Abstract
Dimethyl fumarate (DMF) is a prescribed treatment for multiple sclerosis and has also been used to treat psoriasis. The electrophilicity of DMF suggests that its immunosuppressive activity is related to the covalent modification of cysteine residues in the human proteome. Nonetheless, our understanding of the proteins modified by DMF in human immune cells and the functional consequences of these reactions remains incomplete. Here, we report that DMF inhibits human plasmacytoid dendritic cell (pDC) function through a mechanism of action that is independent of the major electrophile sensor NRF2. Using chemical proteomics, we instead identify cysteine 13 (Cys13) of the innate immune kinase IRAK4 as a principal cellular target of DMF. We show that DMF blocks IRAK4-MYD88 interactions and IRAK4-mediated cytokine production in a Cys13-dependent manner. Our studies thus identify a proteomic hotspot for DMF action that constitutes a druggable protein-protein interface crucial for initiating innate immune responses.
Introduction
Plasmacytoid dendritic cells (pDCs) are a minor dendritic cell subset that produce large amounts of type 1 IFN (IFN-I) in response to viral and endogenous nucleic acids (1, 2). This cell type bridges the innate and adaptive immune systems, a duality reflected in its name – pDCs share a progenitor with the more common conventional dendritic cells (cDCs) and constitutively express the FLT3 receptor; however, pDCs also display a similar morphology as plasma cells, hence the modifier “plasmacytoid” (3, 4). Other hallmarks of pDCs include low MHC class II expression and low (mouse) or negative (human) CD11c expression (1, 5). The pDC population comprises only 0.3–0.5% of human peripheral blood cells, but is implicated in a variety of pathologies (1), where pDCs tend to accumulate in affected tissues to promote local inflammation.
Systemic Lupus Erythematosus patients display low levels of circulating pDCs but accumulate IFN-producing pDCs in the skin (6, 7). Similar results have been found in psoriasis patients, where pDCs infiltrate the skin and become activated, driving the early stages of the disease (8). In a xenograft model of human psoriasis, blockade of IFN-α signaling or production prevented the development of disease (8), suggesting that inhibition of pDC function could have therapeutic value. pDCs have been identified in the cerebrospinal fluid of multiple sclerosis (MS) patients (9) and accumulate in demyelinated lesions of inflamed MS brains (10). It has been reported that depletion of pDCs in chronic and relapsing mouse models of MS results in exacerbation of experimental autoimmune encephalomyelitis (EAE) symptoms and increased CD4+ T-cell activation in the central nervous system, suggesting a negative regulatory role for pDCs in modulating inflammatory T-cell responses (11). Furthermore, pDCs harvested from IFN-β-treated MS patients display lower expression of the activation markers CD83 and CD86 (12), and therefore interferon therapy may act at least in part through modulating pDC function.
Dimethyl fumarate (DMF) is an approved oral therapy for MS and has also been used for the treatment of psoriasis for several decades in an alternative formulation called Fumaderm, with DMF believed to be the active species (13, 14) (15). Despite its long history and use in multiple autoimmune conditions, the mechanism of action of DMF remains poorly understood. DMF can promote an antioxidant response through activation of the NRF2-KEAP1 pathway (16–18). However, DMF has been found to suppress pathology in the EAE model of MS independently of NRF2 (19), indicating that the therapeutic effect of DMF in MS and other autoimmune disorders may involve proteins and pathways beyond the KEAP1-NRF2 system.
DMF is an electrophilic small molecule that can react with nucleophilic cysteine residues in proteins. It is now appreciated that many immune signaling proteins harbor electrophile/oxidation-sensitive cysteines (20–22) and that adduction of reactive cysteines in the proteome by DMF and other electrophilic compounds may constitute a general mechanism for suppressing the activity of immune cells (20, 23, 24). We previously used a quantitative chemical proteomics platform to identify DMF-sensitive cysteines in primary human T cells, which included a CXXC motif in the C2 domain of PKCθ that was found to contribute to CD28 binding and T-cell activation (20). However, the extent to which DMF impacts the function of other immune cells, and whether such effects involve reactivity with specific cysteine residues in the proteome, remains poorly understood. Here, we have examined DMF activity in pDCs, an immune cell type that plays a central role in psoriasis. We first show that DMF, but not structurally-related non-electrophilic analogues, inhibit IFN-α and cytokine/chemokine production from primary human pDCs. DMF-mediated suppression of IFN-α production was independent of either NRF2 activation or global changes in the glutathione content of pDCs. We then used the chemical proteomic method termed isoTOP-ABPP (isotopic tandem orthogonal proteolysis–activity-based protein profiling) to map DMF-sensitive cysteines in the pDC cell line Cal-1. Among the > 4000 total cysteines quantified by isoTOP-ABPP in Cal-1 cells, cysteine-13 (C13) of IRKA4 stood out as being among the most sensitive to DMF. Noting that C13 resides proximal to the interface involved in MyD88 binding (25) and adjacent to a site of mutation in IRAK4 (R12C) implicated in human immunodeficiency (26), we proceeded to show that DMF disrupts both IRAK4-MyD88 interactions and IRAK4-mediated TNF-α production in a C13-dependent manner. Taken together, our data indicate that DMF modifies C13 of IRAK4 in human pDCs and this reaction disrupts MyD88 binding and IRAK4 function. These studies potentially reveal a mechanistic basis for the efficacy of DMF observed in autoimmune syndromes like MS and psoriasis and point to a novel druggable protein-protein interaction for controlling innate immune signaling.
Materials and Methods
Chemical reagents
Assays were performed with the following reagents: dimethyl fumarate (DMF; 242926; Sigma Aldrich), monomethyl fumarate (MMF; 651419; Sigma Aldrich), dimethyl succinate (DMS; W239607; Sigma Aldrich), and buthionine sulfoximine (BSO; 14484; Cayman Chemical). The Nrf2 inhibitor ML385 (SML1833, Sigma Aldrich), Dimethyl maleate (238198; Sigma Aldrich), Diethyl fumarate (D95654: Sigma Alrich), Diethyl maleate (D97703; Sigma Aldrich), Diisobutyl fumarate (7283–69-4; TCI Chemicals) and Diisopropyl fumarate (7283–70-7; TCI Chemicals).
Isolation of human pDCs
All studies with samples from human volunteers followed protocols approved by the TSRI institutional review board. Blood from healthy donors (females aged 30 to 49) were obtained after informed consent. Peripheral blood mononuclear cells (PBMCs) were purified over Histopaque-1077 gradients (10771; Sigma) according to the manufacturer’s instructions. Briefly, blood (20 × 25 ml aliquots) was layered over Histopaque-1077 (12.5 ml) and the samples were then fractionated by centrifugation at 750g for 20 min at 20°C with no brake. PBMCs were harvested from the Histopaque-plasma interface and washed twice with phosphate-buffered saline (PBS). After that time, pDCs were isolated with a CD304 (BDCA-4/Nueropilin-1) cell Isolation Kit (Miltenyi Biotec) according to the manufacturer’s instructions.
Stimulation of pDCs
Synthesized oligodeoxynucleotides (ODNs) were purchased from InvivoGen, Inc. The sequences of the ODNs are as follows: CpG-A (ODN2216), 5′-ggGGGACGA:TCGTCgggggg-3′, and CpG-B (ODN1826), 5′-tccatgacgttcctgacgtt-3′. Bases shown in capital letters are phosphodiester, and those in lower case are phosphorothioate (nuclease resistant). Palindrome is underlined. Influenza virus was propagated and titrated in Madine Darby Canine Kidney (MDCK) cells. Human pDC were stimulated with 1 multiplicity of infection (MOI) of influenza virus overnight and IFN-α levels were measured in the supernatants by ELISA. Isolated human pDCs were stimulated overnight with either 500μM Loxoribine (tlrl-lox, InvivoGen) or 100nM R-848 (Sigma). Supernatant IFN-α levels were measured by ELISA.
Cytokine and chemokine analysis.
ELISAs were performed using the VeriKine™ Human and Mouse IFN-Alpha ELISA Kits (R&D Systems) according to the manufacturer’s instructions.
Cellular analysis and sorting by flow cytometry.
Cells were stained with the following anti-human Abs: Phycoerythrin-conjugated anti-CD123 Ab (clone 6H6; BioLegend), allophycocyanin (APC)-conjugated anti–IFN-α Ab [clone 7N4–1; BD Pharmingen]. Flow cytometry acquisition was performed with a BD FACSDiva-driven BD LSR II flow cytometer (Becton Dickinson). Data were then analyzed with FlowJo software (TreeStar, Inc.).
Generation and culture of IRAK4-deficient human B-EBV cells.
Human IRAK4 deficient cells (patient ID: MB002334) were provided as a gift from the lab of Jean-Laurent Casanova (27). Cells were cultured in RPMI-1640 medium (Life Tech Invitrogen) supplemented with Heat-Inactivated Fetal Bovine Serum and a Penicillin-Streptomycin Solution (Life Tech Invitrogen). Cells were maintained at a concentration of 1 million cells per 1ml of medium with fresh medium added every 2 days.
