Abstract
Allergic eosinophilic asthma is a chronic condition causing airway remodeling resulting in lung dysfunction. We observed that expression of sirtuin 2 (Sirt2), a histone deacetylase, regulates the recruitment of eosinophils after sensitization and challenge with a triple antigen: dust mite, ragweed, and Aspergillus fumigatus (DRA). Our data demonstrate that IL-4 regulates the expression of Sirt2 isoform 3/5. Pharmacological inhibition of Sirt2 by AGK2 resulted in diminished cellular recruitment, decreased CCL17/TARC, and reduced goblet cell hyperplasia. YM1 and Fizz1 expression was reduced in AGK2-treated, IL-4–stimulated lung macrophages in vitro as well as in lung macrophages from AGK2-DRA–challenged mice. Conversely, overexpression of Sirt2 resulted in increased cellular recruitment, CCL17 production, and goblet cell hyperplasia following DRA challenge. Sirt2 isoform 3/5 was upregulated in primary human alveolar macrophages following IL-4 and AGK2 treatment, which resulted in reduced CCL17 and markers of alternative activation. These gain-of-function and loss-of-function studies indicate that Sirt2 could be developed as a treatment for eosinophilic asthma.
Keywords: Immunology, Pulmonology
Keywords: Asthma, Chemokines, Macrophages
An isoform of sirtuin 2, a histone deacetylase, regulates macrophage recruitment and activation in the setting of allergic inflammation.
Introduction
Macrophages are the most abundant immune effector leukocytes found in airways and the parenchyma of the noninflamed lung (1). These cells play an essential role in maintaining homeostasis (2) and protecting against pathogenic and environmental insults. In order to accomplish these important functions, macrophages are differentially activated in response to their milieu and external stimuli. Traditionally, macrophages have been characterized based on their cytokine production profile. Proinflammatory or M1 macrophages are generated in the presence of IFN-γ or LPS (3). Conversely, proasthmatic or M2 macrophages are alternatively activated in response to stimulation by the Th2 cytokines IL-4 and/or IL-13 (4). These differential activation patterns do not reflect terminal differentiation or maturation processes; rather, they represent how macrophages respond to their environment by expressing distinct gene profiles and functional capabilities (5, 6).
Our work and that of other groups has established a link between lung macrophages and type 2 eosinophilic allergic airway inflammation (7–9) and airway remodeling (8, 10) in the setting of allergic asthma. Based on these published data, we hypothesized that the Th2 cytokines IL-4 and/or IL-13 activate macrophages to drive type 2 inflammation and immunity (1, 6). While much of the extant literature has examined the immunoregulatory function of pulmonary macrophages in asthma, the mechanisms that govern the activation of macrophages and their gene expression are still emerging. Previous work from our group has identified transcription factors responsible for macrophage activation during lung injury and inflammation. We have shown that macrophage activation is dependent on the “master myeloid” transcription factor, PU.1 (11, 12). Other work has demonstrated an important role for the transcription factors STAT6, IFN regulatory factor 4 (IRF4), and Krüppel-like factor 4 (KLF4) in promoting alternatively activated, proasthmatic macrophages (13–15). Though transcriptional control of macrophage polarization is the subject of intense investigation, it is thought that the mechanism by which these cells are activated involve combinatorial interactions among transcriptional regulatory proteins, microRNA, and epigenetic mechanisms.
Sirtuins were initially described as transcriptional-silencing histone deacetylases (HDACs) in yeast (16). In mammals, 7 sirtuins (Sirt1–7) all share a similar catalytic domain and use NAD+ as a cosubstrate, but they all have different substrate affinities and subcellular compartmentalization. Although initially identified as deacetylases, sirtuins are now known to regulate a variety of enzymatic activities and biologic processes, including aging, transcription, apoptosis, inflammation, and oxidative stress (16). Class I sirtuins include Sirt1, Sirt2, and Sirt3. Previous work has demonstrated that Sirt2 deacetylates a variety of cytoplasmic and nuclear proteins and is a key modulator of cellular processes, including cell cycle, metabolic homeostasis, apoptosis, and antioxidant defense mechanisms (17–21). For example, during acute inflammation during experimental colitis, Sirt2 has been shown to regulate macrophage polarization. Macrophages isolated from Sirt2-deficient (Sirt2-KO) mice had hyperacetylation on the p65 (Rel A) subunit of NF-κB, which correlated with elevated production of proinflammatory cytokines (22).
There are several different isoforms of Sirt2 found in both human and mouse samples. Previous work has determined that these isoforms result from alternative splicing mechanisms rather than protein degradation (23–25). Sirt2 isoforms 1 and 2 are most similar and are localized in the cytoplasm under resting conditions (23, 24). It has been predicted that Sirt2 isoform 2 could be generated from isoform 1, which may explain differences in isoform expression in different tissue samples. The third Sirt2 isoform has been called isoform 3 in murine studies and isoform 5 in human studies (24). The Sirt2 isoform 3/5 is thought to be primarily localized to the nucleus, since it lacks exon 2–4 and the nuclear export sequence. Although initial reports have determined Sirt2 isoform 5 to be catalytically inactive, its function has yet to be determined.
In this manuscript, we have determined that in vitro treatment of mouse and human macrophages with IL-4 results in elevated gene expression of Sirt2 isoform 3/5. We also observed that pharmacologic ablation of Sirt2 attenuates and genetic overexpression of Sirt2 exaggerates the allergic asthmatic phenotype. These loss- and gain-of-function studies indicate that Sirt2 has an important role in regulating allergic asthmatic response that can be targeted by pharmacologic intervention.
Results
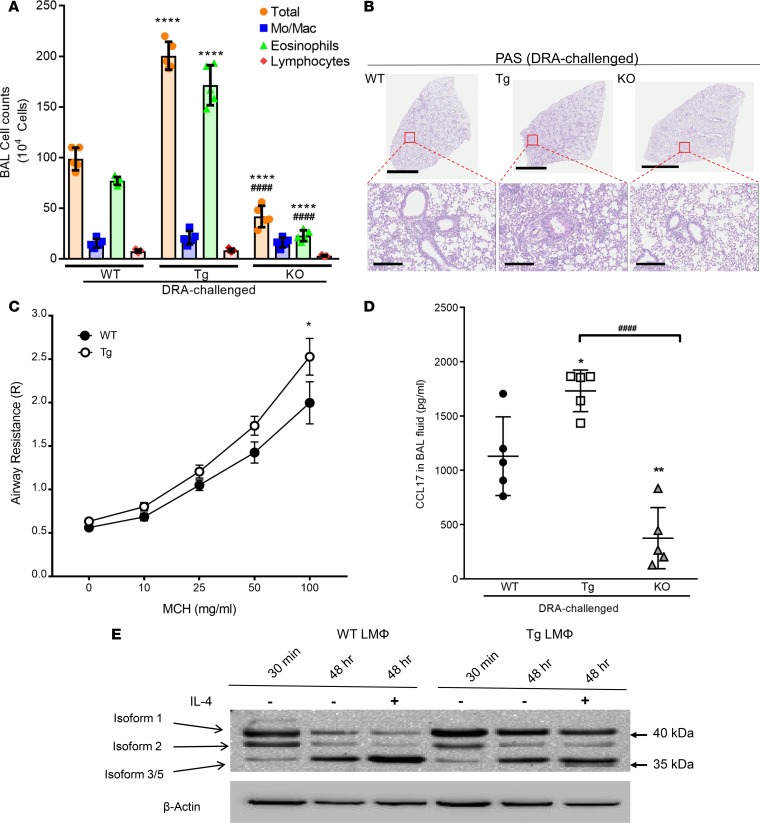
Genetic manipulation of Sirt2 expression regulates allergic inflammation in a murine model of allergen challenge.
