Abstract
Alcohol intake increases the risk of cancer development. Approximately 3.6% human cancers worldwide derive from chronic alcohol drinking, including oral, liver, breast and other organs. Our studies in vivo and in vitro have demonstrated that diluted ethanol increase RNA Pol III gene transcription and promotes cell proliferation and transformation, as well as tumor formation. However, it is unclear about the effect of red wines on the human cancer cells. In present study, we investigated the roles of red wine in human cancer cell growth, colony formation and RNA Pol III gene transcription. Low concentration (12.5mM to 25mM) of ethanol enhances cell proliferation of breast and esophageal cancer lines, whereas its higher concentration (100mM to 200mM) slightly decreases the rates. In contrast, red wines significantly repress cell proliferation of different human cancer lines from low dose to high dose. The results reveal that the red wine also inhibit colony formation of human breast cancer and esophageal carcinoma cells. The effects of repression on different human cancer lines is in a dose-dependent manner. Further analysis indicates that ethanol increases RNA Pol III gene transcription, whereas the red wines significantly reduce transcription of the genes. Interestingly, the effects of mature wine (brick red) on cancer cell phenotypes are much stronger than young wine (intense violet). Together, these new findings suggest that red wines may contain some bioactive components, which are able to inhibit human cancer cell growth and colony formation.
Keywords: Red wine, ethanol, human cancer, cell growth, colony formation, Pol III genes
1. INTRODUCTION
Emerging evidences have indicated that alcohol consumption is consistently associated with increased risk of human cancers (Shi et al, 2017; Yi et al, 2018). Target sites for alcohol-related carcinogenesis include the breast, esophagus, liver and multiple additional organs (Connor J, 2017; Scoccaianti et al, 2013; Allen et al, 2009). Alcohol has been classified as carcinogenic to humans by IARC (the International Agency for Research on Cancer) (Shi et al, 2017; IARC, 2011; Coglian et al, 2011). The epidemiological studies have shown that the relative increase in risk ranges from 5%-10% (up to 1 drink or 10 gram/day) to 40%-50% (3 or more drinks/day) (Singletary et al, 1995; Watabiki et al, 2000). Nucleolar hypertrophy is a consistent cytological feature of cancer cells, where RNA polymerase I and III-dependent genes (RNA Pol III genes) are transcribed (White R, 2004). Upregulation of Pol III genes is tightly linked to cell proliferation, cell transformation, as well as tumor formation (Johnson et al, 2008; Zhong et al, 2009; Zhong et al, 2007; Zhong et al, 2011; Zhang et al, 2013; Zhang et al, 2011). Brf1 (TFIIB-related factor 1) is a transcription factor and specifically regulates Pol III gene transcription. Our studies in vivo and in vitro have demonstrated that diluted ethanol increases Brf1 expression to enhance Pol III gene transcription, resulting in alteration of the cellular phenotypes (Zhong et al, 2009, Zhang et al, 2013, Zhong et al, 2016). This suggests that ethanol-caused upregulation of Brf1 and Pol III genes plays important roles in cell proliferation and cancer development. Our recent studies have demonstrated that liquor spirits (white wine) have the potential inhibiting human cancer growth (Yi et al, 2018). While red wine is more popular alcoholic beverage. However, it is unclear whether red wine affect these phenotypes of human cancer cells.
Red wine is a type of wine made from dark-colored (black) grape varieties. In terms of the actual color of the wine, red wines are divided into two types: mature wines (brick red) and young wines (intense violet). Red wine phenolics contain two major groups: flavonoids and non-flavonoids. The main flavonoid compounds presented in red wine include several classes, such as flavanols [(epi) catechin], flavonols (e.g., myricetin andquercetin) and anthocyanins (e.g., malvidin-3-glucoside), while non-flavonoid compounds presented in red wine comprise phenolic acids, phenols and stilbenes. Renaud et al proposed that the moderate daily consumption of red wine was as a contributing factor to the observed lower incidence of coronary heart disease (Renaud and Lorgeril, 1992). Epidemiological studies have indicated a positive association between red wine ingestion and human health. Studies from diverse populations have revealed that individuals who usually consume moderate amounts of wine experience a 20% to 30% reduction in all-cause mortality, particularly cardiovascular mortality (German and Walzem, 2000; Ruf, 2003). However, a recent study shows that heavy beer and possible sprits consumption is associated with the risk of aggressive prostate cancer, while no dose-dependent relationship was found for red or white wine (Papa et al, 2017). Our studies demonstrated that diluted ethanol-feeding mouse promoted liver tumor development (Zhong et al, 2011). While a previous study indicated that the workers, who tested liquor and sprits to check its quality during the course of production, were not found hepatic fibrosis, cirrhosis and HCC in this special crowd, compared to those of other beverage consumptions (Wu et al, 2002). Our recent studies have demonstrated that liquor spirits are able to repress cell growth and colony formation of human cancer lines (Yi, et al, 2018). These studies imply that wines may play a role in inhibiting cancer development. However, it is known that alcohol intake increases the risk of human cancers (Petri et al, 2004; Singletary and Gapstur, 2001; Chen et al, 2011; Seitz et al, 2012; Dermark-Wahenefien, 2013). These contradictory results mentioned above need to further be studies. To investigate whether red wine plays a role in cancer cell growth, we randomly purchased three commercial red wines, two mature wines and one young wine, from a supermarket. We treated the cells of different human cancer lines with the red wines to observe changes in cell proliferation, colony formation and RNA Pol III gene transcription. The results reveal that all three red wines decrease the rates of cell proliferation, compared to diluted ethanol treatment. The red wines also repress colony formation of human breast cancer and esophageal carcinoma cells and inhibit Pol III gene transcription. The effects of mature wine (brick red) on these cancer cell phenotypes are much stronger than young wine (intense violet). These studies suggest that red wine may contain some bioactive components, which are able to inhibit Pol III gene activity, cell growth and colony formation of human cancer cells.
