Abstract
Immunotherapy has emerged as a promising approach to treat cancer. However, partial responses across multiple clinical trials support the significance of characterizing intertumor and intratumor heterogeneity to achieve better clinical results and as potential tools in selecting patients for different types of cancer immunotherapies. Yet, the type of heterogeneity that informs clinical outcome and patient selection has not been fully explored. In particular, the lack of characterization of immune response–related genes in cancer cells hinders the further development of metrics to select and optimize immunotherapy. Therefore, we analyzed single-cell RNA-Seq data from lung adenocarcinoma patients and cell lines to characterize the intratumor heterogeneity of immune response–related genes and demonstrated their potential impact on the efficacy of immunotherapy. We discovered that IFN-γ signaling pathway genes are heterogeneously expressed and coregulated with other genes in single cancer cells, including MHC class II (MHCII) genes. The downregulation of genes in IFN-γ signaling pathways in cell lines corresponds to an acquired resistance phenotype. Moreover, analysis of 2 groups of tumor-restricted antigens, namely neoantigens and cancer testis antigens, revealed heterogeneity in their expression in single cells. These analyses provide a rationale for applying multiantigen combinatorial therapies to prevent tumor escape and establish a basis for future development of prognostic metrics based on intratumor heterogeneity.
Keywords: Immunology, Oncology
Keywords: Bioinformatics, Immunotherapy
Lung adenocarcinoma exhibits heterogeneity in neoantigen and cancer testis antigen expression and downregulation of MHCII genes and IFN-γ signaling genes correlate with resistance.
Introduction
The recent decade has witnessed the advent and significant advancement of immunotherapy as an effective anticancer strategy. Its demonstrated efficacy against multiple cancer types has attracted more attention to predict the outcomes of various immunotherapies alone or in combination with other therapeutic modalities. One of the most promising approaches for activation of immune responses is immune checkpoint blockade therapy, such as using anti-CTLA4 and anti–PD-1 to block inhibitory molecules on immune cells to unleash antitumor immunity. Moreover, recent studies have begun shedding light on the role of IFN-γ pathway genes on immune checkpoint blockade therapy, demonstrating the effective antitumor immune response induced by IFN-γ when it recognizes its cognate receptor on cancer cells or antigen-presenting cells (1, 2). Another immunotherapy approach gaining more interest is targeting neoantigens or cancer testis antigens (CTAs), whose expressions are cancer cell specific. Both therapeutic vaccines as well as T cell receptor–redirected adoptive cell transfer therapy have been demonstrated to boost T cell responses against tumors expressing these antigenic targets. Several such treatments have entered clinical trials (3–5). However, many of these monotherapies are effective in only a fraction of patients. Although studies have illustrated the intertumor heterogeneity (between patients) of immune signatures (6, 7), as well as intratumor heterogeneity (within the tumor) of immune cells, in multiple cancer types (8–10), it remains elusive how the underlying intratumor heterogeneity of immune response–related gene expression in tumor cells will affect responses and ability to predict outcome. In the current study, we demonstrate that single-cell RNA-Seq (scRNA-Seq) of tumor cells can be used to identify such intratumor heterogeneity.
Lung cancer is one of the most highly mutated cancer types (11), and despite the improved success of immunotherapies in lung cancer, a low response rate (≤20%) is still observed (12). Herein, we used previously published scRNA-Seq data from lung adenocarcinoma (LUAD) patient-derived xenografts (PDXs) and 2 LUAD cell lines, LC2/ad-R (vandetanib resistant) and LC2/ad (vandetanib susceptible), as a test set to demonstrate the functional intratumor heterogeneity of immune response–related genes that might affect immunotherapy responses (13–15). We found that MHCII genes are heterogeneously expressed among tumor cells obtained from LUAD patients and that their expression correlates with a favorable prognosis. Interestingly, MHCII genes are heterogeneously expressed within single cells from individual patients. MHCII genes can be induced by IFN-γ (16). We then sought to identify the intratumor heterogeneity of the IFN-γ signaling pathway and observed coexpression of IFN-γ signaling pathway genes in a fraction of LUAD single cells that had a higher level of MHCII expression. Similar results were found to be enriched in the LC2/ad cell line. Further analysis showed that the opposite trend, where uncoordinated expression of IFN-γ signaling pathway genes was associated with a lower level of MHCII expression, was enriched in the LC2/ad-R cell line that acquired a vandetanib resistance phenotype. This relationship between IFN-γ signaling pathway genes and MHCII genes could also be important in determining resistance to immunotherapy in LUAD. We also uncovered heterogeneity in the expression of predicted cancer neoantigens and CTAs in single cells from both LUAD patients and cell lines. Interestingly, the decrease in the number of neoantigens was also correlated with the acquired resistance phenotype. Our study suggests that using a combinatorial strategy to target multiple tumor antigens in select patients could improve immunotherapy efficacy.
Results
Prognostic prediction of LUAD by the expression pattern of cell cycle and MHCII genes.
