Abstract
Dormant cancer cells known as disseminated tumor cells (DTCs) are often present in bone marrow of breast cancer patients. These DTCs are thought to be responsible for the incurable recurrence of breast cancer. The mechanism underlying the long-term maintenance of DTCs remains unclear, however. Here, we show that Fbxw7 is essential for the maintenance of breast cancer dormancy. Genetic ablation of Fbxw7 in breast cancer cells disrupted the quiescence of DTCs, rendering them proliferative, in mouse xenograft and allograft models. Fbxw7-deficient DTCs were significantly depleted by treatment with paclitaxel, suggesting that cell proliferation induced by Fbxw7 ablation sensitized DTCs to chemotherapy. The combination of Fbxw7 ablation and chemotherapy reduced the number of DTCs even when applied after tumor cell dissemination. Mice injected with Fbxw7-deficient cancer cells survived longer after tumor resection and subsequent chemotherapy than did those injected with wild-type cells. Furthermore, database analysis revealed that breast cancer patients whose tumors expressed FBXW7 at a high level had a poorer prognosis than did those with a low FBXW7 expression level. Our results suggest that a wake-up strategy for DTCs based on Fbxw7 inhibition might be of value in combination with conventional chemotherapy for the treatment of breast cancer.
Keywords: Oncology
Keywords: Breast cancer, Cell cycle, Ubiquitin-proteosome system
Fbxw7 inhibition in combination with chemotherapy is a promising strategy to eradicate DTCs and potentially prolong the overall survival of patients with breast cancer.
Introduction
Cancer is a major public health problem worldwide (1). Although new systemic therapies and improvements in surgery have had a substantial impact on disease outcome in recent years, many cancer patients still develop metastases, the major cause of cancer-related deaths. Control of metastasis is thus one of the most important challenges faced by oncologists (2). Tumor cells that have already arrived at remote organs but not yet formed clinically evident metastatic lesions are known as disseminated tumor cells (DTCs) (3), with most DTCs in patients having been detected as quiescent single cells (4). Among several organs in which DTCs are found, much attention has focused on bone marrow (BM), given that DTCs in BM are largely nonproliferative and persist for long periods (5). Dissemination has already occurred in many patients at the time of diagnosis, with DTCs having been detected in BM of approximately 40% of newly diagnosed breast cancer patients (6). Early dissemination has also been observed in various animal models (7, 8).
Breast cancer patients with DTCs in their BM have a shorter disease-free period and poorer prognosis compared with DTC-negative patients (9, 10). In addition, the presence of DTCs in BM is associated with local recurrence of breast cancer (6), a relationship that has also been suggested experimentally (11). Nevertheless, direct evidence that DTCs in BM are the source of recurrent or metastatic lesions has not been obtained as a result of the inability to monitor individual tumor cells over long periods. On the other hand, several lines of indirect evidence have emerged. For instance, dormant DTCs retrieved from metastasis-free mouse organs were shown to regain tumorigenic and metastatic potency after transplantation into another animal (12). Whole-genome sequencing technology has also suggested that approximately 80% of metastatic lesions are derived from DTCs in breast cancer patients (13). The eradication of dormant DTCs might thus be expected to reduce the incidence of both tumor relapse and metastasis.
Most conventional anticancer drugs target proliferating cells and are therefore ineffective against DTCs as a result of their low proliferation rate (14). Two potential alternative approaches are to attenuate the dormancy of DTCs and thereby convert the quiescent cells into proliferative, chemosensitive cells, or to reinforce the machinery that maintains DTCs dormant (15). An understanding of the mechanisms by which tumor cell dormancy is maintained would thus be expected to lead to the development of therapeutic options for preventing relapse in patients with a history of cancer (16). Although such mechanisms have remained largely obscure, several secreted factors derived from the tumor microenvironment, including TGF-β and BMP, have been shown to maintain the quiescence of DTCs (17). The extracellular matrix also influences the dormant state of DTCs, as evidenced by the finding that DTCs in the lung are reactivated relatively rapidly in response to persistent integrin signaling triggered by the surrounding fibrous microenvironment (16, 18). The quiescence of DTCs in BM is supported by the hematopoietic stem cell niche (19), with various cytokines such as CXCL12 derived therefrom having been shown to contribute to maintenance of the stationary phase of DTCs (20). Indeed, DTCs enter peripheral blood in response to the administration of G-CSF or an inhibitor of CXCL12/CXCR4 signaling in some experimental settings (19). Cell-intrinsic mechanisms such as activation of p38 MAPK or the unfolded protein response (or both) were also shown to promote the survival of dormant DTCs (3). Of note, many molecular programs that promote dormancy of DTCs are thought to be identical to those that regulate self-renewal in adult stem cells (21).
Fbxw7 is the substrate recognition component of a Skp1–Cul1–F box–type (SCF-type) E3 ubiquitin ligase that is responsible for the ubiquitylation and consequent proteasomal degradation of positive regulators of the cell cycle including cyclin E and c-Myc (22). Fbxw7 restrains the cell cycle through the orchestrated degradation of these positive regulators. Fbxw7 is highly expressed in various types of stem cells in which dormancy is required and controls cell cycle entry in vivo (23). In the context of chronic myeloid leukemia (CML), in which leukemia stem cells (LSCs) are maintained dormant like DTCs of breast cancer, we and others previously showed that c-Myc accumulates as a result of Fbxw7 deletion in CML cells and forces quiescent LSCs to enter the cell cycle (24, 25). Fbxw7-null LSCs are sensitive to cytosine arabinoside or imatinib, both of which are widely administered for the treatment of CML, and the combination of Fbxw7 ablation and either of these anticancer drugs resulted in efficient eradication of LSCs and reduced the rate of relapse in mouse models of CML. It has been unclear, however, whether such an approach might also prove effective against cancer stem cells in solid tumors or against DTCs, both of which are also largely quiescent.
In the present study, we aimed to develop a new treatment strategy to target dormant DTCs in breast cancer. We show that expression of the Fbxw7 gene is upregulated in dormant breast cancer cells and that its disruption results in a purge of tumor cells from the quiescent state, rendering them susceptible to chemotherapy. Analysis of clinical data retrieved from The Cancer Genome Atlas (TCGA) also revealed that breast cancer patients with a high level of FBXW7 expression in the primary tumor had a poorer prognosis compared with those with a low level of FBXW7 expression. We propose that inhibition of Fbxw7 in combination with chemotherapy is a promising strategy to eradicate DTCs and thereby to prolong the overall survival of patients with breast cancer.
Results
FBXW7 is highly expressed in quiescent human breast cancer cells.
To identify gene sets most differentially expressed in quiescent DTCs relative to primary breast cancer cells, we exploited a previously published experimental data set (26) in which gene expression patterns were compared at the single-cell level between primary breast tumor cells and quiescent metastatic cancer cells of patient-derived xenograft models. Gene ontology (GO) enrichment analysis of differentially expressed genes in the data set identified “regulation of cell proliferation” (GO: 0042127) as the most enriched gene set in the disseminated cells (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.125138DS1). The expression of CDKN1B, which encodes the cyclin-dependent kinase inhibitor p27Kip1 (27), was increased 7.3-fold in DTCs compared with the primary tumor cells, whereas that of MYC, which encodes a key driver of the cell cycle (28), was markedly suppressed in DTCs (Supplemental Figure 1B). These results suggested that regulation of the cell cycle is pivotal in DTCs. We next focused on Fbxw7, given that its primary function is to degrade multiple proteins, including c-Myc, that promote cell cycle progression (29–31), and that its ablation in LSCs disrupts their quiescence and thereby renders them sensitive to imatinib (24, 25). Given that, like normal hematopoietic stem cells or LSCs, DTCs of breast cancer are maintained in a dormant state and reside in BM (16), we hypothesized that Fbxw7 might be indispensable for maintaining the quiescence of DTCs.