Previous work demonstrated that Sirt2 reduces M2-associated inflammation (22). To determine the role of Sirt2 in regulating allergic inflammation, we sensitized and then challenged WT, Sirt2-KO, and Sirt2-overexpressing transgenic (Sirt2-Tg) mice with 3 combined allergens, dust mite, ragweed, and Aspergillus fumigatus (DRA) extracts (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124710DS1), as previously reported (7, 12). After challenge, there was a doubling of the total bronchoalveolar lavage (BAL) cells in the Sirt2-Tg DRA-challenged mice and an approximately 50% reduction in the total BAL cells in the Sirt2-KO DRA-challenged mice compared with WT mice (Figure 1A). The majority of the cells in all mice, regardless of Sirt2 expression, were eosinophils (Figure 1A). Lung histology demonstrated enhanced periodic acid-Schiff (PAS) staining and goblet cell hyperplasia in the Sirt2-Tg mice and reduced staining in the Sirt2-KO mice (Figure 1B). Airway hyperreactivity (AHR) was assessed in response to nebulized methacholine in WT and Sirt2-Tg DRA-challenged mice. The Sirt2-Tg mice showed a significant increase in airway resistance compared with the WT mice (Figure 1C). Cytokine array analysis demonstrated a marked increase in CCL17 (also known as TARC) in Sirt2-Tg BAL fluid and a decrease in CCL17 in Sirt2-KO BAL fluid following DRA challenge (Supplemental Figure 1B). Overall, there were elevated levels of proinflammatory cytokines, such as IFN-γ, GM-CSF, IL-1, and TNF-α, and antiinflammatory cytokines, such as IL-4 and IL-5, in the Sirt2-Tg and Sirt2-KO mice compared with WT mice. Additionally, there was a decrease in BAL MCP-1 expression in Sirt2-KO mice compared with that in Sirt2 WT and Sirt2-Tg mice. Since the effects of CCL17 vary across the different groups, these findings were validated by ELISA, which confirmed significant differences in CCL17 based on Sirt2 overexpression or deficiency (Figure 1D). Interestingly, overexpression of Sirt2 did not change CCR4 or CC17 receptor on macrophages following DRA challenge (CCR4 expression from WT DRA-challenged, 3.9 ± 1.8-fold increased, and Sirt2-Tg–challenged, 4.5 ± 1.6-fold increase). These gain- and loss-of-function data in Sirt2-Tg and Sirt2-KO mice, respectively, indicate that Sirt2 is an important regulator of allergic inflammation.
Figure 1. Sirt2 regulates allergic inflammation following DRA challenge.
WT, Sirt2-overexpressing transgenic (Tg), and Sirt2-deficient (KO) mice were DRA sensitized and challenged. (A) The total number of cells and cell differentials in BAL fluid after DRA, as determined by cytospin analysis. n = 5 mice/group; analyzed by 1-way ANOVA. (B) Whole lung histological sections were stained with periodic acid-Schiff (PAS) to determine goblet cell hyperplasia. The images are representative of 5 experiments. Scale bar: 3 mm (top); 300 μm (bottom). (C) Airway resistance was measured using increasing doses of methacholine in WT and Tg mice after DRA challenge. n = 5 mice/group; analyzed 2-way ANOVA. (D) CCL7 ELISA. n = 5/group; analyzed by 1-way ANOVA. (E) Lung macrophages from WT or Tg mice were isolated, and expression of Sirt2 isoforms was detected with either N-terminal– or C-terminal–specific antibodies at time 0 and after a 48-hour incubation in the presence or absence of rmIL-4 (20 ng/ml). Isolated macrophage samples collected from 3 mice were combined to evaluate Sirt2 expression; representative blot performed 3 times. *P < 0.05, **P < 0.01, ****P < 0.001 when compared with WT controls; ####P < 0.001 when compared within the groups.
Several different isoforms of Sirt2 have been described previously (18, 24). We sought to determine which isoform was driving the development of allergic airway inflammation in our model. To do this, we isolated lung macrophages via collagenase digestion from WT and Sirt2-Tg DRA-challenged mice and measured the expression of Sirt2 isoforms at the time of isolation or in the presence or absence of IL-4 for 48 hours. In WT lung macrophages, in vitro incubation for 48 hours resulted in a decrease in Sirt2 isoforms 1 and 2. Interestingly, IL-4–stimulated WT lung macrophages had elevated Sirt2 isoform 3/5 expression (Figure 1E). IL-4 stimulation of lung macrophages isolated from Sirt2-Tg mice showed no difference in expression of Sirt2 isoform 1 when compared with control cells. Incubation of lung macrophages from Sirt2-Tg mice for 48 hours resulted in decreased Sirt2 isoform 2 expression and an increase in Sirt2 isoform 3/5 expression (Figure 1E). This appears to be a lung-specific effect, because bone marrow–derived macrophages and peritoneal macrophages isolated from WT mice showed similar expression patterns of Sirt2 isoform 1, 2, and 3/5-4 after overnight IL-4 stimulation (Supplemental Figure 2A).
To verify that the bands on the Western blot corresponded to their respective Sirt2 isoforms, we obtained His-tagged plasmids for Sirt2 isoform 1 and Sirt2 isoform 5 (24). These plasmids were transfected into HEK293 cells, and expression of the specific isoforms was assessed by Western blot analysis using the Sirt2 antibody as well as an antibody against the histidine tag. Using the Sirt2 antibody, a band was found approximately 40 kDa from the Sirt2 isoform 1 plasmid. At 35 kDa, there was a single band from the Sirt2 isoform 3/5 (Supplemental Figure 2B). The same banding pattern was observed when using the histidine-specific antibody.
The mechanism by which Sirt2 isoform 3/5 is generated was examined in lung macrophages isolated from WT mice that were stimulated with IL-4 in vitro in the presence or absence of protein synthesis and degradation inhibitors (Supplemental Figure 2, C and D). IL-4 treatment of WT lung macrophages upregulated Sirt2 isoform 5 (Supplemental Figure 2, C and D). Incubation of lung macrophages with MG-132, a proteasome inhibitor, did not change Sirt2 isoform 1, 2 or 3/5 protein levels at baseline. However, there was a decrease in Sirt2 isoform 3/5 in IL-4–stimulated lung macrophages that were treated with 0.5 μM MG-132 for 24 hours (Supplemental Figure 2C). Incubation of lung macrophages with the protein synthesis inhibitor cycloheximide resulted in reduced Sirt2 isoform 3/5 protein expression at baseline and when stimulated with IL-4 (Supplemental Figure 2D). These data suggest that Sirt2 isoform 3/5 is generated via protein synthesis under homeostatic conditions and possibly via inhibition of proteosomal degradation and accumulated protein synthesis when stimulated.
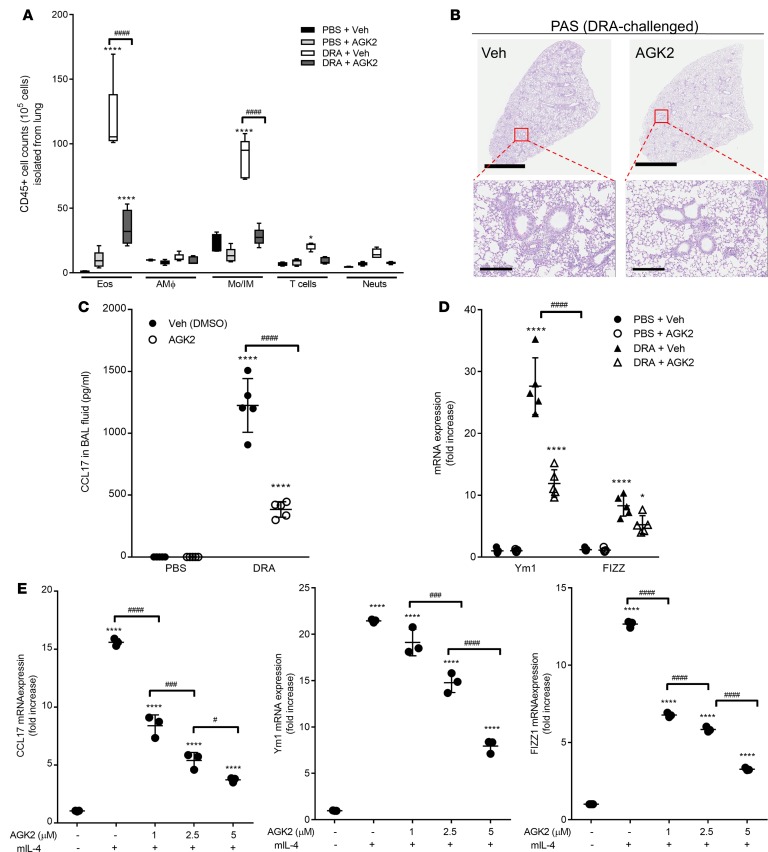
Pharmacologic inhibition of Sirt2 mitigates the development of allergic inflammation in a murine asthma model.
