Abstract
Poly(ADP-ribosyl)ation refers to the covalent attachment of ADP-ribose to protein, generating branched, long chains of ADP-ribose moieties, known as poly(ADP-ribose) (PAR). Poly(ADP-ribose) polymerase 1 (PARP1) is the main polymerase and acceptor of PAR in response to DNA damage. Excessive intracellular PAR accumulation due to PARP1 activation leads cell death in a pathway known as parthanatos. PAR degradation is mainly controlled by poly(ADP-ribose) glycohydrolase (PARG) and ADP-ribose-acceptor hydrolase 3 (ARH3). Our previous results demonstrated that ARH3 confers protection against hydrogen peroxide (H2O2) exposure, by lowering cytosolic and nuclear PAR levels and preventing apoptosis-inducing factor (AIF) nuclear translocation. We identified a family with an ARH3 gene mutation that resulted in a truncated, inactive protein. The 8-year-old proband exhibited a progressive neurodegeneration phenotype. In addition, parthanatos was observed in neurons of the patient’s deceased sibling, and an older sibling exhibited a mild behavioral phenotype. Consistent with the previous findings, the patient’s fibroblasts and ARH3-deficient mice were more sensitive, respectively, to H2O2 stress and cerebral ischemia/reperfusion-induced PAR accumulation and cell death. Further, PARP1 inhibition alleviated cell death and injury resulting from oxidative stress and ischemia/reperfusion. PARP1 inhibitors may attenuate the progression of neurodegeneration in affected patients with ARH3 deficiency.
Keywords: Genetics, Therapeutics
Keywords: Genetic diseases, Neurodegeneration
PARP1 inhibition reduces oxidative stress-induced cell death in ADP-ribose-acceptor hydrolase 3 (ARH3)-deficient murine neurons and patient-derived fibroblasts.
Introduction
ADP-ribosylation is a posttranslational protein modification characterized by the transfer of the ADP-ribose moiety of NAD+ to an acceptor, resulting in the modification of its activity (1, 2). Poly(ADP-ribose) polymerase 1 (PARP1) catalyzes the poly(ADP-ribosyl)ation of acceptor proteins by covalently attaching ADP-ribose to generate a long, branched chain of ADP-ribose moieties, termed poly(ADP-ribose) (PAR), in response to single- and double-stranded DNA breaks (3, 4). Most PAR is attached to PARP1 itself through the automodification domain. Poly(ADP-ribosyl)ation plays a role in a number of crucial cellular functions including mitosis, DNA repair, and cell death (3, 5). PARP1 is the founding member of the PARP family of 17 genes, most of which have poly- and/or mono-ADP-ribosyltransferase activities (5).
Parthanatos, or PARP1-dependent cell death, is a cellular process involved in the pathogenesis of neurological disorders such as ischemic brain injury, Parkinson’s disease, and glutamate excitotoxicity (6–8). Ca2+ overload induced by glutamate receptors and ROS generated by oxidative stress cause single- and double-stranded DNA breaks, which cause PARP1 overactivation (9). Poly(ADP-ribose) glycohydrolase (PARG), the primary enzyme responsible for PAR degradation (10, 11), catalyzes the release of small PAR oligomers from PARP1 and other acceptor proteins (12, 13). PAR then translocates to the cytoplasm (14). Interaction of PAR with mitochondria results in release of a cleaved apoptosis-inducing factor (AIF) from mitochondrial membranes (15). AIF then translocates to the nucleus, facilitated in part by its nuclear localization sequence, leading to activation of endonuclease, which initiates large-scale DNA fragmentation. PAR synthesized by PARP1 serves as a signaling molecule, relaying information related to the extent of DNA damage from the nucleus to mitochondria (15–17).
ADP-ribose-acceptor hydrolase 3 (ARH3) belongs to the ARH gene family, which comprises 3 members encoding 39-kDa proteins (ARH1–3) that are conserved in mouse and human tissues (18–20). ARH3 hydrolyzes the O-glycosidic bond of PAR and O-acetyl ADP-ribose (OAADPr), in both instances generating mono(ADP-ribose) in an Mg2+-dependent manner (21–24). In addition, ARH3 is the main hydrolase responsible for cleaving ADP-ribosylated serine (25, 26). Using ARH3-deficient mouse embryonic fibroblasts (MEFs), ARH3 was shown to regulate PAR content in the nucleus and cytoplasm following oxidative stress (14). ARH3–/– MEFs compared with WT MEFs showed a dramatic increase and sustained elevation of PAR content in the nucleus and cytoplasm in response to hydrogen peroxide (H2O2) exposure. The higher PAR levels in ARH3–/– MEFs induced release of AIF from mitochondria and its accumulation in the nucleus, resulting in chromatin condensation and nuclear shrinkage (14). PAR release into the cytoplasm was dependent on PARP1 to generate the PAR and PARG, which may be required to generate small PAR fragments that serve as substrates for ARH3. Based on these data, the level of PAR is regulated by PARP1, PARG, and ARH3, which catalyze the exoglycosidic cleavage of PAR fragments (14).
Recently, whole-exome or genome sequencing analysis of several families with affected individuals displaying age-dependent, recessive epilepsy-ataxia syndrome, showed that an ARH3 allele carrying various mutations was the top candidate of potentially deleterious genes (27). These mutations, which were located in 5 of the 6 exons of the ARH3 gene, included homozygous mutations introducing a premature stop codon, missense mutations leading to amino acid change, and homozygous deletions resulting in frameshift (27). In this article, we report a new family with ARH3 deficiency, a neurological clinical phenotype, and histological evidence of significant degeneration in the hippocampus, cerebellum, and cortical regions. The identification of this additional ARH3-deficient family supports a causative role of the mutated gene in disease. Genomic analysis of the patient showed homozygous deletion of A and G at positions 340 and 341 in the ARH3 gene, which resulted in the generation of a nonfunctional, truncated ARH3. In this family, in addition to severely affected patients, mild disease was seen in one family member, consistent with additional environmental or genetic causes of this disease. To support the patient data and to develop potential therapeutic options for the affected children, we prepared an ARH3-deficient mouse. Lack of functional ARH3 protein in patient cells and mice enhanced vulnerability to parthanatos based on a model developed earlier. As seen in the ARH3–/– MEFs, the failure of normal PAR degradation in the nucleus and cytoplasm resulted in AIF-mediated DNA fragmentation. In the prior ARH3-deficient families report, a Drosophila model was developed with PARG deficiency in which ARH3 could be replaced by the PARG gene (27). In MEFs, mice, and human fibroblasts, the ARH3 and PARG genes have different and nonredundant roles. In addition, using skin fibroblasts obtained from the patient and ARH3-deficient mice, we show that pharmacological suppression of PARP1 activity inhibited H2O2-induced cell death and ischemia/reperfusion-induced neuronal damage, thus demonstrating the therapeutic potential to inhibit neuronal loss. These data indicate that ARH3 is a PAR-degrading enzyme responsible for the regulation of PAR metabolism under conditions of oxidative stress both in vivo and in vitro, preventing the induction of parthanatos and neuronal cell death.
Results
Case reports.
The proband was an 8-year-old Hispanic male with progressive upper limb girdle neuropathy and severe muscle wasting progressing distal to proximal with autonomic symptoms, dystonic torticollis, and anorexia. He was the youngest and the only male of 4 siblings, of nonconsanguineous parents. 2 older female siblings died at ages 4 years and 8 years of progressive respiratory failure due to severe myopathy and neurodegeneration. The pathology diagnosis, for the affected female who died at age 4, was combined spinocerebellar degeneration and striatal degeneration with hippocampal sclerosis (full clinical details about the patient and pathology diagnosis of one affected sister are provided in Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124519DS1). One older living sibling, a Hispanic female, was relatively mildly affected. When seen in 2012, she was 27 years old and showed moderate intellectual disability (full scale IQ 67), untreated hypothyroidism identified when she was 19 years of age, obesity (BMI, 35.5), premature menopause, postural orthostatic tachycardia, mild sensory neuropathy in a sural distribution only by electromyography/nerve conduction velocity (EMG/NCV), anxiety, and depression. She had normal brain, pituitary, and spine MRI. EEG was consistent with nonspecific mild cerebral dysfunction. Abdominal ultrasound revealed mild hepatomegaly with steatosis. FMR1 (fragile X) genetic mutation testing was negative. A high-resolution karyotype was normal. Hormonal evaluations confirmed hypothyroidism and menopause. All other clinical testing and evaluations were normal.
Truncated, but not the full-length, ARH3 is expressed in the cytoplasm of patient fibroblasts.
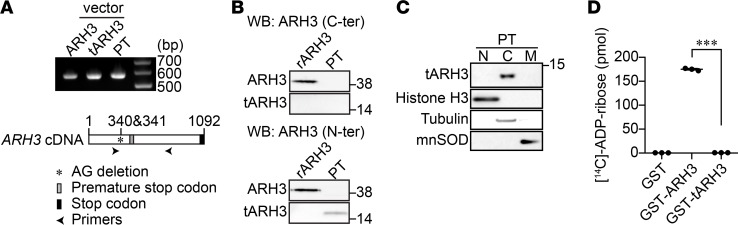
Whole-genome analysis of the family members revealed that the parents each harbored a heterozygous deletion in the ARH3 gene for which the proband was homozygous. One of the deceased siblings from whom DNA was available and the living sibling also had the identical homozygous mutation of ARH3. The patient’s homozygous AG deletions at positions 340 and 341 led to a frameshift that truncates ARH3 from 363 to 135 amino acids. Patient fibroblasts obtained by skin biopsy were examined for ARH3 mRNA expression using RT-PCR. RT-PCR amplified a 566-bp product from patient fibroblasts, similar to the RT-PCR product from expression vectors encoding full-length and truncated ARH3 (Figure 1A). Anti-ARH3 antibody directed against the C-terminal region (355–370 aa) did not detect the truncated ARH3 in patient fibroblasts, while it recognized a recombinant, full-length 39-kDa ARH3 protein (Figure 1B, upper panel). In contrast, anti-ARH3 antibody recognizing the N-terminal region in ARH3 identified a truncated ARH3 (approximately 14.5 kDa) in patient fibroblasts (Figure 1B, lower panel). Subcellular fractionation indicated that the truncated ARH3 was present in the cytoplasm (Figure 1C). The enzymatic activity of ARH3 is Mg2+ dependent (20). Based on crystal structure (23), truncated ARH3 is missing the majority of the amino acid residues essential for catalytic activity and to coordinate with 2 Mg2+ ions. As expected, the recombinant, truncated ARH3 protein failed to hydrolyze [14C]-labeled PAR, whereas WT ARH3 protein efficiently degraded PAR (Figure 1D). These findings indicate that patient fibroblasts express a nonfunctional, truncated cytoplasmic ARH3.