2. MATERIALS AND METHODS
2.1. Cell lines and reagents
Human breast adenocarcinoma cell line MCF-7 (HTB-22), human colon carcinoma cell line RKO (CRL-2577), human colorectal carcinoma cell line HCT-116 (CRL-247), human colorectal adenocarcinoma cell line SW-480 (CCL-228) were from ATCC (Manassas, VA, USA). Human esophageal squamous carcinoma cell line KYSE-510 (ACC 374) was from DSMZ (Braunschweig, Germany). These cell culture media were from Life Technologies Inc (San Diego, CA, USA).
MTT reagent (AR1156) were from Boster Biol. Tech. (Pleasanton, CA, USA). IQ TM SYBR Green (Cat #:7200033) was from Bio-Rad Laboratories (Hercules, CA, USC). The red wines include two mature wines (#1 and #3) and one young wine (#2), which were randomly purchased from a supermarket. The three red wines were labeled with #1 to #3, which contain actual ethanol concentration at 15% (v/v), 13% (v/v) and 13.5 (v/v), respectively. Ethanol (200 proof) was from Sigma-Aldrich (Hayward, CA, USA).
2.2. Cell proliferation and cell death by red wines
Approximately 2 ×103 cells per ml of human cancer cells (MCF-7, KYSE-510, A-549, RKO, HCT-116, SW-480) were seeded in six-well plates in triplicate and grew in the media containing ethanol or red wines for 6 days. Cells were treated with diluted ethanol or red wines (#1, #2 or #3) at corresponding actual ethanol concentration (mM) and cellular morphology was analyzed by microscopy using a Nikon Eclipse TE300 and Metamorph Program (Cell and Tissue Imaging Core of University of Southern California Research Center for Liver Diseases, P30 DK048522). Cells were assayed for viability and counted using a Coulter Counter as described previously and hemacytometer (Zhong et al, 2007, 2009).
The cells of human breast cancer line (MCF-7) and esophageal carcinoma line (KYSE-510) were seeded in six-well plates and grew up to 70% confluence in the culture media without any ethanol and the red wines. Then the cells were treated with the diluted ethanol or the red wines for 48 hours to determine whether cancer death appears by the treatment of red wines. The cell morphology was analyzed and cell numbers were counted as described above.
2.3. Cell Anchorage-independent growth.
MCF-7 cells and KYSE-510 cells (1 × 103 cells/well in 6 well plate) were suspended in 0.35% (w/v) agar in 10% FBS/DMEM/F12 or RIPM1640 media. Cells were fed fresh complete media with diluted ethanol or red wines (#1, #2 or #3) at corresponding actual ethanol concentration twice weekly. Colonies were counted 2-3 weeks or longer after plating as described previously (Zhang et al, 2013; Zhang et al, 2011; Zhong et al, 2016).
2.4. MTT assay
The procedure of MTT assay was followed the instruction of the MTT Kit (AR1156). MCF-7 and KYSE-510 cells were seeded into microplate and treated as mentioned above. Remove the medium and replace it with 100 μl of fresh culture medium for each well of a microplate. Add 10 μl of the MTT labeling reagent to each well including a negative control of 100 μl medium alone.Incubate the microplate at 37°C for 4 hours in the humidified chamber (5% CO2). Add 100 μl of formazan solubilization solution to each well and mix thoroughly and incubate the microplate at 37 °C for 18 hours in the humidified chamber (5% CO2). Measure the absorbance of the samples using Omega microplate reader at 570 nm (Cell Biology Core Laboratory of University of Southern California Research Center for Liver Diseases, P30 DK048522).
2.5. Real time quality PCR (RT-qPCR)
