Abstract
Objective:
Meta-analyses of genome-wide association studies (GWAS) in Europeans have identified >98 loci for Body Mass Index (BMI). We analyze transferability of these established associations in Pima Indians.
Methods:
Among 98 lead SNPs, 82 had minor allele frequency≥0.01 in Pima Indians, and were analyzed for association with the maximum BMI in adulthood (n=3491) and BMI z-score in childhood (n=1958). Common tag SNPs across 98 loci were also analyzed for additional signals.
Results:
Among the lead SNPs, 13 (TMEM18, TCF7L2, MRPS33P4, PRKD1, ZFP64, FTO, TAL1, CALCR, GNPDA2, CREB1, LMX1B, ADCY9, NLRC3) associated with BMI (p≤0.05) in Pima adults. A multi-allelic genetic risk score (GRS), which summed the risk alleles for 82 lead SNPs, showed a significant trend for a positive relationship between GRS and BMI in adulthood (β=0.48% per risk allele, p=1.6×10−9) and BMI z-score in childhood (β=0.024SD, p=1.7×10−7). GRS significantly associated with BMI across all age groups ≥5 years, except for those ≥50 years. The strongest association was seen in adolescence (age 14–16 years, p=1.84×10−9).
Conclusions:
In aggregate, European-derived lead SNPs had a notable effect on BMI in Pima Indians. Polygenic obesity in this population manifests strongly in childhood and adolescence, and persists throughout much of adult life.
Keywords: Established Loci, Body Mass Index, American Indians, Association study, replication
Introduction
Heritable factors are estimated to explain 40–70% of inter-individual variability in body mass index (BMI) (1). Some genes may have a similar impact on BMI across all populations, while others may have ethnic specific differences in effect. Prior meta-analyses of genome-wide association studies (GWAS), predominately from large populations of European ancestry, identified at least 98 loci containing single nucleotide polymorphisms (SNPs) associated with BMI at genome-wide statistical significance (p<5×10−8) (2–4). However, for the most part, it is unknown how these “established” SNPs affect BMI in isolated populations.
The Pima Indians of Arizona are a relatively isolated population with a high prevalence of obesity (5). To identify genetic variants that may contribute to obesity in this population, we previously conducted a GWAS for BMI and have also directly sequenced physiologic candidate genes, such as those in the leptin-melanocortin pathway; these studies have identified both common and rare variants in MC4R, SIM1, LEPR and MAP2K3 that associate with BMI in Pima Indians (6–10). In the current study, we analyze another category of genes, the 98 established GWAS BMI loci, by directly genotyping the lead SNP or a proxy which tags the lead SNP in a longitudinally-studied population based sample of Pima Indians. Furthermore, we also analyze tag SNPs which capture common variation (r2≥0.85 with a minor allele frequency [mAF] ≥0.05) across these GWAS loci in this population to identify potential additional susceptibility variants.
Methods
Subjects
Subjects were derived from a longitudinal study of the etiology of type 2 diabetes (T2D) among the Gila River Indian Community in Arizona, where most residents are of Pima Indian heritage. The study protocols were approved by the institutional review board of the National Institute of Diabetes and Digestive and Kidney Diseases. All residents (age≥5 years) of a geographical section of the community were invited to participate in outpatient biennial exams, which included measures of height and weight for calculation of BMI and well as a 75-g oral glucose tolerance test to determine diabetes according to the criteria of American Diabetes Association (11) (Participants were examined from 1965–2007. The mean±SD number of exams per person was 5.2±3.8. Follow-up time was 19±12 years). Characteristics of the 3491 full-heritage Pima Indians (defined as 8/8 Pima including Tohono O’dham) included in the analysis for each of the established BMI SNPs are shown in Table 1. Maximum BMI in adulthood was defined as the highest recorded BMI from an exam at age ≥15 years. Since diabetes duration and treatment can affect bodyweight, BMI was also analyzed as the highest BMI recorded at an exam at age ≥15 years when the subject was non-diabetic as confirmed by the oral glucose tolerance test. Individuals who did not have a measure of BMI from a non-diabetic exam were excluded from this analysis. To assess susceptibility to obesity in childhood, we analyzed the maximum age and sex adjusted BMI Z-score across examinations during childhood (between the ages of 5–15 years). Median BMI for Pima girls and boys (50th percentile of the Pima population) is at the 95th percentile of the US population at every age (8). Since the distribution of BMI in Pima children is very different from that in standard populations, we used a Pima-specific z-score for these calculations. It was calculated by subtracting the mean from BMI and dividing by its standard deviation in categories of age (1 year) and sex in all research participants. Given the longitudinal nature of the study, many individuals were analyzed as both children and adults. Thus, the analyses of childhood BMI z-score and adulthood BMI were not independent.
Table 1.
Characteristics of full-heritage Pima Indians with longitudinal measures of BMI
| Trait | N | Male % | Age (years) Mean ± SD | BMI (kg/m2) Mean ± SD |
|---|---|---|---|---|
| Maximum Adult BMI (any exam) | 3491 | 42 | 36 ± 13 | 38 ± 9 |
| Maximum Adult BMI (non-diabetic exam) | 2862 | 42 | 32 ± 12 | 36 ± 8 |
| Max Childhood BMI | 1958 | 45 | 11 ± 3 | 24 ± 7 |
Maximum Adult BMI is the maximum lifetime BMI (kg/m2) recorded at age ≥15 years.
Maximum Adult BMI (non-diabetic exam) is the maximum BMI recorded at a non-diabetic exam at age ≥15 years. The maximum childhood BMI was identified between the ages of 5 and 15 years.
Since genetic associations with BMI may vary with age, we further assessed these associations in different age groups. Twelve discrete age groups were analyzed (ages 5–7, 8–10, 11–13, 14–16, 17–19, 20–24, 25–29, 30–34, 35–39, 40–49, 50–59, and ≥60 years), and all individuals who were examined within a given age category were analyzed. If an individual had more than one examination in a given category, the examination closest to the midpoint of the category was used for analysis.
Genotypic data were also available in 3298 additional individuals from the same longitudinal study whose ethnicity was not full-heritage Pima Indian (defined as ≤7/8 Pima Indian; most of these individuals were 4/8 Pima Indian). To identify additional variants in 98 established BMI loci, SNPs associated with BMI in full-heritage Pima Indians, were analyzed for replication in this “mixed-heritage” sample.