Figure 1. Truncated ARH3 expressed in patient fibroblasts lacks PAR-degrading activity.
(A) RT-PCR was performed to detect ARH3 mRNA transcript expression in patient fibroblasts (PTs) using ARH3-specific primers, as described in Supplemental Table 1. Plasmid vectors encoding ARH3 WT and truncated ARH3 were used as expression controls. (B) Expression of truncated ARH3 (tARH3), but not full-length ARH3, in patient fibroblasts. Cells were subjected to Western blotting using anti-ARH3 antibodies recognizing the C- (C-ter) and N-terminal regions of ARH3. Recombinant human ARH3 protein (rARH3) was used as a positive control. (C) Expression of truncated ARH3 in the cytoplasm. Purity of nuclear (N), cytoplasmic (C), and mitochondrial (M) fractions was confirmed using protein markers: histone H3 (nucleus), tubulin (cytoplasm), and manganese superoxide dismutase (MnSOD, mitochondria). (D) PAR-degrading activity of ARH3. [14C]-labeled PAR (52,012 cpm, 245 pmol) was incubated with GST-tagged proteins (200 nM) for 60 minutes. As described in Methods, [14C]-ADP-ribose was separated by HPLC using an LC-18T column; radioactivity was measured with a liquid scintillation counter. Data are mean ± SEM of values obtained from 3 samples. ***P < 0.001 by one-way ANOVA with Tukey’s post-hoc test.
H2O2 exposure induces PAR-mediated parthanatos in patient fibroblasts.
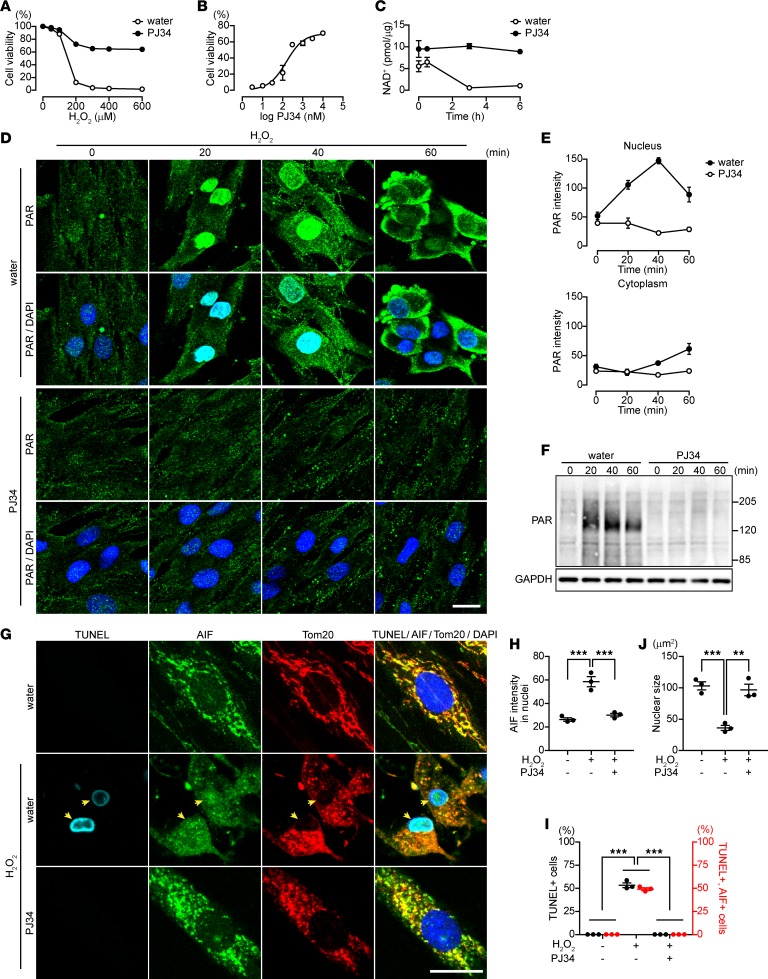
ARH3 in association with PARG catalyzes the hydrolysis of PARP1-generated PAR in the nucleus and cytoplasm following exposure of cells to H2O2, thereby preventing PAR-mediated AIF release from mitochondria and parthanatos (14). We examined the effect of nonfunctional, truncated ARH3 on H2O2-induced cytotoxicity. Exposure of patient fibroblasts to H2O2 for 24 hours was cytotoxic (Figure 2A). Pretreatment of patient fibroblasts with the PARP inhibitors PJ34 (28) and veliparib (29) for 1 hour inhibited H2O2-induced cytotoxicity in a concentration-dependent manner (PJ34: EC50 = 158.2 ± 3.2 nM, Figure 2, A and B; veliparib: EC50 = 35.8 ± 1.3 nM; Supplemental Figure 1, A and B), consistent with the conclusion that the cell death was PARP1 dependent. Cell death in patient fibroblasts was associated with NAD+ consumption, an effect reversed by PJ34 (Figure 2C) and veliparib (Supplemental Figure 1C). All data are consistent with the hypothesis that PARP1 activation following H2O2 exposure is responsible for cell death in patient fibroblasts.
Figure 2. PJ34 inhibits H2O2-induced PAR accumulation and AIF-mediated DNA fragmentation in patient fibroblasts.
(A) H2O2-induced cytotoxicity in patient fibroblasts (PJ34, 10 μM). Data are mean ± SEM of values obtained from 3 experiments conducted in triplicate. Significant differences were observed with 200 μM. (B) Concentration-dependent effect of PJ34 on H2O2-induced cytotoxicity (H2O2, 300 μM). Data represent mean ± SEM of values obtained from 3 experiments conducted in triplicate and were fitted to a sigmoidal (variable slope) curve. IC50= 158.2 ± 3.21 nM. (C) Effect of PARP activation on NAD+ content. NAD+ concentration (pmol/μg) was measured by HPLC and normalized to protein concentration. Data are mean ± SEM of values obtained from 3 experiments. Significant differences were observed at all time points. (D) Effect of PJ34 on time-dependent PAR localization following exposure to H2O2 (300 μM) in patient fibroblasts. DAPI was used as a nuclear marker. (E) Mean PAR fluorescence intensities in nuclei and cytoplasm following exposure to H2O2. Data are mean ± SEM of values obtained from 30–60 cells from three samples. Significant differences in the nucleus and cytoplasm were observed at 20 min and 40 min, respectively. (F) Effect of PJ34 on H2O2-induced PAR accumulation. These representative data were replicated three times with similar results. (G) Effect of PJ34 on AIF translocation to nuclei and DNA fragmentation. After a 16-hour exposure to 300 μM H2O2 without or with PJ34, patient fibroblasts were subjected to TUNEL assay (light blue), DAPI (blue), and immunolabeling with anti-AIF (green) and anti-Tom20 (marker for mitochondria, red) antibodies. Arrowheads indicate nuclei (blue) with DNA fragmentation that are colocalized with AIF. Scale bar: 20 μm (D and G). (H) Nuclear localization of AIF. Data are mean ± SEM of values obtained from 76–164 cells from 3 samples. ***P < 0.001. (I) Percentage of patient fibroblasts with chromatin condensation and AIF translocation to nuclei. Data are mean ± SEM of values obtained from 97–174 cells in 3 samples. ***P < 0.001. (J) Mean nuclear size assessed by DAPI. Data are mean ± SEM of values obtained from 59–221 cells from 3 samples. **P < 0.01, ***P < 0.001, two-way ANOVA with Bonferroni’s post-hoc test (A, C, E, H–J).
Using immunocytochemistry to determine PAR localization, we observed that H2O2 exposure resulted in increased PAR content in the nucleus as early as 20 minutes following H2O2 exposure, with PAR content elevated for more than 1 hour (Figure 2, D–F). After 40 minutes, cytoplasmic PAR content increased, concomitant with decreasing nuclear PAR, consistent with PAR translocation from the nucleus to cytoplasm. Pretreatment of patient fibroblasts with the PARP inhibitor PJ34 prior to H2O2 exposure decreased PAR content in patient fibroblasts (Figure 2, D–F). Following PARP1 activation, PAR production resulted in parthanatos, a caspase-independent cell death pathway that is mediated by PAR-dependent release of AIF from mitochondria (30). Under basal conditions, AIF colocalized exclusively with Tom20, a mitochondrial marker, in patient fibroblasts (Figure 2G). A 16-hour exposure of patient fibroblasts to H2O2 induced AIF translocation to nuclei of patient fibroblasts, along with nuclear shrinkage and apoptosis-related DNA fragmentation, as detected by TUNEL assay (Figure 2, G–J). Approximately 50% of patient fibroblasts became TUNEL positive (Figure 2J), and more than 90% of TUNEL-positive cells also exhibited AIF translocation to nuclei, indicating that parthanatos was responsible for H2O2-induced cell death in patient fibroblasts. Pretreatment of patient fibroblasts with PJ34 for 1 hour before H2O2 exposure counteracted these responses (Figure 2, G–J). All findings are consistent with the conclusion that, in patient fibroblasts, oxidative stress induced by H2O2 increases PAR, leading to AIF-mediated DNA fragmentation. Exposure of patient fibroblasts to the PARP inhibitors PJ34, 3-aminobenzamide, and veliparib for several days did not alter their proliferation (Supplemental Figure 1D).