MCF-7 and KYSE-510 cells grew up to 85% confluence and starved in FBS-free media for 4 hours. The cells were treated with 25mM ethanol or the three red wines at corresponding actual ethanol concentration, respectively. Total RNA was isolated from cultured cells using single step extraction method with TRIzol reagent from Life Technologies Inc (San Diego, CA, USA). RNA samples were quantified and reverse-transcribed in a 20 μl reaction containing 1 x RT (reverse transcription) buffer. After first-strand cDNA synthesis, the cDNAs were diluted in DNase-free water and real time qPCR (RT-qPCR) were performed with specific primers and PCR reagent kits (Bio-Rad Labs) in the ABI prism 7700 Sequence Detection System. Precursor of tRNALeu and 5S rRNA transcripts were measured by RT-qPCR as described previously (Zhong et la, 2011).
3. RESULTS
3.1. Red wines decrease the rate of cell proliferation
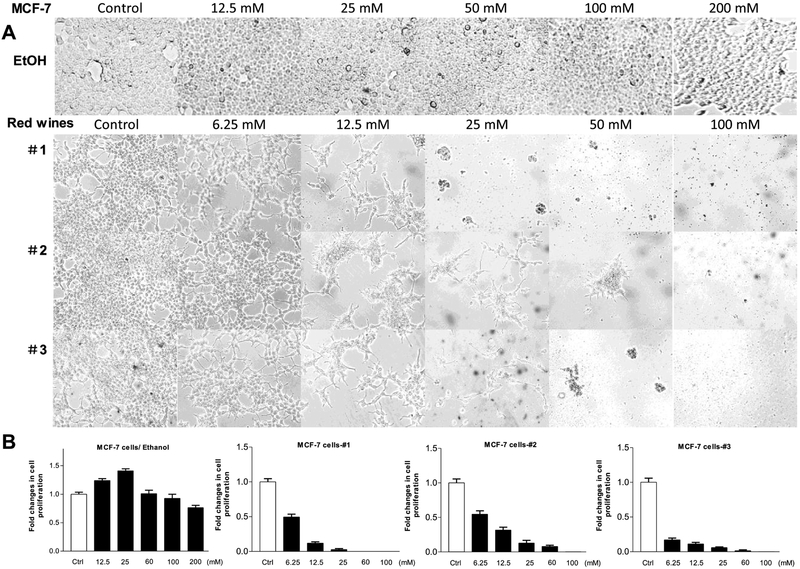
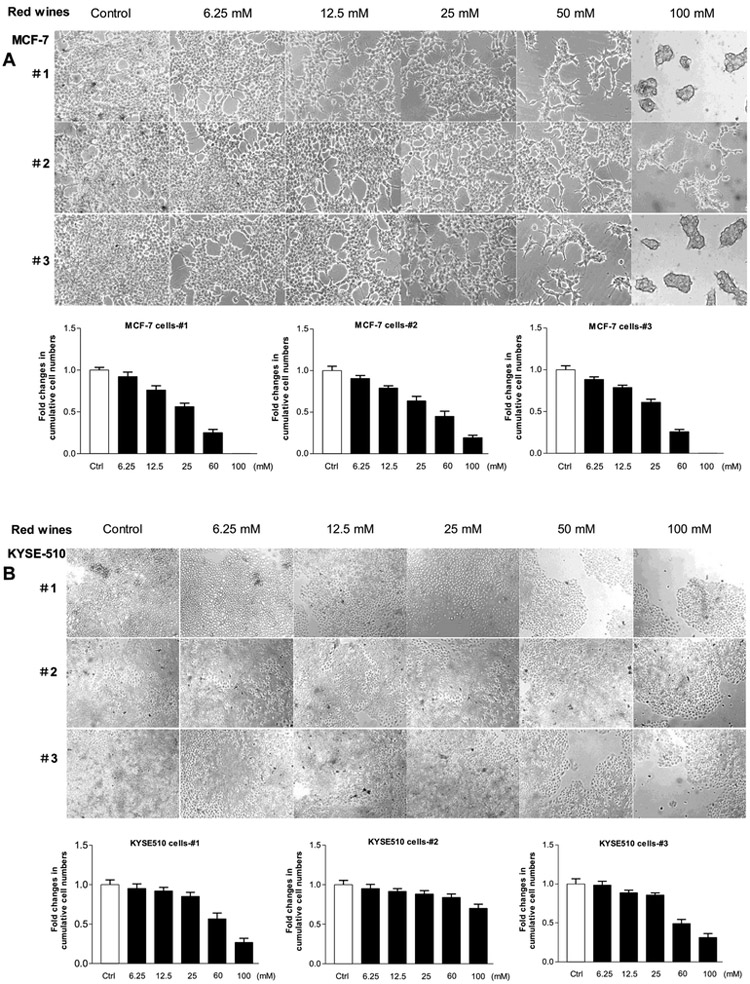
Multiple human cancer cell lines were treated with different amounts of the red wines, calculated the usage volumes of the red wines in same ethanol concentration (mM) referencing their actual ethanol percentages. Figure 1 shows that MCF-7 cells grew up almost full confluence at diluted ethanol-treated group from 12.5 mM to 200mM (Fig, 1A up). At low concentration from 12.5mM to 25mM, ethanol increased accumulative MCF-7 cells. Then, the numbers of cells were then slightly decreased with enhancing ethanol concentration (from 50 mM to 200 mM) (Fig. 1B, Left). In contrast, all three red wines (#1, #2 and #3), either mature wines (#1 and #3) or young wine (#2), reduce the rate of MCF-7 cell growth (Fig. 1A). The repression of the red wines is in a dose-dependent manner (from 6.25 mM to 100 mM). At the concentration of 25 mM, red wines #1 and #3 completely inhibits the cell growth of MCF-7 line, whereas ethanol at same dose increases MCF-7 cell growth (Fig. 1A up and B Left). The results indicate that the red wines are indeed able to inhibit the cell growth of human breast cancer line, MCF-7.
Fig. 1. Red wines repressed cell growth of human breast cancer line, MCF-7.
(A): MCF-7 cells were treated with different amounts of ethanol, red wines #1 (mature wine), #2 (young wine) or #3 (mature wine) for 6 days. The concentrations of the three red wines are equal to ethanol (mM/L) as indicated. The pictures were taken under microscope (Nikon, Eclipse, TE300). Original magnification × 100. (B): the viability and total cell numbers were counted after plated cells 6 days. The cells were treated serially by ethanol, red wine #1, #2 and #3 from left to right as indicated above. The results indicate that ethanol at low concentration increases the cell growth, while red wines dramatically decrease the rate of cell growth. The bars represent Mean ± SE of at least three independent determinations.
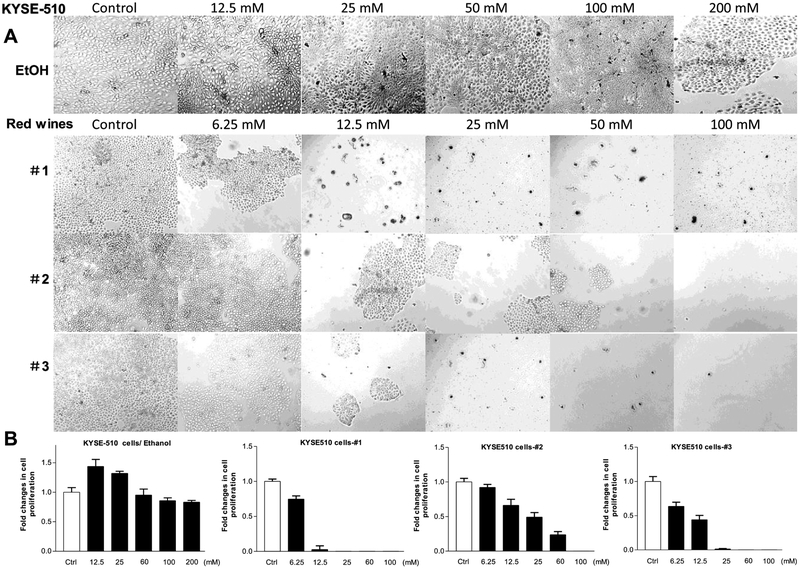
We also determined the effects of the red wines on human esophageal carcinoma cell line, KYSE-510. The results reveal that the accumulation of the KYSE-510 cells is increased by ethanol at 12.5 mM to 25 mM, the high peak is at 12.5 mM ethanol. Whereas higher concentration (up to 200 mM) of ethanol does not further increase the numbers of the KYSE-510 cells (Fig. 2A Up and 2B Left). However, the three red wines dramatically reduce the rate of KYSE-510 cell growth from 6.25 mM to 100 mM (Fig. 2). This cell line to the mature wines (#1 and #3) is much more sensitive than those to the young wine (#2) (Fig. 2).
Fig. 2. Repression of cell growth of human esophageal carcinoma KYSE-510 line.