Genotyping of lead SNPs, identification and genotyping of tag SNPs
Lead SNPs at 30 of the previously established loci were genotyped by TaqMan Allelic Discrimination assay (Life Technologies, CA). Additional data were available from prior genotyping on a custom Axiom array (Affymetrix, Santa Clara, CA), designed to capture common variants (mAF≥0.05 at r2≥0.85 in 300-kb windows) across the genome, identified from whole genome sequence data of 266 full-heritage Pima Indians (12). Genotypes were identified using Analysis Suite Software V1.1.0.6161 (Affymetrix, Santa Clara, CA). These data were used to obtain genotypes on 52 additional lead SNPs which included 12 lead SNPs directly genotyped on the array and 40 proxies (r2≥0.85 with the lead SNP in full-heritage Pima Indians with mean±SD r2=0.97±0.04; r2 between the lead SNP and its proxy is listed in Table S1) on the array. Thus, 82 established variants were analyzed. Sixteen lead SNPs with mAF<0.01 in full-heritage Pima Indians in the whole genome sequence data, were not analyzed for association with BMI. Table S2 lists all 98 lead SNPs, along with their genotyped proxies in the present study. Additional variation across the 98 loci was analyzed using genotypic data for ~6000 tag SNPs derived from the Axiom array. All genotypic data passed quality control metrics of genotype call rate ≥90 %, no deviation from Hardy–Weinberg equilibrium (p≥1×10−4) and discrepant rate ≤2 pairs among 100 blind duplicate pairs (12).
Analyses of cis-acting expression quantitative trait locus (Cis-eQTL) in adipose tissues
Percutaneous abdominal adipose tissue biopsies from 201 non-diabetic Pima Indians were characterized for expression using the Human Exon 1.0 ST Microarray Chips (Affymetrix, Santa Clara, CA) as previously described (13).
Statistical analysis
The association of genotypes with BMI was analyzed by linear regression using a model fitted with a variance components covariance structure to account for genetic relatedness among individuals. The genetic relatedness matrix was estimated as the proportion of the genome shared identical by descent (IBD) between each pair of individuals who had been genotyped (a total of 29,648,850 pairs) (14). Genomic segments shared IBD were identified with the fastIBD function of Beagle package (15) using 482,616 autosomal markers with mAF>0.05. Mixed models were fitted using the SOLAR package (16). The natural logarithm of BMI or the childhood Z-score, was taken as the dependent variable. Results were adjusted for age, sex, birth year and the first five genetic principal components. As the lead SNPs were previously established as associated with BMI at genome-wide statistical significance, the alpha level for significant association of these lead SNPs with BMI in the present study was set at 0.05. For additional variants at each locus, analyses were conducted to identify the most strongly-associated BMI variants in Pima Indians. In addition, conditional analyses were conducted in which the European GWAS lead SNP was included as a covariate in the model to determine whether the signal additionally contributed to the association. In both conditional and unconditional analyses, variants with nominal associations (p<0.05) in full-heritage Pima Indians were assessed for replication in the mixed-heritage individuals. We report all SNPs with nominal evidence for replication in these mixed-heritage individuals (p<0.05). We also note whether any of the new signals for BMI in the combined analysis achieved significance after a Bonferroni-correction for the 6000 variants tested (p<8.3×10−6).
To analyze the effects of all 82 established BMI SNPs in aggregate, we constructed a multi-allelic genetic risk score (GRS). For variants genotyped as part of the Axiom array, missing data for these analyses were imputed with IMPUTE2 using whole genome sequence data of 266 full-heritage Pima Indians (17). For variants genotyped separately, missing data were inferred from genotypes of relatives using a likelihood-based method implemented in MLINK, as previously described (18). The GRS was computed for each subject as the sum of the number of risk alleles observed at each locus (i.e., all variants were given equal weight). The effect size of a 1-unit difference in GRS represents the effect per risk allele at any of the 82 loci. We also conducted analyses in which the GRS was calculated for each subject as the sum of the number of risk alleles observed at each locus weighted by the published effect size in Europeans (taken from the Genetic Investigation of Anthropometric Traits [GIANT], www.broadinstitute.org), divided by the sum of the weights. The results of these analyses (shown in Figure S1) were similar to those with equally weighted effects. Hence, we report the results for the GRS with equal weight for each locus.
To test whether the effects in Pima Indians were consistent with those in Europeans, we compared the regression coefficients observed in Pima Indians with those obtained in GIANT. For these analyses, BMI associations in Pima Indians were analyzed using the inverse Gaussian transformation of the rank of maximum BMI in adulthood to express beta coefficients and SEs in SD units comparable to those used in GIANT. Beta coefficients were compared by the Cochran Q test of homogeneity, and heterogeneity was quantified by the I2 statistic (19). To test for heterogeneity across all 82 lead SNPs analyzed, a combined Z* score was calculated by combining p values for the null hypothesis of homogeneity across all 82 markers, using Stouffer’s method, as described previously (18–20). As presently constructed, if Z* is positive, it indicates that beta coefficients are generally stronger in Pima Indians than in Europeans, whereas if Z* is negative, it indicates that beta coefficients on average are weaker in Pima Indians. To analyze diabetes in the mixed model, a binary linear model was used in which this discrete trait was treated as a continuous (0,1) variable, and these analyses were used for calculating the p value for association. The odds ratio (OR) was calculated following the method of Haggstrom from the regression coefficient (b) and the residual variance (σ2) in the binary linear model: i.e., ln[OR]=b/σ2 (21).
Results
Transferability analysis in Pima Indians of the 98 lead SNPs previously shown to associate with BMI in Europeans
Sixteen of the 98 lead SNPs that associated with BMI in a meta-analysis of Europeans (Supplementary Table 2) had a minor allele frequency <0.01 in Pima Indians; therefore these 16 SNPs were omitted from the statistical analyses. The remaining 82 lead SNPs were genotyped in full-heritage Pima Indians and analyzed for association with the maximum BMI recorded in adulthood (n= 3491), the maximum BMI recorded in adulthood when the subject did not have T2D (n= 2826) and maximum BMI z-score in childhood (n=1958) (Table S2). Table 2 shows the SNPs associated with maximum BMI in adulthood and/or maximum BMI z-score in childhood with p≤0.05 in full-heritage Pima Indians, where the direction of the association was consistent with that observed in Europeans. Thirteen of these lead SNPs associated with maximum BMI in adulthood including those in/near TMEM18, TCF7L2, MRPS33P4, PRKD1, ZFP64, FTO, TAL1, CALCR, GNPDA2, CREB1, LMX1B, ADCY9 and NLRC3 (p≤0.05, Table 2). SNPs with the strongest association with BMI in adulthood were rs2867125 in TMEM18 (p=3.0×10−4, beta=0.03 on loge scale, which corresponds to a 3.0% increase in BMI per copy of the risk allele) and rs7903146 in TCF7L2 (p=4.9×10−4, beta=0.04 or a 4.0% increase in BMI). Most of these SNPs also associated with maximum BMI at a non-diabetic exam in adulthood (p≤0.05, Table 2) and maximum BMI z-score in childhood (p≤0.05, Table 2) in the Pima Indian sample, whereas the SNP with the strongest association with BMI z-score in childhood was rs7193144 in FTO (p=0.003, beta=0.14 SD units per copy of the risk allele).
Table 2.