Next, to confirm the role of the patient’s ARH3 mutation in the response to H2O2, we compared fibroblasts from the patient with those from his father and mother, who are heterozygous for the ARH3 mutation. Parents’ fibroblasts exhibited lower levels of PAR accumulation and cell death in response to H2O2 than did the patient’s fibroblasts. Of note, we observed greater sensitivity to H2O2 of fibroblasts from the patient’s mildly affected sibling, harboring the same homozygous ARH3 mutation (Supplemental Figure 2, A–C). In addition, using MEFs, we demonstrated that heterozygosity, but not homozygosity, of ARH3 had a limited effect on H2O2 response (Supplemental Figure 3, A–C). These results suggest that homozygous mutation of ARH3 is essential for oxidative stress–induced cell death in the patient.
Hippocampal neurons of patient’s affected older sibling showed PAR accumulation, AIF in nuclei, and DNA fragmentation.
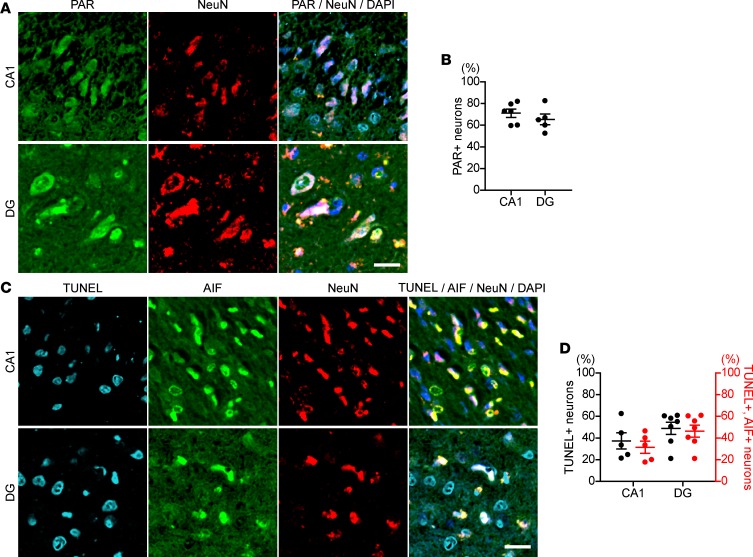
We examined the pathological specimens of the patient’s affected older sibling for potential PAR participation in the pathogenesis of neurological degeneration. Double immunostaining with the antibodies anti-PAR and anti-NeuN, a neuronal marker, revealed that a number of pyramidal neurons in the cornu ammonis 1 (CA1) and granule neurons in the dentate gyrus (DG) accumulated PAR in nuclei (CA1: 70.9% ± 3.6%, DG: 70.1% ± 9.7%; Figure 3, A and B). In addition, most pyramidal and granule neurons also showed AIF accumulation in nuclei and DNA fragmentation, as detected by TUNEL assay (CA1: 37.0% ± 6.7%, DG: 48.7% ± 6.2%; Figure 3, C and D). Approximately 90% of TUNEL-positive neurons in CA1 and DG also showed AIF in nuclei. Combined with the pathological diagnosis, all the data suggested that the neuronal cell loss/death in brain showed characteristics of parthanatos and that the neurodegeneration-induced death was associated with loss of functional ARH3 protein in neurons. Given the circumstances for pathological examination of brain specimens, effects of agonal ischemia cannot be excluded. The family and mouse model presented data for a similar mechanism of cell death.
Figure 3. Hippocampal neurons from proband’s deceased sister show an increase in nuclear PAR and AIF-mediated DNA fragmentation.
(A) PAR accumulation in nuclei of hippocampal neurons in the proband’s deceased sister, with homozygous mutation of ARH3. Hippocampal sections were subjected to immunolabeling with anti-PAR (green) and anti-NeuN (red) antibodies. Nuclei were stained with DAPI (blue). CA1, cornu ammonis 1; DG, dentate gyrus. Scale bar: 20 μm. (B) Percentage of PAR-labeled nuclei of neurons in CA1 and DG. Data are mean ± SEM of values obtained from 980 and 426 neurons in CA1 and DG, respectively. (C) AIF translocation to nuclei and DNA fragmentation in hippocampal neurons of the proband’s sister. Brain sections were subjected to TUNEL assay (right blue) and immunolabeling with anti-AIF (green) and NeuN (red) antibodies. Nuclei were stained with DAPI (blue). Scale bar: 20 μm. (D) Percentage of neurons with DNA fragmentation detected by TUNEL assay and AIF translocation to nuclei. Data are mean ± SEM of values obtained 1571 and 1308 neurons in CA1 and DG, respectively.
ARH3 deficiency enhances PAR content in the nuclei and cytoplasm and neuronal death after cerebral ischemic injury.
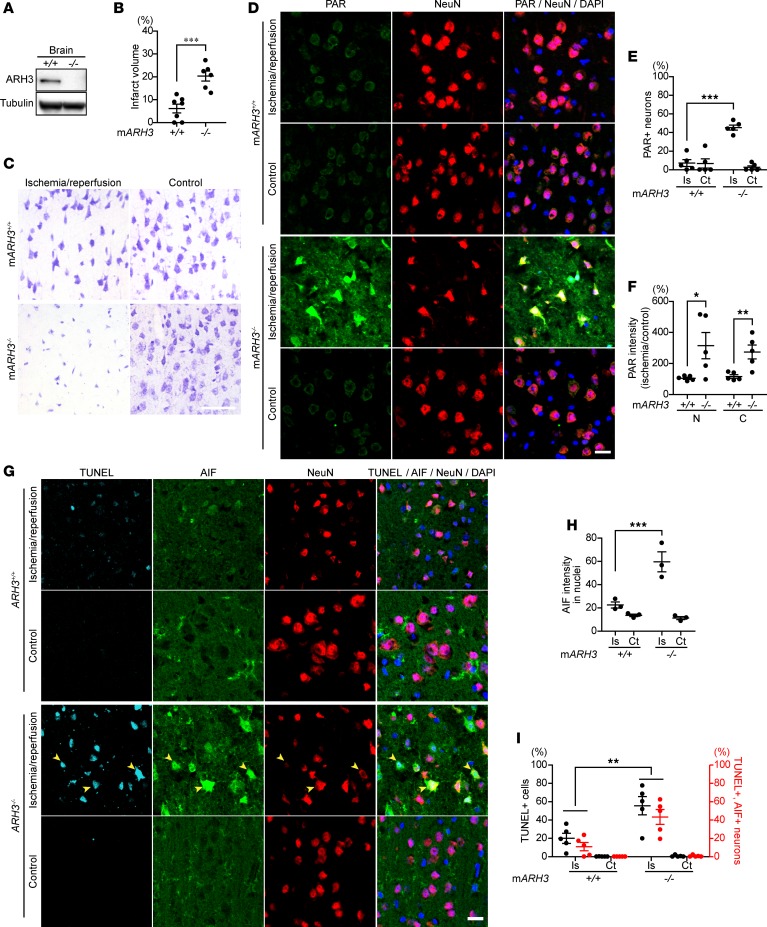
To evaluate the effect of ARH3 deficiency on neuronal death in a murine model of cerebral ischemia, we generated ARH3–/– mice (Figure 4A). Unlike the patients with truncated ARH3, ARH3–/– mice were healthy and fertile and did not display any obvious developmental abnormalities; brain histology and weight appeared normal. When brain ischemia was induced by middle cerebral artery occlusion (MCAO) for 1 hour, all ARH3–/– mice died within 24 hours after ischemia, whereas all WT mice survived (n = 3 in each group), suggesting that ARH3 deficiency may exacerbate ischemic brain injury. After 30-minute ischemia followed by 24-hour reperfusion, ARH3–/– mice survived but had a larger infarct in the cortex, hippocampus, and striatum of the ischemic hemisphere than did WT mice, as evaluated by Nissl staining (WT: 6.1% ± 1.9%, ARH3–/–: 20.3% ± 2.2%; Figure 4, B and C). Nissl bodies were seen in the cortical neurons of WT mice, while they decreased markedly in those of ARH3–/– mice (Figure 4C and Supplemental Figure 4). To assess PAR participation in the pathogenesis of the infarct, we performed immunohistochemistry to determine PAR localization. PAR was localized in perinuclear regions of cells in the control hemisphere of both WT and ARH3–/– mice (Figure 4D). Brain ischemic injury by MCAO caused PAR accumulation in nuclei of cells in the cortex of the ischemic hemisphere of ARH3–/– mice (Figure 4D). The number of cortical neurons that accumulated PAR in nuclei was significantly greater in the ischemic hemisphere of ARH3–/– mice compared with WT mice (WT: 7.3% ± 3.7%, ARH3–/–: 45.3% ± 2.8%; Figure 4E). WT mice did not exhibit an increase in the number of such PAR-positive neurons in the ischemic hemisphere. In addition, nuclear and cytoplasmic PAR levels were 3 times higher in the ischemic hemisphere of ARH3–/– mice compared with that of WT mice (Figure 4F), consistent with the hypothesis that ARH3 deficiency enhanced PAR accumulation in nuclei and its translocation to the cytoplasm after ischemic brain injury.
Figure 4. Enhanced susceptibility of ARH3–/– mice to cerebral ischemia/reperfusion is associated with increased PAR accumulation and AIF translocation to nuclei.