To reconstitute IRAK4 expression within the IRAK4 deficient cells, the IRAK4 open reading frame was cloned from plasmid HsCD00330298 (Harvard PlasmID Repository) and inserted into the pLPC-myc-mCherry lentiviral expression system which was provided as a gift from the Lazzerini Denchi lab. The C13 cysteine mutants were generated using the Q5 Site-Directed Mutagenesis Kit (New England Biolabs).
Protein over-expression
HEK293T cells were seeded 3E6 cells/dish in 10 cm dishes. The next day, to an eppendorf tube was added either 1) 2.5 ug of IRAK4 construct and 2.5 ug of GFP construct or 2) 2.5 ug of IRAK4 construct and 2.5 ug of MyD88 construct. To each tube was added 500 uL of DMEM and 30 uL of PEI. 20 minutes later, the transfection solution was added dropwise to cells. 20 hours later, DMF or DMSO vehicle was added to the cells as indicated.
Protein labeling and click chemistry
HEK293T’s were harvested, lysed by sonication and diluted to a concentration of 2 mg protein/ml. Protein concentrations were measured with the Bio-Rad DC protein assay reagents A and B (5000113, 5000114; Bio-Rad). Proteome sample (500 μl) was treated with 100 μM IA-alkyne probe by adding 5 μl of a 10 mM probe stock (in DMSO). The labeling reactions were incubated at room temperature for 1 hour after which the samples were conjugated to isotopically labeled, TEV-cleavable tags (TEV tags) by copper-catalyzed azide-alkyne cycloaddition (CuACC or “click chemistry”). Heavy click chemistry reaction mixture (60 μl) was added to the DMSO-treated control sample and 60 μl of the light reaction mixture was added to the compound-treated sample. The click reaction mixture consisted of TEV tags [10 μl of a 5 mM stock, light (compound-treated) or heavy (DMSO treated)], CuSO4 (10 μl of a 50 mM stock in water), and TBTA (30 μl of a 1.7 mM stock in 4:1 tBuOH:DMSO). To this was added TCEP (10 μl of a 50 mM stock). The reaction was performed for 1 hour at room temperature. The light- and heavy-labeled samples were then centrifuged at 16,000g for 5 min at 4°C to harvest the precipitated proteins. The resulting pellets were resuspended in 500 μl of cold methanol by sonication, and the heavy and light samples were combined pairwise. Combined pellets were then washed with cold methanol, after which the pellet was solubilized by sonication in PBS, 1.2% SDS. The samples were heated at 90°C for 5 min and subjected to streptavidin enrichment of probe-labeled proteins, sequential on-bead trypsin and TEV digestion, and liquid chromatography-tandem mass spectrometry (LC-MS/MS), according to the published isoTOP-ABPP protocols (28–31).
Peptide and protein identification
RAW Xtractor (version 1.9.9.2; available at http://fields.scripps.edu/downloads.php) was used to extract the MS2 spectra data from the raw files (MS2 spectra data correspond to fragments analyzed during the second stage of mass spectrometry). MS2 data were searched against a reverse concatenated, nonredundant variant of the Human UniProt database (release-2012_11) with the ProLuCID algorithm (publicly available at http://fields.scripps.edu/downloads.php) (32). Cysteine residues were searched with a static modification for carboxyamidomethylation (+57.02146) and up to one differential modification for either the light or heavy TEV tags (+464.28595 or +470.29976, respectively). Peptides were required to have at least one tryptic terminus and to contain the TEV modification. ProLuCID data were filtered through DTASelect (version 2.0) to achieve a peptide false-positive rate below 1% (33).
R value calculation and processing
The quantification of heavy:light ratios (isoTOP-ABPP ratios, R values) was performed by in-house CIMAGE software (30) using default parameters (3 MS1s per peak and a signal-to-noise threshold of 2.5). Site-specific engagement of electrophilic compounds was assessed by blockade of IA-alkyne probe labeling. For peptides that showed a ≥95% reduction in MS1 peak area from the compound-treated proteome (light TEV tag) when compared to the DMSO-treated proteome (heavy TEV tag), a maximal ratio of 20 was assigned. Overlapping peptides with the same labeled cysteine (for example, they had the same local sequence around the labeled cysteines, but had different charge states, MudPIT segment numbers, or tryptic termini) were grouped together, and the median ratio from each group was recorded as the R value of the peptide for that run.
IRAK4-MyD88 Binding Assay:
HEK293Ts (2.5 × 106) were plated in a 10cm plate 24h prior to transfection. For transfection, DNA (5 μg, myc-tagged IRAK4 kinase dead C13C, R12C or other C13 mutants; FLAG-tagged MyD88; or FLAG-tagged MetAP2), serum-free DMEM (500 μL, Corning), and PEI (30 μL) were mixed together and allowed to incubate for 30 min at room teperature. The mixture was then added dropwise to plated cells and cells rested for 48 h. Media from cells transfected with FLAG-tagged protein was aspirated and the cells were washed with PBS (5 mL). Cells were transfered to a 1.5 mL Eppendorf tube and washed with PBS 1x. Freshly-prepared ice-cold lysis buffer was added to the cell pellet (400 μL; 50 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton-X 100, 1 mM DTT with protease (Mini EDTA-free, Roche) and phosphatase (Phos-STOP, Roche) inhibitors. Samples were incubated for 30 min on ice and centrifuged (10 min at 10,000 × g at °C). Protein concentration was normalized to 2 mg/mL. FLAG-tagged protein input samples were generated (100 μg protein in 50 μL sample). FLAG beads (22.5 μL per 1.5 mg protein, Thermo Fisher) were pre-washed with lysis buffer and pelleted by centrifugation (2 min at 1,600 × g at 4 °C). This bead wash protocol was repeated 1x. Pre-washed FLAG beads were then added to 1.5 mg lysate (750 μL) and rotated (3 h at 4 °C). Meanwhile, media from cells transfected with myc-tagged protein was aspirated and cells washed with PBS. Cells were then scraped and transferred to a 1.5 mL Eppendorf tube and subjected to an additional wash with PBS 1x. Freshly-prepared ice-cold lysis buffer was added to each cell pellet (400 μL; 50 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton-X 100, 1 mM DTT with protease (Mini EDTA-free, Roche) and phosphatase (Phos-STOP, Roche) inhibitors. Cell lysates were incubated on ice for 30 min on ice and centrifuged (10 min at 10,000 × g at 4 °C). Protein concentration was normalized to 1 mg/mL. Myc-tagged protein input samples were generated (100 μg protein in 50 μL sample). Upon completion of FLAG enrichment, beads were pelleted by centrifugation (2 min at 1,600 × g at 4 °C). Flow through was aspirated away and ice-cold lysis buffer added. Tubes were inverted gently to wash beads (500 μL). These washes were repeated 2x. FLAG-enrichment was resuspended in lysis buffer (100 μL) and transfered to myc-tagged protein lysate (750 μg, 750 μL). The sample was then incubated on a rotator (2 h at 4 °C). Upon completion, beads were pelleted by centrifugation (2 min at 1,600 × g at 4 °C) and unbound lysate aspirated away. Fresh ice-cold lysis buffer was added, and the tubes inverted gently to wash beads (500 μL). This bead wash was repeated 2x. Beads were resuspended in 1x loading buffer (45 μL lysis buffer and 15 μL 4x loading buffer (40% glycerol, 0.4% bromophenol blue, 2.8% β-mercaptoethanol, pH 6.8)), and samples were boiled to denature proteins and cleave disulfides (5 min, 95 °C). Samples were then loaded onto a 10% Tris-glycine gel for SDS-PAGE separation (30 μL IP, 15 μL input).
