Abstract
Diabetes mellitus is associated with various disorders of the locomotor system including the decline in mass and function of skeletal muscle. The mechanism underlying this association has remained ambiguous, however. We now show that the abundance of the transcription factor KLF15 as well as the expression of genes related to muscle atrophy are increased in skeletal muscle of diabetic model mice, and that mice with muscle-specific KLF15 deficiency are protected from the diabetes-induced decline of skeletal muscle mass. Hyperglycemia was found to upregulate the KLF15 protein in skeletal muscle of diabetic animals, which is achieved via downregulation of the E3 ubiquitin ligase WWP1 and consequent suppression of the ubiquitin-dependent degradation of KLF15. Our results revealed that hyperglycemia, a central disorder in diabetes, promotes muscle atrophy via a WWP1/KLF15 pathway. This pathway may serve as a therapeutic target for decline in skeletal muscle mass accompanied by diabetes mellitus.
Keywords: Metabolism, Muscle Biology
Keywords: Diabetes, Muscle
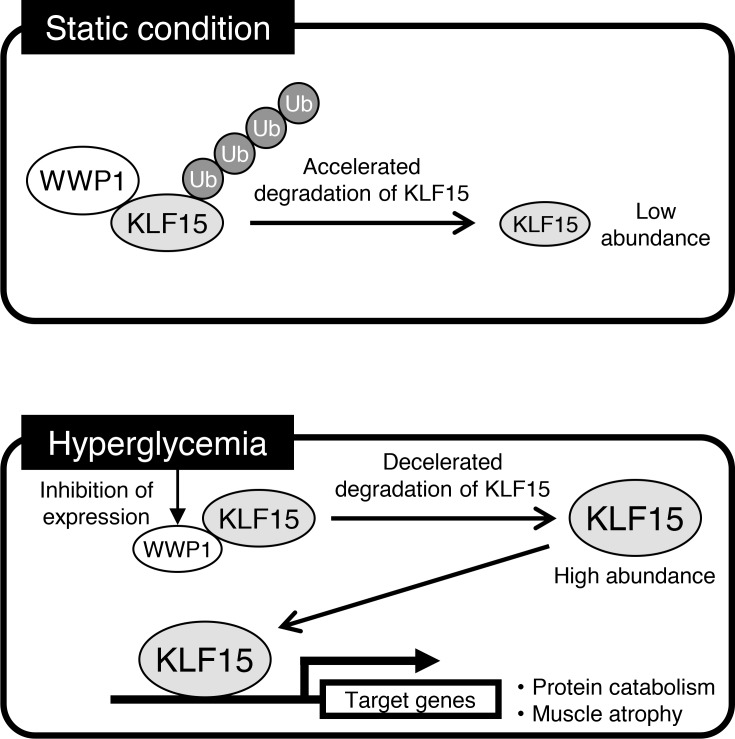
During the development of diabetes-induced decline of muscle mass, hyperglycemia downregulates WWP1, which results in the accumulation of KLF15 and consequent promotion of muscle atrophy.
Introduction
Although the prevention and treatment of micro- and macrovascular complications are major goals of care for diabetes mellitus, this metabolic condition is associated with a variety of diseases other than vascular complications such as cognitive impairment, cancer, and locomotor disorders (1–3). Diabetes mellitus is a well-established risk factor for sarcopenia (4, 5), and is known to accelerate age-dependent decline in skeletal muscle mass and functions (6, 7). A mechanistic link between diabetes mellitus and the decline in skeletal muscle mass remains ambiguous, however. Given that insulin is a potent stimulator of growth and protein synthesis for many cells (8), insufficiency of insulin action in diabetes mellitus likely contributes to muscle mass decline; mouse models in which insulin action is genetically ablated manifest age-dependent decline of muscle mass and physical performance (9, 10). Hyperglycemia, a central disorder of diabetes mellitus, is also implicated in the pathogenesis of decline in mass and function of skeletal muscle on the basis of a population-based cohort study and cross-sectional surveys of individuals with type 1 or type 2 diabetes mellitus (5, 11–13), but the underlying mechanism responsible for this relationship remains unknown.
Here, we investigated the molecular mechanism of diabetes-induced decline of skeletal muscle mass and found that hyperglycemia promotes muscle atrophy in a manner dependent on the E3 ubiquitin ligase WWP1 and the transcription factor KLF15.
Results
To investigate the mechanism underlying how diabetes mellitus promotes skeletal muscle decline, we utilized a mouse model of diabetes induced by streptozotocin (STZ). Intraperitoneal injection of STZ in mice resulted in marked hyperglycemia (~25 mM) accompanied by hypoinsulinemia within 14 days (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124952DS1) as well as a loss of body mass (Supplemental Figure 1C) and skeletal muscle mass (Figure 1A) apparent at 21 days. Given that skeletal muscle mass normalized by body mass was decreased in the treated mice, the loss of muscle mass appears to be specific rather than the result of a generalized increase in catabolism.
Figure 1. Skeletal muscle atrophy associated with diabetes is prevented in mice with skeletal muscle deficiency of KLF15.
(A–D) Ratio of gastrocnemius or extensor digitorum longus (EDL) muscle mass to body mass (A; n = 12), quantitative reverse transcription PCR (RT-PCR) analysis of Klf15 mRNA in gastrocnemius (B; n = 6), immunoblot analysis of KLF15 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH, loading control) in soleus muscle (nuclear fraction from 3 mice was loaded in 1 lane) (C; n = 2), and quantitative RT-PCR analysis of atrophy-related gene expression in gastrocnemius (D; n = 6) for control mice and diabetes-model mice at 21 days after the onset of STZ administration. (E–H) Ratio of muscle mass to body mass (E; n = 12), histological determination of muscle fiber area in EDL (F and G), and atrophy-related gene expression in gastrocnemius (H; n = 6) for WT or M-KLF15KO mice at 21 days after the onset of STZ administration or vehicle (Cont.) injection. In G, the areas of 500 fibers were measured in each condition. All quantitative data are means ± SEM for the indicated numbers of mice. *P < 0.05, **P < 0.01; NS, not significant. Unpaired t test (A, B, and D) or 2-way ANOVA with Bonferroni’s post hoc test (E, G, and H).
Krüppel-like factor 15 (KLF15), a member of the KLF family of transcription factors, regulates carbohydrate, lipid, and protein metabolism (14–18). The expression of KLF15 is upregulated in the liver of diabetic mice and is thought to contribute to their hyperglycemia (15), suggestive of a pathological role for this protein in diabetes. Furthermore, the mRNA abundance of KLF15 is increased by glucocorticoids, and overexpression of KLF15 in muscle cells upregulates genes related to muscle atrophy (19), suggesting that KLF15 is implicated in muscle atrophy induced by glucocorticoids. These findings prompted us to investigate the role of KLF15 in muscle atrophy associated with diabetes.