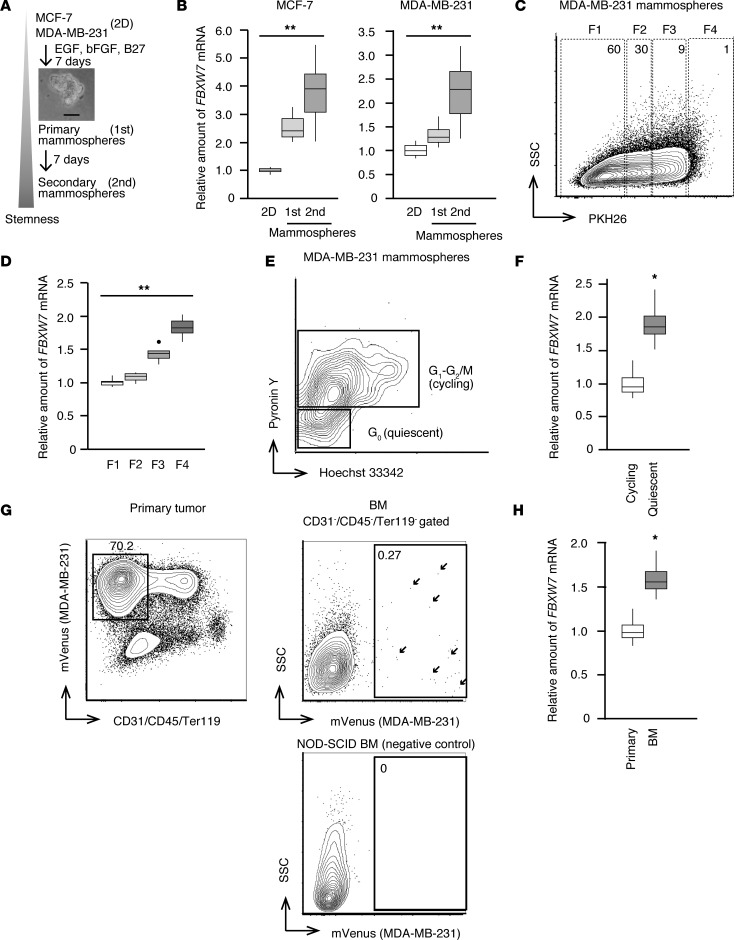
We thus examined FBXW7 expression in quiescent mammary tumor cells. The human breast cancer cell lines MCF-7 and MDA-MB-231 were induced to form mammospheres in order to enrich quiescent stem cells in vitro (32). Primary mammospheres were collected 7 days after the onset of culture and were then either processed for RNA extraction or reseeded for formation of secondary mammospheres during culture for an additional 7 days before RNA extraction (Figure 1A). Reverse transcription (RT) and real-time PCR analysis revealed that FBXW7 expression was upregulated in the primary mammospheres (which reflect both stem cell and progenitor cell states) and further increased in the secondary mammospheres (which reflect the stem cell state) of both cell lines compared with the corresponding cells maintained in 2D culture (Figure 1B).
Figure 1. Preferential expression of FBXW7 in quiescent breast cancer cells.
(A) Scheme for generation of human cell line–derived mammospheres. Cells grown in conventional 2D culture were transferred to ultralow-attachment dishes with the indicated supplements and allowed to form primary (1st) and secondary (2nd) mammospheres. Scale bar indicates 50 μm in the phase-contrast micrograph of primary mammospheres. (B) Primary and secondary mammospheres, as well as cells maintained in the 2D culture condition, were harvested and subjected to RT and real-time PCR analysis of FBXW7 expression (n = 6 independent experiments). (C) PKH26-labeled MDA-MB-231 cells were allowed to form mammospheres for 7 days and then isolated by FACS. Cells in fraction 4 (F4) retain PKH26 to the greatest extent and therefore constitute a slowly dividing population, whereas rapidly growing cells in fraction 1 (F1) had lost most of the PKH26 dye. SSC, side scatter. (D) RT and real-time PCR analysis of FBXW7 mRNA abundance in cell fractions isolated as in C (n = 4 independent experiments). (E) MDA-MB-231–derived mammospheres (day 7) were stained with Hoechst 33342 and pyronin Y for isolation by FACS of quiescent (G0) and cycling (G1-G2/M) populations. (F) RT and real-time PCR analysis of FBXW7 mRNA abundance in quiescent and cycling cell populations isolated as in E (n = 5 independent experiments). (G) MDA-MB-231 cells expressing mVenus were injected into the fourth mammary fat pad of NOD-SCID immunodeficient mice. Primary tumors and BM were isolated from mice when the primary tumor had achieved a diameter of 1 cm and were isolated by FACS as indicated. Arrows indicate DTCs in BM. (H) RT and real-time PCR analysis of FBXW7 mRNA abundance in primary tumor cells and DTCs of BM isolated as in G (n = 4 recipient mice). Box-and-whisker plots show the median, the lower and upper quartiles, 1.5× the interquartile range, and outliers. *P < 0.05, **P < 0.01 by Kruskal-Wallis test (B and D) or Mann-Whitney U test (F and H).
To further enrich quiescent cells in mammospheres, we labeled MDA-MB-231 cells with PKH26, a lipophilic dye that stains slowly dividing cells (33), and then allowed the cells to form mammospheres during culture for 7 days. Label-retaining (quiescent) cells and other cell fractions were isolated by FACS (Figure 1C) and analyzed by quantitative RT-PCR (Figure 1D). The FBXW7 expression level was significantly greater in the slow-cycling fraction (F4) than in rapidly growing populations (F1 and F2). Mammosphere cells were also stained with Hoechst 33342 and pyronin Y (Figure 1E), and those in G0 phase of the cell cycle isolated by FACS were found to express FBXW7 at a higher level than cycling cells (Figure 1F).
We then examined whether FBXW7 is also highly expressed in quiescent breast tumor cells in vivo. Given that MDA-MB-231 cells readily metastasize to various tissues (5) including BM, where DTCs are maintained for long periods in the quiescent state, we established MDA-MB-231 cells that express the fluorescent marker protein mVenus and then injected them orthotopically into NOD-SCID immunodeficient mice in order to allow monitoring of DTCs in vivo. Primary tumor cells as well as DTCs in BM were harvested and analyzed by FACS (Figure 1G). mVenus signals of DTCs were detected in BM of the injected mice, although the frequency of such cells was extremely low. The expression level of FBXW7 was significantly higher in DTCs of BM than in primary tumor cells (Figure 1H). Our observations thus suggested that FBXW7 is highly expressed in quiescent breast cancer cells both in vitro and in vivo.
Fbxw7 ablation induces loss of quiescence in breast tumor cells.
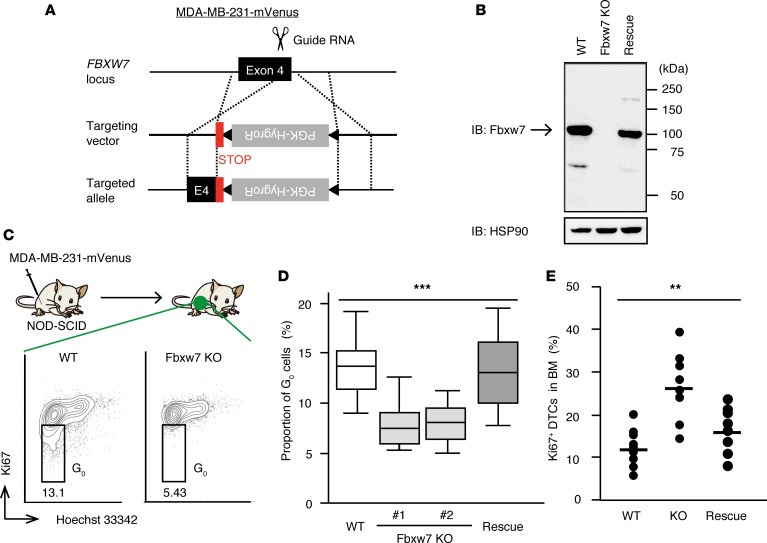
The high expression level of FBXW7 in quiescent breast tumor cells was thus consistent with the notion that Fbxw7 plays a pivotal role in the maintenance of dormancy in these cells. Given that the F-box domain of Fbxw7, which is encoded by exons 4 and 5 of FBXW7, is indispensable for formation of the SCF complex (34), we inserted a 3′ stop-codon cassette into exon 4 of the gene in MDA-MB-231 cells expressing mVenus with the use of the CRISPR-Cas9 system (Figure 2A). The truncated protein encoded by the mutant allele was not detected by immunoblot (IB) analysis with antibodies against Fbxw7 (Figure 2B), suggesting that the mutant protein was unstable. We also infected the Fbxw7-KO cells with a lentivirus containing human FBXW7 cDNA to obtain rescued cells (Figure 2B).