Transferability analysis of established BMI SNPs with maximum BMI in adulthood, maximum BMI recorded at a non-diabetic exam, and maximum BMI z-score in childhood in longitudinally studied full-heritage Pima Indians
| Lead SNP | Locus (in/nearest) | Allele R/N | Full-heritage Pima Indians | Reported Meta-Analysis GIANT | Heterogeneity | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Maximum BMI Adulthood(n=3491) | Maximum BMI (Non-diabetic) Adulthood(n=2862) | Maximum Z-score Childhood(n=1958) | BMIAdulthood (n=339224) | ||||||||||||
| RAF | Beta (SD) | Beta (Loge) | P | Beta (Loge) | P | Beta (SD) | P | RAF | Beta (SD) | P | I2(%) | Phet | |||
| rs2867125 | TMEM18 | C/T | 0.86 | 0.13 | 0.030 | 3.0×10−4 | 0.035 | 6.0×10−5 | 0.131 | 0.005 | 0.87 | 0.06 | 4.4 ×10−52 | 75.2 | 0.045 |
| rs7903146 | TCF7L2 | C/T | 0.92 | 0.16 | 0.039 | 4.9×10−4 | 0.051 | 2.0×10−5 | 0.088 | 0.18 | 0.75 | 0.02 | 1.1×10−12 | 87.6 | 0.004 |
| rs13041126 | MRPS33P4 | T/C | 0.67 | 0.08 | 0.018 | 0.004 | 0.019 | 0.006 | 0.051 | 0.16 | 0.73 | 0.02 | 6.5×10−7 | 78.6 | 0.031 |
| rs12885454 | PRKD1 | C/A | 0.78 | 0.09 | 0.020 | 0.005 | 0.016 | 0.04 | 0.066 | 0.11 | 0.63 | 0.02 | 9.1×10−11 | 78.2 | 0.032 |
| rs6091540 | ZFP64 | C/T | 0.67 | 0.07 | 0.016 | 0.01 | 0.017 | 0.01 | 0.033 | 0.36 | 0.73 | 0.02 | 2.1×10−8 | 70.3 | 0.066 |
| rs7193144 | FTO | C/T | 0.14 | 0.10 | 0.022 | 0.01 | 0.029 | 0.002 | 0.14 | 0.003 | 0.44 | 0.08 | 6.2×10−142 | 0.0 | 0.686 |
| rs977747 | TAL1 | T/G | 0.74 | 0.08 | 0.017 | 0.02 | 0.021 | 0.004 | 0.061 | 0.13 | 0.47 | 0.02 | 2.2×10−8 | 75.2 | 0.044 |
| rs9641123* | CALCR | C/G | 0.09 | 0.11 | 0.023 | 0.02 | 0.021 | 0.05 | 0.059 | 0.32 | 0.39 | 0.01 | 1.8×10−7 | 79.2 | 0.028 |
| rs10938397 | GNPDA2 | G/A | 0.31 | 0.06 | 0.014 | 0.02 | 0.014 | 0.05 | 0.096 | 0.009 | 0.43 | 0.03 | 1.4×10−40 | 27.6 | 0.240 |
| rs17203016 | CREB1 | G/A | 0.15 | 0.08 | 0.017 | 0.03 | 0.016 | 0.06 | 0.073 | 0.12 | 0.20 | 0.02 | 3.4×10−8 | 59.2 | 0.117 |
| rs10733682 | LMX1B | A/G | 0.75 | 0.06 | 0.014 | 0.04 | 0.017 | 0.02 | 0.056 | 0.15 | 0.43 | 0.02 | 2.5×10−10 | 41.1 | 0.192 |
| rs2531995 | ADCY9 | T/C | 0.18 | 0.07 | 0.016 | 0.04 | 0.010 | 0.21 | 0.082 | 0.06 | 0.59 | 0.02 | 7.6×10−10 | 53.5 | 0.142 |
| rs758747 | NLRC3 | T/C | 0.11 | 0.07 | 0.018 | 0.05 | 0.014 | 0.14 | 0.074 | 0.15 | 0.27 | 0.02 | 1.5×10−10 | 37.7 | 0.205 |
| rs1460676* | FIGN | C/T | 0.07 | 0.08 | 0.018 | 0.11 | 0.018 | 0.16 | 0.143 | 0.03 | 0.22 | 0.02 | 5.0×10−8 | 22.2 | 0.257 |
| rs9374842* | LOC285762 | T/C | 0.86 | 0.02 | 0.003 | 0.68 | 0.011 | 0.23 | 0.139 | 0.005 | 0.74 | 0.01 | 7.2×10−9 | 0.0 | 0.749 |
| rs1441264* | MIR548A2 | A/G | 0.73 | 0.01 | 0.002 | 0.78 | 0.003 | 0.62 | 0.114 | 0.003 | 0.55 | 0.01 | 3.0×10−8 | 0.0 | 0.933 |
| rs1555543 | PTBP2 | C/A | 0.54 | −0.01 | −0.001 | 0.93 | −0.001 | 0.89 | 0.076 | 0.03 | 0.57 | 0.02 | 5.5×10−11 | 0.0 | 0.336 |
Data are only given for the SNPs associated with maximum BMI in adulthood and/or maximum BMI z-score in childhood with p≤0.05 in full-heritage Pima Indians (n as indicated). R: risk allele; N: non-risk allele; RAF: risk allele frequency. The risk allele is defined as the allele with higher risk for BMI in Europeans. Beta and p value are adjusted for age, sex, birth year and the first five genetic principal components. BMI is loge-transformed before analyses to approximate a normal distribution. Data for meta-analysis of GIANT in Europeans are derived from the public database and beta coefficients are expressed in SD units by inverse Gaussian transformation of BMI. For comparison with beta coefficients in Europeans, maximum BMI in Pima Indians is also transformed using an inverse Gaussian transformation; results are presented as beta (SD) and used in heterogeneity analyses. I2 represents the percentage of variance in the effect attributable to heterogeneity between Pima Indians and Europeans, and Phet is the p value for the null hypothesis that the two betas are equal.
A proxy SNP was used for genotyping (listed in Table S1). Bold values: p ≤ 0.05.
Heterogeneity of the effect size between full-heritage Pima Indians (n=3491) and Europeans (GIANT, n=339,224) was quantified for all 82 lead SNPs that were analyzed. Six SNPs (in TMEM18, TCF7L2, MRPS33P4, PRKD1, TAL2, CALCR) had significant heterogeneity between the ethnic groups, where the effect of genotype on BMI was greater in Pima Indians as compared to Europeans (Table 2), while three SNPs (in C9orf93, RASA2, SH2B1) had significant heterogeneity where the effect was weaker in Pimas than that in Europeans (Table S2). The largest difference in effect size was observed at rs7903146 (TCF7L2) (β: Pima vs. European: 0.165 vs. 0.024 BMI SD units, I2=87.6%, p=0.004). The overall test for heterogeneity across all 82 SNPs was not statistically significant (Z*=1.21, p=0.23), and this result was not materially different when restricted to the 42 lead SNPs that were directly genotyped (Z*=1.84, p=0.07).
