Abstract
Unlike normal tissues, tumor cells possess a propensity for genomic instability, resulting from elevated oxidant levels produced by oncogenic signaling and aberrant cellular metabolism. Thus, targeting mechanisms that protect cancer cells from the tumor-inhibitory consequences of their redox imbalance and spontaneous DNA-damaging events is expected to have broad-spectrum efficacy and a high therapeutic index. One critical mechanism for tumor cell protection from oxidant stress is the hydrolysis of oxidized nucleotides. Human MutT homolog 1 (MTH1), the mammalian nudix (nucleoside diphosphate X) pyrophosphatase (NUDT1), protects tumor cells from oxidative stress-induced genomic DNA damage by cleansing the nucleotide pool of oxidized purine nucleotides. Depletion or pharmacologic inhibition of MTH1 results in genomic DNA strand breaks in many cancer cells. However, the mechanisms underlying how oxidized nucleotides, thought mainly to be mutagenic rather than genotoxic, induce DNA strand breaks are largely unknown. Given the recent therapeutic interest in targeting MTH1, a better understanding of such mechanisms is crucial to its successful translation into the clinic and in identifying the molecular contexts under which its inhibition is likely to be beneficial. Here we provide a comprehensive perspective on MTH1 function and its importance in protecting genome integrity, in the context of tumor-associated oxidative stress and the mechanisms that likely lead to irreparable DNA strand breaks as a result of MTH1 inhibition.
Keywords: MTH1, DNA repair, 8-oxoguanine, DNA strand breaks, cancer, reactive oxygen species (ROS)
Graphical Abstract

1. Introduction
1.1. Reactive oxygen species & DNA and nucleotide damage
Many tumors sustain high reactive oxygen species (ROS) levels due to aberrant metabolism and constitutive oncogenic signaling [1-3]. ROS are major effectors of cancer-associated DNA damage and concomitant tumor suppression. Therefore, aggressive tumors acquire protective adaptations against oxidative DNA damage. Several studies indicate that the nucleotide pool is more vulnerable to cellular oxidants than chromatin-protected DNA [4-6]. Accordingly, one such important adaptation is the mammalian nucleotide pool-sanitizing enzyme and functional 8-oxodGTPase, MutT Homolog 1 (MTH1, aka NUDT1). It has been reported (from as early on as 2001) [7] that tumors sustain higher MTH1 levels than adjacent normal tissues [8, 9]. This observation has also been made through global gene expression profiling in cancer patient datasets [10, 11]. Indeed, in oncogenic KRAS-driven cancers, such as pancreatic and lung adenocarcinomas, high MTH1 expression increases relapse rates by almost 50% [12]. Thus, tumors associated with elevated oxidant levels [13] and propensity towards DNA damage [14] ostensibly exhibit increased reliance on MTH1. Although MTH1 was initially identified as an anti-mutator protein [15, 16], recent studies indicate it has another important DNA maintenance function as it protects cells under oxidative stress, particularly tumor cells, from genomic DNA breaks and associated anti-viability outcomes [8, 17-19]. The first-in-class MTH1 inhibitors inhibit tumors via production of genomic DNA breaks [19], recapitulating the adverse genomic effects originally reported for MTH1 depletion via shRNA [17]. However, the mechanisms by which MTH1 inhibition-induced oxidation in the nucleotide pool translates to genotoxic damage is not well-established. Here, we survey what is known about MTH1 function in preventing DNA strand breaks in response to oxidative stress, with emphasis on the tumor-relevant molecular contexts that require this function and the cellular mechanisms able to transduce MTH1 functional loss into genomic DNA breaks.
1.2. MTH1 function in normal and cancer cells
MTH1 is an 18 kD nudix pyrophosphatase with substrate specificity for 8-oxodGTP and 8-oxoGTP [15, 20]. MTH1 is also able to hydrolyze 2-OH-dATP and its ribonucleotide analog into their monophosphate counterparts [21, 22]. It has recently been reported that MTH1 can detoxify O6-methylguanine in addition to 8-oxoguanine [23]. The nucleotide pool-sanitizing function of MTH1 is essential for maintaining genomic DNA integrity [5]. Importantly, DNA and RNA polymerases show poor discrimination between the oxidized and non-oxidized nucleotides, potentially leading to G->T, A->C and T->A transversion mutations through 8-oxodG:dA, 2-OHdA:dT or 2-OHdA:dA mispairings. Further, high MTH1 activity can compensate for compromised base excision repair to prevent increased frequency of genomically incorporated oxidized precursors [24].
MTH1 mainly localizes in the cytoplasm, although a mitochondrial isoform has also been identified [25]. Seven alternatively spliced MTH1 variants have been reported, as well as GU or GC polymorphism-carrying variants (at the beginning of exon 2c) that encode longer MTH1 isoforms [25]. The MTH1-KO (germline-null) mouse has no apparent pathology, barring very late onset spontaneous tumorigenesis [26] and somewhat enhanced neurotoxicity under oxidant challenge [27-29].
Although MTH1 is broadly expressed in most tissues, its protein levels are lower in normal tissue when compared to adjacent tumors [8, 9, 30]. Furthermore, MTH1-KO mice do not display increased frequency of the signature 8-oxodG GC -> TA transversion mutations [31]. Coupled with their normal physiology, these observations suggest that, at least in normal murine tissue, oxidized purine nucleotides do not occur at high enough levels for MTH1 loss to produce adverse effects, particularly when DNA base excision repair enzymes such as OGG1 or MUTYH are active. However, MTH1 depletion via shRNA in normal skin and lung fibroblasts induces p53-mediated cell senescence and is accompanied by genomic DNA strand breaks (DSBs) [17]. Conversely, MTH1 transgenic mice show improved functional aging and are protected against ROS-producing chemical mitochondrial toxicants, exhibiting significantly lower levels of 8-oxoG in mitochondrial DNA from heart, muscle and brain relative to their wildtype counterparts [32, 33]. Consistent with this physiologic redox-protective function, overexpression of MTH1 in untransformed human skin fibroblasts prevents oncogene-induced senescence and DSBs [18], and facilitates RAS-induced oncogenic transformation [9].
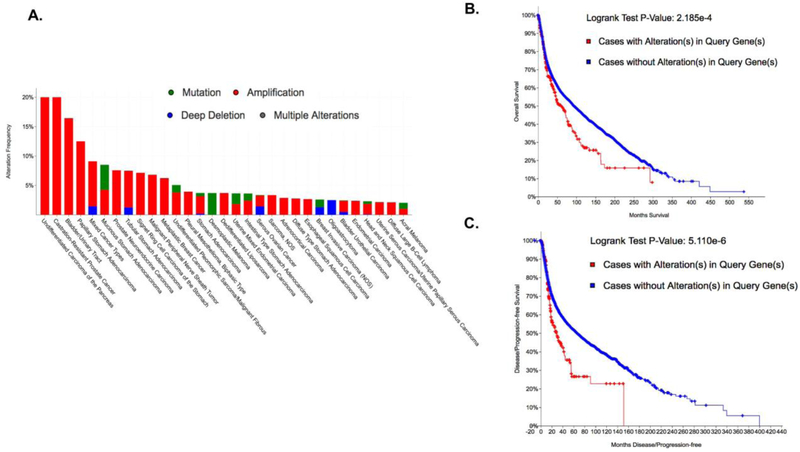
Analysis of large-scale patient tumor datasets (cbioportal.org) [34, 35] shows MTH1 is predominantly amplified in a number of commonly occurring and aggressive cancers (red bars, Fig. 1A) and that this alteration correlates significantly with poor overall (Fig, 1B) as well disease/progression-free survival (Fig. 1C). Furthermore, in the last few years, a plethora of studies reporting on multiple cancer models have implicated MTH1 in various aspects of tumor development and progression. MTH1 is required for optimal malignancy and oncogenic signaling via maintenance of oncogenic ROS levels and inhibition of genotoxic damage in RAS-driven xenograft lung tumors [8]. A recent study reveals that caveolin enforces RAS oncogene-induced senescence (OIS) via direct interaction and inhibition of MTH1 activity [36], supporting the earlier study showing elevated MTH1 levels promote evasion of OIS [18]. MTH1 promotes pro-metastatic cellular traits, such as enhanced migration and invasive ability, in malignant thyroid cells [37] and in lung cancer cells [9]. Skp2 ubiquitin ligase-mediated stabilization of MTH1 is associated with enhanced survival of melanoma cells under oxidative stress [38]. In mismatch repair (MMR)-deficient T-ALL Jurkat cells, both MTH1 and MUTYH are required to protect against apoptosis, suggesting these enzymes coordinately protect cells against the tumor-inhibitory consequences of increased 8-oxodGTP genomic incorporation [39]. MTH1 has also been reported to be important in the maintenance of glioblastoma stem cells, in promoting glioblastoma tumors and in their treatment-refractory behavior [40-42]. Furthermore, a number of studies have shown that MTH1 levels correlate with greater malignancy and poor prognosis in resected human lung tumors [43], colorectal tumors [44], and esophageal squamous cell carcinoma [45], and with greater frequency of ulcerative colitis-associated tumors [46]. Importantly, multiple studies have now shown that MTH1 depletion or inhibition increases genomic instability and DNA damage in cancer cells [8, 19, 40, 47-50]. Yet, recently developed MTH1 inhibitors and deletion of MTH1 by CRISPR/cas9 have been reported to have little, if any, impact on the proliferation of cancer cells in culture [51-53], further complicating our understanding of MTH1’s role in cancer cell growth. However, the mechanism(s) by which functional MTH1 loss induces genomic DNA breaks remain to be fully understood. This knowledge is essential in order to most optimally leverage somatic MTH1 loss for therapeutic purposes.