IRAK4-MyD88 Binding Assay with DMF/MMF/DMF Treatment:
HEK293Ts (2.5 × 106) were plated in a 10cm plate 24h prior to transfection. For transfection, DNA (5 μg, myc-tagged IRAK4 kinase dead C13C, R12C or other C13 mutants; FLAG-tagged MyD88; or FLAG-tagged MetAP2), serum-free DMEM (500 μL, Corning), and PEI (30 μL) were mixed together and allowed to incubate for 30 min at room teperature. The mixture was then added dropwise to plated cells and cells rested for 48 h. Cells expressing myc-tagged proteins were treated with DMSO, DMF, MMF or DMS (1000x stock in DMSO, final concentration 100 μM) and incubated for 4 h. Media from cells transfected with FLAG-tagged protein was aspirated and the cells were washed with PBS (5 mL). Cells were transfered to a 1.5 mL Eppendorf tube and washed with PBS 1x. Freshly-prepared ice-cold lysis buffer was added to the cell pellet (400 μL; 50 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton-X 100, 1 mM DTT with protease (Mini EDTA-free, Roche) and phosphatase (Phos-STOP, Roche) inhibitors. Samples were incubated for 30 min on ice and centrifuged (10 min at 10,000 × g at 4 °C). Protein concentration was normalized to 2 mg/mL. FLAG-tagged protein input samples were generated (100 μg protein in 50 μL sample). FLAG beads (22.5 μL per 1.5 mg protein, Thermo Fisher) were pre-washed with lysis buffer and pelleted by centrifugation (2 min at 1,600 × g at 4 °C). This bead wash protocol was repeated 1x. Pre-washed FLAG beads were then added to 1.5 mg lysate (750 μL) and rotated (3 h at 4 °C). Meanwhile, media from cells transfected with myc-tagged protein was aspirated and cells washed with PBS. Cells were then scraped and transferred to a 1.5 mL Eppendorf tube and subjected to an additional wash with PBS 1x. Freshly-prepared ice-cold lysis buffer was added to each cell pellet (400 μL; 50 mM HEPES, pH 7.4, 150 mM NaCl, 1% Triton-X 100, 1 mM DTT with protease (Mini EDTA-free, Roche) and phosphatase (Phos-STOP, Roche) inhibitors. Cell lysates were incubated on ice for 30 min on ice and centrifuged (10 min at 10,000 × g at 4 °C). Protein concentration was normalized to 1 mg/mL. Myc-tagged protein input samples were generated (100 μg protein in 50 μL sample). Upon completion of FLAG enrichment, beads were pelleted by centrifugation (2 min at 1,600 × g at 4 °C). Flow through was aspirated away and ice-cold lysis buffer added. Tubes were inverted gently to wash beads (500 μL). These washes were repeated 2x. FLAG-enrichment was resuspended in lysis buffer (100 μL) and transfered to myc-tagged protein lysate (750 μg, 750 μL). The sample was then incubated on a rotator (2 h at 4 °C). Upon completion, beads were pelleted by centrifugation (2 min at 1,600 × g at 4 °C) and unbound lysate aspirated away. Fresh ice-cold lysis buffer was added, and the tubes inverted gently to wash beads (500 μL). This bead wash was repeated 2x. Beads were resuspended in 1x loading buffer (45 μL lysis buffer and 15 μL 4x loading buffer (40% glycerol, 0.4% bromophenol blue, 2.8% β-mercaptoethanol, pH 6.8)), and samples were boiled to denature proteins and cleave disulfides (5 min, 95 °C). Samples were then loaded onto a 10% Tris-glycine gel for SDS-PAGE separation (30 μL IP, 15 μL input)
Western Blotting:
Proteins were transferred to nitrocellulose membrane in a wet transfer cell (50 V, 2.5 h). Membrane was blocked in 5% milk for 1 h followed by primary incubation overnight at 4 °C (1:2500 anti-myc (Cell Signaling) or 1:2500 anti-DYKDDDDK (Cell Signaling)). Membranes were washed with TBST (3x, 10 min) and incubated with secondary anti-rabbit antibody (Licor) for 1 h at room temperature.
Kinase Activity Assay:
IRAK4 Kinase activity was evaluated by the IRAK4 Kinase Enzyme System (V9421, Promega) according to the manufactorer’s instructions. Per manufactorer’s instructions, Staurosporine (S6942, Sigma) was administered to generate a kinase inhibitor dose-response curve.
Statistical Analysis:
Significance in Figures 1–3 were determined by 2-tailed unpaired-t-test from at least 2 independent experiments of at least 4 replicates per experiment using Prism 7 (GraphPad). Significance in Figure 4 was determined by paired t-test from three independent experimental replicates using Prism 7 (GraphPad).
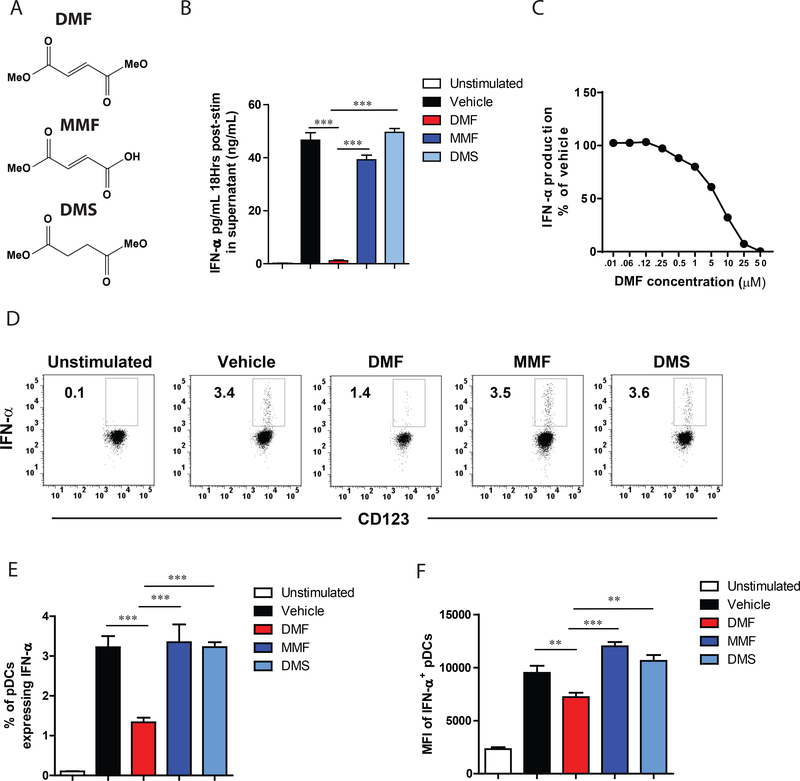
Figure 1. DMF inhibits IFN-α release from human pDCs.
(A) Structures of dimethyl fumarate (DMF), monomethyl fumarate (MMF), and dimethyl succinate (DMS). (B) DMF, but not MMF or DMS, blocks IFN-α release from primary human pDCs. pDCs were isolated from human blood and treated with the indicated compounds (50 μM each) and concomitantly activated with CpGA. After 18 h, IFN-α was measured in the supernatant by ELISA. (C) DMF inhibits IFN-α release in a concentration-dependent manner. pDCs were isolated from human blood and treated with indicated concentrations of DMF for 18 h with concomitant CpGA stimulation. (D) FACS dot plot showing reduced intracellular IFN-α production in DMF-treated human pDCs. pDCs harvested from human blood were stimulated with CpGA and treated concomitantly with DMF (50 μM), MMF (50 μM), or DMS (50 μM) for 6 h. Frequency of IFN-α producing pDC (E) and mean fluorescent intensity of IFN-α levels gated on IFN-α+ pDCs (F). Results are representative of at least two independent experiments and ** P < 0.01, *** P < 0.005 by two-tailed unpaired t test.
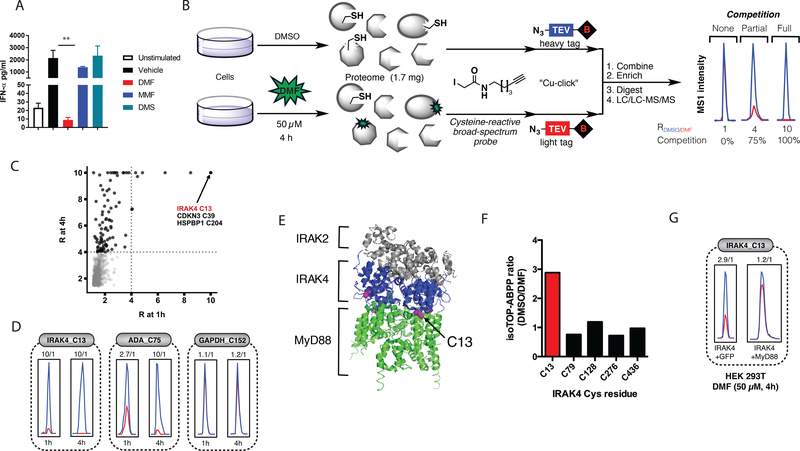
Figure 3. Cysteine-13 (C13) of IRAK4 is a proteomic hot spot for DMF in Cal-1 cells.
(A) IFN-α release from Cal-1 cells stimulated with Sendai virus and treated concomitantly with 50 μM of indicated compounds for 18 h. (B) Schematic diagram depicting competitive IsoTOP for assessing DMF-reactive cysteines. (C) Scatter plot of ratio (R) values (DMSO/DMF) for quantified cysteine residues in isoTOP-ABPP experiments from Cal-1 cells treated for 1 or 4 h with 50 μM DMF. (D) Representative parent ion (MS1) profiles for DMF-hypersensitive (C13 of IRAK4), -moderately sensitive (C75 of ADA), and -insensitive (C152 of GAPDH) cysteines from isoTOP-ABPP experiments in Cal-1 cells. (E) Crystal structure of the Myddosome comprising six MyD88 (green), four IRAK4 (blue), and four IRAK2 (gray) molecules (PDB accession number 3MOP). In magenta is C13 of IRAK4. (F, G) HEK293T cells were transfected with either IRAK4 and GFP or IRAK4 and MyD88. 20 hours later, cells were treated with DMF (50 μM) for 4 hours and lysed. The isoTOP-ABPP protocol was then performed as previously described. The R values (DMSO/DMF) from the isoTOP-ABPP experiment for each quantified cysteine are shown in the bar graph.