Figure 2. Ablation of Fbxw7 induces activation of quiescent breast cancer cells.
(A) Scheme for generation of Fbxw7-KO MDA-MB-231-mVenus cells by CRISPR-Cas9–mediated homology-directed repair. A phosphoglycerate kinase (PGK) gene promoter–hygromycin resistance gene (HygroR) cassette flanked by loxP sequences (triangles) and positioned downstream of 3 stop codons (STOP) was inserted into exon 4 (E4) of FBXW7. (B) IB analysis of Fbxw7 expression in WT MDA-MB-231-mVenus cells, Fbxw7-KO cells, and Fbxw7-KO cells infected with a lentivirus encoding human Fbxw7 (Rescue). HSP90 was examined as a loading control. (C) WT as well as Fbxw7-KO cells (2 independent lines) and Fbxw7-rescued cells were injected separately into the fourth mammary fat pad of NOD-SCID mice. Primary tumors that had achieved a diameter of 1 cm were isolated and stained with Hoechst 33342 and antibodies against Ki67 for FACS analysis of cell cycle status. (D) Quantification of quiescent (G0) cells in primary tumors as in C (n = 8 recipient mice). Box-and-whisker plots show the median, the lower and upper quartiles, and the minimum and maximum values. (E) Proportion of Ki67-positive DTCs in BM of mice as in D. Mean and individual values for the 8 recipient mice are shown. **P < 0.01, ***P < 0.001 by Kruskal-Wallis test.
The Fbxw7-KO and -rescued cells were separately injected into the fourth mammary fat pad of NOD-SCID mice, and the resulting primary tumors were resected for FACS analysis. Such analysis revealed that, compared with WT MDA-MB-231-mVenus cells, the Fbxw7-KO cells contained a significantly reduced proportion of quiescent cells, and this loss of quiescence was reversed by restoration of Fbxw7 expression (Figure 2, C and D). Furthermore, although most DTCs in BM of mice injected with parental MDA-MB-231-mVenus cells were maintained in the dormant state, DTCs derived from Fbxw7-deficient cells showed an increased proportion of cells that had entered the cell cycle and this increase was again attenuated by restoration of Fbxw7 expression (Figure 2E). These results suggested that genetic ablation of Fbxw7 induces loss of quiescence in breast cancer cells.
Fbxw7 ablation allows chemotherapy-induced eradication of DTCs.
Conventional chemotherapeutic agents kill actively cycling tumor cells but spare slow-cycling normal cells. Taxanes are the most active chemotherapeutic agents used for the treatment of metastatic breast tumors (35), with paclitaxel being frequently administered to breast cancer patients (36, 37). However, although such agents eliminate many cancer cells, a small proportion of slowly or rarely cycling tumor cells, including cancer stem cells, remains unaffected and can give rise to relapse (14, 38). Our finding that Fbxw7-deficient breast cancer cells are forced to leave the quiescent state prompted us to examine whether such cancer cells show an increased susceptibility to chemotherapeutic drugs.
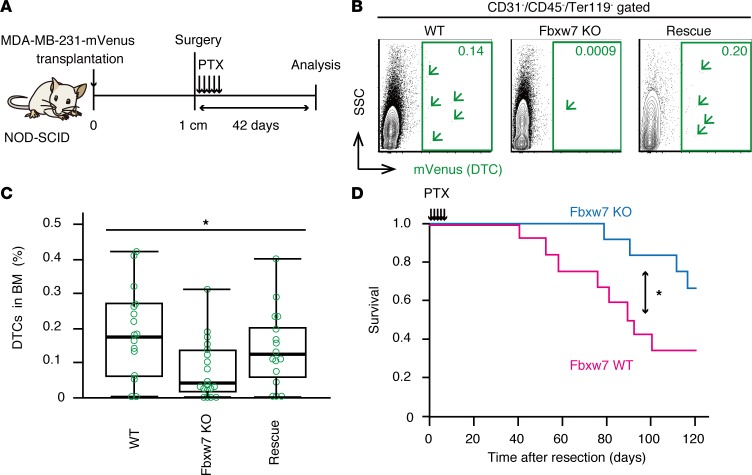
We transplanted WT, Fbxw7-KO, or Fbxw7-rescued MDA-MB-231-mVenus cells into the fourth mammary fat pad of NOD-SCID mice. The resulting primary tumors were resected when they achieved a diameter of 1 cm, and the mice were then injected i.p. with paclitaxel (10 mg/kg per day for 5 consecutive days). Mice with no evidence of recurrence at 42 days after surgery were analyzed by FACS (Figure 3A), as previously described (5). The frequency and absolute number of DTCs in BM were much smaller in recipients of Fbxw7-deficient tumor cells than in those of WT or Fbxw7-rescued cells (Figure 3, B and C). We also found that NOD-SCID mice that received Fbxw7-deficient cells survived longer after surgery and chemotherapy than did those that received WT cells (Figure 3D). These results suggested that Fbxw7-deficient DTCs forced to leave the quiescent state became sensitive to conventional chemotherapy.
Figure 3. Fbxw7-null DTCs are susceptible to conventional chemotherapy.
(A) Experimental strategy for analysis of residual DTCs. Analysis was performed 42 days after surgical resection of the primary tumor and subsequent paclitaxel (PTX) treatment. (B) FACS analysis of BM isolated from recipients of WT, Fbxw7-KO, or Fbxw7-rescued MDA-MB-231-mVenus cells treated as in A. Arrowheads indicate individual DTCs. (C) Quantification of DTCs in BM of mice as in B (n = 16 to 20 recipient mice). Box-and-whisker plots show the median, the lower and upper quartiles, and the minimum and maximum values. (D) Survival of mice after tumor resection and paclitaxel treatment (n = 12 each). *P < 0.05 by Kruskal-Wallis test (C) or log-rank test (D).
Fbxw7-deficient tumor cells are susceptible to chemotherapy in a mouse allograft model.
We also examined the effect of Fbxw7 ablation in a mouse allograft model of breast cancer based on the transplantation of E0771 breast cancer cells into C57BL/6J immunocompetent mice (39). Fbxw7 expression was increased in mammospheres formed by E0771 mouse cells (Figure 4A) and was greater in the quiescent fraction than in the cycling fraction of mammosphere cells (Figure 4, B and C), as was the case for MCF-7 and MDA-MB-231 human cells (Figure 1, B, E, and F). Furthermore, the expression level of Fbxw7 was greater in the fraction of E0771 cells showing high aldehyde dehydrogenase (ALDH) activity, in which breast cancer stem cells are enriched (40), than in those with low ALDH activity (Supplemental Figure 2, A and B).
Figure 4. Fbxw7 ablation combined with chemotherapy eliminates residual tumor cells in a mouse allograft model of breast cancer.
(A) RT and real-time PCR analysis of Fbxw7 mRNA abundance in E0771 breast cancer cells maintained in 2D culture as well as in primary and secondary mammospheres (day 7) formed by these cells (n = 4 independent experiments). (B) FACS analysis of E0771-derived mammospheres (day 7) stained with Hoechst 33342 and pyronin Y. (C) RT and real-time PCR analysis of Fbxw7 mRNA abundance in the cycling and quiescent cell fractions isolated as in B (n = 5 independent experiments). (D) IB analysis of Fbxw7 and c-Myc in Fbxw7-KO E0771 cells generated with the CRISPR-Cas9 system as well as in WT cells. Accumulation of c-Myc in the KO cells was consistent with Fbxw7 ablation. Asterisks indicate nonspecific bands. (E) Experimental strategy for analysis of residual tumor cells in the lung of C57BL/6J mice injected with equal numbers (2.5 × 105 each) of WT E0771 cells expressing mVenus and Fbxw7-KO cells expressing tdTomato into the fourth mammary fat pad. The primary tumors were removed at day 21, and the mice were then treated with paclitaxel (PTX) or vehicle for 5 days and analyzed at day 30. (F) FACS analysis of remaining tumor cells in the lung of mice as in E. Arrows indicate individual residual tumor cells. (G) Proportion of WT (mVenus) or Fbxw7-KO (tdTomato) chemotherapy-resistant cells remaining in the lung. (H) Survival analysis for mice (n = 8 each) injected with WT or Fbxw7-KO E0771 cells (5 × 105) separately and then treated with PTX as in E. Box-and-whisker plots show the median, the lower and upper quartiles, 1.5× the interquartile range, and outliers. *P < 0.05 by Kruskal-Wallis test (A), Mann-Whitney U test (C), or log-rank test (H).