Figure 1. MTH1 (NUDT1) is amplified in multiple cancers and correlates with poor overall as well as disease-free survival.
(A) Data were obtained from cBioportal.org using a minimum 2% alteration cut-off. Note that the most common alteration is amplification (red bars). The metrics in (B) and (C) were compiled by querying 71857 patients / 74247 samples in 240 studies. The p-values (obtained via the Log-Rank test) are shown to the right of the graphs: (B) Overall survival Kaplan-Meier estimate and (C) Disease/progression-free Kaplan-Meier estimate.
2. Tumor-Relevant Oxidative Stressors
The occurrence of MTH1 inhibition-induced DNA breaks depends strongly on cellular oxidant levels. DNA breaks resulting from MTH1 inhibition are found to be lower in cells with low ROS levels and enhanced under conditions of oxidative stress such as oncogene activation [8, 17, 18]. Oncogenic transformation and malignant progression rely on elevated ROS levels (produced by the action of NADPH oxidases or mitochondrial metabolism) to drive pro-mitogenic and pro-survival kinase signaling [54]. Oncogenic ROS can also induce DNA damage and trigger tumor suppressive responses such as cell death or OIS, unless mitigated by protective mechanisms [1].
Mouse embryonic fibroblasts (MEFs) derived from MTH1 germline-null animals exhibit enhanced sensitivity to hydrogen peroxide (H2O2)-induced oxidative DNA damage and cell death relative to wildtype counterparts, despite no reported developmental defects in the MTH1-null animals [55]. Thus, MTH1 function is likely to be physiologically important for protecting cells against enhanced cellular oxidative stress, which is rare in normal, un-diseased tissues but is a hallmark (and vulnerability) of many cancers [54, 56].
Indeed, oncogenic RAS-harboring cells, which sustain elevated ROS [57, 58], possess higher MTH1 expression than their wildtype counterparts, and their proliferation is reduced to a greater extent by MTH1 depletion [5, 18]. Elevated MTH1 expression has been shown to prevent OIS and associated DNA double-strand breaks in oncogenic RAS-driven cells by reducing cellular 8-oxoG levels, and to enhance oncogenic transformation [8, 9, 18]. Furthermore, given the inherent elevated oxidative stress associated with oncogenic signaling, high MTH1 expression is associated with maintaining RAS oncoprotein levels whereas loss of MTH1 selects for tumor cells with lower oncogenic ROS levels and reduced oncogenic signaling, transformation and tumorigenicity [8, 9, 59]. Thus, MTH1 is capable of indirectly affecting the cellular redox state of transformed cells despite having no known ROS detoxifying function.
Other oncogenes such as NFkB, Myc and PI3K also produce ROS and cells expressing these oncogenes are prone to elevated levels of oxidative DNA damage [54]. MTH1-depleted xenograft tumors from KRAS-driven lung cancer cells exhibit reduced levels of Akt signaling, and increasing their Akt signaling through constitutively-activated Akt expression exacerbates MTH1 loss-associated proliferation defects due to elevated Akt signaling-induced ROS [8]. It remains to be investigated whether compromising MTH1 function in tumor cells harboring ROS-producing oncogenic drivers other than PI3K/Akt and RAS also induces DNA strand breaks and tumor-suppressive outcomes.
The metabolic switch from mitochondrial oxidative phosphorylation to glycolysis, produced by many oncogenic drivers, is often associated with adaptive responses to a hypoxic tumor environment [60, 61]. The resulting increase in ROS production under hypoxia stabilizes HIF1α [62-64]. Using a zebrafish embryo model, a recent study [65] found that hypoxia and the accompanying activation of HIF1α promotes oxidation of cellular glutathione pools and thus sensitization of the embryos to MTH1 inhibition. This study further reported that stabilization of HIF1α via the prolyl hydroxylase inhibitor dimethyloxalylglycine (DMOG) or via mutant loss-of-function von Hippel Lindau (VHL) factor leads to elevated MTH1 expression, and induces cytotoxicity by the first-in-class MTH1 inhibitor TH588 [65]. This result could be recapitulated by injecting 8-oxodGTP or 2-OH-dATP into the zebrafish embryos, to increase oxidation in their nucleotide pool. Thus, due to their elevated pro-oxidant state, hypoxic tumors, and particularly those with a dysregulated HIF1α/VHL axis, may respond well to MTH1 inhibition. Whether this sensitization occurs through increased genomic DNA damage or synergizes in some other way with the extant oxidative stress/oxidized nucleotides in the hypoxic milieu needs to be established. More significantly, the collective evidence for reliance of ROS-producing oncogenic signaling on DNA repair mechanisms indicates a complex molecular context underlying MTH1 functional requirement that is yet to be fully elucidated, and likely to be critical for effective clinical use of MTH1 inhibitors.
2.1. DNA maintenance and repair mechanisms
Depletion or chemical inhibition of MTH1 has been reported to elevate 8-oxodG incorporation into the genome of murine as well as human cells. In human cells, particularly cancer cells, several studies show that the acute effect of depleting or inhibiting MTH1 is induction of genomic DNA breaks [9, 17]. The efficacy of the first-in-class MTH1 inhibitors, including Karonudib, which is currently undergoing the first-in-man clinical trial, has been attributed to the ability of these inhibitors to promote the accumulation of DNA double-strand breaks [19, 48]. However, the mechanism(s) by which these genotoxic breaks occur upon MTH1 depletion are still not fully defined. Canonically, 8-oxodG is not known to be a genotoxic lesion as it does not distort the helical structure but rather alters the electrostatic potential of the major groove. As such, the 8-oxodG lesion leads to mispairing of 8-oxodG with dA and thus can give rise to GC -> TA transversions following replication. However, not many studies have assessed the effects of multiple 8-oxodG lesions in close helical proximity or indeed how increased 8-oxodGTP in the nucleotide pool can affect DNA structure, DNA repair capacity/frequency or DNA replication integrity and how this may impact cell viability. Recent studies (discussed below) suggest that MTH1 loss- or inhibition-induced DNA breaks ensue from compromised DNA maintenance mechanisms upon encountering increased 8-oxodG and/or 8-oxodGTP. These include mechanisms of DNA replication, base excision repair, mismatch repair, and telomere maintenance.
2.2. DNA and RNA polymerases
DNA replication and transcription by DNA and RNA polymerases require appropriately balanced dNTP and NTP levels to avoid mutation and catastrophic replication fork stalling [66]. Among the nucleotides, GTP and dGTP are the most prone to oxidation due to the high redox potential of guanines [4, 67]. In mitochondrial DNA isolated from rat tissue, very small amounts of 8-oxodGTP (<1% of the total dGTP pool) are enough to reduce the fidelity of the mitochondrial DNA polymerase (Polγ, DNA polymerase gamma), as measured by the frequency of A->C transversions [68]. The adverse effects on fidelity are most exacerbated in tissues that exhibit already imbalanced nucleotide pools, for instance in the rat cardiac subsarcolemmal tissue where the dGTP pools are abnormally high to begin with and thus the 8-oxodGTP levels are able to effectively compete with dTTP for incorporation opposite dA. However, even in rat liver tissues, which do not exhibit such unbalanced dGTP levels, 8-oxodGTP levels as low as 2.4 nM increase A->C transversions to about 7-fold higher than what was observed in the absence of 8-oxodGTP. Using time-lapse crystallography, a recent study by Freudenthal and colleagues [69] tracked the structural characteristics of 8-oxodGTP insertion opposite dA (a source of G->T transversions) by the base excision repair polymerase DNA polymerase beta (Polβ). These structural studies reveal that the altered charge on 8-oxodGTP vis-à-vis dGTP is accommodated in the Polβ active site during insertion, leading to a nicked DNA repair intermediate. A follow-up study [70] found that 8-oxodGTP insertion by Polβ led to failure of the rate-limiting nick ligation step of BER. This study reported that both MTH1 loss as well as an oxidative stress-inducing agent potassium bromate (KBrO3) sensitized mouse cells to loss of viability and elevated levels of the DNA strand break marker, gamma (γ)-H2AX, contingent on the presence of Polβ. Thus, given the high propensity of tumor cells to nucleotide pool imbalances and both replication and oxidative stress, it could be posited that the addiction of cancer cells to elevated MTH1 levels is required to limit 8-oxodGTP availability to polymerases, enabling DNA replication and repair to proceed without catastrophic strand-break causing errors.