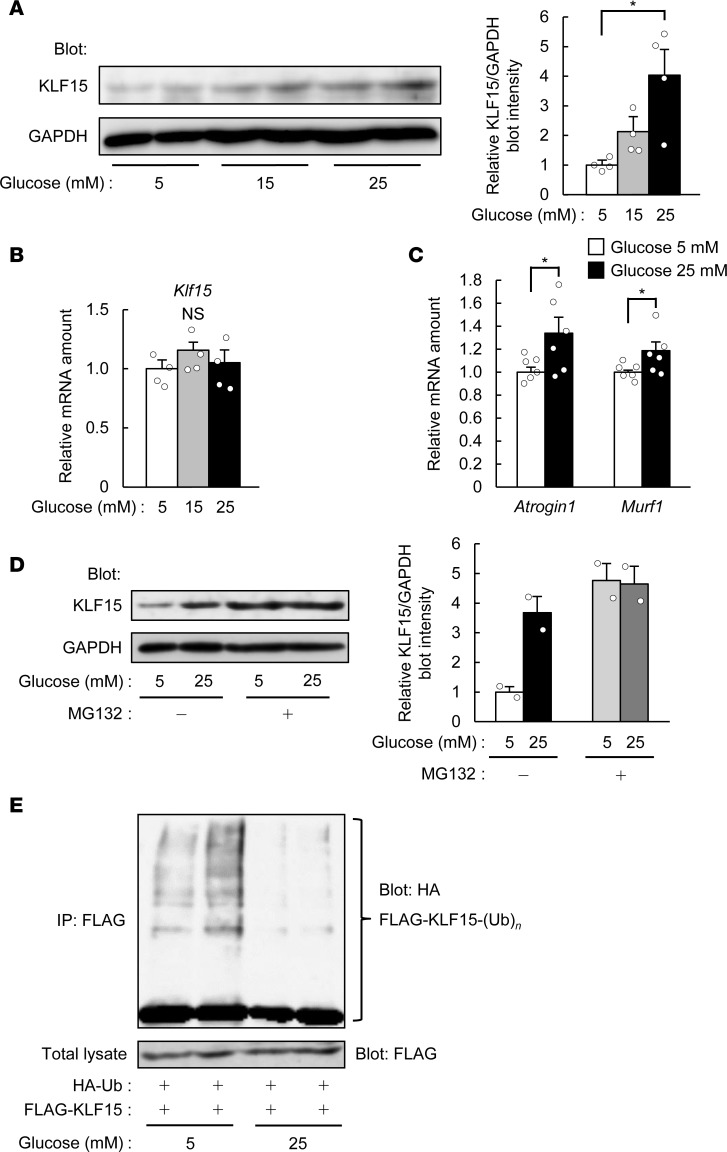
Different from skeletal muscle atrophy induced by glucocorticoids, the amount of Klf15 mRNA in skeletal muscle of mice with STZ treatment was unaltered (Figure 1B). The abundance of KLF15 protein, however, was increased in skeletal muscle of our diabetic model mice at 21 days after the onset of STZ administration (Figure 1C). The expression of genes related to muscle atrophy — Atrogin1, Murf1, Foxo3a, Prodh, and Tdo2 — was also increased by STZ treatment (Figure 1D).
To examine the effect of KLF15 loss on muscle atrophy, we generated mice lacking KLF15 specifically in skeletal muscle (M-KLF15KO mice) by crossing mice harboring a floxed allele of Klf15 (Supplemental Figure 2) with those expressing Cre recombinase under the control of the myosin light chain 1f gene (Mlc1f) promoter (20). Body mass, skeletal muscle mass, and the cross-sectional area of lower limb muscle (Supplemental Figure 3) as well as blood glucose and plasma insulin levels (Supplemental Figure 4, A and B) were unaltered in the mutant mice. Treatment of M-KLF15KO mice with STZ also resulted in hyperglycemia with an extent and time course similar to that in control mice (Supplemental Figure 4C). In contrast, STZ-induced muscle atrophy — as reflected by changes in muscle mass (Figure 1E), the cross-sectional area of lower limb muscle (Supplemental Figure 5A), and muscle fiber area evaluated histologically (Figure 1, F and G) — was prevented in M-KLF15KO mice. Furthermore, whereas STZ treatment increased and decreased the proportions of small and large muscle fibers, respectively, in wild-type (WT) mice, no such effect was apparent in M-KLF15KO mice (Supplemental Figure 5B). In addition, the STZ-induced increase in the expression of muscle atrophy–related genes was abolished in the mutant mice (Figure 1H). The abundance of proteins encoded by muscle atrophy–related genes Atrogin1 and Foxo3a was also increased in the skeletal muscle of STZ-treated mice, and the STZ-induced increase was inhibited in M-KLF15KO mice (Supplemental Figure 6A). Furthermore, muscle function assessed by a passive wire-hang test as well as by the tolerance for maximum speed and the time for exhaustion on a treadmill exercise load test was decreased in STZ diabetic mice and the STZ-induced decline in muscle function was prevented in M-KLF15KO mice (Supplemental Figure 6, B and C). Together, these results thus indicated that KLF15 is responsible for muscle atrophy as well as decline in muscle function in this model of diabetes.
Both hyperglycemia and hypoinsulinemia accompany the STZ-induced diabetes. We have found that exposure of mouse C2C12 myotubes to glucose increased the amount of KLF15 protein in a concentration- and time-dependent manner (Figure 2A and Supplemental Figure 7A), without affecting that of Klf15 mRNA (Figure 2B), as was seen in skeletal muscle of mice treated with STZ. Furthermore, exposure of the cells to glucose increased the expression of muscle atrophy–related genes Atrogin1 and Murf1 (Figure 2C). In contrast, treatment of the myotubes with insulin had no effect on the amount of Klf15 mRNA or the encoded protein (Supplemental Figure 7, B and C), suggesting that hyperglycemia is directly responsible for the upregulation of KLF15 protein in skeletal muscle of diabetic mice.
Figure 2. Glucose decreases the ubiquitination of, and increases the protein abundance of, KLF15.