(A) ARH3 expression in whole brains of WT and ARH3–/– mice. Data are representative of experiments performed 3 times with similar results. (B) Infarct area after MCAO. Coronal sections were stained by the Nissl method to evaluate infarct area of WT and ARH3–/– mice subjected to 30-minute ischemia, followed by a 24-hour reperfusion. Infarct area was assessed by four coronal sections. Data are mean ± SEM of values obtained from WT (n = 7) and ARH3–/– (n = 6) mice. ***P <0.001 by 2-tailed Student’s t test. (C) Nissl-labeled neural cells in cerebral cortex in ischemic and control hemispheres of WT and ARH3–/– mice. Scale bar: 50 μm. (D) PAR localization in cerebral cortex. Brain sections were reacted with anti-PAR (green) and anti-NeuN (red) antibodies, and DAPI (blue). Scale bar: 20 μm. Data are representative of experiments performed 3 times with similar results. (E) Percentage of PAR-labeled nuclei of cortical neurons. Data are mean ± SEM of values obtained from 3182 and 1687 cortical neurons in ischemic (Is) and control (Ct) hemispheres of WT mice (n = 5), respectively; and 2626 and 1831 cortical neurons in ischemic and control hemispheres of ARH3–/– mice (n = 5), respectively. ***P < 0.001. (F) Nuclear and cytoplasmic PAR levels in cortex. Data are mean ± SEM of values obtained from WT (n = 5) and ARH3–/– (n = 5) mice. *P < 0.05, **P < 0.01. (G) DNA fragmentation and AIF translocation to nuclei in cerebral cortex. Brain sections were subjected to TUNEL assay (light blue) and reacted with anti-AIF (green) and anti-NeuN (red) antibodies and DAPI (dark blue). Arrowheads indicate nuclei (blue) with DNA fragmentation that are colocalized with AIF. Scale bar: 20 μm. (H) Nuclear localization of AIF. Data are mean ± SEM of values obtained from 666 and 906 cortical neurons in ischemic and control hemispheres of WT mice (n = 3), respectively; and 1804 and 551 cortical neurons in ischemic and control hemispheres of ARH3–/– mice (n = 3), respectively. ***P < 0.001. (I) Percentage of neurons with DNA fragmentation and AIF translocation to nuclei. Data are mean ± SEM of values obtained from 2443 and 1540 cortical neurons in ischemic and control hemispheres of WT mice (n = 5), respectively; and 2575 and 1772 cortical neurons in ischemic and control hemispheres of ARH3–/– mice (n = 5), respectively. **P < 0.01. Statistical tests done in E, F, H, and I were two-way ANOVA with Bonferroni’s post-hoc test.
Since the parents were heterozygotes for the ARH3 mutation, we next investigated whether the response in mice differed between the WT, heterozygous, and knockout animals. We evaluated cerebral infarction using the 2,3,5-triphenyltetrazolium chloride (TTC) staining technique after temporary MCAO for 30 minutes, followed by 24-hour reperfusion in ARH3–/–, heterozygous (ARH3+/–), and WT mice. Infarct size in ARH3–/– mice (34.58% ± 1.1%) was significantly (P < 0.01) larger than for ARH3+/– and WT mice (13.69% ± 1.1% and 13.73% ± 1.1%, respectively) (Supplemental Figure 5, A and B). During 24 hours of reperfusion after 30 minutes of ischemia, 9 of 15 ARH3–/– mice died. All ARH3+/– and WT mice survived after 30 minutes of ischemia and 24-hour reperfusion (Supplemental Figure 5C). Functional effects of the 30-minute ischemia and 1- or 24-hour reperfusion were evaluated in a blinded manner using neurological scoring of movements by animal facility veterinarians (see Methods). ARH3–/– mice had significantly greater neurological deficits than ARH3+/– and WT mice at both evaluation times (after 1- and 24-hour reperfusion) (Supplemental Figure 5, D and E). In the evaluation of 1-hour reperfusion, all ARH3–/– mice had severe neurological deficits (score of 4 or 5), with deep unconsciousness as a result of severe brain injury (e.g., loss of walking, more than 80% circling). These results suggested that ARH3–/– mice had severe functional outcomes. In contrast, ARH3+/– and WT mice survived and showed the expected effects after 30-minute ischemia and 24-hour reperfusion in both infarct size and neurological evaluation.
We next investigated whether cytoplasmic PAR may be responsible for cleavage and release of AIF from mitochondria (Figure 4G). After 30-minute ischemia followed by 24-hour reperfusion, many cortical neurons in the ischemic hemisphere of ARH3–/– mice showed an increase in nuclear AIF intensity (Figure 4H) and DNA fragmentation, as detected by TUNEL assay, compared with those in the ischemic hemisphere of WT mice (WT: 20% ± 3.3%, ARH3–/–: 55.6% ± 7.1%; Figure 4I). Approximately 80% of TUNEL-positive neurons also showed AIF translocation to nuclei, while AIF was diffusely distributed in the mitochondria of cortical neurons in the control hemisphere of both WT and ARH3–/– mice (Figure 4I). DNA fragmentation was not observed in cortical neurons of the control hemisphere in either WT or ARH3–/– mice. Consistent with the observations following exposure of patient fibroblasts to H2O2, ARH3 deficiency enhanced nuclear and cytoplasmic PAR content after ischemic brain injury, resulting in AIF-mediated DNA fragmentation.
The PARP inhibitor veliparib alleviates brain ischemic injury by preventing PAR and AIF accumulation in ARH3–/– mice.
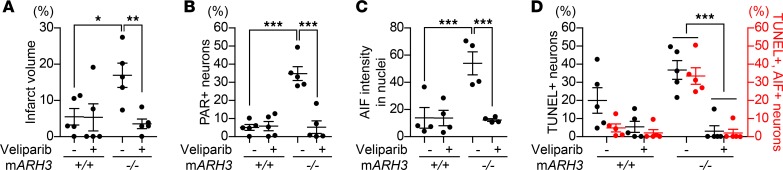
Next, we examined whether a PARP inhibitor, veliparib, alleviates parthanatos-mediated neuronal cell death after ischemic brain injury caused by MCAO. Veliparib resulted in a dramatic reduction of infarct area in ARH3–/– mice (control: 17.0% ± 3.3%, veliparib: 3.5% ± 1.3%), but not in WT mice (control: 5.5% ± 2.3%, veliparib: 5.3% ± 3.7%; Figure 5A). In addition, veliparib prevented PAR accumulation in nuclei of cortical neurons in the ischemic hemisphere of ARH3–/– mice (control: 34.3% ± 2.9%, veliparib: 4.8% ± 1.7%), while it did not affect the PAR accumulation seen in WT mice (control: 5.1% ± 1.2%, veliparib: 5.7% ± 1.6%; Figure 5B). Veliparib reduced the AIF protein level in nuclei of cortical neurons in the ischemic hemisphere of ARH3–/– mice (control: 53.9% ± 7.3%, veliparib: 12.2% ± 0.9%; Figure 5C) and the number of TUNEL-positive cells as well as AIF and TUNEL-double-positive cells in ARH3–/– mice more than in WT mice (TUNEL-positive cells: WT: control: 20.0% ± 3.1%, veliparib: 5.4% ± 1.9%; ARH3–/–: control: 36.9 ± 3.4%, veliparib: 3.0% ± 1.3%, AIF and TUNEL-double positive cells: WT: control: 4.8% ± 1.5%, veliparib: 2.1% ± 1.2%; ARH3–/–: control: 33.5% ± 3.4%, veliparib: 2.0% ± 0.9%; Figure 5D). These results indicate that in ARH3-deficient mice the PARP inhibitor veliparib prevented neuronal cell death after brain ischemic injury with decreased PAR accumulation and AIF-mediated DNA fragmentation.
Figure 5. Exacerbated cerebral ischemic injury and enhanced PAR and AIF accumulation in ARH3–/– mice are prevented by PARP inhibition.
(A) Quantification of infarct volume Data are mean ± SEM of values from WT (n = 5) and ARH3–/– (n = 5) mice. *P < 0. 05, **P < 0.01. (B) Percentage of PAR-labeled of cortical neurons in ischemic cortex. Data are mean ± SEM of values obtained from 1264 and 1783 cortical neurons in control- and veliparib-treated WT mice (n = 5), respectively; and 1453 and 1793 cortical neurons in control- and veliparib-treated ARH3–/– mice (n = 5), respectively. ***P < 0.001. (C) Nuclear localization of AIF. Data are mean ± SEM of values obtained from 349 and 211 cortical neurons in control- and veliparib-treated WT mice (n = 5), respectively; and 394 and 717 cortical neurons in control- and veliparib-treated ARH3–/– mice (n = 5), respectively. ***P < 0.001. (D) Percentage of neurons with DNA fragmentation detected by TUNEL assay and AIF translocation to nuclei. Data are mean ± SEM of values obtained 802 and 847 cortical neurons in control and veliparib-treated WT mice (n = 5), respectively; and 646 and 714 cortical neurons in control and veliparib-treated ARH3–/– mice (n = 5), respectively. ***P < 0.001. Two-way ANOVA with Bonferroni’s post-hoc test used in all panels.
Discussion
In a single-family study, we show that a mutation in the human ARH3 gene is associated with neurological degeneration. The patient and his siblings with homozygous mutations in ARH3 exhibited mild to severe neurodegenerative symptoms, while their parents with heterozygous mutations did not. Using patient fibroblasts and ARH3–/– mice, we confirm that the ARH3 protein participates in an important regulatory pathway to maintain PAR homeostasis in the nucleus and cytoplasm, preventing neurons from undergoing parthanatos — PARP1-dependent, AIF-mediated cell death.
ARH3 truncation produced a catalytically inactive protein.
Our genetic studies found a homozygous deletion of A and G at positions 340 and 341 in the ARH3 gene, which results in the substitution of glycine for arginine at position 114 and a frameshift; this leads to the generation of another 23 amino acids unrelated to the WT ARH3 protein, followed by a premature UGA stop codon. Thus, the base deletions shorten ARH3 from 363 to 136 amino acids. PAR-degrading activity of ARH3 depends on a two-Mg2+ cluster to bind the ADP-ribose moiety of PAR chains (20, 23). Removal of Mg2+ ions or replacement of any of the amino acid residues that coordinate Mg2+ ions (i.e., Glu41, Asp77, Asp78, Asp314, Thr317) resulted in a dramatic reduction in enzymatic activity (20, 23). Truncated ARH3, containing amino acids 1–114 of the ARH3 protein and an additional 23 amino acids, loses amino acids critical to coordinate Mg2+ ions. In an assay quantifying PAR-degradation, the shortened ARH3 did not exhibit enzymatic activity.
