Abstract
Acute cardiorenal syndrome (CRS-1) is a morbid complication of acute cardiovascular disease. Heart-to-kidney signals transmitted by “cardiorenal connectors” have been postulated, but investigation into CRS-1 has been limited by technical limitations and a paucity of models. To address these limitations, we developed a translational model of CRS-1, cardiac arrest and cardiopulmonary resuscitation (CA/CPR), and now report findings from nanoscale mass spectrometry proteomic exploration of glomerular filtrate 2 hours after CA/CPR or sham procedure. Filtrate acquisition was confirmed by imaging, molecular weight and charge distribution, and exclusion of protein specific to surrounding cells. Filtration of proteins specific to the heart was detected following CA/CPR and confirmed with mass spectrometry performed using urine collections from mice with deficient tubular endocytosis. Cardiac LIM protein was a CA/CPR-specific filtrate component. Cardiac arrest induced plasma release of cardiac LIM protein in mice and critically ill human cardiac arrest survivors, and administration of recombinant cardiac LIM protein to mice altered renal function. These findings demonstrate that glomerular filtrate is accessible to nanoscale proteomics and elucidate the population of proteins filtered 2 hours after CA/CPR. The identification of cardiac-specific proteins in renal filtrate suggests a novel signaling mechanism in CRS-1. We expect these findings to advance understanding of CRS-1.
Keywords: Cardiology, Nephrology
Keywords: Cardiovascular disease, Protein traffic, Proteomics
Characterization of the renal filtrate proteome during acute cardiorenal syndrome demonstrates filtration of cardiac proteins, including a cardiac injury–specific protein.
Introduction
Acute cardiorenal syndrome worsens short- and long-term outcomes of patients suffering from myocardial infarction, cardiac arrest, and acute heart failure. Even with improved therapies, including rapid mechanical restoration of flow, the incidence of acute kidney injury (AKI) due to cardiac failure is increasing (1–3). Humoral signals (“cardiorenal connectors”) between the injured heart and the vulnerable kidney have been postulated, and early proteinuria is observed in acute cardiorenal syndrome (4). Since proteins are among the most important signaling molecules, it is possible that cardiorenal connectors could be filtered proteins, which could subsequently modulate tubular cell function after endocytosis via the promiscuous receptor megalin (5–8). It is therefore critical to understand which proteins are in the filtrate in acute cardiorenal syndrome. In contrast to filtrate, when protein is found in urine, it may be filtered or secreted; therefore, urine studies alone cannot establish filtration as the source of protein. We previously found that cardiac arrest and cardiopulmonary resuscitation (CA/CPR) induces glomerular filtration of high-molecular-weight Ficoll, and others working in renal ischemia/reperfusion (I/R) have found concordant results with high-molecular-weight dextran and protein (9–11). However, in vivo assessment of glomerular filtration of large molecules has been limited by reliance on surrogates, such as imaging or recovery of systemically administered single-molecule tracers (12, 13). Discovery of novel protein in the filtrate therefore requires discovery studies of the filtrate itself, which have been difficult to date. Thus, we hypothesized that after CA/CPR, direct assessment of the proteins in Bowman’s urinary space would identify cardiac injury–specific proteins that would be candidate cardiorenal connectors, most likely of high molecular weight, leading to increased mean filtrate protein molecular weight. To overcome limitations in the assessment of the protein content of glomerular filtrate and directly assess the protein content of nanoliter-range samples, we used 2-photon microscopy to perform aspiration of fluid from Bowman’s space in mice and subjected the aspirate to a liquid chromatography/mass spectrometry assay that is orders of magnitude more sensitive than prior technology (14). We then compared the distribution and identity of proteins identified by nanoscale mass spectrometry in the filtrate of mice subjected to CA/CPR or sham procedure in order to determine whether additional high-molecular-weight proteins could be identified in early acute cardiorenal syndrome and inferred protein origins using a proteomic spatial map. Finally, we performed a more conventional mass spectrometry assay on urine from mice with deficient proximal tubular endocytosis and established the filtration of a cardiac protein, which we also determined is elevated in the plasma of CA/CPR-treated mice and human survivors of cardiac arrest.
Results
Multiphoton renal imaging in mice with acute cardiorenal syndrome.
Potassium chloride injection resulted in cardiac arrest in all mice assigned to CA/CPR, as previously described (15). All mice remained normothermic during CA/CPR or sham procedure. 78% of CA/CPR mice and 100% of sham mice survived through renal imaging preparation and were imaged. In a separate cohort, 2 hours postprocedurally, serum urea nitrogen was elevated in mice subjected to CA/CPR, compared with that in sham treated mice (30.4 ± 2.2, n = 4 vs. 46.9 ± 5.4 ng/ml, n = 4, P = 0.03). All animals that started the imaging protocol survived to complete imaging. As compared with sham, filling of superficial cortical vessels with injected FITC-dextran appeared reduced 2 hours after CA/CPR. The number of FITC-fluorescent vessels was reduced, and overall, vessels appeared to be of smaller diameter. Red cell velocity within vessels also appeared reduced, and in some areas, focal vascular dilation with intravascular fluorescence signal voids indicated coagulation. Tubular lumens contained cell debris and blebs. These findings suggest reduced, heterogeneous cortical blood flow in the early reperfusion period due to acute cardiorenal syndrome, consistent with our published studies using laser Doppler measurements (11, 16) and findings from other models of AKI (17). Representative images are presented in Figure 1 and Supplemental Videos 1 and 2 (supplemental material available online with this article; https://doi.org/10.1172/jci.insight.122130DS1).
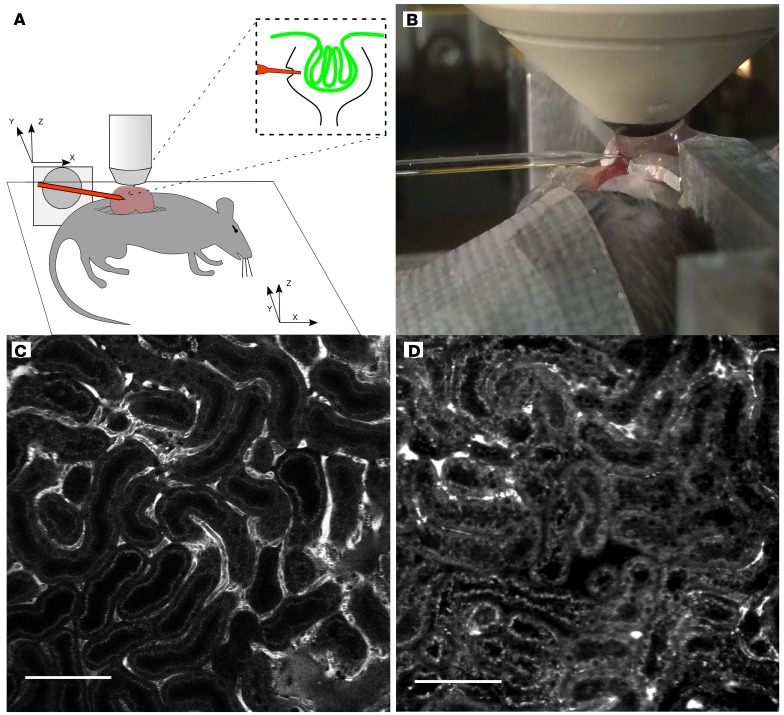
Figure 1. In vivo renal multiphoton imaging acquired after sham treatment and CA/CPR depicts vascular and tubular pathophysiology in early cardiorenal syndrome.
(A) Experimental preparation for multiphoton micropuncture. The kidney was partially exteriorized, immersed in agar, and immobilized with a metal, externally fixated frame. A glass coverslip, overlying the agar, provided a reference point and a stable surface for the water column, which completed the imaging interface to the microscope objective. The pipette tip positioning system was registered in all three planes with the microscope stage positioning system, allowing precision guidance to Bowman’s space of a preidentified glomerulus. (B) Photograph of imaging/accession procedure. The pipette is visible from the left, with the tip 30 μm below the coverslip edge. Partially exteriorized kidney, immobilizing frame, water column, and objective are all visible to the right. (C) Multiphoton image of superficial renal cortex 2 hours after sham procedure. Bright FITC-dextran signal visible in peritubular capillaries with orthogonal signal voids, which are due to red cell movement, indicating intact blood flow. Tubular cell bodies are not opacified, and autofluorescence demonstrates the brush border only. (D) Superficial renal cortex 2 hours after CA/CPR. Fewer FITC-dextran opacified vessels are evident, and they are smaller in caliber. Tubular lumens contain debris, and cell bodies exhibit increased autofluorescence, as does brush border, compared with sham. Images are representative of 7 experiments per condition. Scale bars: 100 μm.
Multiphoton-guided acquisition of samples from Bowman’s space.
Samples were acquired from the Bowman’s space of 14 mice (7 after sham procedure and 7 after CA/CPR) using pipettes with <10 μm opening and 45° bevel, demonstrated by imaging prior to each experiment. The vertical depth (from the capsule directly under the objective to the top of the glomerular tuft) of accessed glomeruli was 64.5 ± 16.7 μm in sham-treated mice and 84.7 ± 20.5 μm (P = 0.07) in CA/CPR-treated mice. The lateral traverse of the micropipette (from contact with the renal capsule to within Bowman’s space) was 554.3 ± 177.0 μm in sham-treated mice and 887.4 ± 312.7 μm (P = 0.08) in CA/CPR-treated mice. Pipettes were observed through access and removal, and no pipette fractured during acquisition procedures. Representative video of an acquisition procedure is provided in Supplemental Video 3. Representative images of pipettes in Bowman’s space are presented in Supplemental Figure 1.
Peptide and protein identification in Bowman’s space aspirate.
Four samples from each group underwent LC/MS analysis. Overall, in the 8-sample cohort 4066 peptide identifications were made. Requiring only 1 unique peptide per protein resulted in identification of 1337 proteins, while a more stringent requirement for 2 unique peptides per protein identified 653 proteins (with a mean of 5.2 ± 5.0 identified peptides/protein). Single-sample spectral counts were low (range 0–54) in accordance with previous estimates of low-protein content in Bowman’s space. MaxQuant label-free quantitation (LFQ), which enables proteomic quantification in small samples (18), was therefore used and yielded a favorable dynamic range (Figure 2, A and B) of over 3 orders of magnitude.