We next examined the effect of Fbxw7 deletion on chemoresistance. The Fbxw7 gene was ablated in E0771 cells with the use of the CRISPR-Cas9 system (Figure 4D), and the WT and KO cells were then infected with lentiviruses encoding mVenus or tdTomato, respectively. We mixed equal numbers (2.5 × 105 cells each) of WT-mVenus and Fbxw7-KO–tdTomato E0771 cells and transplanted them into the fourth mammary fat pad of C57BL/6J mice. Primary tumors were surgically resected 21 days after cell injection, and the mice were then injected i.p. with paclitaxel or vehicle for 5 consecutive days (Figure 4E). At day 30, cells in the lung were examined by FACS (Figure 4F). A substantial number of Fbxw7-KO–tdTomato E0771 cells was detected in the lung of vehicle-treated mice, whereas no or only a few such cells were recovered from the lung of paclitaxel-treated mice (Figure 4G). On the other hand, WT-mVenus cells were detected in the lung of both control and paclitaxel-treated mice, suggesting that the Fbxw7-deficient tumor cells were more susceptible to chemotherapy than were the control cells. Mice injected with Fbxw7-deficient E0771 cells alone and treated with paclitaxel survived significantly longer than did those injected with control cells and treated with the drug (Figure 4H). FACS analysis at day 150 revealed that 3 of the 5 surviving mice injected with Fbxw7-KO–tdTomato cells had no remaining such cells in the lung (Supplemental Figure 2C), indicative of the eradication of Fbxw7-deficient tumor cells by conventional chemotherapy.
Breast cancer patients with a low level of FBXW7 expression survive longer than those with a high level.
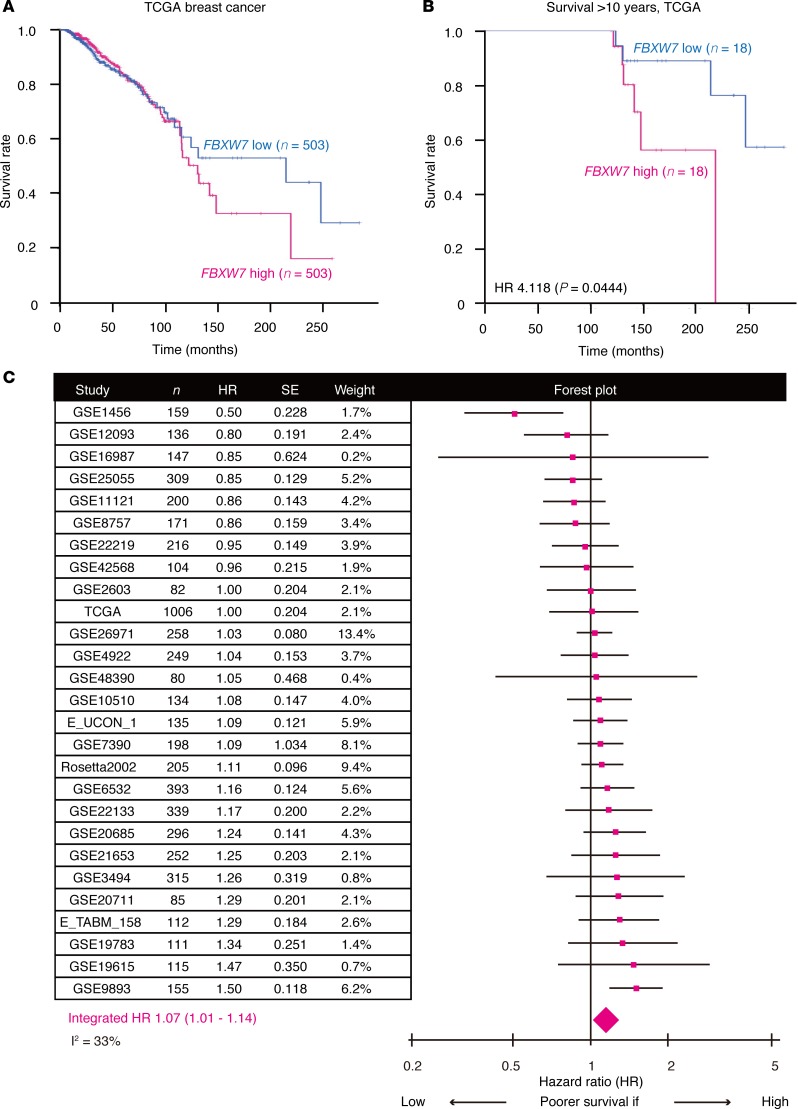
Although Fbxw7 was first identified as a tumor suppressor (41), recent studies have shown that it can also play an oncogenic role depending on the cellular context (42, 43). Our present results showed that Fbxw7 ablation induces quiescent tumor cells to enter the cell cycle and thereby renders them susceptible to conventional chemotherapy in 2 independent mouse models. To extend our investigation to the clinical setting, we retrieved clinical and gene expression data for breast cancer patients from TCGA for analysis of the relationship between FBXW7 expression level and overall survival. Kaplan-Meier curves (Figure 5A) showed that patients with tumors expressing FBXW7 at a level lower than the median (n = 503) tended to survive longer than did those with tumors expressing the gene at a level higher than the median (n = 503). This survival difference was more pronounced between those patients in the top 20% for FBXW7 expression level (n = 202) and the remaining patients (n = 804), with no patient in the FBXW7-high cohort surviving for more than 20 years (Supplemental Figure 3A). Allocation of patients that survived for more than 2.5, 5.0, 7.5, or 10 years to FBXW7-low or FBXW7-high groups based on the median expression level as previously reported (44) revealed a statistically significant difference in survival between the 2 groups for 7.5- and 10-year survivors (Figure 5B and Supplemental Figure 3, B–D), suggesting that expression of FBXW7 is associated with long-term prognosis in breast cancer patients.
Figure 5. FBXW7 expression status is associated with overall prognosis of breast cancer patients.
(A) Kaplan-Meier curves for breast cancer patients in the TCGA data set with tumors expressing FBXW7 at levels lower or higher than the median. (B) Kaplan-Meier curves for 10-year breast cancer survivors in the TCGA data set according to FBXW7 expression level. The P value was determined with the log-rank test. (C) Meta-analysis of breast cancer patients showing a significant impact of FBXW7 expression status on overall survival. The estimated HR (pink squares) and 95% CI are shown for each study. The integrated HR (pink diamond) and 95% CI were 1.07 and 1.01 to 1.14, respectively. SE, standard error.
We also examined 26 additional international breast cancer cohorts published previously (45). Most of these cohorts did not show a significant association between FBXW7 expression status and overall survival in terms of the hazard ratio (HR, Figure 5C), possibly as a result in part of the limited numbers of patients and the fact that FBXW7 expression level did not affect short-term prognosis in the TCGA data set (Figure 5A). However, a meta-analysis (n = 5,962) of these cohorts revealed that the integrated HR was 1.07 (95% CI, 1.01–1.14), indicating that FBXW7-high patients had a significantly poorer prognosis compared with their FBXW7-low counterparts (Figure 5C). This observation further reinforced the notion that FBXW7 expression level affects overall survival of breast cancer patients. We thus conclude that Fbxw7 plays a pivotal role in the maintenance of slow-cycling, chemotherapy-resistant tumor cells not only in mouse models but also in human breast cancer patients.
Wake-up approach to eradication of chemoresistant breast cancer cells.
We finally evaluated the efficacy of a therapy targeting Fbxw7 for breast cancer. Given that inhibitors of Fbxw7 have not been developed to date, we established an MDA-MB-231-mVenus cell line in which FBXW7 can be deleted by Cre recombinase in an inducible manner (Supplemental Figure 4A). Disruption of FBXW7 by Cre induction was confirmed in these cells both in vitro and in vivo (Supplemental Figure 4, B and C). We transplanted the cells into the fourth mammary fat pad of NOD-SCID mice and subsequently removed the primary tumors by surgical resection as well as treated the mice first with tamoxifen (100 mg/kg per day on 5 consecutive days) to delete FBXW7 in DTCs and then with paclitaxel (Figure 6A). At 42 days after surgery, the mice were analyzed for the number of DTCs remaining in BM. Although the number of such cells did not differ significantly between vehicle- and paclitaxel-treated mice not injected with tamoxifen, it was significantly reduced by paclitaxel treatment in tamoxifen-injected mice (Figure 6, B and C). These results suggested that the combination of Fbxw7 inhibition and chemotherapy is a promising approach to improve the long-term survival of breast cancer patients through eradication of DTCs, even after these cells have migrated to BM.