Removal of oxidized guanine ribonucleotides is an important function of MTH1 [71]. Ribonucleotide reductase is unable to convert 8-oxoGTP to 8-oxodGTP, and guanylate kinase cannot phosphorylate 8-oxoGMP [71]. Thus, it is conceivable that MTH1 loss can lead to genomic instability through guanine nucleotide and deoxyribonucleotide pool imbalances. However, very little is known about the effects of MTH1 depletion on RNA quality control or any associated anti-tumor outcomes. Indeed, very little is known in general about the pathophysiological implications of oxidized RNA. RNA is likely to be more susceptible to oxidative damage than DNA given that it is not actively repaired. Furthermore, RNA is not protected by chromatin and is more accessible to mitochondrial oxidants due to its cytoplasmic localization. Consistent with this, RNA from patients with Alzheimer’s disease, Parkinson’s disease, amyotrophic lateral sclerosis (ALS) and dementia exhibit significantly elevated 8-oxoG in their neuronal RNA relative to RNA from healthy controls (reviewed in [72]). Expression of MTH1 is reported to be neuroprotective in hippocampal microglia exposed to kainate, an excitotoxic agent, by limiting 8-oxoG accumulation in mRNA [29]. RNA polymerase II can incorporate 8-oxoGTP in place of GTP at a low but detectable rate (estimated rate for 8-oxoGTP vs. GTP is around 2%) [71], indicating that MTH1 activity limits mutational transcription in addition to protecting against GTP/dGTP imbalances. A recent study by Dai et al. [73] specifically examined the effects of MTH1 depletion (via shRNA) on transcriptional mutagenesis in HeLa cells. This study found that, following addition of 8-oxoGTP to these shMTH1 cells, levels of 8-oxoG- containing mRNA acutely increased to greater than two-fold and then gradually decreased over the next 12 hours, presumably due to preferential degradation. RNA sustaining elevated 8-oxoG levels has been implicated in increased transcriptional mutagenesis [73], ostensibly through RNA polymerase II utilization of 8-oxoGTP, reduced or altered protein expression [72, 74], and/or protein mis-aggregation, including amyloid-like structures [73]. Thus, it is very likely that MTH1 inhibition has deleterious effects on cells through increased incorporation of 8-oxoGTP into RNA. Although the role of RNA oxidation in tumor biology is not well understood, the exponential increase of interest regarding MTH1 function in cancers, in conjunction with the availability of genetic and pharmacologic tools for MTH1 modulation, is likely to facilitate future investigations into this understudied area.
2.3. Telomerase activity and telomere maintenance
There is little understanding with regard to how genomic 8-oxoG lesions can lead to the induction of the genotoxic DNA damage response and the onset of cellular senescence. A leading question is whether there are hotspots for MTH1 inhibition-induced damage, for instance at G-rich DNA sequences. A prime candidate for such a hotspot is the mammalian telomeric repeat, TTAGGG, particularly as DNA damage in this genomic region leads to telomere dysfunction and genomic instability, a major contributing factor to cellular senescence and tumor suppression [75-79]. It has long been suggested that telomeric DNA may be acutely sensitive to oxidative damage [80] and indeed, cells under oxidative stress are unable to be immortalized by the expression of telomerase [81]. A recent study reported that the antioxidant protein, peroxiredoxin-1 (PRDX1), associates with telomeres during S-phase, and that its depletion elevates ROS and 8-oxodGTP, leading to premature chain termination when 8-oxodGTP is incorporated by telomerase [82]. Significantly, this study also reported that terminal 8-oxoG-containing substrates prevent chain extension by telomerase.
Expressing the catalytic domain of telomerase, hTERT, fails to protect BJ skin fibroblasts from premature cellular senescence induced by MTH1 depletion [17], supporting the idea that the accumulation of 8-oxodG at telomeres and/or increased 8-oxodGTP impairs the extension and/or telomere-repairing ability of telomerase. This notion was directly addressed by two recent studies [83, 84], elegantly showing that MTH1 depletion profoundly affects telomerase function under specific molecular contexts that are relevant to cancer cells. Fouquerel and colleagues [83] examined the effects of both elevated 8-oxodGTP levels as well as MTH1 depletion on telomerase activity in HeLa cells with either long or short telomeres. Their results confirm that 8-oxodGTP incorporation is a telomerase chain terminator, although telomerase is able to extend primers with pre-existing terminal 8-oxodG. This would suggest that elevated oxidized nucleotides are more likely to result in telomeric DNA damage via telomerase action vs. in situ generated oxidative lesions. Significantly, following MTH1 depletion, cells with very short telomeres (VST) exhibit elevated markers of irreparable telomere breaks or telomere-induced damage foci (TIFs) and concomitant loss of viability, but the counterpart cells with long telomeres are not affected by MTH1 loss [83]. Considering many tumors display heterogeneity in telomere lengths, including dramatically short telomeres despite the presence of active telomerase [85, 86], this finding has implications for establishing telomeric length as a prognostic marker for the focused and efficacious use of MTH1 inhibition in the clinic. More importantly, this work shows MTH1 function as a previously-unidentified mediator of telomerase efficacy and telomere maintenance.
Tumor cells are known to elevate antioxidant proteins as a protective response against oncogenic oxidative stress. PRDX1, in particular, is known to be elevated in a variety of cancers including lung cancer, bladder cancer, ovarian carcinoma, aggressive esophageal squamous carcinoma, mesothelioma, glioblastoma, cholangiocarcinoma, esophageal squamous cell carcinoma, liver cancer and pancreatic cancer [87]. Lingner and colleagues followed up on their prior work showing the impact of deleting PRDX1 on 8-oxodGTP levels and telomerase function [82] by combining PRDX1 loss with MTH1 loss in HCT116 colon cancer cells [84]. Curiously, single depletion of either gene results in elevated expression of the other gene, suggesting compensatory cross-talk between the expression or function of PRDX1 and MTH1. Co-deletion of both MTH1 and PRDX1 leads to greater telomere shortening than deletion of either single gene. Moreover, MTH1 loss-dependent telomere shortening is highly dependent on the oxygen tension at which the cells were cultured, showing maximal shortening at 40% O2 and none at 5% O2. Significantly, combined loss of MTH1 and PRDX1 reduces elongation rates in a model of rapid telomerase-mediated lengthening produced by a mutant POT1 allele [88] at 20% (but not 5%) O2 culture conditions. Furthermore, introduction of a mutated telomerase variant (TSQ1-hTR) which can incorporate GTTGCC instead of TTAGGG indicates that the PRDX1/MTH1 deleted cells exhibit less telomerase activity at chromosome ends under conditions of oxidative stress. These results provide strong proof-of-principle evidence that MTH1 protects telomeres from shortening under conditions of pro-oxidant stress, prevalent in aggressive tumors, by both maintaining quality control of the deoxyribonucleotide substrates and by preserving telomerase activity. Additional work is required to establish these findings in specific cancer-relevant models and extension into in vivo models of tumorigenesis and MTH1 loss. Nevertheless, these studies point to novel combinatorial approaches (telomerase inhibitors/G-quadruplex inhibitors, targeting of thiol antioxidants) that could enhance MTH1 inhibition as an anti-cancer strategy in the clinical setting.
2.4. Base excision repair and mismatch repair
Genomic 8-oxodG levels reflect the net outcome from incorporation of oxidized deoxyribonucleotides, from in situ oxidation of genomic guanines, and from the removal of 8-oxodG by base excision repair (BER) enzymes. Genomic 8-oxodG lesions are processed by the BER enzyme 8-oxoguanine glycosylase 1 (OGG1) when paired opposite dC and by MUTYH which excises the mis-incorporated dA opposite 8-oxodG [89-91]. It has been established that BER-mediated repair of 8-oxoG involves the following steps: 1) recognition of 8-oxoG lesion by OGG1 (a bifunctional glycosylase), its excision leading to an abasic site and creation of a single-strand break in the DNA backbone, 2) processing of the 3’ end of the break by APE1 to generate a 3’OH group, 3) addition of one nucleotide to the 3′ end of the incised site by DNA polymerase beta (Polβ), simultaneously removing a 5′-sugar phosphate (5′-dRP) via its essential dRP lyase activity, and 4) re-sealing of the nicked DNA backbone by DNA ligase III/XRCC1 or DNA ligase I [92]. Any defect in this tightly coordinated process is likely to generate irreparable DNA strand breaks if BER is initiated on closely located 8-oxoG lesions or clusters of oxidative damage [93, 94]. Essentially, even one such unrepaired DNA double strand break could induce cellular senescence or cell death, if left unrepaired [95].
Previous work suggests that frequent BER initiation at oxidative damage sites, particularly at clustered sites, promotes the accumulation of DSBs [96, 97]. It has also been suggested that adjacent 8-oxoG lesions engage long patch BER where FEN1 is required to remove a > 6 nucleotide stretch. However 8-oxodG clustering can inhibit FEN1 efficiency [98]. Furthermore, AP lyase activity of additional glycosylases such as NTHL1 may promote DNA double strand breaks if OGG1 expression is low [99]. Thus, elevated genomic 8-oxodG, due to MTH1 functional loss, is likely to produce enhanced engagement of repair pathways which, in of themselves, produce DNA nicks as repair intermediates. Thus, the MTH1 addiction observed in a number of cancer cells is plausibly an important protective measure to avoid necessity for such repair processes and their associated risk of DNA breaks, through improved quality control in the nucleotide pool.
Indeed, some tumors such as non-small cell carcinoma (NSCLC) seem to rely largely on MTH1 to limit genomic 8-oxodG and exhibit low or dysfunctional OGG1 levels and activity [24]. Both the germline MTH1-null mice and the OGG1-null mice exhibit spontaneous late-onset lung tumors with a greater frequency of tumors observed in the MTH1-null animals [26] vs. the OGG1-null [100] animals. However the double-null OGG1/MTH1 animals do not develop detectable lung tumors [100]. One interpretation of this finding is that inhibiting OGG1 repair in the context of MTH1 loss could prevent genomic instability-associated malignant events.