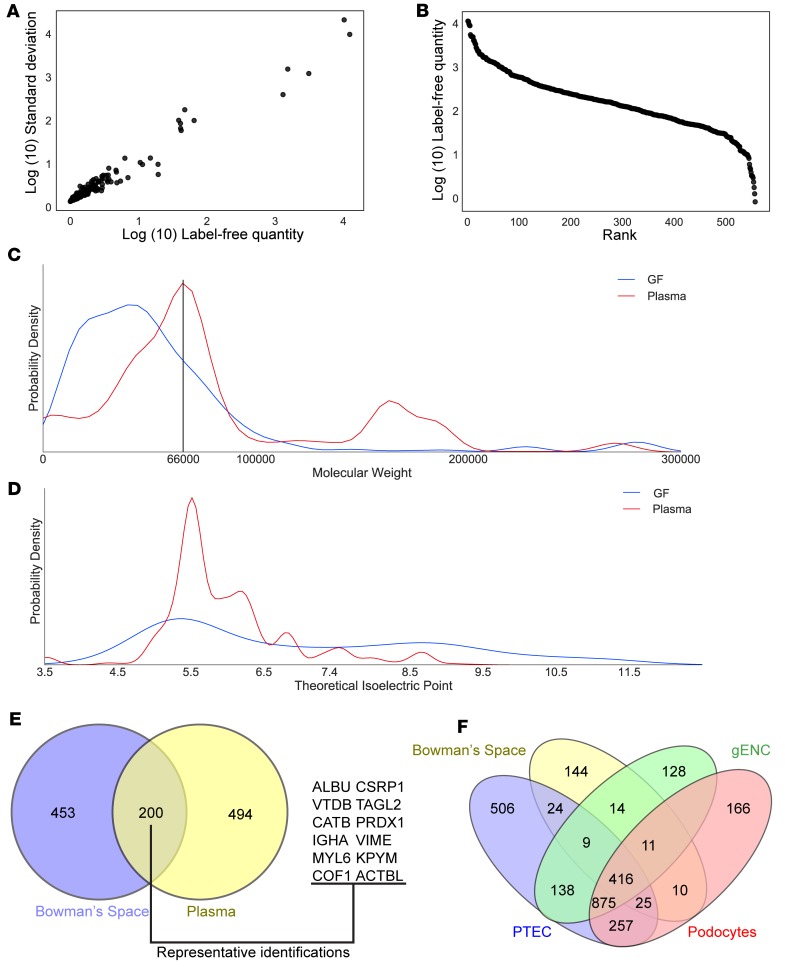
Figure 2. Bowman’s space aspirate contains glomerular filtrate.
(A) Heterogeneity of spectral counts of peptides from Bowman’s space aspiration samples. For each unique protein identification, average spectral count versus standard deviation across 8 samples is displayed. (B) Three-log dynamic range obtained in Bowman’s space aspirate samples using MaxQuant Label-Free Quantification–based (LFQ-based) abundance measurements. For each identification, rank in the total number of identifications is displayed versus log abundance. (C) Kernel density estimate distribution plot of abundance of protein identifications at each molecular weight in Bowman’s space aspirate (GF) and plasma. Mean of 4 experiments from each group. The black vertical line at 66 kDa indicates the molecular weight of albumin, the most prevalent plasma protein. While this is the most prevalent molecular weight region in plasma, and many proteins with greater molecular weight were identified in plasma, in Bowman’s space aspirate very few proteins were identified above this molecular weight. (D) Kernel density estimate plot of abundance of protein identifications at each theoretical isoelectric point in Bowman’s space aspirate and plasma. Mean of 4 experiments from each group. Proteins identified in Bowman’s space demonstrate a greater mean theoretical isoelectric point and are thus predicted to be more positively charged at any given pH. (E) Venn diagram of protein identifications in Bowman’s space and plasma. 200 of the 653 identifications in Bowman’s space aspirates were also identified in plasma. Representative gene symbols of plasma/Bowman’s space mutually identified proteins are shown. (F) Mass spectrometry of Bowman’s space aspirate and of lysate from surrounding cell types (proximal tubular epithelial cells [PTEC], glomerular endothelial cells [gENC], and podocytes) indicates minimal contamination of Bowman’s space aspirate with renal cell proteins. <10% of total identifications in Bowman’s space aspirate were specific to surrounding cell types.
Identification of Bowman’s space aspirate as glomerular filtrate.
Glomerular filtrate is believed to be a size- and charge-exclusion filtrate of plasma. Electrodynamic and electrostatic models of filtration have been proposed that posit neutral or negative charge bias in the filtrate. However, no prior assessment of the distribution of molecular weight and charge of filtered proteins in vivo exists. Accordingly, we assessed the distribution of predicted molecular weight and theoretical isoelectric point (pI) of each protein identified in Bowman’s space aspirate. The mean predicted molecular weight of identified proteins was 55,134 ± 53,738 Da. Figure 2C displays the molecular weight distribution for all proteins identified in Bowman’s space aspirate. The molecular weight distribution peak (single mode) was 41,737 D and demonstrated a sharp cut-off at 60 kDa. As albumin (molecular weight 66 kDa) is the most prevalent plasma protein, though it has a micropuncture-derived filtration coefficient near 0.0005 in rodents (19, 20), this observed distribution and cutoff is consistent with a size-exclusion plasma filtrate.
To determine whether Bowman’s space aspirate is distinct from plasma, we used identical methodology to assess molecular weight and pI of proteins identified in mouse plasma. Plasma protein modal molecular weight was 65,892 Da, mean molecular weight was 60,025 ± 67,842 Da, and mean theoretical was pI 6.41 ± 1.26 m/l. The theoretical pI for Bowman’s space aspirate protein was 7.03 ± 1.88 m/l. Thus, the Bowman’s space aspirate to plasma ratio for modal molecular weight was 0.63, that for molecular weight was 0.92, and that for pI was 1.10, demonstrating that proteins from Bowman’s space aspirate are of lower molecular weight and greater positive charge than those in plasma. Figure 2C depicts the molecular weight distribution found in Bowman’s space aspirate superimposed upon that of plasma, and Figure 2D depicts the pI distribution observed in Bowman’s space aspirate superimposed on that of plasma. To determine whether the low molecular weight, positive charge–biased protein distribution observed comprised a limited set of plasma proteins, we compared identifications from plasma and Bowman’s space; 200 proteins were common to both sets (Figure 2E). Taken together, these data indicate that Bowman’s space aspirate contains size- and charge- restricted proteins in common with plasma.
To determine the extent to which identified proteins were derived specifically from intrarenal cells during pipette passage, we performed mass spectrometry on whole cell lysates of cultured podocytes, tubular epithelial cells, and glomerular endothelial cells, requiring 2 unique peptides for each protein in both of 2 experimental (biological) replicates. In podocytes, 1839 identifications were made. In glomerular endothelial cells, 1670 identifications were made. In proximal tubular epithelial cells (PTECs), 2250 identifications were made. Human orthologs of mouse protein identified in Bowman’s space aspirate were compared with those identified in cell culture, and the results are depicted in Figure 2F. Overall, 416 (6.5% of all) identifications were shared between all 4 assays (Bowman’s space aspirate, glomerular endothelial cell lysate, podocyte lysate, and PTEC lysate). 875 (15.2% of all cell identifications) were shared among the 3 cell culture sets. 257 (4.5% of all cell identifications) identifications shared between PTECs and podocytes represent the largest set of identifications shared between 2 cell types, expected given the shared epithelial lineage of the 2 cell types. 144 proteins (mean molecular weight 55,669 ± 40,432 Da) were exclusive to Bowman’s space aspirate. The number of protein identifications in Bowman’s space aspirate that were exclusive to any single cell type was <10% of the total Bowman aspirate set (14 identifications for glomerular endothelial cells, 10 for podocytes, and 24 for PTECs), indicating that few renal cell–specific proteins were present in the aspirate. Therefore, there was a lack of significant protein contamination of the aspirate samples from cells adjacent to the pipette ingress and egress route.
Thus, the fluid we aspirated from Bowman’s space contained plasma proteins with a size and theoretical pI fraction consistent with glomerular filtrate. Few renal cell–specific protein identifications were made in Bowman’s space aspirate, and substantial numbers of low-molecular-weight plasma proteins were identified in Bowman’s space aspirate. Therefore, the Bowman’s space aspirate distinctively contains glomerular filtrate.
Filtrate proteins in early acute cardiorenal syndrome.
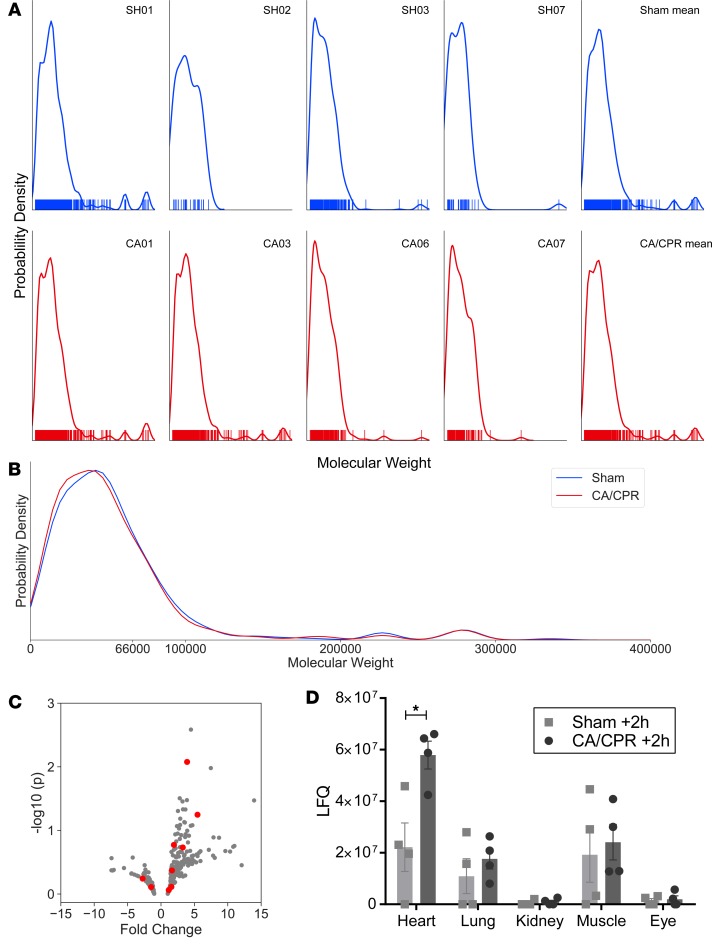
Identified protein molecular weight was reduced after CA/CPR (mean molecular weight 43,123 ± 39,850 Da in sham, 42,093 ± 41,438 Da in CA/CPR, P < 0.0001). No individual protein was significantly regulated by CA/CPR, and no molecular weight–defined band was significantly altered. Since measurement of volume, and therefore concentration, was not possible in these experiments due to modifications required for mass spectrometry as an endpoint (see Methods), this characterization indicates the identification of increased numbers of low-molecular-weight proteins, which reduce the mean molecular weight of filtrate. Since protein content of filtrate is largely derived from nonrenal organs and because CA/CPR exposes the entire organism to I/R, we assessed whether protein from other organs could be differentially regulated in the filtrate after CA/CPR using spatial deconvolution (as described in the Methods). Spatial deconvolution revealed baseline low abundance of kidney-specific protein (an order of magnitude less than cardiac-, brain-, or lung-specific protein), and kidney-specific protein was not upregulated 2 hours after CA/CPR. There was a CA/CPR-mediated difference in cardiac-specific protein (sham 2.2 × 107 vs. 5.8 × 107 LFQ, P = 0.0008, n = 4). Figure 3 illustrates these results. Together, these data indicate that the filtrate is altered after CA/CPR by the inclusion of proteins from organs injured during systemic I/R, those from the heart being most abundant.