(A), KYSE-510 cells were treated with different amounts of ethanol, red wines #1 (mature wine), #2 (young wine) or #3 (mature wine) for 6 days. The concentrations of the three red wines are equal to ethanol (mM/L) as indicated. The pictures were taken under microscope (Nikon, Eclipse, TE300). Original magnification × 100. (B): the viability and total cell numbers were counted after plated cells 6 days. The cells were treated as described in Fig. B. The results indicate that red wines markedly repress KYSE-510 cell growth. The bars represent Mean ± SE of at least three independent determinations.
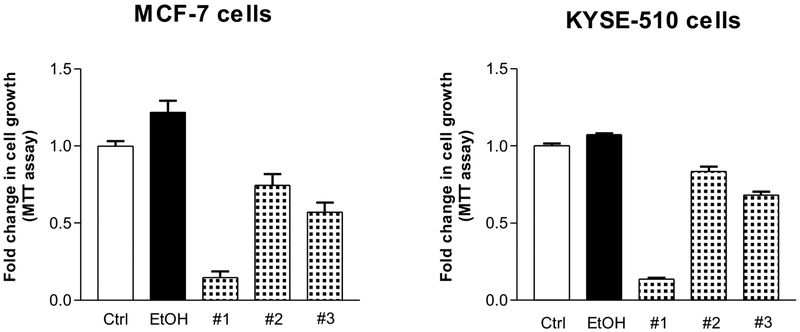
To confirm the effects of red wines on human cancer cells of breast and esophagus, we further performed MTT assay to determine changes in the cell proliferation. The results indicate that mature wine (#1) dramatically decreased both cell growth of human breast (Fig. 3 Left) and esophageal (Fig. 3 Right) cancers. Both red wines (#2 and #3) also markedly reduced the rates of cell growth of the two human cancer cell lines. As mentioned above, these studies indicate that the repression of cancer cell growth by mature wines (#1 and #3) is much stronger than young wine (#2).
Fig. 3. MTT assay of MCF-7 and KYSE-510 cells.
MCF-7 and KYSE-510 cells were seeded into microplate and treated as mentioned above. Perform MTT assays following the procedure of MTT kit (AR1156) from Boster Biol. Tech. (Pleasanton, CA, USA), Measure the absorbance of the samples using Omega microplate reader at 570 nm. Left: MCF- cells; Right: KYSE-510 cells. The results show that red wines #1 (mature wine) and #3 (mature wine) markedly inhibit cancer cell proliferation. The bars represent Mean ± SE of at least three independent determinations.
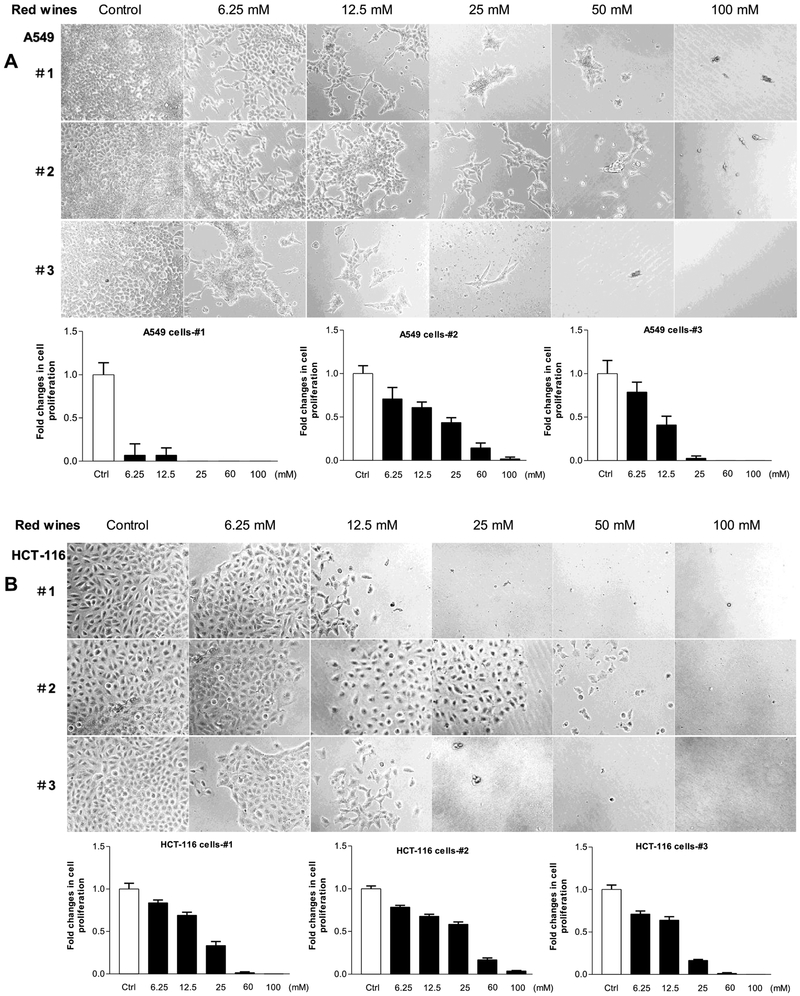
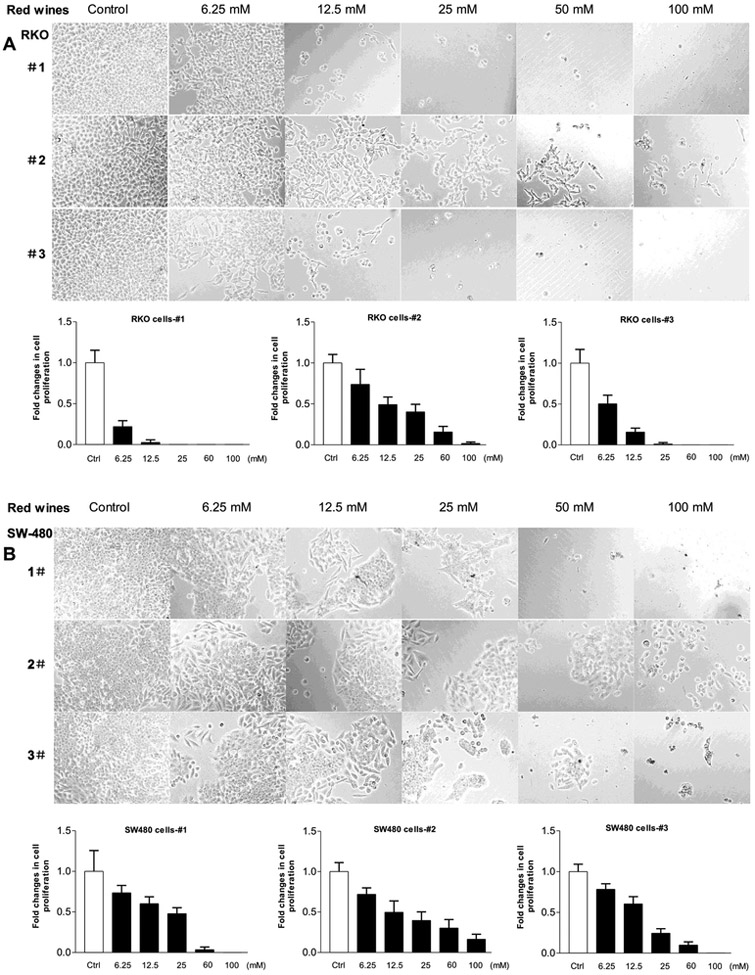
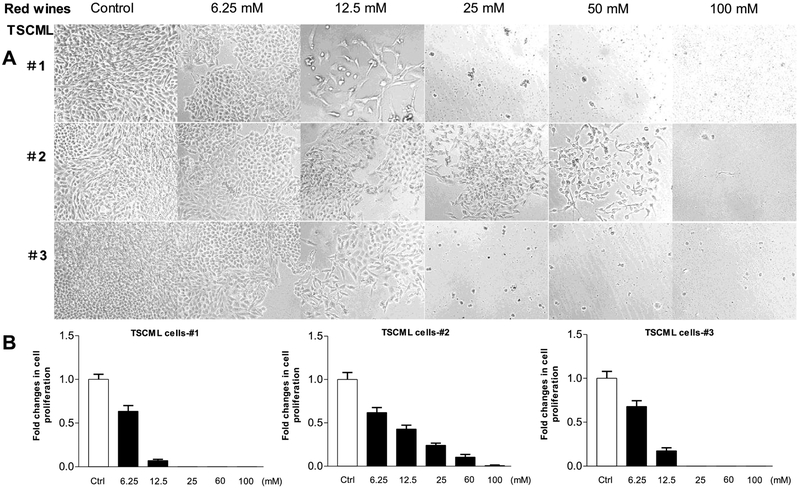
To explore the inhibition effects of the red wines on other human cancer lines, we utilized more human cancer cell lines. We treated human lung carcinoma line (A-549) and human colon cancer lines (HCT-116, SW-480 and RKO) with the red wines. The results indicate that no matter which red wines, all of them significantly decrease the cell growth rates of human lung carcinoma and colon cancer lines (Fig. 4 and Fig. 5). Moreover, the efficiency of repressing cell growth on A549 (Fig.4A) and RKO (Fig. 5A) cells by the red wines are much more significant than those on HCT-116 (Fig.4B) and SW-480 (Fig.5B) cells. Furthermore, we also tested the influence of the red wines in tumor stem cells of mouse liver (TSCML). The results indicate that the three red wines also markedly decrease the rate of cell growth of TSCML (Fig. 6). Together, the studies mentioned above have demonstrated that the red wines display the strong inhibition effects on cell growth of different cancer lines. However, the inhibition role of mature wine (#1 and #3) is much stronger than those of young wine (#2). As we can see, the roles of repressing cancer cell growth by the red wines are in a dose-dependent manner.
