Abstract
Electroporation-based assays were used to test whether the myogenic regulatory factor (MRF) of Ciona intestinalis (CiMRF) interferes with endogenous developmental programs, and to evaluate the importance of its unusual N-terminus for muscle development. We found that CiMRF suppresses both notochord and endoderm development when it is expressed in these tissues by a mechanism that may involve activation of muscle-specific microRNAs. Because these results add to a large body of evidence demonstrating the exceptionally high degree of functional conservation among MRFs, we were surprised to discover that non-ascidian MRFs were not myogenic in Ciona unless they formed part of a chimeric protein containing the CiMRF N-terminus. Equally surprising, we found that despite their widely differing primary sequences, the N-termini of MRFs of other ascidian species could form chimeric MRFs that were also myogenic in Ciona. This domain did not rescue the activity of a Brachyury protein whose transcriptional activation domain had been deleted, and so does not appear to constitute such a domain. Our results indicate that ascidians have previously unrecognized and potentially novel requirements for MRF-directed myogenesis. Moreover, they provide the first example of a domain that is essential to the core function of an important family of gene regulatory proteins, one that, to date, has been found in only a single branch of the family.
Introduction
Myogenic regulatory factors (MRFs) are a well-characterized family of basic-helix-loop-helix (b-hlh) transcription factors that play a central role in metazoan muscle development (reviewed by Baylies and Michelson, 2001; Pownall, et. al., 2002; Buckingham, et. al., 2003; Tajbakhsh, 2005; Berkes and Tapscott, 2005; Tapscott, 2005; Buckingham and Rigby, 2014). Whereas vertebrates have four MRFs with distinct but overlapping functions in myogenesis, invertebrates ordinarily have only one. Despite such differences, MRFs nevertheless are remarkably similar. These transcription factors are typically expressed only in muscle and muscle precursors, and they have the ability to direct myogenesis in non-myogenic cell types. This latter property effectively defines MRFs and distinguishes them from other b-hlh proteins, and has been attributed to the presence of an alanine-threonine dipeptide in the basic domain known as the myogenic code (Brennan, et.al., 1991; Davis and Weintraub, 1992). Most impressive among their similarities is the high level of functional conservation of MRFs, exemplified by their ability to replace each other (Zhang, et.al., 1999; Delfini and Duprez, 2004).
MRFs have also been found to interfere with endogenous programs of development when expressed in non-myogenic cell types. This was shown for MyoD using a variety of cultured cell systems (Weintraub, et.al., 1989; Choi, et.al., 1990; Rosenberg, et.al., 2006), and for the Caenorhabditis elegans MRF, hlh1, in the only in vivo study of this property of MRFs of which we are aware (Fukushige and Krause, 2005). In order to further evaluate to what extent repression of endogenous developmental programs is a shared feature of MRFs, we examined the effect of misexpressing CiMRF, the MRF of Ciona intestinalis (CiMRF is orthologous to vertebrate MRF genes MyoD1, Myf5, MRF4, and myogenin; in the ENSEMBL system it is designated myod) on the development of the ascidian notochord and endoderm. Markers of both tissues were suppressed in these experiments, and stunted tails were observed in embryos expressing CiMRF in the notochord, which is characteristic of mutations that affect the development of this tissue (Nakatani, et.al., 1999; Jiang, et.al., 2005). These findings support the idea that disrupting endogenous developmental programs is another shared feature of MRFs.
Our previous studies of Ci-MRF (Meedel, et.al., 1997; 2002; 2007; Izzi, et.al., 2013), in addition to its ability to suppress endogenous tissue development described here, indicate that Ci-MRF is a typical MRF gene. This conclusion, however, conflicts with the observation that the proteins it encodes have an exceptionally large N-terminus not found in other MRFs (Meedel, et.al., 1997). It is also at odds with the finding that non-ascidian MRFs fail to elicit myogenesis when expressed in Ciona embryos (unpublished results; this communication). Collectively, these observations lead us to suggest that the CiMRF N-terminus is required for the myogenic activity of MRFs in Ciona embryos. Studies presented here support this possibility, and also demonstrate that a large and functionally essential N-terminus is a feature of other, and possibly all, ascidians MRFs. We conclude that myogenesis in ascidians requires an evolutionarily novel type of MRF N-terminus.
Materials and Methods
Plasmid Construction
Plasmids used to direct expression of Ci-MRF (pTCiMRFb) and LacZ (pTLacZ) in the notochord have been described previously (Izzi, et.al., 2013). Plasmids expressed in the endoderm were derived from pTTFLacZ (gift of B. Davidson), which contains cis-regulatory sequences of the TiTf1 gene (Ristoratore, et.al., 1999). BLAST analysis indicated that these regulatory sequences were from Ciona robusta (see Bouchemousse, et.al., 2016 for a discussion of this issue), but they nevertheless drove endoderm expression in our experiments using Ciona intestinalis. pTTFLacZ also served as a negative control in most experiments. All other plasmids designed to be expressed in the endoderm were designated with the prefix “pF” and were assembled in NotI/Acc65I digested pTTFΔSacI, which was created by removing the 2.05kb SacI fragment containing most of the LacZ gene from pTTFLacZ. All synthetic DNA fragments used in this study were supplied by EPOCH Life Sciences (Missouri City, TX) in a blunt-end pBluescript II SK− vector. Several plasmids encoded a thrice-repeated Flag epitope (Flag-tag) that, if deemed necessary, could be used to determine whether the protein encoded by any particular plasmid accumulated in the expected pattern.
pFCiMRFb was used to express CiMRFb (Meedel, et.al., 1997) in the endoderm. It was generated by inserting the 2.6kb Acc65I/NotI fragment of pBSCiMRFb into pTTFΔSacI. pBSCiMRFb was created previously by cloning into PstI/SalI digested pBluescript II KS+ the 0.35kb PstI/SacI fragment of pCiMRF5’ that encoded the 5’ untranslated region and N-terminal coding sequences of CiMRF (Izzi, et.al., 2013) and a 2.3kb SacI/SalI fragment from plasmid pc9m3.5 that contained the remainder of the CiMRFb coding sequence and the 3’ untranslated region (Meedel, et.al., 1997).
pMCiMRFNTf and pFGFPn were plasmids designed to test the fidelity of the Titf1 cis-regulatory sequences used in our experiments. This was done to address concerns about using cis-regulatory sequences from a C. robusta gene in our experiments with C. intestinalis, and because we wanted to determine the potential level of expression of our constructs in the mesenchyme, which is a noted “hot spot” where electroporated plasmids are often misexpressed in Ciona embryos (Harafuji, et.al., 2002). pMCiMRFNTf was driven by promoter sequences of Ci-MRF, which are active in both muscle and mesenchyme (Meedel, unpublished observations and Fig. 2). It was constructed by excising a 1042 bp HindIII/HincII fragment from pCiMRFpro14, a reporter plasmid that contained about 1400 bp of cis-regulatory sequence that is adjacent to the translation start site of Ci-MRF (Meedel and Lee, unpublished). This fragment, together with a 700 bp HincII/SacI fragment of a synthetic DNA (NLSNTSac) was inserted into pBluescript II KS+ that had been linearized with HindIII and SacI. NLSNTSac contained additional sequence of the CiMRF promoter that is immediately upstream of the translation start site (which was excluded by excising the 1042 bp HindIII/HincII fragment of pCiMRFpro14), an SV40 nuclear localization sequence, and approximately 330 bp of CiMRF coding sequence that include a unique SacI site for cloning. The 1740 bp HindIII/SacI insert of this plasmid was isolated and cloned into a plasmid designated pSP72-SE together with an 875 bp SacI/SalI fragment from a previously constructed plasmid (pTCiMRF5NT3Flag3) to create pMCiMRFNTf. pSP72-SE was pSP72 (ProMega) from which the multiple cloning site between the SmaI and EcoRV restriction sites had been deleted. The 875 bp SacI/SalI fragment of pTCiMRF5NT3Flag3 encoded the remainder of the coding sequence of the CiMRF N-terminus (beginning with the SacI site of NLSNTSac), a thrice-repeated Flag-tag and the CiMRFa 3’ UTR. Two synthetic DNA fragments were used to construct pFGFPn. The first (m5GFPCiMRFa) was 936 bp and encoded (5’−3’) a NotI site, the trans-splice leader, 5’ UTR and translation start site of CiMRF, m5GFP (sequence provided by R. Zeller), and CiMRFa sequence between nucleotides 1142–1300 of CiMRFa (Meedel, et.al., 1997). m5GFPCiMRFa was designed to make a second plasmid (pFGFPCiΔNT; see below) as well, and so it contains additional sequence not needed to construct pFGFPn. The second synthetic DNA (DrGFPNLST) was 591 bp and encoded (5’−3’) a DraI site, the C-terminal portion of m5GFP a thrice-repeated SV40 nuclear localization sequence, the translation stop site and 3’ UTR of CiTnI (Ciona intestinalis Troponin I; MacLean, et.al., 1997), and a SacI site. pFGFPn was created by inserting the 441 bp NotI/DraI fragment of m5GFPCiMRFa (this removed sequences encoding the C-terminus of GFP and CiMRF) and DraI/Acc65I digested DrGFPNLST into pTTFΔSacI.
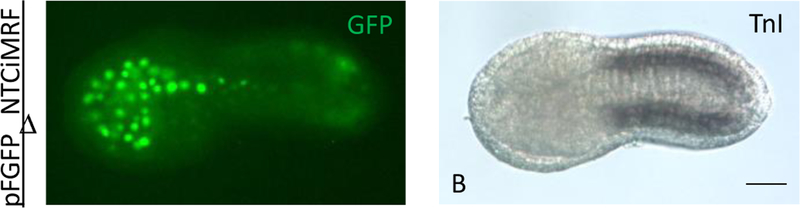
Figure 2.
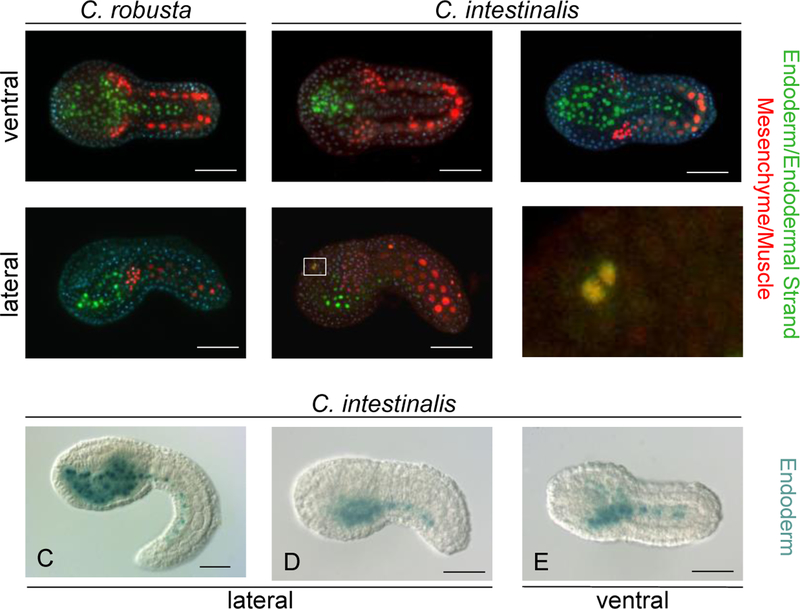
Fidelity of the Titf1 cis-regulatory promoter sequences. Ventral and lateral views respectively of single C. robusta (A and B) and C. intestinalis embryos (A’ and B’) that were co-electroporated with pMCiMRFNTF and pFGFPn and reacted with mouse anti-Flag and rabbit anti-GFP primary antibodies, followed by reaction with goat anti-mouse AF 546 and goat anti-rabbit AF 488 secondary antibodies. A“ is a ventral view of a third C. intestinalis embryo treated the same way and B” is a higher magnification image of the boxed region in B’ showing a rare example of pMCiMRFNTF and pFGFPn expression in the brain. Green signal corresponds to GFP and is restricted to the endoderm of the head and the endodermal strand of the tail, whereas the red signal corresponds to the Flag epitope and is restricted to mesenchyme in the head and muscle in the tail in embryos of both species. Blue signals represent DAPI stained nuclei. C, D, and E showing C. intestinalis embryos that were electroporated with pTTFLacZ and assayed for beta-galactosidase. D and E show the same embryo in lateral and ventral views respectively. Size bars in all photographs are 50μm.
Nautilus plasmids:
Details relevant to the Drosophila melanogaster MRF (Nautilus) correspond to NCBI Reference Sequence: NP_476650.1. pFNau was designed to express the complete Nautilus protein fused to a thrice-repeated Flag-tag. It was constructed by inserting a 1383 bp NotI/BspI fragment of a plasmid (pBS_NautilusF; gift of B. Paterson) that contained a substantial portion of the 5’ UTR and the entire Nautilus coding region, and the 330 bp BspI/Acc65I fragment of a synthetic DNA (CTNauF) encoding the C-terminal domain of Nautilus (amino acids Pro312-Thr332), a thrice-repeated Flag-tag, and the stop codon and 3’ UTR of CiTnI into pTTFΔSacI.
pFCiNTNau was designed to express a fusion protein in which the CiMRF N-terminus replaced the Nautilus N-terminus. It was constructed by inserting a BglII/SalI digested 945 bp synthetic DNA (CiNTNau) into pBSCiMRFb that was also digested with those two enzymes to create pBSCiNTNau. CiNTNau encodes (5’−3’) a portion of CiMRF N-terminus (Ser348-Ser364; Meedel, et.al., 1997) the Cys/His-rich basic-helix-loop-helix domain CH/b-hlh) of Nautilus beginning with His129 and extending to the C-terminus (Thr332), a thrice-repeated Flag-tag, and the stop codon and 3’ UTR of CiTnI. pFCiNTNau was created by inserting the NotI/Acc65I fragment of pBSCiNTNau into pTTFΔSacI.