It should be noted that base excision-associated breaks will also be formed by MUTYH action at sites where 8-oxodG has mispaired with dA. Unlike the single or double MTH1/OGG1 knockout animals, the triple MTH1/OGG1/MUTYH knockout animal does exhibit increased G->T mutagenesis [101]. Thus, MUTYH action alone may be able to compensate for any deficiencies in cellular anti-mutator function produced by loss of MTH1 and/or OGG1. Significantly, it has also been reported that MUTYH depletion reduces both nuclear and mitochondrial DNA strand breaks and the accompanying cytotoxicity [102] produced by treatment with the redox-cycling agent, menadione, which evokes oxidative stress and PARP-dependent cell death [103]. Similar phenomena have been reported by other studies. In MUTYH/MTH1 co-depleted leukemia cells, which are protected against genomic DNA damage accumulation and the resulting cell death, expression of NEIL1 from the Nei family of glycosylases (but neither NEIL2, NEIL3, NTHL1 nor OGG1) is significantly downregulated [39]. The functional overlap in lesion processing [104] among these enzymes suggests it is not abrogation of 8-oxodG or Fapy processing that underlies the observed rescue but possibly prevention of glycosylase activity at guanidinohydantoin (Gh) lesions, which are produced by subsequent oxidation events at 8-oxoG and are specific substrates for NEIL1 [105].
Other secondary oxidative modifications of 8-oxoG can include bulky lesions such as spiroaminodihydantoin or crosslinked guanine-pyrimidines [106, 107]. Accurate cell-physiologic measurements as to the frequency of such lesions in genomic DNA are difficult to establish due to the relatively high background from DNA isolation-related oxidation events. Nevertheless, one might speculate that the high pro-oxidant environment in cancer cells coupled with the lower oxidation potential of 8-oxoG vs. G might increase the frequency of these secondary oxidative guanine lesions in tumors. Therefore, several different guanine lesion-repairing pathways may be responsible for converting incorporated oxidized guanine moieties into DNA strand breaks. Collectively, these studies indicate that suppression of repair-induced DNA strand breaks at sites of genomic 8-oxodG, its mispaired dA or its subsequent oxidation products, is protective against oxidative stress-induced cytotoxicity. Thus, consideration of expression, mutational status and activity levels of the various relevant repair enzymes is likely to be important when evaluating the cytotoxic potential of MTH inhibition on cancer cells.
Previous work has shown MTH1 depletion-dependent DNA strand breaks in KRAS-driven lung cancer cells depend explicitly on wild-type (WT) p53 status [8]. Unlike p53-competent cells, neither p53-null nor p53-mutant cell lines show elevated DNA strand breaks as measured in a comet assay or by changes in the numbers of DSB foci [8]. However, introducing WT p53 into a p53-null cell line causes subsequent MTH1 depletion to lead to elevated DNA strand breaks [8]. Prior studies have implicated p53 in the ligation step of 8-oxodG processing by BER through its stabilization of the complex between Polβ and abasic sites [108]. Thus, one would predict that loss of WT p53 should compromise the rate-limiting BER step that seals 8-oxodG repair-associated DNA nicks and thereby synergize with MTH1 depletion in enhancing DNA strand breaks, which is does not appear to hold true in all cancer lines [8]. However, p53 has also been reported to maintain both OGG1 expression levels [109] and activity [109, 110]. Therefore, it may be that functional p53 loss protects against MTH1 depletion-associated DNA breaks by preventing initiation of BER at 8-oxodG sites through compromised OGG1 levels or activity. In support of this general idea, recent studies have reported that inhibition of OGG1 glycosylase function is protective against DNA break-inducing tumor-suppressive therapies. In pancreatic cancer, depletion of OGG1 protects tumor cells from beta-lapachone-induced DNA damage and PARP1 hyperactivation [111, 112]. Another study similarly reports, following oxidative stress, OGG1-deficient or depleted MEFs and neuroblastoma cells sustain a lower extent of DNA strand lesions and are protected against a form of PARP1 hyperactivation-dependent cell death called parthanatos, relative to their OGG1-functional counterparts [113]. These findings further support that dysfunction in OGG1 (or other 8-oxodG processing mechanisms such as MUTYH) could antagonize the tumor-inhibitory effects of MTH1 inhibition by reducing its ability to produce genomic DNA breaks.
Although MTH1-null animals do not exhibit enhanced mutagenic burden, the dual knockout MTH1/MSH2 double-null animals do exhibit a significantly increased incidence of G->T transversions over their single-null counterparts [31], indicating crosstalk between mismatch repair (MMR) and mechanisms that limit genomic 8-oxodG levels. Several tumors, notably colorectal cancer, exhibit defects in mismatch repair (MMR), manifested in hypermutation rates and microsatellite instability (MSI). Although a priori one might not assume there is a dependence of these phenotypes on endogenous oxidative stress, Russo and colleagues [114] showed that elevated MTH1 levels could alleviate the HPRT mutator phenotype in MMR-deficient MSH2-null mouse cells as well as in colorectal DLD-1 human cells, and also reduce incidences of MSI in the latter. Thus, increased oxidized nucleotides likely contribute to the genomic instability in MMR-deficient cancer cells [39], further increasing the reliance of mismatch-defective tumor cells on high MTH1 expression and function.
Collectively these studies underscore the need for comprehensive consideration of BER and MMR status when evaluating clinical efficacy of MTH1 inhibitors, as these factors will likely impact whether MTH1 inhibition produces genotoxic damage or not.
3. Implications for clinical use of MTH1 inhibitors
Currently, MTH1 as a therapeutic target in cancer is a source of some controversy [12] because, thus far, only the first-in-class inhibitors appear to produce anti-tumor effects in vitro and in vivo. There is also evidence that these inhibitors produce off-target effects on tubulin polymerization [115] and elevate cellular ROS [116], which may partially explain their cytotoxic effects over second-generation MTH1 inhibitors [51-53]. Recently developed MTH1 inhibitors, discovered via fragment-based screening, have yet to be fully evaluated for anti-tumor effects [117, 118]. However, a better understanding of the mechanisms of genotoxic damage induced by MTH1 inhibition (Fig. 2) can promote effective broad-spectrum tumor suppressive outcomes in select cohorts. As discussed above, it is clear this outcome is not one-size-fits-all but rather depends on several molecular contexts including (but likely not limited to) tumor-associated ROS-elevating mechanisms, increased 8-oxoGTP/8-oxodGTP in the nucleotide pool, genomic incorporation of 8-oxodG, 8-oxodG repair processes, p53 status, and telomere length. These contexts are variably represented in different cancers and, thus, the predominant features underlying whether MTH1 inhibition is likely to be efficacious or not will depend on the specific tumors under consideration. As is often the case with chemotherapeutic agents, whereas MTH1 inhibitors may not be successful as single agents, they may become highly efficacious in combination with the right stressor that will alter conditions to favor MTH1-inhibition-induced DNA breaks. Examples of such combinations include ROS-producing therapeutic drugs, targeting antioxidant tumor addictions (for instance, PRDX1), nucleotide metabolism inhibitors, p53 activators or telomere-shortening agents. Conversely, use of agents with antioxidant properties or possibly base excision repair inhibitors may antagonize MTH1 inhibitor efficacy. Similarly, strong damaging agents (hydroxyl radical-producing radiation or topoisomerase II inhibitors) are expected to bypass MTH1 function (or lack thereof) by directly damaging genomic DNA and may not present any combinatorial benefit with MTH1 inhibitors.
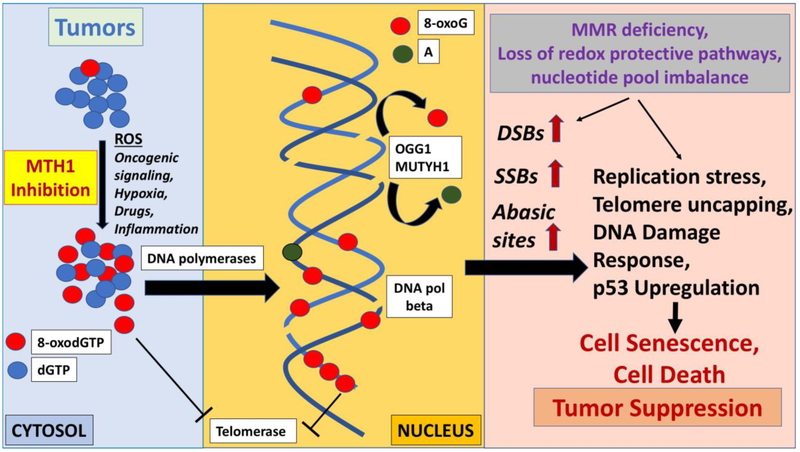
Figure 2. Overall schematic depicting mechanisms through which MTH1 inhibition and concomitant increase in oxidized nucleotides affects DNA maintenance and DNA repair mechanisms.
Cancer-relevant factors that contribute to increased generation of 8-oxodGTP as well as tumor suppression-inducing mechanisms ensuing from unrepaired DNA breaks are noted.
A better understanding of the above-discussed mechanisms is also likely to promote important knowledge regarding resistance mechanisms, which inevitably occur in aggressive tumors and underlie therapeutic failure in the clinic. A recent study evaluated the effects of Karonudib, an MTH1 inhibitor (currently undergoing first-in-man clinical trials in Sweden), in preclinical models of metastatic melanoma [119] and found approximately a third of the patient-derived xenograft (PDX) models exhibited resistance to Karonudib. These resistant tumors exhibited high levels of the multi-drug resistant (MDR) gene ABCB1 and could be partially sensitized to Karonudib through co-treatment with Elacridar, an ATP-dependent drug efflux pump inhibitor [119]. Interestingly, Karonudib efficacy was not correlated with either NRAS or BRAF status, suggesting the sensitizing stress in melanoma may not arise from its oncogenic drivers but some other factor(s).
