Abstract
Despite a dramatic increase in T cell receptor (TCR) sequencing, there are few approaches to biologically parse the data in a fashion that both helps yield new information about immune responses and may guide immunotherapeutic interventions. To address this issue we developed a novel method, ImmunoMap, which utilizes a sequence analysis approach inspired by phylogenetics to examine TCR repertoire relatedness. ImmunoMap analysis of the CD8 T Cell response to self-antigen (Kb-TRP2) or to a model foreign-antigen (Kb-SIY) in naïve and tumor-bearing B6 mice showed unique differences in immune pressures on the T cell repertoire of self versus foreign antigen-specific responses and also detected lymphoid organ-specific differences in TCR repertoires. When used to analyze clinical trial data, ImmunoMap analysis of tumor infiltrating lymphocytes (TILs) from patients on α-PD1 therapy uniquely revealed a clinically predicative signature in pre- and post-therapy samples that were not seen using standard TCR sequence analyses.
Introduction
The advent of immune sequencing has allowed scientists and clinicians to understand antigen-specific responses of the interaction of the immune system with various pathologies in a completely novel way. Its initial applications opened our understanding to the depth and breadth of the T Cell Receptor (TCR) and B Cell Receptor (BCR) repertoire (1–4). Further applications of immune sequencing have contributed to vaccine development and to tracking disease progression in malignancies (5–7). Of recent interest, sequencing efforts have helped to develop our understanding of immune responses to cancer, characterizing the TCR repertoire of circulating as well as of tumor infiltrating lymphocytes (TILs) (8,9).
With the abundance of new “big data” sets, there has arisen a need to develop biologically meaningful techniques for analysis of TCR repertoire sequences. Current methods have failed to provide an intuitive understanding of the immune system repertoire for two reasons. First, as purely mathematical constructs, they focus on diversity defined as a function of the number of different sequences and their respective frequencies, Shannon’s Entropy, and ignore sequence relatedness (10,11). Second, methods that compare different repertoires apply stringent criteria by only comparing exact TCR clonotypes (whether at the nucleotide or amino acid level) to assess similarity (9,12,13).
However, biological sequence similarity, not identity, is the relevant parameter. Ignoring sequence relatedness is a significant omission, since TCR’s with similar, homologous sequences likely recognize related MHC/peptide targets. Recent work by several groups has sought to understand the structural aspects of the underlying TCR repertoire through a variety of techniques which cluster homologous CDR3 sequences, showing that indeed TCR’s seeing the same antigen have highly homologous sequences (14–16). While this work highlights the relevance and rationale behind analyzing TCR sequence repertoire data via clustering methods, we sought to create structural diversity metrics for whole TCR repertoires. To address this, we developed ImmunoMap which visualizes and quantifies immune repertoire diversity in a holistic fashion. ImmunoMap not only enables assessment of similarity between TCR sequences but displays the scope of diversity among different repertoires. Our novel approach combines information about the frequency and relatedness of TCR sequences using a sequence analysis inspired by phylogenetics to determine relatedness among cells of an antigen-specific T cell response as well as the similarities of a particular TCR repertoire to other repertoires.
ImmunoMap was initially trained on and used to analyze T cells responding to Kb-TRP2, a shared self-peptide tumor antigen, and Kb-SIY, a model foreign-antigen, in a model of murine melanoma. In naïve animals, the response to Kb-SIY was highly conserved with many TCR sequences having high sequence homology, a biological observation that was missed by Shannon’s Entropy calculations. In contrast, in the self-antigen response, the bulk of the response came from fewer and distantly related sequences comprising the majority of the response. The presence of tumor had a differential effect on the shaping of the repertoire in the model foreign and self-antigen responses, greatly altering the TCR repertoire of the self-antigen response, with a smaller effect on response to the foreign antigen. To understand the clinical utility of ImmunoMap, we compared ImmunoMap to Shannon’s entropy analysis of TIL from melanoma patients on α-PD1 therapy. While Shannon’s Entropy calculations failed to find any clinically relevant correlates, ImmunoMap uniquely revealed clinically relevant, predicative TCR signatures in patient who responded to α-PD1 therapy after just 4 weeks on therapy. Thus ImmunoMap uniquely revealed a clinically useful parameter that other repertoire analysis techniques failed to show.
Materials and Methods
Mice:
C57BL/6j mice were purchased from Jackson Laboratories (Bar Harbor, ME). All mice were maintained according to Johns Hopkins University’s Institutional Review Board. 4–5 mice (gender and age matched) were used and pooled for each stimulation condition, based on previous T cell expansion experiments, and each stimulation and sequencing run was performed once. Mice were randomly selected for naïve or tumor-bearing treatments and principal investigator was blinded to which mice received tumors. Murine experiments for naïve and tumor-bearing spleens were duplicated in separate pools of animals to demonstrate reproducibility of antigen-specific repertoire characteristics (Suppl. Fig 7).
Preparation of MHC-Ig Dimers and Nano-aAPC:
Soluble MHC-Ig dimers, Kb-Ig, was prepared and loaded with peptides as described (17), see Supplemental Materials. Nano-aAPC were manufactured by direct conjugation of MHC-Ig dimer and anti-CD28 antibody (37.51; Biolegend) to MACS Microbeads (Miltenyi Biotec) as described previously (18).
Lymphocyte Isolation:
Mouse lymphocytes were obtained from homogenized mouse spleens after hypotonic lysis of RBC. Cytotoxic lymphocytes were isolated using a CD8 magnetic enrichment column from Miltenyi Biotec (Cologne, Germany) following the manufacturer’s instructions. Lymphocytes from lymph nodes were obtained from homogenized inguinal lymph nodes and enriched with nano-aAPCs and plated for 7 days. For tumor-bearing animals, murine melanoma cell line B16-SIY, given by Tom Gajewski (The University of Chicago, IL, USA), was injected subcutaneously, measured by calipers and harvested when tumors reach over 50mm2. The B16-SIY cell line is a tumor model modified to express SIY, a completely foreign epitope to the murine B6 background. In naïve mice setting experiments, it was used as a model foreign antigen such as would be a viral epitope and in tumor bearing animals serves as a tumor antigen. Tumor infiltrating lymphocytes were obtained from tumors by manual digestion and washing, a density gradient centrifugation (Lympholyte Cell Separation Media, Mouse, Cedar Lane), and then tumor cells plated for 3 hours at 37°C and lymphocytes washed off and plated with nano-aAPCs (1.25×109 particles/mL). All cell lines underwent testing for mycoplasma contamination.