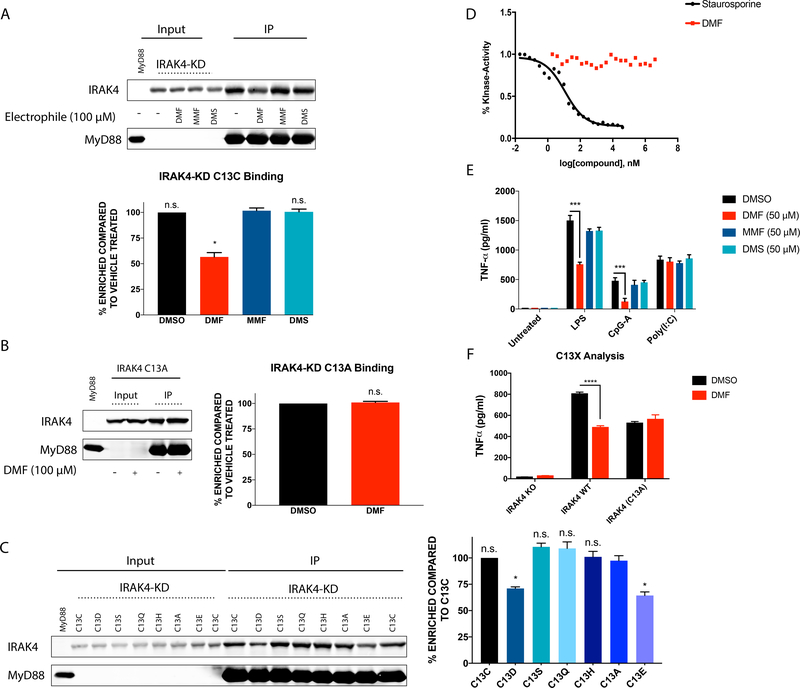
Figure 4. DMF disrupts IRAK4-MyD88 complexes and signaling in a C13-dependent manner.
(A) DMF, but not MMF or DMS, impairs interactions between IRAK4 and MyD88. MyD88-FLAG (immunoprecipitated from HEK293T cells expressing MyD88-FLAG) bound to anti-FLAG beads was exposed to lysate from IRAK4-K2D-expressing HEK293T cells pre-treated with DMF, MMF, or DMS (100 μM compound, 4 h), or DMSO control. Shown Western blot is representative of three replicates used for quantitation. * P = 0.02 DMF vs. MMF as determined by paired t-test. (B) DMF-mediated disruption of the IRAK4-MyD88 interaction depends on C13 of IRAK4. MyD88 was immunoprecipitated from HEK293T cells expressing FLAG-tagged MyD88 and exposed to lysate from WT- or C13A-IRAK4-K2D-expressing HEK293T cells pre-treated with DMF (100 μM, 4 h) or DMSO control. Shown Western blot is representative of three replicates used for quantitation. (C) Impact of mutagenesis of C13 on IRAK4 interactions with Myd88. MyD88 was immunoprecipitated from HEK293T cells expressing FLAG-tagged MyD88 and exposed to lysate from HEK293T cells expressing the indicated C13 mutants of IRAK4. Western blot is representative of three replicates used for quantitation. * C13E vs C13A P = 0.049 * C13D vs C13A P = 0.039 as determined by paired t-test. (D) Impact of DMF on IRAK4 kinase activity. A reaction of purified IRAK4 (50ng/reaction), MBP (100ng/reaction) and ATP (50μM) was was treated with various concentrations of either Staurosporine (40μM to 0.02nM) or DMF (4mM to 2nM) to generate a dose response curve to assess the efficacy of these compounds as inhibitors of IRAK4 kinase activity. ADP-Glo™ Kinase Assay measures the conversion of ATP to ADP, which correlates with overall kinase activity within the reaction. (E) IRAK4-deficient B-EBV cells were reconstituted with human WT IRAK4 by lentiviral transduction, pre-treated with DMF, MMF, or DMS (50 μM, 4 h) or DMSO control, stimulated with LPS, CpG-A or Poly I:C for 24 h, and TNF-α measured by ELISA. *** LPS stimulated cells, DMSO versus DMF Treatment, P = 0.0002. ***CpG-A Treated cells, DMSO versus DMF treatment, P = 0.001. (F) IRAK4-deficient B-EBV cells were reconstituted with WT-IRAK4 or C13A-IRAK4 by lentiviral transduction, pre-treated with DMF (50 μM, 4 h) or DMSO control, stimulated with LPS for 24 hours, and TNF-α levels measured by ELISA. **** DMF treated cells versus DMSO treated cells, P < 0.0001.
Results
DMF inhibits IFN-α release from activated human pDCs
Primary human pDCs isolated from normal human donors were purified, stimulated with CpG-A, and treated concomitantly with DMF, the major DMF metabolite monomethyl fumarate (MMF), or the saturated unreactive analog dimethylsuccinate (DMS) (50 μM each compound; Fig. 1A). DMF, but not MMF or DMS, produced a concentration-dependent inhibition of CpG-A-induced IFN-α release, displaying an IC50 value of ~7 μM (Fig. 1B, C). The inhibitory action of DMF on IFN-α production occurred without an effect on pDC cell viability (Fig. S1A) and was accompanied by the suppression of multiple additional cytokines and chemokines (Fig. S1B). We further found that intracellular IFN-α was also decreased by DMF, but not MMF or DMS (Fig. 1D-F), which pointed to a mechanism of action where DMF blocks the expression, rather than secretion, of IFN-α (Fig. 1D).
We next evaluated additional pDC stimuli and found that, as observed with CpG-A, DMF also blocked IFN-α production following treatment with CpG-B, another TLR9 agonist, or influenza (TLR7 agonist) (Fig. 2A). Interestingly, however, the impact of DMF on other cytokines varied depending on the stimulus, with a broader suppressive effect being observed for DMF with the TLR9 agonists CpG-A and CpG-B, but not following stimulation with influenza virus (Fig. S2). To further investigate the potential role of DMF in blocking TLR7-induced cytokine secretion independent of a viral recognition event, we treated pDCs with the additional TLR7/8 stimulus R848 and the TLR7-specific agonist loxoribine. As we found for influenza virus, DMF treatment impaired cytokine production in response to these additional TLR7 stimuli (Fig. 2A). These data suggest that DMF differentially impairs the activation of innate signaling pathways acting through multiple TLRs.
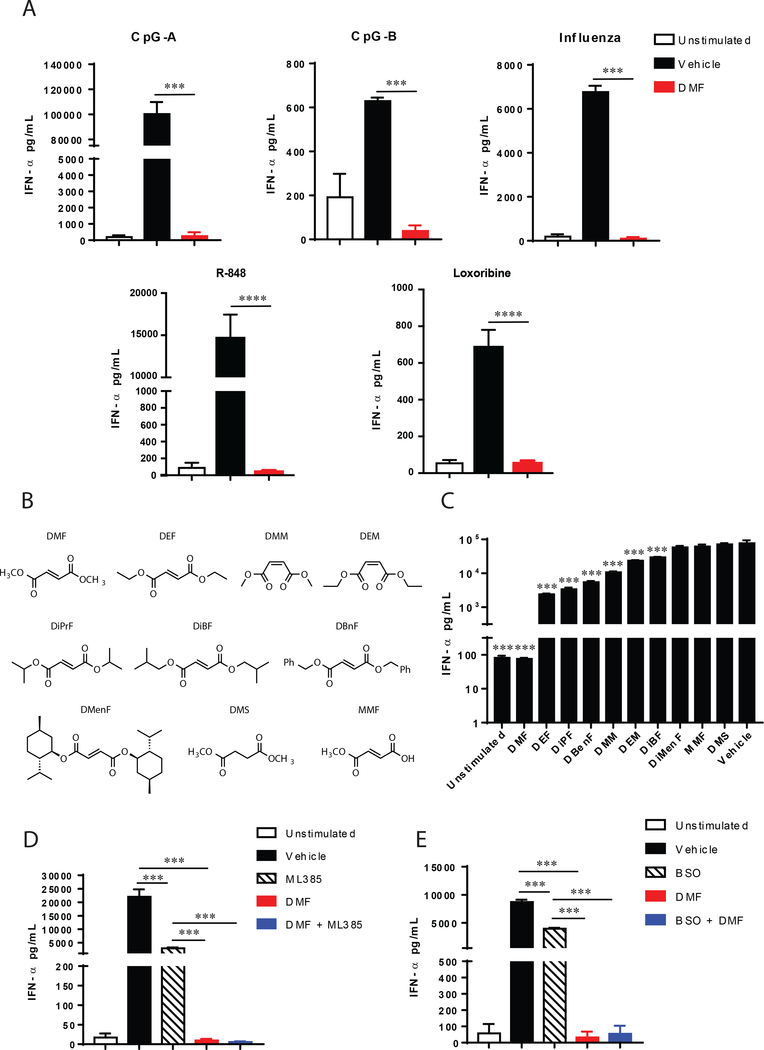
Figure 2. Inhibition of IFN-α production by DMF occurs through multiple TLR signaling pathways and is mostly independent of NRF2 or GSH abundance.