These studies also established that Karonudib does not impair in vivo T cell-mediated cytotoxic effects [119]. Specifically, co-treatment with Karonudib does not alter the anti-tumor response generated in mouse melanoma tumors via treatment with an anti CTLA-4 antibody. Nor does Karonudib impair tumor-suppressive activity produced by injection of autologous tumor-infiltrating T-lymphocytes (TILs) in a PDX model of melanoma. Although further studies are likely needed in this area, these results provide proof-of-principle that MTH1 inhibitors may be combined with immunotherapeutic agents without adversely affecting their efficacy. Furthermore, given that MTH1 loss can increase mutations in conjunction with other cancer-associated DNA repair defects in addition to producing DNA breaks, we speculate MTH1 inhibitors may enhance neoantigen generation and thus promote anti-tumor responses from immune checkpoint inhibitors. Very little is known about this aspect of MTH1 inhibition and we anticipate this will be a fruitful area of study in the coming years.
Further studies are also required to identify additional, possibly tumor-specific, mechanisms of resistance to clinically relevant MTH1 inhibitors. Examples of mechanisms that are likely worthwhile to investigate include enzymes functionally redundant with MTH1, antioxidant/redox protective mechanisms that limit oxidation of dGTP, availability of oxidant-generating labile metals such as iron, factors that increase the efficiency of DNA repair and replication, and nucleotide synthesis/salvage mechanisms. Finally, long-term use of any MTH1 inhibitor-centric treatment needs to account for the necessity of robust MTH1 function in preventing neurotoxicity, age-associated pathology and other systemic issues.
Highlights.
The mammalian 8-oxodGTPase MTH1 is associated with increased tumor malignancy
MTH1 inhibition induces tumor-suppressive genomic DNA breaks via unknown mechanisms
Molecular mechanisms underlying this phenomenon are comprehensively discussed
We provide a perspective on MTH1 function for genome integrity maintenance
Acknowledgements
The authors thank members of the Rai laboratory for helpful discussions. RWS is an Abraham A. Mitchell Distinguished Investigator. This work has been supported by grants from the National Institutes of Health (NIH) to PR [R01CA175086] and to RWS [R01CA148629; R01ES014811; U01ES029518; P01ES028949]. We apologize to those authors whose works could not be cited here due to space constraints.
Footnotes
Conflict of interest
RWS is a scientific consultant for Bio-Techne and Canal House Biosciences, LLC. The authors state that there is no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Gorrini C, Harris IS, Mak TW, Modulation of oxidative stress as an anticancer strategy, Nature reviews. Drug discovery, 12 (2013) 931–947. [DOI] [PubMed] [Google Scholar]
- [2].Trachootham D, Alexandre J, Huang P, Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach?, Nature reviews. Drug discovery, 8 (2009) 579–591. [DOI] [PubMed] [Google Scholar]
- [3].Rai P, Human Mut T Homolog 1 (MTH1): A roadblock for the tumor-suppressive effects of oncogenic RAS-induced ROS, Small GTPases, 3 (2012) 13–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Haghdoost S, Sjolander L, Czene S, Harms-Ringdahl M, The nucleotide pool is a significant target for oxidative stress, Free Radic Biol Med, 41 (2006) 620–626. [DOI] [PubMed] [Google Scholar]
- [5].Rai P, Oxidation in the nucleotide pool, the DNA damage response and cellular senescence: Defective bricks build a defective house, Mutat Res, 703 (2010) 71–81. [DOI] [PubMed] [Google Scholar]
- [6].Kamiya H, Kasai H, Formation of 2-hydroxydeoxyadenosine triphosphate, an oxidatively damaged nucleotide, and its incorporation by DNA polymerases. Steady-state kinetics of the incorporation, The Journal of biological chemistry, 270 (1995) 19446–19450. [DOI] [PubMed] [Google Scholar]
- [7].Iida T, Furuta A, Kawashima M, Nishida J, Nakabeppu Y, Iwaki T, Accumulation of 8-oxo-2'-deoxyguanosine and increased expression of hMTH1 protein in brain tumors, Neuro Oncol, 3 (2001) 73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Patel A, Burton DG, Halvorsen K, Balkan W, Reiner T, Perez-Stable C, Cohen A, Munoz A, Giribaldi MG, Singh S, Robbins DJ, Nguyen DM, Rai P, MutT Homolog 1 (MTH1) maintains multiple KRAS-driven pro-malignant pathways, Oncogene, doi: 10.1038/onc.2014.195 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Giribaldi MG, Munoz A, Halvorsen K, Patel A, Rai P, MTH1 expression is required for effective transformation by oncogenic HRAS, Oncotarget, 6 (2015) 11519–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Buchholz M, Braun M, Heidenblut A, Kestler HA, Kloppel G, Schmiegel W, Hahn SA, Luttges J, Gress TM, Transcriptome analysis of microdissected pancreatic intraepithelial neoplastic lesions, Oncogene, 24 (2005) 6626–6636. [DOI] [PubMed] [Google Scholar]
- [11].Garber ME, Troyanskaya OG, Schluens K, Petersen S, Thaesler Z, Pacyna-Gengelbach M, van de Rijn M, Rosen GD, Perou CM, Whyte RI, Altman RB, Brown PO, Botstein D, Petersen I, Diversity of gene expression in adenocarcinoma of the lung, Proc Natl Acad Sci U S A, 98 (2001) 13784–13789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Samaranayake GJ, Huynh M, Rai P, MTH1 as a Chemotherapeutic Target: The Elephant in the Room, Cancers (Basel), 9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Weinberg F, Hamanaka R, Wheaton WW, Weinberg S, Joseph J, Lopez M, Kalyanaraman B, Mutlu GM, Budinger GR, Chandel NS, Mitochondrial metabolism and ROS generation are essential for Kras-mediated tumorigenicity, Proc Natl Acad Sci U S A, 107 (2010) 8788–8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Grabocka E, Commisso C, Bar-Sagi D, Molecular pathways: targeting the dependence of mutant RAS cancers on the DNA damage response, Clin Cancer Res, 21 (2015) 1243–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sakumi K, Furuichi M, Tsuzuki T, Kakuma T, Kawabata S, Maki H, Sekiguchi M, Cloning and expression of cDNA for a human enzyme that hydrolyzes 8-oxo-dGTP, a mutagenic substrate for DNA synthesis, The Journal of biological chemistry, 268 (1993) 23524–23530. [PubMed] [Google Scholar]
- [16].Furuichi M, Yoshida MC, Oda H, Tajiri T, Nakabeppu Y, Tsuzuki T, Sekiguchi M, Genomic structure and chromosome location of the human mutT homologue gene MTH1 encoding 8-oxo-dGTPase for prevention of A:T to C:G transversion, Genomics, 24 (1994) 485–490. [DOI] [PubMed] [Google Scholar]
- [17].Rai P, Onder TT, Young JJ, McFaline JL, Pang B, Dedon PC, Weinberg RA, Continuous elimination of oxidized nucleotides is necessary to prevent rapid onset of cellular senescence, Proc Natl Acad Sci U S A, 106 (2009) 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Rai P, Young JJ, Burton DG, Giribaldi MG, Onder TT, Weinberg RA, Enhanced elimination of oxidized guanine nucleotides inhibits oncogenic RAS-induced DNA damage and premature senescence, Oncogene, 30 (2011) 1489–1496. [DOI] [PubMed] [Google Scholar]
- [19].Gad H, Koolmeister T, Jemth AS, Eshtad S, Jacques SA, Strom CE, Svensson LM, Schultz N, Lundback T, Einarsdottir BO, Saleh A, Gokturk C, Baranczewski P, Svensson R, Berntsson RP, Gustafsson R, Stromberg K, Sanjiv K, Jacques-Cordonnier MC, Desroses M, Gustavsson AL, Olofsson R, Johansson F, Homan EJ, Loseva O, Brautigam L, Johansson L, Hoglund A, Hagenkort A, Pham T, Altun M, Gaugaz FZ, Vikingsson S, Evers B, Henriksson M, Vallin KS, Wallner OA, Hammarstrom LG, Wiita E, Almlof I, Kalderen C, Axelsson H, Djureinovic T, Puigvert JC, Haggblad M, Jeppsson F, Martens U, Lundin C, Lundgren B, Granelli I, Jensen AJ, Artursson P, Nilsson JA, Stenmark P, Scobie M, Berglund UW, Helleday T, MTH1 inhibition eradicates cancer by preventing sanitation of the dNTP pool, Nature, 508 (2014) 215–221. [DOI] [PubMed] [Google Scholar]
- [20].Maki H, Sekiguchi M, MutT protein specifically hydrolyses a potent mutagenic substrate for DNA synthesis, Nature, 355 (1992) 273–275. [DOI] [PubMed] [Google Scholar]