Enrichment and Expansion:
Nano-aAPC were stored at a concentration of 8.3 nM (5 × 1012 particles/mL), and all volumes refer to particles at this concentration. Ten million CD8-enriched lymphocytes at ~108 cells/mL were incubated with 10 uL of nano-aAPC for 1 hr at 4 °C, for an approximate bead:cell concentration of 5000:1. Cell-particle mixtures were subsequently passed through a magnetic enrichment column, the negative fraction was collected and the positive fraction eluted. Positive fractions were mixed and cultured in 96-well round-bottom plates for 7 days in complete RPMI-1640 medium supplemented l-glutamine, non-essential amino acids, vitamin solution, sodium pyruvate, β-mercaptoethanol, 10% FBS, ciproflaxin, and 1% T cell growth factor, a cytokine cocktail derived from stimulated PBMC as described in the literature (19), in a humidified 5% C02, 37 °C incubator for 1 week. Specificity of CTL was monitored on day 7, by FACS analysis following LIVE/DEAD cell stain (Thermo Fisher), αCD8 (BD Bioscience), and dimeric MHC-Ig staining. The number of antigen-specific cells was calculated by multiplying the number of total cells by the fraction of CD8 and antigen-specific cells; the fraction of antigen-specific cells was calculated after subtracting the non-cognate MHC staining from cognate MHC staining.
Sorting and Sequencing of Antigen-Specific CD8 T Cells:
Following LIVE/DEAD cell stain (Thermo Fisher), αCD8 (BD Bioscience), and dimeric MHC-Ig staining, cells were sorted by gating on cells with cognate Dimeric MHC-Ig staining over non-cognate staining. Antigen-specific CD8 T cells were sent directly for CDR3 β-chain sequencing by Adaptive Biotechnologies.
In vitro nano-aAPC Functionality Assay:
7 days following enrichment and expansion antigen specificity is confirmed by intercellular cytokine staining. Briefly, RMA-S, given by Michael Edidin (Johns Hopkins University, MD, USA) are peptide pulsed (10uM) overnight at room temperature with relevant or no peptide and mixed 1:2 RMAS:T cell ratio with expanded T cells. Unpulsed RMAS cells were used as background stimulation. After 6 hours, cells were washed twice with FACS wash buffer and then stained with viability dye and α-CD8 for 20 minutes. Cells were then fixed and permeabilized with the Cytofix/Cytoperm kit (BD Biosciences) following the manufacturer’s protocol. Anti–TNF-α (Biolegend) was added to the cells and stained for an hour.
Precursor Frequency Assessment:
On day 0 following CD8+ T cell isolation from splenic cells, CD8+ T cells were stained with LIVE/DEAD cell stain (Thermo Fisher), αCD8 (BD Bioscience), and dimeric MHC-Ig staining viability stain with either unloaded or peptide-loaded MHC-Ig. Cells gated on Live cells and anti-CD8a+ staining.
Collection of TILs from Patients undergoing α-PD1 therapy:
Eighty-five patients, providing consent, were accrued to a multi-arm, multi-institutional, institutional-review-board-approved, prospective study (BMS-038) to investigate the pharmacodynamic activity of nivolumab. All patients received nivolumab (3 mg/kg Q2W) until progression for a maximum of 2 years. Tumor samples were collected prior to and four weeks after initiation of Nivolumab therapy. The samples were stored in RNAlater®(Ambion). 34 patients permitted TCR sequencing, and DNA was extracted and submitted to Adaptive Biotechnologies for survey level TCR β-chain sequencing (20,21). Briefly, sequencing libraries were prepared by multiplex PCR targeting all TCR b-chain VJ gene segment combinations. Sequencing was performed using the Illumina HiSeq system. Data for individual TCR sequences, including V and J gene segment identification and CDR3 sequences were obtained from Adaptive Biotechnologies for customized analysis of T-cell repertoire diversity dynamics. Clinical response was assessed via CT scan after 24 weeks of therapy.
Deconvolution Methods:
Due to the fact that animals were pooled together on day 0 prior to expansion of antigen-specific cells, there was only one sequencing run. In order to determine the variance of calculated indicators of the repertoire, the reads from the sequencing file were randomly distributed into the number of bins corresponding to the number of animals that went into the experiment. This method of random deconvolution assured that the variance of the indicator by random chance was not greater than the difference observed between conditions.
Weighted Repertoire Dendrograms:
For the antigen-specific sequencing, productive sequences with a frequency > 0.01% were taken for analysis. For α-PD1 clinical trial analysis, Adaptive files were first filtered to only include sequences with reads greater than or equal to 5 and then top 40% of response was taken for analysis. Sequence distances were calculated based on sequence alignments scores using a PAM10 scoring matrix and gap penalty of 30. Distance matrix was used to create a dendrogram using the Bioinformatics toolbox in MATLAB. Circles were overlaid at the end of the branches corresponding to the CDR3 sequences with diameters proportional to the frequency of the sequence. Furthermore, when using the terminology ‘weighted repertoire dendrogram,’ this does not infer that the distance matrix used to create the dendrogram is weighted; rather, the dendrogram is visually ‘weighted’ by frequency.
Dominant Motif Analysis:
Using the cluster function in MATLAB toolbox, dendrogram was divided into homologous clusters using a homology threshold obtained from analyzing an unexpanded adult CD8 population from a B6 animal (See Supplementary Materials). Clusters whose average sequence distance within cluster <= threshold and met a certain frequency cutoff (3% - See Supplementary Materials) were denoted as ‘Dominant Motifs.’ Cluster frequency was lowered to 1% for α-PD1 clinical trial analysis but held consistent across all patients due to the fact that this was not a single antigen-specific population of cells.
Singular & Novel Clone Analysis:
In order to define singular clones, a matrix was setup to calculate the mapped sequence distance of every unique combination of sequences in the repertoire. Using standard matrix operations within MATLAB, a singular clone was defined as a clone whose frequency was 10x the sum of all other homologous clones. Homologous clones were those who had a sequence distance determined from the dominant motif analysis. In order to define novel clones, the same approach was used, but the matrix was setup in that it calculated the mapped sequence distance of every unique combination of sequences between the two repertoires being compared. A novel clone was defined as a clone whose frequency was 10x the sum of all homologous clones in the other sample.
TCR Diversity Score:
This measurement of diversity was calculated in a similar method as the singular & novel clone analysis. An initial matrix is created where the mapped sequence distance is calculated for every unique combination of sequences in the repertoire. Then the average of the unique combination calculations is taken, weighted by reads, and reported as the TCR Diversity Score. Additional details of the algorithm behind this calculation are shown in Suppl. Fig 6.
Shannon’s Entropy Calculations:
Calculation of Shannon’s Entropy was completed by the following formula where pi represents the frequency of each amino acid sequence and n represents the total number of sequences present in the response:
Statistical Methods:
No specific statistical method was used to determine sample size for the stimulation cohorts. Two-tailed t-tests were used as provided by GraphPad Prism 5 software for all comparative statistics given we expect normal distributions across all experiments.