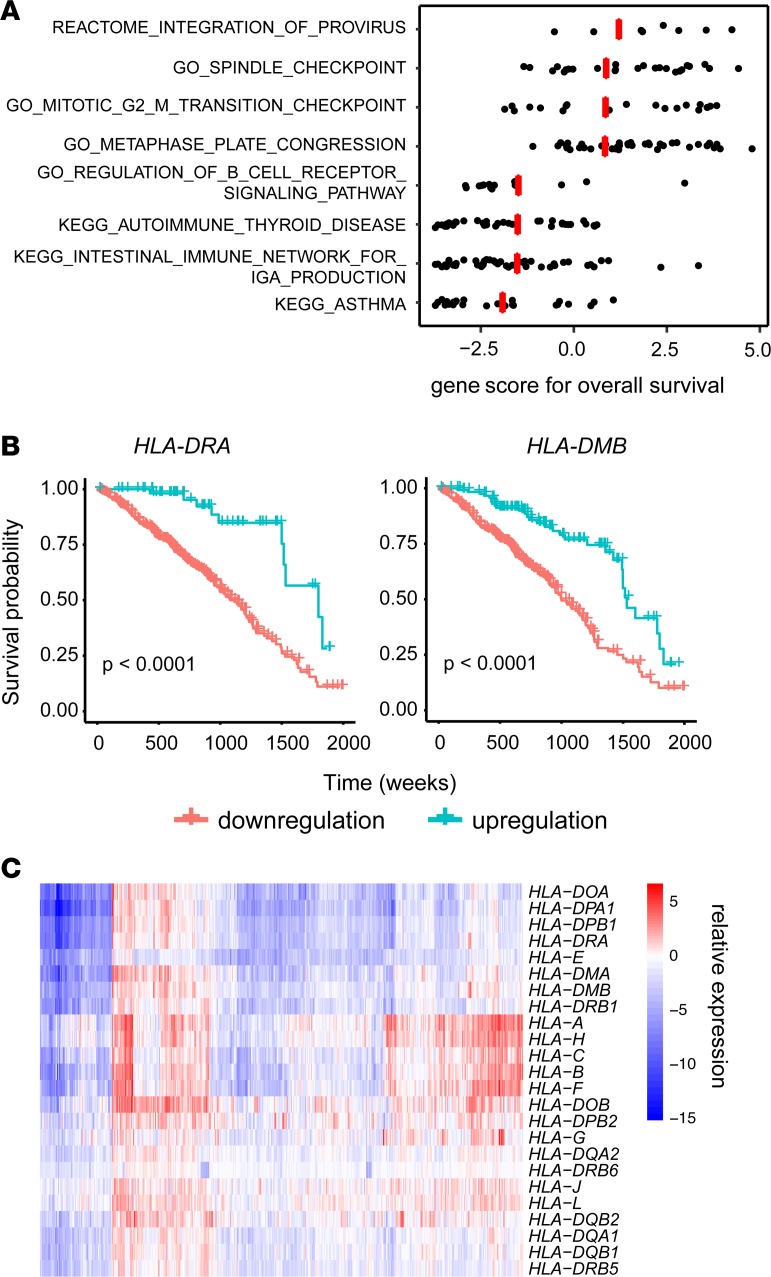
Identifying patients at higher risk of tumor progression or recurrence is crucial for making individualized treatment plans. Despite recently recognized intratumor heterogeneity, there is a lack of understanding of how this is associated with prognosis. In this study, we aimed to characterize the heterogeneity of prognostic predictors in single cancer cells. We first identified pathways that are potential prognostic predictors in LUAD cohorts from The Cancer Genome Atlas Research Network (TCGA) (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.121387DS1). We found that the top pathways associated with an unfavorable prognosis were enriched for cell cycle–related pathways, while the top pathways associated with a favorable prognosis were enriched for immune cell signaling–related pathways (Figure 1A and Supplemental Table 1). Interestingly, the common genes shared by the top favorable prognostic pathways were MHCII genes. Further survival analysis validated the association of upregulated MHCII genes with a better overall survival rate (Figure 1B and Supplemental Table 2). Surprisingly, MHCI genes did not have a significant association with overall survival in LUAD (Supplemental Table 2). Compared with the expression of MHC genes in normal tissues, MHCII genes were more downregulated compared with those of MHCI (Figure 1C). Previously, it has been shown that higher MHCII expression was also associated with better prognosis in multiple other tumor types, such as melanoma and triple-negative breast cancer (17, 18). Especially in melanoma patients, the expression of MHCII can predict response to anti–PD-1/anti–PD-L1 therapy (18).
Figure 1. Cell cycle genes and MHCII genes are potential prognostic predictors of LUAD.
(A) Gene set analysis of TCGA LUAD data to determine the significance of curated canonical pathways with respect to overall patient survival. Each dot represents the individual gene score within the corresponding pathway, and each red line is the score for the gene set calculated from R package GSA. (B) Kaplan-Meier plot showing the 5-year overall survival with respect to HLA-DRA and HLA-DMB expressions for patients in TCGA LUAD cohorts. Log-rank test was performed to determine significance. (C) Heatmap of the relative expression of MHC genes in tumor tissues compared with matched normal tissues for TCGA LUAD data.
Single cancer cells express distinct prognosis-associated gene modules.
Next, we assessed the expression level of prognosis-associated genes, discovered from analyzing bulk cancer sample RNA-Seq data, in LUAD patients at the single-cell level (Supplemental Table 2). We reanalyzed previously reported scRNA-Seq data from LUAD PDXs (14, 15). The LC-PT-45 tumor was taken from a treatment-naive, 60-year-old, male patient with an irregular primary lung lesion, whereas the LC-MBT-15 tumor was taken from a 57-year-old female with a metachronous brain metastasis after standard chemotherapy and erlotinib treatment. A total of 77 and 49 single cells were sequenced for LC-PT-45 and LC-MBT-15, respectively. An average of 5 million reads were sequenced for each single cell, which is needed to saturate the mutation detection (Supplemental Figure 1A). Additional quality control was performed to remove low-quality single cells, which account for a small percentage, based on sequencing metrics, including genome mapping percentage, multimapped read percentage, mitochondrial DNA mapping percentage, cell-to-mean correlation, and transcriptome variance (Supplemental Figure 1B), following a published method (19). High-quality single cells were used in following analyses.