Figure 3. Glomerular filtrate includes cardiac proteins following CA/CPR; comparison of nanoproteomic identifications in glomerular filtrate between sham- and CA/CPR-treated mice.
(A) Kernel density estimate plots of inferred protein abundance (y axis) by inferred molecular weight (x axis) for each mouse. Sham (blue, top row) and CA/CPR (red, bottom row) experiments are shown, with the mean of each group plotted to the far right. Rug plots demonstrate the molecular weight of each protein identification. The molecular weight distribution of identified proteins is remarkably constant between individual animals and across treatments. (B) The mean molecular weight of all identified proteins was reduced after CA/CPR as compared with sham (mean molecular weight 43123 ± 39850 Da in sham, 42093 ± 41438 Da in CA/CPR, P < 0.0001, n = 4 per group). No specific protein or molecular weight band was significantly changed by CA/CPR. (C) Volcano plot demonstrates that proteins identified as cardiac specific (highlighted in red) are primarily upregulated following CA/CPR. (D) Comparison of organ-specific proteins identified in the filtrate, and their sources, in sham- and CA/CPR-treated mice (n = 4 per group, ANOVA). Cardiac-specific protein identifications were significantly upregulated after CA/CPR. Few kidney-specific proteins were identified (comparable to the number of proteins specific to the eye), and they were not regulated 2 hours after CA/CPR (mean ± SEM).
Assessment of systemic protein filtration in mice with deficiency of proximal tubular endocytosis.
To confirm the finding of systemic proteins in the glomerular filtrate and enrichment with cardiac proteins following CA/CPR, we performed quantitative tandem mass-tag mass spectrometry of the urine of mice with deficient proximal tubular endocytosis due to proximal tubule–specific mosaic deletion of the endocytic receptor, megalin (LRP2). We used LRP2fl/fl;apoECre mice and LRP2fl/fl; (Cre–) littermate controls (n = 5), collecting urine for 24 hours before and after CA/CPR. Urine output was 1.0 ± 07 ml/24 hours before CA/CPR and 0.5 ± 0.4 ml/24 hours after CA/CPR (P = 0.43). Mean serum urea nitrogen was 65 ± 69 ml/dl after CA/CPR, comparable to our previously published values in wild-type mice (16). In a separate cohort (n = 12), 24-hour survival after CA/CPR was not different between LRP2fl/fl;apoECre mice and LRP2fl/fl; control mice. Overall, 2029 proteins were quantified in 10 samples with 2 unique peptides required (Figure 4A). There were nearly 6 orders of magnitude difference in reporter ion intensity between the most prevalent species and the least (Supplemental Figure 2). Prior to CA/CPR, LRP2fl/fl;apoECre status conferred reduced urine excretion of LRP2 (normalized intensity 2.2 × 106 vs. 10.7 × 106 in LRP2fl/fl; mice, fold change = –4.9, P = 7.8 × 10–8, FDR 2.0 × 10–6, n = 5) and increased reporter ion intensity for 13 known LRP2 ligands, for example, retinol-binding protein (fold change = 9.5, P < 0.001, FDR = 0.008) and vitamin D–binding protein (fold change = 57.3, P < 0.000001, FDR = 6.0 × 10–10; see Supplemental Table 1). Overall, presence of the apoEcre resulted in upregulated urine excretion of 342 identified proteins primarily of low molecular weight (mean molecular weight 50,676 ± 49,047 Da; Figure 4B).
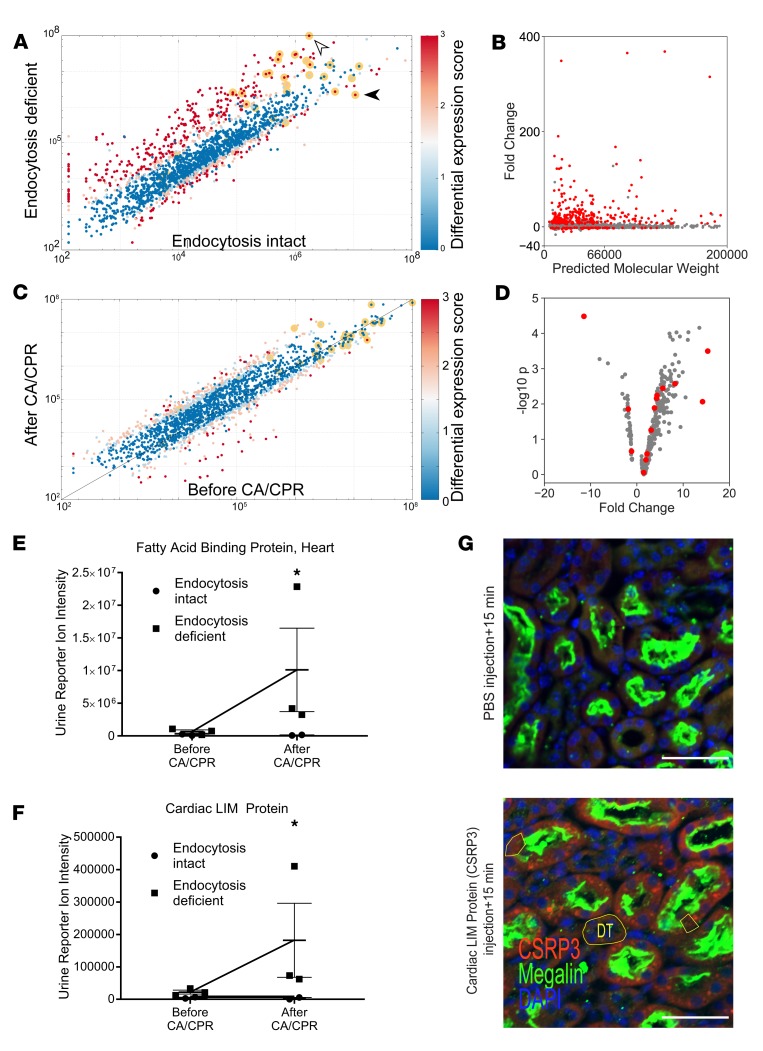
Figure 4. Low-molecular-weight proteinuria in LRP2fl/fl;apoECre mice reveals filtered proteins after cardiac arrest and cardiopulmonary resuscitation.
(A) LRP2fl/fl;apoECre confers proximal tubule–specific mosaic deletion of the critical endocytosis protein, megalin, resulting in proteinuria. Scatterplot compares the log reporter ion intensity of each of 2029 identified urine proteins in mice with megalin interference (“endocytosis deficient,” LRP2fl/fl;apoECre, y axis) with the log intensity of the same protein in the urine of mice without megalin interference (“endocytosis intact,” Cre– littermate controls), prior to cardiac arrest and cardiopulmonary resuscitation (CA/CPR). Color indicates differential expression score, with red indicating FDR < 0.01 and P < 0.001. Known megalin ligands are highlighted with a yellow halo. LRP2fl/fl;apoECre status conferred increased urine content of known megalin ligands, including albumin (white arrowhead) and decreased urine content of megalin itself (black arrowhead). (B) Primarily low-molecular-weight proteins (x axis) are upregulated in endocytosis-deficient mouse urine. Each identified protein is plotted, and those with FDR < 0.01 are highlighted in red. (C) CA/CPR in endocytosis-deficient mice upregulated 70 identified urine proteins. All proteins identified in LRP2fl/fl;apoECre mice are plotted as log10(reporter ion intensity) before (x axis) and 24 hours after (y axis) CA/CPR. (D) Cardiac-specific proteins (highlighted in red) comprise many of the urine proteins upregulated by endocytosis deficiency. (A–D) P values were derived from t tests with Benjamini-Hochberg correction for multiple comparisons after normalization. (E) A plasma biomarker of myocardial injury, fatty acid–binding protein, heart, is selectively upregulated after CA/CPR in the urine of endocytosis-deficient mice but not that of endocytosis-intact mice; megalin interference may allow filtered plasma biomarkers to pass into the excreted urine. (F) Cardiac LIM protein, a myocardiocyte differentiation factor highly specific to cardiomyocytes, is upregulated in the urine of endocytosis-deficient mice after CA/CPR. (E and F) *P < 0.01 by Student’s t test; mean ± SEM. (G) After intravenous administration of CSRP3 to LRP2fl/fl;apoECre mice, CSRP3+ punctae are visualized in megalin+ proximal tubular epithelial cells but are not present in megalin– distal tubule cells (DT) and are attenuated in megalin-null proximal tubule cells (examples outlined in yellow). PBS injection resulted in no CSRP3 signal. Representative of 3 experiments. Scale bars: 50 μm.
In LRP2fl/fl;apoECre mice, CA/CPR was associated with increased urine excretion of 58 proteins (mean fold change = 7.1 ± 2.5, P < 0.01, FDR < 0.05; see Figure 4C, Supplemental Table 2, and Supplemental Figure 3). The third most abundantly upregulated urine protein after CA/CPR in LRP2fl/fl;apoECre mice was troponin I (TNNI2, mean fold change = 15.32), followed by fatty acid–binding protein, heart (FABPH, mean fold change = 15.2); among proteins with organ specificity score (OSS) >4.0, these were the two most abundantly upregulated proteins.