Code Availability:
Upon request, the MATLAB scripts that comprise the ImmunoMap algorithms used to generate the data in this manuscript are available on a GitHub Repository. Additionally, a PC/Mac graphical user interface (GUI) has been developed for ImmunoMap for others to apply their algorithms without a MATLAB license or programming knowledge
Data Availability:
TCR β-chain sequencing raw data is available on Adaptive Biotechnologies’ ImmuneACCESS data portal.
Results
Overview of ImmunoMap Algorithms
Weighted Repertoire Dendrograms:
In order to visualize the immune response, we created weighted dendrograms; combining information about sequence relatedness with information about sequence frequency. We initially applied this analysis to data (taken from the Adaptive Biotechnologies Data Portal(22)) on the response of tetramer-sorted human CD8+ T cells to CMV (Figure 1A). The distance from the end of the dendrogram branches denotes distance in terms of sequence homology; the size of the circles at the ends of the branches denotes frequency of the sequence, and color denotes V-beta usage. Sequence distance is determined as a function of global alignment scores (Needleman-Wunsch(23), PAM10 scoring matrix(24), Gap Penalty = 30) between all unique combination of sequences as follows:
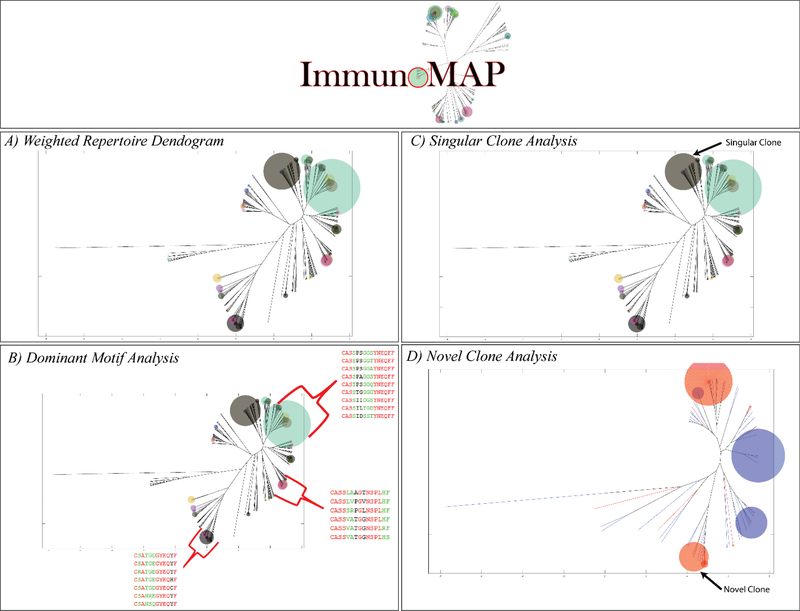
Figure 1. Elements of ImmunoMap Algorithm.
A) Weighted Repertoire Dendogram visualize relatedness of sequences within repertoire along with relative frequency of CDR3 amino acid sequences. B) Dominant Motif Analysis clusters homologous sequences and selects for clusters contributing to significant proportion of the response. 3 Dominant Motifs are shown representing highly represented structural motifs in this individual’s CMV response. C) Singular Clone Analysis defines sequences that expand significantly over the summation of all other homologous sequences D) Novel Clone Analysis is implemented when comparing repertoires from different samples and a novel clone is defined as one that expands significantly over the summation of all homologous sequences in the other sample.
Dominant Motif Analysis:
In order to parse the many sequences that are detected in antigen-specific CTL expansion, we sought to perform hierarchical clustering to determine structural motifs that dominated the response. Thresholds for sequence homology and frequency were set by analyzing the sequences of the naïve B6 CD8+ repertoire, taken from the Adaptive Biotechnologies Data Portal(25), Suppl. Figure 1. We used these thresholds to define homology clusters based on sequence distance and then examined clusters that met a predefined frequency threshold and termed them dominant motifs (Figure 1B).
Singular and Novel Structural Clones Analysis:
In addition to dominant motif analysis, we also defined a singular structural clone as one that has expanded 10x more than the summation of all other homologous clones in a sample, representing a singular solution in ‘sequence space.’ (Figure 1C). Furthermore, when comparing two separate CMV-specific sequencing samples, from different individuals, we define a novel structural clone as one that has expanded 10x more than the summation of all homologous clones to it in another sequencing sample, representing a newly expanded structural clone (Figure 1D).
TCR Diversity Score:
To quantify the diversity of the entire TCR repertoire, we created a metric to quantify the relatedness of an entire sample; defined as the average mapped sequence distance of all unique combinations of sequences in a sample, weighted by number of reads per sequence.
The TCR Diversity score is bounded between 0 and 1, where a score of 0 would correspond to all TCR’s in a response being identical and 1 would correspond to all TCR’s being infinitely different (full details of algorithms to calculate TCR Diversity Score in Suppl. Fig 6).
Naïve TCR Repertoires against Model Tumor Antigens
To understand the clonal diversity of antigen responses, CD8+ T cells pooled naïve B6 mice were expanded against a model foreign-antigen Kb-SIY, or against a self-tumor antigen, Kb-TRP2 (180–188), as previously described (26,27). Briefly, CD8+ T cells were enriched and stimulated with nanoparticle artificial antigen presenting cells (aAPCs) containing peptide-MHC-Ig molecules and cultured in vitro for 7 days (Figure 2A). The resultant CD8+ T cell cultures were shown to be antigen-specific by both peptide-MHC-Ig staining and cytokine analysis confirming their functional specificity (Suppl. Figure 2). Initial precursor frequency was also measured in the endogenous repertoire and even though both antigens can be expanded from naïve animals, Kb-SIY antigen has the higher naïve precursor frequency (Suppl. Figure 2). Antigen-specific populations were sorted and TCR Vβ chain CDR3 sequenced.