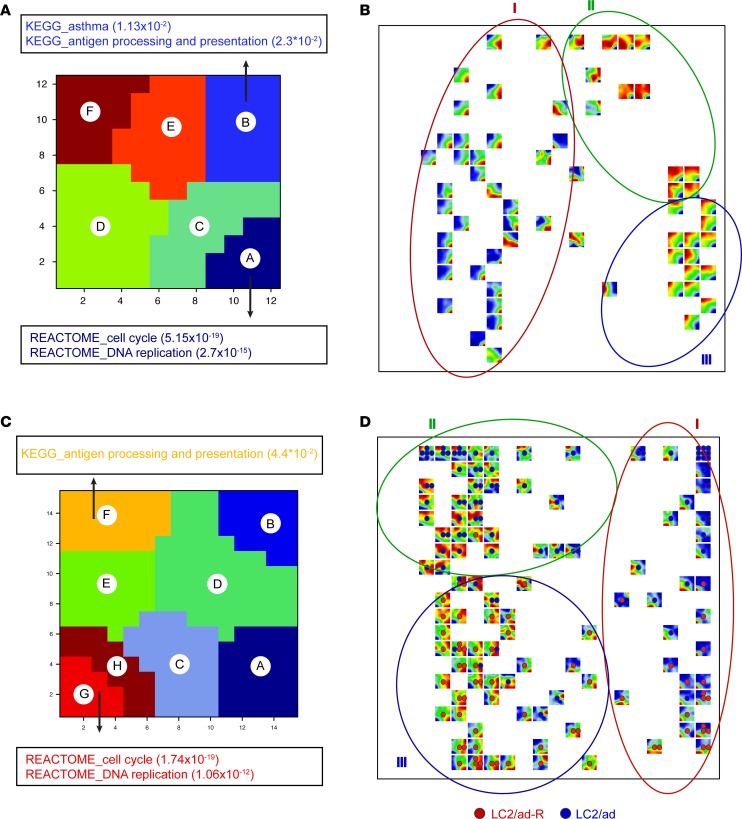
Min et al. previously reported the distinct subpopulations of LUAD single cells with respect to the expression of cell cycle genes (15). Here, we further sought to determine whether prognosis-associated genes and pathways, including cell cycle genes and antigen-presenting pathway genes, are heterogeneously expressed in single LUAD cells and whether there are any expression patterns among these genes. We applied a self-organizing map (SOM), which adopts an unsupervised machine learning approach to map genes into coordinately expressed groups (metagenes) (20). A second-level SOM was then processed by mapping all samples together into a 2-dimensional mosaic pattern based on metagene expression. K-means clustering grouped metagenes into 6 clusters based on coexpression in scRNA-Seq from the LC-PT-45 patient (Figure 2A). Gene set enrichment analysis of genes in these 6 clusters showed that cluster A was enriched in cell cycle genes, while cluster B was enriched for antigen presentation pathways, specifically MHCII presentation (Figure 2A and Supplemental Table 3). We then observed that cells were separated into 3 groups based on their distinct metagene expression patterns: (noted as I) cell cycle pathway high, (noted as II) antigen presentation pathway high, and (noted as III) both pathways low (Figure 2B). We also analyzed the PDX tumor cells from the other LUAD patient (LC-MBT-15). As previously reported, LC-MBT-15 exhibits a less heterogeneous transcriptional profile (14). Nevertheless, SOM analysis revealed that metagenes representing cell cycle and MHCII genes were mapped into different metagene modules (Supplemental Figure 2 and Supplemental Table 3).
Figure 2. Expression of genes in the cell cycle pathway and antigen presentation pathway among single cancer cells are coregulated.
(A and C) Gene pathway clusters from metagene analysis of single cells from PDX LC-PT-45 or from LC2/ad-R and LC2/ad cell lines. K-means clustering of metagenes on SOMs into 6 clusters and 8 clusters, respectively. A hypergeometric test was performed on each cluster of metagenes to determine enrichment of canonical pathways. (A) For PDX LC-PT-45, cluster A was enriched for cell cycle pathways, while cluster B was enriched for antigen presentation pathways. (C) For LC2/ad-R and LC2/ad cell lines, cluster G/H was enriched for cell cycle pathways, while cluster F was enriched for antigen presentation pathways. P values in the parentheses are FDR corrected. (B and D) Second-level clustering of SOMs for (B) 77 single cells from PDX LC-PT-45 or (D) 159 single cells from LC2/ad-R and LC2/ad cell lines. Each square is a unique SOM pattern with the heatmap indicating the gene expression level of metagenes. Cells with the same SOM are collapsed with only 1 representative SOM. SOMs are arranged by mapping all cells together into a 2-dimensional mosaic pattern based on metagene expression. Coexpression of cell cycle pathway and antigen presentation pathway further separates cells into 3 major groups: cell cycle pathway high (I), antigen presentation pathway high (II), and both pathways low (III).
