Abstract
Naïve CD4+ T lymphocytes differentiate into various Th cell subsets following TCR binding to microbial peptide:MHCII (p:MHCII) complexes on dendritic cells (DCs). The affinity of the TCR interaction with p:MHCII plays a role in Th differentiation by mechanisms that are not completely understood. We found that low affinity TCRs biased mouse naïve T cells to become T follicular helper (Tfh) cells while higher affinity TCRs promoted the formation of Type 1 (Th1) or Type 17 (Th17) cells. We explored the basis for this phenomenon by focusing on IL-2 receptor signaling, which is known to promote Th1 and suppress Tfh differentiation. SIRP⍺+ DCs produce abundant p:MHCII complexes and consume IL-2 while XCR1+ DCs weakly produce p:MHCII but do not consume IL-2. We found no evidence, however, of preferential interactions between Th1-prone high affinity T cells and XCR1+ DCs or Tfh-prone low affinity T cells and SIRP⍺+ DCs after infection with bacteria expressing the peptide of interest. Rather, high affinity T cells sustained IL-2 receptor expression longer and expressed two novel Th differentiation regulators, Eef1e1 and Gbp2, to a higher level than low affinity T cells. These results suggest that TCR affinity does not influence Th differentiation by biasing T cell interactions with IL-2-consuming DCs, but instead directly regulates genes in naïve T cells that control the differentiation process.
Introduction
CD4+ T lymphocytes are critical for controlling infections through their ability to provide help to B cells, cytotoxic T cells, or myeloid cells (1). CD4+ T cells provide these diverse functions by differentiating into specialized subsets following TCR recognition of p:MHCII complexes on the surface of DCs and in the context of cytokines from the innate immune system (1–3). Work by our group and others has shown that TCR dwell time on p:MHCII, which strongly correlates with TCR affinity, also influences Th cell differentiation (4–6). In our experiments, increases in TCR affinity corresponding to p:MHCII dwell times of 0.9 to 2.3 s correlated with increased differentiation of macrophage-helping Th1 cells and decreased formation of B cell-helping Tfh cells (4, 5, 7). Although Th1 differentiation fostered by high affinity TCR interactions is related to strong induction of the IRF4 transcription factor (8), additional aspects of the mechanism by which TCR affinity affects T cell differentiation have yet to be determined.
IL-2 receptor signaling promotes Th1 and suppresses Tfh differentiation by driving STAT5 activation and induction of Blimp1, a repressor for the Tfh promoting transcription factor Bcl-6 (9–15). Potentially, TCR affinity regulates Th cell differentiation, in part, by influencing IL-2 signaling. We therefore tested two TCR affinity-regulated IL-2 signaling-based mechanisms, one rooted in dendritic cell Ag presentation and another focused on IL-2 receptor alpha chain (CD25) expression. The two major classical dendritic cells (DCs) in the spleen differ in p:MHCII presentation and IL-2 consumption potential (16–18). XCR1+ DCs are potent producers of the Th1-inducing cytokine IL-12 (18), but are relatively poor producers of p:MHCII complexes, whereas SIRP⍺+ DCs consume IL-2 and are weak IL-12 producers, but are strong producers of p:MHCII complexes (17). Thus, it is possible that high TCR affinity could bias toward Th1 differentiation because only Th cells with high affinity TCRs cells could access the small number of p:MHCII complexes displayed on XCR1+ DCs, while low affinity cells could access the abundant p:MHCII complexes on SIRP⍺+ DCs favoring Tfh differentiation (19). Alternatively, since IL-2 receptor expression is proportional to the strength of TCR signaling and drives Th1-promoting STAT5 activation (9–15), Th cells with high affinity TCRs may be intrinsically more likely to become Th1 cells than Th cells with low affinity TCRs.
We tested these models by examining the influence of TCR affinity on differentiation and T cell-DC interactions using two TCR transgenic (Tg) strains that contain T cells with differing TCR affinities for the same p:MHCII ligand (20). We found that Th cells with low, medium, or high affinity TCRs T cells tended to adopt uncommitted, Tfh, or non-Tfh fates, respectively, after exposure to bacteria or virus expressing the relevant peptide. In all cases, Th cells interacted more frequently with SIRP⍺+ DCs than with XCR1+ DCs, indicating that differential interactions with DCs did not account for TCR affinity-based differences in Th1/Tfh formation. Rather, TCR affinity influenced Th differentiation by controlling the expression of the IL-2 receptor and eukaryotic translation elongation factor 1 epsilon 1 (Eef1e1), which promoted Th1 cells, and guanylate binding protein 2 (Gbp2), which promoted Tfh cells. Our results suggest that TCR affinity-based effects on Th differentiation are related to the capacity of the TCR to induce different genetic programs at different affinity levels.
Materials and Methods
Mice
Six- to eight-wk-old B6 mice were purchased from the Jackson Laboratory or the National Cancer Institute Mouse Repository (Frederick, MD, USA). Cd25−/− mice were purchased from Jackson Laboratory. Rag1−/− B3K506 TCR transgenic, Rag1−/− B3K508 TCR transgenic (20), and Rag1−/− UbcGFP TEa TCR transgenic mice were bred and housed in specific pathogen–free conditions in accordance with guidelines of the University of Minnesota Institutional Animal Care and Use Committee and National Institutes of Health. Clec4a4DTR mice (21) were backcrossed onto the B6 background and used as bone marrow donors. The Institutional Animal Care and Use Committee of the University of Minnesota approved all animal experiments.
Infections and immunization
For intranasal and intravenous infections, mice were injected with 107 colony-forming units of ActA-deficient Listeria monocytogenes (Lm)-3K, Lm-P5R, Lm-P2A, or Lm-Flic bacteria as described previously (5, 22, 23). Mice injected intravenously with Lm-P5R or Lm-P2A bacteria contained similar numbers of organisms in their livers 24 h later (unpublished observation). Lm-3K, Lm-P5R, and Lm-P2A strains were generated by inserting sequences encoding each peptide in frame with the Listeriolysin-O signal sequence and promoter that drives maximal production under in vivo infection conditions, as described previously (22). The P5R epitope was inserted into the PR8 influenza A virus genome as previously described (24, 25). Mice were infected intranasally with 40 plaque-forming units of PR8 influenza A virus expressing P5R.
Cell transfer
Lymph nodes were collected from Rag1−/− B3K506 and Rag1−/− B3K508 (20) TCR transgenic mice and hand-mashed into a single cell suspension. Aliquots were stained with allophycocyanin-labeled CD4 (RM4–5; Tonbo biosciences) antibody and analyzed with Fluorescent AccuCheck counting beads (Invitrogen) to assess CD4+ T cell numbers and purity using a LSR II (BD biosciences) flow cytometer. For imaging experiments, 106 TCR transgenic cells were transferred into B6 mice 24 h before infection. 105 TCR transgenic cells were transferred into B6 mice for flow cytometry experiments examining the initial three d following Lm infection, while 3×103 TCR transgenic cells were transferred for experiments examining the response at seven d post-infection with Lm or influenza.
Cell enrichment and flow cytometry
Single cell suspensions were generated by dissociating spleens with the GentleMACS dissociator (Miltenyi Biotec) or hand-mashed in a petri dish. Single cell suspensions were stained for one h at room temperature with 0.1 μg of FITC-labeled CD90.1 antibody (HIS51; ThermoFisher Scientific) and 2 μg of BV650-labeled (L138D7; Biolegend) or BUV395-labeled (2G8; BD biosciences) CXCR5 antibody. Samples were then enriched for CD90.1 antibody-bound cells using magnetic bead-based enrichment as described previously (26) with the minor modification that EasySep Mouse APC or FITC Positive Selection Kits (Stemcell Technologies) and EasySep magnets (Stemcell Technologies) were used.
For identification of surface markers, the sample was stained on ice with various combinations of the following antibodies: APC-ef780-labeled B220 (RA3–6B2; ThermoFisher Scientific), APC-ef780-labeled CD11b (M1–70; ThermoFisher Scientific), APC-ef780-labeled CD11c (N418; ThermoFisher Scientific), BV786-labeled CD4 (GK1.5; BD biosciences), AF700-labeled CD44 (IM7; ThermoFisher Scientific), FITC-labeled CD45.1 (A20; Biolegend), BUV395-labeled CD45.2 (104; BD biosciences), BUV395-labeled CD25 (PC61; BD biosciences), PE-labeled CD69 (H1.2F3; ThermoFisher Scientific) and FITC-labeled CD90.1 (HIS51; ThermoFisher Scientific). All samples were also stained with a fixable viability dye (Ghost Dye Red 780; Tonbo Biosciences). For transcription factor staining, samples were fixed with the eBioscience Foxp3/transcription factor staining kit (ThermoFisher Scientific) and then stained with BV421-labeled RORɣt (Q31–378; BD biosciences), BV605-labeled or BV421-labeled T-bet (4B10; Biolegend), and AF488-labeled Bcl-6 (K112–91; BD biosciences) antibodies. To calculate cell numbers, Fluorescent AccuCheck counting beads (Invitrogen) were added to each sample after the final wash step. Cells were then analyzed on an LSR II or Fortessa (BD biosciences) flow cytometer. Data were analyzed with FlowJo (TreeStar).