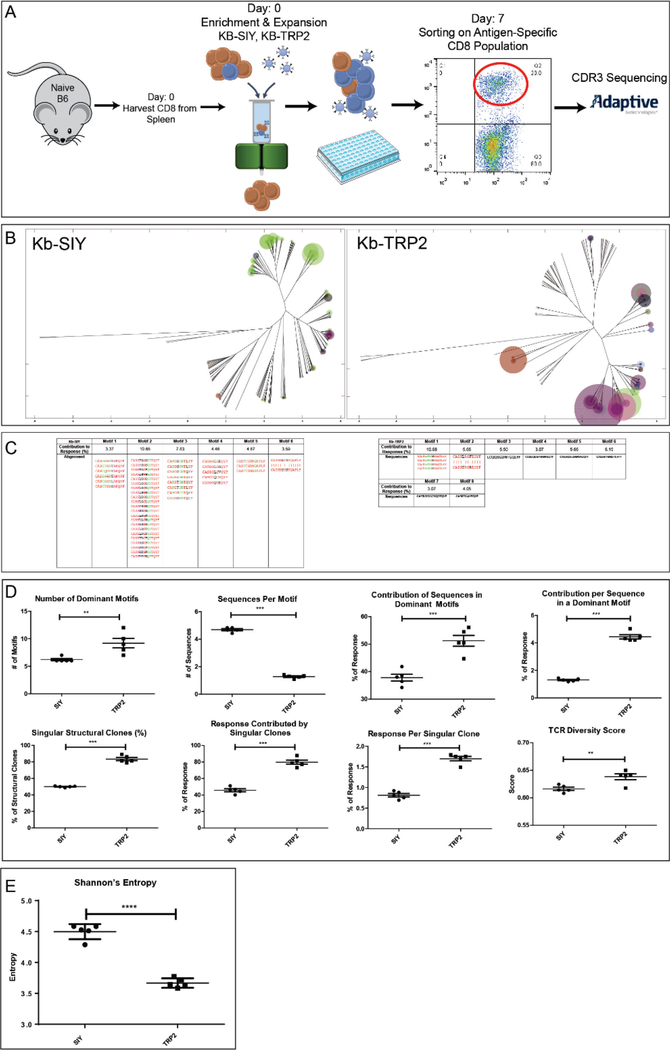
Figure 2. Naïve Repertoire to Kb-SIY vs Kb-TRP2 antigens.
A) Naïve B6 Animals were harvested for CD8 T cells from spleens. CD8 T cells underwent Enrichment & Expansion protocol via nano-aAPC’s and then were cultured ex-vivo for 7 days before undergoing flow-sorting for antigen-specific cells and being sent for TCR β-chain sequencing. B) Weighted Repertoire Dendograms where distance of ends of branches denotes sequence distance, size of circles denotes frequency of sequence, and color of circle denotes specific V beta segment usage. C) Demonstration of dominant motif’s detected for Kb-SIY (left) and Kb-TRP2 (right). Frequency and global sequence alignment is shown (Red = Fully Conserved Amino Acids, Green = Semi-Conserved Amino Acids, Black = Nonconserved Amino Acids). D) Top Row – Quantification of Dominant Motif Analysis comparing the number of dominant motifs, the number of sequences per motif, the contribution of the sequences in the dominant motifs to the response, and the contribution to the response per sequence in a dominant motif. Bottom Row – Singular Structural Clone and TCR Diversity analysis metrics. (n=5). E) Shannon’s Entropy Calculations comparing endogenous Kb-SIY to Kb-TRP2 responses. (N = 5 mice pooled, ** : P-value ≤ 0.01, *** : P-value ≤ 0.001 using the unpaired two-tailed t-test, bar represents mean ± s.e.m)
ImmunoMap analysis of Kb-SIY-specific and Kb-TRP2-specific TCR (Figure 2B) visualizes unique aspects of the polyclonal response for both antigens. Kb-SIY CD8+ T cells consist of clones with homologous TCR sequences; however, the naïve response to Kb-TRP2 is more clonal in nature (more high frequency clones) and uses more unrelated sequences each creating a distinct clonal variant for antigen recognition.
Dominant motif analysis showed that anti-Kb-SIY TCR had fewer yet richer (more sequences per motif) dominant motifs than Kb-TRP2 (Figure 2C, D). Kb-TRP2 specific T cells had a higher percentage of clones representing singular structural T cell expansions and they took up a larger portion of the overall TRP-2 antigen-specific response (Figure 2D – bottom row). Comparing the TCR Diversity Scores, Kb-SIY stimulated a more homologous response while Kb-TRP2 had a more diverse response. Interestingly, the response to Kb-SIY had a more conserved V-beta usage, predominantly using V-beta 13 while response to Kb-TRP2 exhibited a more diverse use of V-beta segments (Suppl. Figure 3).
To demonstrate the novelty of the ImmunoMap analysis over traditional analytic methods, we calculated Shannon’s Entropy for Kb-SIY vs Kb-TRP2 responses (Figure 2E). According to this metric, the diversity of the Kb-SIY response is higher than that of the Kb-TRP2 response. However, since Shannon’s Entropy is largely determined by the number of sequences that are present in the Kb-SIY response and not their relatedness, it missed the important finding that while there are more sequences that respond to Kb-SIY, they are more convergent than the fewer sequences that response to Kb-TRP2. Thus, the ImmunoMap TCR Diversity score and Dominant Motif analyses reflect novel relatedness-information which cannot be seen by conventional Shannon’s Entropy calculations.
Tumor Exerts Differential Expansion Pressure on Antigen-Specific Repertoire
While we know that tumors exert pressure on the immune response, there has been little analysis of how it alters the repertoire of responding T cells. ImmunoMap offers the opportunity to understand the biological impact of tumors on T cell responses and TCR usage by studying TCR repertoire changes in the presence of tumor (B16-SIY) (28). Visualization of the TCR repertoire by ImmunoMap analysis (Figure 3A) demonstrates differential effects of tumor on the repertoire of pooled splenic T cells specific for Kb-SIY or Kb-TRP2. The Kb-SIY CD8 repertoire is largely unaltered in response to tumor, with a high amount of homology between the naïve and tumor-bearing repertoires. In contrast, the Kb-TRP2 response not only exhibits a higher level of clonality but also uses TCR sequences that have minimal sequence homology to the TCR’s seen in the naïve C57Bl/6 response.
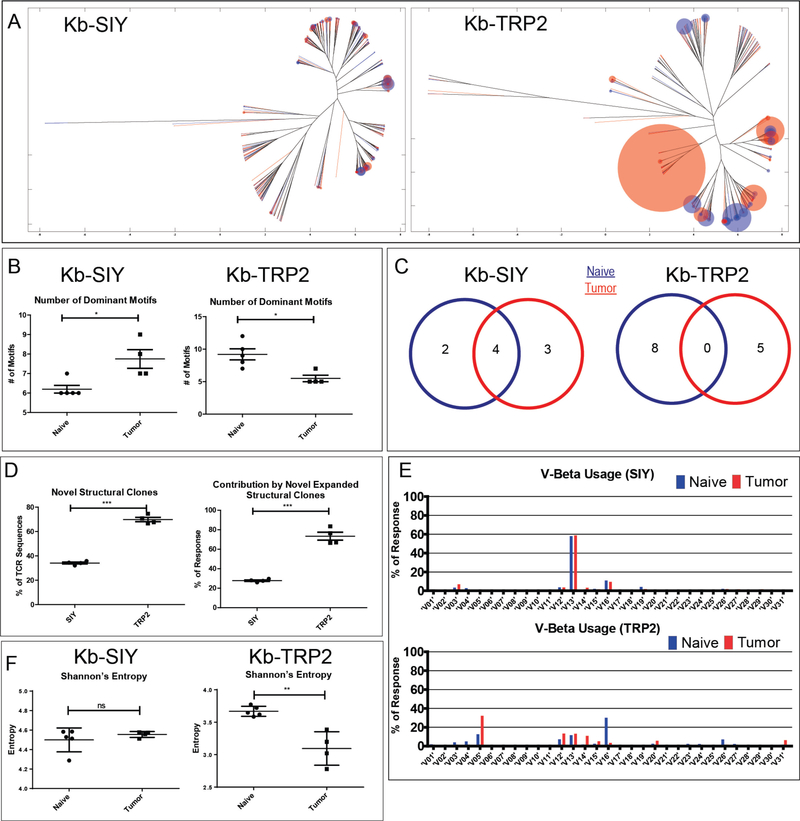
Figure 3. Effects of Tumor on TCR Repertoire.