We further applied the above analysis to scRNA-Seq data from 2 human LUAD cell lines, LC2/ad-R and LC2/ad. LC2/ad-R is a subclone of LC2/ad that has acquired resistance to vandetanib (13). Similar to LUAD patient samples, metagenes representing cell cycle and MHCII genes were also mapped into different metagene modules in the 2 cell lines’ data (Figure 2C and Supplemental Table 3). Additionally, single cells from the 2 cell lines were also separated into 3 groups: (noted as I) cell cycle pathway high, (noted as II) antigen presentation pathway high, and (noted as III) both pathways low (Figure 2D). We noticed that more single cells from the LC2/ad cell line belonged to group II (51 out of 89) compared with single cells from the LC2/ad-R cell line (1 out of 70) (P < 10–5, Fisher’s exact test), which suggests that a significant downregulation of antigen presentation pathway in the LC2/ad-R cell line could render this cell line resistant to immunotherapy in addition to vandetanib.
Heterogeneity of IFN-γ–stimulated genes in single cells.
Because it is known that MHCII genes, a class of genes in the antigen presentation pathway, can be regulated through the IFN-γ signaling pathway (16) and we showed that antigen presentation pathway genes are heterogeneously expressed by LUAD cells, we next examined the expression pattern of IFN-γ signaling pathway genes. Recent works also began to uncover the role of the IFN-γ signaling pathway on cancer immune checkpoint blockade therapies. Gao et al. identified the genomic variations, such as copy number variations and single nucleotide polymorphisms, of IFN-γ pathway genes in melanoma patients as a resistance mechanism to anti-CTLA4 therapy (1). By contrast, Benci et al. suggested an IFN-driven, PD-L1-independent resistance to immune checkpoint blockade (21). These different conclusions on the roles that the IFN-γ signaling pathway plays in cancer resistance to immunotherapy could be due to distinct clinical trial cohorts as well as the underlying intratumor heterogeneity of IFN-γ signaling pathway genes. However, it is not known whether this cancer-immunity interaction network exhibits differential gene expression at the single-cell level, which could be an important prognostic factor and shed light on the mechanism of cancer immune resistance.
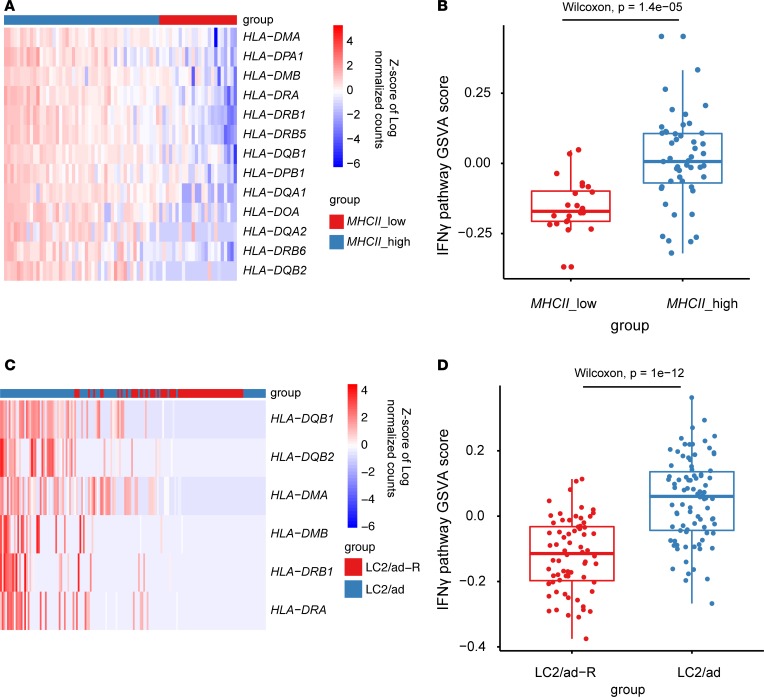
Here, we used the LUAD PDXs and cell line scRNA-Seq data sets to examine the intratumor heterogeneity of the IFN-γ signaling pathway. IFN-γ signaling pathway genes were curated from the Gene Ontology (GO) database and Reactome pathway database. Single cells from LUAD patients were first clustered based on expression of MHCII genes into 2 groups: MHCIIlo and MHCIIhi (Figure 3A and Supplemental Figure 3A). The Gene Set Variation Analysis (GSVA) score of the IFN-γ signaling pathway, which indicates the extent of coordinated expression among pathway genes, was then calculated for each individual cell. We found a significant decrease of IFN-γ signaling pathway GSVA scores in the MHCIIlo group (Figure 3B and Supplemental Figure 3B). We also investigated expression of IFN-γ signaling pathway genes in LC2/ad-R and LC2/ad cell lines. GSVA showed that significantly more single cells in the LC2/ad-R cell line had downregulated IFN-γ signaling pathway genes compared with those in the LC2/ad cell line (Figure 3, C and D, and Supplemental Figure 4).
Figure 3. Coexpression of IFN-γ signaling pathway genes in LC-PT-45 and cell lines.
(A) Heatmap of MHCII gene expression levels for single cells from PDX LC-PT-45. K-means clustering was performed to group all single cells into 2 clusters, MHCIIlo and MHCIIhi. (C) Heatmap of MHCII gene expression levels for single cells in LC2/ad-R and LC2/ad cell lines. LC2/ad-R or LC2/ad were used to label cells. (B and D) Comparison of GSVA scores of IFN-γ signaling pathway genes, calculated for each individual cell analyzed, between (B) MHCIIlo and MHCIIhi groups and (D) LC2/ad-R and LC2/ad groups. Nonparametric Wilcoxon’s test was performed between different groups. The box plots depict the minimum and maximum values (whiskers), the upper and lower quartiles, and the median. The length of the box represents the interquartile range.