(A and B) Immunoblot analysis of KLF15 protein (A) and quantitative RT-PCR analysis of Klf15 mRNA (B; n = 4) in C2C12 myotubes exposed to the indicated concentrations of glucose for 24 hours. In A, a representative blot and quantitative data (n = 4) are shown in the left and right panels, respectively. (C) Quantitative RT-PCR analysis of muscle atrophy–related gene expression for myotubes treated as in A. n = 6. (D) Immunoblot analysis of KLF15 in myotubes exposed to 5 or 25 mM glucose in the absence or presence of 15 μM MG132 for 6 hours. A representative blot and quantitative data (n = 2) are shown in the left and right panels, respectively. (E) C2C12 myoblasts expressing HA-ubiquitin (Ub) and FLAG-KLF15 were incubated with 5 or 25 mM glucose for 24 hours and then subjected to immunoprecipitation (IP) with antibodies against FLAG. The resulting precipitates were analyzed by immunoblot with antibodies against HA to detect polyubiquitinated [-(Ub)n] KLF15, and the original cell lysates were analyzed by immunoblot with antibodies against FLAG. Representative data from 3 independent experiments are shown. All quantitative data are means ± SEM for the indicated numbers of independent experiments. *P < 0.05; NS, not significant. Two-way ANOVA with Bonferroni’s post hoc test (A and B) or unpaired t test (C).
Treatment of C2C12 myotubes with the proteasome inhibitor MG132 also increased the amount of KLF15 protein in the presence of a low glucose concentration (5 mM), and a high concentration of glucose (25 mM) did not further increase KLF15 abundance in the presence of MG132 (Figure 2D). We also found that high glucose inhibited the polyubiquitination of FLAG epitope–tagged KLF15 in C2C12 myotubes also expressing hemagglutinin epitope–tagged (HA-tagged) ubiquitin (Figure 2E), suggesting that glucose upregulates KLF15 protein by inhibiting its ubiquitin-dependent degradation and thereby increases the expression of genes related to muscle atrophy.
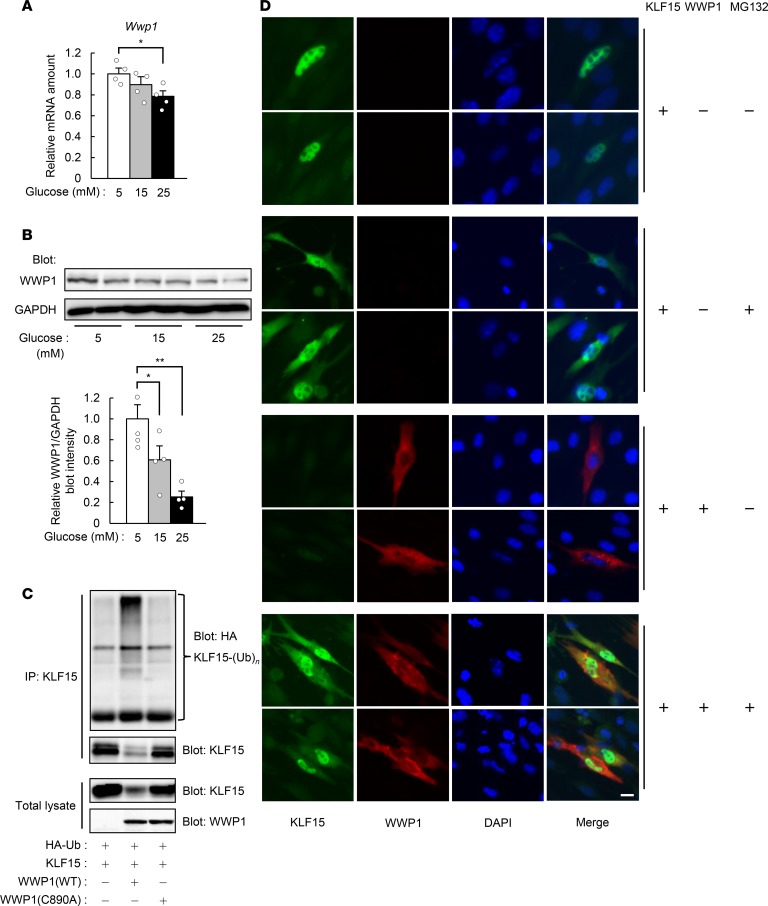
To identify the E3 ubiquitin ligase responsible for the glucose-dependent change in polyubiquitination of KLF15, we investigated the expression of genes encoding E3 ubiquitin ligases in skeletal muscle of mice with STZ-induced diabetes. Microarray analysis showed that the expression of 27 genes encoding E3 ubiquitin ligases was downregulated in skeletal muscle of the diabetic mice (Supplemental Table 1). Analysis of a public database of gene expression revealed that 8 of these 27 genes are highly expressed in skeletal muscle (Supplemental Table 1). Among the proteins encoded by these 8 genes, WW domain–containing E3 ubiquitin protein ligase 1 (WWP1) and neural precursor cell–expressed developmentally downregulated 4 (NEDD4) were previously shown to interact with other members of the KLF family (21, 22). Coimmunoprecipitation analysis revealed that WWP1, but not NEDD4, interacted with KLF15 in transfected COS-7 cells (Supplemental Figure 8A). Furthermore, WWP1 was found to be downregulated at both mRNA and protein levels in skeletal muscle of STZ diabetic mice (Supplemental Figure 8, B and C) as well as in C2C12 myotubes treated with glucose (Figure 3, A and B). Insulin did not affect the amount of Wwp1 mRNA in C2C12 myotubes (Supplemental Figure 8D). The decrease in WWP1 protein (~75% reduction) (Supplemental Figure 8C) in skeletal muscle in response to STZ treatment was greater than that of Wwp1 mRNA (~20% reduction) (Supplemental Figure 8B). Greater reduction in the level of WWP1 protein than that of Wwp1 mRNA was also observed in C2C12 myotubes treated with glucose (Figure 3, A and B). Treatment of MG132 with C2C12 myotubes prevented glucose-induced decrease in the WWP1 protein (Supplemental Figure 8E) without the increase in Wwp1 mRNA (Supplemental Figure 8F), suggesting that glucose stimulates the degradation of WWP1 protein through the proteasome pathway.
Figure 3. WWP1 regulates the polyubiquitination and abundance of KLF15.