A) Overlapped weighted repertoire dendograms of tumor-bearing vs naïve antigen-specific splenic CD8 responses. (Red = Tumor-bearing repertoire. Blue = Naïve repertoire). B) Dominant Motif analysis for Kb-SIY and Kb-TRP2 responses before and after exposure to tumor (n=5 mice). C) Maintenance of Dominant Motifs between Naïve and Tumor-Bearing Repertoire. D) Novel Structural Clone Analysis (n= 5 mice). E) V Beta usage of Kb-SIY and Kb-TRP between Naïve and Tumor-Bearing Repertoire. F) Shannon’s Entropy Calculations comparing endogenous vs tumor-bearing responses to Kb-SIY and Kb-TRP2. (N = 3 mice pooled, * : P-value ≤ 0.05, *** : P-value ≤ 0.001 using the unpaired two-tailed t-test, bar represents mean ± s.e.m)
Dominant motif analysis showed (Figure 3B) that the presence of tumor increased the number of dominant motifs in the Kb-SIY response In contrast, the presence of tumor decreased the number of dominant motifs in the Kb-TRP2 response, suggesting directed immune pressure on the self vs foreign antigens in the context of tumor. Furthermore, there was conservation of dominant Kb-SIY motifs (Figure 3C). In contrast, no common dominant motifs were shared in the Kb-TRP2 response in tumor-bearing animals compared to the naïve response. When examining novel structural clones (Figure 3D), the Kb-TRP2 response had a larger amount of structurally novel sequences that combined took up larger portion of the response as compared to the naïve response. Tumor pressure on the immune response was also seen in analyzing the V-beta usage between naïve and tumor-bearing animals (Figure 3E). We saw a strong pressure on Kb-TRP2 response, eliminating the use of V-beta 16 and an increased use of V-beta 5. In contrast, there was a conservation of V-beta usage in the Kb-SIY response between naïve and tumor-bearing animals.
Additionally, when examining the effect of tumor on Shannon’s Entropy (Figure 3F), we see that while the maintenance of the entropy in the Kb-SIY response and decrease in Entropy in the Kb-TRP2 response generally complement the ImmunoMap Dominant Motif Analysis, they fail to provide important information about the conservation, or lack thereof, of sequence structure caused by the tumor.
Lymphoid Organ-Dependent Differences in TCR Repertoires in Tumor Bearing Mice
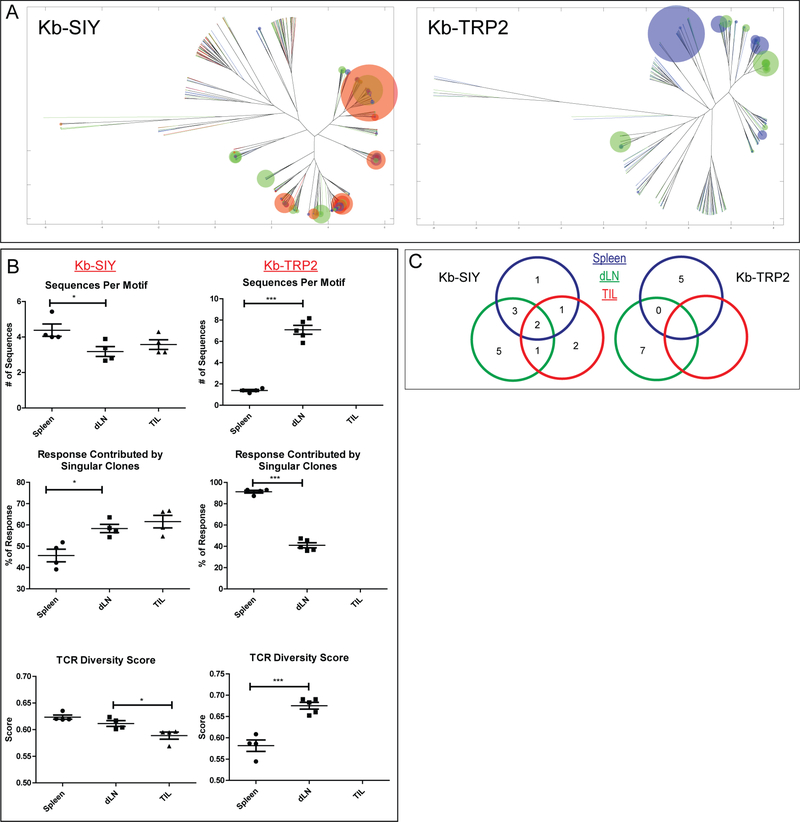
We hypothesized that the influence of tumor on repertoire may also vary dependent on the relationship of the lymphoid organ to the tumor site. This was studied by analyzing antigen-specific TCR repertoires in the spleen versus draining lymph node (dLN), and TIL in pooled tumor-bearing mice lymph nodes and tumor. ImmunoMap analysis revealed that the Kb-SIY repertoire selects for effective structural motifs as one probes compartments closer to the tumor site. This is seen as the richness of dominant motifs decreases, the response contributed by singular clones increases, and the TCR Diversity Score drops as one moves from the spleen towards the tumor (Figure 4B). Additionally, the structural clones expanded in the spleen, dLN, and TIL’s are generally conserved as can be visualized by the dendrograms (Figure 4A) and by tracking dominant motifs in the 3 lymphoid compartments (Figure 4C). In contrast, the opposite trend was seen in the Kb-TRP2 response. Additionally, there was no conservation of dominant motifs between the spleen and draining lymph node; we were unable to expand any Kb-TRP2 specific cells from the TIL’s in multiple experiments (Figure 4 A-C).
Figure 4. Effects of Tumor on TCR Repertoire in Various Lymphoid Organs.