The different pattern we observed in the coordinated expression of IFN-γ signaling pathway genes is consistent with their role in directing antiproliferative and proapoptotic effects in tumor cells (2). However, their roles in activating MHCII expression, enhancing tumor antigen presentation, and inducing the recruitment of other immune cells suggest that the lack of coordinated expression of IFN-γ signaling pathway genes in a subset of cancer cells within a tumor would render these cells resistant to immunotherapy in addition to small-molecule inhibitors (22). Further studies are necessary to demonstrate how the heterogeneity of IFN-γ signaling pathway expression influences the efficacy of immunotherapy.
Heterogeneous expression of predicted cancer neoantigens in single cells.
Neoantigens are a group of promising targets to induce antitumor immunity through recognition of neoantigen-specific T cells and are advantageous in their selective expression in cancer cells without the risk of causing autoimmunity. It has been proposed that targeting multiple neoantigens simultaneously will likely be important to prevent tumor escape by editing of the mutated epitope (5, 11). Here we aimed to analyze whether neoantigens are heterogeneously expressed. If so, then this would warrant further investigation of a new strategy in cancer immunotherapy where a combination of neoantigens could be administered as therapeutic vaccines or a combination of neoantigen-specific T cell receptors could be used to modify patients’ own T cells in adoptive cell transfer therapy. Consequently, the analysis of the heterogeneity of neoantigen expression in single cancer cells would facilitate the development of these therapeutic regimens.
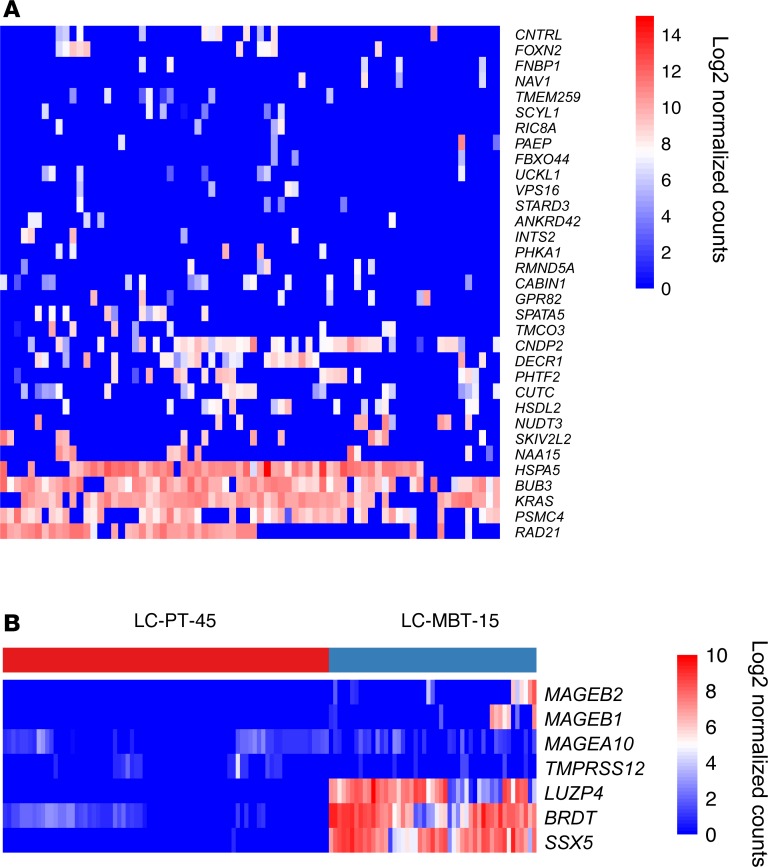
To answer this question, somatic mutations in each single cell were further assessed for neoantigen prediction (see Methods). Only neoantigens detected in more than 3 cells were selected. We found more than half of the neoantigens exhibited a bimodal expression (Figure 4A and Supplemental Figure 5), suggesting the possibility of tumor escape with single neoantigen epitope–based therapy. Surprisingly, we not only found the same bimodal expression of neoantigens in LC2/ad-R and LC2/ad cell lines but also revealed that LC2/ad-R cell lines had a significant decrease of neoantigen load compared with the nonresistant parental cell line (Supplemental Figure 6, A and B). This suggested the degree of neoantigen load might affect cancer cell drug responses. In addition to detecting neoantigens, we identified both wild-type alleles and variant-containing alleles (single nucleotide variants [SNVs] and insertions/deletions) for many genes in many single cells, indicating that cells without neoantigen identified did not mainly result from dropout of corresponding genes (Supplemental Figure 7).
Figure 4. Heterogeneous expression of neoantigens and CTAs in single cancer cells.
(A) Heatmap showing the expression of neoantigens in single cells from LC-PT-45. Log2 normalized counts of 0 represent either no expression or no somatic mutation was detected. Cells were included if a neoantigen was detected regardless of its corresponding wild-type antigen detection status. Only neoantigens detected in more than 3 cells were selected. (B) Heatmap showing the expression of CTAs in single cells from LC-PT-45 and LC-MBT-15. CTAs whose expressions were transcriptionally silent in normal nongermline tissues based on GTEx data and with normalized counts greater than 0 in more than 2 cells were selected.
Heterogeneous expression of CTAs in single cells.