Loss of functional ARH3 led to cell death under oxidative stress conditions via a PAR-dependent, AIF-mediated pathway.
In vitro and in vivo studies using patient-derived fibroblasts and ARH3-deficient mice showed that loss of functional ARH3 protein led to cell death via parthanatos following oxidative stress, i.e., H2O2 exposure and ischemic brain injury induced by MCAO, respectively. PARP1 transfers PAR to PARP1 itself and nuclear acceptor proteins in response to DNA damage; PAR chains are degraded and released by the endoglycosidic activity of PARG (12, 13), with removal of the initial ADP-ribose moiety linked to the acceptor protein by terminal ADP-ribose glycohydrolase (TARG1/C6orf130) (31) or ARH3 (25, 26). The protein-free PAR passes through nuclear pores and then reaches mitochondrial membranes, where it facilitates release of AIF (14). ARH3 protein is located mainly in the cytoplasm, followed by mitochondria and the nucleus (14, 32). PAR in the cytoplasm appears to be a target of ARH3 in neurons, consistent with the observation that the cytoplasmic PAR levels in the cortex were higher in the ischemic hemisphere of ARH3–/– mice than that of WT mice. Moreover, ARH3 is capable of efficiently cleaving serine-ADP-ribose proteins as well as PAR in nucleus in response to H2O2 (26, 33), preventing nucleus PAR accumulation. The above mechanism explains the finding that both in neurons of the patient’s sibling and in mouse brain, PAR accumulated in both cytoplasm and nucleus.
The effect of ARH3, as well as PARG, is likely to be more complex than simply changing the PAR level. For example, ARH3 deficiency may cause suppression of PARP1 activity, due to extensive poly(ADP-ribosyl)ation of PARP1 (34–36), and may downregulate PARP1 gene expression, as PARG knockdown has been shown to do (37). Further study is needed to better understand the function of ARH3.
Oxidative stress is a common feature shared by multiple neurodegenerative diseases, including Parkinson’s disease and Alzheimer’s disease, and is a major cause of enhanced PARP1 activation (38). Parthanatos is seen in neurons of mice with Parkinson’s disease and after ischemic brain injury and glutamate excitotoxicity (6, 39, 40). In the central nervous system, maintaining PAR homeostasis by PAR-degrading enzymes is critical to prevent excessive PAR accumulation. In fact, a Drosophila mutant lacking the catalytic domain of PARG and humans exhibiting homozygous expression of truncated nonfunctional TARG showed severe neurodegeneration (31). ARH3 is expressed in humans and mice, but not in Drosophila. Due to the partial overlap of PARG and ARH3 in PAR degradation activity, expression of ARH3 in PARG–/– Drosophila, rescued the reduced locomotion and life span (27). In contrast, ARH3 has a nonredundant activity with PARG in human and mouse cells. Once DNA is damaged, cytoplasmic PARG isoforms are recruited to sites of nuclear DNA damage (41, 42), while ARH3 is essential for cytoplasmic, nuclear, and perhaps mitochondrial PAR degradation (14, 32). Despite the observation that the PAR-degrading activity of ARH3 is about 2% that of PARG (20), PARG apparently cannot replace ARH3 as a PAR-degrading enzyme (14). Therefore, both PARG and ARH3 are indispensable in regulating PAR metabolism and preventing neuronal cell death via parthanatos (14). Similar to the cell death pathway of ARH3-deficient mice or patient’s and sister’s fibroblasts subjected to oxidative stress, AIF nuclear translocation and DNA damage were present in neurons of the patient’s sibling’s brain slices. Further investigation is necessary to address why a patient with truncated ARH3 develops spontaneous neurodegeneration not seen in ARH3–/– mice. Environmental factors (e.g., fever, infection, stress) should be considered as potential explanations. Environmental factors might also enhance oxidative stress in the central nervous system, resulting in PARP1 activation, leading to neuronal cell death.
The PARP1 inhibitor veliparib, partially attenuated oxidative stress–induced cell death.
More than 90% of PAR is generated by PARP1 activation (43). While the PAR accumulation in cytoplasm is responsible for cell death under oxidative stress in ARH3-deficient mice and in the patient, we speculate that PARP1 inhibition may have therapeutic potential. PARP1 inhibition has proven to be neuroprotective in a transient cerebral ischemia model (44). Indeed, PARP inhibitors significantly reduced cytotoxicity following exposure to H2O2 without having an effect on the proliferation of patient fibroblasts, as well as neuronal cell death after brain ischemic injury in ARH3–/– mice.
PARP1 inhibitors have been approved for treatment of patients with BRCA1/BRCA2-mutated advanced ovarian cancer based on the concept of synthetic lethality (45). For example, veliparib selectively inhibits both PARP-1 and -2, with Ki of 5.2 and 2.9 nmol/l, respectively (29). In a clinical trial, oral veliparib significantly inhibited PARP levels in both tumor tissue and peripheral blood mononuclear cells (46); the effects began 3–6 hours after administration, with recovery at 24 hours in both preclinical models and patients (46). In general, veliparib has an overall acceptable safety profile in patients with breast cancer (47). Further, veliparib was shown to cross the blood-brain barrier, providing a further rationale for its application to treat intracranial diseases (46). PJ34, another PARP1 inhibitor, showed stronger protective effects on H2O2-treated cells than veliparib (Figure 2, A and B, and Supplemental Figure 1, A and B), which might be explained by additional pharmacological effects (48).
In conclusion, our patient and cellular and animal models provide evidence that ARH3 deficiency plays a critical role in PARP1-dependent neuronal cell death through its PAR-degrading activity, which regulates nuclear and cytoplasmic PAR levels. Understanding the precise role of ARH3 may be helpful in understanding the pathogenesis of neurodegenerative disorders. PARP inhibitors might represent a therapeutic option to slow neuronal cell death occurring in an ARH3-deficient patient.
Methods
Antibodies.
Rabbit polyclonal anti-ARH3 antibodies were prepared by immunizing rabbits with a peptide (CTDVLAQSLHRVFQESS) representing amino acids 355–370 of mouse ARH3. Rabbit anti-ARH3 polyclonal antibodies that recognize the N-terminal region of ARH3 (ab102979) were purchased from Abcam; mouse anti-AIF monoclonal antibodies (clone B-9, sc-55519) from Santa Cruz Biotechnology Inc.; rabbit anti-GAPDH (clone 14C10, 2118) and anti–histone H3 (clone D1H2, 4499) monoclonal antibodies from Cell Signaling Technology; anti-tubulin (clone DM1A, T6199) and anti–β-actin (clone AC-15,A5441, A2066) antibodies from Sigma-Aldrich; mouse anti-PAR monoclonal antibodies (clone 10H, ALX-804-220-R100) from Enzo Life Sciences; rabbit anti-NeuN polyclonal antibodies (ABN78) from Millipore; mouse anti-MnSOD monoclonal antibodies (clone 19/MnSOD, 611580) from BD Biosciences; Click-iT TUNEL Alexa Fluor 647 Imaging Assay and fluorescence-conjugated secondary antibodies (Alexa Fluor 488–conjugated goat anti-mouse and 555-conjugated goat anti-rabbit antibodies) from Invitrogen; veliparib from Selleck Chemicals; HRP–goat anti-mouse and anti-rabbit immunoglobulin from Pierce; and IRDye 680RD goat anti-Mouse (no. 926-68070) and 800CW goat anti-rabbit (no. 926-32211) from LI-COR.
Cell culture.
Fibroblasts were generated from a 4-mm punch skin biopsy. Fibroblasts from the patient, his sibling, mother, and father, WT, ARH3+/–, and ARH3–/– MEFs (14) were incubated at 37°C in a humidified atmosphere with 5% CO2 in high-glucose DMEM containing 10% FBS, 100 U penicillin, and 100 μg/ml streptomycin.
RT-PCR.
Total RNA was isolated using an RNeasy Mini Kit (QIAGEN) and reverse transcribed using Ready-to-Go You-Prime first-strand beads (GE Healthcare Life Sciences) and oligo(dT) primer (Thermo Fisher Scientific). RT-PCR was performed using specific ARH3 and GAPDH primers to evaluate mRNA expression level. Sequences of primers are shown in Supplemental Table 3.
Preparation of GST proteins, ARH3 mutant proteins, and GST pull-down assay.
Mutations were generated using the Stratagene QuikChange Site-Directed Mutagenesis Kit (Agilent Technologies), according to the manufacturer’s protocol. Complementary mutant primers used to generate vectors containing the ARH3 mutant are shown in Supplemental Table 3. The entire coding regions were ligated into pGEX-6P-1 expression vector (GE Healthcare Life Sciences) for transfection into Escherichia coli BL21 Rosetta Supercompetent cells. Expression of these GST-proteins was induced by 0.5 mM IPTG (37°C, 6 hours). Proteins were extracted with PBS by sonication, and then 1% Triton X-100 (4°C, 30 minutes) was added. Proteins were purified with Glutathione Sepharose 4B according to the manufacturer’s instructions (GE Healthcare Life Sciences). GST proteins (20 pmol) were incubated with 5 pmol His-tagged AIF (Abcam) and 25 μg glutathione Sepharose 4B beads in 200 μl of 25-mM HEPES containing 10 mM MgCl2 and 5 mM DTT (room temperature [RT], 1 hour). After washing with PBS, 50 μl of 20-mM reduced glutathione in 50 mM Tris-Cl (pH 8.0) was added to the beads. The supernatants were analyzed by Western blot.
Preparation of [14C]-PAR and measurement of ARH3 activity.
