662. [DOI] [PubMed] [Google Scholar]
- [29].Li S, Liu Y-Y, Liu L-J, Hu Q-X, ACS Appl. Mater. Interfaces 2016, 8, 25096. [DOI] [PubMed] [Google Scholar]
- [30].Pimentel CR, Ko SK, Caviglia C, Wolff A, Emnéus J, Keller SS, Dufva M, Acta Biomater. 2018, 65, 174. [DOI] [PubMed] [Google Scholar]
- [31].Chia HN, Wu BM, J. Biol. Eng 2015, 9, 10.1186/s13036-015-0001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Goyanes A, Buanz ABM, Basit AW, Gaisford S, Int. J. Pharm. 2014, 476, 88. [DOI] [PubMed] [Google Scholar]
- [33].Prasad LK, Smyth H, Drug Dev. Ind. Pharm 2016, 42, 1019. [DOI] [PubMed] [Google Scholar]
- [34].Levato R, Visser J, Planell JA, Engel E, Malda J, Mateos-Timoneda MA, Biofabrication 2014, 6, 035020. [DOI] [PubMed] [Google Scholar]
- [35].Sawkins MJ, Mistry P, Brown BN, Shakesheff KM, Bonassar LJ, Yang J, Biofabrication 2015, 7, 035004. [DOI] [PubMed] [Google Scholar]
- [36].Müller AM, Hermanns MI, Skrzynski C, Nesslinger M, Müller K-M, Kirkpatrick CJ, Exp. Mol. Pathol 2002, 72, 221. [DOI] [PubMed] [Google Scholar]
- [37].Maleki M, Ghanbarvand F, Behvarz MR, Ejtemaei M, Ghadirkhomi E, Int. J. Stem Cells 2014, 7, 118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Justice BA, Badr NA, Felder RA, Drug Discovery Today 2009, 14, 102. [DOI] [PubMed] [Google Scholar]
- [39].Tibbitt MW, Anseth KS, Biotechnol. Bioeng 2009, 103, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hughes CS, Postovit LM, Lajoie GA, Proteomics 2010, 10, 1886. [DOI] [PubMed] [Google Scholar]
- [41].Calderon GA, Thai P, Hsu CW, Grigoryan B, Gibson SM, Dickinson ME, Miller JS, Biomater. Sci 2017, 5, 1652. [DOI] [PubMed] [Google Scholar]
- [42].Meng Z, Qiu Y, Lin KC, Kumar A, Placone JK, Fang C, Wang K-C, Lu S, Pan M, Hong AW, Moroishi T, Luo M, Plouffe SW, Diao Y, Ye Z, Park HW, Wang X, Yu F-X, Chien S, Wang C-Y, Ren B, Engler AJ, Guan K-L, Nature 2018, 560, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Qiu Y, Ahn B, Sakurai Y, Hansen CE, Tran R, Mimche PN, Mannino RG, Ciciliano JC, Lamb TJ, Joiner CH, Ofori-Acquah SF, Lam WA, Nat. Biomed. Eng 2018, 2, 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Fleming JM, Miller TC, Meyer MJ, Ginsburg E, Vonderhaar BK, J. Cell. Physiol 2010, 224, 795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kocatürk B, Versteeg HH, J. Visualized Exp 2015, 96, e51967. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.