CiMRF N-terminal replacement plasmids:
pFGFPCiΔNT produces a fusion protein in which m5GFP replaced the N-terminus of CiMRF. It also encoded the 5’ and 3’ UTRs of CiMRFa (Meedel, et.al., 1997). It was created by ligating together a NotI/AflII-digested 936 bp synthetic DNA (m5GFPCiMRFa; see pFGFPn above for details of this DNA), a 537 bp AflII/Acc65I fragment of pBSCiMRFa (Izzi, et.al, 2013), and NotI/Acc65I-digested pTTFΔSacI.
pFHr2Ci produces a fusion protein (Hr2CiMRF) consisting of the N-terminus of the Halocynthia roretzi MRF (HrMRF) and the C/H-bhlh domain and C-terminus of CiMRF. We initially used the sequence of AMD1 (Araki, et.al., 1994) to construct the plasmid pFHrCi. Subsequently, we discovered that this sequence lacked 247 amino acids of the N-terminal domain of HrMRF. pFHr2Ci was created by incorporating the corrected HrMRF sequence that was provided by H. Nishida into one of the original constructs (pBSNTHrCiMRFb.F) containing the incomplete AMD1 sequence as described below.
Two synthetic DNAs were used to construct pBSNTHrCiMRFb.F. The first, NTAMD1, is an 877 bp sequence that contains (5’−3’) a NotI site, sequences encoding the trans-splice leader and 5’ UTR of CiMRF, the N-terminal 242 amino acids of AMD1 (Araki, et.al., 1994), and amino acids 405–432 of CiMRFb (Meedel, et.al., 1997); the internal SphI site that otherwise occurs in this sequence was mutated (GCATGC → GCGTGC) without changing the coding information so that a single SphI site remained at base pair 874 of NTAMD1. This fragment was digested with NotI and SphI and inserted into pBSCiMRFb that was also cut with those two enzymes to produce pBSNTHrCiMRFb. The second synthetic DNA, BCTFlag, is a 483 bp fragment encoding the C-terminal four amino acids of CiMRFb linked to an in-frame thrice-repeated Flag-tag and a portion of the 3’ UTR of CiMRFb that ends with a BsaBI site. pBSNTHrCiMRFb.F was created by inserting the ClaI/BsaBI fragment of BCTFlag into pBSNTHrCiMRFb that was also digested with those two enzymes. As noted above, pBSNTHrCiMRFb.F was designed with incomplete sequence information and so a third 1279 bp sequence, designated Hr404, was synthesized. It consisted of (5’−3’) a NotI site, the trans-splice leader sequence and 5’ UTR of CiMRF, and a sequence encoding the N-terminal 404 amino acids of HrMRF (obtained from the ANISEED database courtesy of H. Nishida). Hr404 was digested with NotI and MluI and inserted into NotI/MluI digested pBSNTHrCiMRFb.F to create pBSNTHr2CiMRFb.F. The NotI/Acc65I insert of pBSNTHr2CiMRFb.F was cloned into pTTFΔSacI to generate pFHr2Ci.
pFPmCi produces a fusion protein (PmCiMRF) consisting of the N-terminus of the Phallusia mammillata MRF (PmMRF; accession number HQ287931; deposited by J. Chenevert) and the C/H-bhlh domain and C-terminus of CiMRF. The sites noted in the following descriptions correspond to the PmMRF sequence. Two synthetic DNAs were used to construct pFPmCi. NTPmMRF contains a 1135 bp DNA sequence containing (5’−3’) a NotI site, the CiMRF trans-spliced leader and 5’ UTR, followed by sequence encoding the N-terminal 356 amino acids of PmMRF (ending with the amino acid sequence HPNG) that extends just beyond an EaeI site at base pair 1079; BCTermFlag is a 483 bp sequence encoding the C-terminal four amino acids of CiMRFb (ERID), a thrice-repeated Flag-tag, the stop codon of CiMRFb and a portion of the 3’ UTR of CiMRFb extending seven nucleotides beyond a BsaBI site. A ClaI site occurs near the 5’ end of this fragment. pBSNTPmCiMRFb was made by ligating NotI-SalI digested pBSIIKS+ with the 1126 bp NotI-EaeI fragment of NTPmMRF and the 1471 bp EaeI/SalI fragment of pBSCiMRFb. pBSNTPmCiMRFFlag was made by cutting pBSNTPmCiMRFb with ClaI and BsaBI and cloning into this site the 396 bp ClaI-BsaBI insert of BCTFlag. pFPmCi was generated by inserting the Acc65I/NotI fragment of pBSNTPmCiMRFFlag into pTTFΔSacI.
pFCsCi produces a fusion protein (CsCiMRF) consisting of the N-terminus of the Ciona savignyi MRF (CsMRF) and the C/H-bhlh domain and C-terminus of CiMRF. The Ciona savignyi genome sequence (Vinson, et.al., 2005) deposited in ENSEMBL was searched using the CiMRF sequence to identify CsMRF. We used this information to make an oligonucleotide, CsMRF01 (CGTGGATCCTGACTATGACTTGTATTTCACTCGAGG), with which to prime cDNA synthesis from RNA isolated from 9-hour C. savignyi embryos. This cDNA was amplified by PCR using CsMRF01 and CsMRF02 (CGTGAGCTCAACCGATCAATCAATTCGTTCCAC), and the resulting product was subcloned into pBluescript II KS+ and sequenced. This plasmid was designated pBSCsMRF and its sequence was used as the basis for synthesizing NTCs357 that encoded (5’−3’) a 5’ NotI site, the CiMRF trans-splice leader and 5’ UTR, the first 357 amino acids of CsMRF, and 58 amino acids of CiMRFb (365–422, beginning with the amino acid sequence HYH and ending with ACA). After digesting with NotI and SphI, NTCs357 was inserted into pBSCiMRFb that also had been digested with those two enzymes to create pBSNTCsCiMRFb. The 396 bp ClaI-BsaBI fragment of BCTFlag (described above) was inserted into pBSNTCsCiMRFb to create pBSNTCsCiMRFb.F, and the NotI-Acc65I insert of this plasmid was isolated and cloned into pTTFΔSacI to produce pFCsCi.
Transactivation Domain Plasmids:
We designed three plasmids to test whether the CiMRF N-terminus could replace the transactivation domain of CiBra, the Ciona Brachyury transcription factor. Information related to CiBra in the following descriptions corresponds to NCBI Reference Sequence: NM_001032487.1. Considering that this sequence was obtained using ascidians from the west coast of the United States, it is most likely from Ciona robusta (Bouchemousse, et.al., 2016).
pFCiBra was designed to express the complete Ciona Brachyrury protein and served as a positive control intended to elicit notochord gene expression in the endoderm. Two synthetic DNAs were used to construct pFCiBra. The first, CiBraL, contains an 839 bp fragment that encodes (5’−3’) a NotI site, and the 5’ UTR and N-terminal 267 amino acids of CiBra. The sequence encoding this part of CiBra contains two HaeII sites; the 5’-most site was mutated without changing the coding information (GGCGTC→GGCCTC) and the 3’-most site was retained for cloning. The second synthetic DNA, CiBraR, contains a 707 bp fragment that encodes (5’−3’) the C-terminal half of CiBra beginning with the HaeII site that occurs in CiBraL, an in-frame, eight-times repeated glycine linker, a thrice-repeated Flag-tag, the stop codon and 3’ UTR of CiBra, and an Acc65I site for cloning. pFCiBra was created by isolating the NotI/HaeII fragment of CiBraL and the HaeII/Acc65I fragment of CiBraR, and ligating those fragments into pTTFΔSacI.
pFCiNTBra was designed to test whether the CiMRF N-terminus could function as a transactivation domain when fused to the DNA binding domain of Brachyury. Two synthetic DNAs were used for its construction: NLS-MRF contains a 1115 bp sequence encoding (5’−3’) a NotI site, the CiMRF trans-splice leader and 5’ UTR, a translation initiation site, the SV40 nuclear localization sequence (SV40NLS), and the N-terminal 349 amino acids of CiMRF. A unique BglII site used for cloning resides near the 3’ end of this sequence. NTBra contains a 937 bp sequence encoding (5’−3’) 49 bp of CiMRF (coding for amino acids 349–364) that include the BglII site found in NLS-MRF, an in-frame linker sequence (GlyGlyGlyGlySer) repeated three times, nucleotides 98–649 coding for the DNA binding domain of CiBra, a thrice-repeated Flag-Tag, the CiTnI stop codon and 3’ UTR, and an Acc65I site. pFCiNTBra was created by isolating the NotI/BglII fragment of NLS-MRF and the BglII/Acc65I fragment of NTBra, and inserting them into pTTFΔSacI.
pFGFPBra was designed to direct endodermal expression of a fusion protein between GFP and the DNA binding domain of CiBra. It served as a negative control because the CiBra DNA binding domain should not drive notochord gene expression in the absence of a transactivation domain. Two synthetic DNAs were used for its construction: L-GFP contains a 766 bp sequence encoding (5’−3’) a NotI site, the CiMRF trans-splice leader and 5’ UTR, a translation initiation site, the SV40NLS, and base pairs 8–686 of m5GFP; GFP-BraR is a 929 bp sequence encoding (5’−3’) bases 681–721 of m5GFP, an in-frame linker sequence (GlyGlyGlyGlySer) repeated three times, nucleotides 98–649 coding for the DNA binding domain of CiBra, a thrice-repeated Flag-tag, the CiTnI stop codon and 3’ UTR, and an Acc65I site. pFGFPBra was made by isolating the NotI/PvuII fragment of L-GFP and the PvuII/Acc65I fragment of GFP-BraR, and inserting them into pTTFΔSacI.
pBSKgmiR-1/133 was the template used to synthesize digoxygenin-labeled RNA transcripts directed against the Ciona miR-1/133 complex. It was supplied by EPOCH Life Sciences in their blunt-ended pBluescript SK− vector and its 345 basepair insert corresponded to the sequence of the Ciona robusta miR-1/133 complex reported by Kusakabe, et.al., 2013. Antisense RNA was prepared from AhdI cut pBSKgmiR-1/133 using T3 RNA polymerase. This produced a transcript of approximately 2 kb in length that consisted of the miR-1/133 target sequences and approximately 1.7 kb of vector sequence. We found that incorporating these vector sequences was necessary to produce a probe with the degree of sensitivity necessary to detect miR-1/133 transcripts.
Animals and Electroporation
Adult Ciona intestinalis were collected from the Sandwich Marina in Sandwich, MA and Point Judith Marina in Snug Harbor, RI. Eggs were obtained by dissection of the oviduct, and fertilized in vitro with sperm of several individuals. Zygotes were dechorionated immediately after fertilization using the methods described by Mita-Miyazawa, et. al. (1985). After electroporation, embryos were reared in 0.2 μm filtered seawater at either 14°C or 18°C in Petri dishes that had been treated with a solution containing 0.1% (w/v) gelatin to prevent dechorionated embryos from sticking to the dish. Adult Ciona robusta were collected from Marina Village in Mission Bay, San Diego, CA, and their gametes and embryos were handled in the same manner as were those from C. intestinalis.
Plasmids were electroporated into Ciona embryos as described by Corbo, et.al. (1997). Fertilized eggs were collected either in 200μL seawater and added to 600μL 0.77 M Mannitol, or they were collected in 200μL of seawater and added to 200μL 0.77 M Mannitol. In either case they were electroporated with approximately 25μg of plasmid between 20 and 30 minutes after fertilization. Embryos for cleavage-arrest experiments were collected beginning at the 8-cell stage and treated with cytochalasin B at a final concentration of 1μg/mL to arrest cleavage at 64 cells (approximately 4.25 hours post-fertilization at 18°C). Normally developing or cleavage-arrested embryos were typically fixed for in situ hybridization during early tail-formation stage; this corresponded to 11–12 hours post fertilization at 18°C and to 14–15 hours post fertilization at 14°C. Occasionally, embryos were fixed at mid-tail formation stage, which occurs about 2 hours later at either temperature.
In Situ Hybridization and Enzyme Histochemistry
Typically, embryos electroporated with a given plasmid were divided into groups containing 15–30 embryos and subjected to in situ hybridization using digoxigenin-labeled antisense RNA probes essentially as described by Wada, et.al. (1995). Incubation times for color development ranged from 3 to 16 hours depending on the probe. This protocol was modified somewhat when probing for miR-1/133 transcripts: hybridization was done for three days, and antibody binding and color development were each done for two days. Probes used for detecting muscle transcripts were described previously (Izzi, et.al., 2013), while probes used for detecting notochord transcripts were designed to be complementary to sequences encoding three notochord-specific proteins: Brachyury (Bra; Corbo, et.al., 1997), Noto1 (Hotta, et.al., 2000) and Tropomyosin-Like (TmL; Di Gregorio and Levine, 1999). Specific accession numbers and/or gene model numbers for each of the genes corresponding to all probes used in the in situ hybridization experiments can be found in Table 1. Except for Troponin I, the source of animals used to make all of the probes was either Japan or the West Coast of the United States; therefore, it is likely that their sequences correspond to Ciona robusta, rather than Ciona intestinalis genes (Bouchemousse, et.al., 2016).
Table 1.