[14C]-PAR was prepared and purified as described by Oka et al. (20). Briefly, 3 μg PARP1 (Enzo Life Sciences) was incubated with 20 μg/ml activated calf thymus DNA, 10 μM unlabeled NAD, and 6 μCi [14C]-NAD (PerkinElmer) in a 150-μl reaction mixture containing 100 mM Tris-HCl, pH 8.0, 10 mM DTT, and 10 mM MgCl2 (37°C, 10 minutes); the reaction was stopped with 20% trichloroacetic acid. After overnight precipitation, the pellet was washed with cold acetone and neutralized. PARP1 was released from [14C]-PAR by incubation at 37°C for 2 hours with 2 M neutral NH2OH in 50 mM MOPS, containing 6 M guanidine HCl. Protein-free [14C]-PAR was purified with dihydroxyboronyl Bio-Rex 70 (100–200 dry mesh, Bio-Rad) affinity chromatography. GST proteins (20 pmol) were incubated with [14C]-PAR (200 nM) in a 100-μl reaction mixture containing 50 mM KPO4, pH 7.5, 5 mM DTT, and 10 mM MgCl2 at 37°C for the indicated times. The reaction was stopped with 2% trifluoroacetic acid. [14C]-ADP-ribose was separated by a reverse-phase HPLC system (series 1100, with a diode array spectrophotometric detector set at 259 nm, Hewlett-Packard) equipped with an LC-18T column (5-μm particle size, 4.6–250 mm, Sigma-Aldrich). Isocratic elution (1 ml/min) with 100% buffer A (50 mM KPO4) from 0 to 2 minutes was followed by a linear gradient to 100% buffer B (methanol) from 2 to 10 minutes, and radioactivity was measured with a lipid scintillation counter.
Cell viability assays.
Fibroblasts from the patient and his family and MEFs (1 × 103 to 3 × 103) were incubated on 96-well plates for 1–2 days before exposure (24 hours) to the indicated concentrations (30–1000 μM) of H2O2. PJ34 (10 μM) or veliparib (0.1–1 μM) was added for 1 hour prior to H2O2 exposure. Cell number was assessed using a Cell Counting Kit-8 (Dojindo) by measuring absorbance at 450 nm according to the manufacturer’s instructions. Cell death was assessed by using a Cytotoxicity LDH Assay kit-WST (Dojindo) by measuring absorbance at 490 nm according to the manufacturer’s instructions. Absorbance was measured using SpectraMax M5 Microplate Reader (Molecular Devices).
Subcellular fractionation.
Subcellular fractionation was performed by a Subcellular Proteome Extraction Kit (Merck), according to the manufacturer’s instructions. Purity of nuclear, cytoplasmic, and mitochondrial fractions was assessed using protein markers: histone H3 (nucleus), tubulin (cytoplasm), and manganese superoxide dismutase (MnSOD, mitochondria).
SDS-PAGE and Western blotting.
Cells were lysed in 20 mM Tris-HCl (pH 7.4) with 2% SDS or RIPA buffer (Sigma-Aldrich) supplemented with proteinase inhibitors. Lysates were subjected to Bis-Tris or Tris-acetate SDS-PAGE (Invitrogen) and then transferred to nitrocellulose or PVDF membranes (Invitrogen). The membranes were blocked with 5% (vol/vol) nonfat dry milk (Bio-Rad) in TBS or Odyssey Blocking Buffer (LI-COR) (4°C, overnight or RT, 1 hour) and then incubated with primary antibody (4°C, overnight, or RT, 1 hour). An ECL system (Fujifilm LAS-3000) or Odyssey Imaging Systems (LI-COR) was used for detection.
TUNEL assay.
Apoptotic cells in patient fibroblasts and brain tissues were visualized by Click-iT TUNEL Assay (Thermo Fisher Scientific), according to the manufacturer’s instructions. The TUNEL assay relies on incorporation of labeled dUTP at sites of DNA breaks.
Immunocytochemistry.
Patient fibroblasts and brain sections were fixed with 4% paraformaldehyde (RT, 20 minutes), permeabilized and blocked with PBS containing 0.5% Triton X-100, 10% FBS, and 1% BSA (RT, 1 hour) and then incubated with primary antibodies (overnight, 4°C). After washing with PBS, samples were treated with either Alexa Fluor 488–conjugated goat anti-mouse IgG or Alexa Fluor 555–conjugated goat anti-mouse IgG (1:500) (1 hour, RT) and then incubated with 300 nM DAPI (5 minutes, RT) to stain nuclei. Brain sections were imaged with a confocal microscope (Zeiss LSM 510 UV) equipped with a Plan NeoApo objective (20′, numerical aperture, 0.75), an argon laser (488 nm) to excite Alexa Fluor 488, a 561 diode (561 nm) to excite Alexa Fluor 555, a 405 diode (405 nm) to excite DAPI, and a 633 diode to excite Alexa Fluor 647. Fluorescence data were processed with ImageJ 1.37a (NIH).
Measurement of NAD+ concentrations.
Patient fibroblasts seeded in 10-cm dishes were incubated in DMEM with 10% FBS (2 days, 37°C). After washing with PBS, 1 M HClO4 (300 μl) was added, and the incubation continued on ice for 10 minutes. The resulting solution was centrifuged at 14,000 g, and the supernatant (240 μl) was neutralized with 3 M K2CO3 (80 μl). After centrifugation, samples were loaded on an HPLC equipped with an LC-18T column. NAD (Sigma-Aldrich) was used to generate a standard curve. Isocratic elution (1 ml/min) with 100% buffer A (50 mM KPO4) from 0 to 2 minutes was followed by a linear gradient to 100% buffer B (methanol) from 2 to 10 minutes.
Measurement of infarct size.
After 24-hour reperfusion, the mice were euthanized by exsanguination under general anesthesia (1%–3% isoflurane). The brain was collected and kept on dry ice. Then, the brains were sliced into 2-mm coronal sections and stained with 1% TTC (Sigma-Aldrich) solution for 30 minutes at 37°C and fixed in 4% paraformaldehyde at 4°C overnight. After fixation, the brain slices were scanned using a flatbed scanner (Epson V700 Photo scanner). Infarct size of each brain slice was quantified with NIH ImageJ software. Infarct size was calculated by summing the infarct areas measured in 6 brain slices (2-mm thickness) and was expressed as the percentage of whole area of cross-sectional slices of mouse brain (49).
Functional consequences of focal cerebral ischemia/reperfusion injury in ARH3-deficient mice.
Neurological deficits were assessed at 1 and 24 hours after reperfusion using a 6-point scale; 0: normal; 1: mild circling behavior with or without inconsistent rotation when picked up by the tail, <50% attempts to rotate to the contralateral side; 2: mild consistent circling, >50% attempts to rotate to the contralateral side; 3: consistent strong and immediate circling, the mouse holds a rotation position for more than 1–2 seconds, with its nose almost reaching its tail; 4: severe rotation progressing into barreling, loss of walking or righting reflex; 5: comatose or moribund (50). Mice were assessed in a blinded manner by animal facility veterinarians.
Targeting construct and generation of ARH3–/– mice.
A BAC (RPCI 22) clone containing an ARH3 gene was obtained from a mouse spleen-129s6/SvEv BAC library (Thermo Fisher Scientific). A 2.2-kb fragment containing part of intron 2 was amplified from the BAC clone by PCR and subcloned into the 5′ arm of the pNTK loxp/FrtII vector. The pNTK loxp/FrtII vector was provided by Warren Leonard (NHLBI, NIH). A 4.6-kb fragment containing part of introns 2–5 and exons 3-5 was amplified from BAC clone by PCR and subcloned into the 3′ arm of the pNTK loxp/FrtII vector. The targeted 0.5-kb fragment was flanked by 2 loxP sites (Supplemental Figure 6A, shaded triangle), allowing Cre-mediated excision of the exon 3 region of the ARH3 locus. The deleted region included D77 and D78, which are critical for catalytic activity (20). The targeting vector also contained the neomycin resistance gene (Neo) driven by the phosphoglycerate kinase (PGK) promoter and flanked by 2 Frt sites (Supplemental Figure 6A, closed triangles), allowing Flp-mediated excision of the PGK-Neo cassette. The unique Not I site was used to linearize the pNTK loxp/FrtII vector before electroporation of embryonic stem (ES) cells. ES cells were electroporated, and transformed cells were selected by standard techniques. Clones resistant to both G418 and ganciclovir were analyzed by Southern blot analysis. WT and targeted ARH3 alleles were found in, respectively, 8.7- and 5.7-kb bands, and 8.7- and 4.7-kb bands by Southern blot analysis of the ES cell genomic DNA digested with HindIII and hybridized with 5′ probe (including exon 1) or 3′ probe (including exon 5) (Supplemental Figure 6B). Chimeric mice (129/SvJ-derived ES cells in blastocysts of C57BL/6J mice) were generated by standard procedures. Chimeric mice were identified by coat color and then were bred to C57BL/6J mice. Transmission of the germline was identified by PCR and confirmed by Southern blot analysis. Heterozygous germline-transmitted mice (ARH3T/+) were bred with ACT-Flpe transgenic mice (The Jackson Laboratory) to produce ARH3F/+ mice. To delete the target region (exon 3), ARH3F/+ mice were mated with EIIa-Cre transgenic mice ubiquitously expressing Cre recombinase (The Jackson Laboratory) to generate ARH3D/+ (ARH3+/–) mice. Genotypes of these mice were confirmed by PCR (Supplemental Figure 6, C and D).
Southern blot analysis.
Southern blot analysis was carried out as described previously (51). Briefly, samples (5–10 μg) of genomic DNA prepared from ES cells or mouse tails using a Wizard genomic DNA purification kit according to standard protocols were digested with HindIII (Roche), subjected to electrophoresis in 1.0% agarose gel, and transferred to nylon membranes. The 5′ (670 bp) and 3′ (320 bp) cDNA probes corresponding to bases in the mouse ARH3 genomic DNA were amplified from the BAC clone. Membranes were incubated for 1 hour at 68°C in ExpressHyb (Takara) with [α-32P]dATP–labeled (PerkinElmer) probes located outside the 5′ and 3′ regions of the targeting vector. Sequences of primer sets to amplify 5′ and 3′ probes are shown in Supplemental Table 3. Membranes were washed twice for 20 minutes at RT with 2× SSC containing 0.1% SDS and twice for 30 minutes at 60°C with the same solution before exposure to film. [32P] was detected by exposure of X-ray film (Kodak).