Cell sorting and co-culture
Spleens were harvested from mice 48 h after intravenous infection with Lm-P5R bacteria. The spleens were chopped into small pieces and digested with collagenase P (Millipore Sigma) for 20 min at 37°C prior to hand-mashing in a petri dish to generate a single cell suspension. The single cell suspension was enriched for CD11c expressing cells using a CD11c positive selection kit (Miltenyi Biotec). The enriched samples were stained for 30 min at room temperature with BV421-labeled XCR1 (ZET; Biolegend), PE-labeled CD64 (X54–5/7.1; Biolegend), APC-labeled SIRP⍺ (P84; Biolegend), APC-Cy7-labeled Ly-6G (1A8; Biolegend), APC-Cy7-labeled Siglec F (E50–2440; BD biosciences), APC-eF780-labeled NKp46 (29A1.4; ThermoFisher Scientific), APC-eF780-labeled CD90.2 (53–2.1; ThermoFisher Scientific), PE-Cy7-labeled Ly-6C (HK1.4; ThermoFisher Scientific), FITC-labeled CD11c (N418; ThermoFisher Scientific), and a fixable viability dye (Ghost Dye Red 780; Tonbo Biosciences). The cells were sorted with a FACS Aria II (BD biosciences) to isolate live XCR1+ (CD64− NKp46− Ly-6C− CD90.2− Ly-6G− SIRP⍺− CD11c+ XCR1+) and SIRP⍺+ (CD64− NKp46− Ly-6C− CD90.2− Ly-6G− XCR1− CD11c+ SIRP⍺+) DCs.
For co-culture, naïve B3K508 CD4+ T cells were isolated from Rag1−/− B3K508 TCR transgenic mice as described in the cell transfer section. B3K508 cells were co-cultured with the sort-purified XCR1+ or SIRP⍺+ DCs at 1 to 1, 1 to 3, and 1 to 10 ratios of DCs to T cells in complete IMDM for 24 h at 37°C. Some T cells were also cultured without DCs to serve as negative staining controls for markers of activation. After culture, T cells were stained with antibody and analyzed on a flow cytometry as described in the cell enrichment and flow cytometry section.
Confocal microscopy
Confocal microscopy was performed using a Leica SP5 confocal microscope with 2 HyD detectors and two PMT detectors as well as 405, 458, 488, 514, 543, 594 and 633 lasers. Twenty μm paraformaldehyde-fixed splenic sections from naive or Lm-P5R infected mice were imaged with a 63X oil immersion objective lens with 1.4NA. The splenic sections were stained with F4/80 BV421-labeled (BM8; Biolegend), Pacific Blue-labeled B220 (RA3–6B2; Biolegend), CF405L-labeled CD8⍺ (53–6.7; Biolegend), AF488-labeled phospho-S6 kinase (pS6) (2F9; Cell Signaling Technologies), CF555-labeled CD86 (GL-1; Biolegend), AF647-labeled CD45.2 (104, Biolegend), AF700-labeled MHCII (M5/114.15.2; Biolegend), CF514-labeled CD11c (N418; Biolegend), BV480-labeled CD3 (17A2; BD biosciences), and AF594-labeled SIRP⍺ (P84; Biolegend) antibodies. CF-labeled antibodies were generated by conjugating purified antibodies from Biolegend to CF405L, CF514, or CF555 with Biotium Mix-n-Stain labelling kits (Biotium). The mark and find feature in the Leica Application Suite was used to image 12 T cell zones in each spleen with each image consisting of a 20 μm z-stack acquired at a 0.5 μm step size. Additionally, the Leica SP5 microscope was used to image single color-stained Ultracomp eBeads (ThermoFisher Scientific) for generating a compensation matrix.
Image processing and histo-cytometry analysis
Image analysis was performed using Chrysalis (27). In brief, a compensation matrix was created by automatic image-based spectral measurements on calibration samples in ImageJ by using Generate Compensation Matrix (27). The compensation matrix was used to perform linear unmixing on 3D images and movies with Chrysalis. Chrysalis was also used for further automated image processing, including merging images from the same tissue into a single stack, generating preview time-lapse sequences to identify informative fields, and generating new channels based on mathematical and morphological operations on existing channels. Imaris 8.3, 8.4, 9.0, and 9.1 (Bitplane) were used for surface creation to identify cells in images and to run the Sortomato, XTCreateSurfaces, and XTChrysalis Xtensions (27). These Xtensions were used for creating surfaces based on protein expression, quantifying cell-cell interactions, and exporting cell surface statistics. These exported statistics were imported into FlowJo for quantitative image analysis.
Quantitative PCR
106 B3K506 or B3K508 TCR transgenic cells were transferred into B6 mice 24 h before intravenous infection with Lm-P5R bacteria. The spleen and lymph nodes were harvested 48 h after infection and stained with FITC-labeled CD90.1 (HIS51; ThermoFisher Scientific), APC-ef780-labeled B220 (RA3–6B2; ThermoFisher Scientific), APC-ef780-labeled CD11b (M1–70; ThermoFisher Scientific), APC-ef780-labeled CD11c (N418; ThermoFisher Scientific), BV786-labeled CD4 (GK1.5; BD biosciences), AF700-labeled CD44 (IM7; ThermoFisher Scientific), and a fixable viability dye (Ghost Dye Red 780; Tonbo Biosciences). The TCR transgenic cells were then enriched with the EasySep Mouse FITC Positive Selection Kit (Stemcell Technologies) and EasySep magnets (Stemcell Technologies). Live TCR transgenic CD4+ T cells were sorted from the enriched sample into Trizol (ThermoFisher Scientific) using a FACS Aria II (BD biosciences). RNA was extracted using chloroform (ThermoFisher Scientific) and purified with the RNeasy micro kit (Qiagen). RNA quantities were measured with a Qubit 2.0 (ThermoFisher Scientific) and 41 ng RNA of each sample was converted to cDNA with SuperScript IV reverse transcriptase (ThermoFisher Scientific). Primers (Supplementary Table 1) were designed with the NCBI Primer-Blast tool (28). Quantitative PCR was performed with FastStart SYBR Green (Millipore Sigma) in a thermocycler (Eppendorf). The resulting data was analyzed by normalizing to the housekeeping gene Gapdh and then calculating fold change in gene expression for B3K506 cells relative to B3K508 cells with the comparative CT method (29).
CRISPR/Cas9 Screen
A CRISPR/Cas9 system utilizing two MSCV-based gamma retroviral vectors was used to target genes of interest. One virus encoded Cas9 and the fluorescent protein mNeongreen (30) and the other encoded gRNAs and the fluorescent protein mAmetrine (31). These vectors were created by modifying the LMP-Amt vector (32), which was a gift from S. Crotty (La Jolla Institute). The vector encoding Cas9 and mNeongreen was generated by replacing the shRNA-encoding segment, PGK promoter, and mAmetrine gene with Cas9-P2A and mNeongreen (Allele Biotechnology) through In-Fusion cloning (Takara Bio USA). The Cas9-P2A fragment was cloned from the lentiCRISPRv2 puro plasmid which was a gift from B. Stringer (Addgene plasmid # 98290).
To generate the gRNA vector, the LMP-Amt vector (32) was modified by removing the SapI site and replacing the shRNA-encoding segment with the bacterial toxin gene CCDB flanked by AarI sites using In-Fusion cloning (Takara Bio USA). This CCDB containing fragment was cloned from the pMOD_B2303 plasmid, which was a gift from D. Voytas (University of Minnesota). This modified LMP-Amt plasmid served as the recipient for Golden Gate cloning reactions that replaced the CCDB gene with a tRNA-gRNA array (33–35) using AarI (ThermoFisher Scientific), SapI (New England BioLabs), and T4 (New England BioLabs) enzymes. The gRNAs used in the tRNA-gRNA arrays were designed in Benchling (Benchling) to target functional motifs identified in InterPro (36) with two gRNA designed for each target gene. Primers encoding the gRNAs were then generated using the Voytas laboratory toolkit (37) and the cloning of the tRNA-gRNA array was performed as described previously (37).
Retrovirus was prepared as described previously(38), with minor alterations. In brief, Platinum-E cells (Cell Biolabs) were grown in complete DMEM media (Gibco) prior to transfection with Polyethylenimine, Linear, MW 25,000 (Polysciences), pCL-Eco and retroviral plasmids encoding Cas9 or tRNA-gRNA arrays. After transfection, the media was supplemented with ViralBoost (Alstem Cell Advancements) and 30 μM water soluble cholesterol (Millipore Sigma). The virus containing supernatant was collected 24 and 48 h after transfection and filtered with a 0.45 μm Nylon 25 mm Syringe Filter (Fisher). Aliquots were stored at −80°C for up to three mo.