Fig. 4. Red wines decrease the rates of cell growth of lung and colorectal carcinoma lines.
Human lung cancer A-549 cells (A) and colorectal carcinoma HCT-116 (B) cells were treated with different amounts of ethanol and red wines as described above. The viability and total cell numbers were counted after the cells plated 6 days.
Fig. 5. Cell growth of colorectal carcinoma lines repressed by the red wines.
Human colorectal carcinoma RKO (A) and SW-480 (B) cells were treated with different amounts of ethanol and red wines as described above. The viability and total cell numbers were counted after the cells plated 6 days. The bars represent Mean ± SE of at least three independent determinations.
Fig. 6. Red wines repressed cell growth of mouse TSCML line.
(A): TSCML (tumor stem cells of mouse liver) MCF-7 cells were treated with different amounts of red wines #1 (mature wine), #2 (young wine) or #3 (mature wine) for 6 days. The concentrations of the three red wines are equal to actual ethanol (mM/L) as indicated. The pictures were taken under microscope (Nikon, Eclipse, TE300). Original magnification × 100. (B): the viability and total cell numbers were counted after plated cells 6 days. The cells were treated serially by red wines: #1, #2 and #3 from left to right as indicated. The results indicate that red wines dramatically decrease the rate of cell growth of TSCML line.
3.2. The cancer cell death was caused by the treatment of the red wines
Above studies have demonstrated that the red wines indeed decrease the cell growth rates of human cancer lines. Next, we explore whether the red wines are able to kill the grown-up human cancer cells. The MCF-7 cells and KYSE-510 cells grew to 70% confluence and then were treated with the three red wines for 48 hours, respectively, to determine accumulative cells. Figure 7 indicates that higher concentration (100 mM) of mature wines (#1 and #3) completely caused the MCF-7 cell death (Fig. 7A), while the mature wines at same dose also dramatically abate KYSE-510 cells (Fig. 7B). However, the young wine (2#) has less inhibition role in the same concentration of ethanol to the cancer lines (Fig. 7A and7B). These results show that the effect of the red wines on human breast cancer cells (MCF-7 line) is much stronger than those of human esophageal carcinoma cells (KYSE-510 line). But, the capacity of the red wines killing cancer cells is less than their inhibition role in cancer cell growth.
Fig. 7. Human breast and esophageal carcinoma cells killed by red wines.
MCF-7 cells (A) and KYSE-510 cells (B) were seeded in six-well plates and grew to 70% confluence in media without the red wines and then the cells were treated with different amounts of the red wines #1, #2 or #3 in media for 48 hours. The viability and total cell numbers were counted after red wine treatment 48 hours. The results indicate that red wines are able to kill the grown-up cells of breast cancer and esophageal carcinoma. The bars represent Mean ± SE of at least three independent determinations.