A) Overlapped weighted repertoire dendograms (blue = spleen, green = draining lymph node, red = TILs). B) Dominant Motif and TCR Diversity Metrics (Kb-SIY n=4 mice, Kb-TRP2 Spleen n=4 mice, Kb-TRP2 dLN n=5 mice). C) Maintenance of Dominant Motifs between various lymphoid organs. (N = 4 mice pooled by organ, * : P-value ≤ 0.05, *** : P-value ≤ 0.001 using the unpaired two-tailed t-test, bar represents mean ± s.e.m)
Analysis of α-PD1 Clinical Trial Data Reveals Indicators of Response
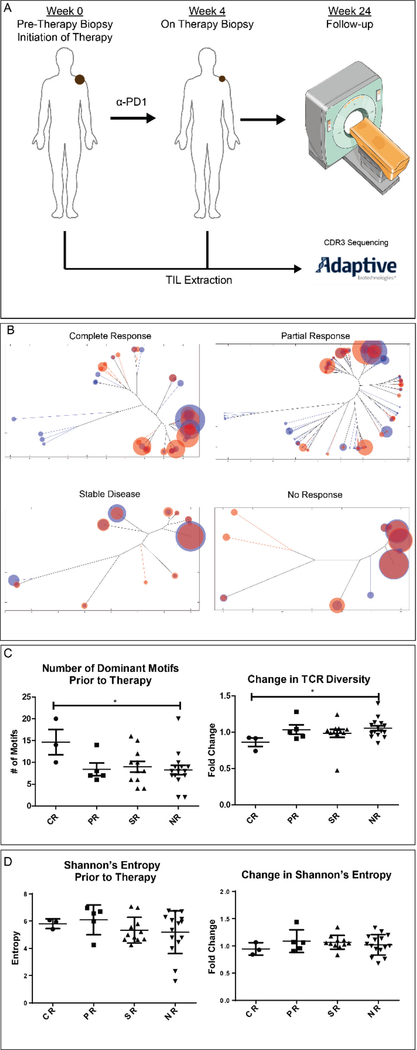
Recent studies have implicated changes in T cell responses as important in clinical outcomes to checkpoint blockade. We therefore applied ImmunoMap analysis to clinical trial data (BMS-038) from patients with metastatic melanoma undergoing α-PD1 therapy (Nivolumab). For this analysis, FFPE scrapings were taken from 34 patients, %TIL was estimated as per Adaptive protocol (See Materials and Methods) and CDR3 β-chains sequenced pre- and on-therapy (Figure 5A). In all samples analyzed there was no statistically significant difference in number of T cells sequenced (Suppl. Figure 5).
Figure 5. TCR Repertoire Analysis of Patients Undergoing α-PD1 (Nivolumab) Therapy.
A) Clinical Protocol for sample collection and response stratification. Pre-therapy biopsies were taken from tumor sites prior to initiation of therapy. 4 weeks after initiation of α-PD1 therapy, on-therapy biopsies were taken from same tumor sites. TIL extraction was completed sent to Adaptive Biotechnologies for CDR3 β-chain sequencing. B) Token weighted repertoire dendograms for each of the cohorts of responders. C) Dominant Motif and TCR Diversity analysis (CR = 3, PR=5, SR=11, NR=15).D) Shannon’s Entropy Calculations for responses prior and after initiation of α-PD1. (* : P-value ≤ 0.05 using the Mann Whitney test, bar represents mean ± s.e.m)
ImmunoMap was used to compare TCR repertoire prior to and after 4 weeks of α-PD1 therapy (all ImmunoMap metrics in BMS038Results.xlsx). Weighted repertoire dendrograms (Figure 5B) revealed distinct differences between responders and non-responders. Dominant motif analysis (Figure 5C) showed that patients who had more dominant motifs prior to initiation of therapy had more favorable responses to therapy. Additionally, those patients who had a decrease in their TCR Diversity Score (Figure 5C) on therapy had more favorable outcomes to therapy. In contrast no clinically relevant signature could be found by Shannon entropy calculations (Figure 5D). Thus ImmunoMap analysis uniquely revealed repertoire characteristics predicting response to therapy after just four weeks of treatment.
Discussion
Here we show a novel bioinformatics approach to analyze TCR repertoire sequence data, ImmunoMap, and used the approach to characterize repertoire changes in responses to model murine tumors and in patients undergoing immunotherapy for melanoma. By combining information about sequence relatedness and frequency, ImmunoMap allows an intuitive appreciation of TCR repertoire characteristics that reconciles the structure and function of the repertoire.
ImmunoMap analysis of foreign (Kb-SIY) to self (Kb-TRP2) antigens, showed distinct differences in the naïve repertoire to these two different antigens. Our observations show interesting differences in the foreign vs self-antigen repertoire, the conclusions of this analysis cannot be expanded to all foreign vs self-antigens. The presence of more dominant motifs in the Kb-TRP2 response in combination with its higher level of clonality suggests that central and peripheral tolerance mechanisms limited clonal responses with more distinct clones occupying a larger portion of the TRP2-specific repertoire. Self-reactive clones, with TRP2, TCR’s would be either removed during central thymic development or tolerized in the periphery, explaining the inability to find more numerous TCR sequences per dominant motif (29–31). Since our analysis was conducted on expanded antigen-specific populations, our results demonstrate the ‘expansion potential’ of the antigen-specific T cell repertoire for a model foreign and shared tumor antigen in the setting of both naïve and tumor-bearing animals. It is possible that the limited TCR relatedness of TRP2 responses could be due to the lower precursor frequency in naïve animals, and the T cells that have the ability to expand do not cluster in the same dominant motif due to lower initial cell frequency. Furthermore, it is important to also consider the impact of pooling animals prior to expansion and sequencing. In this scenario, one could be selecting for ‘public’ clones and possibly enriching for these parts of the repertoire over ‘private’ clones, unique to each animal. While individual mice are genetically identical, VDJ recombination occurs as an independent process in each animal and the primary TCR repertoire capable of responding to a given antigen could vary between individual animals. Therefore, the effects on shaping of the repertoire may be most relevant to ‘public’ or conserved sequences. Finally, the higher TCR diversity score of Kb-TRP2 alongside with the higher number of dominant motifs suggests that the immune system has to reach further to find solutions to bind the cognate antigen/MHC complex.