- [21].Fujikawa K, Kamiya H, Yakushiji H, Nakabeppu Y, Kasai H, Human MTH1 protein hydrolyzes the oxidized ribonucleotide, 2-hydroxy-ATP, Nucleic acids research, 29 (2001) 449–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Fujikawa K, Kamiya H, Yakushiji H, Fujii Y, Nakabeppu Y, Kasai H, The oxidized forms of dATP are substrates for the human MutT homologue, the hMTH1 protein, The Journal of biological chemistry, 274 (1999) 18201–18205. [DOI] [PubMed] [Google Scholar]
- [23].Jemth AS, Gustafsson R, Brautigam L, Henriksson L, Vallin KSA, Sarno A, Almlof I, Homan E, Rasti A, Warpman Berglund U, Stenmark P, Helleday T, MutT homologue 1 (MTH1) catalyzes the hydrolysis of mutagenic O6-methyl-dGTP, Nucleic acids research, 46 (2018) 10888–10904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Speina E, Arczewska KD, Gackowski D, Zielinska M, Siomek A, Kowalewski J, Olinski R, Tudek B, Kusmierek JT, Contribution of hMTH1 to the maintenance of 8-oxoguanine levels in lung DNA of non-small-cell lung cancer patients, J Natl Cancer Inst, 97 (2005) 384–395. [DOI] [PubMed] [Google Scholar]
- [25].Nakabeppu Y, Molecular genetics and structural biology of human MutT homolog, MTH1, Mutat Res, 477 (2001) 59–70. [DOI] [PubMed] [Google Scholar]
- [26].Tsuzuki T, Egashira A, Igarashi H, Iwakuma T, Nakatsuru Y, Tominaga Y, Kawate H, Nakao K, Nakamura K, Ide F, Kura S, Nakabeppu Y, Katsuki M, Ishikawa T, Sekiguchi M, Spontaneous tumorigenesis in mice defective in the MTH1 gene encoding 8-oxo-dGTPase, Proc Natl Acad Sci U S A, 98 (2001) 11456–11461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nakabeppu Y, Ohta E, Abolhassani N, MTH1 as a nucleotide pool sanitizing enzyme: Friend or foe?, Free Radic Biol Med, 107 (2017) 151–158. [DOI] [PubMed] [Google Scholar]
- [28].Yamaguchi H, Kajitani K, Dan Y, Furuichi M, Ohno M, Sakumi K, Kang D, Nakabeppu Y, MTH1, an oxidized purine nucleoside triphosphatase, protects the dopamine neurons from oxidative damage in nucleic acids caused by 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine, Cell Death Differ, 13 (2006) 551–563. [DOI] [PubMed] [Google Scholar]
- [29].Kajitani K, Yamaguchi H, Dan Y, Furuichi M, Kang D, Nakabeppu Y, MTH1, an oxidized purine nucleoside triphosphatase, suppresses the accumulation of oxidative damage of nucleic acids in the hippocampal microglia during kainate-induced excitotoxicity, J Neurosci, 26 (2006) 1688–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Kennedy CH, Pass HI, Mitchell JB, Expression of human MutT homologue (hMTH1) protein in primary non-small-cell lung carcinomas and histologically normal surrounding tissue, Free Radic Biol Med, 34 (2003) 1447–1457. [DOI] [PubMed] [Google Scholar]
- [31].Egashira A, Yamauchi K, Yoshiyama K, Kawate H, Katsuki M, Sekiguchi M, Sugimachi K, Maki H, Tsuzuki T, Mutational specificity of mice defective in the MTH1 and/or the MSH2 genes, DNA repair, 1 (2002) 881–893. [DOI] [PubMed] [Google Scholar]
- [32].De Luca G, Ventura I, Sanghez V, Russo MT, Ajmone-Cat MA, Cacci E, Martire A, Popoli P, Falcone G, Michelini F, Crescenzi M, Degan P, Minghetti L, Bignami M, Calamandrei G, Prolonged lifespan with enhanced exploratory behavior in mice overexpressing the oxidized nucleoside triphosphatase hMTH1, Aging Cell, 12 (2013) 695–705. [DOI] [PubMed] [Google Scholar]
- [33].Ventura I, Russo MT, De Nuccio C, De Luca G, Degan P, Bernardo A, Visentin S, Minghetti L, Bignami M, hMTH1 expression protects mitochondria from Huntington's disease-like impairment, Neurobiol Dis, 49 (2013) 148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N, Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal, Sci Signal, 6 (2013) pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, Antipin Y, Reva B, Goldberg AP, Sander C, Schultz N, The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data, Cancer Discov, 2 (2012) 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Volonte D, Vyas AR, Chen C, Dacic S, Stabile LP, Kurland BF, Abberbock SR, Burns TF, Herman JG, Di YP, Galbiati F, Caveolin-1 promotes the tumor suppressor properties of oncogene-induced cellular senescence, The Journal of biological chemistry, 293 (2018) 1794–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Arczewska KD, Stachurska A, Wojewodzka M, Karpinska K, Kruszewski M, Nilsen H, Czarnocka B, hMTH1 is required for maintaining migration and invasion potential of human thyroid cancer cells, DNA repair, 69 (2018) 53–62. [DOI] [PubMed] [Google Scholar]
- [38].Wang JY, Liu GZ, Wilmott JS, La T, Feng YC, Yari H, Yan XG, Thorne RF, Scolyer RA, Zhang XD, Jin L, Skp2-Mediated Stabilization of MTH1 Promotes Survival of Melanoma Cells upon Oxidative Stress, Cancer Res, 77 (2017) 6226–6239. [DOI] [PubMed] [Google Scholar]
- [39].Eshtad S, Mavajian Z, Rudd SG, Visnes T, Bostrom J, Altun M, Helleday T, hMYH and hMTH1 cooperate for survival in mismatch repair defective T-cell acute lymphoblastic leukemia, Oncogenesis, 5 (2016) e275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Pudelko L, Rouhi P, Sanjiv K, Gad H, Kalderen C, Hoglund A, Squatrito M, Schuhmacher AJ, Edwards S, Hagerstrand D, Berglund UW, Helleday T, Brautigam L, Glioblastoma and glioblastoma stem cells are dependent on functional MTH1, Oncotarget, 8 (2017) 84671–84684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Versano Z, Shany E, Freedman S, Tuval-Kochen L, Leitner M, Paglin S, Toren A, Yalon M, MutT homolog 1 counteracts the effect of anti-neoplastic treatments in adult and pediatric glioblastoma cells, Oncotarget, 9 (2018) 27547–27563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tu Y, Wang Z, Wang X, Yang H, Zhang P, Johnson M, Liu N, Liu H, Jin W, Zhang Y, Cui D, Birth of MTH1 as a therapeutic target for glioblastoma: MTH1 is indispensable for gliomatumorigenesis, Am J Transl Res, 8 (2016) 2803–2811. [PMC free article] [PubMed] [Google Scholar]
- [43].Fujishita T, Okamoto T, Akamine T, Takamori S, Takada K, Katsura M, Toyokawa G, Shoji F, Shimokawa M, Oda Y, Nakabeppu Y, Maehara Y, Association of MTH1 expression with the tumor malignant potential and poor prognosis in patients with resected lung cancer, Lung Cancer, 109 (2017) 52–57. [DOI] [PubMed] [Google Scholar]
- [44].Li J, Yang CC, Tian XY, Li YX, Cui J, Chen Z, Deng ZL, Chen FJ, Hayakawa H, Sekiguchi M, Cai JP, MutT-related proteins are novel progression and prognostic markers for colorectal cancer, Oncotarget, 8 (2017) 105714–105726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Akiyama S, Saeki H, Nakashima Y, Iimori M, Kitao H, Oki E, Oda Y, Nakabeppu Y, Kakeji Y, Maehara Y, Prognostic impact of MutT homolog-1 expression on esophageal squamous cell carcinoma, Cancer Med, 6 (2017) 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Kumagae Y, Hirahashi M, Takizawa K, Yamamoto H, Gushima M, Esaki M, Matsumoto T, Nakamura M, Kitazono T, Oda Y, Overexpression of MTH1 and OGG1 proteins in ulcerative colitis-associated carcinogenesis, Oncol Lett, 16 (2018) 1765–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].van der Waals LM, Laoukili J, Jongen JMJ, Raats DA, Borel Rinkes IHM, Kranenburg O, Differential anti-tumour effects of MTH1 inhibitors in patient-derived 3D colorectal cancer cultures, Sci Rep, 9 (2019) 819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Warpman Berglund U, Sanjiv K, Gad H, Kalderen C, Koolmeister T, Pham T, Gokturk C, Jafari R, Maddalo G, Seashore-Ludlow B, Chernobrovkin A, Manoilov A, Pateras IS, Rasti A, Jemth AS, Almlof I, Loseva O, Visnes T, Einarsdottir BO, Gaugaz FZ, Saleh A, Platzack B, Wallner OA, Vallin KS, Henriksson M, Wakchaure P, Borhade S, Herr P, Kallberg Y, Baranczewski P, Homan EJ, Wiita E, Nagpal V, Meijer T, Schipper N, Rudd SG, Brautigam L, Lindqvist A, Filppula A, Lee TC, Artursson P, Nilsson JA, Gorgoulis VG, Lehtio J, Zubarev RA, Scobie M, Helleday T, Validation and development of MTH1 inhibitors for treatment of cancer, Annals of oncology : official journal of the European Society for Medical Oncology, 27 (2016) 2275–2283. [DOI] [PubMed] [Google Scholar]