Retroviral transduction was performed as described previously (38), with minor alterations. Specifically, naïve B3K508 CD4+ T cells or naïve Cas9+ B3K508 CD4+ T cells were isolated from Rag1−/− B3K508 TCR transgenic or Rag1−/− Cas9+ B3K508 TCR transgenic mice, respectively, as described in the cell transfer section. These B3K508 T cells were grown in a 96 well plate with complete IMDM media (Millipore Sigma) containing IL-7 (Tonbo biosciences). Cells were activated in plates coated with anti-CD3 (2C11; BioXcell) and anti-CD28 (37.51; BioXcell). The cells were transduced with retroviral supernatant and polybrene (Millipore Sigma) 24 and 40 h after activation. B3K508 T cells were transduced with retrovirus encoding Cas9 and retrovirus encoding tRNA-gRNA arrays, while the Cas9+ B3K508 T cells were only transduced with tRNA-gRNA array encoding retrovirus. For transductions, plates were spun at 1500 rpm for two h at 37°C and then the media was exchanged for complete IMDM media containing IL-2 (Peprotech). After three d of culture, the cells were moved to plates that were not coated with anti-CD3 or anti-CD28 and two d later the media was switched to complete IMDM media containing IL-7 (Tonbo biosciences). After 24 h, cells were either directly transferred into B6 mice or stained with BV786-labeled CD4 (RM4–5; BD biosciences) antibody and a fixable viability dye (Ghost Dye Red 780; Tonbo Biosciences). The stained cells were sorted with a FACS Aria II (BD biosciences) to isolate live Cas9+ gRNA+ CD4+ T cells, which were then transferred into B6 mice. Each B6 mouse received 2500 T cells. Four d after transfer, mice were infected with Lm-3K bacteria and the spleens were harvested seven d post-infection for flow cytometric analysis.
Statistical analysis
Statistical significance was determined using Prism (Graphpad) software for unpaired two-tailed Student’s t test, one-way and two-way ANOVA tests. Prism (Graphpad) was also used to calculate linear correlations and R squared.
Code availability
All of the custom code generated for image processing or analysis can be downloaded at https://histo-cytometry.github.io/Chrysalis/.
Results
TCR affinity biases Th cell differentiation
We utilized B3K506 and B3K508 TCR Tg strains (20) to examine the relationship between TCR affinity and T cell differentiation. B3K506 and B3K508 mice contain monoclonal populations of CD4+ T cells with TCRs that bind to a peptide called P5R complexed with the I-Ab MHCII molecule of C57BL/6 (B6) mice. The B3K506 TCR, however, binds more strongly to this ligand than B3K508 TCR (KDs of 11 and 93 μM) while the B3K508 TCR binds to a related ligand P2A:I-Ab more strongly than the B3K506 TCR (KDs of 175 and 278 μM). The KDs of 11 and 93 μM will be considered high and medium affinities, respectively, while KDs of 175 and 278 μM will be considered to be low affinities. The effect of these affinity differences on differentiation was examined by adoptive transfer of CD90.1+ B3K506 or B3K508 T cells into CD90.2+ mice (39) that were then infected intravenously with Lm bacteria engineered to express P5R (Lm-P5R) or P2A (Lm-P2A) peptide.
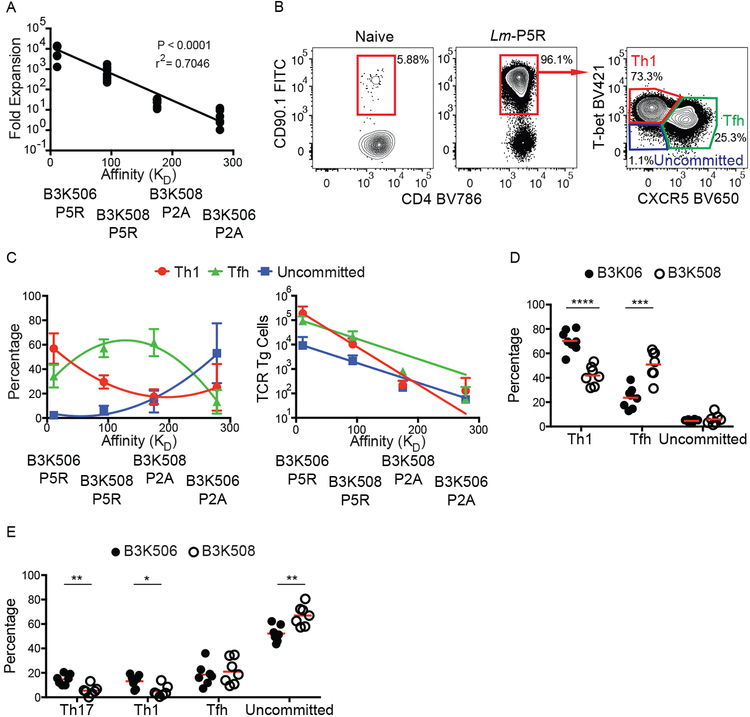
As expected from the work of Huseby and colleagues (20), the expansion of B3K506 and B3K508 T cells in response to Lm-P5R and Lm-P2A infection correlated positively with TCR affinity (Fig. 1A). Previous work (9) indicated that Lm infection drives the differentiation of Th1 cells, which express the T-bet transcription factor (40), and Tfh cells, which express the CXCR5 chemokine receptor (41–43). B3K506 and B3K508 cells also adopted these fates (Fig. 1B), although high TCR affinity (lower KD) biased differentiation to the Th1 fate and medium affinity towards the Tfh fate as we reported previously (5), while lower affinity T cells predominantly remained uncommitted as defined by a lack of T-bet or CXCR5 (Fig. 1C). Despite having a lower percentage of Tfh cells than the medium TCR affinity population, the high TCR affinity T cell population had a greater absolute number of Tfh cells due to greater proliferation (Fig. 1C). The TCR affinity-mediated differentiation bias was recapitulated in mice infected with influenza expressing P5R (PR8-P5R). After PR8-P5R infection, T cells with a higher TCR affinity were again biased towards the Th1 fate while lower TCR affinity biased cells towards the Tfh fate, demonstrating that the influence of TCR affinity on T cell differentiation is not restricted to Lm infection (Fig. 1D).
Figure 1. TCR affinity differences between B3K506 and B3K508 cells influence T cell expansion and bias T cell differentiation.
(A) Fold expansion of B3K506 or B3K508 cells seven d after Lm-P5R (n=7) or Lm-P2A (B3K506 n=7, B3K508 n=8) infection plotted in relation to the affinity of the relevant TCR-p:MHCII interaction. (B) Representative flow cytometric identification of TCR Tg cells (red gates in the left and middle panels) in naive or Lm-P5R infected mice. Th1 (T-bet+), Tfh (CXCR5+), or uncommitted (T-bet− CXCR5−) cells were identified in the right panel in the red, green and purple gates, respectively. (C) Differentiation (Th1 in red, Tfh in green, and uncommitted in blue) of B3K506 or B3K508 cells seven d after Lm-P5R (n=7) or Lm-P2A (B3K506 n=7, B3K508 n=8) infection plotted in relation to the affinity of the relevant TCR-p:MHCII interaction. (D) Frequency of Th1, Tfh, or uncommitted cells among B3K506 (filled circle, n=8) or B3K508 (empty circle, n=7) effector cell populations seven d after infection with influenza PR8-P5R. (E) Frequency of Th17, Th1, Tfh, or uncommitted cells among B3K506 (filled circle, n=7) or B3K508 (empty circle, n=7) cells seven d after intranasal infection with Lm-P5R. The bars in D and E represent the mean. Pooled data from two or three independent experiments are shown. Spleens were analyzed for the depicted experiments. Multiple t tests were used to determine significance for D and E. A linear regression was used to calculate significance for A and a nonlinear fit was used to calculate the r2. * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
The effect of TCR affinity on T cell differentiation was also tested after intranasal Lm-P5R infection, which induces Th17 cells along with Th1 and Tfh cells (23). Th17 differentiation was identified by expression of the lineage-defining transcription factor RORɣt (44). B3K506 cells generated larger fractions of Th17 and Th1 cells, an equal fraction of Tfh cells, and a lower fraction of uncommitted cells than B3K508 cells (Fig. 1E), suggesting that high TCR affinity also promotes the Th17 fate. The large percentage of uncommitted Th cells observed after this infection was likely a consequence of low Ag presentation due to poor adaptation of this enteric bacterium to the intranasal route.
XCR1+ and SIRP⍺+ DCs differ in Ag presentation and effect on T cell differentiation
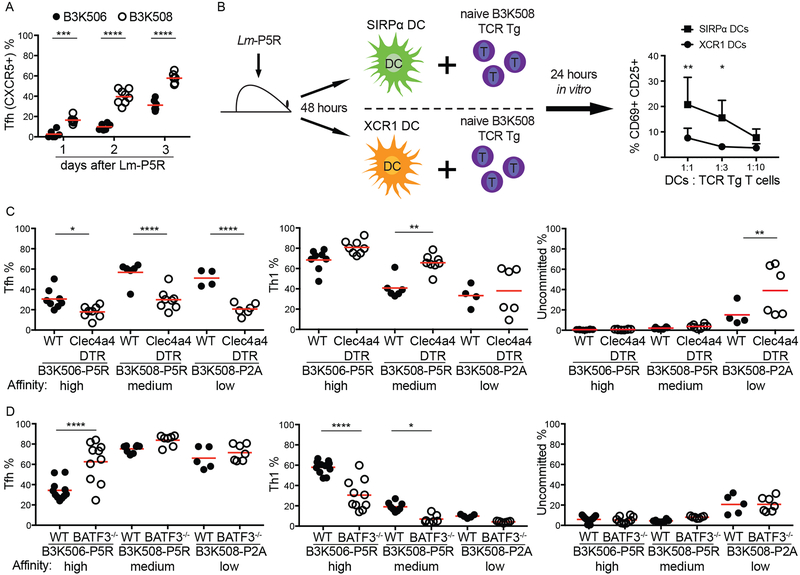
Before testing the differential Ag presentation hypothesis, we first examined the kinetics of Th differentiation to identify the time when critical T-DC interactions occur. B3K506 and B3K508 cell differentiation was examined over the first three d after Lm infection because Th1/Tfh bifurcation occurred in this time frame in another system (11, 38). A larger fraction of the B3K508 effector T cell population consisted of CXCR5+ cells than the B3K506 population on d one, two, and three after infection (Fig. 2A) and did not change after d three (Fig. 1C and2A). These results suggest that the Th1/Tfh cell bifurcation occurs within the first three d following Lm infection.