(A and B) Quantitative RT-PCR analysis of Wwp1 mRNA (A; n = 4) and immunoblot analysis of WWP1 protein (B) in C2C12 myotubes exposed to the indicated concentrations of glucose for 24 hours. In B, a representative blot and quantitative data (n = 4) are shown in the left and right panels, respectively. (C) COS-7 cells transfected with vectors for HA-Ub, KLF15, and either WT or C890A mutant forms of WWP1 were subjected to immunoprecipitation with antibodies against KLF15. The resulting precipitates and the original cell lysates were analyzed by immunoblot as indicated. (D) Immunofluorescence analysis of KLF15 and WWP1 in C2C12 myoblasts transfected with vectors for these proteins and exposed to 15 μM MG132 for 6 hours as indicated. Nuclei were stained with DAPI. Scale bar: 10 μm. In C and D, representative data from at least 3 independent experiments are shown. All quantitative data are means ± SEM for the indicated numbers of independent experiments. *P < 0.05, **P < 0.01. Two-way ANOVA with Bonferroni’s post hoc test (A and B).
Forced expression of WWP1 in COS-7 cells reduced the abundance (Supplemental Figure 9A) and increased the polyubiquitination (Figure 3C) of KLF15, whereas expression of a catalytically inactive mutant (C890A) of WWP1 had no effect on KLF15 polyubiquitination (Figure 3C). Immunofluorescence analysis of transfected C2C12 myoblasts revealed that KLF15 immunoreactivity was localized predominantly to the nucleus, with smaller amounts also being detected in the cytosol (Figure 3D). Treatment of the cells with MG132 increased the amount of KLF15 immunoreactivity in the cytosol. In contrast, WWP1 was localized exclusively to the cytosol, and its forced expression resulted in the complete or partial loss of KLF15 in the cytosol and nucleus, respectively, with these effects being prevented in the presence of MG132 (Figure 3D). These results thus suggested that WWP1 interacts with KLF15 in the cytosol, which results in the downregulation of KLF15 in both the cytosol and the nucleus.
We next transfected a vector expressing both enhanced green fluorescent protein (EGFP) and WWP1 or a control vector only expressing EGFP into C2C12 myotubes, and evaluated the effect of glucose on the size of transfected myotubes (detected by EGFP fluorescence). Treatment with glucose decreased the diameters of the myotubes transfected only with EGFP, whereas the effect of glucose was inhibited in myotubes transfected with EGFP plus WWP1 (Supplemental Figure 9, B and C). These results suggest that glucose induces atrophy of the myotubes in a WWP1-dependent manner.
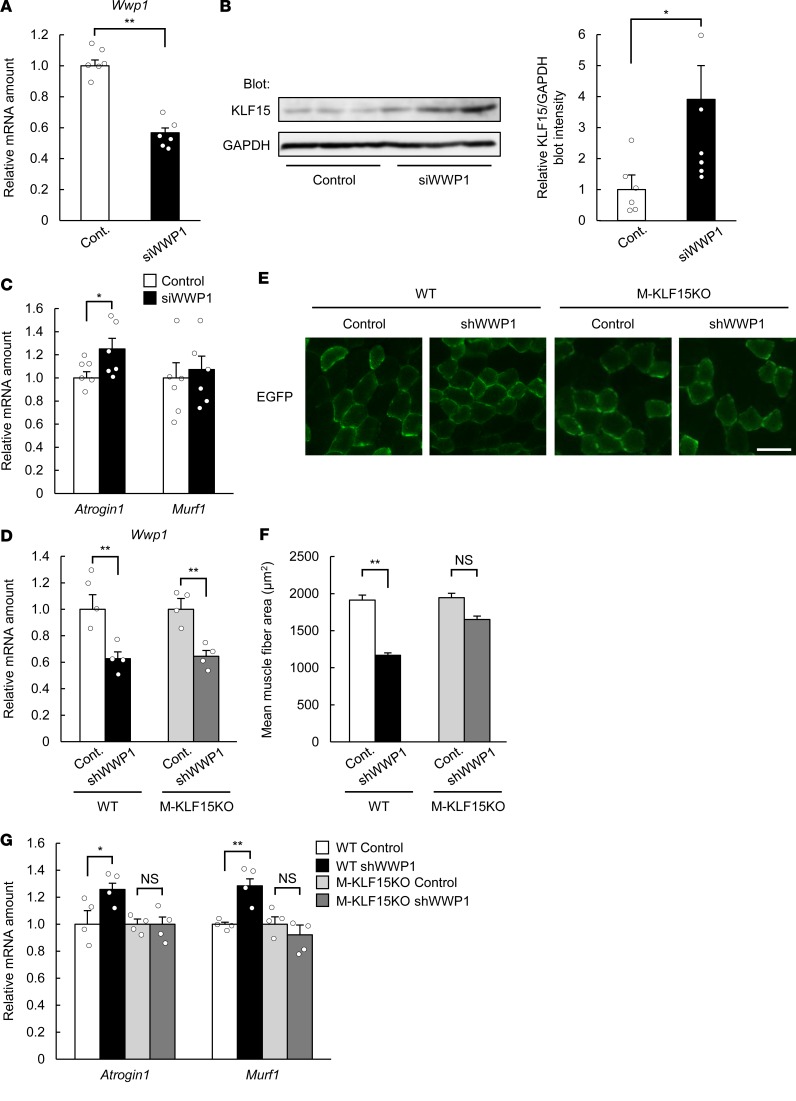
Depletion of WWP1 in C2C12 myotubes by transfection with small interfering RNA (siRNA) (Figure 4A) increased the amount of KLF15 protein (Figure 4B) as well as that of Atrogin1 mRNA (Figure 4C), indicating that the downregulation of WWP1 leads to the upregulation of KLF15 protein. To deplete WWP1 in skeletal muscle of mice, we constructed adeno-associated virus (AAV) vectors encoding both a short hairpin RNA (shRNA) specific for Wwp1 mRNA (Supplemental Figure 9D) and EGFP to visualize virus-infected muscle fibers. Whereas injection of skeletal muscle (tibialis anterior) with the AAV vector led to only an approximately 40% reduction of Wwp1 mRNA in whole-muscle extracts in both WT and M-KLF15KO mice (Figure 4D), fluorescence microscopic analysis revealed that only a part of the muscle fibers was infected with the virus (positive for GFP) (Figure 4E), suggesting that the reduction of the amount of Wwp1 mRNA in the infected muscle fibers was greater than the extent evaluated with the whole-muscle extracts. The cross-sectional area of virus-infected muscle fibers (detected by EGFP fluorescence) was significantly smaller in WT mice injected with the AAV encoding WWP1 shRNA than in those injected with a control AAV (Figure 4, E and F). In contrast, such depletion of WWP1 did not significantly decrease the cross-sectional area of skeletal muscle fibers in M-KLF15KO mice. Infection with the AAV encoding WWP1 shRNA also increased the expression of Atrogin1 and Murf1 in whole-muscle extracts of WT mice but not of M-KLF15KO mice (Figure 4G). Collectively, these results thus indicated that downregulation of WWP1 promotes skeletal muscle atrophy in live animals in a KLF15-dependent manner.