If interference with tubular protein endocytosis allows transit of filtered proteins to end-urine, there should be overlap between glomerular filtrate (GF) and urine from LRP2fl/fl;apoECre mice. Accordingly, we compared protein identifications in GF (653 total) with proteins upregulated in the urine of endocytosis-deficient LRP2fl/fl;apoECre mice. 36 of 653 GF proteins, including vitamin D–binding protein, retinol-binding protein, gelsolin, lactotransferrin, galectin 1, galectin 3, and cathepsin B, were upregulated in endocytosis-deficient mice. This confirms that urine from LRP2fl/fl;apoECre mice may be used to make conditional inferences regarding filtration of selected proteins.
Finally, we performed spatial deconvolution on protein identified after CA/CPR in LRP2fl/fl;apoECre mice. Prior to CA/CPR, heart-specific (having OSS >4.0) protein intensity was not different from the overall mean intensity for all urine identifications (all: 1.9 × 106 ± 3.4 × 107, organ specific: 12,900 ± 224,400, P = 0.72). However, 24 hours after CA/CPR, heart-specific proteins were significantly upregulated (fold change from before to after CA/CPR = 3.6 ± 6.6 for heart-specific proteins vs. 2.0 ± 3.0 for all identified proteins, P = 0.0008; Figure 4D). Cardiac proteins that were upregulated were all of low molecular weight (n = 11, molecular weight 23,520 ± 17,442 Da), compared with those that were not upregulated or downregulated (n = 3, P = 0.02, molecular weight 103,330 ± 109,845 Da), suggesting the role of filtration. Accordingly, the high-molecular-weight cardiac biomarker myosin-binding protein C, cardiac (MyBPC3, molecular weight 140.6 kDa), which was not detectable in any filtrate samples, was detectable in plasma 2 hours after CA/CPR (mean concentration 14.9 ± 4.7 ng/ml, n = 4). The third-most upregulated heart-specific protein after CA/CPR in LRP2fl/fl;apoECre mice was cardiac LIM protein (CSRP3, see Supplemental Table 3). Cardiac LIM protein is a myocardial differentiation factor, which has previously been identified in mass spectrometry study of coronary sinus blood in an I/R model and proposed as a potential biomarker of myocardial infarction (21). We therefore tested whether cardiac LIM protein could be a novel plasma marker of myocardial injury that undergoes renal filtration. First, we assessed the urine reporter ion intensity for FABPH, a known plasma myocardial injury marker. FABPH was sharply upregulated by CA/CPR in the urine of LRP2fl/fl;apoECre mice only, suggesting that after CA/CPR plasma cardiac biomarkers may be filtered and revealed in the end-urine of endocytosis-deficient mice (Figure 4E). Accordingly, we assessed the urine reporter ion intensity for cardiac LIM protein, which was specifically upregulated only after CA/CPR in the urine of endocytosis-deficient mice (Figure 4F).
To confirm filtration of cardiac LIM protein, we injected recombinant cardiac LIM protein into healthy LRP2fl/fl;apoECre mice. Renal immunofluorescence images following intravenous injection demonstrated concentration of cardiac LIM protein signal at the tubular brush border and within tubular epithelial cells, as compared with images following injection of PBS, which demonstrated no cardiac LIM protein signal (Figure 4G). Taken together, these findings confirm that CA/CPR specifically upregulates cardiac protein in the glomerular filtrate and that cardiac LIM protein is renally filtered.
Plasma and renal cardiac LIM protein after cardiac arrest.
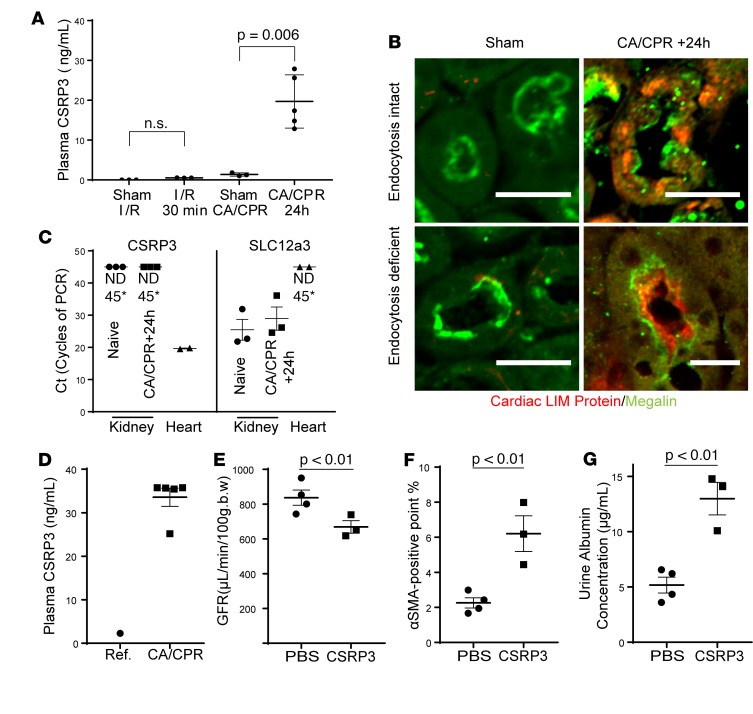
To determine whether cardiac LIM protein specifically filters after CA/CPR, we interrogated plasma cardiac LIM protein after CA/CPR (which leads to renal injury with cardiac injury), renal I/R (which leads to renal injury without acute cardiac injury), and respective sham procedures. Mice subjected to CA/CPR demonstrated 15 times more serum cardiac LIM protein than mice subjected to sham procedure, while mice subjected to renal I/R injury did not demonstrate elevated plasma cardiac LIM protein (sham CA/CPR: 1.37 ± 0.4, CA/CPR: 20.9 ± 4.82, P = 0.016; sham I/R: 0 ± 0 ng/ml, I/R: 0.5 ± 0.01 ng/ml, P = 0.9, n = 3/group; Figure 5A). Elevated plasma cardiac LIM protein was reflected in renal tissue; immunofluorescence demonstrated the presence of cardiac LIM protein in punctae within tubular epithelial cells and in tubular lumens after CA/CPR but not sham procedure (Figure 5B). Next, to confirm the source of renal cardiac LIM protein was not the kidney itself, we tested gene translation in the kidney 24 hours after CA/CPR, subjecting renal lysates from sham and CA/CPR mice to quantitative PCR. mRNA encoding CSRP3, the gene encoding cardiac LIM protein, was not detectable at 45 cycles of PCR in renal tissue after sham or CA/CPR, while the gene encoding sodium-chloride cotransporter (SLC12a3), a positive control, was readily detected in all renal lysates (Figure 5C).Since very limited data suggest cardiac LIM protein may appear in human plasma after myocardial injury (21), we further explored this finding in intensive care unit patients following CA/CPR. Etiologies associated with CA included myocardial infarction (n = 2), hemorrhagic pulseless electrical activity (n = 2), and unknown etiologies (n = 1). Plasma samples used for measurement were obtained 23.7 ± 5.9 hours after CA. Mean cardiac troponin I (a sensitive biomarker of myocardial injury) was 9.4 ± 10.4 ng/ml (more than 10 times the upper limit of normal, 0.8 ng/ml) in 3 patients for whom this value was available. The mean plasma cardiac LIM protein concentration was 54.4 ± 22.1 ng/ml. As the normative plasma cardiac LIM protein concentration is nearly 0 (22), these patients demonstrate very elevated plasma cardiac LIM protein concentration, concordant with that of mice subjected to CA/CPR (Figure 5D). Taken together, these data indicate that plasma cardiac LIM protein rises acutely after CA.
Figure 5. Cardiac LIM protein originates outside the kidney in cardiac arrest and transits from plasma to tubular epithelial cells.
(A) Plasma cardiac LIM protein (CSRP3) levels are greatly increased 24 hours after CA/CPR but not 24 hours after renal-only ischemia, induced by renal pedicle clamping. (B) Tubular detection of cardiac LIM protein by immunohistochemistry is specific to CA/CPR. Cardiac LIM protein was not detected after sham procedure but was present in tubular epithelial cells 24 hours after CA/CPR in wild-type, endocytosis-intact mice; it concentrated at the brush border and within some tubular cells in endocytosis-deficient mice. Representative of 3 experiments per genotype. Scale bars: 50 μm. (C) CSRP3 is not expressed in the kidney in surgically naive mice or 24 hours after CA/CPR. Shown are the number of cycles of PCR required to detect PCR product of CSRP3- or thiazide-sensitive sodium/chloride cotransporter (NCC, SCL12a3), a kidney-specific gene. Lower numbers of cycles denote increased levels of expression. CSRP3 was not detected in kidney (*ND45, not detected after 45 cycles of PCR), while SLC12a3 was easily detected in kidney. CSRP3 was readily detected in heart tissue, while SLC12a3 was not. (D) Critically ill human survivors of cardiac arrest and cardiopulmonary resuscitation, 23.9 ± 5.9 hours after CA, demonstrate mean plasma CSRP3 comparable to that of CA/CPR mice, which was elevated compared with reference human value (22). (E) 28 days after administration of CSRP3 to healthy, wild-type mice, GFR was reduced. (F) Tubulointerstitial αSMA was increased and (G) urine albumin was also increased, indicating that CSRP3 administration negatively affects renal function in a manner consistent with early chronic kidney disease. (A and E–F) Mean ± SEM displayed. (A) P value was calculated ANOVA with Sidak’s test. (E and F) P values were calculated using Student’s t test.
Effect of administration of cardiac LIM protein to mice.