Because of the relative abundance of the different sirtuin proteins, we utilized a specific pharmacological inhibitor of Sirt2 in order to dissect compensatory changes that would occur by genetic overexpression or deficiency. Previous work has identified that AGK2 is a potent and highly selective inhibitor of Sirt2 (26). WT mice were sensitized and challenged with either saline or DRA and then treated with either AGK2 or vehicle control 30 minutes prior to DRA challenge on days 12, 13, and 14 (Supplemental Figure 1A). The relative number of leukocytes was quantified in the lungs at day 15 (Figure 2A) using the flow cytometric gating strategy shown in Supplemental Figure 3. As previously demonstrated, DRA challenge resulted in a marked increase in eosinophils (CD45+CD3–SigF+Ly6G–CD11c–) (Supplemental Figure 3). Interestingly, there was also a substantial increase in monocytes and interstitial macrophages (CD45+CD3–SigF–Ly6G–CD11b+), but no change in the total number of resident alveolar macrophages (CD45+CD3–Ly6G–SigF+) following DRA challenge. There was a slight increase in the total number of T cells (CD45+CD3+) and neutrophils (CD45+CD3–SigF–LyG6+CD11b+) in mice that received DRA challenge compared with saline alone. Treatment of the saline-challenged mice with AGK2 did not result in any difference in the CD45+ cells in the lung. However, DRA-challenged mice treated with AGK2 had markedly fewer eosinophils, monocytes/interstitial macrophages, T cells, and neutrophils in their lungs compared with DRA-challenged mice treated with vehicle. In addition, AGK2-treated DRA-challenged mice had less cellular infiltration and reduced PAS staining and goblet cell hyperplasia (Figure 2B). Furthermore, there was significantly less CCL17 in the BAL fluid of AGK2-treated DRA-challenged WT mice compared with vehicle treated mice (Figure 2C). These data demonstrate that pharmacological inhibition of Sirt2 reduced allergic inflammation following DRA challenge and mirror the changes seen in Sirt2-KO mice (Figure 1).
Figure 2. Administration of AGK2, a selective Sirt2 inhibitor, attenuates allergic inflammation by modulating alternative activation of macrophages and CCL17 production.
AGK2 (10 mg/kg; i.p.) was administered 30 minutes prior to DRA challenge on days 12, 13, and 14. (A) Total lung cells were labeled with surface markers for CD45, CD3, CD11c, CD11b, Ly6G, and Siglec F, gated on CD45+ cells, and the percentage of eosinophils, alveolar macrophages, monocytes/interstitial macrophages, T cells, and neutrophils was determined. n = 5 mice/group; analyzed by 1-way ANOVA. (B) Whole lung histological sections were PAS stained. The images are representative of 5 experiments. Scale bar: 3 mm (top); 300 μm (bottom). (C). CCL17 production in BAL fluid was measured by ELISA. n = 5 mice/group; analyzed by 1-way ANOVA. (D) Alternative activation macrophage markers were assessed in whole lung tissue after DRA challenge by qPCR. n = 5 mice/group; analyzed by 1-way ANOVA. (E) Lung macrophages were isolated from WT mice and incubated with rmIL-4 (20 ng/ml) for 24 hours in the presence of AGK2, and expression of CCL17 and alternative activation markers was assessed by qPCR. n = 3 mice/group; analyzed by 1-way ANOVA. *P < 0.05, ****P < 0.001 when compared with WT control. #P < 0.05, ###P < 0.001, ####P < 0.0001 when compared within the groups.
Because we observed differences in macrophage/monocyte populations in the lung following DRA challenge, we were interested in determining the activation phenotype of these cells. We measured expression of alternative activation markers Ym1 and Fizz1 in whole lung tissue from saline- and DRA-challenged mice treated with AGK2 or vehicle control. DRA-challenged mice had significant increases in YM1 and Fizz1 expression; however, inhibition of Sirt2 following DRA challenge resulted in a significant reduction in Ym1 and Fizz1 expression (Figure 2D). In order to determine the role of Sirt2 in regulating macrophage activation, lung macrophages were isolated from WT mice and incubated with IL-4 and increasing doses of AGK2 in vitro. After 48 hours, IL-4–treated macrophages showed a significant increase in the expression of CCL17, Ym1, and Fizz1 mRNA transcripts, as measured by qPCR (Figure 2E). There was a dose-dependent decrease of alternative activation markers when treated with AGK2 in the presence of IL-4 stimulation (Figure 2E). These data suggest that the Sirt2 inhibitor AGK2 alters allergic inflammation by DRA challenge by regulating the macrophage activation phenotype.
Sirt2 expression in bone marrow–derived cells regulates the development of allergic airway inflammation.
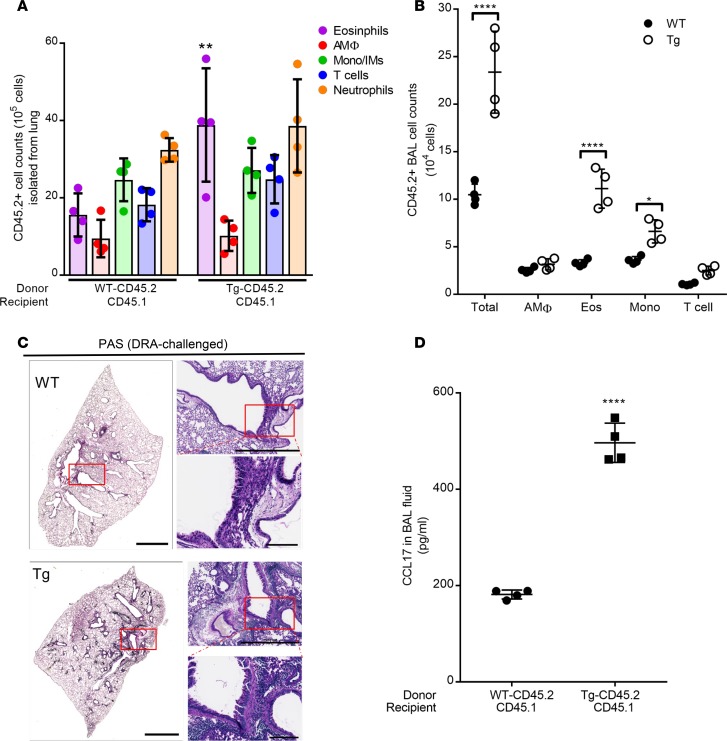
In order to determine the relative effects of Sirt2 expression of bone marrow–derived cells compared with structural cells, we developed a bone marrow transplantation (BMT) chimera model. Bone marrow was isolated from WT or Sirt2-Tg mice and transplanted into lethally irradiated WT mice. In order to validate complete reconstitution of the BMT chimera model, we utilized the congenic markers to track CD45.2 donor (WT and Sirt2-Tg) and CD45.1 recipient cells. After reconstitution, the mice had approximately 95% donor-derived (CD45.2) cells (Supplemental Figure 4). Interestingly, additional flow cytometric gating determined that the remaining recipient-derived cells present in the lung following irradiation and reconstitution were positive for CD3 (Supplemental Figure 4). These data demonstrate that, following BMT, the majority of the macrophages present in the lung were donor derived.
Overexpression of Sirt2, specifically in the bone marrow compartment, resulted in a significant increase in total cells in the whole lung and BAL fluid (Figure 3, A and B). There were also significantly more eosinophils in both the whole lung as well as the BAL fluid in Sirt2-Tg BMT mice compared with WT BMT mice. In addition, there was a significant increase in the number of monocytes/interstitial macrophages in the BAL fluid of Sirt2-Tg BMT mice compared with those that received WT bone marrow. Histology demonstrated increased PAS staining in Sirt2-Tg BMT mice compared with WT BMT (Figure 3C). There was a significant increase in CCL17 in the BAL fluid as well as whole lung of Sirt2-Tg BMT mice compared with WT BMT mice (Figure 3D). Together, these data demonstrate that overexpression of Sirt2 only in bone marrow–derived cells enhances DRA-induced allergic inflammation, resulting in increased eosinophilic infiltration and goblet cell hyperplasia in the lungs.
Figure 3. Bone marrow–derived cells overexpressing Srit2 have enhanced susceptibility to DRA challenge.
Bone marrow chimeras were generated by transplanting WT or Tg Sirt2 bone marrow cells (CD45.2) into lethally irradiated CD45.1 mice. After completion of bone marrow reconstitution (6 weeks), mice were sensitized and challenged. Left lung tissue was isolated for histology, and lung cells from right lobes were isolated via collagenase digestion. (A) The total number of CD45.2 cells was quantified in the lung tissue. n = 4 mice/group; analyzed by 1-way ANOVA. (B) Cellularity of the BAL was determined by flow cytometric staining and graphed as the percentage of donor-derived CD45.2 cells. n = 4 mice/group; analyzed by 1-way ANOVA. (C) Whole lung histological sections were PAS stained. The images are representative of 4 experiments. Scale bar: 2 mm (left); 1 mm (top right); 200 μM (bottom right). (D) Production of CCL17 was assessed in BAL fluid by ELISA. n = 4 mice/group; analyzed by unpaired t test. *P < 0.05, **P < 0.01, ****P < 0.005 when compared with WT control.