Figure 2. Effect of genetic ablation of XCR1+ or SIRP⍺+ DCs on Th differentiation.
(A) Tfh differentiation by B3K506 (filled circle, n=6) and B3K508 (empty circle, d 1 d 2 n=6, d 3 n=8) cells in the initial three d following Lm-P5R infection. (B) Experimental model and resulting mean frequencies ± SD of CD69+ CD25+ B3K508 cells after one d of co-culture with SIRP⍺+ (square, n=3) or XCR1+ (circle, n=3) DCs from Lm-P5R infected mice two d post-infection. (C) Frequency of Tfh, Th1, or uncommitted cells among B3K506 or B3K508 TCR Tg cells in DT-treated WT (filled circle) or Clec4a4DTR (empty circle) mice three d after Lm-P5R (B3K506 WT n=8, Clec4a4DTR n=8; B3K508 WT n=6, Clec4a4DTR n=8) or Lm-P2A (B3K508 WT n=4, Clec4a4DTR n=6) infection. (D) Frequency of Tfh, Th1, or uncommitted cells among B3K506 or B3K508 cells in WT (filled circle) or Batf3−/− (empty circle) mice three d after Lm-P5R (B3K506 WT n=11, Batf3−/− n=7; B3K508 WT n=8, Batf3−/− n=7) or Lm-P2A (B3K508 WT n=5, Batf3−/− n=7) infection. The bars in A, C, and D represent the mean. Pooled data from two or three independent experiments are shown. Spleens were analyzed for the depicted experiments. Two-way ANOVA was used to determine significance for B and one-way ANOVA with Sidak’s multiple comparison test was used to determine significance for A, C and D. * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
We also confirmed that SIRP⍺+ DCs produce more p:MHCII complexes than XCR1+ DCs after Lm infection as reported in another system (17). The amount of p:MHCII presentation was determined by measuring the capacity of purified splenic DCs from d two intravenously Lm-P5R-infected mice to stimulate CD69 and CD25 expression by cognate Th cells in vitro (10, 17) (Fig. 2B). SIRP⍺+ DCs were more potent activators of B3K508 cells than XCR1+ DCs in this assay (Fig. 2B), indicating that SIRP⍺+ DCs display more P5R:I-Ab complexes than XCR1+ DCs as expected.
We then tested how the different DC types affected Th differentiation using a cell ablation strategy. The majority of SIRP⍺+ DCs, which express Clec4a4 (DCIR2), were ablated by administering diphtheria toxin (DT) to radiation chimeras generated with Clec4a4DTR bone marrow (21). The effect of XCR1+ DCs was studied with Batf3−/− mice, which lack these cells (45). A 60% reduction in SIRP⍺+ DCs led to an increase in Th1 and a decrease in Tfh differentiation by B3K506 and B3K508 cells following Lm-P5R infection irrespective of TCR affinity (Fig. 2C). Ablation of XCR1+ DCs had the opposite effect - increased Tfh and decreased Th1 formation for high and medium affinity TCR T cells (Fig. 2D). These results indicate that XCR1+ DCs are critical for optimal Th1 and SIRP⍺+ DCs for optimal Tfh differentiation after Lm infection for high and low affinity T cells. Thus, this approach indicated that differential Ag presentation by XCR1+ and SIRP⍺+ DCs cannot explain the tendency of high affinity cells to produce Th1 cells and low affinity cells to produce Tfh cells.
TCR affinity does not influence T cell-DC interactions following Listeria infection
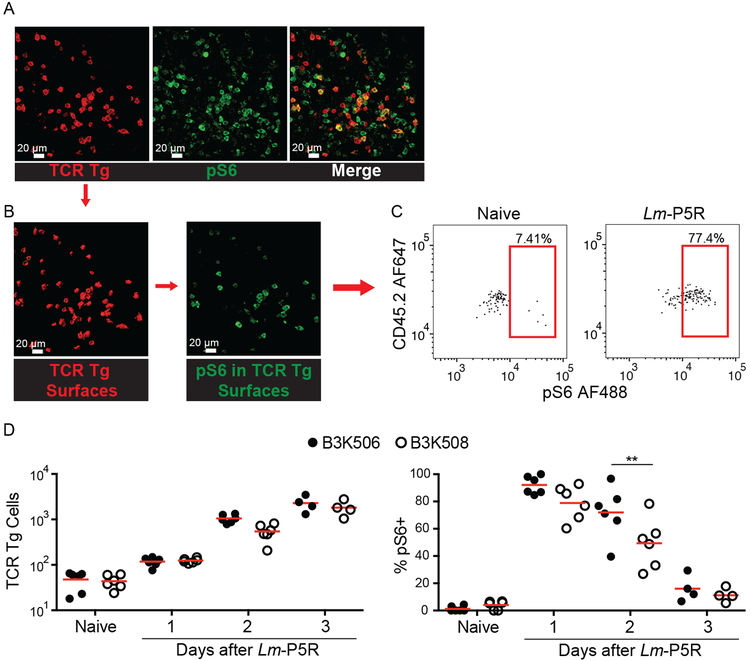
Because cell ablation can result in compensatory effects (46), we also tested the DC preference model by direct observation of T-DC interactions in mice with a normal complement of DCs. Histo-cytometry was used to assess B3K506 and B3K508 T cell interactions with XCR1+ and SIRP⍺+ DC because two-photon microscopy cannot simultaneously resolve all the T cell and DC populations of interest. The analysis focused on T cell zones of splenic sections from Lm-P5R infected and naive CD45.1+ mice that received CD45.2+ B3K506 or B3K508 cells because the T cells were primarily located in this location (39). TCR Tg cells were identified based on the congenic marker, CD45.2, which was used to generate digital surfaces corresponding to these cells (Fig. 3A and 3B). TCR Tg Th cells receiving acute TCR signals and thus likely engaged in productive interactions with DCs were identified by staining pS6, a T cell activation-induced phosphorylation event associated with the mTORC1 pathway (47) (Fig. 3B and 3C). Although B3K506 and B3K508 cells had equal pS6 levels one d after infection, B3K508 cells had significantly lower values one d later (Fig. 3D). This result suggests that higher TCR affinity B3K506 cells had more prolonged TCR signaling than lower TCR affinity B3K508 cells.
Figure 3. Histo-cytometry analysis of the effect of TCR affinity on T cell activation.
(A) Representative images of TCR Tg cells identified with CD45.2 (red) and pS6 (green) in a spleen one d post-Lm-P5R infection. (B) Digital surfaces of the cells were created and analyzed with histo-cytometry. (C) TCR Tg cell expression of pS6 was then quantified in Flowjo. (D) Histo-cyotmetric quantification of B3K506 (filled circle) and B3K508 (empty circle) cell numbers and their pS6 expression in naïve mice and mice during the initial three d following Lm-P5R infection (naïve, d 1, d 2 n=6; d 3 n=4). The bars in B represent the mean. Scale bar, 20 μm. Pooled data from three independent experiments are shown. Spleens were analyzed for the depicted experiments. One-way ANOVA was used to determine significance for D. ** = p < 0.01.