Figure 4. WWP1 regulates skeletal muscle atrophy in a KLF15-dependent manner.
(A) Quantitative RT-PCR analysis of Wwp1 mRNA in C2C12 myotubes transfected with control or WWP1 siRNA. n = 6. (B and C) Immunoblot analysis of KLF15 (B) and quantitative RT-PCR analysis of Atrogin1 and Murf1 mRNAs (C; n = 6) in C2C12 myotubes transfected as in A. In B, a representative blot and quantitative data (n = 6) are shown in the left and right panels, respectively. (D) Quantitative RT-PCR analysis of Wwp1 mRNA in tibialis anterior muscle of WT or M-KLF15KO mice injected with AAVs encoding EGFP with or without (Cont.) WWP1 shRNA. n = 4. (E) Fluorescence microscopic detection of EGFP-positive muscle fibers of mice infected as in D. Scale bar: 100 μm. (F) Cross-sectional area of EGFP-positive muscle fibers determined from images as in E. The areas of 100 fibers were measured in each condition. (G) Quantitative RT-PCR analysis of Atrogin1 and Murf1 mRNAs in tibialis anterior muscle of mice infected as in D. n = 4. All quantitative data are means ± SEM for the indicated numbers of independent experiments (A–C) or mice (D, F, and G). *P < 0.05, **P < 0.01; NS, not significant. Unpaired t test (A–C) or 2-way ANOVA with Bonferroni’s post hoc test (D, F, and G).
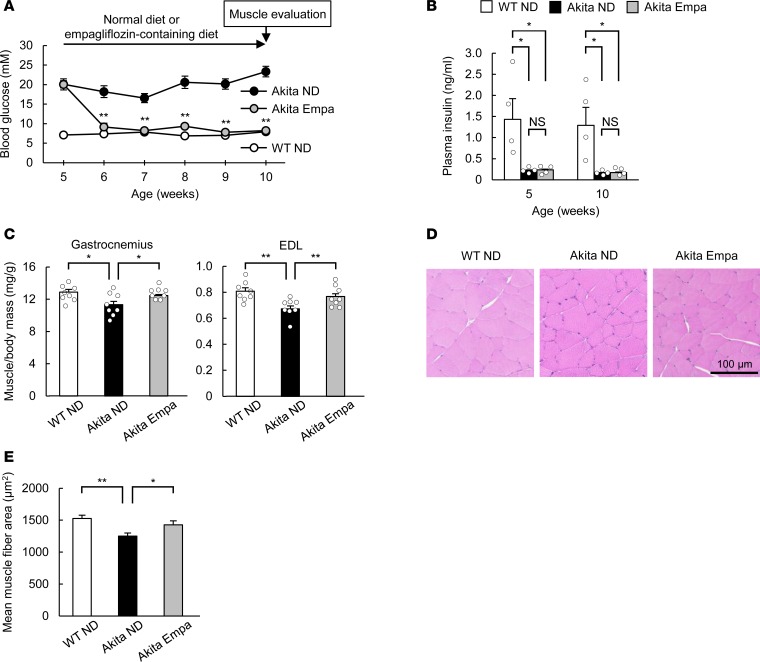
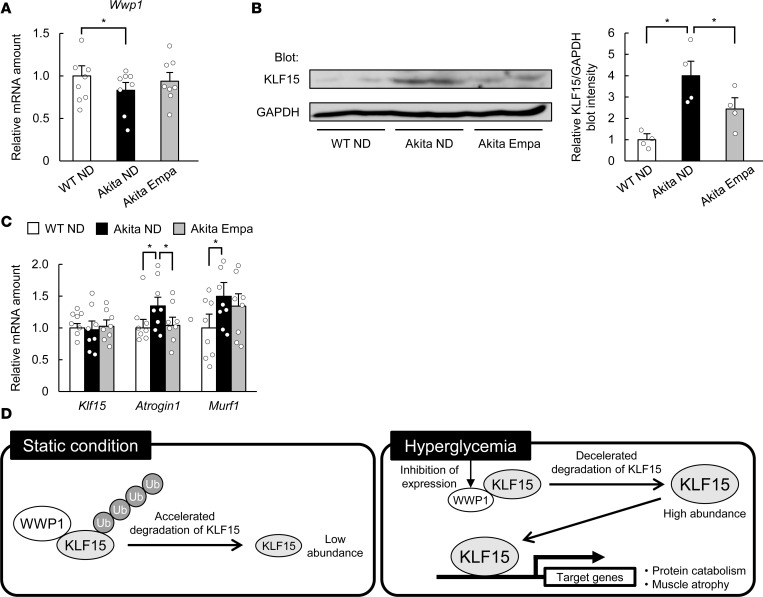
We lastly examined whether hyperglycemia is responsible for declines of skeletal muscle mass in a chronic model of diabetes. Akita mice, which develop diabetes as a result of pancreatic β cell dysfunction (23), manifested hyperglycemia as early as 5 weeks of age (Figure 5A). Although skeletal muscle mass at 6 weeks of age was similar in Akita and WT mice, that at 10 weeks was smaller in the former animals (Supplemental Figure 10), indicating that this chronic model of diabetes develops age-dependent skeletal muscle atrophy. Inhibitors of sodium-glucose cotransporter 2 (SGLT2) such as empagliflozin lower blood glucose levels in an insulin-independent manner (24) and thus are a useful tool for the evaluation of the pathological significance of blood glucose. Administration of empagliflozin thus lowered the blood glucose concentration of Akita mice to a level similar to that of WT mice within 1 week, without affecting the plasma insulin level (Figure 5, A and B). Empagliflozin prevented the reduction in skeletal muscle mass and muscle fiber area apparent in control Akita mice (Figure 5, C–E). The amount of Wwp1 mRNA in skeletal muscle of Akita mice was smaller than that in WT mice but was increased by empagliflozin treatment (Figure 6A). The amount of KLF15 protein was increased in skeletal muscle of Akita mice, but this increase was attenuated by the administration of empagliflozin (Figure 6B). The amount of Klf15 mRNA in skeletal muscle was similar in Akita and WT mice and was not affected by empagliflozin (Figure 6C). The expression of muscle atrophy–related genes was also increased in skeletal muscle of Akita mice in a manner sensitive to empagliflozin treatment (Figure 6C). Together, these results suggested that a mechanism similar to that operative in STZ diabetic mice contributes to the age-dependent muscle atrophy in this chronic model of diabetes.