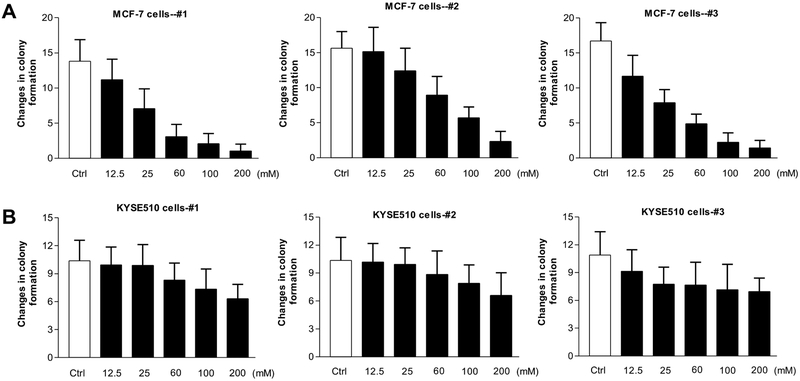
3.3. The red wines repress colony formation of human cancer cells
Our previous studies have indicated that inhibiting Pol III gene transcription is able to repress cell transformation (Zhong et al, 2011; Zhang et al, 2013; Zhang et al, 2011; Zhong et al, 2016). Early studies have demonstrated that ethanol promotes cell transformation of normal breast and liver cells and increases the rate of colony formation (Zhong et al, 2011; Zhang et al, 2013; Zhang et al, 2011; Zhong et al, 2016). Therefore, we further determine whether the red wines affect the colony formation of human cancer cells. The human breast cancer cells, MCF-7 line and human esophageal carcinoma cells, KYSE-510 line were seeded in soft agar and treated with the three red wines, respectively. The results indicate that the red wines markedly decrease the rates of colony formation of the human breast cancer cells (Fig.8A), and they also inhibit the anchorage-independent growth of human esophageal carcinoma cells, KYSE-510 line (Fig.8B). However, the efficiency repressing colony formation on breast cancer cells is more significant than those on esophageal carcinoma cells.
Fig. 8. Inhibition of colony formation of human cancer cells by China spirits.
MCF-7 cells (A) and KYSE-510 cells (B) were poured in triplicate into 6-well plate with 0.35% agar containing in turn red wines #1 (Left), #2 (Middle) or #3 (Right) in the media as indicated. The cells were analyzed for colony formation in soft agar after plated 2-3 weeks. The mature wines (#1 and #3) and young wine (#2) reduce the rates of colony formation of MCF-7 (A) and KYSE-510 (B) cells. The bars represent Mean ± SE of at least three independent determinations.
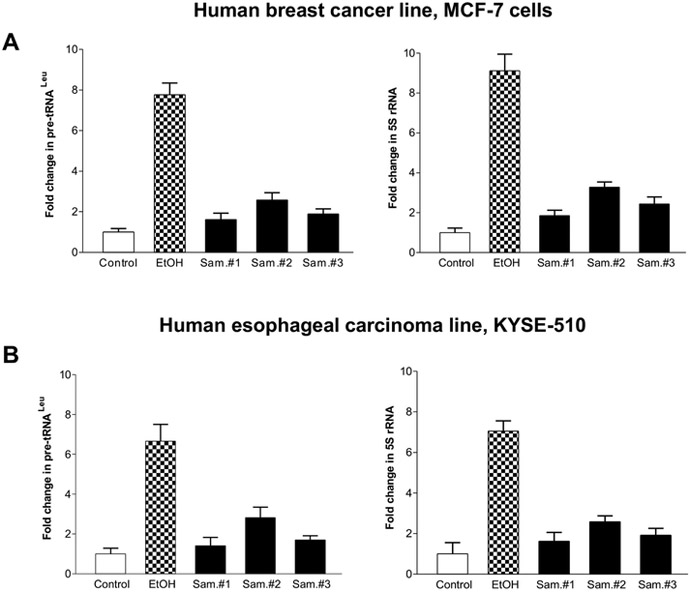
3.4. Transcription of RNA Pol III genes was inhibited by the red wines
Our studies have demonstrated that ethanol increased RNA Pol III-dependent transcription in vitro and in vivo by using cell culture model and animal model (Zhong et al, 2011; Zhang et al, 2013; Zhang et al, 2011). To investigate whether red wines affect Pol III gene transcription. MCF-7 (Fig. 9A) and KYSE-510 (Fig. 9B) cells were treated red wines, the amounts of precursor tRNALeu (Fig. 9A, Left) and 5S rRNA (Fig. 9B Right) transcript were measured by RT-qPCR. As we can see, diluted ethanol treatment resulted in augment of pre-tRNALeu (Fig. 9A Left) and 5S rRNA (Fig. 9B Right) transcription at the condition of 25mM ethanol for 60min. In contrast, at the same concentration of actual ethanol, red wines dramatically inhibit pre-tRNALeu (Fig. 9A Left) and 5S rRNA (Fig. 9B Right) transcription, either MCF-7 cells or KYSE-510 cells. These results reveal that induction of Pol III genes by diluted ethanol is able to be inhibited by bioactive components in red wines.
Fig. 9. The red wines inhibit RNA Pol III-dependent transcription.
MCF-7 (A) and KYSE-510 (B) cells were starved in Free/FBS media for 4 hours. Cells were treated with 25mM ethanol and similar amount and three red wines which amounts are equal to ethanol (mM/L) as indicated. RNA was isolated from these cells and RT-qPCR was performed to measure the amounts of pre-tRNALeu (A and B, Left panel) and 5S rRNA (A and B, Right Panel). The fold change was calculated by normalizing to the amount of GAPDH mRNA. The bars represent Mean ± SE of at least three independent determinations.