CTAs are a group of tumor antigens with normal expression restricted to male germ cells in the testis. In cancer, alteration of gene regulation results in aberrant expression of CTAs in various tumor types (23). Previous studies have demonstrated the extensive heterogeneity of CTAs among patients of different cancer types (6, 7). However, little is known about the heterogeneity of all possible CTAs expressed within each patient. Thus, we examined the expression of all possible CTAs in LUAD single cells and demonstrated the extensive heterogeneity of their expression at the single-cell level. To restrict our analysis to only CTAs expressed in tumors, we selected a subset of 276 known CTAs that are transcriptionally silent in normal nongermline tissues based on Genotype-Tissue Expression (GTEx) data (7). Only CTAs with normalized counts greater than 0 in more than 2 cells were selected. In both LUAD patients, we observed both substantial intertumor heterogeneity and intratumor heterogeneity of expressed CTAs (Figure 4B). In addition, compared with neoantigens, CTAs exhibited a lower expression level (Figure 4 and Supplemental Figure 5). Further analysis of CTAs in cell lines revealed that LC2/ad-R and LC2/ad cell lines can also be separated based on CTA expression. Expression of melanoma-associated antigen-A6 (MAGE-A6) and MAGE-A2 was significantly higher in LC2/ad-R compared with LC2/ad (Supplemental Figure 6C). These results suggest that CTAs could be therapeutic targets for cancers that are resistant to small-molecule inhibitor therapy.
Discussion
In this study, we applied both LUAD and cell line scRNA-Seq to characterize the intratumor heterogeneity of immune response–related genes. We identified that (a) prognosis-related genes, especially cell cycle pathway and antigen presentation pathway genes, were independently coexpressed among single cells; (b) IFN-γ signaling pathways were heterogeneously expressed within cancer cells and downregulation was correlated to a drug-resistant phenotype; and (c) promising cancer vaccination targets, such as neoantigens and CTAs, were also heterogeneously expressed.
Previous studies showed that IFN-γ induced the expression of MHCII genes in multiple myeloma cells (24) and normal epidermal melanocytes (25). Because MHCII expression on cancer cells is important for CD4+ T cell immunity as well as T cell exhaustion (26), our findings on the heterogeneity of expression in cancer cells, especially its lost coordinated expression with IFN-γ signaling pathway genes in the LC2/ad-R cell line, could have important implications for cancer immunotherapy. Although it is yet to be determined whether cell cycle pathways and MHCII genes in tumor cells are mechanistically associated, recent findings that CDK4/6 inhibitors not only induce cell cycle arrest, but also promote antitumor immunity through activation of IFN signaling and suppression of Tregs, indicates a connection between these 2 gene modules (27). Besides, a recent study investigating resistance to immune checkpoint blockade in melanoma revealed a cancer resistance program, which is enriched for upregulation of E2F targets and downregulation of antigen presentation, is associated with T cell exclusion (28). Although it is known that IFN-γ can induce MHCII expression (16), the analysis of immune response–related genes in single cancer cells revealed that genes of the IFN-γ signaling pathway are coexpressed, including many IFN-γ–stimulated genes in addition to MHCII. One of the coexpressed IFN-γ–stimulated genes, IDO1, is responsible for the conversion of tryptophan and other indole derivatives to kynurenine and has been widely studied as an important suppressor of antitumor immunity. Several IDO inhibitors have entered clinical trials as monotherapies and in combination with CTLA-4 and PD-1 immune checkpoint blockade therapy (29). Our analysis also indicated that heterogeneity in IFN-γ signaling pathways might affect the responses of IFN-γ pathway–directed therapies.
Another aspect of intratumor heterogeneity unveiled by our analysis was in cancer vaccine targets, including neoantigens and CTAs. Many completed clinical trials have failed to observe significant responses to CTA vaccines. For example, clinical trials in non–small cell lung carcinoma identified no significant difference in MAGE-A3 compared with control groups (30). Yet another recent clinical trial study in melanoma patients reported 4 out of 6 patients had no recurrence after neoantigen vaccination (3). Our study further demonstrates the individuality and intratumor heterogeneity of neoantigens and CTAs. The analysis of heterogeneous expression of neoantigens and CTAs together highlights the challenge of cancer vaccine monoepitope therapy to elicit effective antitumor immunity and supports the possible advantages of targeting multiple epitopes in neoantigen vaccine– or neoantigen-specific T cell–based immunotherapy.
In summary, we demonstrated that the intratumor heterogeneity of immune module expression will help develop better prognoses of immunotherapies. Examining the gene expression in single cancer cells not only provides a rationale for combinatorial immunotherapies, in particular neoantigen/CTA-directed therapies, but also paves the road for future analyses on how intratumor heterogeneity affects immunotherapy efficacy.
Methods
Single-cell RNA-Seq data set.
Raw reads of scRNA-Seq and whole-exome sequencing were all downloaded from the National Center for Biotechnology Information Gene Expression Omnibus DataSets website under accession number GSE69405 (https://www.ncbi.nlm.nih.gov/geo). Two PDX samples were processed for scRNA-Seq, LC-PT-45 and LC-MBT-15 (14). Raw reads of scRNA-Seq data from LC2/ad-R and LC2/ad cell lines were downloaded from the DNA Data Bank of Japan under accession number DRA001287. Raw reads of scRNA-Seq were first mapped to human genome reference GRCh37 with RSEM (31) and then normalized using SCnorm (32). Genes with read counts fewer than 2 in more than 10% of all single cells were filtered out.
TCGA data set.
Quantile normalized read counts of RNA-Seq from LUAD patients and corresponding clinical data were downloaded from the Broad Institute TCGA Genome Data Analysis Center Firehose website (http://gdac.broadinstitute.org/). Aggregated somatic mutation data processed by MuTect2 was downloaded from National Cancer Institute Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/).
Prognostic gene and pathway analysis of LUAD.