(A) DMF inhibits TLR7 and TLR9-induced pDC IFN-α production. pDCs were isolated from human blood and treated with 50μM DMF for 18 h along with 1 μM CpG-A, 1 μM CpG-B, one multiplicity of infection with Influenza virus, 100nM R-848 or 500uM Loxoribine and the levels of IFN-α were quantified by ELISA. Results are representative of at least two independent experiments and *** P < 0.005, **** P < 0.0001 by two-tailed unpaired t test. (B) Structures of dimethyl fumarate (DMF) analogs. DEF: diethyl fumarate; DMM: dimethyl maleate; DEM: diethyl maleate; DiPrF: diisopropyl fumarate; DBnF: dibenzyl fumarate; DMenF: dimenthyl fumarate; TMA: tetramethyl fumaramide; DMS: dimethyl succinate; MMF: monomethyl fumarate. (C) IFN-α release from purified human pDCs stimulated with CpG-A and treated concomitantly with 50 μM of indicated compounds in (B) for 18 h. (D) IFN-α release measured by ELISA from purified human pDCs stimulated with CpG-A and treated with either the NRF2 inhibitor ML385 (5 μM), DMF (50 μM), or ML385 and DMF. (E) IFN-α release measured by ELISA from purified human pDCs stimulated with CpG-A and treated with either the glutathione inhibitor BSO (2 mM), DMF (50 μM), or BSO and DMF. Results are representative of at least two independent experiments and *** P < 0.005 by two-tailed unpaired t test.
We further explored the structure-activity relationship (SAR) for blockade of IFN-α production by treating pDCs with a set of fumarate ester analogues with varied stereoelectronic properties (Fig. 2B). While several compounds with increased steric bulk (e.g., diethyl fumarate (DEF), diisopropyl fumarate (DiPrF), diisobutyl fumarate (DiBF), dibenzyl fumarate (DBnF)) suppressed IFN-α production, none exhibited comparable activity to DMF (50 μM of each compound; Fig. 2C). We also tested the impact of double bond geometry and found that dimethyl maleate (DMM) and diethyl maleate (DEM) partially blocked IFN-α production, but neither compound displayed equivalent activity to DMF (Fig. 2C). To the extent that the Z-olefin arrangement in DMM and DEM would be expected to increase electrophilicity, these data suggest a more complex, non-linear relationship between the intrinsic reactivity and immunosuppressive effects of fumarate esters.
DMF blockade of pDC IFN-α production involves NRF2-independent mechanisms
The pharmacological effects of DMF have been proposed to occur, at least in part, through activation of the KEAP1-NRF2 pathway (16), which is major mediator of cellular responses to electrophilic/oxidative stress (34). We found, however, that DMF fully suppressed IFN-α production in pDCs co-treated with the NRF2 inhibitor ML385 (35) (Fig. 2D). ML385 treatment alone produced a partial reduction in IFN-α, but this effect was much less dramatic than the complete blockade of IFN-α production by DMF (Fig. 2D). DMF has also been shown to deplete intracellular concentrations of glutathione (GSH) (14, 17, 18, 36), and we found that the GSH synthesis inhibitor buthionine sulfoximine (BSO) modestly decreased IFN-α release from pDCs (Fig. 2E). Again, this change was much lower in magnitude than the IFN-α suppression caused by DMF (Fig. 2E). These data, taken together, indicate that the primary mechanism by which DMF suppresses IFN-α production is independent of NRF2 and involves biochemical pathways beyond the reduction of GSH content in pDCs.
Mapping DMF-sensitive cysteines in the pDC cell line model Cal-1
We next sought to identify candidate protein targets that might contribute to the IFN-α suppressing activity of DMF in pDCs. We previously used the chemical proteomic method isoTOP-ABPP to globally map DMF-sensitive cysteines in primary human T cells (20). The sample requirements for such chemical proteomic experiments are, however, quite high (milligrams of protein) and beyond the scale that could be prepared from primary human pDCs. We therefore turned to Cal-1 cells as a pDC model cell line (37) for the discovery of DMF-sensitive cysteines.
We first confirmed that DMF maintains IFN-α-suppressing activity in Cal-1 cells (Fig. 3A) and then applied isoTOP-ABPP to map DMF-sensitive cysteines in these cells. In brief, Cal-1 cells were treated with DMF (50 μM, 1 or 4 h) or DMSO control, lysed and exposed to the general cysteine-reactive probe iodoacetamide (IA)-alkyne, followed by click chemistry-mediated conjugation (38) to isotopically differentiated azide-biotin tags containing a tobacco etch virus (TEV) protease–cleavable linker. Samples were then subject to streptavidin enrichment and protease cleavage, after which heavy and light samples were combined, and analyzed by LC-MS to quantify IA-alkyne-labeled cysteines using an LTQ-Orbitrap Velos instrument (Fig. 3B) (20, 21, 30)
Among the 4408 cysteines quantified by isoTOP-ABPP in Cal-1 cells, ~170 residues showed substantial sensitivity to DMF, as reflected in > 4-fold changes in IA-alkyne reactivity in DMF-treated cells (Fig. 3C). Most of these DMF-sensitive cysteines showed greater change in IA-reactivity at 4 h (Fig. 3C), likely reflecting a time-dependent increase in modification by DMF. A representative example of such time-dependent change is shown for C75 of adenosine deaminase (ADA) in Fig. 3D. A select subset of cysteines, on the other hand, showed near-complete loss of IA-alkyne reactivity following only 1 h of DMF treatment. These “DMF-hypersensitive” cysteines included C13 of IRAK4 (Fig. 3C, D), a protein kinase that plays a central role in Toll-like receptor (TLR)-mediated innate immune cell signaling pathways that produce IFN-α (39). The immunological functions of IRAK4 have inspired the development of several ATP-competitive inhibitors of this kinase as potential drugs to treat autoimmune and autoinflammatory disorders (40, 41). Interestingly, however, C13 is not found in the IRAK4 active site, but rather located at the interface of IRAK4 that binds to the adaptor protein MyD88 (Fig. 3E). We next sought to understand if DMF modification of C13 in IRAK4 affects interactions with MyD88 and downstream signaling.
DMF disrupts IRAK4-MYD88 interactions
A primary function of pDCs is to produce IFN-α in response to viral or endogenous nucleic acids (1, 2). The receptors for these nucleic acids are TLR7/8 and TLR9 (5), which in turn signal through the “Myddosome” complex comprising myeloid differentiation primary response gene 88 (MyD88), interleukin-1 receptor-associated kinase 2 (IRAK2), and IRAK4 (42). The location of the DMF-hypersensitive cysteine in IRAK4 – C13 – is at the interface of the IRAK4-MyD88 protein-protein interaction (Fig. 3E), suggesting a potential functional role for this residue. This hypothesis is also supported by human genetics, as mutation of the adjacent residue Arg12 (R12C) produces an immunodeficiency syndrome that manifests as increased susceptibility to pyogenic infections in children (27, 43, 44). The R12C mutation has been shown to disrupt binding between IRAK4 and MyD88 (45), presumably hampering pDC responses to exogenous nucleic acids. We found that recombinantly expressed IRAK4 (produced by transient transfection in HEK293T cells) maintained site-specific sensitivity to DMF at C13 (Fig. 3F), and this interaction was disrupted by co-expression of MyD88 in HEK293T cells (Fig. 3G).
We next established an in vitro binding assay for measuring the IRAK4-MyD88 interaction. We immobilized FLAG-tagged MyD88 on anti-FLAG beads for incubation with lysate from HEK293T cells expressing various R12/C13 mutants in the context of a kinase-dead variant of IRAK4 (K213A/K214A; termed IRAK4-K2D). We chose to utilize the kinase-dead IRAK4 because this form of IRAK4 has been shown to interact more stably with MyD88, likely indicating that IRAK4 and MyD88 engage in a dynamic signaling complex that can be disassembled by autophosphorylation of IRAK4 (46–48). Consistent with expectations based on past work (45), we found that the R12C mutant of IRAK4-K2D blocked co-precipitation of this protein with FLAG-tagged MyD88 (Fig. S3A). We next treated IRAK4-K2D-transfected cells with DMF (100 μM, 4 h) and found that this compound, but not structurally related analogues that lack IFN-α-suppressing activity (MMF, DMS), substantially blocked the interaction of IRAK4-KD with MyD88 in vitro (Fig. 4A). This antagonistic effect of DMF on the IRAK4-K2D-MYD88 interaction was not observed for a C13A-mutant of IRAK4-K2D (Fig. 4B), supporting that DMF reactivity with C13 is responsible for disrupting IRAK4-K2D binding to MyD88. Interestingly, we found that mutation of C13 to negatively charged residues glutamate or aspartate, but not other amino acids, including glutamine, mimicked the inhibitory effect of DMF on the IRAK4-KD-MYD88 interaction (Fig. 4C). One interpretation of these findings is that DMF, once reacted with C13 of IRAK4-KD, may undergo esterolysis to present a carboxylate group that, like the C13D and C13E mutants, impairs binding to MyD88.