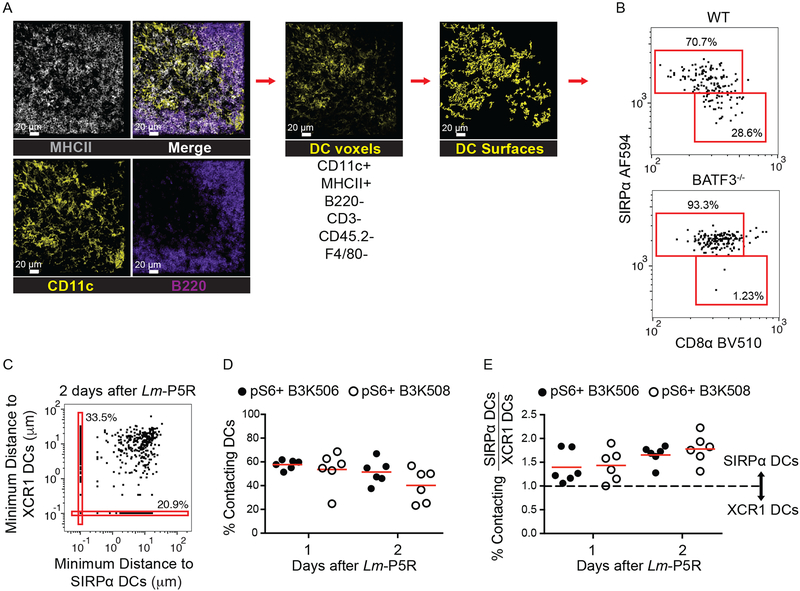
Th cell interactions with the DCs subsets were then assessed. DCs were identified by generating surfaces on a DC channel that contained voxels with strong MHCII and CD11c, but weak B220, CD3, F4/80, and CD45.2 signal (Fig. 4A). XCR1+ DCs and SIRP⍺+ DCs were identified based on CD8⍺ and SIRP⍺ expression, respectively (16, 18, 48, 49) (Fig. 4B). Identification of XCR1+ and SIRP⍺+ DCs was validated by comparing the DC gating strategy in WT and Batf3−/− mice, which lack XCR1+ DCs, 1 d after Lm-P5R infection (Fig. 4B). DCs in the WT mice consisted of approximately 30% XCR1+ DCs and 70% SIRP⍺+ DCs, while over 90% of the DCs were XCR1+ DCs in Batf3−/− mice (Fig. 4B). Productive T cell-DC interactions were identified as T cells undergoing T cell activation (pS6+) and touching a DC(s) (Fig. 4C). B3K506 and B3K508 cells interacted equally with DCs one and two d after Lm-P5R infection (Fig. 4D). The ratio of T cell contacts with SIRP⍺+ or XCR1+ DCs was analyzed to determine whether low or high affinity T cells interacted preferentially with one DC subset or the other. The analysis revealed that high affinity B3K506 and medium affinity B3K508 cells interacted 1.5 times more frequently with SIRP⍺+ than with XCR1+ DCs one and two d after infection (Fig. 4E). Thus, TCR affinity-mediated bias in T cell differentiation was not associated with preferential access of low or high affinity T cells to a particular DC subset.
Figure 4. Histo-cytometry analysis of DCs and T cell-DC interactions.
(A) Representative images of MHCII (grey), CD11c (yellow), and B220 (purple) staining from a spleen one d post-Lm-P5R infection. Histo-cytometry was used to generate surfaces for DCs (CD11c+ MHCII+ B220− CD3− CD45.2− F4/80−). XCR1+ or SIRP⍺+ surfaces were identified among the DC surfaces based on the indicated gates. (B) Histo-cytometry identification of XCR1+ and SIRP⍺+ DCs in WT and Batf3−/− mice infected with Lm-P5R for one d. (C) Representative histo-cytometry analysis displaying the percentage of pS6+ TCR Tg cells interacting with XCR1+ or SIRP⍺+ DCs in mice infected with Lm-P5R for one or two d. Interactions are defined as TCR Tg cells that are within 0 μm of either DC subset with 10−1 μm added to each cell for logarithmic visualization. (D) Histo-cytometry-identified interactions between pS6+ B3K506 (filled circle, n=6) or B3K508 (empty circle, n=6) T cells with XCR1+ or SIRP⍺+ DCs in mice infected with Lm-P5R for one or two d. (E) Ratio of T cell interactions with SIRP⍺+ DC to XCR1+ DC interactions by pS6+ B3K506 (filled circle, n=6) or B3K508 (empty circle, n=6) T cells. The bars in D and E represent means. Scale bar, 20 μm. Pooled data from three independent experiments are shown. Spleen was analyzed for the depicted experiments. One-way ANOVA was used to determine significance for D and E. No significant differences were detected.
CD25, Eef1e1, and Gbp2 are TCR regulated proteins that bias helper T cell differentiation
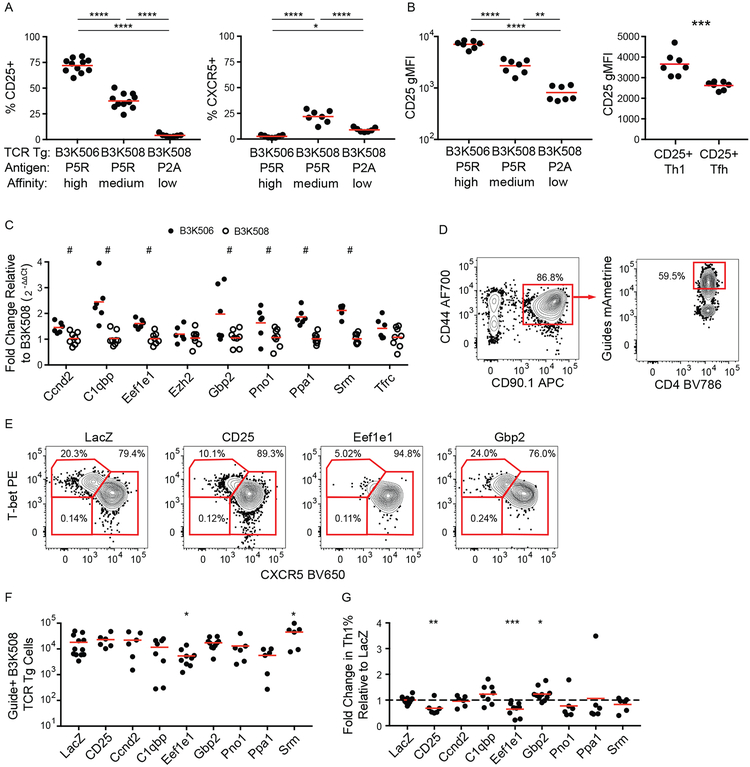
The lack of impact on T cell-DC interactions suggested that TCR affinity regulates differentiation in a T cell intrinsic manner. We, therefore, tested a model in which increasing TCR affinity for p:MHCII drives greater IL-2 receptor expression, STAT5 signaling, and Th1 differentiation. As described previously (4), B3K506 and B3K508 cells expressed CD25 (IL-2 receptor α) in a TCR affinity dependent manner two d after infection with Lm-P5R, with higher TCR affinity correlating with higher CD25 expression (Fig. 5A and 5B). This difference in CD25 expression inversely correlated with Tfh differentiation for high and medium TCR affinity T cells, in line with IL-2 receptor signal transduction suppressing Tfh differentiation (12) (Fig. 5A). T cells with a low affinity TCR had the lowest CD25 expression, but less Tfh differentiation than T cells with a medium affinity TCR, which could be attributed to a larger proportion of uncommitted cells (Fig. 5A). Moreover, CD25+ Th1 cells expressed more CD25 than CD25+ Tfh cells (Fig. 5B) adding support to the hypothesis that CD25 signaling represses Tfh differentiation. These results suggest that high affinity TCR-p:MHCII interactions promote Th1 differentiation by maintaining CD25, which represses the Tfh fate.
Figure 5. CD25, Eef1e1, and CD25 are upregulated by TCR signaling and bias helper T cell differentiation.
(A) Frequency of CD25+ and CXCR5+ B3K506 and B3K508 T cells two d after infection with Lm-P5R (CD25% n=11, CXCR5% n=7) or Lm-P2A (n=7). (B) CD25 geometric MFI (gMFI) for B3K506 and B3K508 T cells two d after infection with Lm-P5R (n=7) or Lm-P2A (n=7) and for CD25+ Th1 (T-bet+) and Tfh (CXCR5+) B3K508 T cells two d after infection with Lm-P5R (n=9). (C) Fold change in gene expression relative to B3K508 T cells based on qPCR analysis for B3K506 (filled circle, n=6) and B3K508 (empty circle, n=8) T cells two d after infection with Lm-P5R. (D) Representative flow cytometric identification of gRNA+ Cas9+ B3K508 T cells (red squares) seven d after Lm-3K infection and (E) their differentiation into Th1 (T-bet+), Tfh (CXCR5+), or uncommitted (T-bet− CXCR5−) lineages when targeting LacZ, CD25, Eef1e1, or Gbp2. (F) Quantification of gRNA+ Cas9+ B3K508 T cells and (G) their fold change in Th1% relative to the LacZ group seven d after Lm-3K infection. The bars in A, B, C, F, and G represent means. Pooled data from two or three independent experiments are shown. Spleen was analyzed for the depicted experiments, except (C), which represents data from spleen and lymph nodes. Multiple t tests were used to determine discoveries based on a false discovery rate of 5% for C and significance for A, B, F, and G. # = discovery, * = p < 0.05, ** = p < 0.01, *** = p < 0.001, **** = p < 0.0001.
TCR signaling induces many proteins in addition to CD25, some of which could be mediators of TCR affinity-based effects on Th differentiation. We focused on nine candidates from a published list of genes induced in naïve TCR transgenic T cells in vitro to a greater extent by a large amount of agonist peptide than by a lower amount (10). The candidate genes were identified by excluding genes with less than 10 Reads Per Kilobase of transcript, per Million mapped for the no peptide condition, less than a 4-fold increase in expression in the high versus no peptide conditions, and less than a 2-fold increase in expression in the high versus low peptide conditions. Of the 12 most abundantly expressed genes, Nr4a1 and Irf4 were excluded from further analysis due to their well-established roles in T cell differentiation (8, 50–55), while CD25 was used as a positive control.