4. DISCUSSION
In present study, we determined the effects of red wines on cell growth, colony formation and Pol III gene transcription of different human cancer lines. The results indicate that diluted ethanol promotes cell proliferation at low concentration (12.5 mM and 25 mM). In contrast, red wines at same ethanol concentration dramatically decrease the rates of cell growth. The inhibition of cell growth by red wines is in a dose-dependent manner. Further analysis reveals that the mature wines (#1 and #3) are able to cause death of human breast cancer cells, as well as esophageal carcinoma cells. The results of soft agar assay indicate that the red wines markedly repress colony formation of MCF-7 and KYSE-510 cells. The red wines also dramatically inhibit RNA Pol III gene transcription, compared to diluted ethanol. The effects of the mature wines (#1 and #3) on cell growth, colony formation and Pol III gene transcription of human cancer cells are much stronger than young wine (#2). It is the first report on the effects of red wines to inhibit cell growth of different human cancer lines. These studies demonstrate that intaking suitable amounts of red wines may play a role in preventing cancer development.
Studies from our and other laboratories have demonstrated that increase in RNA Pol III gene transcription boosts cell proliferation and promotes cell transformation and tumor development (Johnson et al, 2008; Zhong and Johnson, 2009; Zhong et al, 2007). While inhibition of Pol III gene transcription decreases the rates of cell proliferation and colony formation (Zhong et al, 2009; Zhong et al, 2011; Zhang et al, 2013; Zhang et al, 2011). Therefore, repressing the cellular phenotypic changes stand for decrease in Pol III gene transcription. Here, our data further support this idea that inhibition of Pol III gene transcription (Fig. 9) by the red wines results in repression of cell growth (Fig. 1-7) and colony formation (Fig. 8). Our early studies indicated that 25~50 mM ethanol increases Pol III gene transcription in liver and breast cancer cells (Zhong et al, 2009; Zhong et al, 2011; Zhang et al, 2013; Zhang et al, 2011; Feng et al, 2017; Zhong et al, 2014). Studies have demonstrated that ethanol promoted liver tumor development and enhanced aggressiveness of breast cancer of mice (Zhong et al, 2011; Xu et al, 2016). Here, the results indicate that 12.5 mM to 25 mM ethanol also increases the cell proliferation of MCF-7 and KYSE-510 lines. Emerging studies have shown that alcohol consumption is associated with human cancers (Shi and Zhong, 2017). However, the mechanism is still unclear. Diluted ethanol (200 proof) has widely been using to explore molecular mechanisms of alcohol-caused human health issues, including tumor development. However, persons intake wines, but not ethanol. Red wines are made from dark-colored (black) grapes and a class of liquid mixtures. Although red wine contains ethanol, but also includes other components, such as anthocyanin from fluits which is of protective property against cancer (Xu et al, 2010). Therefore, the wine is not equal to ethanol. Yi and their colleagues reported that the liquor spirits (white wine) repressed DEN-induced liver tumor formation of mice (Yi et al, 2014). Our recent study reveals that the liquor spirits (white wine) indeed inhibit cell growth of human cancer lines (Yi et al, 2018). At the present study, our results further demonstrate that the red wines more dramatically decrease the rates of cell growth and colony formation than liquor spirits (Yi et al, 2018) on multiple human cancer lines. This indicates that the red wine indeed plays some extent roles in inhibiting human cancer cell growth. However, more studies in vivo by using animals need to be carried out to elucidate the role of red wine in preventing cancer development. For further studies in vivo, we can utilize mouse model mouse model of liver cancer caused by DEN and then feed the mice with the red wines in drinking water to determine whether the red wines are able to repress liver tumor growth; we can also feed the mice injected with DEN to observe whether red wines are able to against tumor development.
In summary, our studies reveal that the red wines, particularly mature wines (#1 and #3), dramatically decrease the rates of human cancer cell growth and colony formation, while diluted ethanol at same concentration boosts cell growth. The red wines also cause death of the grew up cancer cells and inhibit Pol III gene transcription. It implies that the red wine may contain some bioactive components and function potential to repress cancer development. Thus, identifying the bioactive components in red wine and enhancing their yielding ratio during producing processes will enhance the quality of red wine, which will benefit people with red wine consumption.
Highlights:
Red wines repressed cell proliferation and colony formation of different human cancer cells;
Red wines inhibit RNA Pol III gene transcription, while ethanol increases the activity of Pol III genes;
Diluted ethanol enhances these cellular phenotypic changes in human breast cancer and esophageal carcinoma lines;
The repressing effects on cell growth, colony formation and Pol III genes by mature wines are much stronger than ones by young wine;
Acknowledgements
We want to thank Drs. M. R. Stallcup, Danial Levy and Neil Kaplowitz (University of Southern California) for scientific discussions. This work was supported by NIAAA/NIH grants AA017288, AA021114, AA023247 and AA04169 to S.Z. and CNJ14C007 in China to Y.Y.