Transient focal cerebral ischemia by MCAO.
Six-month-old male C57BL/6 WT and ARH3–/– mice were anesthetized for induction with 3%–5% isoflurane and maintained in 1%–3% isoflurane using a vaporizer. The body temperature of the mice during surgery was maintained at 36.5 ± 0.5°C with a heating plate. The left MCA was occluded by inserting an 8-0 silk monofilament suture with rounded tip into the internal carotid artery from a small hole in the external carotid artery. After appropriate periods (30 and 60 minutes) of ischemia, the mice were anesthetized again to remove the suture and start the reperfusion. The time interval between the end of the ischemia and reperfusion was less than 10 minutes, including time for anesthesia and reperfusion procedures. The PARP inhibitor veliparib at 25 mg/kg or vehicle buffer (0.5% methylcellulose/0.2% Tween 80) was administrated by intraperitoneal injection 3 times at indicated time points: 10 minutes before ischemia, 10 minutes before reperfusion, and 30 minutes after reperfusion. After 24-hour reperfusion, mice were anesthetized with isoflurane and were perfused with heparinized 0.9% saline followed by 4% paraformaldehyde in PBS. The entire brain was removed from the skull and immersed in 4% paraformaldehyde (4°C, 2 hours). The fixed brain was cryoprotected by immersing in 20% sucrose in PBS for (4°C, 1 day) and was then embedded in Jung tissue-freezing medium (Leica). Coronal sections (20 μm) were prepared using a cryostat microtome (Leica) at –22°C. Sham-treated animals underwent identical surgical procedures, except that the left middle cerebral artery (MCA) was not occluded. For evaluation of infarct area, Nissl staining was performed. Sections were treated with acetone (RT, 30 minutes) and rehydrated in a descending series of ethanol (100%, 95%, and 75% ethanol and water). The sections were treated with 0.2% cresyl violet (Sigma-Aldrich) solution (RT, 30 minutes). Then, the sections were dehydrated in an ascending series of water/ethanol (water, 75% and 95% ethanol), and treated with xylene. Infarct volume was calculated from infarct area of 4 coronal sections.
Statistics.
Statistical analysis was performed using GraphPad Prism or Excel software (Microsoft). Significance was determined using paired t tests, Student’s t test for pairwise comparison, or 1- or 2-way ANOVA with Bonferroni’s post hoc test. Data are mean ± SEM of values from the indicated number of experiments. P values less than 0.05 were considered significant. All representative experiments were repeated 3 times.
Study approval.
One family with 3 affected siblings was studied. The proband, a participant in the NIH Undiagnosed Diseases Program (REFS A-C), was admitted to the NIH Clinical Center (NIH-CC) and enrolled in protocol 76-HG-0238, “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders” (ClinicalTrials.gov, NCT00369421), approved by the National Human Genome Research Institute (NHGRI) IRB (52–54). The family was also seen under protocols 96-H-0100 and 82-H0032, approved by the NHLBI IRB. The parents gave written informed consent. Animal studies were approved by the NHLBI Animal Care and Use Committee (protocols H-0128 and H-0271). ARH3 mice were backcrossed 11 times using C57BL6/J mice (The Jackson Laboratory).
Author contributions
JM conceived, designed, and coordinated the study, and served as the primary author of the manuscript. MM, XB, and KA designed, performed, and analyzed the majority of the experiments. JK prepared the ARH3 recombinant protein and conducted activity assays. JK and AK generated ARH3 knockout mice. JK, HIE, and LAS performed and analyzed experiments. WAG, LAW, CT, and DA performed genomic analysis, and provided patient fibroblasts and case reports. MM, XB, KA, and JM prepared the figures and wrote the manuscript. JM, TM, and WAG edited the manuscript.
Supplementary Material
Acknowledgments
We thank Martha Vaughan (now deceased) for helpful discussions and critical review of the manuscript. We appreciate the professional skills and advice of Randy Clevenger and Karen Keeran (Animal Surgery and Resources Core, NHLBI, NIH), who performed the cerebral ischemia and reperfusion surgery. We appreciate the professional skills and advice of Christian A. Combs and Daniela Malide (Light Microscopy Core, NHLBI, NIH) regarding microscopy-related experiments. We thank Chengyu Liu, NHLBI Transgenic Core, for expertise in generating ARH3-deficient mice; Zu-Xi Yu, NHLBI Pathological Core, for support in performing immunohistochemistry of human and mouse brain; and Danielle A. Springer, NHLBI Murine Phenotyping Core, for support in the functional analysis of ARH3-deficient mice. This study was supported by the NIH Intramural Research Program, NHLBI and National Human Genome Research Institute.
Version 1. 02/21/2019
Electronic publication
Funding Statement
This study was supported by the Intramural Research Program, National Institutes of Health, National Heart, Lung, and Blood Institute and National Human Genome Research Institute
Footnotes
MM’s present address is: Department of Pharmacology, Faculty of Pharmaceutical Sciences, Doshisha Women’s College of Liberal Arts, Kyotanabe, Kyoto, Japan.
AK’s present address is: Department of Oral Science, Graduate School of Medicine, Chiba University, Chuo-ku, Chiba, Japan.
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(4):e124519. https://doi.org/10.1172/jci.insight.124519.
Contributor Information
Masato Mashimo, Email: mmashimo@dwc.doshisha.ac.jp.
Xiangning Bu, Email: xiangningbu@gmail.com.
Kazumasa Aoyama, Email: aoyamakazumasafepsg@gmail.com.
Jiro Kato, Email: katoj@nhlbi.nih.gov.
Hiroko Ishiwata-Endo, Email: endohiro@mail.nih.gov.
Linda A. Stevens, Email: stevensla61@gmail.com.
Atsushi Kasamatsu, Email: kasamatsua@faculty.chiba-u.jp.
Lynne A. Wolfe, Email: lynne.wolfe@nih.gov.
Camilo Toro, Email: toroc@mail.nih.gov.
David Adams, Email: dadams1@mail.nih.gov.
Thomas Markello, Email: markellot@mail.nih.gov.
William A. Gahl, Email: gahlw@mail.nih.gov.
Joel Moss, Email: mossj@nhlbi.nih.gov.
References
- 1.Palazzo L, Mikoč A, Ahel I. ADP-ribosylation: new facets of an ancient modification. FEBS J. 2017;284(18):2932–2946. doi: 10.1111/febs.14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaughan M, Moss J. Mono (ADP-ribosyl)transferases and their effects on cellular metabolism. Curr Top Cell Regul. 1981;20:205–246. doi: 10.1016/B978-0-12-152820-1.50010-9. [DOI] [PubMed] [Google Scholar]
- 3.Cohen MS, Chang P. Insights into the biogenesis, function, and regulation of ADP-ribosylation. Nat Chem Biol. 2018;14(3):236–243. doi: 10.1038/nchembio.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crawford K, Bonfiglio JJ, Mikoč A, Matic I, Ahel I. Specificity of reversible ADP-ribosylation and regulation of cellular processes. Crit Rev Biochem Mol Biol. 2018;53(1):64–82. doi: 10.1080/10409238.2017.1394265. [DOI] [PubMed] [Google Scholar]
- 5.Gupte R, Liu Z, Kraus WL. PARPs and ADP-ribosylation: recent advances linking molecular functions to biological outcomes. Genes Dev. 2017;31(2):101–126. doi: 10.1101/gad.291518.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrabi SA, et al. Iduna protects the brain from glutamate excitotoxicity and stroke by interfering with poly(ADP-ribose) polymer-induced cell death. Nat Med. 2011;17(6):692–699. doi: 10.1038/nm.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee Y, et al. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat Neurosci. 2013;16(10):1392–1400. doi: 10.1038/nn.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Niimura M, et al. Prevention of apoptosis-inducing factor translocation is a possible mechanism for protective effects of hepatocyte growth factor against neuronal cell death in the hippocampus after transient forebrain ischemia. J Cereb Blood Flow Metab. 2006;26(11):1354–1365. doi: 10.1038/sj.jcbfm.9600287. [DOI] [PubMed] [Google Scholar]
- 9.Yu SW, Wang H, Dawson TM, Dawson VL. Poly(ADP-ribose) polymerase-1 and apoptosis inducing factor in neurotoxicity. Neurobiol Dis. 2003;14(3):303–317. doi: 10.1016/j.nbd.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Bonicalzi ME, Haince JF, Droit A, Poirier GG. Regulation of poly(ADP-ribose) metabolism by poly(ADP-ribose) glycohydrolase: where and when? Cell Mol Life Sci. 2005;62(7-8):739–750. doi: 10.1007/s00018-004-4505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hatakeyama K, Nemoto Y, Ueda K, Hayaishi O. Purification and characterization of poly(ADP-ribose) glycohydrolase. Different modes of action on large and small poly(ADP-ribose) J Biol Chem. 1986;261(32):14902–14911. [PubMed] [Google Scholar]
- 12.Barkauskaite E, et al. Visualization of poly(ADP-ribose) bound to PARG reveals inherent balance between exo- and endo-glycohydrolase activities. Nat Commun. 2013;4:2164. doi: 10.1038/ncomms3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Braun SA, Panzeter PL, Collinge MA, Althaus FR. Endoglycosidic cleavage of branched polymers by poly(ADP-ribose) glycohydrolase. Eur J Biochem. 1994;220(2):369–375. doi: 10.1111/j.1432-1033.1994.tb18633.x. [DOI] [PubMed] [Google Scholar]