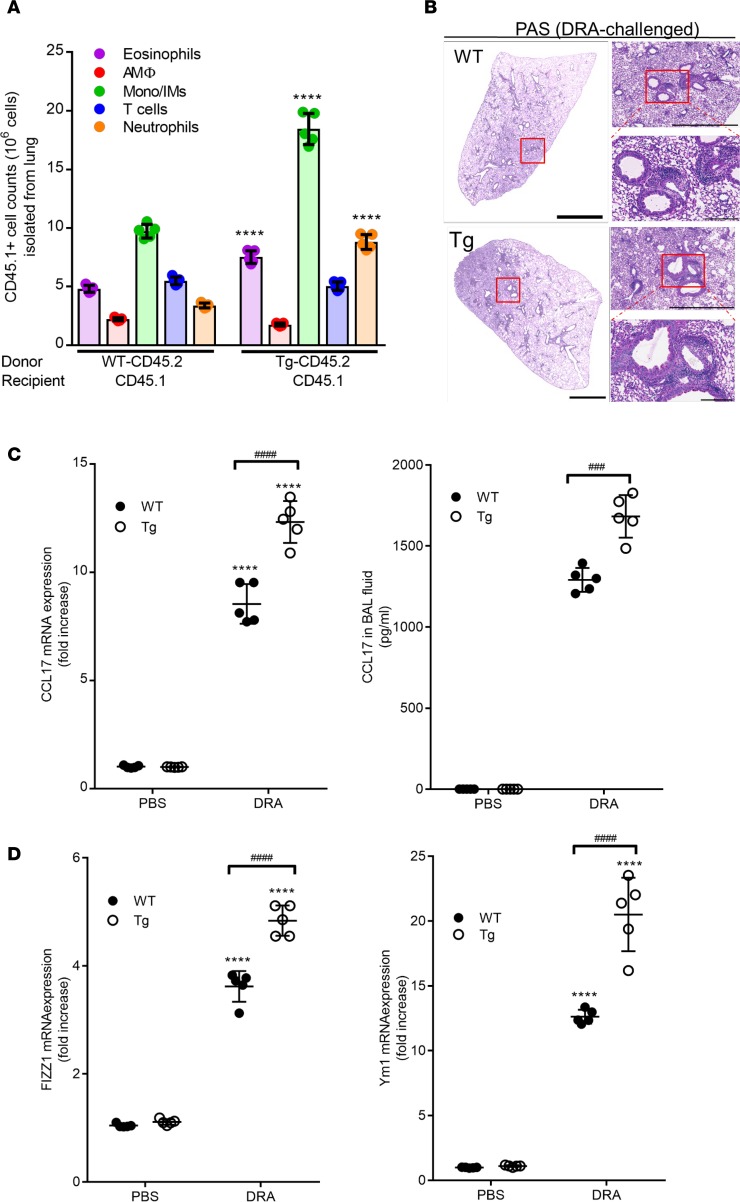
Adoptive transfer of Sirt2-Tg lung macrophages exacerbates allergic airway inflammation.
To validate the role of macrophages in regulating the allergic inflammatory response in the lung following DRA challenge, naive primary macrophages were isolated from WT or Sirt2-Tg mice (expressing the congenic CD45.2 marker) and adoptively transferred into CD45.1 recipient mice on day 8 (Supplemental Figure 1A). On day 15 following DRA challenge, the total number of recipient-derived cells (CD45.1) was determined by flow cytometric analysis (Supplemental Figure 4). As shown in Figure 4A, there was a significant increase in eosinophils and alveolar macrophages in the mice that received the lung macrophages isolated from Sirt2-Tg mice compared with those receiving macrophages isolated from WT mice. Similar to the findings in the BMT experiment, histologic analysis revealed enhanced PAS staining in the CD45.1 recipient mice that received the Sirt2-Tg macrophages (Figure 4B). In addition, there was elevated expression of CCL17 in whole lung tissue as well as BAL fluid in mice that received the adoptive transfer of Sirt2-Tg lung macrophages (Figure 4C). Finally, adoptive transfer of lung macrophages from Sirt2-Tg mice significantly increased the expression of markers of alternative macrophage activation in whole lung tissue (Figure 4D). Together, these data demonstrate that adoptive transfer of lung macrophages that overexpress of Sirt2 enhances DRA-induced allergic airway inflammation.
Figure 4. Adoptive transfer of Sirt2-overexpressing transgenic lung macrophages exacerbates DRA challenge.
Lung macrophages from WT or Tg Sirt2 mice were transferred i.n. into sensitized CD45.1 mice on day 8. Both groups of mice were then challenged with DRA on day 12–14. On day 15, left lung tissue was isolated for histology, and lung cells from right lobes were isolated via collagenase digestion. (A) Relative percentages of different subpopulations of lung immunological cells were assessed by flow cytometry, as described in Supplemental Figure 2. The number of cells was quantified based on the number of recipient CD45.1+ cells. n = 5 mice/group; analyzed by 1-way ANOVA. (B) Lung histological slides were stained with PAS to determine goblet cell hyperplasia. The images are representative of 5 experiments. Scale bar: 2 mm (left); 1 mm (top right); 200 μM (bottom right). (C) Expression of CCL17 was assessed in whole lung tissue by qPCR, and production of CCL17 in BAL fluid was measured by ELISA. n = 5 mice/group; analyzed by 1-way ANOVA. (D) Expression of markers of alternatively activated macrophages, Fizz1 and YM1, was determined in lung tissue after DRA challenge was assessed by qPCR. n = 5 mice/group; analyzed by 1-way ANOVA. ****P < 0.001 when compared with WT control; ###P < 0.001, ####P < 0.0001 when compared within the groups.
Sirt2 expression regulates CCL17 and macrophage alternative activation markers in human lung macrophages.
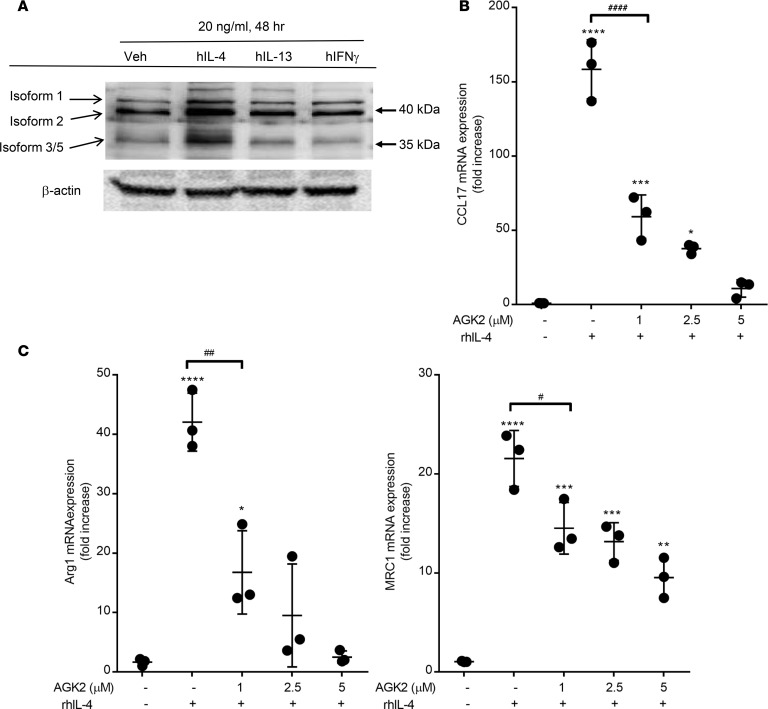
Primary human alveolar macrophages were isolated via ex vivo lavage from donor lungs deemed not viable for transplantation. Expression of Sirt2 isoforms was measured after stimulation with macrophage-polarizing cytokines (Figure 5A). Stimulation of human lung macrophages with IL-4, IL-13, or IFN-γ did not alter expression of Sirt2 isoforms 1 and 2. Interestingly, stimulation with IL-4 for 48 hours resulted in a marked increase in Sirt2 isoform 3/5 expression (Figure 5A), similar to our findings in murine macrophages (Figure 1E). Incubation of human alveolar macrophages with IL-4 resulted in a significant increase in CCL17, Arg1, and mannose receptor C-type (MRC1) expression, as measured by qPCR (Figure 5, B and C). Inhibition of Sirt2 by AGK2 significantly reduced expression of alternatively activated macrophage markers in a dose-dependent manner. These data demonstrate that the Sirt2 isoform 3/5 is an IL-4–regulated protein in human and murine macrophages that regulates the development of allergic inflammation in the lung.