Aggregate analysis of the lead SNPs previously shown to associate with BMI in Europeans
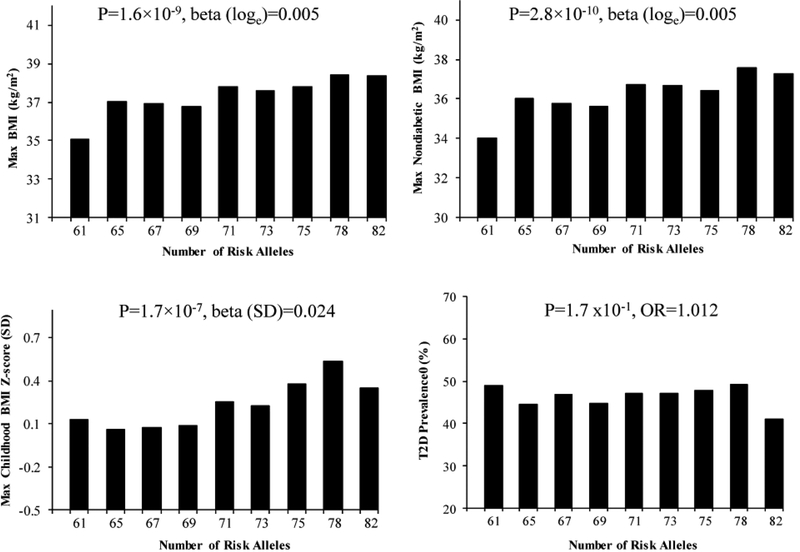
To assess whether the European-derived risk alleles contribute in aggregate to obesity in Pima Indians, a multi-allelic genetic risk score was created by summing the number of the risk alleles of all 82 SNPs with a mAF ≥ 0.01 with equal weight for each locus. The GRS showed a significant trend for the relationship between increasing number of risk alleles in Pima Indians and maximum BMI in adulthood (Figure 1, β=0.0048 on loge scale, which corresponds to a 0.48% increase in BMI per unit increase of the GRS, p=1.6× 10−9), maximum BMI at a non-diabetic exam in adulthood (β=0.0054 or a 0.54% increase per unit increase of the GRS, p=2.8× 10−10) and maximum BMI z-score in childhood (β=0.024 SD per unit increase of the GRS, p=1.7× 10−7). The GRS was also strongly associated with BMI in 3298 mixed-heritage Pima Indians (Figure S2). Despite BMI being a risk factor for development of T2D, the GRS for BMI was not associated with diabetes status (Figure 1, OR=1.012 [95% confidence interval= 0.9951.028], p=0.17). When analyzed individually, 4 SNPs rs7193144 in FTO, rs12286929 in CADM1, rs1441264 in MIR548A2 and rs10938397 in GNPDA2 nominally associated with T2D in 3747 Pima Indians (OR=1.21–1.15 per risk allele for BMI, p=0.008–0.05, Table S3), where the BMI risk allele associated with higher risk of diabetes.
Figure 1.
The aggregate effect of 82 lead SNPs on maximum BMI in adulthood (n=3491, age ≥15), maximum BMI recorded at a non-diabetic exam (n=2862, age ≥15), maximum BMI z-score in childhood (n=1958, age 5–15) and T2D (n=3747) in full-heritage Pima Indians. The GRS was created by summing the number of the risk alleles of all 82 SNPs with a mAF ≥ 0.01 with equal weight for each locus. The p values were adjusted for age, sex, birth year and the first five genetic principal components. Individuals were divided into 9 categories based on the GRS: ≤63, >63- ≤66, >66- ≤68, >68- ≤70, >70- ≤72, >72- ≤74, >74- ≤76, >76- ≤79, >79 risk alleles. In the figure, the value on the x-axis reflects the mean GRS for individuals within each category.
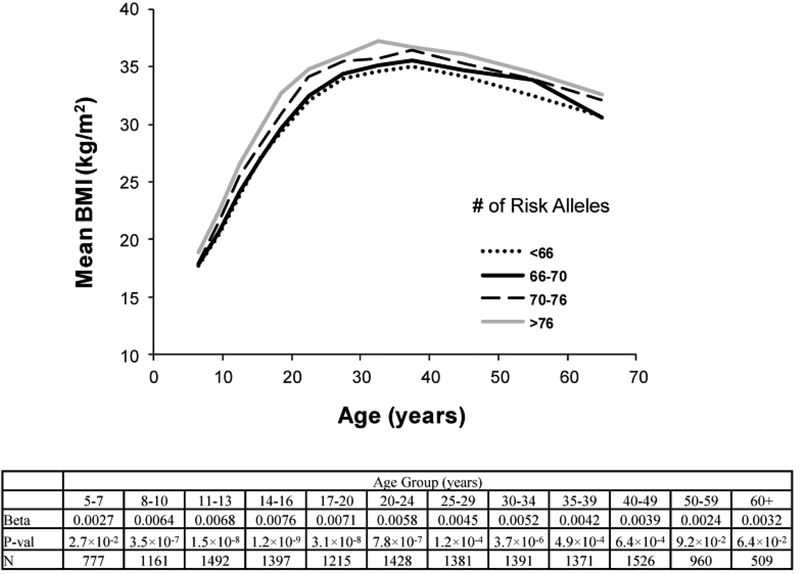
The aggregate effect of 82 lead SNPs on BMI at various ages was also assessed in analyses of the longitudinal measures for BMI in Pima children and adults. The GRS associated with higher BMI in most age groups, except for those >50 years old, and effects tended to be stronger at younger ages. The strongest effect, in terms of the regression coefficient, was observed in those 14–16 years old (β=0.0076, corresponding to a 0.76% increase in BMI per unit increase of the GRS, p=1.2 × 10−9). (Figure 2).
Figure 2.
The aggregate effect of 82 lead SNPs on BMI at various ages in Pima Indians. For the life-time assessment of BMI with the genetic risk score within discrete age groups which cross childhood and adulthood, the logarithm of BMI was uniformly analyzed in all age groups to avoid discontinuity of units across childhood and adulthood. Within each discrete group, the analyses are adjusted for age, sex, birth year and the first five genetic principal components. The beta coefficient [loge(BMI) per unit GRS], p value and number of subject for each age group were listed.