While prior work on TCR clustering has focused on understanding the structural aspects that confer antigen-specificity (14–15), the effects of perturbations to the immune system on antigen-specific responses has not been studied. With ImmunoMap, we studied the changes in repertoire in response to tumor, and observed that the effects of tumor on the anti-self-Kb-TRP 2 peptide repertoire indicates that tumors exert greater pressure on the self- than on the foreign-antigen. Not only did the tumor increase the clonality of the response to self by decreasing the number of dominant motifs and increasing their contribution to the net response but it was able to shift the response to entirely novel, presumably suboptimal, motifs. Additionally, the differences in repertoire characteristics among various lymphoid organs for the two different model antigens indicates that tumor effectively eliminates the expansion of certain clones from its microenvironment. The consequences of these findings are relevant to both antigen-discovery and targeting for immune therapies related to treating cancer. Due to limitations of personalized antigen-specific therapy, targeting shared antigens like MART1, a self-antigen specific for melanocytes has been a mainstay of antigen-specific cancer immunotherapy (32–34). This approach has typically relied on TCR transgenic models where a single TCR clone is chosen as the source for the antigen-specific receptor (35–37). Given our analysis, several problems with this approach are apparent: (1) antigen-specific expansion not only generates a diversity of TCR sequences but one that spans the entire sequence distance of the naïve repertoire, (2) self-antigen expansion represents a limited repertoire and arsenal against a given epitope due to effects of tolerance, and (3) the tumor is able to exert pressure on the self-antigen specific immune response in a more profound way than in the case of a foreign antigen. Our findings call into question the approach of using self or over-expressed antigens as targets for immune therapy and highlight the importance of exploring responses to neoantigens, novel MHC-specific epitopes that arise from mutations in a patient’s individual malignancy (38–42).
Furthermore, we also used ImmunoMap algorithms to understand mechanisms of successful immune responses to cancer against 4T1, a murine breast cancer model. In that model when analyzing TIL’s from animals treated with α-CTLA4, radiation, or the combination of these therapies, we found that TCR structural repertoire prior to therapy from TIL’s was highly conserved, seemingly targeting a single antigen and following combination therapy, there was a broadening of the structural response within the TIL’s and each individual animal developed its own uniquely expanded repertoire (43).
Finally, we used ImmunoMap to study TIL’s from clinical trial specimens to determine if structural diversity is an important parameter in determining successful immune responses to cancer immunotherapy. Our analysis revealed that patients who had more dominant motifs prior to therapy responded more favorably to therapy. Additionally, the change in TCR diversity suggest that patients who respond to therapy converge on a solution of successful TCR sequences and thus their repertoire is actually less diverse after therapy. In contrast to previous work by Madi et. al that demonstrated a structural broadening of the peripheral repertoire to α-CTLA4 therapy in melanoma patients but did not correlate this finding with response, we focused our analysis on studying changes in the repertoire within the TIL’s and were able to determine structural signatures of response (16). While our findings are significant, we note the scope of the clinical trial was limited which impacted of the distribution of clinical responses. Nevertheless, taken together, ImmunoMap analysis revealed that patients with a broader repertoire prior to therapy have a higher probability of expanding effective TCR sequences and converging on them.
ImmunoMap not only has the potential to have clinical implications for monitoring patients on therapy, knowing their likelihood to respond, but also has opened new paths of biological inquiry around antigen-specific immune responses that could alter current immune therapies.
Supplementary Material
Statement of Significance:
TCR sequencing helps understand antigen-specific immune responses. While there has been an increase in sequencing data, there has been little innovation in parsing it into biologically meaningful conclusions. ImmunoMap is a bioinformatics tool that reconciles TCR repertoire function and structure by quantifying the ‘relatedness’ of the response by sequence homology.
Acknowledgements
The authors thank the JHU-Coulter Translational Partnership, the TEDCO Maryland Innovation Initiative, The Troper Wojcicki Foundation, the NIH (R01- AI44129,CA108835, and U01 – AI113315), and sponsored research agreements with Miltenyi Biotec and NexImmune for support of this research. Animal and clinical protocol images reproduced under a Creative Commons License from Servier Medical Art (http://www.servier.com/Powerpoint-image-bank). Additionally, we thank each patient and their families for participation in this study and for their willingness to accept on-treatment biopsies to facilitate scientific advancement in cancer immunotherapy. This work was funded by Bristol Myers-Squibb, the Pershing Square Sohn Cancer Research Foundation (T.A.C.), the PaineWebber Chair (T.A.C.), Stand Up 2 Cancer (T.A.C.), and the STARR Cancer Consortium (T.A.C.). Partial Support was provided by the MSK Core Grant (P30 CA008748). Editorial assistance was provided by Dr. Michael Edidin and Dr. Drew Pardoll.
Footnotes
Disclosure of Potential Conflicts of Interest
Under a licensing agreement between NexImmune and The Johns Hopkins University, Jonathan Schneck is entitled to a share of royalty received by the university on sales of products described in this article. He was a founder of NexImmune and currently serves as a member of NexImmune’s Scientific Advisory Board and Board of Directors and has equity in NexImmune. Timothy A. Chan is the co-founder of Gritstone Oncology. Jonathan Havel’s wife is a full-time employee of Regeneron Pharmaceuticals.
In order to use the ImmunoMap algorithms, we have developed a MATLAB-based Graphical User Interface (GUI) that can be found along with the source code at https://github.com/sidhomj/ImmunoMap. Supplementary Figure 8 demonstrates the use of the GUI.
References
- 1.Boyd SD, Marshall EL & Merker JD. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel VDJ pyrosequencing. Science translational … (2009). at <http://stm.sciencemag.org/content/1/12/12ra23.short> [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vollmers C, Sit RV & Weinstein JA. Genetic measurement of memory B-cell recall using antibody repertoire sequencing. Proceedings of the … (2013). doi: 10.1073/pnas.1312146110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Robins HS, Campregher PV, Srivastava SK & Wacher A Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood (2009). at <http://www.bloodjournal.org/content/114/19/4099?variant=long> [DOI] [PMC free article] [PubMed]