Because cardiac LIM protein is a transcriptional coactivator that induces cardiomyocyte differentiation (23) and, through activation of MyoD, may upregulate expression of α smooth muscle actin (αSMA) in epithelial cells (24), we next tested whether exogenous cardiac LIM protein could alter renal function and αSMA deposition. One mouse died 2 hours after the second injection of cardiac LIM protein; necropsy revealed no clear cause. Twenty-hour hours after completing injections, the urine cardiac LIM protein was not different in PBS- and cardiac LIM protein–injected mice (1.3 ± 0.1 ng/ml vs. 1.3 ± 0.2 ng/ml, n = 3–4, P = 0.94), consistent with the hypothesis that intact megalin in wild-type mice excludes cardiac LIM protein from the urine. At 28 days, although urine output was not different between groups (0.19 ± 0.03 ml/g/24 hours vs. 0.16 ± 0.05 ml/g/24 hours, n = 3–4), GFR was reduced in mice treated with cardiac LIM protein (837 ± 44 μl/min/100 g in PBS-injected group vs. 669 ± 36 μl/min/100 g in cardiac LIM protein–injected mice, P = 0.03, n = 3–4, Figure 5E). Quantitative stereologic evaluation of tubulointerstitial αSMA revealed a cardiac LIM protein–mediated increase (2.3% ± 0.3% positive points, n = 4 vs. 6.2% ± 1.0% positive points, P < 0.01, n = 3–4; Figure 5G), and urine albumin concentration was also increased in cardiac LIM–treated mice (5.2 ± 0.7 μg/ml vs. 13.0 ± 1.5 μg/ml, P < 0.01, n = 3–4; Figure 5F). The reduction in GFR combined with relative albuminuria and increased tubulointerstitial αSMA due to injection of cardiac LIM protein strongly suggest that cardiac LIM protein alters renal function.
Discussion
We characterized acute change in the filtrate proteome in early acute cardiorenal syndrome within hours of CA/CPR. Our complementary approach of direct nanoproteomic assessment of Bowman’s space aspirate paired with quantitative mass spectrometry on urine from mice with defective tubular endocytosis enabled what we believe to be novel findings and mitigated the considerable challenges of identifying filtered proteins using urine proteomics alone. We found that cardiac proteins appear in the filtrate hours after CA/CPR, and we identified what we believe to be a previously unknown, cardiac injury–specific component of the filtrate, cardiac LIM protein, in the urine of endocytosis-deficient mice. We confirmed the filtration of cardiac LIM protein with renal immunofluorescence following injection of recombinant protein and determined that cardiac LIM protein is specifically elevated in the plasma and after CA/CPR in mice and in human cardiac arrest survivors.
Cardiac LIM protein is a transcription coactivator, found on the cardiomyocyte z-disk, which translocates from the cytosol to the nucleus to initiate cardiomyocyte differentiation (25, 26). Current understanding of the role of this protein in human disease is limited to the heart, where gene association studies in humans and animal models have demonstrated a role in hypertrophic and dilated cardiomyopathy (27–30). In vitro, cardiac LIM protein is released into culture media by necrotic but not apoptotic cardiomyocytes (31). Ex vivo, release into myocardial perfusate is specific to I/R injury, with kinetics comparable to that of troponin I, a well-described cardiac biomarker (21). Our study is the first to our knowledge to associate elevation in plasma cardiac LIM protein with CA/CPR. Given the prior studies, we interpret the rise in cardiac LIM protein after cardiac arrest as an indication of myocardial I/R injury, which occurs during arrest and return of circulation. Although, with our model, it is not possible to eliminate myocardial mechanical injury from chest compression as a potential cause of biomarker release.
Whether cardiac LIM protein has a role in renal disease is unknown. However, three findings of our study suggest this may be worthy of additional investigation. First, cardiac LIM protein was only present in the urine after CA/CPR in mice with impaired renal protein endocytosis due to mosaic deletion of proximal tubular megalin. Since proximal tubule–specific deletion of megalin is very unlikely to eliminate cardiac release of cardiac LIM protein due to cardiac injury, this finding suggests that cardiac LIM protein interacts with megalin, a major mediator of protein endocytosis in the PTEC. Known megalin interactors, such as vitamin D–binding protein, which undergo endocytosis in the proximal tubule and exit the endosome to perform their critical roles (6, 32), were similarly upregulated in the urine by the proximal tubule–specific deletion of megalin (above and Supplemental Table 1). Second, the appearance of cardiac LIM protein signal within tubular epithelial cell bodies after injection suggests the possibility that this protein undergoes endocytosis in the proximal tubule. Third, injection of 5 μg recombinant cardiac LIM protein into healthy wild-type mice reduced glomerular filtration rate and increased tubulointerstitial deposition of αSMA compared with vehicle.
To our knowledge, this is the first report demonstrating highly sensitive mass spectrometry analysis of glomerular filtrate. We identified >600 proteins with a 3-log dynamic range. Our findings are consistent with models of glomerular filtration and micropuncture measurements of filtrate protein content in rodents, as total protein abundance is very low, and the abundance of albumin is much less than that in plasma (19, 20). In addition to application of what we believe to be a highly novel mass spectrometry assay, a strength of our study is aspiration of fluid from Bowman’s space, where filtrate is formed and may be less modified by endocytosis or secretion than in the proximal tubule. Consistent with this, we found few proteins from tubular epithelial or other renal cells in our filtrate samples, and spatial deconvolution of the filtrate demonstrated very low abundance of renal-specific protein compared with protein specific to other organs. Micropuncture studies have primarily reported protein concentrations from the early proximal tubule, in part because, with conventional microscopy, only surface glomeruli (rare in mice) are accessible (33). Use of 2-photon microscopy in the present study rendered subsurface glomeruli (several to many in each mouse) accessible, allowing micropuncture of Bowman’s space in wild-type mice.
We hypothesized that identifications of high-molecular-weight protein would be increased in the renal filtrate after CA/CPR compared with sham treatment, increasing the mean molecular weight. To our surprise, we found the mean molecular weight of filtered protein was reduced, not increased, after CA/CPR. This finding is in contrast to our prior finding that sieving of Ficoll-70 is increased at the same time point after CA/CPR (10) and to the findings of Andersson and Rippe (34, 35) in an ex vivo preparation. A difference between the technique of the current investigation and the prior investigation into ischemic glomerular barrier properties is that the present results were obtained without an exogenous filtration marker. Polysaccharide markers may be deformed or cleaved in an unpredictable fashion in the plasma prior to filtration (potentially leaving the fluorescent tag with a fraction rather than the intact molecule), but in the present study proteins were identified using mass spectrometry. The requirement for multiple peptides demonstrating predictable trypsin cleavages for each identified protein robustly predicts the presence of intact protein. It is unlikely that the observed molecular weight distribution would be obtained in the case of filtration of precleaved (i.e., in the plasma) peptides, as this would equally affect high- and low-molecular-weight plasma proteins, producing a greater abundance of high-molecular-weight protein identifications than we observed. Therefore, we are confident that the observed proteins largely reflect proteins present in the filtrate samples.
To better understand findings from Bowman’s aspirate, we performed tandem mass tag–labeled quantification of proteins in the urine of mice with defective proximal tubular protein endocytosis, in which urine contains filtrate proteins ordinarily not present due to endocytosis in the proximal tubule (36, 37). Accordingly, we found low-molecular-weight proteins in the urine of these mice with our assay. A proximal tubule–specific, inducible megalin deletion was recently used in a similar fashion to demonstrate increased filtration of albumin in a diabetic nephropathy model (38); we chose the lifelong mosaic deletion model due to its demonstrated stability in injury models (39). In our model, ApoEcre status did not affect survival after CA/CPR but sharply increased the urine excretion of low-molecular-weight proteins, including many known megalin ligands, before CA/CPR.
Because the origin of filtered protein is spatially convoluted, we used a mouse proteomic spatial map to deconvolute the filtrate, leading to the observation that proteins with specificity to the heart appear in the filtrate in acute cardiorenal syndrome, which we then followed up with specific investigation. It is important to note that the spatial deconvolution method is inferential, and, as the OSS was derived from healthy SILAC mice (40), it is possible that protein derivation may be less specific in an injury model. Our experimental design using complementary data sets 2 and 24 hours after CA/CPR and controls mitigates this limitation. Further, we confirmed the finding of cardiac LIM protein in the filtered urine with immunofluorescence and plasma studies.
Our finding of cardiac protein in the renal filtrate in early acute cardiorenal syndrome may advance the search for cardiorenal connectors (4, 41) in this common sequela of acute cardiovascular disease. Cardiac LIM protein is a candidate for this cardiorenal connector status and is the focus of further investigations in our laboratory. Additionally, aberrant protein filtration correlates with outcome in the most common causes of chronic kidney disease, hypertensive, and diabetic nephropathies; we therefore expect the techniques developed for our investigation will have wide applicability in renal investigation.
Methods
CA/CPR and sham procedure.
Eight-minute normothermic CA/CPR leading to AKI has previously been described by our group and was conducted identically to prior experiments (10, 42–45). Following induction of anesthesia with isoflurane, mice (male C57BL/6, 8–12 weeks old, n = 16) were orotracheally intubated with a 22-gauge catheter. A PE-10 catheter was placed in the jugular vein. Transcutaneous electrodes were used to monitor electrocardiography, and the anterior chest was depilated to monitor the visible point of maximal cardiac impulse. Core temperature was maintained within normothermic limits (36.5°C–37.5°C) with a rectal temperature monitor, heating lamp, temperature controller, and insulating blanket. Eight minutes after potassium-induced cardiac arrest, resuscitation was begun with chest compressions (300/min) and administration of intravenous epinephrine (8–16 μg). Efforts to resuscitate ceased if return of spontaneous circulation was not observed 180 seconds after starting resuscitation. After return of spontaneous respiration, mice were orotracheally extubated, decannulated, and moved to a recovery cage. Sham procedures were identical, except that cardiac arrest was not induced, and neither chest compressions nor epinephrine were administered.
In vivo renal microscopy.
Following sham or CA/CPR, mice were prepared for in vivo microscopy, which was performed as described previously (46). Briefly, under isoflurane general anesthesia, a left flank incision was made and the kidney was partly extruded through the muscle and subcutaneous tissue and immobilized in 10% agar. An aluminum frame within a custom polysiloxane support (Sugru) was applied to support a cover glass, and gaps filled with 1% agar. The mouse was then immobilized on the imaging platform, and a heating pad applied to maintain normothermia. 100 μl FITC-dextran (2000-kDa molecular weight) was injected via the retro-orbital sinus immediately before imaging. Imaging was carried out using a Zeiss LSM 7MP microscope with 525/50 and 593/35 band-pass filters and excitation set at 810 nm, with power between 20 and 40 μA. In order to reduce energy transfer to tissue, which might lead to functional change, live imaging was primarily employed. When laser scanning was employed for recording images, the laser energy, laser dwell time, and the number of images per unit of tissue were kept to the absolute minimum required to document position. The experimental apparatus is detailed in Figure 1.