Muscle and notochord genes assayed.
| Gene Name | Probe Name* |
Normal Site of Expression |
KH Gene Model |
|---|---|---|---|
| Muscle Actin (MA3) | Actin | Muscle | KH.C7.67 |
| SET MYND Domain (QZF438) |
SMYD1 | Muscle | KH.S423.6 |
| Tropomyosin 2 | TPM2 | Muscle | KH.C3.814 |
| Troponin I | Tnl | Muscle | KH.C 11.673 |
| Brachyury (T) | Bra | Notochord | KH.S 1404.1 |
| Notol | Notol | Notochord | KH.S643.6 |
| Tropomyosin-Like | TmL | Notochord | KH.C7.260 |
| microRNA-1/133 | miR-1/133 | Muscle | KhCl :8,386,031 ..8,401,030† |
Probe Name refers to the name used in this study to describe the probe. Except for Troponin I whose sequence corresponds to the Ciona intestinalis gene (MacLean et al., 1997), all probe sequences correspond to Ciona robusta genes.
Indicates the KH genomic coordinates of the miR-1/133 probe sequence, which falls within a large intron of the Mindbomb E3 Ligase gene (mib; KH.C1.94). KH refers to Kyoto Hoya gene models.
Embryos for alkaline phosphatase activity were fixed for 30–40 minutes on ice in seawater containing 4% paraformaldehyde and 0.1 % Tween 80. Alkaline phosphatase was localized by staining fixed embryos in a solution containing 50mM TrisHCl pH 9.5, 0.5M NaCl, 10mM MgCl2, 0.1% Tween 80, nitro blue tetrazolium chloride (50mg/ml), and 5-bromo-4-chloro-3’ indolylphosphate p-toluidine salt (100mg/ml) for 1 hour at room temperature. Embryos assayed for β-galactosidase activity were fixed for 30 minutes on ice in seawater containing 1.5% paraformaldehyde and 0.1% Tween 80; they were then washed in phosphate buffered saline (PBS) containing 0.1% Tween 80 and incubated in staining solution (0.04% XGal, 2mM MgCl2, 0.06 M Na2HPO4, 0.04 M NaH2PO4, 4mM potassium ferrocyanide, 4mM potassium ferricyanide, 0.1% Tween 80) at room temperature for 1–4 hours.
Quantitative Assay of Alkaline Phosphatase Activity
Embryos were lysed in a homogenization buffer consisting of 20mM Tris-HCl pH7.5, 1mM MgCl2, 1μM ZnCl2, 1% Triton X-100, and 0.5% sodium deoxycholate. Typically, the volume of homogenization buffer was 0.5–1.0 μl of buffer/embryo; in order to ensure complete lysis, embryos stored on ice were subjected to intermittent vortex mixing over a period of 0.5–1 hour and the resulting homogenate was inspected visually using a dissecting microscope. Alkaline phosphatase activity was determined by mixing equal volumes of embryo homogenate and a solution containing 1mg/ml of p-nitrophenyl phosphate dissolved in 1M Tris-HCl pH9.5. Quantification of the colored product, p-nitrophenol, was determined at 405 nm using a BioRad Smartspec spectrophotometer.
Immunofluorescence Assays
Embryos were fixed in 2% paraformaldehyde, 0.1% Tween 20 in seawater for 12 minutes at room temperature, washed twice with PBT (phosphate buffered saline containing 0.1% Tween 20), rinsed quickly with −20°C methanol, and washed four times for a total of 40 minutes in PBT. Blocking was done for 10 minutes using a 1% solution of BSA dissolved in PBS. Embryos were exposed to diluted primary antibodies either for 2 hours at room temperature or overnight at 4°C. They were then washed four times for a total of 40 minutes in PBT and blocked as above. Secondary antibody incubations were done in the dark either for 2 hours at room temperature or overnight at 4°C. Embryos were again washed four times for a total of 40 minutes in PBT. Typically, embryos were imaged in 50% glycerol, but for confocal microscopy they were first incubated in 50% glycerol containing DAPI and then imaged in 70% glycerol. Primary antibodies were used at 1:2000 dilutions and were mouse anti-Flag (Sigma; F1804-M2) and rabbit anti-GFP (Thermo; A11122). Secondary antibodies were used at 1:500 dilutions and were goat anti-mouse AF546 (Thermo; A11003) and goat anti-rabbit AF 488 (Thermo; A11034).
Photography
Brightfield images were taken using either an Olympus BHS Model microscope equipped with a Pixelink 6.6 megapixel camera, a Zeiss AxioPlan 2e microscope equipped with a AxioCam ICc1 color camera, or with a Leica dissecting microscope, equipped with a Leica DFC290 HD digital camera with 3 Megapixel standard resolution. Fluorescence images were taken using a Nikon Eclipse E-660 microscope equipped with wide field epifluorescence and a Retiga 2000R camera, and confocal images were taken with a Zeiss LSM 780 microscope. Minor editing of photographs was done using ImageJ software.
Results
CiMRF Suppresses Notochord Development
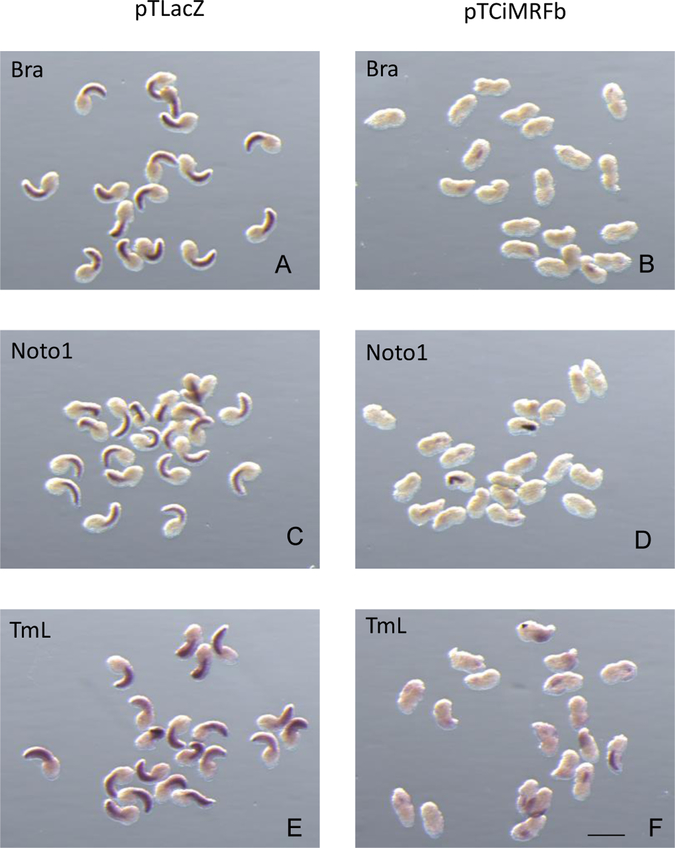
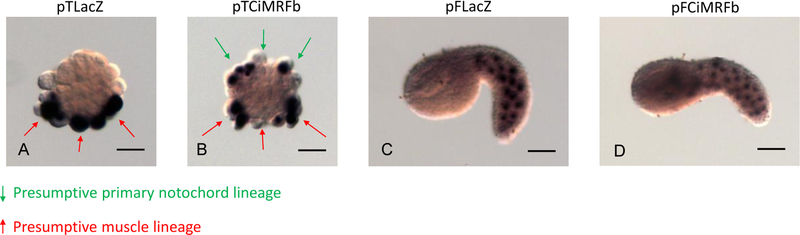
Previous studies in our laboratory showed that expression of Ci-MRF in the notochord led to the activation of several muscle-specific genes in that tissue (Izzi, et.al, 2013). We also demonstrated in those experiments that the tails of electroporated embryos were very often abnormal. Tail development depends on normal notochord morphogenesis, which in turn depends on the activity of Brachyury (Bra), a notochord-specific regulatory gene in ascidians (Yasuo and Satoh, 1994). Therefore, we reasoned that such abnormalities could be due to suppression of Bra activity by CiMRF. To investigate this possibility, we subjected embryos expressing CiMRF in the notochord to in situ hybridization (ISH) using a Bra probe as well as probes for two other genes (Noto1 and Tropomyosin Like [TmL]) that are characteristic of differentiating notochord and whose activity is most likely regulated by Bra (Takahashi, et.al., 1999; Di Gregorio and Levine, 1999; Hotta, et.al., 2000). Such embryos are shown in Figure 1 and it is apparent that the intensity of ISH reactions, which is a measure of the relative level of target transcript abundance, was reduced for all three markers in embryos electroporated with pTCiMRFb (see Meedel et al., 1997 for a description of CiMRFb versus CiMRFa) compared to those electroporated with pTLacZ. Figure 1 (B, D, F) also illustrates the severe tail abnormalities that are typical of embryos expressing CiMRF in the notochord, that we noted previously (Izzi, et al., 2013).
Figure 1.
CiMRF expression in the notochord suppresses gene activity in this tissue. Control embryos in A, C, and E were electroporated with pTLacZ and assayed for expression of Bra, Noto1, or TmL respectively. Embryos in B, D, and F were electroporated with pTCiMRFb and assayed for expression of Bra, Noto1, or TmL respectively. Size bar is 200μm, and all embryos shown are C. intestinalis.
In order to better quantify the effect of CiMRF on notochord gene expression, we determined the number of Bra positive cells in zygotes electroporated with either pTLacZ or pTCiMRFb that were subsequently cleavage-arrested at the 64-cell stage. The mean number of cells expressing Bra in embryos electroporated with pTCiMRFb was significantly less than in those electroporated with pTLacZ ( = 1.50 vs. 3.75). In addition, almost one-third of the pTCiMRFbelectroporated embryos had no cells expressing Bra, whereas all pTLacZ-electroporated embryos had at least some cells expressing this gene (Table 2). Collectively, our results demonstrate that CiMRF activity in the notochord interferes with the development of this tissue by a mechanism that probably involves inhibiting Bra activity.
Table 2.
Quantification of Bra expression.
| Bra+ Cells/Embryo | pTLacZ Bra+ Embryos |
pTCiMRFb Bra+ Embryos |
|---|---|---|
| 0 | 0 | 46 |
| 1 | 4 | 25 |
| 2 | 6 | 45 |
| 3 | 21 | 14 |
| 4 | 151 | 15 |
| 5 or 6 | 0 | 0 |
| Total | 182 | 145 |
| Mean | 3.75±0.62 | 1.50±1.31 |
Embryos electroporated with either pTLacZ or pTCiMRFb were cleavage-arrested at the 64-cell stage (approximately 4.25 hours post-fertilization) and assayed for Bra activity by ISH at 11 hours post-fertilization.
CiMRF Directs Muscle Gene Expression in the Endoderm
We next studied the effect of expressing CiMRF in a tissue other than notochord by constructing plasmids that relied on cis-regulatory sequences of the Ciona robusta Titf1 gene to drive expression in the endoderm lineage (Ristoratore, et.al., 1999). In a series of experiments designed to assess the fidelity of the Titf1 promoter sequences in both C. intestinalis and C. robusta embryos, we co-electroporated pMCiMRFNTFlag and pTTFGFPn and immunostained against Flag and GFP. Examination of over 100 embryos per experiment revealed GFP expression to be restricted to the endoderm (Fig. 2) [in rare cases this activity was seen in cells of the brain (Fig. 2 B’, B”), which are known to express Titf1 during tail formation stages (Imai, et.al., 2004)]. Particularly notable was the absence of Titf1-directed activity in the mesenchyme, which has been termed a “hot spot” of reporter plasmid misexpression in electroporated Ciona embryos (Harafuji, et.al., 2002). These results indicate that the Titf1 sequences used in this study are exceptionally faithful in replicating the expected endodermal pattern of expression in both Ciona species tested.
We next examined whether expressing CiMRF in the endoderm could trans-differentiate this tissue to muscle by electroporating a plasmid (pFCiMRFb) that expressed CiMRFb (Meedel, et.al., 1997) under the control of Titf1 cis-regulatory sequences. As a negative control, we used pTTFLacZ, which expresses β-galactosidase in the endoderm. Figure 2C shows that embryos electroporated with pTTFLacZ exhibited β-galactosidase activity in both head endoderm and in the endodermal strand of the tail. These embryos exhibited the morphology typical of this stage of development and muscle marker gene expression occurred only in their tail muscle cells (Fig. 3A) demonstrating that pTTFLacZ does not disrupt the normal pattern of embryogenesis and therefore, serves as an appropriate negative control. In many instances, we also observed β-galactosidase activity only in lateral regions of the posterior head and the endodermal strand of embryos electroporated with pTTFLacZ (Fig. 2D and E). Given the experiments described above demonstrating the fidelity of the Titf1 promoter used in our experiments, we suspect that this activity in the lateral posterior of the head is in the B-line endoderm. However, because we cannot be absolutely certain that none of it was in the mesenchyme, expression of muscle genes in this region will be described as occurring in the “lateral posterior head” (LPH).