Additional variants in established BMI loci associate with BMI in American Indians
To determine whether additional variants in these 98 loci associate with BMI in Pima Indians in a stronger fashion than the lead SNPs, the genotypes previously generated for ~6000 SNPs which tag (r2≥0.85) common variation (mAF ≥0.05) across all 98 loci were analyzed for BMI associations in full-heritage Pima Indians. Those SNPs with p<0.05 were further analyzed in the replication sample of 3298 non-full heritage Pima Indians from the same longitudinal study. Thirty-eight tag SNPs, which map to 18 loci, had nominal associations with BMI (p≤0.05) in both full-heritage and non-full heritage Pima Indians (Table 3). The strongest signal for BMI in adulthood was rs3751837 in NLRC3 (p=5.8×10−5 in the combined analysis of full-heritage and non-full heritage Pima Indians), whereas SNPs rs12462812 in PGPEP1, rs2531982 in ADCY9, rs149906922 in CADM1 and rs17602834 in LRP1B had associations with p values <10−3. In addition, to determine whether additional variants contribute to BMI, independent of the lead SNP, those SNPs with p<0.05 after conditioning on the established lead SNPs were replicated in non-full heritage Pima Indians. Nine tag SNPs in 7 loci showed BMI associations in both samples (p≤0.05 after conditioning on the established SNP, Table 4). For 5 of these 7 loci (PGPEP1, CADM1, CLIP1, LRP1B, EHBP1), the previously established SNP did not significantly associate with BMI in Pima Indians, whereas for ZFP64 and TCF7L2, there was evidence of both the established SNP and new SNP affecting BMI. The strongest potential new signals for BMI in adulthood were rs6021702 in ZFP64 (p=8.4×10−4 after conditioning on the lead SNP rs6091540) and rs12462812 in PGPEP1 (p=8.8×10−4 after conditioning on the lead SNP rs17724992). However, no variant, in either the unconditional or conditional analyses, achieved statistical significance after correction for testing ~6000 SNPs (p<8.3×10−6). Identification of Cis-eQTL in the established or putative BMI-associated variants
Table 3.
Associations of additional SNPs with BMI in 98 established BMI loci in an expanded population-based study of American Indians
| Locus | SNP | Maximum BMI Full-heritage (n=3491) | Maximum BMI Mixed-heritage (n=3298) | Maximum BMI Combined (n=6789) | BMI GIANT (n=339224) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RS# | Allele 1/2 | Freq Alelle1 American Indian | Freq Allele1 European | Beta (Loge) | P | Beta (Loge) | P | Beta (Loge) | P | Beta (SD) | P | |
| NLRC3 | rs3751837 | T/C | 0.11 | 0.22 | 0.023 | 0.02 | 0.028 | 0.002 | 0.028 | 5.8×10−5 | - | - |
| NLRC3 | rs11646156 | C/G | 0.06 | 0.03 | 0.025 | 0.05 | 0.025 | 0.04 | 0.029 | 0.001 | 0.028 | 0.003 |
| NLRC3 | rs758747 | T/C | 0.13 | 0.27 | 0.018 | 0.05 | 0.019 | 0.03 | 0.019 | 0.004 | 0.023 | 1.5×10−10 |
| PGPEP1 | rs12462812 | A/G | 0.19 | 0.01 | −0.016 | 0.03 | −0.022 | 0.007 | −0.019 | 0.0005 | - | - |
| ADCY9 | rs2531982 | A/G | 0.86 | 0.70 | −0.023 | 0.01 | −0.022 | 0.01 | −0.022 | 0.0005 | −0.020 | 5.0×10−7 |
| ADCY9 | rs2531995 | T/C | 0.21 | 0.59 | 0.016 | 0.04 | 0.017 | 0.02 | 0.017 | 0.002 | 0.024 | 7.6×10−10 |
| ADCY9 | rs35384844 | T/C | 0.86 | 0.95 | −0.017 | 0.04 | −0.020 | 0.02 | −0.018 | 0.004 | - | - |
| CADM1 | rs149906922 | A/T | 0.38 | 0 | 0.019 | 0.001 | 0.014 | 0.03 | 0.016 | 0.0007 | - | - |
| CADM1 | rs718484 | A/G | 0.28 | 0.09 | 0.019 | 0.003 | 0.020 | 0.005 | 0.016 | 0.001 | 0.007 | 0.29 |
| CADM1 | rs77550241 | A/G | 0.25 | 0.01 | 0.018 | 0.008 | 0.020 | 0.007 | 0.015 | 0.003 | - | - |
| CADM1 | rs17118342 | T/C | 0.78 | 1 | −0.020 | 0.005 | −0.017 | 0.03 | −0.014 | 0.007 | - | - |
| CADM1 | rs11215485 | T/C | 0.30 | 0.12 | 0.019 | 0.003 | 0.013 | 0.05 | 0.013 | 0.008 | −0.001 | 0.85 |
| CADM1 | rs72306674 | −/TCTC | 0.25 | 0.01 | 0.016 | 0.01 | 0.017 | 0.02 | 0.013 | 0.01 | - | - |
| LRP1B | rs17602834 | T/C | 0.60 | 0.48 | −0.013 | 0.04 | −0.015 | 0.02 | −0.015 | 0.0009 | 0.007 | 0.10 |
| LRP1B | rs17576955 | A/G | 0.91 | 0.99 | 0.023 | 0.02 | 0.022 | 0.03 | 0.021 | 0.005 | 0.012 | 0.32 |
| ZFP64 | rs139447701 | A/C | 0.18 | 0 | −0.019 | 0.008 | −0.016 | 0.05 | −0.019 | 0.001 | - | - |
| ZFP64 | rs6021702 | T/C | 0.17 | 0.37 | 0.022 | 0.02 | 0.016 | 0.04 | 0.017 | 0.005 | −0.003 | 0.56 |
| ZFP64 | rs11086366 | A/C | 0.54 | 0.88 | 0.012 | 0.05 | 0.016 | 0.01 | 0.013 | 0.005 | 0.001 | 0.88 |
| FTO | rs1861869 | C/G | 0.70 | 0.47 | −0.013 | 0.04 | −0.015 | 0.02 | −0.015 | 0.001 | −0.030 | 9.9×10−17 |
| FTO | rs3751814 | A/G | 0.15 | 0.41 | 0.021 | 0.009 | 0.017 | 0.05 | 0.019 | 0.002 | - | - |
| FTO | rs7206790 | C/G | 0.81 | 0.49 | −0.019 | 0.01 | −0.016 | 0.04 | −0.017 | 0.002 | −0.066 | 3×10−95 |
| TCF7L2 | rs146479796 | T/C | 0.80 | 1 | −0.017 | 0.02 | −0.017 | 0.03 | −0.017 | 0.002 | - | - |
| TCF7L2 | rs7895657 | A/G | 0.29 | 0.30 | 0.013 | 0.05 | 0.013 | 0.05 | 0.012 | 0.01 | - | - |
| ZBTB10 | rs183925020 | A/C | 0.13 | 0 | 0.019 | 0.02 | 0.021 | 0.04 | 0.021 | 0.002 | - | - |