Bowman’s space micropuncture.
Planned analysis with mass spectrometry required 3 modifications to the usual micropuncture practice. First, to reduce potential confounding of this highly sensitive analytic technique by tubular secretion of protein, we acquired samples from Bowman’s space, rather than the proximal tubule. Second, to ensure pipette positioning within Bowman’s space, stereotactic techniques were coupled with 3D imaging provided by 2-photon microscopy. Third, to prevent overwhelming signal from petrochemical sample contamination, mineral oil was not used, and the sample volume was therefore not measured. Prior to micropuncture, a quantum dot–coated (QDot 594, Thermo Fisher) borosilicate glass micropipette (47) with a 45° beveled, tapered 5- to 10-μm orifice was secured in an x-y-z–controlled headstage (WPI). The pipette was attached to a precision syringe microinjector (WPI), with tubing and pipette filled with perfluorodecalin, a biologically inert, hydrophobic single-molecule fluid (MilliporeSigma). The pipette was introduced into the water column above the coverslip, and the coordinates of the microscope stage and that of the controller were registered to allow guidance to intrarenal structures.
Glomeruli were identified as FITC-enhanced spheroid vascular tufts, with a diameter of between 70 and 100 μM and visible red cell motion. Based on preliminary experiments, we chose favorably located and oriented glomeruli, defined as having their top-most urinary space within 80 μm of the cover glass in the Z dimension, and their most lateral urinary space within 700 μm of the lateral surface of the kidney (the direction of pipette access). After identifying a target glomerulus, stage coordinates were recorded, and the pipette coordinates of the glomerulus were calculated based on the registration procedure. The micropipette was introduced under live imaging, and an image of the pipette tip in the urinary space was obtained. We then injected 150 nl perfluorodecalin over 3 minutes and confirmed placement by observing expansion of the urinary space in the immediate vicinity of the pipette. After waiting 3 minutes for additional filtration, we set the precision microinjector to aspirate 300 nl over 6 minutes, disconnected the pipette from negative pressure, and withdrew it from the kidney. While withdrawing the pipette, we looked for extravasation of FITC-dextran and for FITC signal within the pipette, which could indicate plasma contamination of the sample. After removal from the kidney, the sample was immediately diluted in 1 μl 8 M urea, 100 mM Tris(hydroxymethyl)aminomethane buffer (pH 8.0) within the pipette to prevent protein interaction prior to flash-freezing. Volume of the sample was not determined to prevent contamination with organic chemicals from mineral oil, which is typically used for this application in micropuncture experiments.
Cell culture.
Human cells were used. HK2 cells (human PTECs) were purchased from ATCC and cultured in accordance with extant practice. Conditionally immortalized human podocytes were a gift from Moin Saleem, University of Bristol, Bristol, United Kingdom. Conditionally immortalized human glomerular endothelial cells were a gift from Simon Satchel, also of the University of Bristol.
Comparison with data from mouse experiments was facilitated by conversion to species orthologs using the biodbnet:dbOrtho tool (48, 49), followed by hand curation for missing elements.
Sample handling for online simple nanoproteomic platform proteomics.
Frozen capillary tubes containing Bowman’s space aspirate were inverted into low-retention LC-MS vials (Waters Corp.). To prevent contamination from the outside of the tube, the sharp end was removed prior to recovery. Capillaries were then warmed, facilitating transfer to the vial, and rinsed with 25 μl of 8 M Tris (pH 8), with 10 mM dithiothreitol, to total volume of approximately 26 μl. The aqueous layer was transferred to a fresh vial and incubated at 60°C for 30 minutes to denature and reduce the proteins. The entire sample volume was then injected for simple nanoproteomic platform (SNaPP) analysis.
A 20-μl aliquot of mouse plasma (Equitech-Bio Inc.) was used for SNaPP analysis. The same procedure was used to process cell pellets from culture and mouse plasma. Samples were denatured and reduced by adding 100 μl of 8 M urea, 10 mM DTT in 50 mM Tris(hydroxymethyl)aminomethane buffer (pH 8.0), followed by 30 seconds of sonication and 30 minutes of incubation at 37°C. Samples were then centrifuged at 21,130 g, and the supernatant transferred to a fresh tube. Protein concentration was assessed by Coomassie assay. Protein isolates were then diluted with fresh 50 mM Tris(hydroxymethyl)aminomethane (pH 8) to a concentration of 0.02 μg/μl for SNaPP analysis.
Nanoscale sample processing and mass spectrometric analysis.
SNaPP sample processing and LC-MS/MS analysis was performed as previously described (50, 51). Briefly, the sample was passed through a Poroszyme (Thermo Fisher) immobilized trypsin column, 150 μm ID × 2 cm fused silica capillary, at 0.5 μl/min for digestion. Resultant peptides were desalted inline by trapping on a 150 μm ID × 4 cm C18 column. After washing, peptides were separated on an in-house packed 50 μm ID × 75 cm C18 analytical column. The SNaPP system was coupled to a Qexactive HF mass spectrometer (Thermo Scientific) operated in a top-12 data-dependent acquisition mode, with a 200-millisecond MS2 maximum IT to increase sensitivity.
SNaPP mass spectrometry data analysis.
Spectral identification of peptides from SNaPP analysis was carried out using MSGF+ (52, 53) and MaxQuant software tools (54, 55). MSGF searches were carried out with a 20-ppm mass error tolerance, with methionine oxidation as a dynamic modification, against the UniProt Mus musculus protein database (UniProtKB, downloaded February 2016). Unique peptide identifications were filtered to an optimized FDR of 1% against a reversed decoy database, as previously described (56). Raw data were processed using MaxQuant, version 1.5.3.30, with an FDR of 0.01, against the same Mus musculus Uniprot database. N-terminal acetylation and methionine oxidation were included as dynamic modifications. For protein quantification, the match-between-runs feature was enabled using default settings as well as LFQ normalization for protein abundances. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (57) with the data set identifier PXD012148. Following urine mass spectrometry, proteins were identified using Proteome Discoverer v1.4 against the SPROT_Mouse_2015.07 proteome, and protein abundance was expressed as reporter ion intensity after normalization.
Spatial deconvolution of filtered proteins.
Proteins in plasma, and therefore those in glomerular filtrate, originate in many cell types and in all organs of the body. Therefore, the set of proteins identified in the filtrate contains convoluted information about organs that are distant from the kidney. In order to make inferences about protein origin, spatial deconvolution was performed. We applied proteomic spatial mapping (40) to protein identifications. We applied an OSS, defined as follows, to each protein in the spatial map: OSS = ((Abundanceorgan tissue) – (Abundancemean)/σ(Abundance))
Each protein therefore was evaluated by organ specificity to each organ system. Protein-organ pairs with OSS >3 were considered specific to single organs. These highly specific protein-organ pairs were then mapped to protein identifications from Bowman’s space and urine to infer organ system origination.
Identification of filtered proteins in urine.
To confirm the presence of proteins in the filtrate, we performed urine proteomics before and after CA/CPR in mice with a deficiency of proximal tubular endocytosis. LRP2fl/fl;apoECre mice (a gift of Thomas Willnow, Max Delbruck Institute, Berlin, Germany) exhibit mosaic, proximal tubule–specific deletion of the primary protein endocytosis receptor for the proximal tubule, megalin. LRP2fl/fl;apoECre mice demonstrate characteristic low-molecular-weight proteinuria, including filtered serum chaperones with known tubular uptake, such as vitamin D–binding protein and retinol-binding protein(37). Urine from LRP2fl/fl;apoECre and LRP2fl/fl; (Cre– control) mice was collected onto dry ice in metabolic cages with protease inhibition (Roche) for 24 hours before and 24 hours after CA/CPR (n = 10 samples from 5 mice) to ensure adequate quantity for tandem mass tag label mass spectrometry. Equal volumes were subjected to trichloroacetic acid precipitation, washed with acetone, centrifuged,and air dried. The pellet was dissolved in 8 M urea, 0.1 M ammonium bicarbonate, and the protein concentration was determined by Bradford Coomassie assay. 50 μg isolated protein from each sample was reduced, alkylated, and digested with trypsin overnight. Digests were then solid-phase extracted in a C18 column, eluted, and dried. 25 μg per sample was tandem mass tag–labeled, pooled, and analyzed on an Orbitrap Fusion (Thermo Scientific). These proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (57) with the data set identifier PXD012193.
Evaluation of cardiac LIM protein filtration by immunofluorescence.
During brief isoflurane general anesthesia, 20 μg recombinant human cardiac LIM protein (Origine, TP325217) diluted to total volume in 100 μl PBS or 100 μl PBS was injected into the retroorbital plexus of LRP2fl/fl;apoECre mice (n = 3/group). 15 minutes later, under deep isoflurane general anesthesia, mice were killed and perfusion fixed as described above. The right kidney was removed, fixed in 4% paraformaldehyde, paraffin embedded, and cut such that 4 equidistant 6-μm-thick sections were placed on a glass slide. Slides were stained with DAPI and the following antibodies and concentrations: megalin (P-20,Santa Cruz, sc-16478, 1:200) and CSRP3 (Abcam, ab173301, 1:100).
Renal I/R injury model.
Adult male C57BL/7 mice (Charles River) were subjected to 30-minute renal ischemia or to sham surgery as described previously (58). Plasma were collected 24 hours after renal IR injury and stored at –80°C prior to analysis.
Determination of plasma myocardiocyte protein levels.
Stored plasma samples from C57BL/6 mice subjected to sham or CA/CPR (n = 6) were assessed for cardiac LIM protein concentration using a commercially available ELISA (MyBioSource, MBS7211399). We assessed plasma MyBPC3 in an additional cohort of wild-type mice (n = 4) 2 hours after they were subjected to CA/CPR using a commercially available ELISA kit (MyBioSource, MBS9321524). For the study of human patients, 9 consecutive cardiac arrest patients were screened and 5 were enrolled. Four patients were excluded for predefined criteria: (a) prearrest dialysis dependence (which might impair cardiac LIM protein excretion) (n = 1), (b) because no plasma sample was available (n = 1), and (c) because they were screened after the planned enrollment goal was met (n = 2). Plasma samples were obtained as aliquots from the clinical laboratory and preserved for analysis at –80°C. After all samples were acquired, the concentration of cardiac LIM protein was determined using a commercially available kit (MyBioSource, MBS7230380).