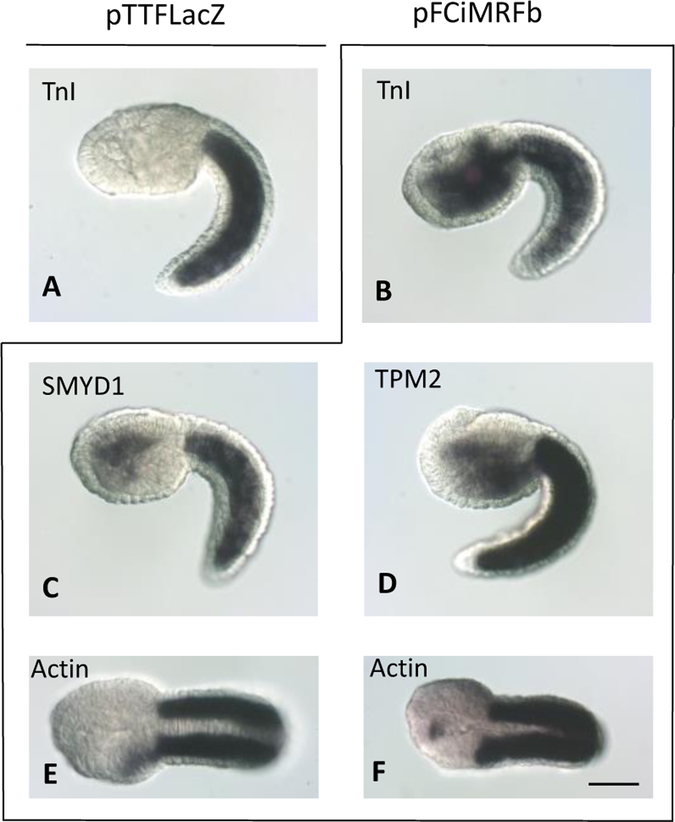
Figure 3.
CiMRF expression in the endoderm activates muscle-specific genes. Embryo in A was electroporated with pTTFLacZ and assayed for TnI; all other embryos were electroporated with pFCiMRFb and assayed for TnI (B), SMYD1 (C), TPM2 (D), or Actin (E, F). Embryos A-D are shown in lateral view, and embryos in E and F are shown in ventral view. Size bar is 50μm, and all embryos shown are C. intestinalis.
We limited our analysis of the effect of CiMRF to head endoderm, because any induced muscle gene activity in the endodermal strand was usually obscured by normal muscle gene activity in the adjacent tail muscle cells. Expression of three single-copy muscle-specific genes, Troponin I (TnI), SET-MYND-1 (SMYD-1), and Tropomyosin-2 (TPM-2), was clearly evident in the anterior endoderm in the head of embryos electroporated with pFCiMRFb (Fig. 3B–D), and was often observed in the posterior head of such embryos as well (e.g. Fig. 3B and D). In addition, a probe representing the Actin gene family often yielded a signal (20–40% of pFCiMRFb electroporated embryos) that was limited to the LPH (Fig. 3E), although in a small number of embryos this signal also was observed in the anterior of the head, which is clearly endoderm (< 10%; Fig. 3F). Probes against five other genes or gene families (Myosin Binding Protein [MBP], Myosin Light Chain [MLC], Myosin Heavy Chain [MHC], Myosin Regulatory Light Chain [MRLC], and Troponin T [TnT]) did not yield a signal anywhere in the head in response to electroporated pFCiMRFb (data not shown). Most notably, our results show that CiMRF activates three, and probably four, of the muscle-specific genes that we tested in the embryonic endoderm. Because of their robust response to CiMRF, their well-defined expression in the endoderm, and because they are single copy genes, all further analyses of muscle gene expression in the endoderm were restricted to TnI, SMYD-1, and TPM-2.
CiMRF Suppresses Endoderm Development
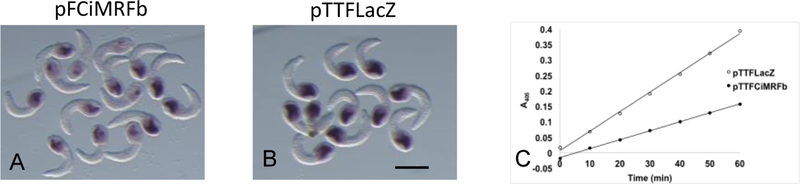
We used alkaline phosphatase as a marker to assess the effect of CiMRF on endoderm development. Although this enzyme does not occur uniquely in the endoderm of Ciona embryos, its level of activity there is so much greater than in all other embryonic tissues combined that it is effectively endoderm specific (Whittaker and Meedel, 1989). Histochemical assays indicated that alkaline phosphatase activity in embryos electroporated with pFCiMRFb was typically less than in that of control embryos electroporated with pTTFLacZ (compare Fig. 4A and 4B). Quantitative colorimetric assays showing that the level of alkaline phosphatase activity in pFCiMRFb-electroporated embryos was about one-half of that in pTTFLacZ-electroporated embryos confirmed this observation (Fig. 4C). These results demonstrate that CiMRF expression in the endoderm suppresses the differentiation of that tissue.
Figure 4.
CiMRF expression in the endoderm suppresses embryonic alkaline phosphatase activity. Embryos were electroporated either with pFCiMRFb (A) or pTTFLacZ (B) and examined for alkaline phosphatase activity using a histochemical assay. Enzyme activity is proportional to the intensity of the purple colored precipitate in the heads of the embryos and appears to be reduced by about 50% in embryos electroporated with pFCiMRFb (A) relative to those electroporated with pTTFLacZ (B). This was confirmed by a quantitative colorimetric assay that showed the ratio of alkaline phosphatase activity in embryos electroporated with pFCiMRFb relative to those electroporated with pTTFLacZ was 0.54 (C). In two other experiments this ratio was 0.43 and 0.57. Size bar in (B) is 100μm, and all embryos shown are C. intestinalis.
Misexpressing CiMRF Activates the miR-1/133 Complex
Others have shown that muscle-specific micro RNAs (miRNAs) including miR-1, miR-133, and/or miR-206 can be activated by expressing vertebrate MRFs in mouse embryonic fibroblasts and in chick neural tubes (Rosenberg, et.al., 2006; Sweetman, et al., 2008). Additionally, the study of Rosenberg, et al. (2006) demonstrated that expression of endogenous cell fate markers was suppressed by MyoD-activated miR-206. While Ciona do not have the miR-206 gene, they do have the muscle-specific miR-1/miR-133 complex (Kusakabe, et.al., 2013) that likely was duplicated in the lineage leading to vertebrates to give rise to the closely related miR-206 gene (Tani, et.al., 2013). Given the strong conservation between CiMRF and other MRFs, we speculated that CiMRF also elicits muscle-specific miRNA activity. Here we show that in addition to its expected expression in muscle, the miR-1/miR-133 complex was activated in both notochord and endoderm in response to CiMRF expression in those tissues (Fig. 5). In three separate experiments, cleavage-arrested embryos that were electroporated with pTCiMRFb had 2–3 more miR-1/133 positive cells/embryo (attributed to expression in the notochord) than those electroporated with pTLacZ (Fig. 5). Similarly, over 40% of embryos electroporated with pFCiMRFb exhibited miR-1/133 expression in the head, whereas only one of 149 embryos electroporated with pFLacZ did so (Table 3; Fig. 5). These findings indicate that like vertebrate MRFs, CiMRF activates muscle-specific miRNA gene expression, and lead us to suggest that like mouse MyoD, and possibly other MRFs, it may suppress endogenous developmental programs via a pathway involving miRNA genes.
Figure 5.
CiMRF expression leads to activation of the miR-1/133 complex. Embryos were subjected to in situ hybridization with a probe complementary to miR-1/133 complex transcripts. Zygotes electroporated with pTLacZ (A) or pTCiMRFb (B) were cleavage-arrested at the 64-cell stage and subjected to in situ hybridization at the equivalent of early-mid tail formation stage. Embryos in panels A and B are shown with the presumptive primary notochord lineage at the top and the presumptive muscle lineage at the bottom. Zygotes electroporated with pFLacZ (C) or pFCiMRFb (D) were allowed to develop normally and subjected to in situ hybridization at early-mid tail formation stage. Size bars are 50μm, and all embryos shown are C. robusta.
Table 3.
Ectopic CiMRF elicits miR-1/133 expression in the notochord and endoderm.
| Exp.# | pTLacZ | pTCiMRFb | pFLacZ | pFCiMRFb |
|---|---|---|---|---|
| 1 | 4.1 ± 0.9 (31) | 6.3 ± 1.4 (28) | 0/51 | 33/63 |
| 2 | 5.6 ± 0.9 (22) | 7.6 ± 1.6 (36) | 0/39 | 42/93 |
| 3 | 5.2 ± 1.3 (44) | 8.1 ± 2.1 (74) | 1/58 | 34/96 |
Results of three experiments showing the mean number of miR-1/133 positive cells per cleavage arrested embryo in zygotes electroporated with pTLacZ or pTCiMRFb (number of embryos examined is shown in parentheses). The number of embryos positive for miR-1/133 transcripts in the head endoderm /total number of embryos examined is shown for zygotes electroporated with pFLacZ and pFCiMRFb. In the third experiment with embryos electroporated with pFLacZ, three of the 58 embryos were scored as miR-1/133 positive in the posterior-lateral region of the head.
The CiMRF N-terminus is Essential for Myogenesis
Our interest in the functional evolution of the MRF family led us to construct a series of plasmids to determine whether other members of this family were myogenic in our assays. Other than CiMRF, of the five MRFs we tested in the notochord – mouse (Mus musculus; MyoD), sea urchin (Lytechinus variegatus; SUM1), fruit fly (Drosophila melanogaster; Nautilus), nematode (Caenorhabditis elegans; hlh1) and ascidian (Ciona savignyi; CsMRF), only the ascidian MRF was myogenic (unpublished results). The one obvious difference among the MRFs that were or were not myogenic in this assay was the presence of an exceptionally large N-terminal domain preceding the Cys-His rich/basic helix-loop helix domain (CH/b-hlh domain). N-termini of the MRFs of the two Ciona species are approximately 360 amino acids in length, whereas those of the four other MRFs tested range from approximately 80 to 120 amino acids. To test whether this domain might possess elements that are necessary for MRF activity in Ciona embryos, we created a series of plasmids encoding chimeric MRFs in which the CH/b-hlh domain of the MRFs of the species mentioned above (or just the b-hlh domain of the nematode MRF, which lacks a CH domain) replaced the CiMRF CH/b-hlh domain. In support of a potentially important role(s) of the Ciona MRF N-terminus in myogenesis, all chimeric constructs were found to direct muscle gene expression in the notochord (unpublished results).
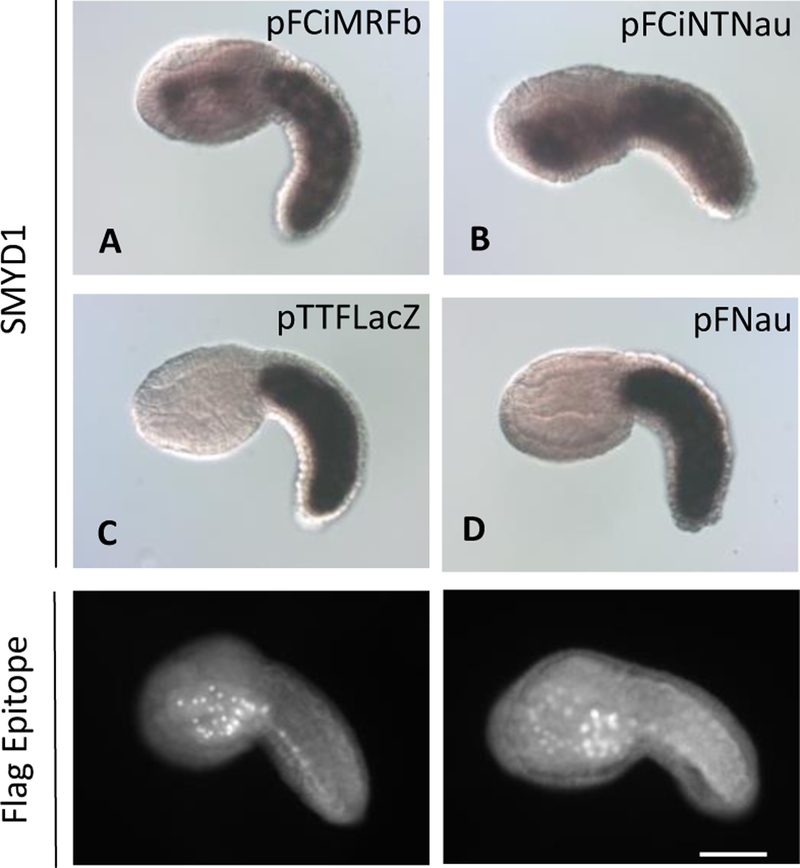
Here, two additional studies using the endoderm assay are presented that indicate the CiMRF N-terminus plays an essential functional role(s) in muscle development during Ciona embryogenesis. In these studies, only embryos electroporated with chimeric plasmids containing the CiMRF N-terminus expressed muscle markers in the head, although, for reasons described above, embryos that exhibited such activity only in the lateral posterior head were not counted as positive for muscle expression in the endoderm. We first created a plasmid (pFGFPΔNTCiMRF) to express a GFP-CiMRF fusion protein in the endoderm in which the CiMRF N-terminus was replaced with GFP. Despite detecting GFP signal in the endodermal cell nuclei of embryos expressing GFP-CiMRF (Fig. 6A), this fusion protein was not myogenic in any of the embryos examined (Table 4; Fig. 6B) demonstrating that the lack of myogenic activity of this protein was not due to its absence from the endoderm, or from its failure to become properly localized within endodermal cells. Next, we constructed two plasmids to test the effect of replacing the N-terminus of Nautilus with the CiMRF N-terminus: pFNau expressed the complete Nautilus protein, and pFCiNTNau expressed a chimeric protein in which the CiMRF N-terminus replaced the Nautilus N-terminus. pFCiNTNau was designed to rule out possible complications arising from the presence of the CiMRFb C-terminus in the chimeric proteins that we previously tested in the notochord, which contained the C-terminal domain of CiMRFb as well as its N-terminus. In addition, unlike the chimeric proteins expressed in the notochord, both constructs encoded a Flag epitope tag, so that we could determine if they produced stable, properly localized proteins. In three separate experiments, only the chimeric fusion protein produced by pFCiNTNau was myogenic (Fig. 7 and Table 5). Notably, the Flag epitope was detected in endodermal cell nuclei of embryos electroporated with pFNau (Fig. 7), indicating that the failure of Nautilus protein to stimulate myogenesis was not due to its absence or to its failure to be localized in nuclei. These findings further support our assertion that the CiMRF N-terminus contains features that are essential for myogenesis in Ciona embryos.