- [49].Abbas HHK, Alhamoudi KMH, Evans MD, Jones GDD, Foster SS, MTH1 deficiency selectively increases non-cytotoxic oxidative DNA damage in lung cancer cells: more bad news than good?, BMC Cancer, 18 (2018) 423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Huber KV, Salah E, Radic B, Gridling M, Elkins JM, Stukalov A, Jemth AS, Gokturk C, Sanjiv K, Stromberg K, Pham T, Berglund UW, Colinge J, Bennett KL, Loizou JI, Helleday T, Knapp S, Superti-Furga G, Stereospecific targeting of MTH1 by (S)-crizotinib as an anticancer strategy, Nature, 508 (2014) 222–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kettle JG, Alwan H, Bista M, Breed J, Davies NL, Eckersley K, Fillery S, Foote KM, Goodwin L, Jones DR, Kack H, Lau A, Nissink JW, Read J, Scott JS, Taylor B, Walker G, Wissler L, Wylot M, Potent and Selective Inhibitors of MTH1 Probe Its Role in Cancer Cell Survival, J Med Chem, 59 (2016) 2346–2361. [DOI] [PubMed] [Google Scholar]
- [52].Petrocchi A, Leo E, Reyna NJ, Hamilton MM, Shi X, Parker CA, Mseeh F, Bardenhagen JP, Leonard P, Cross JB, Huang S, Jiang Y, Cardozo M, Draetta G, Marszalek JR, Toniatti C, Jones P, Lewis RT, Identification of potent and selective MTH1 inhibitors, Bioorg Med Chem Lett, 26 (2016) 1503–1507. [DOI] [PubMed] [Google Scholar]
- [53].Ellermann M, Eheim A, Rahm F, Viklund J, Guenther J, Andersson M, Ericsson U, Forsblom R, Ginman T, Lindstrom J, Silvander C, Tresaugues L, Giese A, Bunse S, Neuhaus R, Weiske J, Quanz M, Glasauer A, Nowak-Reppel K, Bader B, Irlbacher H, Meyer H, Queisser N, Bauser M, Haegebarth A, Gorjanacz M, Novel Class of Potent and Cellularly Active Inhibitors Devalidates MTH1 as Broad-Spectrum Cancer Target, ACS Chem Biol, 12 (2017) 1986–1992. [DOI] [PubMed] [Google Scholar]
- [54].Liou GY, Storz P, Reactive oxygen species in cancer, Free radical research, 44 (2010) 479–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Yoshimura D, Sakumi K, Ohno M, Sakai Y, Furuichi M, Iwai S, Nakabeppu Y, An oxidized purine nucleoside triphosphatase, MTH1, suppresses cell death caused by oxidative stress, The Journal of biological chemistry, 278 (2003) 37965–37973. [DOI] [PubMed] [Google Scholar]
- [56].Nogueira V, Hay N, Molecular pathways: reactive oxygen species homeostasis in cancer cells and implications for cancer therapy, Clin Cancer Res, 19 (2013) 4309–4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Irani K, Xia Y, Zweier JL, Sollott SJ, Der CJ, Fearon ER, Sundaresan M, Finkel T, Goldschmidt-Clermont PJ, Mitogenic signaling mediated by oxidants in Ras-transformed fibroblasts, Science, 275 (1997) 1649–1652. [DOI] [PubMed] [Google Scholar]
- [58].Lee AC, Fenster BE, Ito H, Takeda K, Bae NS, Hirai T, Yu ZX, Ferrans VJ, Howard BH, Finkel T, Ras proteins induce senescence by altering the intracellular levels of reactive oxygen species, The Journal of biological chemistry, 274 (1999) 7936–7940. [DOI] [PubMed] [Google Scholar]
- [59].Burton DG, Rai P, MTH1 counteracts oncogenic oxidative stress, Oncoscience, 2 (2015) 785–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Gatenby RA, Gillies RJ, Why do cancers have high aerobic glycolysis?, Nat Rev Cancer, 4 (2004) 891–899. [DOI] [PubMed] [Google Scholar]
- [61].Eales KL, Hollinshead KE, Tennant DA, Hypoxia and metabolic adaptation of cancer cells, Oncogenesis, 5 (2016) e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT, Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing, The Journal of biological chemistry, 275 (2000) 25130–25138. [DOI] [PubMed] [Google Scholar]
- [63].Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT, Mitochondrial reactive oxygen species trigger hypoxia-induced transcription, Proc Natl Acad Sci U S A, 95 (1998) 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Bonello S, Zahringer C, BelAiba RS, Djordjevic T, Hess J, Michiels C, Kietzmann T, Gorlach A, Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site, Arterioscler Thromb Vasc Biol, 27 (2007) 755–761. [DOI] [PubMed] [Google Scholar]
- [65].Brautigam L, Pudelko L, Jemth AS, Gad H, Narwal M, Gustafsson R, Karsten S, Carreras Puigvert J, Homan E, Berndt C, Berglund UW, Stenmark P, Helleday T, Hypoxic Signaling and the Cellular Redox Tumor Environment Determine Sensitivity to MTH1 Inhibition, Cancer Res, 76 (2016) 2366–2375. [DOI] [PubMed] [Google Scholar]
- [66].Ganai RA, Johansson E, DNA Replication-A Matter of Fidelity, Mol Cell, 62 (2016) 745–755. [DOI] [PubMed] [Google Scholar]
- [67].Neeley WL, Essigmann JM, Mechanisms of formation, genotoxicity, and mutation of guanine oxidation products, Chem Res Toxicol, 19 (2006) 491–505. [DOI] [PubMed] [Google Scholar]
- [68].Pursell ZF, McDonald JT, Mathews CK, Kunkel TA, Trace amounts of 8-oxodGTP in mitochondrial dNTP pools reduce DNA polymerase gamma replication fidelity, Nucleic acids research, 36 (2008) 2174–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Freudenthal BD, Beard WA, Perera L, Shock DD, Kim T, Schlick T, Wilson SH, Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide, Nature, 517 (2015) 635–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Caglayan M, Horton JK, Dai DP, Stefanick DF, Wilson SH, Oxidized nucleotide insertion by pol beta confounds ligation during base excision repair, Nat Commun, 8 (2017) 14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Hayakawa H, Hofer A, Thelander L, Kitajima S, Cai Y, Oshiro S, Yakushiji H, Nakabeppu Y, Kuwano M, Sekiguchi M, Metabolic fate of oxidized guanine ribonucleotides in mammalian cells, Biochemistry, 38 (1999) 3610–3614. [DOI] [PubMed] [Google Scholar]
- [72].Simms CL, Zaher HS, Quality control of chemically damaged RNA, Cell Mol Life Sci, 73 (2016) 3639–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Dai DP, Gan W, Hayakawa H, Zhu JL, Zhang XQ, Hu GX, Xu T, Jiang ZL, Zhang LQ, Hu XD, Nie B, Zhou Y, Li J, Zhou XY, Li J, Zhang TM, He Q, Liu DG, Chen HB, Yang N, Zuo PP, Zhang ZX, Yang HM, Wang Y, Wilson SH, Zeng YX, Wang JY, Sekiguchi M, Cai JP, Transcriptional mutagenesis mediated by 8-oxoG induces translational errors in mammalian cells, Proc Natl Acad Sci U S A, 115 (2018) 4218–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Kong Q, Lin CL, Oxidative damage to RNA: mechanisms, consequences, and diseases, Cell Mol Life Sci, 67 (2010) 1817–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Feldser DM, Greider CW, Short Telomeres Limit Tumor Progression In Vivo by Inducing Senescence, Cell, 11 (2007) 461–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Harley CB, Futcher AB, Greider CW, Telomeres shorten during ageing of human fibroblasts, Nature, 345 (1990) 458–460. [DOI] [PubMed] [Google Scholar]
- [77].Takai H, Smogorzewska A, de Lange T, DNA Damage Foci at Dysfunctional Telomeres, Current Biology, 13 (2003) 1549–1556. [DOI] [PubMed] [Google Scholar]
- [78].Fagagna F.d.A.d., Reaper PM, Clay-Farrace L, Fiegler H, Carr P, von Zglinicki T, Saretzki G, Carter NP, Jackson SP, A DNA damage checkpoint response in telomere-initiated senescence, Nature, 426 (2003) 194–198. [DOI] [PubMed] [Google Scholar]
- [79].Herbig U, Jobling WA, Chen BP, Chen DJ, Sedivy JM, Telomere shortening triggers senescence of human cells through a pathway involving ATM, p53, and p21(CIP1), but not p16(INK4a), Mol Cell, 14 (2004) 501–513. [DOI] [PubMed] [Google Scholar]
- [80].von Zglinicki T, Role of oxidative stress in telomere length regulation and replicative senescence, Ann N Y Acad Sci, 908 (2000) 99–110. [DOI] [PubMed] [Google Scholar]
- [81].Forsyth NR, Evans AP, Shay JW, Wright WE, Developmental differences in the immortalization of lung fibroblasts by telomerase, Aging Cell, 2 (2003) 235–243. [DOI] [PubMed] [Google Scholar]
- [82].Aeby E, Ahmed W, Redon S, Simanis V, Lingner J, Peroxiredoxin 1 Protects Telomeres from Oxidative Damage and Preserves Telomeric DNA for Extension by Telomerase, Cell Rep, 17 (2016) 3107–3114. [DOI] [PubMed] [Google Scholar]
- [83].Fouquerel E, Lormand J, Bose A, Lee HT, Kim GS, Li J, Sobol RW, Freudenthal BD, Myong S, Opresko PL, Oxidative guanine base damage regulates human telomerase activity, Nat Struct Mol Biol, 23 (2016) 1092–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ahmed W, Lingner J, PRDX1 and MTH1 cooperate to prevent ROS-mediated inhibition of telomerase, Genes Dev, 32 (2018) 658–669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].der-Sarkissian H, Bacchetti S, Cazes L, Londono-Vallejo JA, The shortest telomeres drive karyotype evolution in transformed cells, Oncogene, 23 (2004) 1221–1228. [DOI] [PubMed] [Google Scholar]
- [86].Londono-Vallejo JA, Telomere instability and cancer, Biochimie, 90 (2008) 73–82. [DOI] [PubMed] [Google Scholar]