Figure 5. Sirt2 plays important role on alternative macrophage activation and CCL17 expression.
Primary human alveolar macrophages were isolated via ex vivo lavage from healthy donors. (A) Alveolar macrophages were stimulated with IL-4, IL-13 or IFN-γ for 48 hours and expression of Sirt2 isoforms were detected with either N-terminus or C-terminus specific antibodies. Blot represents 1 set of patient samples, but performed on 3 separate patients. (B) Human AMs macrophages were incubated with rhIL-4 (20 ng/ml) for 24 hours in the presence of AGK2 and expression of CCL17, and (C) alternative activation macrophage markers Arg1 and MRC1 were assessed by qPCR. n = 3 samples/group; analyzed by 1-way ANOVA. *P < 0.05, **P > 0.01, ***P < 0.005, ****P < 0.001 when compared with untreated; ##P < 0.01, ####P < 0.001 when compared across groups.
Discussion
Asthma is a major health concern; despite increasing prevalence there are relatively few new classes of therapeutics developed to treat or control this disease. Using a murine model of allergic airway inflammation, our data demonstrate that expression of Sirt2 regulates eosinophilic inflammation in response to allergen challenge. We observed a substantial upregulation in Sirt2 isoform 3/5 in lung macrophages following in vitro IL-4 stimulation or in vivo DRA challenge. In addition, our data indicate that overexpression or inhibition of Sirt2 regulates expression of CCL17 and recruitment of monocytes and macrophages into the airways following allergen sensitization and challenge. We demonstrate that Sirt2 isoform 3/5 is regulated by IL-4 stimulation and protein synthesis in lung macrophages. Interestingly, this effect appears to be lung specific, since there was no difference in Sirt2 isoform 3/5 expression in bone marrow–derived or peritoneal macrophages after IL-4 stimulation. Pharmacologic inhibition of Sirt2 reduced cellular recruitment, diminished CCL17 expression, and decreased expression of alternatively activated macrophage markers. Conversely, overexpression of Sirt2 using bone marrow chimeras or adoptive transfer of lung macrophages led to an exacerbated allergic response, with elevated CCL17 production and enhanced goblet cell hyperplasia. Importantly, we also show that IL-4 regulates Sirt2 isoform 3/5 in human alveolar macrophages, suggesting that this pathway may be relevant to human asthma. Finally, incubation of IL-4–skewed macrophages with a Sirt2 inhibitor resulted in reduced expression of CCL17 as well as macrophage markers of alternative activation. Together, these data shed light on the growing body of literature suggesting that macrophage activation plays an important role in regulating the development of allergic inflammation in asthma and identify a role for Sirt2 in regulating a proasthmatic macrophage phenotype.
The incidence of asthma and allergy has been on the rise in the US and is of significant public health concern because of rising health care costs as well as loss of productivity and reduced quality of life. Previous work has determined that exposure to common indoor allergens, such as dust mites and those generated from animals and mold, are considered a risk factor for asthma. Previous work has demonstrated that at least half of the over 800 housing units surveyed across 75 different locations in the US had one allergen and 45.8% of them had at least 3 allergens (27). These data are reinforced by a recent study highlighting the fact that multicomponent interventions, but not single interventions, of removing indoor allergens showed improved asthma outcomes (28). In order to understand the complex interactions which occur in the setting of allergic inflammation, several groups have developed multiple allergen murine models (29, 30). Our work seeks to extend these results and examine the role of macrophages in regulating the acute allergic inflammatory phenotype by using a triple-antigen model. However, we acknowledge that use of 3 antigens, as well as the use of alum during the sensitization phase, does complicate the system and is a limitation in the current model.
Despite the fact that macrophages are the most abundant immune cell in the homeostatic lung, the role of macrophages in regulating the development of allergic inflammation has been controversial (31). Previous work done by our group and others has demonstrated that macrophages play an important role in the development of the allergic inflammatory response (7, 32–34). In contrast, other reports have suggested that these innate immune cells may only play a minor role in the development of an experimental allergic response (35, 36). One important consideration when examining the body of literature in regards to the role of macrophages in allergic inflammation is there are genetic differences between alternatively activated mouse and human macrophages (37). Another aspect is the role of other cells in regulating monocyte trafficking in response to an allergen challenge as well as macrophage differentiation and activation. We acknowledge that our study did not focus on dendritic cells or CD4 T cells and their role in producing cytokines and chemokines; rather, our work demonstrates that lung macrophages collected following DRA challenge have a differential activation phenotype and produce altered cytokines and chemokines.
Because macrophages are a heterogeneous population of cells, the discrepancy among the conclusions in these reports may be different due to the lack of consideration of the derivation and the overall activation phenotype of the different macrophage subpopulations. Exciting new work has emerged that has begun to identify derivation markers to distinguish between embryonic and bone marrow–derived macrophage subpopulations in the lung (38, 39); however, additional work is needed to fully understand the implications of this macrophage heterogeneity and diversity in the setting of allergic inflammation. It is also important to consider that allergic asthma presents with different endotypes characterized by a predominance of eosinophils or neutrophils in the airways. These endotypes can be modeled in the laboratory by manipulating the breed of mouse, allergen, timeline, or route of administration of the challenge, and macrophages may play different roles during inflammation, depending on the prevalent granulocytic cell type found in the model. This current study aimed to delineate the role of macrophage activation in regulating eosinophilic allergic airway inflammation.
Our work has demonstrated an important role of Sirt2 in regulating the production of CCL17 in the setting of allergic inflammation. Chemokines, such as CCL17 and CCL22, are pivotal in regulating allergic inflammation, and increased levels of CCL17 have been found in plasma and BAL fluid from asthmatic patients (40). A variety of different cell types have been shown to produce CCL17, including structural cells, such as bronchial epithelial cells, and several leukocyte populations, such as T cells, dendritic cells, and macrophages. Several reports have demonstrated that IL-4 induced CCL17 production from monocytes and macrophages (41, 42). Although it has been suggested that CCL17 is a marker of alternative macrophage activation, other work has suggested that they function more to recruit cells to the site of inflammation (43). CCL17, as well as CCL22, signals via the CCR4 receptor, and recent studies have demonstrated that targeting CCR4 resulted in reduced AHR and controlled allergic airway inflammation (44, 45). Although there was a marked increase in the mRNA expression of CCR4 in macrophages isolated from the lung after DRA challenge, there was no difference in the upregulation of CCR4 in Sirt2-Tg mice compared with Sirt2 WT mice. Taken together, these data demonstrate an important role for CCL17 in regulating macrophage activation as well as allergic inflammation in the lung.
Previous work has demonstrated that epigenetic changes contribute to the development of allergic asthmatic inflammation (46). Sirtuins are a class of proteins that possess mono-ADP-ribosyltransferase activity and are considered a HDAC capable of epigenetic modifications. Several classes of sirtuin have been characterized by subcellular localization, enzymatic activity, and function (47). Our work focuses on the role of Sirt2, which is a member of the class I sirtuins that have been shown to be key modulators of cell signaling processes involved in homeostatic regulation, cell division, cell death, and production of defense mechanisms. In particular, it has previously been shown that the pan–class I sirtuin inhibitor, sirtinol, was effective in reversing allergic inflammation in murine model of OVA exposure (48). While the role of Sirt2 has not been investigated previously during allergic asthmatic inflammation, treatment of mice with the Sirt2 inhibitor AGK2 in the setting of experimental sepsis and occlusive cerebrovascular ischemia resulted in both a survival and an antiinflammatory benefit (49–51). In contrast to pan-class 1 Sirt inhibitors, AGK2 has been previously shown to be highly specific for Sirt2. AGK2 has a calculated IC50 for Sirt2 of 3.5 μM and only slightly inhibits Sirt1 and Sirt3 at concentrations over 40 μM (26). Together, these data suggest that upregulation of Sirt2 could be a necessary mechanism by which allergic inflammation occurs.