Figure 6. Wake-up therapy is a promising strategy to prolong overall survival of breast cancer patients.
(A) Experimental strategy for analysis of residual DTCs at 42 days after surgical resection of primary tumors formed in NOD-SCID mice by Fbxw7 conditional KO MDA-MB-231-mVenus cells and treatment with tamoxifen (TAM) to induce FBXW7 deletion and with paclitaxel (PTX). (B and C) Representative FACS analysis (B) and quantification (C) of remaining DTCs in BM of mice (n = 9 or 10) as in A. Box-and-whisker plots show the median, the lower and upper quartiles, and the minimum and maximum values. *P < 0.05 by 1-way ANOVA. NS, not significant.
Discussion
Efforts to treat metastatic disease are hindered by the fact that metastatic cells often remain dormant for periods of years or even decades (46). Dormancy confers on tumor cells the ability to resist conventional therapies that target rapidly cycling cells and to eventually give rise to overt metastases (47). In the present study, we found that the Fbxw7 gene is highly expressed in quiescent breast cancer cells and that its deletion forced DTCs to exit the dormant state and thereby rendered them susceptible to conventional chemotherapy in both immunodeficient xenograft and immunocompetent allograft mouse models. We also found that breast cancer patients whose tumors express FBXW7 at a low level survive longer than their counterparts with a high FBXW7 expression level, with this difference in survival being especially pronounced for long-term survivors. Furthermore, we showed that the combination of Fbxw7 ablation and chemotherapy reduced the number of DTCs even when applied after tumor cell dissemination.
Although ubiquitin ligases including SCFFbxw7 have been considered to be undruggable, they have the potential to become drug targets as a result of various recent technological innovations (48). For example, a biplanar dicarboxylic acid compound has been shown to inhibit substrate recognition by Cdc4, a yeast ortholog of Fbxw7 (49). The combination of inhibitors of human Fbxw7, when developed, with conventional anticancer drugs would be expected to reduce the number of existing DTCs and thereby to improve the overall survival of breast cancer patients.
From a biological point of view, the upstream regulator of Fbxw7 expression in DTCs remains to be identified. Expression of p53 was previously shown to be increased in quiescent cells (50), and the Fbxw7 gene is a target for transcriptional regulation by p53 (41). To determine whether p53 functions as an upstream regulator of FBXW7 expression in breast cancer, we examined the amount of p53 mRNA in primary tumor cells and DTCs isolated from the MDA-MB-231 xenograft model. RT and real-time PCR analysis revealed that the abundance of p53 mRNA was indeed increased in DTCs compared with the primary breast tumor cells (Supplemental Figure 5A). This result is consistent with public data GSE70555 (26). We next treated mammospheres derived from MCF-7 or MDA-MB-231 cells with the p53 inhibitor pifithrin-α (51, 52). As a positive control, we confirmed that pifithrin-α significantly reduced the expression of CDKN1A (Supplemental Figure 5B), a well-established p53 target gene (53). In contrast, the upregulation of FBXW7 mRNA in mammospheres was not significantly affected by this drug (Supplemental Figure 5C). These results thus suggested that p53 is not a direct upstream regulator of FBXW7 expression in this context. Fbxw7 exists in several isoforms: Fbxw7β, Fbxw7γ, and the major isoform Fbxw7α (42). Various stress conditions have been shown to specifically increase Fbxw7β expression, largely in a p53-dependent manner (54). Consistent with this finding, inhibition of the Mdm2-p53 interaction resulted in marked upregulation of Fbxw7β and Fbxw7γ, but not of Fbxw7α, in HCT116 cells (55). Ablation of each Fbxw7 isoform separately in cultured cells revealed that Fbxw7α mediates most Fbxw7 functions (56). Together, these various findings suggest that Fbxw7α may play the dominant role in dormant breast cancer cells.
Expression of the Fbxw7 gene has also been shown to be regulated by miRNA-mediated silencing (57). The miRNA miR-223 has been implicated in human cancer and is commonly repressed in various tumors (58–60). Forced expression of miR-223 was found to suppress Fbxw7 expression and thereby to increase cyclin E abundance and activity, whereas depletion of miR-223 had the opposite effects in primary mouse embryonic fibroblasts, indicating that the expression of Fbxw7 can be modulated directly by miR-223 (61). A similar inverse relationship between miR-223 and FBXW7 expression was previously detected in breast cancer (62). We also examined the relationship between miR-223 expression and overall survival of breast cancer patients in TCGA with the use of PROGmiR, an internet-based bioinformatics tool that calculates and plots Kaplan-Meier curves (63). We found that miR-223–high patients tended to survive longer than did miR-223–low patients (Supplemental Figure 5D), consistent with the better prognosis of FBXW7-low breast cancer patients. We thus speculate that miR-223 might be an upstream regulator of Fbxw7 expression in DTCs.
We previously showed that c-Myc is the key substrate of Fbxw7 with regard to maintenance of the quiescence of leukemia-initiating cells (25). It is of note that the p38 MAPK signaling pathway has been shown to be activated in dormant tumor cells (3, 64, 65), and that p38 activation in turn increases c-Myc expression (66, 67). It is thus possible that upregulation of Fbxw7 in dormant cells serves to degrade c-Myc protein that has been produced in a p38-dependent manner and thereby to maintain quiescence. The c-Myc protein might therefore also be the most critical substrate of Fbxw7 in DTCs.
We also extended our survival analysis of TCGA patients to clear cell renal cell carcinoma, in which the importance of dormancy has been demonstrated in the clinical setting (68). Renal cell carcinoma patients with a high level of FBXW7 expression had a poorer prognosis compared with those with a low level of FBXW7 expression (Supplemental Figure 5E), suggesting that Fbxw7 might play a role in this tumor type similar to that in breast cancer.
Although reactivation of DTCs is a promising approach to eradication of metastatic tumor lesions, there are several potential problems with this wake-up strategy. One risk is that it might accelerate disease progression by promoting the proliferation of DTCs. This unwanted consequence might be avoided by administration of agents that inhibit Fbxw7 together with or before (or both) standard care regimens. Clinical trials of wake-up therapies for CML were performed after initial chemotherapy and in combination with such chemotherapy, with regrowth of cancer cells not having been detected under such conditions (69–71), suggesting that this strategy may not be risky when combined with currently available chemotherapeutics. Another potential risk of the wake-up strategy is the acquisition of novel mutations by the newly proliferating cells. In addition, this approach might damage not only DTCs but also normal tissue stem cells, given that these cells are also maintained in the quiescent state in most cases. Such a risk may be mitigated by the transient administration of agents that induce cell cycle entry. Clinical studies of hematologic malignancies have not revealed excessive toxicity in patients receiving a cell cycle inducer together with standard chemotherapy (69–71), suggesting that it might be possible to identify a therapeutic window at least for some such regimens.
In contrast to wake-up therapy, several agents to maintain DTC quiescence, known as hibernation therapy, have been tested for their efficacy (15). Patients must continue to use such drugs for their entire lives, however, which may result in side effects and other problems (15) as well as become a financial burden. Indeed, such problems have been recognized with regard to HAART (highly active antiretroviral therapy) for HIV infection (72), and strategies for activation and targeting of latently infected cells are under consideration (73). A combination of wake-up therapy with hibernation therapy is a potential approach that makes use of the advantages of both strategies. For example, after an initial wake-up treatment, remaining DTCs that are not eradicated with anticancer agents could be immediately induced to hibernate. Repeated cycles of wake-up and hibernation therapy might then eventually lead to the complete elimination of residual DTCs.
In summary, we have shown that Fbxw7 depletion forces breast tumor cells to exit the quiescent state and thereby sensitizes DTCs to conventional chemotherapy. Further development of this wake-up therapy, alone or in combination with hibernation therapy, may eradicate DTCs and prolong the survival of breast cancer patients.
Methods
Mice.
C57BL/6J and NOD-SCID mice were obtained from The Jackson Laboratory. Mice were injected i.p. with paclitaxel (Sigma-Aldrich) and tamoxifen (Sigma-Aldrich) at a dose of 10 mg/kg and 100 mg/kg, respectively.