Expression of the candidate genes was validated by quantitative PCR (qPCR) analysis of B3K506 and B3K508 T cells two d after Lm-P5R infection. Of the nine genes analyzed, seven were induced to a greater extent in B3K506 than B3K508 T cells indicating a dependence on TCR signaling (Fig. 5C). The role of these seven genes in Th cell differentiation was determined by disrupting each gene in B3K508 cells using CRISPR/Cas9-mediated gene targeting. Cas9-expressing B3K508 cells were transduced with a retrovirus encoding the fluorescent protein mAmetrine and guide RNAs (gRNA) targeting the candidate genes or the bacterial gene LacZ as a control. The transduced T cells were then transferred into B6 mice before intravenous infection of the mice with Lm bacteria expressing 3K peptide (Lm-3K). The transferred gRNA+ T cells were identified based on CD90.1 and mAmetrine expression and evaluated for adoption of the Th1, Tfh, and uncommitted cell lineages (Fig. 5D and 5E). The expansion of gRNA+ B3K508 cells in the recipient mice was unaffected in the majority of cases, with the exceptions of Eef1e1 and Srm, loss of which resulted in reduced or enhanced expansion, respectively (Fig. 5F). As expected, targeting the Il2ra gene encoding CD25 reduced Th1 differentiation relative to the LacZ control population (Fig. 5G). Of the candidate genes, only Eef1e1 (encoding eukaryotic translation elongation factor 1 epsilon-1 (Eef1e1), also known as AIMP3 and p18) and Gbp2 (encoding GTP-binding protein 2 (Gbp2)) had significant effects on T cell differentiation. Ablation of Eef1e1 reduced Th1 differentiation (Fig. 5G) while targeting Gbp2 led to enhanced Th1 differentiation. These results demonstrate that TCR-driven Eef1e1 promotes Th1 differentiation whereas Gbp2 inhibits this process.
Discussion
Our results indicate that uncommitted effector Th cells are induced by low affinity TCR-p:MHCII interactions. Tfh cells then become the preferred effector cells as TCR affinity increases, until Th1 and Th17 cells become predominant at higher affinities. The facts that XCR1+ DCs are potent producers of IL-12 (18) but relatively poor producers of p:MHCII complexes, while IL-2 consuming SIRP⍺+ DCs have the opposite properties suggested an extrinsic explanation of these TCR affinity-based effects on Th cell differentiation. XCR1+ DCs could display fewer p:MHCII complexes than SIRP⍺+ DCs creating a situation where Th cells with low affinity TCRs would be unable to interact with XCR1+ DCs thereby receive IL-12 and become Th1 cells, but could still interact with SIRP⍺+ DCs to become Tfh cells. There was potential for this model to be correct because we confirmed that XCR1+ DCs present fewer P5R:I-Ab complexes than SIRP⍺+ DCs two d after Lm-P5R infection, the time frame when Tfh differentiation is initiated. However, we found medium and high affinity Th cells equally interacted with each DC subset, 20% with SIRP⍺+ DCs and 15% with XCR1+ DCs. Thus, the frequency of interaction with XCR1+ DCs cannot explain the greater tendency of high affinity Th cells to produce non-Tfh cells in this system.
We found better evidence in favor of a Th cell intrinsic mechanism. The CD25 component of the IL-2 receptor was an attractive mediator of such an effect because its expression is proportional to TCR signaling (7, 10, 13, 14) and biases differentiation by inhibiting Tfh formation through Blimp1 induction (12). We found that high TCR affinity positively correlated with stronger TCR signaling, maintenance of CD25 expression, and Th1 differentiation. Longer expression of the IL-2 receptor would provide more time for IL-2 binding and induction of the STAT5-regulated Tfh repressor Blimp1 (11, 12). DiToro et al. (56) recently showed that the capacity to produce IL-2 is also a key factor in the Tfh/Th1 bifurcation process. They found that Th1 cells are derived from precursors that express CD25 and experience strong STAT5 signaling but do not produce their own IL-2, while the Tfh cells are derived from precursors that produce the IL-2 used by the CD25hi Th1 precursors but do not respond to IL-2 because of low IL-2 receptor expression. Our result that Th1 cells express more CD25 than Tfh cells is consistent with this model. TCR affinity likely influences the non-Tfh/Tfh balance by determining how many precursor cells respond to IL-2. This process may also operate during Type 2 immune responses since the population of house dust mite allergen p:MHCII-specific Th cells induced by allergen inhalation is comprised of Th2 and Tfh cells (57) and the Th2 cells are CD25-dependent (58–60).
Our results indicate that the Eef1e1 is another TCR signal strength responsive driver of the Th1 fate. Eef1e1 binds to Lamin A (61) and overexpression studies performed in cell lines demonstrated that Eef1e1 facilitates Lamin A ubiquitination and degradation via the ubiquitin ligase, Siah1 (62). Interestingly, Lamin A-deficient T cells have reduced in vivo Th1 differentiation (63), while Siah1 promotes Th17 cell differentiation in vitro (64). Thus, strong TCR signaling may favor the differentiation of non-Tfh cells by engaging a pathway involving Eef1e1, Lamin A, and Siah1. The Th1-promoting effects of Eef1e1 may also be related to the role that we identified for this molecule in proliferation. Craft and colleagues showed that IL-2-mediated activation pathways associated with cellular proliferation promote Th1 over Tfh cell differentiation (65). Eef1e1, which interacts with ATM (66), a protein critical for TCR signaling-induced proliferation (67) may contribute to this non-Tfh-promoting process.
Increased expression of Eef1e1, CD25, and IRF4 and induction of downstream pathways is a plausible explanation for the increase in Th1 formation that occurs as TCR affinity increases from moderate to high levels. This mechanism, however, cannot explain the increase in Tfh formation that occurs as TCR affinity increases from low to moderate levels. Our results indicate that Gbp2, a TCR-induced protein that promoted Tfh cell formation, could be involved in this transition. Gbp2 is associated with reduced mitochondrial fission (68), which is important for aerobic glycolysis (69–71) and operates at low levels in Tfh cells (65, 72). Therefore, Gbp2 may promote Tfh formation by keeping aerobic glycolysis in check. An alternative hypothesis is that Gbp2 represses Th1 differentiation and high TCR affinity allows T cells to overcome this inhibition.
Together, our results suggest a general model in which low TCR affinity and signaling induces Gbp2 thereby promoting Tfh differentiation, while high TCR affinity and signaling drives Eef1e1 and CD25, which represses the Tfh fate and promotes non-Tfh cell formation. The type of non-Tfh cells would then depend on the type of cytokines produced by the innate immune system. For example, intranasal infection, which elicits IL-6 and TGF-β production by cervical lymph node DCs (73), would favor Th17 differentiation, while intravenous infection, which stimulates IL-12 production by splenic XCR1+ DCs (18), would bias towards Th1 differentiation. This mechanism would ensure that polyclonal populations of epitope-specific naïve Th cells, which always contain a spectrum TCR affinities (26), generate effector cells capable of promoting humoral and cellular immunity suited to clearance of the microbe. The tendency of moderate affinity T cells to become Tfh cells may foster affinity maturation by only allowing B cells with the highest affinity immunoglobulins to garner T cell help, while diversion of high affinity T cells to become myeloid cell-helpers would ensure that the helped cells achieve the optimal state of microbicidal activity.
Supplementary Material
Acknowledgments
We thank J. Walter, C. Ellwood, the University of Minnesota Center for Immunology Imaging Core, the University of Minnesota Imaging Centers and the University of Minnesota Flow Cytometry Resource for technical assistance, D.F. Voytas and J.J. Belanto for helping generate the CRISPR/Cas9 system, and M.Y. Gerner for suggestions on histo-cytometry.
This work was supported by US National Institutes of Health grants R01 AI039614 and P01 AI35296 to M.K.J., T32 AI083196 and T32 AI007313 to D. I. K., and R01 AI106791 and P01 AI35296 to B.T.F.