- [87].Nicolussi A, D'Inzeo S, Capalbo C, Giannini G, Coppa A, The role of peroxiredoxins in cancer, Mol Clin Oncol, 6 (2017) 139–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Loayza D, De Lange T, POT1 as a terminal transducer of TRF1 telomere length control, Nature, 423 (2003) 1013–1018. [DOI] [PubMed] [Google Scholar]
- [89].Oka S, Leon J, Tsuchimoto D, Sakumi K, Nakabeppu Y, MUTYH, an adenine DNA glycosylase, mediates p53 tumor suppression via PARP-dependent cell death, Oncogenesis, 4 (2015) e142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sobol RW, For MutY, it's all about the OG, Chem Biol, 19 (2012) 313–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Brinkmeyer MK, Pope MA, David SS, Catalytic contributions of key residues in the adenine glycosylase MutY revealed by pH-dependent kinetics and cellular repair assays, Chem Biol, 19 (2012) 276–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Svilar D, Goellner EM, Almeida KH, Sobol RW, Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage, Antioxid Redox Signal, 14 (2011) 2491–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Budworth H, Matthewman G, O'Neill P, Dianov GL, Repair of Tandem Base Lesions in DNA by Human Cell Extracts Generates Persisting Single-strand Breaks, Journal of Molecular Biology, 351 (2005) 1020–1029. [DOI] [PubMed] [Google Scholar]
- [94].David-Cordonnier MH, Boiteux S, O'Neill P, Excision of 8-oxoguanine within clustered damage by the yeast OGG1 protein, Nucleic acids research, 29 (2001) 1107–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Huang L.-c., Clarkin KC, Wahl GM, Sensitivity and selectivity of the DNA damage sensor responsible for activating p53-dependent G1 arrest, PNAS, 93 (1996) 4827–4832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Blaisdell JO, Harrison L, Wallace SS, Base excision repair processing of radiation-induced clustered DNA lesions, Radiat Prot Dosimetry, 97 (2001) 25–31. [DOI] [PubMed] [Google Scholar]
- [97].Ensminger M, Iloff L, Ebel C, Nikolova T, Kaina B, Lbrich M, DNA breaks and chromosomal aberrations arise when replication meets base excision repair, The Journal of cell biology, 206 (2014) 29–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Budworth H, Dianova II, Podust VN, Dianov GL, Repair of clustered DNA lesions. Sequence-specific inhibition of long-patch base excision repair be 8-oxoguanine, The Journal of biological chemistry, 277 (2002) 21300–21305. [DOI] [PubMed] [Google Scholar]
- [99].Yang N, Chaudhry MA, Wallace SS, Base excision repair by hNTH1 and hOGG1: a two edged sword in the processing of DNA damage in gamma-irradiated human cells, DNA repair, 5 (2006) 43–51. [DOI] [PubMed] [Google Scholar]
- [100].Sakumi K, Tominaga Y, Furuichi M, Xu P, Tsuzuki T, Sekiguchi M, Nakabeppu Y, Ogg1 knockout-associated lung tumorigenesis and its suppression by Mth1 gene disruption, Cancer Res, 63 (2003) 902–905. [PubMed] [Google Scholar]
- [101].Ohno M, Sakumi K, Fukumura R, Furuichi M, Iwasaki Y, Hokama M, Ikemura T, Tsuzuki T, Gondo Y, Nakabeppu Y, 8-oxoguanine causes spontaneous de novo germline mutations in mice, Sci Rep, 4 (2014) 4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Oka S, Ohno M, Tsuchimoto D, Sakumi K, Furuichi M, Nakabeppu Y, Two distinct pathways of cell death triggered by oxidative damage to nuclear and mitochondrial DNAs, The EMBO journal, 27 (2008) 421–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Loor G, Kondapalli J, Schriewer JM, Chandel NS, Vanden Hoek TL, Schumacker PT, Menadione triggers cell death through ROS-dependent mechanisms involving PARP activation without requiring apoptosis, Free Radic Biol Med, 49 (2010) 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Jacobs AL, Schar P, DNA glycosylases: in DNA repair and beyond, Chromosoma, 121 (2012) 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Zhao X, Krishnamurthy N, Burrows CJ, David SS, Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts, Biochemistry, 49 (2010) 1658–1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].AbdulSalam SF, Thowfeik FS, Merino EJ, Excessive Reactive Oxygen Species and Exotic DNA Lesions as an Exploitable Liability, Biochemistry, 55 (2016) 5341–5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Henderson PT, Neeley WL, Delaney JC, Gu F, Niles JC, Hah SS, Tannenbaum SR, Essigmann JM, Urea lesion formation in DNA as a consequence of 7,8-dihydro-8-oxoguanine oxidation and hydrolysis provides a potent source of point mutations, Chem Res Toxicol, 18 (2005) 12–18. [DOI] [PubMed] [Google Scholar]
- [108].Zhou J, Ahn J, Wilson SH, Prives C, A role for p53 in base excision repair, The EMBO journal, 20 (2001) 914–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Chatterjee A, Mambo E, Osada M, Upadhyay S, Sidransky D, The effect of p53-RNAi and p53 knockout on human 8-oxoguanine DNA glycosylase (hOgg1) activity, Faseb J, 20 (2006) 112–114. [DOI] [PubMed] [Google Scholar]
- [110].Achanta G, Huang P, Role of p53 in sensing oxidative DNA damage in response to reactive oxygen species-generating agents, Cancer Res, 64 (2004) 6233–6239. [DOI] [PubMed] [Google Scholar]
- [111].Chakrabarti G, Silvers MA, Ilcheva M, Liu Y, Moore ZR, Luo X, Gao J, Anderson G, Liu L, Sarode V, Gerber DE, Burma S, DeBerardinis RJ, Gerson SL, Boothman DA, Tumor-selective use of DNA base excision repair inhibition in pancreatic cancer using the NQO1 bioactivatable drug, beta-lapachone, Sci Rep, 5 (2015) 17066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Huang X, Motea EA, Moore ZR, Yao J, Dong Y, Chakrabarti G, Kilgore JA, Silvers MA, Patidar PL, Cholka A, Fattah F, Cha Y, Anderson GG, Kusko R, Peyton M, Yan J, Xie XJ, Sarode V, Williams NS, Minna JD, Beg M, Gerber DE, Bey EA, Boothman DA, Leveraging an NQO1 Bioactivatable Drug for Tumor-Selective Use of Poly(ADP-ribose) Polymerase Inhibitors, Cancer Cell, 30 (2016) 940–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Wang R, Li C, Qiao P, Xue Y, Zheng X, Chen H, Zeng X, Liu W, Boldogh I, Ba X, OGG1-initiated base excision repair exacerbates oxidative stress-induced parthanatos, Cell Death Dis, 9 (2018) 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Russo MT, Blasi MF, Chiera F, Fortini P, Degan P, Macpherson P, Furuichi M, Nakabeppu Y, Karran P, Aquilina G, Bignami M, The oxidized deoxynucleoside triphosphate pool is a significant contributor to genetic instability in mismatch repair-deficient cells, Mol Cell Biol, 24 (2004) 465–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Kawamura T, Kawatani M, Muroi M, Kondoh Y, Futamura Y, Aono H, Tanaka M, Honda K, Osada H, Proteomic profiling of small-molecule inhibitors reveals dispensability of MTH1 for cancer cell survival, Sci Rep, 6 (2016) 26521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Wang JY, Jin L, Yan XG, Sherwin S, Farrelly M, Zhang YY, Liu F, Wang CY, Guo ST, Yari H, La T, McFarlane J, Lei FX, Tabatabaee H, Chen JZ, Croft A, Jiang CC, Zhang XD, Reactive Oxygen Species Dictate the Apoptotic Response of Melanoma Cells to TH588, J Invest Dermatol, 136 (2016) 2277–2286. [DOI] [PubMed] [Google Scholar]
- [117].Rudling A, Gustafsson R, Almlof I, Homan E, Scobie M, Warpman Berglund U, Helleday T, Stenmark P, Carlsson J, Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space, J Med Chem, 60 (2017) 8160–8169. [DOI] [PubMed] [Google Scholar]
- [118].Rahm F, Viklund J, Tresaugues L, Ellermann M, Giese A, Ericsson U, Forsblom R, Ginman T, Gunther J, Hallberg K, Lindstrom J, Persson LB, Silvander C, Talagas A, Diaz-Saez L, Fedorov O, Huber KVM, Panagakou I, Siejka P, Gorjanacz M, Bauser M, Andersson M, Creation of a Novel Class of Potent and Selective MutT Homologue 1 (MTH1) Inhibitors Using Fragment-Based Screening and Structure-Based Drug Design, J Med Chem, 61 (2018) 2533–2551. [DOI] [PubMed] [Google Scholar]
- [119].Einarsdottir BO, Karlsson J, Soderberg EMV, Lindberg MF, Funck-Brentano E, Jespersen H, Brynjolfsson SF, Bagge RO, Carstam L, Scobie M, Koolmeister T, Wallner O, Stierner U, Berglund UW, Ny L, Nilsson LM, Larsson E, Helleday T, Nilsson JA, A patient-derived xenograft pre-clinical trial reveals treatment responses and a resistance mechanism to karonudib in metastatic melanoma, Cell Death Dis, 9 (2018) 810. [DOI] [PMC free article] [PubMed] [Google Scholar]