Administration of recombinant cardiac LIM protein to mice.
As plasma kinetics for cardiac LIM protein are unknown, we modeled them, using the value measured 24 hours after CA/CPR in mice and known kinetics of myocardial injury biomarker troponin I to estimate an AUC, which had mean value 2.8 μg. To account for protein binding and expected technical loss, we chose a total dose of 5 μg. Recombinant cardiac LIM protein (Origene, TP325217) was diluted to 1 μg/150 μl in PBS (pH 7.4). Under brief isoflurane anesthesia, 1 μg cardiac LIM protein or an equivalent volume of PBS was injected retro-orbitally to mice in an order determined by a random number generator, and this order was followed on consecutive days to a total dose of 5 μg in the treatment group and the equivalent volume in the PBS (vehicle) group. For 24 hours after the last injection, urine was collected in metabolic cages. Twenty-five days after the last injection, mice were housed in metabolic cages for 24 hours for urine collection and then underwent measurement of glomerular filtration rate followed by euthanasia and perfusion-fixation via the left ventricular apex. Kidneys were preserved for analysis in 4% paraformaldehyde.
Measurement of renal function in mice.
GFR was measured by transcutaneous measurement of FITC-sinistrin fluorescence as described previously (59). To reduce movement-induced measurement variation, this was performed under light isoflurane anesthesia (1%–1.2% in air), with isoflurane titrated to mouse respiratory rate to achieve a rate of approximately 120 breaths per minutes, which is just below the threshold of spontaneous movement. Urine volume was recorded after 24 hours collection in metabolic cages, and urine protein was measured using the Bradford Coomassie assay.
Kidney immunofluorescence and quantitative determination of tubulointerstitial αSMA.
Following perfusion-fixation, paraffin-embedded kidneys were sagittally sectioned at 60-μm intervals, starting at a random point from the inferior pole. Six consecutive sections were mounted per slide. For immunofluorescence qualitative visualization of cardiac LIM protein and megalin, sections were incubated for 2 hours at 20°C in primary antibodies (megalin, Santa-Cruz goat anti mouse P-20; cardiac LIM protein, Abcam rabbit anti-mouse, 173301) in 1% bovine serum albumin/PBS, followed by Cy2 and Alexa Fluor 594–conjugated secondary antibodies (1:500, Thermo Fisher) for 1 hour at room temperature. For quantitative visualization of tubulointerstitial αSMA, sections were stained with FITC-conjugated anti-αSMA antibody (MilliporeSigma, F3777). Three random high-power field images (×400) were taken per section, and an 8 × 10 grid of points was superimposed. Each point was assessed for (a) tubulointerstitial location and (b) αSMA stain positivity, and perivascular αSMA was excluded. For each image, the number of positive points/80 × 100 represented the percentage of positive points, and the mean of percentage of positive points for each mouse was calculated.
Statistics.
Statistical methods used to identify peptides from mass spectra and proteins from peptides are described above. Statistical analysis for hypothesis testing was performed in R (using the EdgeR Bioconductor package), scientific Python (using packages Scipy.stats and statsmodels), and Prism (version 7). All t tests were 2 tailed.
Analysis of differential expression of proteins identified with label-free quantification (Bowman’s space aspirate) was performed using the Benjamini-Hochberg correction for multiple t tests (FDR), with the family-wise error rate controlled for α = 0.01. Statistical significance was inferred if P < 0.05 and FDR < 0.05. Differential protein abundances between groups in TMT-labeled mass spectrometry conducted on urine samples were determined by comparing the total reporter ion intensities using the Bioconductor package edgeR. Additional data normalizations, multiple testing corrections, and calculation of FDRs were performed within edgeR. Only results with FDR < 0.1 were considered significant (60, 61). Multiple group analysis of differences in organ-specific protein filtration was performed by ANOVA with post-hoc correction using Sidak’s test. Two-group comparisons for nominal variables (serum creatinine and urine output) were conducted with Student’s t test.
Study approvals.
All studies conducted in mice were conducted at Oregon Health & Science University and were approved by the Oregon Health & Science University Institutional Animal Care and Use Committee, except for renal I/R, which was conducted at Columbia University and approved by the Columbia University Institutional Animal Care and Use Committee. Animal experiments were conducted in accordance with the NIH Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011). The study of waste plasma from human survivors of cardiac arrest, including waiver of informed consent, was approved by the Oregon Health & Science University Institutional Review Board.
Author contributions
RW performed animal surgeries and experiments and codeveloped the imaging preparation and protocols. KM performed critical assays and helped author the manuscript. KG performed animal experimental surgeries and codeveloped the imaging preparation and imaging protocols. SA conceived the experimental design, guided and critiqued experimental execution, analyzed data, and critiqued the manuscript. MBE performed critical assays. DJO contributed to experimental design and statistical analysis and performed analysis and critical assays. SJH conducted critical experiments. HTL conducted critical experiments and authored the manuscript. RDS led the nanoproteomics team, contributed to experimental design and statistical analysis, and performed analysis. KDR contributed to experimental design and statistical analysis, performed analysis, and authored the manuscript. PDP conceived and executed the nanoproteomic assay, contributed to experimental design, performed critical assays, and authored the manuscript. MPH conceived the program of research, assembled and directed the research team, performed experiments, and wrote the manuscript.
Supplementary Material
Acknowledgments
This study was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (K08DK090754 to MPH) and the National Institute of General Medical Sciences (P41 GM103493 to RDS). Mass spectrometric analysis was performed by the Oregon Health & Science University Proteomics Shared Resource, with partial support from NIH grants P30EY010572, P30CA069533, and S10OD012246. The authors wish to express their gratitude to Phillip Wilmarth and Ashok Reddy, of the Oregon Health & Science University Proteomics Shared Resource, for helpful assistance. This material is the result of work (by MPH) that was supported with resources and the use of facilities at the Portland Veterans Affairs Medical Center. The contents do not represent the views of the US Department of Veterans Affairs or the United States Government.
Version 1. 02/21/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(4):e122130. https://doi.org/10.1172/jci.insight.122130.
Contributor Information
Rumie Wakasaki, Email: darumie68@gmail.com.
Katsuyuki Matsushita, Email: mysql@ohsu.edu.
Kirsti Golgotiu, Email: kgolgo@gmail.com.
Sharon Anderson, Email: anderssh@ohsu.edu.
Daniel J. Orton, Email: daniel.orton@pnnl.gov.
Sang Jun Han, Email: sh3740@cumc.columbia.edu.
Richard D. Smith, Email: dick.smith@pnnl.gov.
Paul D. Piehowski, Email: Paul.Piehowski@pnnl.gov.
References
- 1.Pimienta González R, et al. Incidence, mortality and positive predictive value of type 1 cardiorenal syndrome in acute coronary syndrome. PLoS ONE. 2016;11(12):e0167166. doi: 10.1371/journal.pone.0167166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ronco C, Di Lullo L. Cardiorenal syndrome in Western countries: epidemiology, diagnosis and management approaches. Kidney Dis (Basel) 2017;2(4):151–163. doi: 10.1159/000448749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vandenberghe W, et al. Acute kidney injury in cardiorenal syndrome type 1 patients: a systematic review and meta-analysis. Cardiorenal Med. 2016;6(2):116–128. doi: 10.1159/000442300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Braam B, Joles JA, Danishwar AH, Gaillard CA. Cardiorenal syndrome--current understanding and future perspectives. Nat Rev Nephrol. 2014;10(1):48–55. doi: 10.1038/nrneph.2013.250. [DOI] [PubMed] [Google Scholar]
- 5.Tenten V, et al. Albumin is recycled from the primary urine by tubular transcytosis. J Am Soc Nephrol. 2013;24(12):1966–1980. doi: 10.1681/ASN.2013010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nykjaer A, et al. An endocytic pathway essential for renal uptake and activation of the steroid 25-(OH) vitamin D3. Cell. 1999;96(4):507–515. doi: 10.1016/S0092-8674(00)80655-8. [DOI] [PubMed] [Google Scholar]
- 7.Sousa MM, et al. Evidence for the role of megalin in renal uptake of transthyretin. J Biol Chem. 2000;275(49):38176–38181. doi: 10.1074/jbc.M002886200. [DOI] [PubMed] [Google Scholar]
- 8.Zhai XY, et al. Cubilin- and megalin-mediated uptake of albumin in cultured proximal tubule cells of opossum kidney. Kidney Int. 2000;58(4):1523–1533. doi: 10.1046/j.1523-1755.2000.00314.x. [DOI] [PubMed] [Google Scholar]
- 9.Ware LB, Johnson AC, Zager RA. Renal cortical albumin gene induction and urinary albumin excretion in response to acute kidney injury. Am J Physiol Renal Physiol. 2011;300(3):F628–F638. doi: 10.1152/ajprenal.00654.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hutchens MP, Fujiyoshi T, Komers R, Herson PS, Anderson S. Estrogen protects renal endothelial barrier function from ischemia-reperfusion in vitro and in vivo. Am J Physiol Renal Physiol. 2012;303(3):F377–F385. doi: 10.1152/ajprenal.00354.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda M, et al. Determination of renal function and injury using near-infrared fluorimetry in experimental cardiorenal syndrome. Am J Physiol Renal Physiol. 2017;312(4):F629–F639. doi: 10.1152/ajprenal.00573.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo LM, et al. The normal kidney filters nephrotic levels of albumin retrieved by proximal tubule cells: retrieval is disrupted in nephrotic states. Kidney Int. 2007;71(6):504–513. doi: 10.1038/sj.ki.5002041. [DOI] [PubMed] [Google Scholar]
- 13.Tanner GA. Glomerular sieving coefficient of serum albumin in the rat: a two-photon microscopy study. Am J Physiol Renal Physiol. 2009;296(6):F1258–F1265. doi: 10.1152/ajprenal.90638.2008. [DOI] [PubMed] [Google Scholar]