Our studies pointed to a kinase activity-independent mechanism by which covalent DMF modification of IRAK4 disrupts Myddosome function (i.e., disruption of IRAK4-MyD88 interactions). To more directly assess whether DMF impacts IRAK4 kinase activity, we treated purified IRAK4 (50ng/reaction) with DMF (4mM-2nM) or the pan-kinase inhibitor staurosporine (40μM-0.02nM) and measured residual IRAK4 activity using a substrate assay. DMF did not alter IRAK4 kinase activity in this assay, in contrast to staurosporine, which showed clear concentration-dependent inhibition of IRAK4 (Fig. 4D).
DMF inhibits IRAK4-MyD88 signaling and cytokine production through engagement of IRAK4 Cysteine 13
One of the downstream consequences of Myddosome complex formation is activation of the NF-κB signaling pathway. DMF has been reported to inhibit NF-κB signaling in a variety of cell types (49–53). Consistent with these past findings and with disruption of the Myddosome complex, we found that DMF blocks phosphorylation of the NF-κB pathway member p65 in pDCs stimulated with CpG-B (Fig. S3B).
We next examined the impact of DMF on IRAK4-induced cytokine production in human immune cells. We employed Epstein-Barr Virus immortalized B cells (B-EBV) that were generated from peripheral blood mononuclear cells of a patient with a large deletion in the IRAK4 gene that renders it non-functional (27). We used the B-EBV cells as a source of IRAK4-null cells amenable to genetic complementation studies. IRAK4-deficient B-EBV cells were then reconstituted with either wild-type or the C13A mutant of IRAK4 using lentiviral transduction. Reconstitution of IRAK4-deficient B-EBV cells with wild-type IRAK4 resulted in significantly greater TNF-α production following LPS and CpG stimulation compared to IRAK4-deficient B-EBV cells, while the TLR3/RIG-I/MDA5 agonist Poly I:C, which signals independent of MyD88/IRAK4, induced TNF-α production in both wild-type IRAK4 and IRAK4-deficient cells (Fig. S3C). We next discovered that DMF, but not MMF or DMS, suppressed TNF-α production following stimulation with LPS and CpG in the B-EBV cells reconstituted with wild type IRAK4 (Fig. 4E). In contrast, DMF did not affect TNF-α production following Poly I:C stimulation (Fig. 4E), suggesting that DMF acts to suppress pDC cytokine production predominantly through IRAK4/MyD88 signaling. Finally, we confirmed that DMF does not decrease CpG-induced TNF-α production in IRAK4-deficient cells reconstituted with the C13A-IRAK4 mutant (Fig. 4F). We should note that B-EBV cells expressing the C13A-IRAK4 mutant showed a lower TNF-α induction than WT-IRAK4-reconstituted cells, but, regardless, the TNF-α induction in C13A-IRAK4 mutant cells was still much larger than that of IRAK4-deficient cells (which showed a negligible response; see Fig. S3C) and DMF treatment did not further reduce TNF-α production in the C13A-IRAK4 cells.
Discussion
Plasmacytoid DCs were first definitively characterized in 1999 and have since been implicated in the initiation of psoriasis (8, 54) and the progression of MS (9, 10, 12). Despite their low abundance in the blood, pDCs are the primary producers of IFN-α and can thus drive local inflammation (1). The immunomodulatory drug DMF is a widely prescribed and effective treatment for MS and psoriasis, both of which are inflammatory diseases. More recently, DMF has also been reportedas a potential therapy for cutaneous T-cell lymphoma and is at times prescribed off-label to bone marrow transplant patients due to its immunomodulatory properties (55). The mechanism of DMF is not well understood, but this drug has been shown to impair the activation of T cells (19, 20) and conventional dendritic cells (56). However, little is known about the effect of DMF on pDCs. Our studies indicate that pDCs are sensitive to DMF, and that this pharmacological effect is largely independent of NRF2 and, at least in part, through the NF-κB signaling pathway, with covalent adduction of C13 of IRAK4 residing at the top of the signaling cascade. Given that DMF is relatively well tolerated and capable of suppressing pro-inflammatory signaling in both activated T-cells and now pDCs, a more detailed understanding of its mechanisms of action may reveal potential new therapeutic applications in hematological malignancies and autoimmune disorders.
We have shown here that DMF and more elaborated electrophilic analogs, but not the poorly reactive hydrolytic metabolite MMF or DMS, inhibit IFN-α secretion from pDCs. This modulation depends primarily on proteins and pathways beyond the conventional electrophile-sensing NRF2-KEAP1 pathway or glutathione depletion. Additionally, DMF inhibits multiple modes of pDC stimulation, consistent with the drug targeting a central signaling pathway underlying different types of immune cell activation. TLR7 and TLR9 are the primary innate immune receptors on pDCs (3). Their activation promotes the assembly of the Myddosome complex, which is comprised of MyD88, IRAK2, and IRAK4. Using a global chemical proteomic method that quantified > 4000 cysteine residues in the human pDC cell line Cal-1, we identified C13 of IRAK4 as a “hot spot” for modification of DMF. IRAK4 signaling underlies each of the different modes of pDC activation shown to be inhibited by DMF. That not only DMF, but also mutation of C13 to aspartate or glutamate, disrupted IRAK4-MYD88 interactions suggests a potential mechanism where DMF reacts with C13 on IRAK4 and is subsequently hydrolyzed to unveil a negatively charged appendage that disrupts MyD88 binding. Importantly, but not entirely surprising, given the distal location of the modified residue to the active site, DMF adduction of C13 does not affect IRAK4 kinase activity, suggesting that disruption of the protein-protein interaction (PPI) of IRAK4 and MyD88 is the primary mechanism by which DMF impairs Myddosome function in pDCs.
While PPIs play numerous and critical roles in cell signaling, targeting this class of interactions with small-molecule drugs has historically proven challenging, as many PPI interfaces are flat and cover large surface areas (57). Our discovery that DMF can disrupt the IRAK4-MyD88 interaction through cysteine reactivity demonstrates the potential of covalent small molecules as chemical probes and therapeutics to target disease-relevant PPIs. Further elaboration of the methyl fumarate ester still suppressed IFNα production, but to a much lesser degree than DMF. This suggests that the small size of DMF is critical to its pharmacological activity. It is possible that some of this effect reflects a greater vulnerability of the methyl-succinylated cysteine on DMF-adducted proteins to undergo hydrolysis (enzymatic or solvolytic) to furnish a negatively charged group.
It has also been reported that endogenous fumarate production through the citric acid cycle is associated with oncogenesis (58, 59). We have demonstrated that other methyl fumarate-bearing small molecules (60) are vulnerable to enzymatic hydrolysis by carboxylesterases, but it is unclear whether DMF can be hydrolyzed twice to reveal the free-acid fumarate oncometabolite in appreciable concentrations. We have previously reported that the singly-hydrolyzed metabolic product of DMF, MMF does not appreciably react with proteinaceous cysteines at treatment relevant concentrations or suppress immune cell function (20). Nonetheless, we cannot rule out the possibility of an increase of free fumarate, or a pharmacological effect of this possible outcome, as a result of DMF treatment.
Supplementary Material
Key Points.
Dimethylfumarate (DMF) suppresses cytokine production in pDCs
DMF blocks Myddosome formation by disrupting the MyD88-IRAK4 interaction
DMF blocks the MyD88-IRAK4 interaction by targeting C13 in IRAK4
Acknowledgments
This work was supported by the NIH (CA231991), the Life Science Research Foundation (E.V.V.), the American Cancer Society PF-15-142-01-CDD (B.W.Z.), the National Science Foundation DGE-1346837 (M.M.B.), The Donald E. and Delia B. Baxter Foundation Faculty Scholar Grant (J.R.T.).