Abbreviation
- Pol III genes
RNA polymerase III-dependent genes
- RT-PCR
Real time-polymerase chain reaction
Footnotes
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference:
- Allen NE, Beral V, Casabonne D, Kan SW, Reeves GK, Brown A, and Green J, 2009. Moderate alcohol intake and cancer incidence in women. J Natl Cancer Inst 101,296–305. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang N, Ling Y, Wakai T, He Y, and Wei L 2011. Alcohol Consumption as a Risk Factor for Esophageal Adenocarcinoma in North China. The Tohoku Journal of Experimental Medicine, 224, 21–27. [DOI] [PubMed] [Google Scholar]
- Cogliani VJ, Baan R, Straif K, Crosse Y, Lauby-Secretan B, El Ghissassi F, Bouvard V, Benbrahim-Tallaa L, Guha N, Freeman C, Galichet L, Wild CP., 2011. Preventable exposures associated with human cancers. J Natl. Cancer Inst 103: 1827–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor J (2017). Alcohol consumption as a cause of cancer. Addiction, 112, 222–228. [DOI] [PubMed] [Google Scholar]
- Demark-Wahenefien W, &Goodwin PJ, 2013. To your health: how does the latest research on alcohol and breast cancer inform clinical practice? Journal of Clinical Oncology, 31, 1917–1919. [DOI] [PubMed] [Google Scholar]
- Fang Z, Yi Y, Shi G, Li S, Chen S, Lin Y, Li Z, He Z, Li W, Zhong S (2017). Role of Brf1 interaction with ERα, and significance of its overexpression in human breast cancer. Molecular Oncology 11: 1752–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German JB, & Walzem RL 2000. The health benefits of wine. Annual Review of Nutrition, 20, 561–593. [DOI] [PubMed] [Google Scholar]
- Johnson SA, Dubeau L, Johnson DL., 2008. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 283:19184–19191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IARC. 2011. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol 100, A Review of Human Carcinogens. Lyon, France: International Agency for Research on Cancer; 2011. [Google Scholar]
- Papa NP, Maclnnis RJ, Jayasekara H, English DR, Bolton D, and Davis ID, 2017. Total and beverage-specific alcohol intake and the risk of aggressive prostate cancer: a case- control study. Prostate Cancer and Prostatic Diseases, 20, 305–310. [DOI] [PubMed] [Google Scholar]
- Petri AL, Anne T, Michael G, Ditte J, Susanne H, Thorkild S, and Moeten G. (2004) Alcohol intake, type of beverage, and risk of breast cancer in pre-and postmenopausal women, Alcohol Clin Exp Res 2004; 28: 1084–1090. [DOI] [PubMed] [Google Scholar]
- Renaud S, & Lorgeril M 11-13]D. 1992. Wine, alcohol, platelets, and the French paradox for coronary heart disease. Lancet, 339, 1523–1526. [DOI] [PubMed] [Google Scholar]
- Ruf JC, 2003. Overview of epidemiological studies on wine, health and mortality. Drugs Under Experimental and Clinical Research, 29, 173–179. [PubMed] [Google Scholar]
- Seitz HK, Pelucchi C, Bagnardi V, and Vecchia C, 2012. Epidemiology and pathophysiology of alcohol and breast cancer: Update 2012, Alcohol 2012; 47: 204–12. [DOI] [PubMed] [Google Scholar]
- Scoccianti C, Straif K, and Romieu I, 2013. Recent evidence on alcohol and cancer epidemiology. Future Oncol 9:1315–1322. [DOI] [PubMed] [Google Scholar]
- Shi GG, and Zhong SP. (2017) Alcohol-associated cancer and deregulation of Pol III genes. Gene 612: 25–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singletary KW. and Gapstur SM, 2001. Alcohol and breast cancer: review of epidemiologic and experimental evidence and potential mechanisms, JAMA 286: 2143–2151. [DOI] [PubMed] [Google Scholar]
- Singletary KM Nelshoppen J and Wallig M, 1995. Enhancement by chronic ethanol intake of N-methyl Nitrosourea-induced rat mammary tumorigenesis, Carcinogenesis 16: 959–964. [DOI] [PubMed] [Google Scholar]
- Watabiki T, Okii Y, Tokiyasu T, Yoshimura S, Yoshida M, Akane A, Shikata N, and Tsubura A. 2000. Long term ethanol consumption in ICR mice causes mammary tumor in females and liver fibrosis in males. Alcohol Clin Exp Res 2000; 24: 117S–22S. [PubMed] [Google Scholar]
- White RJ., 2004. RNA polymerase III transcription and cancer. Oncogene. 23, 3208–3016. [DOI] [PubMed] [Google Scholar]
- Wu J, Cheng ML, Zhang GH, Zhai RW, Huang NH,&Li CX, 2002. Epidemiological and histopathological study of relevance of Guizhou Maotai liquor and liver diseases. World Journal of Gastroenterology, 8, 571–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Bower KA, Wang S, Frank JA, Chen G, Ding M, Wang S, Shi X, Ke Z, Luo J, 2010. Cyanidin-3-glucoside inhibits ethanol-induced invasion of breast cancer cells overexpressing ErbB2. Mol. Cancer. 9: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Ren ZH, Wang X, Comer A, Frank JA, Ke ZJ, Huang Y, Zhang Z, Shi X, Wang S, and Luo J, 2016. ErbB2 and p38γ MAPK mediate alcohol-induced increase in breast cancer stem cells and metastasis. Molecular Cancer 15: 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi X, Li L, Yang CZ, Lu YY, and Cheng LM, 2014. Maotai Ameliorates Diethylnitrosamine- Initiated Hepatocellular Carcinoma Formation in Mice. PLoS One. 2014; 9: e93599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi YF, Huang CH, Zhang YM, Tian SK, Lei JX, Chen SL, Shi GG, Wu ZD, Xia NS, and Zhong S, 2017. Exploring a common mechanism of alcohol-induced deregulation of RNA Pol III genes in liver and breast cells. Gene. 626:309–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Y, Lei J, Shi G, Chen S, Zhang Y, Hong Z, He Z and Zhong S., 2018. The effects of liquor spirits on RNA Pol III genes and cell growth of human cancer lines. Food and Nutrition Sciences. 9(3): 208–220, 2018. [Google Scholar]
- Zhang Q, Jin J, Zhong Q, Yu XL, Levy D, and Zhong S, 2013. ERα mediates alcohol-induced deregulation of Pol III genes in breast cancer cells. Carcinogenesis 2013; 34: 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Zhong Q, Evans AG, Levy D, and Zhong S, 2011. Phosphorylation of Histone H3 serine 28 modulates RNA polymerase III-dependent transcription. Oncogene, 30, 3943–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Shi G, Zhang Q, Lu L, Levy D, Zhong S, 2014. Tamoxifen represses alcohol-induced transcription of RNA polymerase III-dependent genes. Oncotarget 5: 12410–12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Q, Xi SY, Liang JZ, Shi GG, Huang Y, Zhang YM, Levy D, and Zhong SP (2016). The significance of Brf1 overexpression in human hepatocellular carcinoma. Oncotarget 2016; 7: 6243–6254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Fromm J, and Johnson DL (2007) TBP is differentially regulated by JNK1 and JNK2 through Elk-1, controlling c-Jun expression and cell proliferation Mol. Cell. Biol 27: 54–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Johnson DL (2009) The JNKs differentially regulate RNA polymerase III transcription by coordinately modulating the expression of all TFIIIB subunits. Proc Natl Acad Sci USA. 106: 12682–12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Machida K, Tsukamoto H, Johnson DL (2011) Alcohol induces RNA Pol III-dependent transcription through c-jun by coregulating TBP and Brf1 expression. J Biol Chem 286: 2393–2401. [DOI] [PMC free article] [PubMed] [Google Scholar]