Figure 5. SGLT2 inhibitor ameliorates age-dependent muscle atrophy in Akita mice.
(A and B) Blood glucose (A; n = 8) and plasma insulin (B; n = 4) levels of WT or Akita mice fed a normal diet (ND) or a diet containing empagliflozin (Empa) beginning at 5 weeks of age. (C–E) Skeletal muscle/body mass ratios (C; n = 8) as well as histological determination of muscle fiber area in tibialis anterior muscle (D and E) of mice as in A at 10 weeks of age. In E, the areas of 200 fibers were measured in each condition. All quantitative data are means ± SEM for the indicated numbers of mice. *P < 0.05, **P < 0.01 for Akita Empa versus Akita ND (A) or for the indicated comparisons. NS, not significant. Two-way ANOVA with Bonferroni’s post hoc test (A–C, and E).
Figure 6. SGLT2 inhibitor prevents the alteration of gene expression in Akita mice.
(A–C) Quantitative RT-PCR analysis of Wwp1 (A; n = 8) or Klf15, Atrogin1, and Murf1 (C; n = 8) mRNAs, and immunoblot analysis of KLF15 (B) in gastrocnemius (A and C) or soleus muscle (nuclear fraction from 2 mice was loaded on one lane) (B) of mice as in Figure 5A at 10 weeks of age. In B, a representative blot and quantitative data (n = 4) are shown in the left and right panels, respectively. All quantitative data are means ± SEM for the indicated numbers of mice. *P < 0.05. Two-way ANOVA with Bonferroni’s post hoc test (A–C). (D) Proposed role for the WWP1/KLF15 axis in hyperglycemia-induced muscle atrophy.
Discussion
In the present study, our data have revealed that hyperglycemia upregulates KLF15, which then contributes to the decline in skeletal muscle mass, in mouse models of diabetes. The increase in KLF15 abundance induced by hyperglycemia results from downregulation of the E3 ubiquitin ligase WWP1 and the consequent suppression of ubiquitin-dependent degradation of KLF15 (Figure 6D). The decrease in anabolic effects of insulin has been implicated in the muscle mass decline in diabetes mellitus (9, 10). Our results showed, for the first time to our knowledge, that hyperglycemia directly exerts catabolic effects on skeletal muscle. The STZ-induced loss of muscle mass was significantly, but not completely, attenuated in M-KLF15KO mice. The residual decline in muscle mass in the mutant mice might therefore be attributable to insufficiency of insulin action.
We have identified the E3 ubiquitin ligase WWP1 as a regulator of hyperglycemia-dependent change in the abundance of KLF15. Although WWP1 has been shown to be involved in various physiological and pathological processes including differentiation and tumorigenesis, as well as infectious and neural diseases (25), to the best of our knowledge its contribution to the pathology of diabetes has not been reported. Whether WWP1 contributes to other pathological processes triggered by hyperglycemia warrants further study. Our data suggest that glucose downregulates the abundance of WWP1 both through the suppression of mRNA and the stimulation of protein degradation. However, a complete molecular mechanism by which glucose downregulates the abundance of WWP1 remains to be elucidated. Intriguingly, a missense mutation in a Wwp1 ortholog was identified in a chicken model of muscular dystrophy (26). It would also be interesting to investigate whether the mechanism identified in this study is related to the pathogenesis of the chicken model of the muscle disease.
In conclusion, we have shown that hyperglycemia, a central disorder in diabetes, promotes skeletal muscle atrophy via a WWP1/KLF15 pathway. This pathway may thus serve as a therapeutic target for muscle atrophy triggered by diabetes.
Methods
Animals.
Mice were maintained on a 12-hour light/dark cycle and allowed free access to food (CE-2, CLEA) and water. A Klf15-floxed mouse line was generated by insertion of loxP sites flanking exon 2 of the Klf15 gene through collaboration with the RIKEN Center (Supplemental Figure 2). For generation of Klf15 FRT–floxed mice, the targeting construct was introduced into embryonic stem cells by electroporation. The Klf15 FRT–floxed mice were then crossed with FLP-transgenic mice to remove the neo cassette from between the FRT sites, and Klf15-floxed mice were selected and backcrossed for 7 generations with C57BL/6J mice (CLEA). We generated skeletal muscle–specific KLF15-knockout (M-KLF15KO) mice by crossing Klf15-floxed mice and Mlc1f-Cre mice (20). Klf15-floxed, Cre-negative littermates served as controls. Akita mice (with a mutation in the Ins2 gene) on the C57BL/6J background were obtained from Japan SLC and were compared with age-matched WT controls.
For intraperitoneal glucose tolerance tests, male mice at 20 weeks of age were deprived of food for 18 hours and then injected intraperitoneally with glucose (2 g/kg). Blood samples were collected, and blood glucose concentration was measured with a glucometer (Sanwa Kagaku). Plasma insulin concentration was measured with an enzyme-linked immunosorbent assay kit for mouse insulin (Shibayagi).
Male WT and M-KLF15KO mice at 10 weeks of age were injected intraperitoneally with sodium citrate (control) or 50 mg/kg STZ (Wako), dissolved in 50 mM sodium citrate (pH 4.5) on 5 consecutive days. After 2 weeks, mice with a blood glucose level of greater than 20 mM were considered to be diabetic and used for experiments. Diabetic mice were euthanized for final analysis at 1 week after the establishment of diabetes.
An empagliflozin-containing diet (0.045%) was obtained from Boehringer Ingelheim Pharma. This drug concentration was estimated to correspond to a dose of approximately 90 mg/kg per day based on data for daily food intake. Female Akita mice at 5 weeks of age received either a normal diet or the empagliflozin-containing diet until 10 weeks of age.
Computed tomography was performed with an RmCT2 system (Rigaku) to evaluate the cross-sectional area of lower limb muscle. The maximum diameter of lower limb muscle was quantified with the manufacturer’s software.
For passive wire-hang test, mice were placed on the cage top, which was then inverted and suspended above the home cage. The latency to when the animals fell was recorded. A single data point for an individual mouse represents the mean of 2 separate trials. For treadmill exercise test, mice were placed on a treadmill MK-680 (Muromachi), and allowed to warm up at 10 m/min with the slope at 9% for 4 minutes. Then, the speed was increased by 1 m/min every 1 minute. Mice were considered to be exhausted when the hindlimbs remained on the electric grid for more than 5 seconds.
Cell culture.