| ZBTB10 | rs575452 | A/G | 0.75 | 0.87 | 0.020 | 0.003 | 0.014 | 0.05 | 0.015 | 0.004 | 0.000 | 0.95 |
| RARB | rs186133817 | C/G | 0.75 | 0.87 | −0.016 | 0.01 | −0.015 | 0.04 | −0.016 | 0.002 | - | - |
| CLIP1 | rs34383196 | T/C | 0.29 | 0.24 | 0.013 | 0.04 | 0.018 | 0.01 | 0.015 | 0.002 | - | - |
| LMX1B | rs3814120 | A/G | 0.30 | 0.07 | 0.015 | 0.02 | 0.014 | 0.03 | 0.015 | 0.002 | 0.022 | 0.0008 |
| LMX1B | rs10733682 | A/G | 0.75 | 0.43 | 0.014 | 0.04 | 0.019 | 0.01 | 0.016 | 0.003 | 0.019 | 2.5×10−10 |
| LMX1B | rs16929203 | T/C | 0.32 | 0.08 | 0.016 | 0.01 | 0.013 | 0.05 | 0.014 | 0.003 | 0.022 | 0.0006 |
| LMX1B | rs28687510 | T/G | 0.32 | 0.11 | 0.015 | 0.01 | 0.013 | 0.05 | 0.014 | 0.004 | - | - |
| LMX1B | rs10739682 | T/C | 0.89 | 0.63 | 0.023 | 0.03 | 0.018 | 0.05 | 0.018 | 0.01 | - | - |
| LOC284260 | rs9304270 | T/C | 0.12 | 0.10 | 0.018 | 0.04 | 0.022 | 0.02 | 0.020 | 0.003 | 0.001 | 0.89 |
| GPRC5B | rs1292635 | A/G | 0.10 | 0.23 | 0.020 | 0.05 | 0.021 | 0.04 | 0.021 | 0.005 | −0.001 | 0.82 |
| MC4R | rs1943217 | T/G | 0.47 | 0.71 | −0.013 | 0.03 | −0.013 | 0.03 | −0.012 | 0.005 | −0.011 | 0.05 |
| MC4R | rs8092350 | A/C | 0.55 | 0.66 | −0.012 | 0.04 | −0.012 | 0.04 | −0.011 | 0.01 | −0.032 | 1.3×10−22 |
| CALCR | rs12666730 | T/C | 0.49 | 0.86 | 0.012 | 0.05 | 0.014 | 0.02 | 0.012 | 0.008 | 0.006 | 0.19 |
| EHBP1 | rs147306320 | T/C | 0.08 | 0 | −0.020 | 0.04 | −0.027 | 0.03 | −0.018 | 0.03 | - | - |
Data are given for the analysis of full-heritage Pima Indians, mixed-heritage Pima Indians and combined samples (n as indicated). Allele 1 is defined as the reference allele; allele 2 is as the altered allele. Beta and p value are adjusted for age, sex, birth year and the first five genetic principal components. SNPs which were not available in the meta-analysis of GIANT are denoted with “-“.
Table 4.
Additional SNPs independent of the lead SNPs in 7 of 98 established BMI loci associated with BMI in an expanded population-based study of American Indians
| Locus | SNP | Maximum BMI Full-heritage (n=3491) | Maximum BMI Mixed-heritage (n=3298) | Maximum BMI Combined (n=6789) | BMI GIANT (n=339224) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RS# | Allele 1/2 | Freq Allele 1 American Indian | Freq Allele 1European | Beta (Loge) | P after conditional analysis for lead SNP | Beta (Loge) | P after conditional analysis for lead SNP | Beta (Loge) | P after conditional analysis for lead SNP | Beta (SD) | P | |
| ZFP64 | rs6021702 | T/C | 0.17 | 0.36 | 0.024 | 0.01 | 0.020 | 0.009 | 0.021 | 0.0008 | −0.003 | 0.56 |
| PGPEP1 | rs12462812 | A/G | 0.19 | 0.01 | −0.019 | 0.01 | −0.018 | 0.03 | −0.019 | 0.0009 | - | - |
| TCF7L2 | rs146479796 | T/C | 0.80 | 1.00 | −0.020 | 0.01 | −0.016 | 0.05 | −0.018 | 0.001 | - | - |
| CADM1 | rs718484 | A/G | 0.28 | 0.09 | 0.019 | 0.003 | 0.018 | 0.01 | 0.015 | 0.002 | 0.007 | 0.29 |
| CADM1 | rs77550241 | A/G | 0.25 | 0.01 | 0.018 | 0.009 | 0.018 | 0.02 | 0.014 | 0.006 | - | - |
| CADM1 | rs72306674 | -/TCTC | 0.25 | 0.01 | 0.016 | 0.02 | 0.015 | 0.05 | 0.012 | 0.02 | - | - |
| CLIP1 | rs34383196 | T/C | 0.29 | 0.24 | 0.013 | 0.05 | 0.018 | 0.01 | 0.015 | 0.003 | - | - |
| EHBP1 | rs147306320 | T/C | 0.08 | 0.00 | −0.025 | 0.02 | −0.025 | 0.05 | −0.018 | 0.03 | - | - |
Data are given for the analysis of full-heritage Pima Indians, mixed-heritage Pima Indians and combined samples (n as indicated). Allele 1 is defined as the reference allele; allele 2 is as the altered allele. Conditional analyses were conducted in which the European GWAS lead SNP was included as a covariate in the model to determine whether the signal additionally contributed to the BMI association. Beta and p value are also adjusted for age, sex, birth year and the first five genetic principal components. SNPs which were not available in the meta-analysis of GIANT are denoted with “-“.
To determine whether any of the 82 previously established or 9 newly identified putative SNPs that associate with BMI may function as a cis-eQTL, genotypic data were merged with expression data from adipose tissue biopsies collected from 201 Pima Indians. Four established lead SNPs (rs2531995 in ADCY9, rs2176598 in HSD17B12, rs657452 in AGBL4, rs10150332 in NRXN3) correlated with RNA levels of the respective gene with p<0.05 (Table 5). The strongest correlation was observed between the 3-UTR SNP rs2531995 in ADCY9 and its expression (p=5.3×10−6), where the allele for higher BMI in Europeans (T at rs2531995) associated with reduced ADCY9 expression in adipose tissue. The established intronic SNP rs2176598 in HSD17B12 also had evidence of being a cis-eQTL; it correlated with HSD17B12 expression in adipose tissue (p=0.006) where the BMI risk allele T had a lower RNA level in Pima Indians. This correlation replicated in 298 adipose tissues reported in the GTEx database (p=2.2×10−35, GTEx Analysis Release V6p) in a direction consistent with that in Pima Indians.
Table 5.