Cell culture and genetic manipulation.
MDA-MB-231 cells (National Institute of Biomedical Innovation) and MCF-7 cells (ATCC) were maintained in DMEM (Sigma-Aldrich). E0771 cells (CH3 BioSystems) were cultured as described previously (39). For stable expression of mVenus, MDA-MB-231 or E0771 cells were infected with a lentivirus encoding mVenus for 2 days, after which mVenus-expressing cells were isolated with the use of a FACSAria II instrument (BD). For generation of Fbxw7-deficient MDA-MB-231 or E0771 cells with the use of the CRISPR-Cas9 system, complementary oligonucleotides corresponding to the target sequence of single guide RNAs for human FBXW7 (5′-CACCGTGCTGACCCCAAGAAGGAAT-3′ and 5′-AAACATTCCTTCTTGGGGTCAGCAC-3′) or mouse Fbxw7 (5′-CACCGCGGCTCAGACTTGTCGATAC-3′ and 5′-AAACGTATCGACAAGTCTGAGCCGC-3′) were annealed and subcloned into the BbsI site of pX330 (Addgene). Both 5′ and 3′ arms of the template vector for homology-directed repair were amplified by PCR with MDA-MB-231 genomic DNA and subcloned into pBlueScript II SK+ (Addgene) with the use of appropriate primers and restriction enzymes. The resulting template vector was introduced into cells together with the appropriate pX330-derived vector and an expression vector containing either a hygromycin resistance gene (for MDA-MB-231 cells) or a puromycin resistance gene (for E0771 cells) with the use of Lipofectamine LTX reagent (Roche), and the cells were then selected in medium containing hygromycin (1,000 μg/ml) or puromycin (5 μg/ml), respectively. The cells were subsequently transferred at low dilution to 96-well plates in order to obtain single cell–derived clones that no longer expressed Fbxw7. Fbxw7-deficient MDA-MB-231 cells were infected with a lentivirus encoding human Fbxw7 in order to rescue Fbxw7 expression. For generation of Fbxw7 conditional KO cells, we infected Fbxw7-deficient MDA-MB-231-mVenus cells with retroviruses for CreERT2 or containing FBXW7 cDNA flanked by loxP sites. The cells were treated with 10 μM 4-hydroxytamoxifen (Sigma-Aldrich) for 48 hours to induce ablation of Fbxw7.
Mammosphere formation.
Mammospheres were formed by culture of MDA-MB-231, MCF-7, or E0771 cells (2 × 103 per well) for 7 days in 6-well ultralow-attachment dishes (Corning) containing DMEM-F12 (Invitrogen) supplemented with recombinant human EGF (20 ng/ml, BD), recombinant human basic FGF (bFGF, 20 ng/ml, Corning), and B27 supplement (Thermo Fisher Scientific). Primary mammospheres were collected for generation of secondary mammospheres as described previously (32). Mammospheres were treated with pifithrin-α (Merck) at 10 or 50 μM.
PKH26 and ALDEFLUOR assays.
PKH26-positive cells and ALDHhi cells were isolated with the use of PKH26GL (Sigma-Aldrich) and ALDEFLUOR (Veritas) kits, respectively.
Tumor cell transplantation and surgery.
Tumor cells (5 × 106 MDA-MB-231 cells with Matrigel [Corning], or 5 × 105 E0771 cells) were injected into the fourth mammary fat pad of 7- to 8-week-old female NOD-SCID or C57BL/6J mice, respectively. The resulting primary tumor was removed together with the surrounding normal fat pad by surgical resection when the tumor had achieved a diameter of 1 cm, as described previously (5).
FACS sample preparation and analysis.
Tumor dissociation was performed as described previously (74), with minor modifications. In brief, the tumor tissue was mechanically dissociated with razor blades and digested by incubation for 1 hour at 37°C with collagenase IV (0.2 mg/ml, Worthington), dispase (2 mg/ml, Gibco), hyaluronidase (4 mg/ml, Sigma-Aldrich), and DNase I (0.1 mg/ml, Roche) in RPMI 1640 medium supplemented with 10% FBS. Cells in BM were isolated as described previously (75). Both cell suspensions were washed with PBS, and red blood cells therein were lysed with hemolysis buffer (10 mM Tris-HCl [pH 7.5], 140 mM NH4Cl). The isolated cells were then stained for 30 minutes at 4°C with antibodies against mouse CD31 (BD, 553371), CD45 (eBioscience, 13-0451-82), and Ter119 (BD, 553672). Dead cells were excluded with the use of a LIVE/DEAD Fixable Near-IR Dead Cell Stain Kit (Invitrogen). For assay of quiescent cells, cells were stained at 37°C first with Hoechst 33342 (Sigma-Aldrich) and 5 μM verapamil (Sigma-Aldrich) for 40 minutes and then in the additional presence of pyronin Y (Sigma-Aldrich) at 1 μg/ml for 20 minutes. For Ki67 staining, cells were fixed with 4% paraformaldehyde for 10 minutes and then stained for 30 minutes at 4°C with antibodies against Ki67 (BioLegend, 652407) in permeabilization buffer (PBS containing 0.5% Triton X-100). All stained cells were analyzed with a FACSVerse flow cytometer (BD) or sorted with a FACSAria II instrument.
RT and real-time PCR analysis.
Total RNA extracted from cells with the use of Isogen (Nippon Gene) was purified and then subjected to RT with random hexanucleotide primers with the use of a QuantiTect RT kit (Qiagen). The resulting cDNA was analyzed by real-time PCR as described previously (25). The sequences of the primers (sense and antisense, respectively) were 5′-CCACTGGGCTTGTACCATGTT-3′ and 5′-CAGATGTAATTCGGCGTCGTT-3′ for human FBXW7, 5′-CAGCACATGACGGAGGTTGT-3′ and 5′-TCATCCAAATACTCCACACGC-3′ for human TP53, 5′-TGTCCGTCAGAACCCATGC-3′ and 5′-AAAGTCGAAGTTCCATCGCTC-3′ for human CDKN1A, 5′-ATGGACAGGACTGAACGTCTTGCT-3′ and 5′-TTGAGCACACAGAGGGCTACAATG-3′ for human hypoxanthine phosphoribosyltransferase gene (HPRT), 5′-TGCAAAGTCTCAGATTATACC-3′ and 5′-ACTTCTCTGGTCCGCTCCAGC-3′ for mouse Fbxw7, and 5′-ATGCTCCCCGGGCTGTAT-3′ and 5′-CATAGGAGTCCTTCTGACCCATTC-3′ for mouse β-actin gene. Results were normalized by the corresponding amount of HPRT (human) or β-actin (mouse) mRNA.
Immunofluorescence analysis.
Bone tissue was fixed with 4% paraformaldehyde in PBS, decalcified in 10% EDTA, embedded in 30% sucrose overnight, sectioned at a thickness of 10 μm with a cryostat, and stained as described previously (39). Antibodies against MKI67 were from Thermo Fisher Scientific (RM-9106-S1). Immune complexes were detected with secondary antibodies labeled with Alexa Fluor 546 (Molecular Probes, A-11035) at a dilution of 1:2,000. The sections were mounted in VECTASHIELD medium (Vector Laboratories) and examined with a laser-scanning confocal microscope (LSM700, Carl Zeiss).
IB analysis.
Total protein extracts were prepared from MDA-MB-231 and E0771 cells and were analyzed by IB as previously described (25). Antibodies against Fbxw7 were obtained from Bethyl Laboratories (A301-720A), those against c-Myc were from Abcam (ab32072), and those against HSP90 (loading control) were from BD Biosciences (610419).
GO analysis.
The experimental data set GSE70555 was downloaded and analyzed as described previously (26). Differentially expressed genes were uploaded to the GO database (http://www.geneontology.org), and the most significant GO terms for all 3 categories were identified.
Human clinical information.
The breast cancer and renal cell carcinoma data sets were obtained from TCGA cBioPortal (http://www.cbioportal.org). Kaplan-Meier curves were plotted with the use of the survival R package (CRAN repository). HRs were recalculated for previously described studies (45) with the use of the survival R package, and the integrated HR was calculated with the use of RevMan5 software (Cochrane Community). We used the PROGmiR internet tool (63) for miRNA analysis.
Statistics.