C2C12 myoblasts as well as COS-7 and HEK 293 cells were maintained in growth medium (Dulbecco’s modified Eagle’s medium [DMEM] supplemented with 10% heat-inactivated fetal bovine serum and 1% penicillin-streptomycin) at 37°C under a mixture of 95% air and 5% CO2. C2C12 myoblasts were passaged before they achieved 80% confluence in order to maintain their undifferentiated state. The differentiation of C2C12 myoblasts was induced by replacement of growth medium with differentiation medium (DMEM supplemented with 2% horse serum and 1% penicillin-streptomycin) at 100% confluence. All experiments with C2C12 myotubes were performed with cells in the fully differentiated state after exposure to differentiation medium for 4 days. The glucose concentration of the medium was changed to the low condition (5 mM) for at least 18 hours after full differentiation; it was then changed to the high condition (15 or 25 mM) for the indicated times. MG132 was obtained from Merck.
Plasmids and transfection.
Mammalian expression plasmids encoding HA-tagged ubiquitin or mouse KLF15 with or without the FLAG tag were described previously (15). Mammalian expression plasmids for human WWP1 or NEDD4-2 were provided by K. Miyazono (University of Tokyo). For transient transfection, the culture medium of C2C12 myoblasts, COS-7 cells, or HEK 293 cells at 40% confluence was replaced with Opti-MEM I (Invitrogen) and vectors were introduced into the cells with the use of the FuGENE HD reagent (Promega). The total amount of plasmid DNA was maintained constant by the addition of control plasmids. The WWP1 and negative control siRNAs were obtained from Invitrogen and were delivered into cells with the use of the Lipofectamine RNAiMAX transfection reagent (Invitrogen).
RNA isolation and quantitative RT-PCR analysis.
Total RNA was extracted from mouse skeletal muscle and C2C12 myotubes with the use of the RNeasy Mini Kit (Qiagen). Quantitative reverse transcription PCR (RT-PCR) analysis was performed with the Real-Time PCR system (Applied Biosystems). Data were normalized by the amount of 36B4 mRNA. Primer sequences are listed in Supplemental Table 2.
Immunoprecipitation and immunoblot analysis.
Lysates of C2C12 myoblasts, myotubes, and COS-7 cells were prepared with radioimmunoprecipitation (RIPA) buffer containing 0.1% SDS, and lysates of skeletal muscle were prepared with RIPA buffer containing 1.0% SDS. Nuclear fractions were prepared with a fractionation kit (BioVision). The protein concentration of each sample was measured with a BCA protein assay kit (Thermo Fisher Scientific), and GAPDH or α-tubulin served as a loading control. Immunoprecipitation was performed with the use of Dynabeads Protein G (Invitrogen). Rabbit polyclonal antibodies against mouse KLF15 (15) and those against mouse WWP1 (27) were generated as described previously. The following primary antibodies were purchased: anti-KLF15 (ab2647, Abcam), anti-Atrogin1 (ab168372, Abcam), anti-Foxo3a (2497, Cell Signaling Technology), anti-FLAG (F3165, Sigma-Aldrich), anti-HA (12CA5, 11583816001, Roche Diagnostics), anti-Myc (2278, Cell Signaling Technology). See complete unedited blots in Supplemental Figure 11.
Histological analysis.
Skeletal muscle sections were stained with hematoxylin and eosin to quantify myofiber area. The areas of muscle fibers were quantified per image with the use of ImageJ software (NIH). For immunohistochemical analysis in vivo, freshly harvested muscle tissue was mounted in OCT compound and frozen in isopentane cooled with liquid nitrogen. The frozen samples were subsequently sectioned with a cryostat microtome, the sections were fixed in ice-cold acetone, and EGFP-expressing myofibers were directly detected with a BZ-X710 fluorescence microscope (Keyence). For immunofluorescence staining in vitro, cultured myoblasts were fixed in 4% paraformaldehyde, and permeabilization solution (0.4% saponin, 1% BSA, and 2% normal goat serum) was used for cell membrane permeabilization prior the overnight hybridization of primary antibodies. FITC- or Cy3-conjugated secondary antibodies were used to visualize the myoblast structure, and nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). The stained myoblasts were detected with a BZ-X710 fluorescence microscope.
Recombinant AAV vectors.
For generation of an AAV6 vector encoding WWP1 shRNA, oligonucleotides including the hairpin loop sequence for the target site (5′-GTCCTTGATGGATTAGTGATTGA-3′) were subcloned into the pAAV-U6-GFP vector. The recombinant AAV6 vector was produced with the use of an AAV Helper-Free system (Cell Biolabs). For local delivery, 1 × 1011 vector genomes for the AAV6 vector with or without (control) the shRNA-coding sequence were injected in 50 μl of saline directly into the tibialis anterior muscle. Muscle tissue was collected for analysis at 2 weeks after the injection.
DNA microarray analysis.
Total RNA was extracted from gastrocnemius muscle of STZ diabetic or control mice and was hybridized with an Affymetrix Mouse Gene 2.0 ST Array. All original microarray data were deposited in the NCBI’s Gene Expression Omnibus database (GEO GSE123540). The data were analyzed by DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/). The expression of E3 ubiquitin ligase genes in skeletal muscle was determined by analysis of the public database BioGPS (http://biogps.org/).
Statistics.
Quantitative data are presented as means ± SEM. P values were calculated by 2-tailed unpaired Student’s t test or 2-way analysis of variance (ANOVA) with Bonferroni’s post hoc test. A P value of <0.05 was considered statistically significant.
Study approval.
All animal experiments were approved by the animal experimentation committee of Kobe University Graduate School of Medicine (no. P171012).
Author contributions
YH, KN, and WO conceived the study and analyzed the data. YO and TH contributed to animal experiments. YS contributed to cell experiments. KK, SO, and YM contributed to the production of AAV vectors. MI and ST contributed to skeletal muscle analyses. TH contributed to the generation of genetically engineered mice. WO, YH, KN, and TH contributed to discussion. YH, KN, and WO wrote the manuscript.
Supplementary Material
Acknowledgments
We thank K. Miyazono and S.J. Burden for providing vectors for WWP1 and Mlc1f-Cre mice, respectively. This work was supported in part by a grant-in-aid for scientific research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (16H06970 to YO).
Version 1. 02/21/2019
Electronic publication
Footnotes
Conflict of interest: WO has received a research grant and lecture fees from Boehringer Ingelheim.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(4):e124952. https://doi.org/10.1172/jci.insight.124952.
Contributor Information
Yu Hirata, Email: yuhirata@med.kobe-u.ac.jp.
Kazuhiro Nomura, Email: knomura@med.kobe-u.ac.jp.
Yoko Senga, Email: ysenga4@gmail.com.