- 14.Huang EL, et al. SNaPP: simplified nanoproteomics platform for reproducible global proteomic analysis of nanogram protein quantities. Endocrinology. 2016;157(3):1307–1314. doi: 10.1210/en.2015-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Veeneman JM, de Jong PE, Huisman RM, Reijngoud DJ. Re: Adey et al. Reduced synthesis of muscle proteins in chronic renal failure. Am J Physiol Endocrinol Metab 278: E219-E225, 2000. Am J Physiol Endocrinol Metab. 2001;280(1):E197–E198. doi: 10.1152/ajpendo.2001.280.1.E197. [DOI] [PubMed] [Google Scholar]
- 16.Hutchens MP, et al. Estrogen is renoprotective via a nonreceptor-dependent mechanism after cardiac arrest in vivo. Anesthesiology. 2010;112(2):395–405. doi: 10.1097/ALN.0b013e3181c98da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharfuddin AA, et al. Soluble thrombomodulin protects ischemic kidneys. J Am Soc Nephrol. 2009;20(3):524–534. doi: 10.1681/ASN.2008060593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics. 2014;13(9):2513–2526. doi: 10.1074/mcp.M113.031591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tojo A, Endou H. Intrarenal handling of proteins in rats using fractional micropuncture technique. Am J Physiol. 1992;263(4 Pt 2):F601–F606. doi: 10.1152/ajprenal.1992.263.4.F601. [DOI] [PubMed] [Google Scholar]
- 20.Oken DE, Flamenbaum W. Micropuncture studies of proximal tubule albumin concentrations in normal and nephrotic rats. J Clin Invest. 1971;50(7):1498–1505. doi: 10.1172/JCI106635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cordwell SJ, et al. Release of tissue-specific proteins into coronary perfusate as a model for biomarker discovery in myocardial ischemia/reperfusion injury. J Proteome Res. 2012;11(4):2114–2126. doi: 10.1021/pr2006928. [DOI] [PubMed] [Google Scholar]
- 22.Farrah T, et al. A high-confidence human plasma proteome reference set with estimated concentrations in PeptideAtlas. Mol Cell Proteomics. 2011;10(9):M110.006353. doi: 10.1074/mcp.M110.006353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kong Y, Flick MJ, Kudla AJ, Konieczny SF. Muscle LIM protein promotes myogenesis by enhancing the activity of MyoD. Mol Cell Biol. 1997;17(8):4750–4760. doi: 10.1128/MCB.17.8.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, Holtzer H. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc Natl Acad Sci USA. 1990;87(20):7988–7992. doi: 10.1073/pnas.87.20.7988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boateng SY, Senyo SE, Qi L, Goldspink PH, Russell B. Myocyte remodeling in response to hypertrophic stimuli requires nucleocytoplasmic shuttling of muscle LIM protein. J Mol Cell Cardiol. 2009;47(4):426–435. doi: 10.1016/j.yjmcc.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vafiadaki E, Arvanitis DA, Sanoudou D. Muscle LIM Protein: Master regulator of cardiac and skeletal muscle functions. Gene. 2015;566(1):1–7. doi: 10.1016/j.gene.2015.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arber S, et al. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 1997;88(3):393–403. doi: 10.1016/S0092-8674(00)81878-4. [DOI] [PubMed] [Google Scholar]
- 28.Geier C, et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum Mol Genet. 2008;17(18):2753–2765. doi: 10.1093/hmg/ddn160. [DOI] [PubMed] [Google Scholar]
- 29.Huby AC, et al. Disturbance in Z-disk mechanosensitive proteins induced by a persistent mutant myopalladin causes familial restrictive cardiomyopathy. J Am Coll Cardiol. 2014;64(25):2765–2776. doi: 10.1016/j.jacc.2014.09.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boateng SY, et al. Cardiac dysfunction and heart failure are associated with abnormalities in the subcellular distribution and amounts of oligomeric muscle LIM protein. Am J Physiol Heart Circ Physiol. 2007;292(1):H259–H269. doi: 10.1152/ajpheart.00766.2006. [DOI] [PubMed] [Google Scholar]
- 31.Marshall KD, Edwards MA, Krenz M, Davis JW, Baines CP. Proteomic mapping of proteins released during necrosis and apoptosis from cultured neonatal cardiac myocytes. Am J Physiol, Cell Physiol. 2014;306(7):C639–C647. doi: 10.1152/ajpcell.00167.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marinó M, Andrews D, Brown D, McCluskey RT. Transcytosis of retinol-binding protein across renal proximal tubule cells after megalin (gp 330)-mediated endocytosis. J Am Soc Nephrol. 2001;12(4):637–648. doi: 10.1681/ASN.V124637. [DOI] [PubMed] [Google Scholar]
- 33.Lorenz JN. Micropuncture of the kidney: a primer on techniques. Compr Physiol. 2012;2(1):621–637. doi: 10.1002/cphy.c110035. [DOI] [PubMed] [Google Scholar]
- 34.Rippe C, Rippe A, Larsson A, Asgeirsson D, Rippe B. Nature of glomerular capillary permeability changes following acute renal ischemia-reperfusion injury in rats. Am J Physiol Renal Physiol. 2006;291(6):F1362–F1368. doi: 10.1152/ajprenal.00123.2006. [DOI] [PubMed] [Google Scholar]
- 35.Andersson M, Nilsson U, Hjalmarsson C, Haraldsson B, Nyström JS. Mild renal ischemia-reperfusion reduces charge and size selectivity of the glomerular barrier. Am J Physiol Renal Physiol. 2007;292(6):F1802–F1809. doi: 10.1152/ajprenal.00152.2006. [DOI] [PubMed] [Google Scholar]
- 36.Leheste JR, et al. Megalin knockout mice as an animal model of low molecular weight proteinuria. Am J Pathol. 1999;155(4):1361–1370. doi: 10.1016/S0002-9440(10)65238-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leheste JR, et al. Hypocalcemia and osteopathy in mice with kidney-specific megalin gene defect. FASEB J. 2003;17(2):247–249. doi: 10.1096/fj.02-0578fje. [DOI] [PubMed] [Google Scholar]
- 38.Mori KP, et al. Increase of total nephron albumin filtration and reabsorption in diabetic nephropathy. J Am Soc Nephrol. 2017;28(1):278–289. doi: 10.1681/ASN.2015101168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Theilig F, et al. Abrogation of protein uptake through megalin-deficient proximal tubules does not safeguard against tubulointerstitial injury. J Am Soc Nephrol. 2007;18(6):1824–1834. doi: 10.1681/ASN.2006111266. [DOI] [PubMed] [Google Scholar]
- 40.Geiger T, et al. Initial quantitative proteomic map of 28 mouse tissues using the SILAC mouse. Mol Cell Proteomics. 2013;12(6):1709–1722. doi: 10.1074/mcp.M112.024919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bongartz LG, et al. Target organ cross talk in cardiorenal syndrome: animal models. Am J Physiol Renal Physiol. 2012;303(9):F1253–F1263. doi: 10.1152/ajprenal.00392.2012. [DOI] [PubMed] [Google Scholar]
- 42.Ikeda M, Swide T, Vayl A, Lahm T, Anderson S, Hutchens MP. Estrogen administered after cardiac arrest and cardiopulmonary resuscitation ameliorates acute kidney injury in a sex- and age-specific manner. Crit Care. 2015;19:332. doi: 10.1186/s13054-015-1049-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hutchens MP, Fujiyoshi T, Koerner IP, Herson PS. Extracranial hypothermia during cardiac arrest and cardiopulmonary resuscitation is neuroprotective in vivo. Ther Hypothermia Temp Manag. 2014;4(2):79–87. doi: 10.1089/ther.2014.0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hutchens MP, et al. Estrogen-mediated renoprotection following cardiac arrest and cardiopulmonary resuscitation is robust to GPR30 gene deletion. PLoS ONE. 2014;9(6):e99910. doi: 10.1371/journal.pone.0099910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hutchens MP, et al. Estrogen is renoprotective via a nonreceptor-dependent mechanism after cardiac arrest in vivo. Anesthesiology. 2010;112(2):395–405. doi: 10.1097/ALN.0b013e3181c98da9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsushita K, et al. Micropuncture of Bowman’s space in mice facilitated by 2 photon microscopy. J Vis Exp. 2018;(140):e58206. doi: 10.3791/58206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrásfalvy BK, et al. Quantum dot-based multiphoton fluorescent pipettes for targeted neuronal electrophysiology. Nat Methods. 2014;11(12):1237–1241. doi: 10.1038/nmeth.3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ortholog conversions. Advanced Biomedical Computing Center Frederick National Laboratory for Cancer Research, National Cancer Institute at Frederick. https://biodbnet-abcc.ncifcrf.gov/db/dbOrtho.php Accessed January 17, 2019.
- 49.Mudunuri U, Che A, Yi M, Stephens RM. bioDBnet: the biological database network. Bioinformatics. 2009;25(4):555–556. doi: 10.1093/bioinformatics/btn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Clair G, et al. Spatially-resolved proteomics: rapid quantitative analysis of laser capture microdissected alveolar tissue samples. Sci Rep. 2016;6:39223. doi: 10.1038/srep39223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang EL, et al. SNaPP: simplified nanoproteomics platform for reproducible global proteomic analysis of nanogram protein quantities. Endocrinology. 2016;157(3):1307–1314. doi: 10.1210/en.2015-1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim S, Gupta N, Pevzner PA. Spectral probabilities and generating functions of tandem mass spectra: a strike against decoy databases. J Proteome Res. 2008;7(8):3354–3363. doi: 10.1021/pr8001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S, Pevzner PA. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol. 2008;26(12):1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 55.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 56.Piehowski PD, et al. STEPS: a grid search methodology for optimized peptide identification filtering of MS/MS database search results. Proteomics. 2013;13(5):766–770. doi: 10.1002/pmic.201200096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vizcaíno JA, et al. 2016 update of the PRIDE database and its related tools. Nucleic Acids Res. 2016;44(D1):D447–D456. doi: 10.1093/nar/gkv1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Han SJ, Li H, Kim M, Shlomchik MJ, Lee HT. Kidney proximal tubular TLR9 exacerbates ischemic acute kidney injury. J Immunol. 2018;201(3):1073–1085. doi: 10.4049/jimmunol.1800211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schock-Kusch D, et al. Reliability of transcutaneous measurement of renal function in various strains of conscious mice. PLoS ONE. 2013;8(8):e71519. doi: 10.1371/journal.pone.0071519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Plubell DL, et al. Extended multiplexing of tandem mass tags (TMT) labeling reveals age and high fat diet specific proteome changes in mouse epididymal adipose tissue. Mol Cell Proteomics. 2017;16(5):873–890. doi: 10.1074/mcp.M116.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.