Figure 6.
Loss of the N-terminus eliminates myogenicity of CiMRF. Zygotes electroporated with pFGFPΔNTCiMRF were raised to the early-mid tail formation stage and examined for GFP fluorescence (A). Embryos exhibiting GFP fluorescence were then examined by ISH for TnI expression, which was only detected in the muscle cells (B). The same C. intestinalis embryo is shown in ventral view in both panels. Size bar is 50μm.
Table 4.
GFP does not possess the myogenic properties of the CiMRF N-terminus.
| Marker | |||
|---|---|---|---|
| Plasmid | SMYD1 | TPM2 | TnI |
| pTTFLacZ | 0/85 | 0/76 | 0/74 |
| pFCiMRFb | 55/79 | 52/74 | 73/89 |
| pFGFPCiMRFANT | 1/82 | 0/76 | 0/106 |
Zygotes were electroporated with the indicated plasmid and reared to early-mid tail formation stage when they were fixed and processed for ISH. Data are expressed as the number of embryos that expressed a given muscle marker in the endoderm/total number of embryos assayed. Results reflect the combined totals of three independent experiments.
Figure 7.
The CiMRF N-terminus confers myogenicity on Nautilus. Embryos electroporated with pFCiMRFb (A) and pFCiNTNau (B) express SMYD1 in the endoderm, whereas embryos electroporated with pTTFLacZ (C) and pFNau (D) do not. Flag-tagged Nau protein and Flag-tagged CiMRF-Nau chimeric protein is present in endodermal cell nuclei of embryos electroporated with pFNau (E) and pFCiNTNau (F). Size bar is 50μm, and all embryos shown are C. intestinalis.
Table 5.
The CiMRF N-terminus confers myogenic potential on Nautilus.
| Marker | |||
|---|---|---|---|
| Plasmid | SMYD1 | TPM2 | TnI |
| pTTFLacZ | 0/126 | 1/180 | 0/183 |
| pFCiMRFb | 62/122 | 122/144 | 133/149 |
| pFNau | 0/69 | 0/79 | 0/65 |
| pFCiNTNau | 41/66 | 69/78 | 83/126 |
Zygotes were electroporated with the indicated plasmid and reared to early-mid tail formation stage when they were fixed and processed for ISH. Data are shown as the number of embryos expressing a given muscle marker in the endoderm/total number of embryos examined. Results reflect three independent experiments.
Functional Conservation of Ascidian MRF N-termini
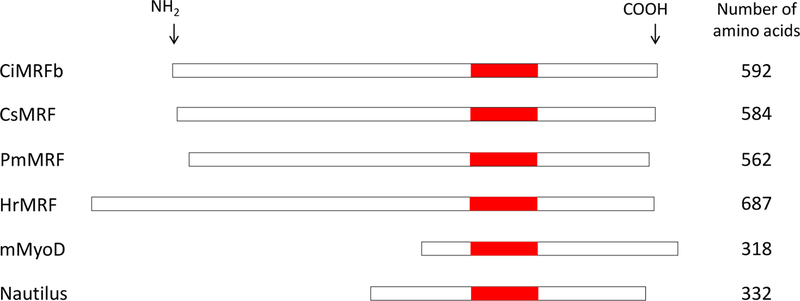
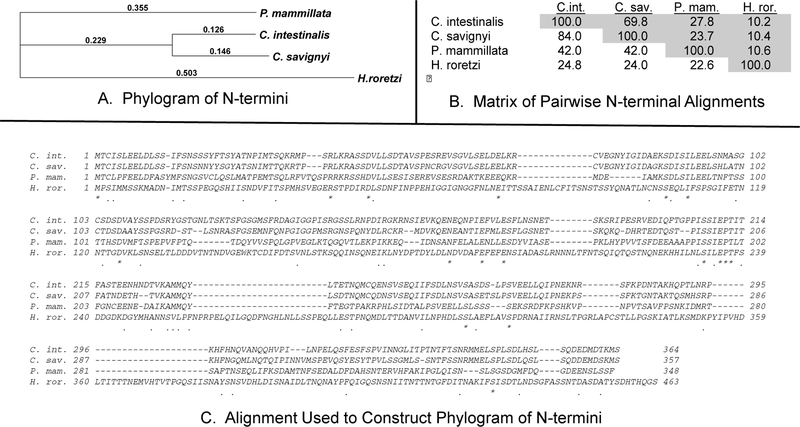
The results described above raised the question of whether domains similar to the CiMRF N-terminus are unique to Ciona species or whether they might also be present in MRFs of other animals. Database searches revealed large N-termini in several species of Platyhelminthes (e.g. 560 amino acids in the putative MRF of Schistosoma mansoni [Accession XP_018645980]), but none exhibited sequence similarity with the CiMRF N-terminus and no very large N-termini were identified in the MRFs of other chordates or deuterostomes. We also identified large N-termini in two other ascidian species whose MRFs have been characterized (see below). An alignment of these four ascidian MRFs together with mouse MyoD, and D. melanogaster Nautilus is shown in Figure 8. The ascidian proteins are clearly much larger than either of the other two MRFs, and this difference is attributable to the dramatic increase in the number of amino acids preceding the CH/b-hlh domain, which we refer to as the N-terminal domain.
Figure 8.
MRF Alignment. Scale drawings of MRFs of four ascidian species (C. intestinalis, C. savignyi, P. mammillata, and H. roretzi), mouse MyoD (Mus musculus; Ensembl:ENSMUSG00000009471), and Nautilus (Drosophila melanogaster; FLYBASE:FBgn0002922) illustrate the relatively large N-termini of ascidian MRFs. The amino acid content of each MRF is shown on the right of the diagram, and MRFs are aligned on their C/H-bhlh domains, which are shown in red.
In order to assess whether the large N-termini of ascidian MRFs are functionally conserved we chose to test these domains of the MRFs of Ciona savignyi (CsMRF), Phallusia mammillata (PmMRF), and Halocynthia roretzi (HrMRF) in our endoderm assay. These species were chosen based on the availability and reliability of their MRF sequences and their phylogenetic relationship to Ciona intestinalis. C. savignyi (family Cionidae) is a sister species of C. intestinalis, P. mammillata (family Ascidiidae) is more distantly related, and H. roretzi (family Pyuridae) is considered to be very distantly related to the other three species (Shenkar, et.al., 2018). Comparison of the N-terminal sequences of these four ascidian MRFs was consistent with the above classification that is based on extensive classic criteria and molecular data, and showed that the sequences of the other ascidian MRF N-termini differ from that of CiMRF as a function of their phylogenetic position relative to Ciona intestinalis. (Fig 9). The alignment of Figure 9C also illustrates that there are very few amino acid sequence similarities that occur in all four N-termini. However, pairwise alignments (Fig. 9B) of these MRF sequences (Fig. S1) reveal extensive similarities between the N-termini of the C. intestinalis and C. savignyi proteins and several, though fewer, regions of similarity between either the C. intestinalis or C. savignyi N-termini and the P. mammillata MRF N-terminus, and very few similarities between the N-terminus of the H. roretzi MRF and those of any of the other three ascidian MRFs. In fact, pairwise comparison of the H. roretzi MRF N-terminus with those of each of the other three ascidian species indicated sequence similarities no greater than would be expected by chance alone. For example, sequence identity and sequence similarity between the CiMRF and HrMRF N-termini were 9.9% and 15.8% respectively, which were no higher than the values obtained when we compared ten random sequences generated from the HrMRF N-terminus to the CiMRF N-terminus (in fact, they were slightly lower; unpublished result). We conclude that the N-termini of CiMRF and HrMRF are unrelated with respect to primary sequence.
Figure 9.
Comparison of the N-termini of four ascidian MRFs. Data were generated using the Multiple Sequence Alignment tool of the Clustal W Phylogeny software in MacVector 16.0.8. The phylogram in panel A (Method: Neighbor joining; Best Tree; tie-breaking = random) was based on the alignment shown in panel C, which used the following multiple alignment parameters: Open Gap Penalty = 10.0; Extended Gap Penalty = 0.2; Delay Divergent = 30%; Gap Distance = 4; Similarity Matrix: gonnet. Distances presented for each branch were uncorrected and gaps were distributed proportionally. Identical amino acids (*) and similar amino acids (.) are indicated below the alignments. Panel B shows the percent identity (above the diagonal) and percent similarity (below the diagonal) scores of pairwise alignments of the four N-termini. Similarity scores reflect the percentage of amino acid identities plus the percentage of amino acid similarities. Alignments were done in the slow mode and alignment parameters were: Open Gap Penalty = 10.0; Extend Gap Penalty = 0.1; Similarity Matrix: gonnet.
By comparing the ability of the different ascidian MRF N-termini to support myogenesis as described below, we believed it might be possible to distinguish whether sequence similarities encoding secondary structural motifs or a shared property such as enrichment of certain amino acids (e.g. the serine content of the four ascidian MRF N-termini is high [12–17%] as is the level of acidic amino acids [Glu + Asp =13–17%]) is key to their role(s). In order to do this, we designed chimeric fusion proteins in which the N-terminus of each of the three ascidian MRFs replaced this domain of CiMRF. Fusion proteins were expressed from plasmids designated pFCsCi, pFPmCi, and pFHr2Ci, where the third and fourth characters of each plasmid name correspond to the first letters of the genus and species names from which the N-terminal coding sequences were derived (e.g. in pFCsCi the N-terminus of the Ciona savignyi MRF replaced the N-terminus of CiMRF).
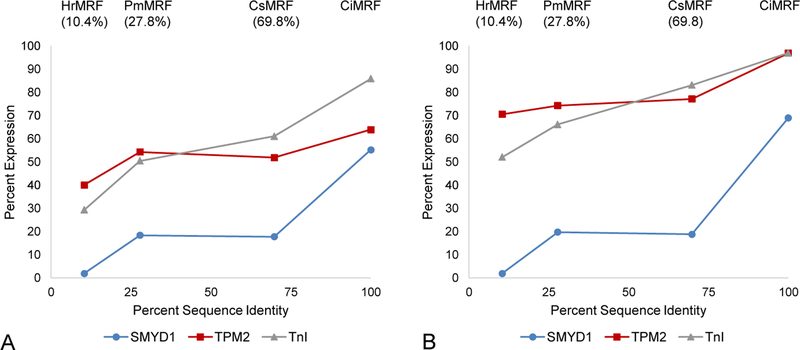
Electroporated embryos were scored as either endoderm positive (End+), lateral posterior head positive (LPH+), or negative for ectopic expression of TnI, TPM-2, or SMYD-1 in these experiments. This scoring system resulted in undercounting embryos that were positive for muscle marker expression in the LPH because all embryos expressing a muscle marker in the anterior endoderm were scored as End+, regardless of whether they were also LPH+, as were most of the End+ embryos. Embryos scored as LPH+ expressed muscle markers only in the lateral posterior head. With the exception of pFCsCi and pFPmCi probed for TPM2, the three chimeric plasmids were not as effective as pFCiMRFb at eliciting muscle gene activity in the endoderm (Fig 10A; Table 6; Table S1). Including LPH+ data in our analysis, under the assumption that such expression was in the B-line endoderm, as we believe is likely, indicated that neither pFCsCi nor pFPmCi were as effective as pFCiMRFb at directing ectopic TPM-2 expression (Fig. 10B; Table 6; Table S1). In addition, only pFHr2Ci was unable to stimulate SMYD-1 expression anywhere in the head relative to the negative control to an extent that was judged statistically significant (Fig. 10 A and B; Table 6; Table S2). These results demonstrate that all three chimeric proteins can direct muscle gene expression in the endoderm, although not as effectively as CiMRF.
Figure 10.
Relative myogenic activity of ascidian MRF N-termini. Data from Table 6 are expressed as the percentage of muscle-specific gene activity as a function of the percent identity (shown in parenthesis) of the indicated N-termini versus the CiMRF N-terminus. (A) Relative expression in End+ embryos. (B) Relative expression in End+ plus LPH+ embryos.
Table 6.