References
- 1.Reizis B, Bunin A, Ghosh HS, Lewis KL, and Sisirak V 2011. Plasmacytoid dendritic cells: recent progress and open questions. Annu Rev Immunol 29: 163–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gilliet M, Cao W, and Liu YJ 2008. Plasmacytoid dendritic cells: sensing nucleic acids in viral infection and autoimmune diseases. Nat Rev Immunol 8: 594–606. [DOI] [PubMed] [Google Scholar]
- 3.Reizis B, Colonna M, Trinchieri G, Barrat F, and Gilliet M 2011. Plasmacytoid dendritic cells: one-trick ponies or workhorses of the immune system? Nat Rev Immunol 11: 558–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Colonna M, Trinchieri G, and Liu YJ 2004. Plasmacytoid dendritic cells in immunity. Nat Immunol 5: 1219–1226. [DOI] [PubMed] [Google Scholar]
- 5.Swiecki M, and Colonna M 2015. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 15: 471–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Farkas L, Beiske K, Lund-Johansen F, Brandtzaeg P, and Jahnsen FL 2001. Plasmacytoid dendritic cells (natural interferon- alpha/beta-producing cells) accumulate in cutaneous lupus erythematosus lesions. Am J Pathol 159: 237–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ronnblom L, Alm GV, and Eloranta ML 2009. Type I interferon and lupus. Curr Opin Rheumatol 21: 471–477. [DOI] [PubMed] [Google Scholar]
- 8.Nestle FO, Conrad C, Tun-Kyi A, Homey B, Gombert M, Boyman O, Burg G, Liu YJ, and Gilliet M 2005. Plasmacytoid predendritic cells initiate psoriasis through interferon-alpha production. J Exp Med 202: 135–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pashenkov M, Huang YM, Kostulas V, Haglund M, Soderstrom M, and Link H 2001. Two subsets of dendritic cells are present in human cerebrospinal fluid. Brain 124: 480–492. [DOI] [PubMed] [Google Scholar]
- 10.Serafini B, Rosicarelli B, Franciotta D, Magliozzi R, Reynolds R, Cinque P, Andreoni L, Trivedi P, Salvetti M, Faggioni A, and Aloisi F 2007. Dysregulated Epstein-Barr virus infection in the multiple sclerosis brain. J Exp Med 204: 2899–2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey-Bucktrout SL, Caulkins SC, Goings G, Fischer JA, Dzionek A, and Miller SD 2008. Cutting edge: central nervous system plasmacytoid dendritic cells regulate the severity of relapsing experimental autoimmune encephalomyelitis. J Immunol 180: 6457–6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lande R, Gafa V, Serafini B, Giacomini E, Visconti A, Remoli ME, Severa M, Parmentier M, Ristori G, Salvetti M, Aloisi F, and Coccia EM 2008. Plasmacytoid dendritic cells in multiple sclerosis: intracerebral recruitment and impaired maturation in response to interferon-beta. J Neuropathol Exp Neurol 67: 388–401. [DOI] [PubMed] [Google Scholar]
- 13.Mrowietz U, Christophers E, and Altmeyer P 1999. Treatment of severe psoriasis with fumaric acid esters: scientific background and guidelines for therapeutic use. The German Fumaric Acid Ester Consensus Conference. Br J Dermatol 141: 424–429. [DOI] [PubMed] [Google Scholar]
- 14.Mrowietz U, and Asadullah K 2005. Dimethylfumarate for psoriasis: more than a dietary curiosity. Trends Mol Med 11: 43–48. [DOI] [PubMed] [Google Scholar]
- 15.Nieboer C, de Hoop D, Langendijk PN, van Loenen AC, and Gubbels J 1990. Fumaric acid therapy in psoriasis: a double-blind comparison between fumaric acid compound therapy and monotherapy with dimethylfumaric acid ester. Dermatologica 181: 33–37. [DOI] [PubMed] [Google Scholar]
- 16.Scannevin RH, Chollate S, Jung MY, Shackett M, Patel H, Bista P, Zeng W, Ryan S, Yamamoto M, Lukashev M, and Rhodes KJ 2012. Fumarates promote cytoprotection of central nervous system cells against oxidative stress via the nuclear factor (erythroid-derived 2)-like 2 pathway. J Pharmacol Exp Ther 341: 274–284. [DOI] [PubMed] [Google Scholar]
- 17.Lin SX, Lisi L, Dello Russo C, Polak PE, Sharp A, Weinberg G, Kalinin S, and Feinstein DL 2011. The anti-inflammatory effects of dimethyl fumarate in astrocytes involve glutathione and haem oxygenase-1. ASN neuro 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrecht P, Bouchachia I, Goebels N, Henke N, Hofstetter HH, Issberner A, Kovacs Z, Lewerenz J, Lisak D, Maher P, Mausberg AK, Quasthoff K, Zimmermann C, Hartung HP, and Methner A 2012. Effects of dimethyl fumarate on neuroprotection and immunomodulation. J Neuroinflammation 9: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schulze-Topphoff U, Varrin-Doyer M, Pekarek K, Spencer CM, Shetty A, Sagan SA, Cree BA, Sobel RA, Wipke BT, Steinman L, Scannevin RH, and Zamvil SS 2016. Dimethyl fumarate treatment induces adaptive and innate immune modulation independent of Nrf2. Proc Natl Acad Sci U S A 113: 4777–4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Blewett MM, Xie J, Zaro BW, Backus KM, Altman A, Teijaro JR, and Cravatt BF 2016. Chemical proteomic map of dimethyl fumarate-sensitive cysteines in primary human T cells. Sci Signal 9: rs10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Backus KM, Correia BE, Lum KM, Forli S, Horning BD, Gonzalez-Paez GE, Chatterjee S, Lanning BR, Teijaro JR, Olson AJ, Wolan DW, and Cravatt BF 2016. Proteome-wide covalent ligand discovery in native biological systems. Nature 534: 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sobotta MC, Liou W, Stocker S, Talwar D, Oehler M, Ruppert T, Scharf AN, and Dick TP 2015. Peroxiredoxin-2 and STAT3 form a redox relay for H2O2 signaling. Nat Chem Biol 11: 64–70. [DOI] [PubMed] [Google Scholar]
- 23.Kornberg MD, Bhargava P, Kim PM, Putluri V, Snowman AM, Putluri N, Calabresi PA, and Snyder SH 2018. Dimethyl fumarate targets GAPDH and aerobic glycolysis to modulate immunity. Science 360: 449–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bambouskova M, Gorvel L, Lampropoulou V, Sergushichev A, Loginicheva E, Johnson K, Korenfeld D, Mathyer ME, Kim H, Huang LH, Duncan D, Bregman H, Keskin A, Santeford A, Apte RS, Sehgal R, Johnson B, Amarasinghe GK, Soares MP, Satoh T, Akira S, Hai T, de Guzman Strong C, Auclair K, Roddy TP, Biller SA, Jovanovic M, Klechevsky E, Stewart KM, Randolph GJ, and Artyomov MN 2018. Electrophilic properties of itaconate and derivatives regulate the IkappaBzeta-ATF3 inflammatory axis. Nature 556: 501–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De S, Karim F, Kiessu E, Cushing L, Lin LL, Ghandil P, Hoarau C, Casanova JL, Puel A, and Rao VR 2018. Mechanism of dysfunction of human variants of the IRAK4 kinase and a role for its kinase activity in interleukin-1 receptor signaling. J Biol Chem 293: 15208–15220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hoarau C, Gerard B, Lescanne E, Henry D, Francois S, Lacapere JJ, El Benna J, Dang PM, Grandchamp B, Lebranchu Y, Gougerot-Pocidalo MA, and Elbim C 2007. TLR9 activation induces normal neutrophil responses in a child with IRAK-4 deficiency: involvement of the direct PI3K pathway. J Immunol 179: 4754–4765. [DOI] [PubMed] [Google Scholar]
- 27.Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, Yang K, Soudais C, Dupuis S, Feinberg J, Fieschi C, Elbim C, Hitchcock R, Lammas D, Davies G, Al-Ghonaium A, Al-Rayes H, Al-Jumaah S, Al-Hajjar S, Al-Mohsen IZ, Frayha HH, Rucker R, Hawn TR, Aderem A, Tufenkeji H, Haraguchi S, Day NK, Good RA, Gougerot-Pocidalo MA, Ozinsky A, and Casanova JL 2003. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science 299: 2076–2079. [DOI] [PubMed] [Google Scholar]
- 28.Weerapana E, Speers AE, and Cravatt BF 2007. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat. Protoc 2: 1414–1425. [DOI] [PubMed] [Google Scholar]
- 29.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, and Cravatt BF 2010. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468: 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang C, Weerapana E, Blewett MM, and Cravatt BF 2014. A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles. Nat Methods 11: 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weerapana E, Speers AE, and Cravatt BF 2007. Tandem orthogonal proteolysis-activity-based protein profiling (TOP-ABPP)--a general method for mapping sites of probe modification in proteomes. Nat Protoc 2: 1414–1425. [DOI] [PubMed] [Google Scholar]