Analyses of cis-eQTL variants in the established BMI loci in adipose tissue biopsies of 201 Pima Indians
| SNP | Gene | Allele (R/N) | RAF | Location | Mean (R/R) | Mean (R/N) | Mean (N/N) | Beta | P |
|---|---|---|---|---|---|---|---|---|---|
| rs2531995 | ADCY9 | T/C | 0.18 | 3’UTR | −1.30 | −0.25 | 0.21 | −0.24 | 5.3 ×10−6 |
| rs2176598 | HSD17B12 | T/C | 0.52 | intron | −0.28 | 0.018 | 0.29 | −0.29 | 0.006 |
| rs657452 | AGBL4 | A/G | 0.39 | intron | −0.34 | −0.04 | 0.18 | −0.26 | 0.007 |
| rs10150332 | NRXN3 | C/T | 0.36 | intron | 0.13 | 0.16 | −0.22 | 0.24 | 0.02 |
Adipose tissue gene expression levels were determined using Human Exon 1.0 ST Microarray Chips (Affymetrix, Santa Clara, CA, USA) and expressed as batch- and sex-standardized values (SD units). R: risk allele; N: non-risk allele. RAF: risk allele frequency. The risk allele is defined as the allele with higher risk for BMI in Europeans. Beta and p value are adjusted for age at time of biopsy and the first genetic principal component.
Discussion
Recent meta-analyses of genome-wide association studies have identified many genetic variants that associate with BMI across multiple studies of European populations (2–4); however, the extent to which these variants contribute to the high rate of obesity found in more isolated populations is not well understood. Therefore, in the current study, we determined the effect of 98 established SNPs on BMI measured in Pima Indians of Arizona. Our analysis showed that 13 established SNPs associated with maximum BMI in adulthood and 7 SNPs associated with maximum BMI z-score in childhood with p ≤0.05, where the direction of the association was consistent with that observed in Europeans. The p values for the association between SNP and BMI were much more significant among the 339,224 Caucasians as compared to the 3,491 Pima Indians; no SNP in the Pima Indian analysis achieved genome-wide statistical significance. This is not surprising given the relatively small sample size of Pima Indians. Nonetheless, some of SNPs had significantly stronger effects in Pima Indians than that in Europeans. For example, the lead SNP rs2867125 in TMEM18 had a 2-fold larger effect size (β per copy of risk allele) in Pima Indians as compared with Europeans (0.13 and 0.06 SD units respectively, p=0.04 for difference in effect size), while the lead SNP rs7903146 in TCF7L2 had the largest difference in effect size between Pima Indians and Europeans (0.165 and 0.024 SD units respectively, p=0.005). Some variants, on the other hand, had significantly weaker effects in Pima Indians (in C9orf93, RASA2, SH2B1). Overall, however, the differences in effect sizes between Pima Indians and Europeans were not statistically different; this suggests that, in general, these established obesity variants have similar effects in both populations. Some of the variants analyzed in the present study were proxies that tag the lead SNPs identified in Europeans, and incomplete concordance between the tag and lead SNP could introduce heterogeneity. However, the overall test for heterogeneity was still not statistically significant when restricted to the 42 lead SNPs that had been directly genotyped. Although most BMI-associated variants identified in Europeans are also associated with BMI in other populations, there are some ethnic-specific associations. In the current study, we did not assess variants that were primarily identified in studies of non-European ethnic groups (22).
Our longitudinal data with measures of BMI across multiple ages, spanning both childhood and adulthood, allow for a comprehensive assessment for the effect of established BMI loci on obesity risk. For example, when all 82 SNPs with a mAF≥0.01 were considered in aggregate, the GRS was statistically significant in relation to maximum BMI during adulthood as well as childhood in Pima Indians. In general, the effects, in terms of the strength of the (logarithmic) regression coefficient, were stronger in childhood and adolescence than in adulthood. When analyzed individually, SNPs in FTO, MC4R, GNPDA2, TMEM18, SEC16B, FAIM2, TFAP2B, TNNI3K and LMX1B associated with childhood obesity in a meta-analysis of 47541children from 33 studies predominately of European ancestry (23). We have previously assessed 36 Pima specific BMI SNPs for their role in BMI gain during life time, and also found stronger genetic effects in childhood than in adulthood (24). In the present study, the GRS derived from established obesity variants was significantly associated with BMI in Pima Indians in most age groups, except for the oldest individuals. The strongest effects were observed in adolescence (ages of 14–16 years). This suggests that obesity conferred by these loci is established in childhood and adolescence in this population, and persists throughout adulthood, with some attenuation at older ages.
Although obesity is a major risk factor for development of T2D, the lead BMI SNPs showed little or no evidence for association with T2D when analyzed individually or as a GRS in 3747 Pima Indians. Statistical power for individual variants in the present study is limited due to a relatively small sample size. However, the effect of the GRS on T2D was very modest and not statistically significant, and this suggests that on average, the effect of these established obesity loci on T2D is small.
The current study included a comprehensive assessment of the 98 established loci for new or additional signals for BMI in Pima Indians. Thirty-eight tag SNPs, which map to 18 loci, had nominal associations with BMI in both full-heritage and non-full heritage Pima Indians (p≤0.05). The strongest signal for BMI was rs3751837 in NLRC3 (p=5.8×10−5). Additional independent signals were identified in 9 tag SNPs, that mapped to 7 loci and had slightly stronger associations with BMI than the previously reported lead SNP. Two SNPs (rs6021702 in ZFP64; rs12462812 in PGPEP1) which were distinct from the lead SNPs, provided the strongest evidence for a new independent signal for BMI. The previously established SNP in ZFP64 significantly associated with BMI, whereas the established SNP in PGPEP1 did not associate with BMI in the Pima Indian sample. Given that causal variants at most of these loci have not been identified, different signals between Pima Indians and Europeans might be expected due to differences in linkage disequilibrium. Nevertheless, none of these additional signals achieved statistical significance after correction for testing ~6000 SNPs (p<8.3×10−6).
Determining the function of SNPs identified in large meta-analyses can be challenging. Our finding that the 3’-UTR SNP rs2531995 in ADCY9 significantly associated with reduced ADCY9 expression (p=5.3×10−6) in 201 adipose biopsies of Pima Indians indicates that this SNP functions as (or tags) a cis-eQTL. Although our evidence that the intronic SNP rs2176598 affects HSD17B12 expression in adipose biopsies from Pima Indians was somewhat weaker (p=0.006), this finding was supported by data provided by the GTEx consortium where the risk allele T at rs2176598 had a reduced HSD17B12 expression level in adipose tissues (p=2.2×10−35, GTEx Analysis Release V6p). ADCY9 (Adenylyl cyclase 9) is part of the signaling pathway of the β2adrenergic receptor (ADRB2) (25–26), whereas HSD17B12 (Hydroxysteroid 17-Beta Dehydrogenase 12) gene expression is dysregulated in BDNF knock-out mice (27). Our results suggest that these variants may affect obesity through an effect on expression of ADCY9 and HSD17B12.