Yuko Okada, Email: yokada@med.kobe-u.ac.jp.
Kenta Kobayashi, Email: kobaya@nips.ac.jp.
Shiki Okamoto, Email: shiki@med.u-ryukyu.ac.jp.
Yasuhiko Minokoshi, Email: minokosh@nips.ac.jp.
Michihiro Imamura, Email: imamura@ncnp.go.jp.
Tetsuya Hosooka, Email: thosooka@med.kobe-u.ac.jp.
Wataru Ogawa, Email: ogawa@med.kobe-u.ac.jp.
References
- 1.Kodl CT, Seaquist ER. Cognitive dysfunction and diabetes mellitus. Endocr Rev. 2008;29(4):494–511. doi: 10.1210/er.2007-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Calle EE, Kaaks R. Overweight, obesity and cancer: epidemiological evidence and proposed mechanisms. Nat Rev Cancer. 2004;4(8):579–591. doi: 10.1038/nrc1408. [DOI] [PubMed] [Google Scholar]
- 3.Wong E, et al. Diabetes and risk of physical disability in adults: a systematic review and meta-analysis. Lancet Diabetes Endocrinol. 2013;1(2):106–114. doi: 10.1016/S2213-8587(13)70046-9. [DOI] [PubMed] [Google Scholar]
- 4.Monaco CMF, Perry CGR, Hawke TJ. Diabetic myopathy: current molecular understanding of this novel neuromuscular disorder. Curr Opin Neurol. 2017;30(5):545–552. doi: 10.1097/WCO.0000000000000479. [DOI] [PubMed] [Google Scholar]
- 5.Kalyani RR, Corriere M, Ferrucci L. Age-related and disease-related muscle loss: the effect of diabetes, obesity, and other diseases. Lancet Diabetes Endocrinol. 2014;2(10):819–829. doi: 10.1016/S2213-8587(14)70034-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park SW, et al. Accelerated loss of skeletal muscle strength in older adults with type 2 diabetes: the health, aging, and body composition study. Diabetes Care. 2007;30(6):1507–1512. doi: 10.2337/dc06-2537. [DOI] [PubMed] [Google Scholar]
- 7.Park SW, et al. Excessive loss of skeletal muscle mass in older adults with type 2 diabetes. Diabetes Care. 2009;32(11):1993–1997. doi: 10.2337/dc09-0264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.James HA, O’Neill BT, Nair KS. Insulin regulation of proteostasis and clinical implications. Cell Metab. 2017;26(2):310–323. doi: 10.1016/j.cmet.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Neill BT, et al. Insulin and IGF-1 receptors regulate FoxO-mediated signaling in muscle proteostasis. J Clin Invest. 2016;126(9):3433–3446. doi: 10.1172/JCI86522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luo J, et al. Loss of class IA PI3K signaling in muscle leads to impaired muscle growth, insulin response, and hyperlipidemia. Cell Metab. 2006;3(5):355–366. doi: 10.1016/j.cmet.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 11.Kalyani RR, Metter EJ, Egan J, Golden SH, Ferrucci L. Hyperglycemia predicts persistently lower muscle strength with aging. Diabetes Care. 2015;38(1):82–90. doi: 10.2337/dc14-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mori H, et al. Advanced glycation end-products are a risk for muscle weakness in Japanese patients with type 1 diabetes. J Diabetes Investig. 2017;8(3):377–382. doi: 10.1111/jdi.12582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yoon JW, et al. Hyperglycemia is associated with impaired muscle quality in older men with diabetes: The Korean Longitudinal Study on Health and Aging. Diabetes Metab J. 2016;40(2):140–146. doi: 10.4093/dmj.2016.40.2.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray S, et al. Regulation of gluconeogenesis by Krüppel-like factor 15. Cell Metab. 2007;5(4):305–312. doi: 10.1016/j.cmet.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takashima M, et al. Role of KLF15 in regulation of hepatic gluconeogenesis and metformin action. Diabetes. 2010;59(7):1608–1615. doi: 10.2337/db09-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takeuchi Y, et al. KLF15 enables rapid switching between lipogenesis and gluconeogenesis during fasting. Cell Rep. 2016;16(9):2373–2386. doi: 10.1016/j.celrep.2016.07.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mori T, et al. Role of Krüppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J Biol Chem. 2005;280(13):12867–12875. doi: 10.1074/jbc.M410515200. [DOI] [PubMed] [Google Scholar]
- 18.Jeyaraj D, et al. Klf15 orchestrates circadian nitrogen homeostasis. Cell Metab. 2012;15(3):311–323. doi: 10.1016/j.cmet.2012.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shimizu N, et al. Crosstalk between glucocorticoid receptor and nutritional sensor mTOR in skeletal muscle. Cell Metab. 2011;13(2):170–182. doi: 10.1016/j.cmet.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 20.Bothe GW, Haspel JA, Smith CL, Wiener HH, Burden SJ. Selective expression of Cre recombinase in skeletal muscle fibers. Genesis. 2000;26(2):165–166. doi: 10.1002/(SICI)1526-968X(200002)26:2<165::AID-GENE22>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 21.Zhang X, Srinivasan SV, Lingrel JB. WWP1-dependent ubiquitination and degradation of the lung Krüppel-like factor, KLF2. Biochem Biophys Res Commun. 2004;316(1):139–148. doi: 10.1016/j.bbrc.2004.02.033. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, et al. Human Kruppel-like factor 5 is a target of the E3 ubiquitin ligase WWP1 for proteolysis in epithelial cells. J Biol Chem. 2005;280(50):41553–41561. doi: 10.1074/jbc.M506183200. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, et al. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. J Clin Invest. 1999;103(1):27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vallon V, Thomson SC. Targeting renal glucose reabsorption to treat hyperglycaemia: the pleiotropic effects of SGLT2 inhibition. Diabetologia. 2017;60(2):215–225. doi: 10.1007/s00125-016-4157-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhi X, Chen C. WWP1: a versatile ubiquitin E3 ligase in signaling and diseases. Cell Mol Life Sci. 2012;69(9):1425–1434. doi: 10.1007/s00018-011-0871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsumoto H, et al. The ubiquitin ligase gene (WWP1) is responsible for the chicken muscular dystrophy. FEBS Lett. 2008;582(15):2212–2218. doi: 10.1016/j.febslet.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 27.Imamura M, Nakamura A, Mannen H, Takeda S. Characterization of WWP1 protein expression in skeletal muscle of muscular dystrophy chickens. J Biochem. 2016;159(2):171–179. doi: 10.1093/jb/mvv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.