A structural analysis of Sirt2 indicates that it contains a NAD-binding domain and a catalytic domain with deacetylase activity (52). Initial characterization of different Sirt2 variants revealed that isoforms 1 and 2 both contain a nuclear export signal and predominantly localize to the cytoplasm (18). Although a predicted Sirt2 isoform 4 variant is in the GenBank database, the predicted protein structure and functionality of this isoform have yet to be determined. Several groups have identified a third variant of Sirt2, which has been referred to as isoform 3, found in mice and rats (23, 25), as well as isoform 5, found in humans (24). Because we examine expression of the Sirt2 isoforms using both human and murine samples, we have chosen to refer to the third isoform as Sirt2 isoform 3/5. Our data show that in vivo exposure to allergen and in vitro treatment with IL-4 increases Sirt2 isoform 3/5 in lung macrophages. Based on our data, we hypothesize that Sirt2 isoform 3/5 is the IL-4–dependent variant, which regulates allergic inflammation. The mechanism by which Sirt2 isoform 3/5 regulates this response is somewhat perplexing since it does not contain a nuclear export sequence and is therefore stuck in the nucleus and initial reports failed to demonstrate in vitro catalytic activity, despite an intact catalytic domain (24). It is known that nuclear Sirt2 has specific histone H3K18 deacetylation activity (53), and, recently, it has been shown that Sirt2 has increased enzymatic activity when incubated with nucleosomal-linked substrates over naked protein (54). This is an important finding, because previous work has demonstrated that cytoplasmic Sirt2 functions during mitosis as a tubulin deacetylase (18, 55, 56), whereas nuclear Sirt2 has specific HDAC activity (52, 53) for histone 3 and histone 4 when engaged with an acetylated nucleosome (54). Additional work to determine the target and mechanism by which IL-4–stimulated Sirt2 isoform 3/5 can regulate macrophage activation in the setting of allergic inflammation is an area of ongoing research in our laboratory.
Taken together, these results demonstrate that Sirt2 isoform 3/5 influences the development of experimental allergic asthmatic inflammation. Our data show a mechanism by which expression of a HDAC, Sirt2, regulates the production of CCL17 and recruitment of myeloid-derived cells into the lung following sensitization and challenge with DRA. Using both pharmacologic inhibitors and genetic manipulation, we demonstrate changes in cellular composition and airway resistance. We believe that these data can have direct implications to human allergic airway disease, because of similarities between Sirt2 isoform 3/5 expression in both primary mouse and human lung macrophages after in vitro IL-4 stimulation. In summary, our findings provide additional support for the growing hypothesis that macrophage activation regulates the development of allergic inflammation.
Methods
Mice.
Sirt2-Tg and Sirt2-KO mice were a gift from Fen Xia (The Ohio State University). WT C57BL/6 mice were obtained from The Jackson Laboratory. Male and female mice were used at 8–12 weeks of age in this study.
Allergic inflammation model and Sirt2 inhibition in vivo.
The allergic asthma model was performed as previously described (12, 57). The DRA triple-allergen mixture was composed of extracts of dust mite (Dermatophagoides farina), ragweed (Ambrosia artemisiifolia), and A. fumigatus (Greer Laboratories). Briefly, on days 0 and 5, mice were i.p. sensitized with the DRA allergen mixture (5 μg dust mite, 50 μg ragweed, and 5 μg A. fumigatus) in 200 μl alum as an adjuvant (Imject, Alum; Thermo Scientific). Mice were then i.n. challenged with 50 μl saline or DRA (8.3 μg dust mite, 83.4 μg ragweed, and 8.3 μg A. fumigatus) on days 12, 13, and 14. Control mice were sensitized as above, but were i.n. challenged with saline in place of DRA. Mice were sacrificed on day 15, and BAL fluid and lung tissues were collected for further analysis. AGK2 (10 mg/kg) was administered i.p. with 2% tween20 and 10% PEG-400 in PBS 30 minutes prior to DRA challenge at days 12, 13 and 14. The timeline of the DRA and AGK2 administration is shown in Supplemental Figure 1A.
BAL differential cell count.
BAL fluid was collected as previously described (12, 57). Briefly, airways were lavaged with PBS and 0.6 mM EDTA and analyzed for total cell counts using the Countess automated cell counter (Life Technologies). Cells isolated from the BAL fluid were stained with HEMA 3 (Thermo Scientific), and the number of macrophages and eosinophils was quantitated by microscopy.
Measurement of cytokines.
The Proteome Profiler mouse cytokine array panel A (R&D Systems) was used to detect the cytokine expression profile in mouse BAL fluid. In addition, BAL fluid and cytokine secretion in culture supernatants was analyzed by an ELISA specific for mouse CCL17 (R&D Systems) following the protocols supplied by the manufacture.
Isolation of macrophages.
Macrophages were isolated from the whole lungs of mice via a collagenase digestion, as previously described (58). Cells were adherence purified for 30 minutes in serum-free media, nonadherent cells (such as lymphocytes) were washed away, and complete media were replaced for overnight incubation. Lung macrophages were incubated in the presence or absence of rmIL-4 (20 ng/ml). Mouse bone marrow cells were isolated from the femurs and tibias of mice and incubated with rmM-CSF for 7 days in order to differentiate bone marrow–derived macrophages (59). Peritoneal macrophages were isolated from the peritoneal cavity and adherence purified before use.
Lung histology and airway mucus expression.
Mouse lung tissue was prepared using pressurized low-melting agarose as described previously (7). Briefly, low-melting agarose was infused through the tracheostomy and then the tube was tied and the lung was removed and placed into 10% formalin. Both H&E staining and PAS staining were conducted by the Comparative Pathology and Mouse Phenotyping Shared Resource at The Ohio State University. Whole lung images were analyzed by Aperio or Gen5 software.
Measurements of airway hyperresponsiveness.
Mice were anesthetized with pentobarbital (100 mg/kg; i.p.), pancuronium bromide was administered (0.8 ml/kg; i.p.), and a tracheostomy tube was inserted. Mechanical ventilation was initiated using a FlexiVent small-animal ventilator (Sireq). Continuous EKG and pulse oximetry monitoring was performed. Airway resistance was measured after sequentially increasing doses of aerosolized methacholine using the FlexiVent system, as described previously (60).
Western blot analysis.
Cells were lysed in RIPA buffer (MilliporeSigma) in the presence of protease inhibitor (Cell Signaling Technology). Cell lysates containing equal amounts of protein were electrophoresed and immunoblotted using the following antibodies: Sirt2 (C-term, MilliporeSigma), β-actin (Cell Signaling; 8H10D10), and histidine tag (AbD Serotec; AD1.1.10). All secondary antibodies were purchased from Cell Signaling. See complete unedited blots in the supplemental material.
Sirt2 overexpression vectors and transfections.
Sirt2 isoform 1 and isoform 5 plasmid vectors were generated by in-house (24). HEK 293 cells were transfected with pcDNA3.1/V5-His-Sirt2 isoform 1 or isoform 5 by Lipofectamine 3000 (Thermo Fisher) for 3 hours and harvested at 48 hours. Expression of isoforms 1 and 5 was confirmed with anti-His and anti–Sirt2 C-terminal antibodies by immunoblotting.
Flow cytometry.
Cells collected from BAL fluid or isolated from collagenase digestion of lungs were fixed with 2% PFA for flow cytometric analysis. Cells were incubated with Fc-blocking anti-mouse CD16/32 antibody (BD Bioscience; 2.4G2), followed by antibodies against pan-CD45 (BioLegend; 30-F11), CD45.1 (BioLegend; A20), CD45.2 (BioLegend;104), CD11c (BioLegend; N418), CD11b (BioLegend; M1/70), CD3 (e Bioscience; 14A2 and 145-2C11), Ly6G (BioLegend; 1A8), and Siglec F (BS Biosciences; E50-2440). Cells were analyzed on a BD LSR II (BD Bioscience) where gating was based on respective unstained cell populations and isotype matching control antibodies. The data were analyzed with FlowJo software (TreeStar). Flow cytometric gating scheme is presented in Supplemental Figure 2.
BMT.
Mice received 13-Gy total body irradiation delivered in 2 fractions separated by 3 hours as previously described (58). Bone marrow was harvested from the femurs of donor mice, and bone marrow was transplanted into the lethally irradiated recipients via tail vein infusion. All experiments with BMT mice were performed 6 weeks after BMT, and complete reconstitution was determined by measuring donor versus recipient cells (Supplemental Figure 2). Total lung cells were isolated by collagenase digestion from the right lobes, BAL fluid was collected from left lobe, and whole lung tissue was collected for cell isolation and histological samples.
Adoptive transfer experiments.
CD45.1 mice were sensitized by DRA and alum at days 0 and 5. WT or Sirt2-Tg mouse lung macrophages were isolated as described above and adherence purified for 1 hour in serum-free media. The adherent cells were collected using 0.05% trypsin, and the percentage of live cells was quantified. Cells were resuspended at a concentration of 4 × 106 cells in 50 μl PBS and then transferred to the sensitized CD45.1 mice i.n. at day 8.