Evaluation of the myogenic activity of chimeric ascidian MRFs and CiMRF.
| SMYD1 | TPM2 | TnI | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Plasmid | End+ | LPH+ | Total | End+ | LPH+ | Total | End+ | LPH+ | Total |
| pTTFLacZ | 0 | 0 | 64 | 0 | 0 | 51 | 1 | 1 | 113 |
| pFCiMRFb | 32 | 8 | 58 | 62 | 32 | 97 | 85 | 11 | 99 |
| pFHr2Ci | 2 | 0 | 109 | 38 | 29 | 95 | 36 | 28 | 123 |
| pFPmCi | 13 | 1 | 71 | 38 | 14 | 70 | 61 | 19 | 121 |
| pFCsCi | 17 | 1 | 96 | 43 | 21 | 83 | 47 | 17 | 77 |
Data are the combined totals of at least three independent experiments and are displayed as the number of embryos positive for the expression of the indicated muscle marker anywhere in the head endoderm (End+) or in the lateral posterior of the head only (LPH+). Total refers to the total number of embryos examined, and differs from the sum of End+ and LPH+ by the number of embryos that were negative for expression of the indicated marker in the head. Using Fisher’s exact test (two tailed) to calculate p-values, there were two instances where chimeric MRF plasmids acted similarly to pFCiMRFb (pFPmCi and pFCsCi assayed for TPM2 expression vs. pFCiMFRb; P = 0.2628 and 0.1293, respectively). In all other cases examined, none were deemed as effective as pFCiMRFb at eliciting muscle gene activity in the endoderm (P < 0.01). If LPH+ embryos were included in this analysis of TPM2 expression the differences between embryos electroporated with pFPmCi or pFCsCi and pFCiMRFb were significant (P < 0.0001 for both plasmids vs. pFCiMRFb). Conversely, except for pFHr2Ci, which like the negative control failed to elicit SMYD1 activity anywhere in the head (P = 0.5311 vs. pTTFLacZ), all plasmids stimulated the expression of all muscle markers to an extent greater than pTTFLacZ (P < 0.01).
Does the CiMRF N-terminus Act as a Transactivation Domain?
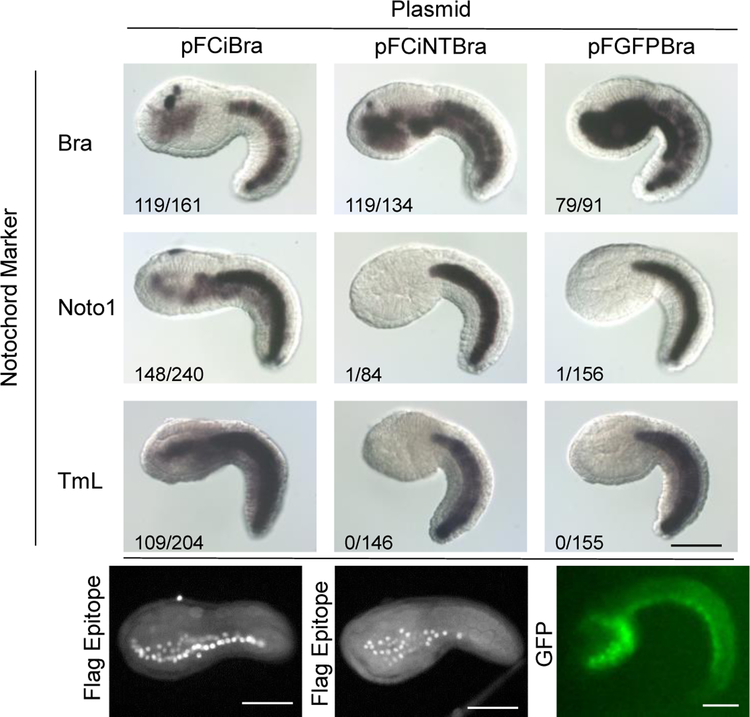
Except for revealing that the CiMRF N-terminus is intrinsically disordered, bioinformatics analyses gave no clues about its structure that might provide insight into its function and did not identify any similar protein sequences, other than those of ascidian MRFs (above and unpublished results). We were, however, able to test the possibility that the CiMRF N-terminus acts as a transcriptional activation domain, albeit one that must have unusual properties since non-ascidian MRFs, which contain their own such domains, were not myogenic in our assay. We did this by constructing a series of plasmids driven by the Titf1 promoter encoding intact Brachyury (pFCiBra) or chimeric proteins in which either the CiMRF N-terminus (pFCiNTBra) or GFP (pFGFPCiBra) replaced the transactivation domain of Brachyury. As expected, electroporation of pFCiBra resulted in the accumulation of notochord-specific transcripts in the endoderm (e.g. Takahashi et al., 1999), but neither pFCiNTBra nor pFGFPCiBra elicited this response (Fig. 11). Detection of the Flag epitope signal in embryos electroporated with pFCiNTBra or of the GFP signal in pFGFPBra indicated that the failure of these plasmids to elicit notochord gene activity was not due to the absence of their fusion proteins in endodermal nuclei (Fig. 11). These findings are not consistent with the idea that the CiMRF N-terminus functions as a transcriptional activation domain, but indicate instead that it, and presumably those of other ascidian MRFs, provide some additional function(s) that is required for these transcription factors to direct myogenesis in Ciona embryos.
Figure 11.
The CiMRF N-terminus cannot replace the transcriptional activation domain of Bra. The upper three rows show examples of notochord marker gene expression in embryos electroporated with the indicated plasmids. Numbers in each figure represent the number of embryos expressing a given marker/total number of embryos examined for each plasmid that was electroporated. Bottom row: fluorescent detection of proteins expressed from pFCiBra and pFCiNTBra by immunostaining the Flag epitope (A & B, respectively) and GFP fluorescence in a live embryo expressing pFGFPBra (C). Size bars are 100μm; all embryos shown are C. intestinalis.
Discussion
Cell Fate Suppression
Our studies provide the first in vivo demonstration that a chordate MRF can suppress endogenous developmental programs: alkaline phosphatase activity was reduced in the endoderm, as was expression of Bra, Noto-1 and Tm-L in the notochord. Features of both of the tissues that we studied persisted in treated embryos, and while the reasons for this might be as simple as mosaic expression of CiMRF in targeted cells or failure to accumulate sufficient levels of CiMRF, numerous precedents exist for co-expression of multiple programs of differentiation within a single cell. For example, cleavage-arrested ascidian zygotes accumulated features of more than one differentiated cell type (Crowther and Whittaker, 1986; Whittaker and Meedel, 1989), and forced expression of mouse MyoD resulted in cells in which endogenous developmental programs coexisted with induced myogenesis (Weintraub, et.al., 1989). Additionally, heterokaryons between rat neural cells and chick skeletal myocytes or between mouse adrenal cells and chick skeletal myocytes exhibited differentiation characters of both partners (Wright, 1984 a, b). Conversely, cleavage-arrest of nematode embryos (Cowan and McIntosh, 1985) or cell fusions (e.g. Weiss and Chaplain, 1971; Killary and Fournier, 1984; Miller, et.al., 1988; Choi, et.al., 1990) yielded only a single developmental program. Collectively, these studies indicate that cellular responses to interventions of this kind are complex and variable, and likely depend on conditions specific to the cells in question.
Together with studies of vertebrate MRFs (Rosenberg, et.al., 2006 and references therein) and the C. elegans MRF (Fukushige and Krause, 2005), our findings indicate that suppression of cell fates is another conserved property of MRFs. As pointed out elsewhere (e.g. Rosenberg, et.al., 2006), this ability is likely to be shared by many, if not all, fate-determining transcription factors because it would allow them to shut down early developmental programs of the cells in which they are normally activated, while also initiating programs that lead to terminal differentiation. In addition, Rosenberg, et.al. (2006) showed that this property of mouse MyoD was mediated by a muscle-specific miRNA (miR 206), which is related to the muscle-specific miR-1/miR-133 complex that we showed was activated in the notochord and endoderm of Ciona embryos when CiMRF was active in those tissues. This finding is consistent with the possibility that miRNA-mediated suppression of endogenous tissue development is a mechanism shared by MRFs, and provides further support for the idea that these proteins exhibit a high level of functional conservation.
Myogenic Potential of CiMRF in the Endoderm
Three genes (SMYD-1, TnI, and TPM-2) were clearly activated in the endoderm in response to CiMRF and transcripts of a fourth representing the Actin gene family were observed in this tissue as well, albeit at a much lower frequency and at levels that were substantially less robust. No other muscle markers we tested were expressed in the endoderm in response to CiMRF. These results differ from a previous study in which electroporation was used to express CiMRF in the notochord of ascidian embryos (Izzi, et.al., 2013). In those experiments, transcripts representing two other gene families (MHC and MRLC) were detected in the notochord in addition to SMYD-1, TnI, and TPM-2; also, the frequency of expression of the Actin family was much higher and its levels were more robust in the notochord than we saw in the current study of the endoderm. We conclude from these results that CiMRF is more effective at eliciting muscle gene activity in a mesodermal tissue like notochord, than in the endoderm. Additionally, when mRNAs encoding CiMRF were injected into Ciona eggs, no muscle gene expression was detected in ectodermal cells of the resulting embryos (Meedel, et.al., 2007). Therefore, it appears that mesodermal and endodermal cells, while differing from one another, are permissive for trans-differentiation to a muscle phenotype by CiMRF, but that ectodermal cells are not. This interpretation supports the existence of a distinct “mesendodermal” regulatory state in Ciona (Rodaway and Patient, 2001; Hudson, et.al., 2016).
The N-terminal Domain is Essential for Myogenesis
Removing the CiMRF N-terminus and replacing it with GFP resulted in a stable, nuclear-localized protein (GFPCiMRF) that was unable to elicit myogenesis. Conversely, replacing the N-terminus of an MRF (Nautilus) that was not myogenic in our assay, but whose presence was detected in endodermal cell nuclei, with the N-terminus of CiMRF resulted in a chimeric protein that was myogenic. Notably, both of these proteins that failed to direct myogenesis possessed authentic MRF b-hlh domains, which in the case of the MyoD b-hlh domain could activate myogenesis by itself (Tapscott, et.al., 1988). Although it is conceivable that GFPCiMRF and Nautilus were inactive as a result of misfolding or because of some other abnormality that was unrelated to the absence of the CiMRF N-terminus, the fact that aberrant proteins tend to form aggregates (Johnston, et.al., 1998), which we did not see, and/or to be unstable (Comyn, et.al., 2014), which was not the case, argue against this possibility. Moreover, their proper localization within target cells, and the fluorescence signal of GFPCiMRF further support our assertion that these proteins assumed normal conformations, leading us to conclude that the most plausible explanation of our results is that the CiMRF N-terminus is essential for myogenesis in Ciona.
Whatever role the ascidian MRF N-terminus plays in myogenesis, it is not likely to function as a transcriptional activation domain, because a chimeric protein in which it replaced the Brachyury transcriptional activation domain did not direct notochord gene expression. Also, a Flag-tagged chimeric protein in which the Herpes simplex VP-16 transcriptional activation replaced the CiMRF N-terminus was not myogenic, but we did not report these results because we could not detect this fusion protein in embryos.
Demonstration that the large N-terminus of ascidian MRFs is necessary for these transcription factors to direct myogenesis in Ciona implies that requirements for MRF function in ascidian embryos differ from those of other organisms and that those requirements are somehow fulfilled by this domain. The intrinsic disorder of the ascidian MRF N-terminus may be relevant to understanding what those requirements are, since disordered domains often interact with other proteins to carry out their activities (Dyson and Wright, 2005; Wright and Dyson, 2015). Assuming that such interactions are necessary for MRF-regulated myogenesis in ascidian embryos, then an approach similar to the one that we used to demonstrate the importance of the CiMRF N-terminus could also be exploited to validate its potential protein partners, which we predict would have the capacity to enable CiMRF to trans-differentiate ectoderm to muscle, which it cannot do otherwise (see above).
Origin of the Ascidian MRF N-terminus
Several mechanisms have been proposed for domain gain in proteins, including gene fusion via joining of exons from adjacent genes, retrotransposition, and exon insertion into introns (Buljan, et.al., 2010; Nagy and Patthy, 2011; Bornberg-Bauer and Mar Albà, 2013). Its N-terminal position is consistent with the origin of the ascidian MRF N-terminus by gene fusion, which is also the mechanism that is generally considered to be the most common pathway for domain gain (Buljan, et.al., 2010; Bornberg-Bauer and Mar Albà, 2013; for an alternative view see Nagy and Patthy, 2011). Gene fusion normally relies on gene duplication (Buljan, et.al., 2010) and while ascidians are known for their compact genomes, gene duplication has occurred in these animals (Dehal, et.al., 2002). On the other hand, the entire CiMRF N-terminus and CH/b-hlh domain reside on a single exon (Meedel and Lee, unpublished results). This argues against exon insertion into an intron (which would require a second exon fusion), but because the absence of introns is often a signature of retrotransposition (Buljan, et.al., 2010; Nagy and Patthy, 2011; Bornberg-Bauer and Mar Albà, 2013), we do not exclude this mechanism as another possibility. Ascidians have also acquired genes by horizontal gene transfer (e.g. a bacterial cellulose synthase; Dehal, et.al., 2002) making it conceivable that such an event was involved in the origin of the N-terminus of their MRFs. As with retrotransposition, gene transfer would presumably require additional processes for domain gain such as fusion of exons from adjoining genes. Finally, because the ascidian N-terminus does not resemble any other identified protein domain, it may have arisen de novo in the tunicates through the stepwise accumulation of many small changes. Such a mechanism is consistent with the relatively rapid evolution of tunicate protein sequences (Tsagkogeorga et al., 2012; Berná and Alvarez-Valin, 2014), and probably would have occurred in concert with changes in the sequence of a predicted binding partner.
The high level of sequence variability and intrinsic disorder of ascidian MRF N-termini have combined to limit the utility of available bioinformatics tools (e.g. sequence comparisons, structure prediction, etc.) for clarifying the origin of the CiMRF N-terminus. However, experimental approaches such as those mentioned above that are designed to further define the role of this evolutionary novelty may provide insight into this intriguing question as well. For instance, N-terminal domain binding partners may have different binding partners in other organisms that could provide clues about its origin.