- 14.Mashimo M, Kato J, Moss J. ADP-ribosyl-acceptor hydrolase 3 regulates poly (ADP-ribose) degradation and cell death during oxidative stress. Proc Natl Acad Sci USA. 2013;110(47):18964–18969. doi: 10.1073/pnas.1312783110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Y, et al. Poly(ADP-ribose) (PAR) binding to apoptosis-inducing factor is critical for PAR polymerase-1-dependent cell death (parthanatos) Sci Signal. 2011;4(167):ra20. doi: 10.1126/scisignal.2000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu SW, et al. Apoptosis-inducing factor mediates poly(ADP-ribose) (PAR) polymer-induced cell death. Proc Natl Acad Sci USA. 2006;103(48):18314–18319. doi: 10.1073/pnas.0606528103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu SW, et al. Mediation of poly(ADP-ribose) polymerase-1-dependent cell death by apoptosis-inducing factor. Science. 2002;297(5579):259–263. doi: 10.1126/science.1072221. [DOI] [PubMed] [Google Scholar]
- 18.Glowacki G, et al. The family of toxin-related ecto-ADP-ribosyltransferases in humans and the mouse. Protein Sci. 2002;11(7):1657–1670. doi: 10.1110/ps.0200602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss J, et al. Molecular and immunological characterization of ADP-ribosylarginine hydrolases. J Biol Chem. 1992;267(15):10481–10488. [PubMed] [Google Scholar]
- 20.Oka S, Kato J, Moss J. Identification and characterization of a mammalian 39-kDa poly(ADP-ribose) glycohydrolase. J Biol Chem. 2006;281(2):705–713. doi: 10.1074/jbc.M510290200. [DOI] [PubMed] [Google Scholar]
- 21.Kasamatsu A, et al. Hydrolysis of O-acetyl-ADP-ribose isomers by ADP-ribosylhydrolase 3. J Biol Chem. 2011;286(24):21110–21117. doi: 10.1074/jbc.M111.237636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mashimo M, Kato J, Moss J. Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair (Amst) 2014;23:88–94. doi: 10.1016/j.dnarep.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mueller-Dieckmann C, et al. The structure of human ADP-ribosylhydrolase 3 (ARH3) provides insights into the reversibility of protein ADP-ribosylation. Proc Natl Acad Sci USA. 2006;103(41):15026–15031. doi: 10.1073/pnas.0606762103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono T, Kasamatsu A, Oka S, Moss J. The 39-kDa poly(ADP-ribose) glycohydrolase ARH3 hydrolyzes O-acetyl-ADP-ribose, a product of the Sir2 family of acetyl-histone deacetylases. Proc Natl Acad Sci USA. 2006;103(45):16687–16691. doi: 10.1073/pnas.0607911103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abplanalp J, et al. Proteomic analyses identify ARH3 as a serine mono-ADP-ribosylhydrolase. Nat Commun. 2017;8(1):2055. doi: 10.1038/s41467-017-02253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fontana P, Bonfiglio JJ, Palazzo L, Bartlett E, Matic I, Ahel I. Serine ADP-ribosylation reversal by the hydrolase ARH3. Elife. 2017;6:e28533. doi: 10.7554/eLife.28533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh SG, et al. Biallelic mutations in ADPRHL2, encoding ADP-ribosylhydrolase 3, lead to a degenerative pediatric stress-induced epileptic ataxia syndrome. Am J Hum Genet. 2018;103(3):431–439. doi: 10.1016/j.ajhg.2018.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jagtap P, et al. Novel phenanthridinone inhibitors of poly (adenosine 5’-diphosphate-ribose) synthetase: potent cytoprotective and antishock agents. Crit Care Med. 2002;30(5):1071–1082. doi: 10.1097/00003246-200205000-00019. [DOI] [PubMed] [Google Scholar]
- 29.Donawho CK, et al. ABT-888, an orally active poly(ADP-ribose) polymerase inhibitor that potentiates DNA-damaging agents in preclinical tumor models. Clin Cancer Res. 2007;13(9):2728–2737. doi: 10.1158/1078-0432.CCR-06-3039. [DOI] [PubMed] [Google Scholar]
- 30.Hong SJ, Dawson TM, Dawson VL. Nuclear and mitochondrial conversations in cell death: PARP-1 and AIF signaling. Trends Pharmacol Sci. 2004;25(5):259–264. doi: 10.1016/j.tips.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 31.Sharifi R, et al. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J. 2013;32(9):1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Niere M, et al. ADP-ribosylhydrolase 3 (ARH3), not poly(ADP-ribose) glycohydrolase (PARG) isoforms, is responsible for degradation of mitochondrial matrix-associated poly(ADP-ribose) J Biol Chem. 2012;287(20):16088–16102. doi: 10.1074/jbc.M112.349183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palazzo L, Leidecker O, Prokhorova E, Dauben H, Matic I, Ahel I. Serine is the major residue for ADP-ribosylation upon DNA damage. Elife. 2018;7:e34334. doi: 10.7554/eLife.34334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Formentini L, et al. Mono-galloyl glucose derivatives are potent poly(ADP-ribose) glycohydrolase (PARG) inhibitors and partially reduce PARP-1-dependent cell death. Br J Pharmacol. 2008;155(8):1235–1249. doi: 10.1038/bjp.2008.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rapizzi E, Fossati S, Moroni F, Chiarugi A. Inhibition of poly(ADP-ribose) glycohydrolase by gallotannin selectively up-regulates expression of proinflammatory genes. Mol Pharmacol. 2004;66(4):890–898. doi: 10.1124/mol.104.000968. [DOI] [PubMed] [Google Scholar]
- 36.Ying W, Sevigny MB, Chen Y, Swanson RA. Poly(ADP-ribose) glycohydrolase mediates oxidative and excitotoxic neuronal death. Proc Natl Acad Sci USA. 2001;98(21):12227–12232. doi: 10.1073/pnas.211202598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uchiumi F, Watanabe T, Ohta R, Abe H, Tanuma S. PARP1 gene expression is downregulated by knockdown of PARG gene. Oncol Rep. 2013;29(5):1683–1688. doi: 10.3892/or.2013.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martire S, Mosca L, d’Erme M. PARP-1 involvement in neurodegeneration: a focus on Alzheimer’s and Parkinson’s diseases. Mech Ageing Dev. 2015;146-148:53–64. doi: 10.1016/j.mad.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Lee Y, Kang HC, Lee BD, Lee YI, Kim YP, Shin JH. Poly (ADP-ribose) in the pathogenesis of Parkinson’s disease. BMB Rep. 2014;47(8):424–432. doi: 10.5483/BMBRep.2014.47.8.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang X, Cheng J, Gao Y, Ding J, Ni X. Downregulation of Iduna is associated with AIF nuclear translocation in neonatal brain after hypoxia-ischemia. Neuroscience. 2017;346:74–80. doi: 10.1016/j.neuroscience.2017.01.010. [DOI] [PubMed] [Google Scholar]
- 41.Haince JF, Ouellet ME, McDonald D, Hendzel MJ, Poirier GG. Dynamic relocation of poly(ADP-ribose) glycohydrolase isoforms during radiation-induced DNA damage. Biochim Biophys Acta. 2006;1763(2):226–237. doi: 10.1016/j.bbamcr.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Hanai S, et al. Loss of poly(ADP-ribose) glycohydrolase causes progressive neurodegeneration in Drosophila melanogaster. Proc Natl Acad Sci USA. 2004;101(1):82–86. doi: 10.1073/pnas.2237114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schuhwerk H, et al. Kinetics of poly(ADP-ribosyl)ation, but not PARP1 itself, determines the cell fate in response to DNA damage in vitro and in vivo. Nucleic Acids Res. 2017;45(19):11174–11192. doi: 10.1093/nar/gkx717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teng F, et al. Neuroprotective Effects of poly(ADP-ribose)polymerase inhibitor olaparib in transient cerebral ischemia. Neurochem Res. 2016;41(7):1516–1526. doi: 10.1007/s11064-016-1864-6. [DOI] [PubMed] [Google Scholar]
- 45.Ohmoto A, Yachida S. Current status of poly(ADP-ribose) polymerase inhibitors and future directions. Onco Targets Ther. 2017;10:5195–5208. doi: 10.2147/OTT.S139336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner LM. Profile of veliparib and its potential in the treatment of solid tumors. Onco Targets Ther. 2015;8:1931–1939. doi: 10.2147/OTT.S69935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pahuja S, et al. A phase I study of veliparib (ABT-888) in combination with weekly carboplatin and paclitaxel in advanced solid malignancies and enriched for triple-negative breast cancer (TNBC) J Clin Oncol. 2015;33(15):1015 [Google Scholar]
- 48.Nicolescu AC, Holt A, Kandasamy AD, Pacher P, Schulz R. Inhibition of matrix metalloproteinase-2 by PARP inhibitors. Biochem Biophys Res Commun. 2009;387(4):646–650. doi: 10.1016/j.bbrc.2009.07.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ishiwata-Endo H, et al. Role of a TRIM72 ADP-ribosylation cycle in myocardial injury and membrane repair. JCI Insight. 2018;3(22):e97898. doi: 10.1172/jci.insight.97898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jiang SX, et al. Chlortetracycline and demeclocycline inhibit calpains and protect mouse neurons against glutamate toxicity and cerebral ischemia. J Biol Chem. 2005;280(40):33811–33818. doi: 10.1074/jbc.M503113200. [DOI] [PubMed] [Google Scholar]
- 51.Kato J, Zhu J, Liu C, Moss J. Enhanced sensitivity to cholera toxin in ADP-ribosylarginine hydrolase-deficient mice. Mol Cell Biol. 2007;27(15):5534–5543. doi: 10.1128/MCB.00302-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gahl WA, Tifft CJ. The NIH Undiagnosed Diseases Program: lessons learned. JAMA. 2011;305(18):1904–1905. doi: 10.1001/jama.2011.613. [DOI] [PubMed] [Google Scholar]
- 53.Gahl WA, Boerkoel CF, Boehm M. The NIH Undiagnosed Diseases Program: bonding scientists and clinicians. Dis Model Mech. 2012;5(1):3–5. doi: 10.1242/dmm.009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gahl WA, et al. The NIH Undiagnosed Diseases Program and Network: applications to modern medicine. Mol Genet Metab. 2016;117(4):393–400. doi: 10.1016/j.ymgme.2016.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.