RNA extraction and qPCR.
RNA was extracted from cells or lung tissues homogenates by using a Direct-zol RNA miniPrep Plus Kit (Zymo Research) according to the manufacture’s instructions. cDNA synthesis was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher). Gene expression was measured by quantitative PCR in an Roche LightCycler 480 (Roche). Data were analyzed by the 2–ΔΔCt method with GAPDH expression as an endogenous control.
Human cells.
Alveolar macrophages were isolated via ex vivo BAL by instilling PBS and 0.6 mM EDTA into the airways of the lungs and then removing the lavage fluid by suction. Cells were adherence purified by incubating for 1 hour in serum-free media, and overall macrophage purity was >95%, as determined by modified Wright-Giemsa staining. Cells were either utilized immediately or stimulated with rhIL-4 (20 ng/ml), rhIL-13 (20 ng/ml), and rhIFN-γ (20 ng/ml) to induce macrophage activation.
Statistics.
Data are expressed as mean ± SEM. Differences between groups of mice were evaluated with 2-way ANOVA between indicated groups. Two-tailed Student’s t test was used for other analyses. The statistical software Sigmaplot 12.0 was used for the analysis. P < 0.05 was considered statistically significant.
Study approval.
All experiments involving mice were conducted with protocols approved by the Institutional Animal Care and Use Committee of The Ohio State University. Human cells were isolated from lungs deemed not viable for transplantation based on a panel of specific donor characteristics. These lungs were procured from LifeLine of Ohio, which is the organ procurement agency in central Ohio.
Author contributions
YGL, JAE, MZ, GYP, JWC, and MNB designed the research studies. YGL, BFR, DH, AS, SC, MK, and MNB conducted the research studies. YGL, BFR, DH, AS, SC, and MK acquired the data. YGL, DH, AS, JAE, GYP, JWC, and MNB analyzed the data. MZ and GYP provided the reagents. YGL, JAE, JWC, and MNB wrote the manuscript.
Supplementary Material
Acknowledgments
We would like to thank analytical flow cytometry core (P30CA016058) for their technical assistance and the Davis Heart and Lung Research Institute. This work was supported by grants from the National Institutes of Health (R01HL075557, R01HL137224, K08GM102695, and 5R01HL126852 to JWC, JAE and GYP), American Heart Association/Scientist Development Grant grant (to MNB and MK), American Lung Association (to SC), and the American Thoracic Society Research Foundation (to JAE and MNB).
Version 1. 01/22/2019
In-Press Preview
Version 2. 02/21/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(4):e124710. https://doi.org/10.1172/jci.insight.124710.
Contributor Information
Yong Gyu Lee, Email: Yonggyu.Lee@osumc.edu.
Brenda F. Reader, Email: brenda.cuson@osumc.edu.
Derrick Herman, Email: derrick.herman@osumc.edu.
Adam Streicher, Email: streicherae@gmail.com.
Joshua A. Englert, Email: Joshua.Englert@osumc.edu.
Mathias Ziegler, Email: Mathias.Ziegler@uib.no.
Sangwoon Chung, Email: sangwoon.chung@osumc.edu.
Manjula Karpurapu, Email: manjula.karpurapu@osumc.edu.
Gye Young Park, Email: parkgy@uic.edu.
John W. Christman, Email: John.Christman@osumc.edu.
Megan N. Ballinger, Email: megan.ballinger@osumc.edu.
References
- 1.Balhara J, Gounni AS. The alveolar macrophages in asthma: a double-edged sword. Mucosal Immunol. 2012;5(6):605–609. doi: 10.1038/mi.2012.74. [DOI] [PubMed] [Google Scholar]
- 2.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14(2):81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 3.Murray PJ, et al. Macrophage activation and polarization: nomenclature and experimental guidelines. Immunity. 2014;41(1):14–20. doi: 10.1016/j.immuni.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moreira AP, Hogaboam CM. Macrophages in allergic asthma: fine-tuning their pro- and anti-inflammatory actions for disease resolution. J Interferon Cytokine Res. 2011;31(6):485–491. doi: 10.1089/jir.2011.0027. [DOI] [PubMed] [Google Scholar]
- 5.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wynn TA, Vannella KM. Macrophages in tissue repair, regeneration, and fibrosis. Immunity. 2016;44(3):450–462. doi: 10.1016/j.immuni.2016.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YG, et al. Recruited alveolar macrophages, in response to airway epithelial-derived monocyte chemoattractant protein 1/CCl2, regulate airway inflammation and remodeling in allergic asthma. Am J Respir Cell Mol Biol. 2015;52(6):772–784. doi: 10.1165/rcmb.2014-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moon KA, et al. Allergen-induced CD11b+ CD11c(int) CCR3+ macrophages in the lung promote eosinophilic airway inflammation in a mouse asthma model. Int Immunol. 2007;19(12):1371–1381. doi: 10.1093/intimm/dxm108. [DOI] [PubMed] [Google Scholar]
- 9.Park GY, et al. Autotaxin production of lysophosphatidic acid mediates allergic asthmatic inflammation. Am J Respir Crit Care Med. 2013;188(8):928–940. doi: 10.1164/rccm.201306-1014OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mautino G, Henriquet C, Jaffuel D, Bousquet J, Capony F. Tissue inhibitor of metalloproteinase-1 levels in bronchoalveolar lavage fluid from asthmatic subjects. Am J Respir Crit Care Med. 1999;160(1):324–330. doi: 10.1164/ajrccm.160.1.9808087. [DOI] [PubMed] [Google Scholar]
- 11.Karpurapu M, et al. Functional PU.1 in macrophages has a pivotal role in NF-κB activation and neutrophilic lung inflammation during endotoxemia. Blood. 2011;118(19):5255–5266. doi: 10.1182/blood-2011-03-341123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qian F, et al. The transcription factor PU.1 promotes alternative macrophage polarization and asthmatic airway inflammation. J Mol Cell Biol. 2015;7(6):557–567. doi: 10.1093/jmcb/mjv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satoh T, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nat Immunol. 2010;11(10):936–944. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 14.Liao X, et al. Krüppel-like factor 4 regulates macrophage polarization. J Clin Invest. 2011;121(7):2736–2749. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson SM, Lei X, Prabhu KS. Selenium levels affect the IL-4-induced expression of alternative activation markers in murine macrophages. J Nutr. 2011;141(9):1754–1761. doi: 10.3945/jn.111.141176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wątroba M, Dudek I, Skoda M, Stangret A, Rzodkiewicz P, Szukiewicz D. Sirtuins, epigenetics and longevity. Ageing Res Rev. 2017;40:11–19. doi: 10.1016/j.arr.2017.08.001. [DOI] [PubMed] [Google Scholar]
- 17.Buechler N, Wang X, Yoza BK, McCall CE, Vachharajani V. Sirtuin 2 regulates microvascular inflammation during sepsis. J Immunol Res. 2017;2017:2648946. doi: 10.1155/2017/2648946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.North BJ, Verdin E. Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS ONE. 2007;2(8):e784. doi: 10.1371/journal.pone.0000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang X, et al. Sirtuin-2 regulates sepsis inflammation in ob/ob mice. PLoS ONE. 2016;11(8):e0160431. doi: 10.1371/journal.pone.0160431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomes P, Fleming Outeiro T, Cavadas C. Emerging role of sirtuin 2 in the regulation of mammalian metabolism. Trends Pharmacol Sci. 2015;36(11):756–768. doi: 10.1016/j.tips.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 21.Nie H, et al. SIRT2 mediates oxidative stress-induced apoptosis of differentiated PC12 cells. Neuroreport. 2014;25(11):838–842. doi: 10.1097/WNR.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 22.Lo Sasso G, et al. SIRT2 deficiency modulates macrophage polarization and susceptibility to experimental colitis. PLoS ONE. 2014;9(7):e103573. doi: 10.1371/journal.pone.0103573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maxwell MM, et al. The Sirtuin 2 microtubule deacetylase is an abundant neuronal protein that accumulates in the aging CNS. Hum Mol Genet. 2011;20(20):3986–3996. doi: 10.1093/hmg/ddr326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rack JG, VanLinden MR, Lutter T, Aasland R, Ziegler M. Constitutive nuclear localization of an alternatively spliced sirtuin-2 isoform. J Mol Biol. 2014;426(8):1677–1691. doi: 10.1016/j.jmb.2013.10.027. [DOI] [PubMed] [Google Scholar]