- 4.Wang C, Sanders CM & Yang Q High throughput sequencing reveals a complex pattern of dynamic interrelationships among human T cell subsets. Proceedings of the … (2010). doi: 10.1073/pnas.0913939107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jackson K, Liu Y, Roskin KM, Glanville J & Hoh RA. Human responses to influenza vaccination show seroconversion signatures and convergent antibody rearrangements. Cell host & …(2014). doi: 10.1016/j.chom.2014.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Logan AC, Gao H, Wang C & Sahaf B High-throughput VDJ sequencing for quantification of minimal residual disease in chronic lymphocytic leukemia and immune reconstitution assessment.Proceedings of the … (2011). doi: 10.1073/pnas.1118357109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grupp SA, Kalos M, Barrett D & Aplenc R Chimeric antigen receptor–modified T cells for acute lymphoid leukemia. … England Journal of … (2013). doi: 10.1056/NEJMoa1215134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carreno BM et al. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science (New York, N.Y.) 348, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sherwood AM, Emerson RO & Scherer D Tumor-infiltrating lymphocytes in colorectal tumors display a diversity of T cell receptor sequences that differ from the [DOI] [PMC free article] [PubMed]
- 10.Jost L Partitioning diversity into independent alpha and beta components.Ecology (2007). doi: 10.1890/06-1736.1 [DOI] [PubMed] [Google Scholar]
- 11.Stewart JJ et al. A Shannon entropy analysis of immunoglobulin and T cell receptor.Molecular immunology 34, 1067–1082 (1997). [DOI] [PubMed] [Google Scholar]
- 12.Venturi V, Kedzierska K, Tanaka MM & Turner SJ. Method for assessing the similarity between subsets of the T cell receptor repertoire. Journal of … (2008). doi: 10.1016/j.jim.2007.09.016 [DOI] [PubMed] [Google Scholar]
- 13.Victor T-SC, Rech AJ, Maity A & Rengan R Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature (2015). doi: 10.1038/nature14292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glanville J, Huang H, Nau A, Hatton O, Wagar LE, Rubelt F, ... & Haas, N. (2017). Identifying specificity groups in the T cell receptor repertoire. Nature, 547(7661), 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dash P, Fiore-Gartland AJ, Hertz T, Wang GC, Sharma S, Souquette A, ... & La Gruta NL (2017). Quantifiable predictive features define epitope-specific T cell receptor repertoires. Nature, 547(7661), 89–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madi A, Poran A, Shifrut E, Reich-Zeliger S, Greenstein E, Zaretsky I, ... & Sun PD (2017). T cell receptor repertoires of mice and humans are clustered in similarity networks around conserved public CDR3 sequences. Elife, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oelke M et al. Ex vivo induction and expansion of antigen-specific cytotoxic T cells by HLA-Ig-coated artificial antigen-presenting cells. Nat. Med 9, 619–24 (2003). [DOI] [PubMed] [Google Scholar]
- 18.Perica K et al. Nanoscale artificial antigen presenting cells for T cell immunotherapy.Nanomedicine 10, 119–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oelke M, Moehrle U, Chen JL, Behringer D, Cerundolo V, Lindemann A, & Mackensen A (2000). Generation and purification of CD8+ melan-A-specific cytotoxic T lymphocytes for adoptive transfer in tumor immunotherapy. Clinical Cancer Research, 6(5), 1997–2005. [PubMed] [Google Scholar]
- 20.Robins HS, Campregher PV, Srivastava SK, Wacher A, Turtle CJ, Kahsai O, ... & Carlson CS (2009). Comprehensive assessment of T-cell receptor β-chain diversity in αβ T cells. Blood, 114(19), 4099–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlson CS et al. Using synthetic templates to design an unbiased multiplex PCR assay. Nat Commun 4, 2680 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Suessmuth Y et al. CMV reactivation drives posttransplant T-cell reconstitution and results in defects in the underlying TCRβ repertoire. Blood 125, 3835–3850 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Needleman SB & Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of molecular biology(1970). at <http://www.sciencedirect.com/science/article/pii/0022283670900574> [DOI] [PubMed]
- 24.Altschul SF. Amino acid substitution matrices from an information theoretic perspective. Journal of molecular biology(1991). at <http://www.sciencedirect.com/science/article/pii/002228369190193A> [DOI] [PMC free article] [PubMed]
- 25.Carey AJ et al. Rapid Evolution of the CD8+ TCR Repertoire in Neonatal Mice.The Journal of Immunology 196, 2602–2613 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oelke M et al. Ex vivo induction and expansion of antigen-specific cytotoxic T cells by HLA-Ig-coated artificial antigen-presenting cells. Nat. Med 9, 619–24 (2003). [DOI] [PubMed] [Google Scholar]
- 27.Perica K et al. Enrichment and Expansion with Nanoscale Arti fi cial Antigen Presenting Cells for Adoptive Immunotherapy. 6861–6871 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blank C et al. PD-L1/B7H-1 inhibits the effector phase of tumor rejection by T cell receptor (TCR) transgenic CD8+ T cells.Cancer research 64, 1140–1145 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Kyewski B & Klein L A central role for central tolerance. Annu. Rev. Immunol(2006). doi: 10.1146/annurev.immunol.23.021704.115601 [DOI] [PubMed] [Google Scholar]
- 30.Hogquist KA, Baldwin TA & Jameson SC. Central tolerance: learning self-control in the thymus. Nature Reviews Immunology(2005). doi: 10.1038/nri1707 [DOI] [PubMed] [Google Scholar]
- 31.Piccirillo CA & Thornton AM. Cornerstone of peripheral tolerance: naturally occurring CD4+ CD25+ regulatory T cells. Trends in immunology(2004). doi: 10.1016/j.it.2004.04.009 [DOI] [PubMed] [Google Scholar]
- 32.Chodon T et al. Adoptive transfer of MART-1 T-cell receptor transgenic lymphocytes and dendritic cell vaccination in patients with metastatic melanoma.Clin. Cancer Res 20, 2457–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Bade E, Kuniyoshi C, Spears L & Jeffery G Phase I trial of a MART-1 peptide vaccine with incomplete Freund’s adjuvant for resected high-risk melanoma.Clinical Cancer … (1999). at <http://clincancerres.aacrjournals.org/content/5/10/2756.short> [PubMed] [Google Scholar]
- 34.Rosenberg SA, Zhai Y & Yang JC. Immunizing patients with metastatic melanoma using recombinant adenoviruses encoding MART-1 or gp100 melanoma antigens. Journal of the …(1998). doi: 10.1093/jnci/90.24.1894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanson HL, Donermeyer DL, Ikeda H & White JM. Eradication of established tumors by CD8+ T cell adoptive immunotherapy. Immunity (2000). doi: 10.1016/S1074-7613(00)00026-1 [DOI] [PubMed] [Google Scholar]
- 36.Morgan RA, Dudley ME & Wunderlich JR. Cancer regression in patients after transfer of genetically engineered lymphocytes. … (2006). doi: 10.1126/science.1129003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vatakis DN, Koya RC & Nixon CC. Antitumor activity from antigen-specific CD8 T cells generated in vivo from genetically engineered human hematopoietic stem cells. Proceedings of the … (2011). doi: 10.1073/pnas.1115050108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dudley ME & Rosenberg SA. Adoptive-cell-transfer therapy for the treatment of patients with cancer. Nature Reviews Cancer(2003). doi: 10.1038/nrc1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Speiser DE, Miranda R & Zakarian A Self antigens expressed by solid tumors do not efficiently stimulate naive or activated T cells: implications for immunotherapy.The Journal of … (1997). doi: 10.1084/jem.186.5.645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schumacher TN & Schreiber RD. Neoantigens in cancer immunotherapy. Science (2015). doi: 10.1126/science.aaa4971 [DOI] [PubMed] [Google Scholar]
- 41.Lennerz V, Fatho M & Gentilini C The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proceedings of the … (2005). doi: 10.1073/pnas.0500090102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gros A et al. Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat. Med (2016). doi: 10.1038/nm.4051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rudqvist NP et al. Radiotherapy and CTLA-4 blockade shape the TCR repertoire of tumor-infiltrating cells. Cancer Immunology Research (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TCR β-chain sequencing raw data is available on Adaptive Biotechnologies’ ImmuneACCESS data portal.