In conclusion, although many individual SNPs associated with BMI in large European studies do not show significant evidence for association in the smaller Pima Indian sample, calculation of genetic risk scores to analyze the SNPs in aggregate shows strong association between number of risk alleles and BMI in Pima Indian adults and children. This suggests that obesity loci identified in Europeans generally also affect BMI in Pima Indians.
Supplementary Material
Answer the Study Important Questions.
What is already known about this subject?
Heritable/genetic factors are estimated to explain 40–70% of inter-individual variability in BMI.
Prior meta-analyses of genome-wide association studies (GWAS) have identified at least 98 established (p<5 × 10−8) loci for Body Mass Index (BMI) predominately from populations of European ancestry.
What does your study add?
Although many individual SNPs associated with BMI in large European studies do not show significant evidence for association in the smaller full-heritage Pima Indian sample (n=3491), calculation of genetic risk scores to analyze the lead SNPs in aggregate shows strong association between number of risk alleles and BMI in Pima Indian adults and children. This suggests that obesity loci identified in Europeans also affect BMI in Pima Indians with a high prevalence of obesity.
Our longitudinal data with measures of BMI across multiple ages, spanning both childhood and adulthood, allow for a comprehensive assessment for the effect of established BMI loci on obesity risk. The strongest effects were observed in adolescence. Our study suggests that obesity conferred by these loci is established in childhood and adolescence in this population, and persists throughout adulthood, with some attenuation at older ages.
Our study also included a comprehensive assessment of the 98 established loci for new or additional signals for BMI in Pima Indians. The strongest signal for BMI in adulthood was rs3751837 in NLRC3 (p=5.8×10−5). Two SNPs (rs6021702 in ZFP64; rs12462812 in PGPEP1) which were distinct from the lead SNPs, provided the strongest independent evidence (p=8.4×10−4, p=8.8×10−4 after conditioning on the lead SNP, respectively) for a new signal for BMI.
Acknowledgements
We thank the clinical staff of the Phoenix Epidemiology and Clinical Research Branch for conducting the study. We also thank all study participants.
Funding: This work was supported by the intramural research programme of the National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health.
Footnotes
Disclosure Statement: The authors have nothing to disclose.
References
- 1.Barsh GS, Farooqi IS, O’Rahilly S. Genetics of body-weight regulation. Nature 2000; 404:644–651. [DOI] [PubMed] [Google Scholar]
- 2.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature 2015;518:197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet 2010;42:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berndt SI, Gustafsson S, Mägi R, et al. Genome-wide meta-analysis identifies 11 new loci for anthropometric traits and provides insights into genetic architecture. Nat Genet 2013;45:501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knowler WC, Pettitt DJ, Saad MF, Bennett PH. Diabetes mellitus in the Pima Indians: incidence, risk factors and pathogenesis. Diabetes Metab Rev 1990;6:1–27. [DOI] [PubMed] [Google Scholar]
- 6.Traurig M, Mack J, Hanson RL, et al. Common variation in SIM1 is reproducibly associated with BMI in Pima Indians. Diabetes 2009,58:1682–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller YL, Thearle MS, Piaggi P, et al. Common genetic variation in and near the melanocortin 4 receptor gene (MC4R) is associated with body mass index in American Indian adults and children. Hum Genet 2014;133:1431–1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thearle MS, Muller YL, Hanson RL, et al. Greater impact of melanocortin-4 receptor deficiency on rates of growth and risk of type 2 diabetes during childhood compared with adulthood in Pima Indians. Diabetes 2012;61:250–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Traurig MT, Perez JM, Ma L, et al. Variants in the LEPR gene are nominally associated with higher BMI and lower 24-h energy expenditure in Pima Indians. Obesity (Silver Spring) 2012;20:2426–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bian L, Traurig M, Hanson RL, et al. MAP2K3 is associated with body mass index in American Indians and Caucasians and may mediate hypothalamic inflammation. Hum Mol Genet 2013;22:4438–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Expert Committee on the Diagnosis and Classification of Diabetes Mellitus: Report of the Expert Committee on the Diagnosis and Classification of Diabetes Mellitus Diabetes Care 1997;20:1183–1197. [DOI] [PubMed] [Google Scholar]
- 12.Muller YL, Piaggi P, Chen P, et al. Assessing Variation across Eight Established East Asian Loci for Type 2 Diabetes in American Indians: Suggestive Evidence for New Sex-specific Diabetes Signals in GLIS3 and ZFAND3. Diabetes Metab Res Rev. 2017;33:doi: 10.1002/dmrr.2869. [DOI] [PubMed] [Google Scholar]
- 13.Mason CC, Hanson RL, Ossowski V, et al. Bimodal distribution of RNA expression levels in human skeletal muscle tissue. BMC Genomics. 2011;12:98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsueh WC, Nair AK, Kobes S, et al. Identity-by-Descent Mapping Identifies Major Locus for Serum Triglycerides in Amerindians Largely Explained by an APOC3 Founder Mutation. Circ Cardiovasc Genet 2017;10: doi: 10.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Browning BL, Browning SR. A fast, powerful method for detecting identity by descent. Am J Hum Genet 2011;88:173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998;62:1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanson RL, Rong R, Kobes S, et al. Role of Established Type 2 Diabetes-Susceptibility Genetic Variants in a High Prevalence American Indian Population. Diabetes 2015;64:2646–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Higgins JPT, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in metaanalyses. BMJ 2003;327:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stouffer SA, Suchman EA, DeVinney LC, Star SA, Williams RM. How the volumes were produced In the American Soldier, Volume 1: Adjustment During Army Life. Princeton, NJ, Princeton University Press; 1949. pp.45. [Google Scholar]
- 21.Haggstrom GW. Logistic regression and discriminant analysis by ordinary least squares. J Bus Econ Statist 1983;1:229–238. [Google Scholar]
- 22.Wen W, Zheng W, Okada Y. Meta-analysis of genome-wide association studies in East Asian-ancestry populations identifies four new loci for body mass index. Hum Mol Genet. 2014;23:5492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Felix JF, Bradfield JP, Monnereau C, et al. Genome-wide association analysis identifies three new susceptibility loci for childhood body mass index. Hum Mol Genet 2016;25:389–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hohenadel MG, Baier LJ, Piaggi P, et al. The impact of genetic variants on BMI increase during childhood versus adulthood. Int J Obes 2016;40:1301–1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prior SJ, Goldberg AP, Ryan AS. ADRB2 haplotype is associated with glucose tolerance and insulin sensitivity in obese postmenopausal women. Obesity (Silver Spring) 2011;19:396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pearen MA, Muscat GE. Minireview: Nuclear hormone receptor 4A signaling: implications for metabolic disease. Mol Endocrinol 2010;24:1891–1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glorioso C, Sabatini M, Unger T, et al. Specificity and timing of neocortical transcriptome changes in response to BDNF gene ablation during embryogenesis or adulthood. Mol Psychiatry 2006;11:633–648. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.