Non-standard Abbreviations:
- B6
C57BL/6
- DC
dendritic cell
- DT
diphtheria toxin
- Lm
Listeria monocytogenes
- p:MHCII
peptide bound to MHCII
- pS6
phosphorylated S6 kinase
- Tfh
T follicular helper cell
- Tg
transgenic
- Th1
Type 1 Th cell
- Th17
Type 17 Th cell
- WT
wild-type
- gMFI
geometric MFI
References
- 1.van Panhuys N 2016. TCR Signal Strength Alters T-DC Activation and Interaction Times and Directs the Outcome of Differentiation. Front. Immunol 7: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meredith MM, Liu K, Darrasse-Jeze G, Kamphorst AO, Schreiber HA, Guermonprez P, Idoyaga J, Cheong C, Yao KH, Niec RE, and Nussenzweig MC. 2012. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med 209: 1153–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mempel TR, Henrickson SE, and Von Andrian UH. 2004. T-cell priming by dendritic cells in lymph nodes occurs in three distinct phases. Nature 427: 154–159. [DOI] [PubMed] [Google Scholar]
- 4.Keck S, Schmaler M, Ganter S, Wyss L, Oberle S, Huseby ES, Zehn D, and King CG. 2014. Antigen affinity and antigen dose exert distinct influences on CD4 T-cell differentiation. Proc. Natl. Acad. Sci. U.S.A 111: 14852–14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tubo NJ, Pagan AJ, Taylor JJ, Nelson RW, Linehan JL, Ertelt JM, Huseby ES, Way SS, and Jenkins MK. 2013. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell 153: 785–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Panhuys N, Klauschen F, and Germain RN. 2014. T-cell-receptor-dependent signal intensity dominantly controls CD4(+) T cell polarization In Vivo. Immunity 41: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Snook JP, Kim C, and Williams MA. 2018. TCR signal strength controls the differentiation of CD4(+) effector and memory T cells. Sci. Immunol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krishnamoorthy V, Kannanganat S, Maienschein-Cline M, Cook SL, Chen J, Bahroos N, Sievert E, Corse E, Chong A, and Sciammas R. 2017. The IRF4 Gene Regulatory Module Functions as a Read-Write Integrator to Dynamically Coordinate T Helper Cell Fate. Immunity 47: 481–497 e487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pepper M, Pagan AJ, Igyarto BZ, Taylor JJ, and Jenkins MK. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity 35: 583–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Allison KA, Sajti E, Collier JG, Gosselin D, Troutman TD, Stone EL, Hedrick SM, and Glass CK. 2016. Affinity and dose of TCR engagement yield proportional enhancer and gene activity in CD4+ T cells. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, Lao C, and Crotty S. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity 34: 932–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnston RJ, Choi YS, Diamond JA, Yang JA, and Crotty S. 2012. STAT5 is a potent negative regulator of TFH cell differentiation. J. Exp. Med 209: 243–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tkach KE, Barik D, Voisinne G, Malandro N, Hathorn MM, Cotari JW, Vogel R, Merghoub T, Wolchok J, Krichevsky O, and Altan-Bonnet G. 2014. T cells translate individual, quantal activation into collective, analog cytokine responses via time-integrated feedbacks. Elife 3: e01944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Voisinne G, Nixon BG, Melbinger A, Gasteiger G, Vergassola M, and Altan-Bonnet G. 2015. T Cells Integrate Local and Global Cues to Discriminate between Structurally Similar Antigens. Cell Rep. 11: 1208–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ballesteros-Tato A, Leon B, Graf BA, Moquin A, Adams PS, Lund FE, and Randall TD. 2012. Interleukin-2 inhibits germinal center formation by limiting T follicular helper cell differentiation. Immunity 36: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilliams M, Dutertre CA, Scott CL, McGovern N, Sichien D, Chakarov S, Van Gassen S, Chen J, Poidinger M, De Prijck S, Tavernier SJ, Low I, Irac SE, Mattar CN, Sumatoh HR, Low GHL, Chung TJK, Chan DKH, Tan KK, Hon TLK, Fossum E, Bogen B, Choolani M, Chan JKY, Larbi A, Luche H, Henri S, Saeys Y, Newell EW, Lambrecht BN, Malissen B, and Ginhoux F. 2016. Unsupervised High-Dimensional Analysis Aligns Dendritic Cells across Tissues and Species. Immunity 45: 669–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, Steinman RM, and Nussenzweig MC. 2007. Differential antigen processing by dendritic cell subsets in vivo. Science 315: 107–111. [DOI] [PubMed] [Google Scholar]
- 18.Alexandre YO, Ghilas S, Sanchez C, Le Bon A, Crozat K, and Dalod M. 2016. XCR1+ dendritic cells promote memory CD8+ T cell recall upon secondary infections with Listeria monocytogenes or certain viruses. J. Exp. Med 213: 75–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briseno CG, Satpathy AT, Davidson J. T. t., Ferris ST, Durai V, Bagadia P, O’Connor KW, Theisen DJ, Murphy TL, and Murphy KM. 2018. Notch2-dependent DC2s mediate splenic germinal center responses. Proc. Natl. Acad. Sci. U.S.A.115: 10726–10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huseby ES, White J, Crawford F, Vass T, Becker D, Pinilla C, Marrack P, and Kappler JW. 2005. How the T cell repertoire becomes peptide and MHC specific. Cell 122: 247–260. [DOI] [PubMed] [Google Scholar]
- 21.Muzaki AR, Tetlak P, Sheng J, Loh SC, Setiagani YA, Poidinger M, Zolezzi F, Karjalainen K, and Ruedl C. 2016. Intestinal CD103(+)CD11b(−) dendritic cells restrain colitis via IFN-gamma-induced anti-inflammatory response in epithelial cells. Mucosal Immunol. 9: 336–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ertelt JM, Rowe JH, Johanns TM, Lai JC, McLachlan JB, and Way SS. 2009. Selective priming and expansion of antigen-specific Foxp3- CD4+ T cells during Listeria monocytogenes infection. J. Immunol 182: 3032–3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pepper M, Linehan JL, Pagan AJ, Zell T, Dileepan T, Cleary PP, and Jenkins MK. 2010. Different routes of bacterial infection induce long-lived TH1 memory cells and short-lived TH17 cells. Nat. Immunol 11: 83–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heaton NS, Sachs D, Chen CJ, Hai R, and Palese P. 2013. Genome-wide mutagenesis of influenza virus reveals unique plasticity of the hemagglutinin and NS1 proteins. Proc. Natl. Acad. Sci. U.S.A. 110: 20248–20253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Waring BM, Sjaastad LE, Fiege JK, Fay EJ, Reyes I, Moriarity B, and Langlois RA. 2018. MicroRNA-Based Attenuation of Influenza Virus across Susceptible Hosts. J. Virol 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moon JJ, Chu HH, Pepper M, McSorley SJ, Jameson SC, Kedl RM, and Jenkins MK. 2007. Naive CD4(+) T cell frequency varies for different epitopes and predicts repertoire diversity and response magnitude. Immunity 27: 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kotov DI, Pengo T, Mitchell JS, Gastinger MJ, and Jenkins MK. 2019. Chrysalis: A New Method for High-Throughput Histo-Cytometry Analysis of Images and Movies. J. Immunol 202: 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, and Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13: 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schmittgen TD, and Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 30.Shaner NC, Lambert GG, Chammas A, Ni Y, Cranfill PJ, Baird MA, Sell BR, Allen JR, Day RN, Israelsson M, Davidson MW, and Wang J. 2013. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nat. Methods 10: 407–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ai HW, Hazelwood KL, Davidson MW, and Campbell RE. 2008. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat. Methods 5: 401–403. [DOI] [PubMed] [Google Scholar]
- 32.Chen R, Belanger S, Frederick MA, Li B, Johnston RJ, Xiao N, Liu YC, Sharma S, Peters B, Rao A, Crotty S, and Pipkin ME. 2014. In vivo RNA interference screens identify regulators of antiviral CD4(+) and CD8(+) T cell differentiation. Immunity 41: 325–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie K, Minkenberg B, and Yang Y. 2015. Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. Proc. Natl. Acad. Sci. U.S.A. 112: 3570–3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu L, Zhao L, Gao Y, Xu J, and Han R. 2017. Empower multiplex cell and tissue-specific CRISPR-mediated gene manipulation with self-cleaving ribozymes and tRNA. Nucleic Acids Res. 45: e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dong F, Xie K, Chen Y, Yang Y, and Mao Y. 2017. Polycistronic tRNA and CRISPR guide-RNA enables highly efficient multiplexed genome engineering in human cells. Biochem Biophys. Res. Commun 482: 889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finn RD, Attwood TK, Babbitt PC, Bateman A, Bork P, Bridge AJ, Chang HY, Dosztanyi Z, El-Gebali S, Fraser M, Gough J, Haft D, Holliday GL, Huang H, Huang X, Letunic I, Lopez R, Lu S, Marchler-Bauer A, Mi H, Mistry J, Natale DA, Necci M, Nuka G, Orengo CA, Park Y, Pesseat S, Piovesan D, Potter SC, Rawlings ND, Redaschi N, Richardson L, Rivoire C, Sangrador-Vegas A, Sigrist C, Sillitoe I, Smithers B, Squizzato S, Sutton G, Thanki N, Thomas PD, Tosatto SC, Wu CH, Xenarios I, Yeh LS, Young SY, and Mitchell AL. 2017. InterPro in 2017-beyond protein family and domain annotations. Nucleic Acids Res. 45: D190–D199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cermak T, Curtin SJ, Gil-Humanes J, Cegan R, Kono TJY, Konecna E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, and Voytas DF. 2017. A Multipurpose Toolkit to Enable Advanced Genome Engineering in Plants. Plant Cell 29: 1196–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choi YS, Yang JA, Yusuf I, Johnston RJ, Greenbaum J, Peters B, and Crotty S. 2013. Bcl6 expressing follicular helper CD4 T cells are fate committed early and have the capacity to form memory. J. Immunol 190: 4014–4026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kearney ER, Pape KA, Loh DY, and Jenkins MK. 1994. Visualization of peptide-specific T cell immunity and peripheral tolerance induction in vivo. Immunity 1: 327–339. [DOI] [PubMed] [Google Scholar]