Evolution of the Ascidian MRF N-terminus
Animals share a highly conserved set of regulatory genes that govern their development (Davidson, 2001; Wilkins, 2002; Carroll, et.al., 2005). Change in the cis-regulatory sequences of these genes leading to their differential deployment is believed to be a key driver of evolutionary diversification (Carroll, 2008; Chan, et.al., 2010; Wittkopp and Kalay, 2012), although it is also apparent that changes in the protein coding regions of regulatory genes are important to evolutionary change as well (Marcellini, et.al., 2003; Lynch and Wagner, 2008; Cheatle Jarvela and Hinman, 2015; Weinberger, et.al., 2017). While MRFs are certainly members of this “toolkit” of important developmental regulatory genes, unlike most other members of this group they exhibit an unusually high degree of evolutionary stasis in their expression pattern and their role during development (see Introduction and references therein). As shown here and elsewhere (Meedel, et.al., 1997; 2002; 2007; Izzi, et.al., 2013), CiMRF is comparable to other MRFs in these respects, and yet together with other ascidian MRFs it has acquired a large N-terminus that appears not to alter its function as an MRF, but instead is required to maintain the ancestral myogenic activity that is the hallmark of this gene family. To our knowledge this is the first example of such a remarkable occurrence and it raises several issues that are addressed below.
Large MRF N-termini have been identified in what are most likely to be MRFs of Platyhelminthes such as S. mansoni. Could the MRF N-termini of this group of animals be homologous with the N-termini of the ascidian MRFs that we characterized? Sequence comparison between the S. mansoni MRF N-terminus and that of ascidian MRFs revealed no significant regions of similarity (unpublished result). This result was not unexpected considering that no such regions of similarity were apparent between the CiMRF N-terminus and the N-terminus of the MRF of another ascidian, H. roretzi. However, the observation that large MRF N-termini are not only extremely rare, but have not been found in any chordates or deuterostomes other than ascidians, indicate that these domains of Platyhelminthes and ascidian MRFs are not homologous. Instead, the most parsimonious explanation is that these two groups of animals acquired these N-termini independently. Nevertheless, it would be interesting to test whether a chimeric protein in which the N-terminus of a Platyhelminthes MRF replaced that of CiMRF is myogenic in our assay. Perhaps even more revealing, and as a way to further distinguish between the importance of primary sequence versus amino acid composition in its activity, would be to test whether a CiMRF protein containing a randomized N-terminus is myogenic in our assay. Because intrinsically disordered protein domains often acquire secondary structural motifs that are functionally important when they interact with their binding partners (Dyson and Wright, 2005; Wright and Dyson, 2015), if this were the case with CiMRF, then we would expect that altering its primary sequence by randomizing its N-terminus would have a deleterious effect on myogenic activity.
Within the tunicates, the large MRF N-terminus is found in all of the ascidians that we examined, but it does not occur in the putative MRF of Oikopleura dioica (Denoeud, et.al., 2010; Accession: CBY08250.1), which is a member of the class Appendicularia. Thus, within this chordate subphylum, large MRF N-termini may be limited to the class Ascidiacea. No MRF sequences are available currently from the Thaliacea (traditionally considered to be the third class of tunicates consisting of the doliolids, pyrosomes, and salps; Holland, 2016). It will be interesting to determine whether this group possesses MRFs with large N-termini since phylogenomic studies place thaliaceans within the Ascidiacea (e.g. Kocot, et al., 2018).
Other than the acquisition of an unusual and essential N-terminus, the most remarkable finding of our studies is that despite the high degree of amino acid sequence variation of this domain in ascidian MRFs, its ability to facilitate myogenesis in Ciona is conserved to some extent. This conclusion is best illustrated by the myogenic activity of the chimeric protein Hr2CiMRF, which despite having an N-terminus whose sequence is no more related to that of CiMRF than is expected by chance, was able to stimulate endodermal expression of both TnI and TPM-2 although to a lesser extent than was a plasmid encoding full-length CiMRFb. Together with the ability to direct muscle gene expression of the other two ascidian MRF fusion proteins that we studied, these findings indicate that a remarkable degree of sequence variation is allowed in the ascidian MRF N-terminus. This is in striking contrast to the relatively minor variability seen when comparing their CH/b-hlh domains, and demonstrates that different selection pressures must be acting on these two parts of ascidian MRFs.
All of the ascidian MRF fusion proteins were able to stimulate muscle gene expression, but except for two situations (CsCiMRF and PmCiMRF electroporated End+ embryos assayed for TPM-2 expression), none were as effective as CiMRF. Additionally, Hr2CiMRF was completely ineffective at eliciting SMYD-1 expression in our assay. Collectively, these results point to a general but imperfect trend in which the ability of a given ascidian MRF N-terminus to facilitate muscle gene expression in Ciona as part of a fusion protein with CiMRF correlates with the similarity of its sequence to the CiMRF N-terminus.
Our studies also demonstrate that different regulatory requirements exist for muscle-specific genes whose activities are regulated by MRFs in ascidians that depend on the properties of the N-terminus. For example, Hr2CiMRF drives expression of TnI and TPM-2 in the Ciona embryo assay, but not SMYD-1. This result indicates that as MRF N-termini diverged in ascidians, other components of the myogenic control system that interact with this domain undoubtedly diverged as well to maintain a robust regulatory state in the different species.
We have shown that the unusual N-terminus of ascidian MRFs is necessary for their function as myogenic regulatory factors. The discovery of an essential domain of a gene regulatory protein that has a highly conserved function (i.e. promoting myogenesis), but that occurs only in a subset of the organisms in which those proteins direct a particular developmental process (i.e. myogenesis) appears to be an evolutionary novelty. We expect that as more in-depth studies are undertaken, particularly of non-model systems, other examples of this phenomenon will be identified. Perhaps the large N-termini of the putative MRFs of Platyhelminthes represent another such case.
Supplementary Material
Highlights.
Ascidians have evolved novel requirements for myogenesis
Ascidian Myogenic Regulatory Factors (MRFs) possess large and unusual N-termini
Misexpressing ascidian MRFs activates muscle genes in Ciona endoderm and notochord
Non-ascidian MRFs are myogenic only when fused to the Ciona MRF N-terminus
Endogenous developmental programs are suppressed in response to the Ciona MRF
Acknowledgments:
We thank Bob Zeller for his insightful comments that improved this manuscript and Chris Hemme for his help with the phylogenetic analysis. This research was supported by Student Training Pilot Awards made available by RI-INBRE Grants 8P20GM103430 and 2P0GM103430 and by Awards from the National Institutes of Health (2R15HD047357–02; 3R15HD047357–02S1; 3R15HD47357–02S2; 3R15HD047357–02S3) to THM.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This paper is dedicated to the memory of Jamie Lee, an exceptional scientist, an outstanding mentor, and an even better friend.
References
- Araki S, Saiga H, Makabe KW, Satou N 1994. Expression of AMD 1, a gene for a MyoD 1-related factor in the ascidian Halocynthia roretzi. Roux’s Arch. Dev. Biol 203, 320–327. doi: 10.1007/BF00457803 [DOI] [PubMed] [Google Scholar]
- Baylies MK, Michelson AM, 2001. Invertebrate myogenesis: looking back to the future of muscle development. Current Opinion in Genetics & Development 11, 431–439. 10.1016/S0959-437X(00)00214-8 [DOI] [PubMed] [Google Scholar]
- Berkes CA and Tapscott SJ 2005. MyoD and the transcriptional control of myogenesis. Semin. Cell Dev. Biol 16, 585–595. doi: 10.1016/j.semcdb.2005.07.006 [DOI] [PubMed] [Google Scholar]
- Berná L and Alvarez-Valin F 2014. Evolutionary genomics of fast evolving tunicates. Genome Biol. Evol 6, 1724–1738. 10.1093/gbe/evu122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornberg-Bauer E and Mar Albà M 2013. Dynamic and adaptive benefits of modular protein evolution. Curr. Opin. Struc. Biol 23, 459–466. doi: 10.1016/j.sbi.2013.02.012 [DOI] [PubMed] [Google Scholar]
- Bouchemousse S, Bishop JDD, and Viard F 2016. Contrasting global genetic patterns in two biologically similar, widespread and invasive Ciona species (Tunicata, Ascidiacea). Scientific Reports | 6:24875 | doi: 10.1038/srep24875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan TJ, Chakraborty T, Olson EN 1991. Mutagenesis of the myogenin basic region identifies an ancient protein motif critical for activation of myogenesis. Proc. Natl. Acad. Sci 88, 5675–5679. 10.1073/pnas.88.13.5675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Bajard L, Chang T, Daubas P, Hadchouel J, Meilhac S, Montarras D, Rocancourt D, and Relaix F, 2003. The formation of skeletal muscle: from somite to limb. J. Anat 202, 59–68. doi: 10.1046/j.1469-7580.2003.00139.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M and Rigby PWJ 2014. Gene regulatory networks and transcriptional mechanisms that control myogenesis Dev. Cell 28, 225–238. doi: 10.1016/j.devcel.2013.12.020 [DOI] [PubMed] [Google Scholar]
- Buljan M, Frankish A, and Bateman A 2010. Quantifying the mechanisms of domain gain in animal proteins. Genome Biol 11, R74. doi: 10.1186/gb-2010-11-7-r74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134, 25–36. doi: 10.1016/j.cell.2008.06.030 [DOI] [PubMed] [Google Scholar]
- Carroll SB, Grenier JK and Weatherbee SD 2005. From DNA to Diversity. Molecular Genetics and the Evolution of Animal Design, 2nd ed., Blackwell Publishing, Malden, MA, USA: ISBN-13: 978–1405119504 [Google Scholar]
- Chan YF, Marks ME, Jones FC Villarreal G, Shapiro MD, Brady SD, Southwick AM, Absher DM, Grimwood J, Schmutz J, Myers RM, Petrov D, Jonsson B, Schluter D, Bell MA, and Kingsley DM 2010. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science 327, 302–305. doi: 10.1126/science.1182213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheatle Jarvela A.M. and Hinman VF 2015. Evolution of transcription factor function as a mechanism for changing metazoan developmental gene regulatory networks. Evo. Devo 6, 3. doi: 10.1186/2041-9139-6-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Costa ML, Mermelstein CS, Chagas C, Holtzer S, and Holtzer H 1990. MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc. Natl. Acad. Sci. USA 87, 7988–7992. 10.1073/pnas.87.20.7988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comyn SA, Chan GT, and Mayor T 2014. False start: Cotranslational protein ubiquitination and cytosolic protein quality control. J. Proteomics 100, 92–101. doi: 10.1016/j.jprot.2013.08.005 [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW 1997. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development 124, 589–602. PMID: [DOI] [PubMed] [Google Scholar]
- Cowan A and McIntosh R 1985. Mapping the distribution of differentiation potential for intestine, muscle, and hypodermis during early development in Caenorhabditis elegans. Cell 41, 923–932. 10.1016/S0092-8674(85)80073-8 [DOI] [PubMed] [Google Scholar]
- Crowther RJ and Whittaker JR 1986. Differentiation without Cleavage: Multiple Cytospecific Ultrastructural Expression in Individual One-Celled Ascidian Embryos. Dev. Biol 117, 114–126. 10.1016/0012-1606(86)90354-4 [DOI] [PubMed] [Google Scholar]
- Davidson EH 2001. Genomic Regulatory Systems: Academic Press, San Diego, CA, USA: ISBN 9780122053511; eBook ISBN: 9780080525594 [Google Scholar]
- Davis RL and Weintraub H 1992. Acquisition of myogenic specificity by replacement of three amino acid residues from MyoD into E12. Science 256, 1027–1030. doi: 10.1126/science.1317057 [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, Harafuji N, Hastings KE, Ho I, Hotta K, Huang W, Kawashima T, Lemaire P, Martinez D, Meinertzhagen IA, Necula S, Nonaka M, Putnam N, Rash S, Saiga H, Satake M, Terry A, Yamada L, Wang HG, Awazu S, Azumi K, Boore J, Branno M, Chin-Bow S, DeSantis R, Doyle S, Francino P, Keys DN, Haga S, Hayashi H, Hino K, Imai KS, Inaba K, Kano S, Kobayashi K, Kobayashi M, Lee BI, Makabe KW, Manohar C, Matassi G, Medina M, Mochizuki Y, Mount S, Morishita T, Miura S, Nakayama A, Nishizaka S, Nomoto H, Ohta F, Oishi K, Rigoutsos I, Sano M, Sasaki A, Sasakura Y, Shoguchi E, Shin-i T, Spagnuolo A, Stainier D, Suzuki MM, Tassy O, Takatori N, Tokuoka M, Yagi K, Yoshizaki F, Wada S, Zhang C, Hyatt PD, Larimer F, Detter C, Doggett N, Glavina T, Hawkins T, Richardson P, Lucas S, Kohara Y, Levine M, Satoh N, Rokhsar DS 2002. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157–2167. doi: 10.1126/science.1080049 [DOI] [PubMed] [Google Scholar]