Each gene in the TCGA data set was classified into “high expression” and “low expression” by comparison with its mean expression. Then “survival,” an R package, was used to calculate the Cox proportional hazards regression hazard ratio (HR) and P value of the log-rank test. R package “survminer” was used to plot Kaplan-Meier plots. The adjusted P value of the log-rank test was applied using false discovery rate (FDR) correction method. R package “GSA” was used to identify pathways that were potential prognosis predictors of overall survival in LUAD patients (33). Curated canonical pathways were downloaded from Molecular Signatures Database version 6.1 (http://software.broadinstitute.org/gsea/msigdb/index.jsp).
Somatic mutation detection.
Mapping of whole-exome sequencing reads and preprocessing of mappable reads were processed as described previously (14). Then somatic mutations in both PDX samples were called using MuTect2 with default settings.
Somatic mutations of LC2/ad cell lines were curated from the Catalogue Of Somatic Mutations In Cancer database. Over 97% of somatic mutations were overlapping between LC2/ad-R and LC2/ad cell lines (13).
SOM analysis in scRNA-Seq.
Prognostic genes with FDR less than 0.05 were selected, and genes with mean normalized counts less than 2 were filtered. Gene expression data were then log transformed, centralized, and clustered using SOMs. Genes were clustered onto a 12 × 12 grid or 15 × 15 grid for LUAD scRNA-Seq and cell line scRNA-Seq, respectively. R package “oposSOM” was implemented for SOM processing and downstream analysis, including k-means clustering of metagenes and the second-level SOM (20).
Supervised analysis of IFN-γ signaling pathways in scRNA-Seq.
IFN-γ signaling pathway genes were curated from the GO and Reactome pathway database, under GO:0034341 and R-HAS-877300.1, respectively. GSVA was used to calculate the sample-wise gene set enrichment score of IFN-γ signaling pathways in each individual single cell (34). K-means clustering was used to group single cells into MHCIIlo and MHCIIhi groups based on MHCII gene expression. Nonparametric Wilcoxon’s test was used to perform significance testing of GSVA scores between different groups. R package “pheatmap” was used to generate heatmaps.
Tumor-specific HLA typing and HLA-binding neoepitope prediction and expression in scRNA-Seq.
Raw reads of whole-exome sequencing were processed with OptiType (35). Then VCF files of each individual single cell generated by GATK HaplotypeCaller and MHC class I alleles (HLA-A, HLA-B, and HLA-C) predicted by OptiType were used to predict neoepitopes with topiary (https://github.com/hammerlab/topiary). NetMHCpan was selected in topiary as the MHC binding predictor. Cells were included if a neoantigen was detected regardless of its corresponding wild-type antigen detection status. Only genes that had predicted neoepitopes in more than 3 cells were examined for expression.
CTA expression in scRNA-Seq.
CTA genes were selected as previously reported (7). Briefly, known CTAs with negligible expression in GTEx normal tissues (95th percentile value < 1 TPM in all somatic tissue types) were analyzed for scRNA-Seq data.
Statistics.
All P values were FDR corrected, and FDR less than 0.05 was treated as significant.
Author contributions
KYM wrote the scripts, performed the analysis, interpreted the results, and wrote the manuscript. AAS helped with SNV analysis; AB helped interpret the results. CVDB helped interpret the results. SGE helped design the study. ZL helped design study. NJ designed the study, interpreted the results, and wrote the manuscript with KYM and with input from all authors.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R00AG040149 and R33CA225539 (to NJ), National Science Foundation CAREER Award 1653866 (to NJ), the Welch Foundation grant F1785 (to NJ), and the Cancer Prevention and Research Institute of Texas grant RR160093 (to SGE). NJ is a Cancer Prevention & Research Institute of Texas (CPRIT) Scholar and a Damon Runyon-Rachleff Innovator. SGE is also a CPRIT Scholar.
Version 1. 02/21/2019
Electronic publication
Footnotes
Conflict of interest: NJ is a scientific advisor for ImmuDX LLC and Immune Arch Inc.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(4):e121387. https://doi.org/10.1172/jci.insight.121387.
Contributor Information
Ke-Yue Ma, Email: keyue.ma@gmail.com.
Alexandra A. Schonnesen, Email: schonnesen@utexas.edu.
Amy Brock, Email: amy.brock@utexas.edu.
Carla Van Den Berg, Email: carla.vandenberg@austin.utexas.edu.
S. Gail Eckhardt, Email: gail.eckhardt@austin.utexas.edu.
Zhihua Liu, Email: liuzhihualzh@hotmail.com.
Ning Jiang, Email: jiang@austin.utexas.edu.