- 40.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, and Glimcher LH. 2000. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 100: 655–669. [DOI] [PubMed] [Google Scholar]
- 41.Breitfeld D, Ohl L, Kremmer E, Ellwart J, Sallusto F, Lipp M, and Forster R. 2000. Follicular B helper T cells express CXC chemokine receptor 5, localize to B cell follicles, and support immunoglobulin production. J. Exp. Med 192: 1545–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Forster R, Emrich T, Kremmer E, and Lipp M. 1994. Expression of the G-protein--coupled receptor BLR1 defines mature, recirculating B cells and a subset of T-helper memory cells. Blood 84: 830–840. [PubMed] [Google Scholar]
- 43.Schaerli P, Willimann K, Lang AB, Lipp M, Loetscher P, and Moser B. 2000. CXC chemokine receptor 5 expression defines follicular homing T cells with B cell helper function. J. Exp. Med 192: 1553–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, and Littman DR. 2006. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell 126: 1121–1133. [DOI] [PubMed] [Google Scholar]
- 45.Bachem A, Hartung E, Guttler S, Mora A, Zhou X, Hegemann A, Plantinga M, Mazzini E, Stoitzner P, Gurka S, Henn V, Mages HW, and Kroczek RA. 2012. Expression of XCR1 Characterizes the Batf3-Dependent Lineage of Dendritic Cells Capable of Antigen Cross-Presentation. Front. Immunol 3: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tussiwand R, Lee WL, Murphy TL, Mashayekhi M, Kc W, Albring JC, Satpathy AT, Rotondo JA, Edelson BT, Kretzer NM, Wu X, Weiss LA, Glasmacher E, Li P, Liao W, Behnke M, Lam SS, Aurthur CT, Leonard WJ, Singh H, Stallings CL, Sibley LD, Schreiber RD, and Murphy KM. 2012. Compensatory dendritic cell development mediated by BATF-IRF interactions. Nature 490: 502–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katzman SD, O’Gorman WE, Villarino AV, Gallo E, Friedman RS, Krummel MF, Nolan GP, and Abbas AK. 2010. Duration of antigen receptor signaling determines T-cell tolerance or activation. Proc. Natl. Acad. Sci. U.S.A. 107: 18085–18090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Lu E, Yi T, and Cyster JG. 2016. EBI2 augments Tfh cell fate by promoting interaction with IL-2-quenching dendritic cells. Nature 533: 110–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krishnaswamy JK, Gowthaman U, Zhang B, Mattsson J, Szeponik L, Liu D, Wu R, White T, Calabro S, Xu L, Collet MA, Yurieva M, Alsen S, Fogelstrand P, Walter A, Heath WR, Mueller SN, Yrlid U, Williams A, and Eisenbarth SC. 2017. Migratory CD11b(+) conventional dendritic cells induce T follicular helper cell-dependent antibody responses. Sci. Immunol 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brustle A, Heink S, Huber M, Rosenplanter C, Stadelmann C, Yu P, Arpaia E, Mak TW, Kamradt T, and Lohoff M. 2007. The development of inflammatory T(H)-17 cells requires interferon-regulatory factor 4. Nat. Immunol 8: 958–966. [DOI] [PubMed] [Google Scholar]
- 51.Huber M, Brustle A, Reinhard K, Guralnik A, Walter G, Mahiny A, von Low E, and Lohoff M. 2008. IRF4 is essential for IL-21-mediated induction, amplification, and stabilization of the Th17 phenotype. Proc. Natl. Acad. Sci. U.S.A 105: 20846–20851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Man K, Miasari M, Shi W, Xin A, Henstridge DC, Preston S, Pellegrini M, Belz GT, Smyth GK, Febbraio MA, Nutt SL, and Kallies A. 2013. The transcription factor IRF4 is essential for TCR affinity-mediated metabolic programming and clonal expansion of T cells. Nat. Immunol 14: 1155–1165. [DOI] [PubMed] [Google Scholar]
- 53.Mahnke J, Schumacher V, Ahrens S, Kading N, Feldhoff LM, Huber M, Rupp J, Raczkowski F, and Mittrucker HW. 2016. Interferon Regulatory Factor 4 controls TH1 cell effector function and metabolism. Sci. Rep 6: 35521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fassett MS, Jiang W, D’Alise AM, Mathis D, and Benoist C. 2012. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion. Proc. Natl. Acad. Sci. U.S.A 109: 3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Caton AJ, Kropf E, Simons DM, Aitken M, Weissler KA, and Jordan MS. 2014. Strength of TCR signal from self-peptide modulates autoreactive thymocyte deletion and Foxp3(+) Treg-cell formation. Eur. J. Immunol 44: 785–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.DiToro D, Winstead CJ, Pham D, Witte S, Andargachew R, Singer JR, Wilson CG, Zindl CL, Luther RJ, Silberger DJ, Weaver BT, Kolawole EM, Martinez RJ, Turner H, Hatton RD, Moon JJ, Way SS, Evavold BD, and Weaver CT. 2018. Differential IL-2 expression defines developmental fates of follicular versus nonfollicular helper T cells. Science 361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hondowicz BD, An D, Schenkel JM, Kim KS, Steach HR, Krishnamurty AT, Keitany GJ, Garza EN, Fraser KA, Moon JJ, Altemeier WA, Masopust D, and Pepper M. 2016. Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma. Immunity 44: 155–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhu J, Cote-Sierra J, Guo L, and Paul WE. 2003. Stat5 activation plays a critical role in Th2 differentiation. Immunity 19: 739–748. [DOI] [PubMed] [Google Scholar]
- 59.Cote-Sierra J, Foucras G, Guo L, Chiodetti L, Young HA, Hu-Li J, Zhu J, and Paul WE. 2004. Interleukin 2 plays a central role in Th2 differentiation. Proc. Natl. Acad. Sci. U.S.A. 101: 3880–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao W, Schones DE, Oh J, Cui Y, Cui K, Roh TY, Zhao K, and Leonard WJ. 2008. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat. Immunol 9: 1288–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tao Y, Fang P, Kim S, Guo M, Young NL, and Marshall AG. 2017. Mapping the contact surfaces in the Lamin A:AIMP3 complex by hydrogen/deuterium exchange FT-ICR mass spectrometry. PLoS One 12: e0181869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oh YS, Kim DG, Kim G, Choi EC, Kennedy BK, Suh Y, Park BJ, and Kim S. 2010. Downregulation of lamin A by tumor suppressor AIMP3/p18 leads to a progeroid phenotype in mice. Aging Cell 9: 810–822. [DOI] [PubMed] [Google Scholar]
- 63.Toribio-Fernandez R, Zorita V, Rocha-Perugini V, Iborra S, Martinez Del Hoyo G, Chevre R, Dorado B, Sancho D, Sanchez-Madrid F, Andres V, and Gonzalez-Granado JM. 2018. Lamin A/C augments Th1 differentiation and response against vaccinia virus and Leishmania major. Cell Death Dis. 9: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matsui-Hasumi A, Sato Y, Uto-Konomi A, Yamashita S, Uehori J, Yoshimura A, Yamashita M, Asahara H, Suzuki S, and Kubo M. 2017. E3 ubiquitin ligases SIAH1/2 regulate hypoxia-inducible factor-1 (HIF-1)-mediated Th17 cell differentiation. Int. Immunol 29: 133–143. [DOI] [PubMed] [Google Scholar]
- 65.Ray JP, Staron MM, Shyer JA, Ho PC, Marshall HD, Gray SM, Laidlaw BJ, Araki K, Ahmed R, Kaech SM, and Craft J. 2015. The Interleukin-2-mTORc1 Kinase Axis Defines the Signaling, Differentiation, and Metabolism of T Helper 1 and Follicular B Helper T Cells. Immunity 43: 690–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Park BJ, Kang JW, Lee SW, Choi SJ, Shin YK, Ahn YH, Choi YH, Choi D, Lee KS, and Kim S. 2005. The haploinsufficient tumor suppressor p18 upregulates p53 via interactions with ATM/ATR. Cell 120: 209–221. [DOI] [PubMed] [Google Scholar]
- 67.Bagley J, Singh G, and Iacomini J. 2007. Regulation of oxidative stress responses by ataxia-telangiectasia mutated is required for T cell proliferation. J. Immunol 178: 4757–4763. [DOI] [PubMed] [Google Scholar]
- 68.Zhang J, Zhang Y, Wu W, Wang F, Liu X, Shui G, and Nie C. 2017. Guanylate-binding protein 2 regulates Drp1-mediated mitochondrial fission to suppress breast cancer cell invasion. Cell Death Dis. 8: e3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hagenbuchner J, Kuznetsov AV, Obexer P, and Ausserlechner MJ. 2013. BIRC5/Survivin enhances aerobic glycolysis and drug resistance by altered regulation of the mitochondrial fusion/fission machinery. Oncogene 32: 4748–4757. [DOI] [PubMed] [Google Scholar]
- 70.Salabei JK, and Hill BG. 2013. Mitochondrial fission induced by platelet-derived growth factor regulates vascular smooth muscle cell bioenergetics and cell proliferation. Redox Biol. 1: 542–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Guido C, Whitaker-Menezes D, Lin Z, Pestell RG, Howell A, Zimmers TA, Casimiro MC, Aquila S, Ando S, Martinez-Outschoorn UE, Sotgia F, and Lisanti MP. 2012. Mitochondrial fission induces glycolytic reprogramming in cancer-associated myofibroblasts, driving stromal lactate production, and early tumor growth. Oncotarget 3: 798–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Oestreich KJ, Read KA, Gilbertson SE, Hough KP, McDonald PW, Krishnamoorthy V, and Weinmann AS. 2014. Bcl-6 directly represses the gene program of the glycolysis pathway. Nat. Immunol 15: 957–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Linehan JL, Dileepan T, Kashem SW, Kaplan DH, Cleary P, and Jenkins MK. 2015. Generation of Th17 cells in response to intranasal infection requires TGF-beta1 from dendritic cells and IL-6 from CD301b+ dendritic cells. Proc. Natl. Acad. Sci. U.S.A 112: 12782–12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.