- 32.Xu T, Park SK, Venable JD, Wohlschlegel JA, Diedrich JK, Cociorva D, Lu B, Liao L, Hewel J, Han X, Wong CC, Fonslow B, Delahunty C, Gao Y, Shah H, and Yates JR 3rd. 2015. ProLuCID: An improved SEQUEST-like algorithm with enhanced sensitivity and specificity. J. Proteomics 129: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tabb DL, McDonald WH, and Yates JR 3rd. 2002. DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome. Res. 1: 21–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taguchi K, Motohashi H, and Yamamoto M 2011. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells 16: 123–140. [DOI] [PubMed] [Google Scholar]
- 35.Singh A, Venkannagari S, Oh KH, Zhang YQ, Rohde JM, Liu L, Nimmagadda S, Sudini K, Brimacombe KR, Gajghate S, Ma J, Wang A, Xu X, Shahane SA, Xia M, Woo J, Mensah GA, Wang Z, Ferrer M, Gabrielson E, Li Z, Rastinejad F, Shen M, Boxer MB, and Biswal S 2016. Small Molecule Inhibitor of NRF2 Selectively Intervenes Therapeutic Resistance in KEAP1-Deficient NSCLC Tumors. ACS Chem Biol 11: 3214–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehmann JC, Listopad JJ, Rentzsch CU, Igney FH, von Bonin A, Hennekes HH, Asadullah K, and Docke WD 2007. Dimethylfumarate induces immunosuppression via glutathione depletion and subsequent induction of heme oxygenase 1. J Invest Dermatol 127: 835–845. [DOI] [PubMed] [Google Scholar]
- 37.Maeda T, Murata K, Fukushima T, Sugahara K, Tsuruda K, Anami M, Onimaru Y, Tsukasaki K, Tomonaga M, Moriuchi R, Hasegawa H, Yamada Y, and Kamihira S 2005. A novel plasmacytoid dendritic cell line, CAL-1, established from a patient with blastic natural killer cell lymphoma. Int J Hematol 81: 148–154. [DOI] [PubMed] [Google Scholar]
- 38.Rostovtsev VV, Green LG, Fokin VV, and Sharpless KB 2002. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew Chem Int Ed Engl 41: 2596–2599. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki N, and Saito T 2006. IRAK-4--a shared NF-kappaB activator in innate and acquired immunity. Trends Immunol 27: 566–572. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Wesche H, Stevens T, Walker N, and Yeh WC 2009. IRAK-4 inhibitors for inflammation. Curr Top Med Chem 9: 724–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McElroy WT, Tan Z, Ho G, Paliwal S, Li G, Seganish WM, Tulshian D, Tata J, Fischmann TO, Sondey C, Bian H, Bober L, Jackson J, Garlisi CG, Devito K, Fossetta J, Lundell D, and Niu X 2015. Potent and Selective Amidopyrazole Inhibitors of IRAK4 That Are Efficacious in a Rodent Model of Inflammation. ACS Med Chem Lett 6: 677–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gay NJ, Gangloff M, and O’Neill LA 2011. What the Myddosome structure tells us about the initiation of innate immunity. Trends Immunol 32: 104–109. [DOI] [PubMed] [Google Scholar]
- 43.Medvedev AE, Lentschat A, Kuhns DB, Blanco JC, Salkowski C, Zhang S, Arditi M, Gallin JI, and Vogel SN 2003. Distinct mutations in IRAK-4 confer hyporesponsiveness to lipopolysaccharide and interleukin-1 in a patient with recurrent bacterial infections. J Exp Med 198: 521–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picard C, von Bernuth H, Ghandil P, Chrabieh M, Levy O, Arkwright PD, McDonald D, Geha RS, Takada H, Krause JC, Creech CB, Ku CL, Ehl S, Marodi L, Al-Muhsen S, Al-Hajjar S, Al-Ghonaium A, Day-Good NK, Holland SM, Gallin JI, Chapel H, Speert DP, Rodriguez-Gallego C, Colino E, Garty BZ, Roifman C, Hara T, Yoshikawa H, Nonoyama S, Domachowske J, Issekutz AC, Tang M, Smart J, Zitnik SE, Hoarau C, Kumararatne DS, Thrasher AJ, Davies EG, Bethune C, Sirvent N, de Ricaud D, Camcioglu Y, Vasconcelos J, Guedes M, Vitor AB, Rodrigo C, Almazan F, Mendez M, Arostegui JI, Alsina L, Fortuny C, Reichenbach J, Verbsky JW, Bossuyt X, Doffinger R, Abel L, Puel A, and Casanova JL 2010. Clinical features and outcome of patients with IRAK-4 and MyD88 deficiency. Medicine (Baltimore) 89: 403–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamamoto T, Tsutsumi N, Tochio H, Ohnishi H, Kubota K, Kato Z, Shirakawa M, and Kondo N 2014. Functional assessment of the mutational effects of human IRAK4 and MyD88 genes. Mol Immunol 58: 66–76. [DOI] [PubMed] [Google Scholar]
- 46.Wesche H, Henzel WJ, Shillinglaw W, Li S, and Cao Z 1997. MyD88: an adapter that recruits IRAK to the IL-1 receptor complex. Immunity 7: 837–847. [DOI] [PubMed] [Google Scholar]
- 47.Li S, Strelow A, Fontana EJ, and Wesche H 2002. IRAK-4: a novel member of the IRAK family with the properties of an IRAK-kinase. Proc Natl Acad Sci U S A 99: 5567–5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loiarro M, Gallo G, Fanto N, De Santis R, Carminati P, Ruggiero V, and Sette C 2009. Identification of critical residues of the MyD88 death domain involved in the recruitment of downstream kinases. J Biol Chem 284: 28093–28103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gillard GO, Collette B, Anderson J, Chao J, Scannevin RH, Huss DJ, and Fontenot JD 2015. DMF, but not other fumarates, inhibits NF-kappaB activity in vitro in an Nrf2-independent manner. J Neuroimmunol 283: 74–85. [DOI] [PubMed] [Google Scholar]
- 50.Kastrati I, Siklos MI, Calderon-Gierszal EL, El-Shennawy L, Georgieva G, Thayer EN, Thatcher GR, and Frasor J 2016. Dimethyl Fumarate Inhibits the Nuclear Factor kappaB Pathway in Breast Cancer Cells by Covalent Modification of p65 Protein. J Biol Chem 291: 3639–3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loewe R, Holnthoner W, Groger M, Pillinger M, Gruber F, Mechtcheriakova D, Hofer E, Wolff K, and Petzelbauer P 2002. Dimethylfumarate inhibits TNF-induced nuclear entry of NF-kappa B/p65 in human endothelial cells. J Immunol 168: 4781–4787. [DOI] [PubMed] [Google Scholar]
- 52.Loewe R, Pillinger M, de Martin R, Mrowietz U, Groger M, Holnthoner W, Wolff K, Wiegrebe W, Jirovsky D, and Petzelbauer P 2001. Dimethylfumarate inhibits tumor-necrosis-factor-induced CD62E expression in an NF-kappa B-dependent manner. J Invest Dermatol 117: 1363–1368. [DOI] [PubMed] [Google Scholar]
- 53.Peng H, Guerau-de-Arellano M, Mehta VB, Yang Y, Huss DJ, Papenfuss TL, Lovett-Racke AE, and Racke MK 2012. Dimethyl fumarate inhibits dendritic cell maturation via nuclear factor kappaB (NF-kappaB) and extracellular signal-regulated kinase 1 and 2 (ERK1/2) and mitogen stress-activated kinase 1 (MSK1) signaling. J Biol Chem 287: 28017–28026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lande R, Gregorio J, Facchinetti V, Chatterjee B, Wang YH, Homey B, Cao W, Wang YH, Su B, Nestle FO, Zal T, Mellman I, Schroder JM, Liu YJ, and Gilliet M 2007. Plasmacytoid dendritic cells sense self-DNA coupled with antimicrobial peptide. Nature 449: 564–569. [DOI] [PubMed] [Google Scholar]
- 55.Nicolay JP, Muller-Decker K, Schroeder A, Brechmann M, Mobs M, Geraud C, Assaf C, Goerdt S, Krammer PH, and Gulow K 2016. Dimethyl fumarate restores apoptosis sensitivity and inhibits tumor growth and metastasis in CTCL by targeting NF-kappaB. Blood 128: 805–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ghoreschi K, Bruck J, Kellerer C, Deng C, Peng H, Rothfuss O, Hussain RZ, Gocke AR, Respa A, Glocova I, Valtcheva N, Alexander E, Feil S, Feil R, Schulze-Osthoff K, Rupec RA, Lovett-Racke AE, Dringen R, Racke MK, and Rocken M 2011. Fumarates improve psoriasis and multiple sclerosis by inducing type II dendritic cells. J Exp Med 208: 2291–2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arkin MR, Tang Y, and Wells JA 2014. Small-molecule inhibitors of protein-protein interactions: progressing toward the reality. Chem Biol 21: 1102–1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang M, Soga T, Pollard PJ, and Adam J 2012. The emerging role of fumarate as an oncometabolite. Front Oncol 2: 85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kulkarni RA, Bak DW, Wei D, Bergholtz SE, Briney CA, Shrimp JH, Alpsoy A, Thorpe AL, Bavari AE, Crooks DR, Levy M, Florens L, Washburn MP, Frizzell N, Dykhuizen EC, Weerapana E, Linehan WM, and Meier JL 2019. A chemoproteomic portrait of the oncometabolite fumarate. Nat Chem Biol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zaro BW, Whitby LR, Lum KM, and Cravatt BF 2016. Metabolically Labile Fumarate Esters Impart Kinetic Selectivity to Irreversible Inhibitors. J Am Chem Soc 138: 15841–15844. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.