Statistical analysis was performed with the nonparametric Mann-Whitney U test or ANOVA, with the exception that survival curves were compared with the log-rank nonparametric test. All statistical analysis was performed with R software. P values were adjusted by the Holm method for multiple comparisons. A P value of <0.05 was considered statistically significant.
Study approval.
All mouse experiments were approved by the Animal Ethics Committee of Kyushu University, and animal care was in accordance with institutional guidelines.
Author contributions
HS, ST, and KIN designed the study. HS performed all experiments. HS, ST, HN, and KIN interpreted the data. HS and KIN wrote the manuscript. KIN contributed to supervision of the entire study.
Supplementary Material
Acknowledgments
We thank T. Higa, K. Yumimoto, A. Niihara, K. Tsunematsu, and E. Koba for technical assistance; Y. Yamauchi for statistical advice; laboratory members for comments on the manuscript; and A. Ohta for help in preparation of the manuscript. This work was supported in part by a Japan Society for the Promotion of Science (JSPS) Fellowship to HS and by KAKENHI grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan to KIN (18H05215, 17H06301, and 25221303).
Version 1. 02/21/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(4):e125138. https://doi.org/10.1172/jci.insight.125138.
Contributor Information
Hideyuki Shimizu, Email: h_shimizu@bioreg.kyushu-u.ac.jp.
Shoichiro Takeishi, Email: shoichiro.takeishi@einstein.yu.edu.
Hirokazu Nakatsumi, Email: nakatsumi@phar.nagoya-cu.ac.jp.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging biological principles of metastasis. Cell. 2017;168(4):670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kang Y, Pantel K. Tumor cell dissemination: emerging biological insights from animal models and cancer patients. Cancer Cell. 2013;23(5):573–581. doi: 10.1016/j.ccr.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naumov GN, et al. Persistence of solitary mammary carcinoma cells in a secondary site: a possible contributor to dormancy. Cancer Res. 2002;62(7):2162–2168. [PubMed] [Google Scholar]
- 5.Ghajar CM, et al. The perivascular niche regulates breast tumour dormancy. Nat Cell Biol. 2013;15(7):807–817. doi: 10.1038/ncb2767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braun S, et al. A pooled analysis of bone marrow micrometastasis in breast cancer. N Engl J Med. 2005;353(8):793–802. doi: 10.1056/NEJMoa050434. [DOI] [PubMed] [Google Scholar]
- 7.Hüsemann Y, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13(1):58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Rhim AD, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–361. doi: 10.1016/j.cell.2011.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Janni W, et al. Persistence of disseminated tumor cells in the bone marrow of breast cancer patients predicts increased risk for relapse--a European pooled analysis. Clin Cancer Res. 2011;17(9):2967–2976. doi: 10.1158/1078-0432.CCR-10-2515. [DOI] [PubMed] [Google Scholar]
- 10.Hartkopf AD, et al. Disseminated tumor cells from the bone marrow of patients with nonmetastatic primary breast cancer are predictive of locoregional relapse. Ann Oncol. 2015;26(6):1155–1160. doi: 10.1093/annonc/mdv148. [DOI] [PubMed] [Google Scholar]
- 11.Kim MY, et al. Tumor self-seeding by circulating cancer cells. Cell. 2009;139(7):1315–1326. doi: 10.1016/j.cell.2009.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki M, Mose ES, Montel V, Tarin D. Dormant cancer cells retrieved from metastasis-free organs regain tumorigenic and metastatic potency. Am J Pathol. 2006;169(2):673–681. doi: 10.2353/ajpath.2006.060053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosseini H, et al. Early dissemination seeds metastasis in breast cancer. Nature. 2016;540(7634):552–558. doi: 10.1038/nature20785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun S, et al. Lack of effect of adjuvant chemotherapy on the elimination of single dormant tumor cells in bone marrow of high-risk breast cancer patients. J Clin Oncol. 2000;18(1):80–86. doi: 10.1200/JCO.2000.18.1.80. [DOI] [PubMed] [Google Scholar]
- 15.Ghajar CM. Metastasis prevention by targeting the dormant niche. Nat Rev Cancer. 2015;15(4):238–247. doi: 10.1038/nrc3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aguirre-Ghiso JA, Bragado P, Sosa MS. Metastasis awakening: targeting dormant cancer. Nat Med. 2013;19(3):276–277. doi: 10.1038/nm.3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao H, et al. The BMP inhibitor Coco reactivates breast cancer cells at lung metastatic sites. Cell. 2012;150(4):764–779. doi: 10.1016/j.cell.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao H, et al. Multi-organ site metastatic reactivation mediated by non-canonical discoidin domain receptor 1 signaling. Cell. 2016;166(1):47–62. doi: 10.1016/j.cell.2016.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shiozawa Y, et al. Human prostate cancer metastases target the hematopoietic stem cell niche to establish footholds in mouse bone marrow. J Clin Invest. 2011;121(4):1298–1312. doi: 10.1172/JCI43414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Domanska UM, et al. A review on CXCR4/CXCL12 axis in oncology: no place to hide. Eur J Cancer. 2013;49(1):219–230. doi: 10.1016/j.ejca.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Giancotti FG. Mechanisms governing metastatic dormancy and reactivation. Cell. 2013;155(4):750–764. doi: 10.1016/j.cell.2013.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Onoyama I, Nakayama KI. Fbxw7 in cell cycle exit and stem cell maintenance: insight from gene-targeted mice. Cell Cycle. 2008;7(21):3307–3313. doi: 10.4161/cc.7.21.6931. [DOI] [PubMed] [Google Scholar]
- 23.Takeishi S, Nakayama KI. Role of Fbxw7 in the maintenance of normal stem cells and cancer-initiating cells. Br J Cancer. 2014;111(6):1054–1059. doi: 10.1038/bjc.2014.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reavie L, et al. Regulation of c-Myc ubiquitination controls chronic myelogenous leukemia initiation and progression. Cancer Cell. 2013;23(3):362–375. doi: 10.1016/j.ccr.2013.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takeishi S, Matsumoto A, Onoyama I, Naka K, Hirao A, Nakayama KI. Ablation of Fbxw7 eliminates leukemia-initiating cells by preventing quiescence. Cancer Cell. 2013;23(3):347–361. doi: 10.1016/j.ccr.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 26.Lawson DA, et al. Single-cell analysis reveals a stem-cell program in human metastatic breast cancer cells. Nature. 2015;526(7571):131–135. doi: 10.1038/nature15260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Susaki E, Nakayama KI. Functional similarities and uniqueness of p27 and p57: insight from a knock-in mouse model. Cell Cycle. 2009;8(16):2497–2501. doi: 10.4161/cc.8.16.9330. [DOI] [PubMed] [Google Scholar]
- 28.Bretones G, Delgado MD, León J. Myc and cell cycle control. Biochim Biophys Acta. 2015;1849(5):506–516. doi: 10.1016/j.bbagrm.2014.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Yada M, et al. Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 2004;23(10):2116–2125. doi: 10.1038/sj.emboj.7600217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welcker M, Orian A, Grim JE, Grim JA, Eisenman RN, Clurman BE. A nucleolar isoform of the Fbw7 ubiquitin ligase regulates c-Myc and cell size. Curr Biol. 2004;14(20):1852–1857. doi: 10.1016/j.cub.2004.09.083. [DOI] [PubMed] [Google Scholar]
- 31.Nakayama KI, Nakayama K. Ubiquitin ligases: cell-cycle control and cancer. Nat Rev Cancer. 2006;6(5):369–381. doi: 10.1038/nrc1881. [DOI] [PubMed] [Google Scholar]
- 32.Dontu G, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17(10):1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punzel M, Ho AD. Divisional history and pluripotency of human hematopoietic stem cells. Ann N Y Acad Sci. 2001;938:72–81. doi: 10.1111/j.1749-6632.2001.tb03576.x. [DOI] [PubMed] [Google Scholar]