- Delfini M and Duprez D 2004. Ectopic Myf5 or MyoD prevents the neuronal differentiation program in addition to inducing skeletal muscle differentiation, in the chick neural tube. Development 131, 713–723. doi: 10.1242/dev.00967 [DOI] [PubMed] [Google Scholar]
- Denoeud F, Henriet S, Mungpakdee S, Aury JM, Da Silva C, Brinkmann H, Mikhaleva J, Olsen LC, Jubin C, Canestro C, Bouquet JM, Danks G, Poulain J, Campsteijn C, Adamski M, Cross I, Yadetie F, Muffato M, Louis A, Butcher S, Tsagkogeorga G, Konrad A, Singh S, Jensen MF, Huynh Cong E, Eikeseth-Otteraa H, Noel B, Anthouard V, Porcel BM, Kachouri-Lafond R, Nishino A, Ugolini M, Chourrout P, Nishida H, Aasland R, Huzurbazar S, Westhof E, Delsuc F, Lehrach H, Reinhardt R, Weissenbach J, Roy SW, Artiguenave F, Postlethwait JH, Manak JR, Thompson EM, Jaillon O, Du Pasquier L, Boudinot P, Liberles DA, Volff JN, Philippe H, Lenhard B, Roest Crollius H, Wincker P and Chourrout D 2010. Plasticity of animal genome architecture unmasked by rapid evolution of a pelagic tunicate. Science 330, 1381–1385. doi: 10.1126/science.1194167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Gregorio A and Levine M 1999. Regulation of Ci-tropomyosin-like, a Brachyury target gene in the ascidian Ciona intestinalis. Development 126, 5599–5609. PMID: [DOI] [PubMed] [Google Scholar]
- Dyson JH and Wright PE 2005. Intrinsically unstructured proteins and their functions. Nature Reviews Mol. Cell Biol 6, 197–208. doi: 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- Fukushige T and Krause M 2005. The myogenic potency of HLH-1 reveals wide-spread developmental plasticity in early C. elegans embryos. Development 132, 1795–1805. doi: 10.1242/dev.01774 [DOI] [PubMed] [Google Scholar]
- Harafuji N, Keys D, Levine M 2002. Genome-wide identification of tissue-specific enhancers in the Ciona tadpole. Proc. Nat. Acad. Sci 99, 6802–6805. doi: 10.1073/pnas.052024999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland LZ 2016. Tunicates. Current Biology 26, R146–R152. 10.1016/j.cub.2015.12.024 [DOI] [PubMed] [Google Scholar]
- Hotta K, Takahashi H, Asakura T, Saitoh B, Takatori N, Satou Y, and Satoh N 2000. Characterization of Brachyury-downstream notochord genes in the Ciona intestinalis embryo. Dev. Biol 224, 69–80. doi: 10.1006/dbio.2000.9765 [DOI] [PubMed] [Google Scholar]
- Hudson C, Sirour C, and Yasuo H 2016. Co-expression of Foxa.a, Foxd and Fgf9/16/20 defines a transient mesendoderm regulatory state in ascidian embryos. Elife 2016 Jun 28;5. doi: 10.7554/elife.14692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai K, Hino K, Yagi K, Satoh N and Satou Y 2004. Gene expression profiles of transcription factors and signaling molecules in the ascidian embryo: towards a comprehensive understanding of gene networks. Development 131, 4047–4058 doi: 10.1242/dev.01270 [DOI] [PubMed] [Google Scholar]
- Izzi S, Colantuono B, Sullivan K, Khare P, Meedel TH 2013. Functional studies of the Ciona intestinalis myogenic regulatory factor reveal conserved features of chordate myogenesis. Dev. Biol 376, 213–223. doi: 10.1016/j.ydbio.2013.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang D, Munro EM, and Smith WC 2005. Ascidian prickle regulates both mediolateral and anterior-posterior cell polarity of notochord cells. Curr. Biol 15, 79–85. 10.1016/j.cub.2004.12.041 [DOI] [PubMed] [Google Scholar]
- Johnston JA, Ward CL, and Kopito RR 1998. Aggresomes: A cellular response to misfolded proteins. J. Cell Biol 143, 1883–1898. doi: 10.1083/jcb.143.7.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killary AM and Fournier K 1984. A genetic analysis of extinction: trans-dominant loci regulate expression of liver-specific traits in hepatoma hybrid cells. Cell 38, 523–534. 10.1016/0092-8674(84)90507-5 [DOI] [PubMed] [Google Scholar]
- Kocot KM, Tassia MG, Halanych KM, and Swalla BJ 2018. Phylgenomics offers resolution of major tunicate relationships. Mol. Phylogenet. Evol 121, 166–173. doi: 10.1016/j.ympev.2018.01.005 [DOI] [PubMed] [Google Scholar]
- Kusakabe R, Tani S, Nishitsuji K, Shindo M, Kamura K, Miyamoto Y, Nakai K, Suzuki Y, Kusakabe G, and Inoue K 2013. Characterization of the compact bicistronic microRNA precursor, miR-1/miR-133, expressed specifically in Ciona muscle tissues. Gene Exp. Patterns 13, 43–50. doi: 10.1016/j.gep.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Lynch VJ and Wagner GP 2008. Resurrecting the role of transcription factor change in developmental evolution. Evolution 62, 2131–2154. doi: 10.1111/j.1558-5646.2008.00440.x [DOI] [PubMed] [Google Scholar]
- MacLean D, Meedel TH, Hastings KEM, 1997. Tissue-specific alternative splicing of ascidian troponin I isoforms: redesign of a protein isoform-generating mechanism during chordate evolution. J. Biol. Chem 272, 32115–32120. doi: 10.1074/jbc.272.51.32115 [DOI] [PubMed] [Google Scholar]
- Marcellini S, Technau U, Smith JC, and Lemaire P 2003. Evolution of Brachyury proteins: identification of a novel regulatory domain conserved within bilateria. Dev. Biol 260, 352–361. 10.1016/S0012-1606(03)00244-6 [DOI] [PubMed] [Google Scholar]
- Meedel TH, Farmer SC, and Lee JJ 1997. The single MyoD family gene of Ciona intestinalis encodes two differentially expressed proteins: implications for the evolution of chordate muscle gene regulation. Development 124, 1711–1721. PMID: [DOI] [PubMed] [Google Scholar]
- Meedel TH, Lee JJ, and Whittaker JR 2002. Muscle development and lineage-specific expression of CiMDF, the MyoD-family gene of Ciona intestinalis. Dev. Biol 241, 238–246. doi: 10.1006/dbio.2001.0511 [DOI] [PubMed] [Google Scholar]
- Meedel TH, Chang P, Yasuo H 2007. Muscle development in Ciona intestinalis requires the b-HLH myogenic regulatory factor gene Ci-MRF. Dev. Biol 302, 333–344. doi: 10.1016/jydbio.2006.09.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller SC, Pavlath GK, Blakely BT, and Blau HM 1988. Muscle cell components dictate hepatocyte gene expression and the distribution of the Golgi apparatus in heterokaryons. Genes & Dev 2, 330–340. doi: 10.1101/gad.2.3.330 [DOI] [PubMed] [Google Scholar]
- Mita-Miyazawa I, Ikegami S, Satoh N, 1985. Histospecific acetylcholinesterase development in the presumptive muscle cells isolated from 16-cell-stage ascidian embryos with respect to the number of DNA replications. J. Embryol. Exp. Morphol 87, 1–12. PMID: [PubMed] [Google Scholar]
- Nagy A and Patthy L 2011. Reassessing domain architecture evolution of metazoan proteins: the contribution of different evolutionary mechanisms. Genes 2, 578–598. doi: 10.3390/genes2030578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatani Y, Moody R, and Smith WC 1999. Mutations affecting tail and notochord development in the ascidian Ciona savignyi. Development 126, 3293–3301. PMID: [DOI] [PubMed] [Google Scholar]
- Pownall ME, Gustafsson MK, Emerson CP Jr., 2002. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annual Review of Cell and Developmental Biology 18, 747–783. doi: 10.1146/annurev.cellbio.18.012502.105758 [DOI] [PubMed] [Google Scholar]
- Ristoratore F, Spagnuolo A, Aniello F, Branno M, Fabbrini F, and Di Lauro R 1999. Expression and functional analysis of Cititf1, an ascidian NK-2 class gene, suggests its role in endoderm development. Development 126, 5149–5159. PMID: [DOI] [PubMed] [Google Scholar]
- Rodaway A and Patient R, 2001. Mesendoderm: An Ancient Germ Layer? Cell 105, 169–172. 10.1016/S0092-8674(01)00307-5 [DOI] [PubMed] [Google Scholar]
- Rosenberg MI, Georges SA, Asawachaicharn A, Analau E, and Tapscott SJ 2006. MyoD inhibits Fst1 and Utrn expression by inducing transcription of miR-206. J. Cell Biol 175, 77–85. doi: 10.1083/jcb.200603039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenkar N, Gittenberger A, Lambert G, Rius M, Moreira Da Rocha R, Swalla BJ, Turon X 2018. Ascidiacea World Database Accessed at http://www.marinespecies.org/ascidiacea. [Google Scholar]
- Sweetman D, Goljanek K, Rathjen T, Oustanina S, Braun T, Dalmay T, and Munsterberg A 2008. Specific requirements of MRFs for the expression of muscle specific microRNAs, miR-1, miR-206 and miR-133. Dev. Biol 321, 491–499. doi: 10.1016/j.ydbio.2008.06.019 [DOI] [PubMed] [Google Scholar]
- Tajbakhsh S 2005. Skeletal muscle stem and progenitor cells: Reconciling genetics and lineage. Exp. Cell Res 306, 364–372. doi: 10.1016/j.yexcr.2005.03.033 [DOI] [PubMed] [Google Scholar]
- Takahashi H, Hotta K, Erives A, Di Gregorio A, Zeller R, Levine M, Satoh N 1999. Brachyury downstream notochord differentiation in the ascidian embryo. Genes & Dev 13, 1519–1523. doi: 10.1101/gad.13.12.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tani S, Kuraku S, Sakamoto H, Inoue K, and Kusakabe R 2013. Developmental expression and evolution of muscle-specific microRNAs conserved in vertebrates. Evol. & Dev 15, 293–304. doi: 10.1111/ede.12039 [DOI] [PubMed] [Google Scholar]
- Tapscott SJ, Davis RL, Thayer MJ, Cheng P-F, Weintraub H, and Lassar AB 1988. MyoD1: A nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science 242, 405–411. doi: 10.1126/science.3175662 [DOI] [PubMed] [Google Scholar]
- Tapscott SJ 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development 132, 2685–2695. doi: 10.1242/dev.01874 [DOI] [PubMed] [Google Scholar]
- Tsagkogeorga G, Cahais V, and Galtier N 2012. The population genomics of a fast evolver: high levels of diversity, functional constraint, and molecular adaptation in the tunicate Ciona intestinalis. Genome Biol. Evol 4, 852–861. doi: 10.1093/gbe/evs054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinson JP, Jaffe DB, O’Neill K, Karlsson EK, Stange-Thomann N, Anderson S, Mesirov JP, Satoh N, Satou Y, Nusbaum C, Birren B, Galaan JE, and Lander ES 2005. Assembly of polymorphic genomes: algorithms and application to Ciona savignyi. Genome Res 15, 1127–1135. doi: 10.1101/gr.3722605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada S, Katsuyama Y, Yasugi S, Saiga H 1995. Spatially and temporally regulated expression of the LIM class homeobox gene Hrlim suggests multiple distinct functions in development of the ascidian, Halocynthia roretzi. Mech. of Dev 51, 115–126. 10.1016/0925-4773(95)00359-9 [DOI] [PubMed] [Google Scholar]
- Weinberger S, Topping MP, Yan J, Claeys A, De Geest N, Ozbay D, Hassan T, He X, Albert JT, Bassem A, Hassan BA, and Ramaekers A 2017. Evolutionary changes in transcription factor coding sequence quantitatively alter sensory organ development and function. eLife; 6:e26402. doi: 10.7554/eLife.26402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H, Tapscott SJ, Davis RL, Thayer MJ, Adam MA, Lassar AB, and Miller AD 1989. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci 86, 5434–5438. 10.1073/pnas.86.14.5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss MC and Chaplain M 1971. Expression of differentiated functions in hepatoma cell hybrids: reappearance of tyrosine aminotransferase inducibility after the loss of chromosomes. Proc. Natl. Acad. Sci 68, 3026–3030. PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker JR and Meedel TH 1989. Two histospecific enzyme expressions in the same cleavage-arrested one-celled ascidian embryos. J. Exp. Zool 250, 168–175. doi: 10.1002/jez.1402500208 [DOI] [PubMed] [Google Scholar]
- Wilkins AS 2002. The Evolution of Developmental Pathways Sinauer Associates, Inc., Sunderland, MA, USA: ISBN: 0878939164 [Google Scholar]
- Wittkopp PJ and Kalay G 2012. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Gen, 13, 59–69. doi: 10.1038/nrg3095 [DOI] [PubMed] [Google Scholar]
- Wright PE and Dyson HJ 2015. Intrinsically disordered proteins in cellular signalling and regulation. Nat. Rev. Mol. Cell Biol 16, 18–29. doi: 10.1038/nrm3920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright WE 1984a. Expression of differentiated functions in heterokaryons between skeletal myocytes, adrenal cells, fibroblasts and glial cells. Exp. Cell. Res 151, 55–69. 10.1016/0014-4827(84)90355-0 [DOI] [PubMed] [Google Scholar]
- Wright WE 1984b. Induction of muscle genes in neural cells. J. Cell Biol 98, 427–435. doi: 10.1083/jcb.98.2.427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuo H and Satoh N 1994. An ascidian homolog of the mouse Brachyury (T) gene is expressed exclusively in notochord cells at the fate restricted stage. Dev. Growth Differ 36, 9–18. doi: 10.1111/j.1440-169X.1994.00009.x [DOI] [PubMed] [Google Scholar]
- Zhang JM, Chen L, Krause M, Fire A, Paterson BM 1999. Evolutionary conservation of MyoD function and differential utilization of E proteins. Dev. Biol 208, 465–472. doi: 10.1006/dbio.1999.9218 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.