References
- 1.Gao J, et al. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell. 2016;167(2):397–404.e9. doi: 10.1016/j.cell.2016.08.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zaretsky JM, et al. Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med. 2016;375(9):819–829. doi: 10.1056/NEJMoa1604958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ott PA, et al. An immunogenic personal neoantigen vaccine for patients with melanoma. Nature. 2017;547(7662):217–221. doi: 10.1038/nature22991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robbins PF, et al. A pilot trial using lymphocytes genetically engineered with an NY-ESO-1-reactive T-cell receptor: long-term follow-up and correlates with response. Clin Cancer Res. 2015;21(5):1019–1027. doi: 10.1158/1078-0432.CCR-14-2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tran E, et al. T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med. 2016;375(23):2255–2262. doi: 10.1056/NEJMoa1609279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li B, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi: 10.1186/s13059-016-1028-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1–2):48–61. doi: 10.1016/j.cell.2014.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chevrier S, et al. An immune atlas of clear cell renal cell carcinoma. Cell. 2017;169(4):736–749.e18. doi: 10.1016/j.cell.2017.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lavin Y, et al. Innate immune landscape in early lung adenocarcinoma by paired single-cell analyses. Cell. 2017;169(4):750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng C, et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169(7):1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- 11.Schumacher TN, Schreiber RD. Neoantigens in cancer immunotherapy. Science. 2015;348(6230):69–74. doi: 10.1126/science.aaa4971. [DOI] [PubMed] [Google Scholar]
- 12.Nishino M, Ramaiya NH, Hatabu H, Hodi FS. Monitoring immune-checkpoint blockade: response evaluation and biomarker development. Nat Rev Clin Oncol. 2017;14(11):655–668. doi: 10.1038/nrclinonc.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suzuki A, et al. Single-cell analysis of lung adenocarcinoma cell lines reveals diverse expression patterns of individual cells invoked by a molecular target drug treatment. Genome Biol. 2015;16:66. doi: 10.1186/s13059-015-0636-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim KT, et al. Single-cell mRNA sequencing identifies subclonal heterogeneity in anti-cancer drug responses of lung adenocarcinoma cells. Genome Biol. 2015;16:127. doi: 10.1186/s13059-015-0692-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Min JW, et al. Identification of distinct tumor subpopulations in lung adenocarcinoma via single-cell RNA-seq. PLoS One. 2015;10(8):e0135817. doi: 10.1371/journal.pone.0135817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steimle V, Siegrist CA, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-γ mediated by the transactivator gene CIITA. Science. 1994;265(5168):106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 17.Forero A, et al. Expression of the MHC class II pathway in triple-negative breast cancer tumor cells is associated with a good prognosis and infiltrating lymphocytes. Cancer Immunol Res. 2016;4(5):390–399. doi: 10.1158/2326-6066.CIR-15-0243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson DB, et al. Melanoma-specific MHC-II expression represents a tumour-autonomous phenotype and predicts response to anti-PD-1/PD-L1 therapy. Nat Commun. 2016;7:10582. doi: 10.1038/ncomms10582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ilicic T, et al. Classification of low quality cells from single-cell RNA-seq data. Genome Biol. 2016;17:29. doi: 10.1186/s13059-016-0888-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löffler-Wirth H, Kalcher M, Binder H. oposSOM: R-package for high-dimensional portraying of genome-wide expression landscapes on bioconductor. Bioinformatics. 2015;31(19):3225–3227. doi: 10.1093/bioinformatics/btv342. [DOI] [PubMed] [Google Scholar]
- 21.Benci JL, et al. Tumor interferon signaling regulates a multigenic resistance program to immune checkpoint blockade. Cell. 2016;167(6):1540–1554.e12. doi: 10.1016/j.cell.2016.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castro F, Cardoso AP, Gonçalves RM, Serre K, Oliveira MJ. Interferon-γ at the crossroads of tumor immune surveillance or evasion. Front Immunol. 2018;9:847. doi: 10.3389/fimmu.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scanlan MJ, Gure AO, Jungbluth AA, Old LJ, Chen YT. Cancer/testis antigens: an expanding family of targets for cancer immunotherapy. Immunol Rev. 2002;188:22–32. doi: 10.1034/j.1600-065X.2002.18803.x. [DOI] [PubMed] [Google Scholar]
- 24.Zhao M, et al. MHC class II transactivator (CIITA) expression is upregulated in multiple myeloma cells by IFN-γ. Mol Immunol. 2007;44(11):2923–2932. doi: 10.1016/j.molimm.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsujisaki M, Igarashi M, Sakaguchi K, Eisinger M, Herlyn M, Ferrone S. Immunochemical and functional analysis of HLA class II antigens induced by recombinant immune interferon on normal epidermal melanocytes. J Immunol. 1987;138(4):1310–1316. [PubMed] [Google Scholar]
- 26.Maruhashi T, et al. LAG-3 inhibits the activation of CD4+ T cells that recognize stable pMHCII through its conformation-dependent recognition of pMHCII. Nat Immunol. 2018;19(12):1415–1426. doi: 10.1038/s41590-018-0217-9. [DOI] [PubMed] [Google Scholar]
- 27.Goel S, et al. CDK4/6 inhibition triggers anti-tumour immunity. Nature. 2017;548(7668):471–475. doi: 10.1038/nature23465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jerby-Arnon L, et al. A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. 2018;175(4):984–997.e24. doi: 10.1016/j.cell.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ott PA, Hodi FS, Kaufman HL, Wigginton JM, Wolchok JD. Combination immunotherapy: a road map. J Immunother Cancer. 2017;5:16. doi: 10.1186/s40425-017-0218-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vansteenkiste JF, et al. Efficacy of the MAGE-A3 cancer immunotherapeutic as adjuvant therapy in patients with resected MAGE-A3-positive non-small-cell lung cancer (MAGRIT): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2016;17(6):822–835. doi: 10.1016/S1470-2045(16)00099-1. [DOI] [PubMed] [Google Scholar]
- 31.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bacher R, et al. SCnorm: robust normalization of single-cell RNA-seq data. Nat Methods. 2017;14(6):584–586. doi: 10.1038/nmeth.4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Efron B, Tibshirani R. On testing the significance of sets of genes. Ann Appl Stat. 2007;1(1):107–129. doi: 10.1214/07-AOAS101. [DOI] [Google Scholar]
- 34.Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics. 2014;30(23):3310–3316. doi: 10.1093/bioinformatics/btu548. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.