- 34.Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8(2):83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- 35.Ghersi D, Willson ML, Chan MM, Simes J, Donoghue E, Wilcken N. Taxane-containing regimens for metastatic breast cancer. Cochrane Database Syst Rev. 2015;6:Cd003366. doi: 10.1002/14651858.CD003366.pub3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sachdev JC, Jahanzeb M. Use of cytotoxic chemotherapy in metastatic breast cancer: putting taxanes in perspective. Clin Breast Cancer. 2016;16(2):73–81. doi: 10.1016/j.clbc.2015.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Sparano JA, et al. Weekly paclitaxel in the adjuvant treatment of breast cancer. N Engl J Med. 2008;358(16):1663–1671. doi: 10.1056/NEJMoa0707056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oskarsson T, Batlle E, Massagué J. Metastatic stem cells: sources, niches, and vital pathways. Cell Stem Cell. 2014;14(3):306–321. doi: 10.1016/j.stem.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yumimoto K, et al. F-box protein FBXW7 inhibits cancer metastasis in a non-cell-autonomous manner. J Clin Invest. 2015;125(2):621–635. doi: 10.1172/JCI78782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Davis RJ, Welcker M, Clurman BE. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26(4):455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu W, Taranets L, Popov N. Regulating Fbw7 on the road to cancer. Semin Cancer Biol. 2016;36:62–70. doi: 10.1016/j.semcancer.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 43.Robinson DR, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. doi: 10.1038/nature23306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reddy A, et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell. 2017;171(2):481–494.e15. doi: 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Abdel-Fatah TMA, et al. SPAG5 as a prognostic biomarker and chemotherapy sensitivity predictor in breast cancer: a retrospective, integrated genomic, transcriptomic, and protein analysis. Lancet Oncol. 2016;17(7):1004–1018. doi: 10.1016/S1470-2045(16)00174-1. [DOI] [PubMed] [Google Scholar]
- 46.Sosa MS, Bragado P, Aguirre-Ghiso JA. Mechanisms of disseminated cancer cell dormancy: an awakening field. Nat Rev Cancer. 2014;14(9):611–622. doi: 10.1038/nrc3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gomis RR, Gawrzak S. Tumor cell dormancy. Mol Oncol. 2017;11:62–78. doi: 10.1016/j.molonc.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kitagawa K, Kitagawa M. The SCF-type E3 ubiquitin ligases as cancer targets. Curr Cancer Drug Targets. 2016;16(2):119–129. doi: 10.2174/1568009616666151112122231. [DOI] [PubMed] [Google Scholar]
- 49.Orlicky S, et al. An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase. Nat Biotechnol. 2010;28(7):733–737. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dai Y, et al. Activation of anaphase-promoting complex by p53 induces a state of dormancy in cancer cells against chemotherapeutic stress. Oncotarget. 2016;7(18):25478–25492. doi: 10.18632/oncotarget.8172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Komarov PG, et al. A chemical inhibitor of p53 that protects mice from the side effects of cancer therapy. Science. 1999;285(5434):1733–1737. doi: 10.1126/science.285.5434.1733. [DOI] [PubMed] [Google Scholar]
- 52.Mukhopadhyay S, Antalis TM, Nguyen KP, Hoofnagle MH, Sarkar R. Myeloid p53 regulates macrophage polarization and venous thrombus resolution by inflammatory vascular remodeling in mice. Blood. 2017;129(24):3245–3255. doi: 10.1182/blood-2016-07-727180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livneh Z. Keeping mammalian mutation load in check: regulation of the activity of error-prone DNA polymerases by p53 and p21. Cell Cycle. 2006;5(17):1918–1922. doi: 10.4161/cc.5.17.3193. [DOI] [PubMed] [Google Scholar]
- 54.Sionov RV, Netzer E, Shaulian E. Differential regulation of FBXW7 isoforms by various stress stimuli. Cell Cycle. 2013;12(22):3547–3554. doi: 10.4161/cc.26591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grinkevich VV, et al. Ablation of key oncogenic pathways by RITA-reactivated p53 is required for efficient apoptosis. Cancer Cell. 2009;15(5):441–453. doi: 10.1016/j.ccr.2009.03.021. [DOI] [PubMed] [Google Scholar]
- 56.Grim JE, et al. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J Cell Biol. 2008;181(6):913–920. doi: 10.1083/jcb.200802076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang L, Ye X, Liu Y, Wei W, Wang Z. Aberrant regulation of FBW7 in cancer. Oncotarget. 2014;5(8):2000–2015. doi: 10.18632/oncotarget.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pulikkan JA, et al. Cell-cycle regulator E2F1 and microRNA-223 comprise an autoregulatory negative feedback loop in acute myeloid leukemia. Blood. 2010;115(9):1768–1778. doi: 10.1182/blood-2009-08-240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stamatopoulos B, et al. microRNA-29c and microRNA-223 down-regulation has in vivo significance in chronic lymphocytic leukemia and improves disease risk stratification. Blood. 2009;113(21):5237–5245. doi: 10.1182/blood-2008-11-189407. [DOI] [PubMed] [Google Scholar]
- 60.Wong QW, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135(1):257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 61.Xu Y, Sengupta T, Kukreja L, Minella AC. MicroRNA-223 regulates cyclin E activity by modulating expression of F-box and WD-40 domain protein 7. J Biol Chem. 2010;285(45):34439–34446. doi: 10.1074/jbc.M110.152306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pinatel EM, et al. miR-223 is a coordinator of breast cancer progression as revealed by bioinformatics predictions. PLoS One. 2014;9(1):e84859. doi: 10.1371/journal.pone.0084859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Goswami CP, Nakshatri H. PROGmiR: a tool for identifying prognostic miRNA biomarkers in multiple cancers using publicly available data. J Clin Bioinforma. 2012;2(1):23. doi: 10.1186/2043-9113-2-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sosa MS, Avivar-Valderas A, Bragado P, Wen HC, Aguirre-Ghiso JA. ERK1/2 and p38α/β signaling in tumor cell quiescence: opportunities to control dormant residual disease. Clin Cancer Res. 2011;17(18):5850–5857. doi: 10.1158/1078-0432.CCR-10-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bragado P, et al. TGF-β2 dictates disseminated tumour cell fate in target organs through TGF-β-RIII and p38α/β signalling. Nat Cell Biol. 2013;15(11):1351–1361. doi: 10.1038/ncb2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muranen T, et al. ERK and p38 MAPK activities determine sensitivity to PI3K/mTOR inhibition via regulation of MYC and YAP. Cancer Res. 2016;76(24):7168–7180. doi: 10.1158/0008-5472.CAN-16-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frank SB, Berger PL, Ljungman M, Miranti CK. Human prostate luminal cell differentiation requires NOTCH3 induction by p38-MAPK and MYC. J Cell Sci. 2017;130(11):1952–1964. doi: 10.1242/jcs.197152. [DOI] [PubMed] [Google Scholar]
- 68.Uhr JW, Pantel K. Controversies in clinical cancer dormancy. Proc Natl Acad Sci USA. 2011;108(30):12396–12400. doi: 10.1073/pnas.1106613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Löwenberg B, et al. Effect of priming with granulocyte colony-stimulating factor on the outcome of chemotherapy for acute myeloid leukemia. N Engl J Med. 2003;349(8):743–752. doi: 10.1056/NEJMoa025406. [DOI] [PubMed] [Google Scholar]
- 70.Uy GL, et al. A phase 1/2 study of chemosensitization with the CXCR4 antagonist plerixafor in relapsed or refractory acute myeloid leukemia. Blood. 2012;119(17):3917–3924. doi: 10.1182/blood-2011-10-383406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ohno R, et al. A double-blind controlled study of granulocyte colony-stimulating factor started two days before induction chemotherapy in refractory acute myeloid leukemia. Kohseisho Leukemia Study Group. Blood. 1994;83(8):2086–2092. [PubMed] [Google Scholar]
- 72.Richman DD, Margolis DM, Delaney M, Greene WC, Hazuda D, Pomerantz RJ. The challenge of finding a cure for HIV infection. Science. 2009;323(5919):1304–1307. doi: 10.1126/science.1165706. [DOI] [PubMed] [Google Scholar]
- 73.Archin NM, et al. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487(7408):482–485. doi: 10.1038/nature11286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nakatsumi H, Matsumoto M, Nakayama KI. Noncanonical pathway for regulation of CCL2 expression by an mTORC1-FOXK1 axis promotes recruitment of tumor-associated macrophages. Cell Rep. 2017;21(9):2471–2486. doi: 10.1016/j.celrep.2017.11.014. [DOI] [PubMed] [Google Scholar]
- 75.Houlihan DD, et al. Isolation of mouse mesenchymal stem cells on the basis of expression of Sca-1 and PDGFR-α. Nat Protoc. 2012;7(12):2103–2111. doi: 10.1038/nprot.2012.125. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.