- 25.Thangaraj MP, et al. RNA-binding protein quaking stabilizes Sirt2 mRNA during oligodendroglial differentiation. J Biol Chem. 2017;292(13):5166–5182. doi: 10.1074/jbc.M117.775544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Outeiro TF, et al. Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity in models of Parkinson’s disease. Science. 2007;317(5837):516–519. doi: 10.1126/science.1143780. [DOI] [PubMed] [Google Scholar]
- 27.Salo PM, Arbes SJ, Crockett PW, Thorne PS, Cohn RD, Zeldin DC. Exposure to multiple indoor allergens in US homes and its relationship to asthma. J Allergy Clin Immunol. 2008;121(3):678–684.e2. doi: 10.1016/j.jaci.2007.12.1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leas BF, et al. Effectiveness of indoor allergen reduction in asthma management: A systematic review. J Allergy Clin Immunol. 2018;141(5):1854–1869. doi: 10.1016/j.jaci.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 29.DiGiovanni FA, Ellis R, Wattie J, Hirota JA, Southam DS, Inman MD. Concurrent dual allergen exposure and its effects on airway hyperresponsiveness, inflammation and remodeling in mice. Dis Model Mech. 2009;2(5-6):275–282. doi: 10.1242/dmm.001859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarpong SB, Zhang LY, Kleeberger SR. A novel mouse model of experimental asthma. Int Arch Allergy Immunol. 2003;132(4):346–354. doi: 10.1159/000074902. [DOI] [PubMed] [Google Scholar]
- 31.Draijer C, Peters-Golden M. Alveolar macrophages in allergic asthma: the forgotten cell awakes. Curr Allergy Asthma Rep. 2017;17(2):12. doi: 10.1007/s11882-017-0681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Role of C/EBP homologous protein and endoplasmic reticulum stress in asthma exacerbation by regulating the IL-4/signal transducer and activator of transcription 6/transcription factor EC/IL-4 receptor α positive feedback loop in M2 macrophages. J Allergy Clin Immunol. 2017;140(6):1550–1561.e8. doi: 10.1016/j.jaci.2017.01.024. [DOI] [PubMed] [Google Scholar]
- 33.Kawasaki T, Ito K, Miyata H, Akira S, Kawai T. Deletion of PIKfyve alters alveolar macrophage populations and exacerbates allergic inflammation in mice. EMBO J. 2017;36(12):1707–1718. doi: 10.15252/embj.201695528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bang BR, et al. Alveolar macrophages modulate allergic inflammation in a murine model of asthma. Exp Mol Med. 2011;43(5):275–280. doi: 10.3858/emm.2011.43.5.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wiese AV, et al. The C5a/C5aR1 axis controls the development of experimental allergic asthma independent of LysM-expressing pulmonary immune cells. PLoS ONE. 2017;12(9):e0184956. doi: 10.1371/journal.pone.0184956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staples KJ, Hinks TS, Ward JA, Gunn V, Smith C, Djukanović R. Phenotypic characterization of lung macrophages in asthmatic patients: overexpression of CCL17. J Allergy Clin Immunol. 2012;130(6):1404–12.e7. doi: 10.1016/j.jaci.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez FO, et al. Genetic programs expressed in resting and IL-4 alternatively activated mouse and human macrophages: similarities and differences. Blood. 2013;121(9):e57–e69. doi: 10.1182/blood-2012-06-436212. [DOI] [PubMed] [Google Scholar]
- 38.McCubbrey AL, et al. Deletion of c-FLIP from CD11bhi macrophages prevents development of bleomycin-induced lung fibrosis. Am J Respir Cell Mol Biol. 2018;58(1):66–78. doi: 10.1165/rcmb.2017-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misharin AV, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med. 2017;214(8):2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartl D, Lee CG, Da Silva CA, Chupp GL, Elias JA. Novel biomarkers in asthma: chemokines and chitinase-like proteins. Curr Opin Allergy Clin Immunol. 2009;9(1):60–66. doi: 10.1097/ACI.0b013e32831f8ee0. [DOI] [PubMed] [Google Scholar]
- 41.Hsu AT, et al. Epigenetic and transcriptional regulation of IL4-induced CCL17 production in human monocytes and murine macrophages. J Biol Chem. 2018;293(29):11415–11423. doi: 10.1074/jbc.RA118.002416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liddiard K, Welch JS, Lozach J, Heinz S, Glass CK, Greaves DR. Interleukin-4 induction of the CC chemokine TARC (CCL17) in murine macrophages is mediated by multiple STAT6 sites in the TARC gene promoter. BMC Mol Biol. 2006;7:45. doi: 10.1186/1471-2199-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rajaiah R, Perkins DJ, Polumuri SK, Zhao A, Keegan AD, Vogel SN. Dissociation of endotoxin tolerance and differentiation of alternatively activated macrophages. J Immunol. 2013;190(9):4763–4772. doi: 10.4049/jimmunol.1202407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Honjo A, et al. Targeted reduction of CCR4(+) cells is sufficient to suppress allergic airway inflammation. Respir Investig. 2013;51(4):241–249. doi: 10.1016/j.resinv.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y, et al. A new antagonist for CCR4 attenuates allergic lung inflammation in a mouse model of asthma. Sci Rep. 2017;7(1):15038. doi: 10.1038/s41598-017-11868-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Harb H, Renz H. Update on epigenetics in allergic disease. J Allergy Clin Immunol. 2015;135(1):15–24. doi: 10.1016/j.jaci.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 47.Kupis W, Pałyga J, Tomal E, Niewiadomska E. The role of sirtuins in cellular homeostasis. J Physiol Biochem. 2016;72(3):371–380. doi: 10.1007/s13105-016-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim SR, et al. Involvement of sirtuin 1 in airway inflammation and hyperresponsiveness of allergic airway disease. J Allergy Clin Immunol. 2010;125(2):449–460.e14. doi: 10.1016/j.jaci.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Krey L, et al. Knockout of silent information regulator 2 (SIRT2) preserves neurological function after experimental stroke in mice. J Cereb Blood Flow Metab. 2015;35(12):2080–2088. doi: 10.1038/jcbfm.2015.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee AS, et al. SIRT2 ameliorates lipopolysaccharide-induced inflammation in macrophages. Biochem Biophys Res Commun. 2014;450(4):1363–1369. doi: 10.1016/j.bbrc.2014.06.135. [DOI] [PubMed] [Google Scholar]
- 51.Zhao T, et al. Selective Inhibition of SIRT2 improves outcomes in a lethal septic model. Curr Mol Med. 2015;15(7):634–641. doi: 10.2174/156652401507150903185852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Finnin MS, Donigian JR, Pavletich NP. Structure of the histone deacetylase SIRT2. Nat Struct Biol. 2001;8(7):621–625. doi: 10.1038/89668. [DOI] [PubMed] [Google Scholar]
- 53.Eskandarian HA, et al. A role for SIRT2-dependent histone H3K18 deacetylation in bacterial infection. Science. 2013;341(6145):1238858. doi: 10.1126/science.1238858. [DOI] [PubMed] [Google Scholar]
- 54.Hsu WW, Wu B, Liu WR. Sirtuins 1 and 2 are universal histone deacetylases. ACS Chem Biol. 2016;11(3):792–799. doi: 10.1021/acschembio.5b00886. [DOI] [PubMed] [Google Scholar]
- 55.Inoue T, et al. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26(7):945–957. doi: 10.1038/sj.onc.1209857. [DOI] [PubMed] [Google Scholar]
- 56.Skoge RH, Ziegler M. SIRT2 inactivation reveals a subset of hyperacetylated perinuclear microtubules inaccessible to HDAC6. J Cell Sci. 2016;129(15):2972–2982. doi: 10.1242/jcs.187518. [DOI] [PubMed] [Google Scholar]
- 57.Lee YG, et al. Recruited alveolar macrophages, in response to airway epithelial-derived monocyte chemoattractant protein 1/CCl2, regulate airway inflammation and remodeling in allergic asthma. Am J Respir Cell Mol Biol. 2015;52(6):772–784. doi: 10.1165/rcmb.2014-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ballinger MN, et al. IRAK-M promotes alternative macrophage activation and fibroproliferation in bleomycin-induced lung injury. J Immunol. 2015;194(4):1894–1904. doi: 10.4049/jimmunol.1402377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Karpurapu M, et al. Krüppel like factor 4 promoter undergoes active demethylation during monocyte/macrophage differentiation. PLoS ONE. 2014;9(4):e93362. doi: 10.1371/journal.pone.0093362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lai PS, et al. Chronic endotoxin exposure produces airflow obstruction and lung dendritic cell expansion. Am J Respir Cell Mol Biol. 2012;47(2):209–217. doi: 10.1165/rcmb.2011-0447OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.