ABSTRACT
CRISPR-Cas systems provide adaptive immunity against mobile genetic elements, but employment of this resistance mechanism is often reported with a fitness cost for the host. Whether or not CRISPR-Cas systems are important barriers for the horizontal spread of conjugative plasmids, which play a crucial role in the spread of antibiotic resistance, will depend on the fitness costs of employing CRISPR-based defences and the benefits of resisting conjugative plasmids. To estimate these costs and benefits we measured bacterial fitness associated with plasmid immunity using Escherichia coli and the conjugative plasmid pOX38-Cm. We find that CRISPR-mediated immunity fails to confer a fitness benefit in the absence of antibiotics, despite the large fitness cost associated with carrying the plasmid in this context. Similar to many other conjugative plasmids, pOX38-Cm carries a CcdAB toxin–anti-toxin (TA) addiction system. These addiction systems encode long-lived toxins and short-lived anti-toxins, resulting in toxic effects following the loss of the TA genes from the bacterial host. Our data suggest that the lack of a fitness benefit associated with CRISPR-mediated defence is due to expression of the TA system before plasmid detection and degradation. As most antibiotic resistance plasmids encode TA systems this could have important consequences for the role of CRISPR-Cas systems in limiting the spread of antibiotic resistance.
Keywords: CRISPR, TA, toxin, adaptive immunity, plasmid, bacteria
A cost of CRISPR immunity due to addiction systems.
INTRODUCTION
Prokaryotes often carry multiple immune systems (Labrie, Samson and Moineau 2010; Doron et al. 2018), including a highly sophisticated adaptive immune system known as CRISPR-Cas (Clustered Regularly Interspaced Short Palindromic Repeats—CRISPR-associated), reviewed in (Marraffini 2015). This system functions by integrating sequences of viruses, plasmids and transposable elements (Barrangou et al. 2007; Bikard et al. 2012; Lopez-Sanchez et al. 2012) (known as spacers) into CRISPR loci, which subsequently provide immunity against re-infection (Barrangou, et al. 2007; Brouns et al., 2008; Tyson and Banfield, 2008; Datsenko et al. 2012; Swarts et al. 2012; Yosef, Goren and Qimron 2012; van Houte, Buckling and Westra 2016a). Conjugative plasmids often carry antibiotic resistance genes and therefore play a crucial role in the spread of antibiotic resistance (Maiden 1998; Dionisio et al. 2002; Svara and Rankin, 2011; Carattoli, 2013). Whether CRISPR-dependent immunity to plasmids is important in limiting the spread of antibiotic resistance (Marraffini and Sontheimer, 2008 and Palmer and Gilmore, 2010; Gophna et al. 2015) depends on the efficacy of the CRISPR-Cas immune system (Hullahalli et al. 2018), and on the fitness cost associated with carrying the plasmid (in the absence of antibiotics) and the cost of resistance associated with CRISPR-immunity. Experimental observations (Jiang et al. 2013; Vercoe et al. 2013) and theory predicts that CRISPR-Cas systems can degenerate if they carry a cost (Levin 2010; Weinberger, Wolf and Lobkovsky 2012; Iranzo et al. 2013). While large costs are likely when the CRISPR-Cas system behaves maladaptively, such as autoimmunity (Stern et al. 2010; Vercoe et al. 2013) and the prevention of beneficial infection (Bikard et al. 2012; Jiang et al. 2013), there may also be costs when the system prevents infection by costly genetic elements, for example due to immunopathological effects or energetic costs of immune activation (Vale et al. 2015; Westra et al. 2015; Westra et al. 2016; van Houte et al. 2016b). Here, we investigate this possibility using Escherichia coli and the conjugative F-plasmid pOX38-Cm. Our data show that CRISPR-mediated immunity against this costly plasmid is associated with a fitness cost under non-selective conditions. Our data further suggest that this cost of immunity may not only result from energetic costs, but is caused by a plasmid-encoded CcdAB toxin–antitoxin (TA) addiction system (Ogura and Hiraga 1983; Jaffe, Ogura and Hiraga 1985; Bahl, Hansen and Sorensen 2009), which plays a critical role in avoiding plasmid curing (Hayes 2003). Hence, TA systems may limit the evolution of bacterial adaptive immunity against plasmids, which could have important consequences for the spread of antibiotic resistance.
METHODS
Bacterial strains
Escherichia coli K12 Δhns (BW25113) strains, which have a constitutively active CRISPR-Cas system (Pul et al. 2010; Westra et al. 2010) were used as recipient cells. These strains, which were obtained from the KEIO collection, were cured from the kanamycin resistance cassette using FLP recombinase (Datsenko and Wanner 2000) and were engineered to carry synthetic CRISPR loci, sequences of which can be found in Table S2 (Supporting Information). Escherichiacoli MC4100 carrying pOX38-Cm was used as the donor strain.
Cloning of spacers, lacZ and CcdA
Spacers, lacZ and ccdA were cloned into the recombination cassette located on the previously described plasmid pRECOMB-Cr2.1(Westra et al. 2010). Spacer-containing DNA fragments from plasmid pWUR693 and pWUR700 (Westra et al. 2013) were cloned using the BamHI and EcoRI restriction sites of pRECOMB-Cr2.1. The lacZ and ccdA genes were PCR amplified from the E. coli K12 W3110 genome and plasmid pOX38-Cm, respectively (Figure S1, Tables S1 and S2, Supplementary Information) and cloned using restriction enzymes NotI and KpnI. Resulting plasmids were used as a template for PCR amplification using primers BG4452 and BG4453 (Figure S1, Tables S1 and S2, Supplementary Information) and the amplicon was subsequently gel purified. Escherichia coli Δhns cells containing the plasmid pKD46 were transformed with the amplicon (Datsenko and Wanner 2000), after which the bacteria were plated on LB agar containing kanamycin (50 mg/L) to select for recombinants. Plated bacteria were grown at 37°C overnight to cure the cells from pKD46. Recombination was confirmed using colony PCR and Sanger sequencing (GATC Biotech, Germany).
Growth measurements
Growth curves of E. coli K12 Δhns (BW25113) and E. coli K12 Δhns (BW25113) carrying pOX38-Cm were measured as follows. Bacteria were inoculated 1:100 in 1 L fresh LB medium from overnight cultures containing the same optical density and grown at 37°C while shaking at 180 rpm (four replicas per treatment). The optical density (OD600) of the cultures was measured every 30 min. At each of these time points a sample of 10 or 20 mL was taken. Cells were washed with Millipore water and dried overnight at 130°C. The dry weight of the bacteria was measured of every sample to determine the specific growth rate (in gram new cells·gram cell−1·hr−1). The specific growth rate was determined for the log phase of the growth curve and used as a measurement of bacterial fitness.
Competition experiments
Competition experiments were inoculated from overnight cultures grown at 37°C with equal optical densities. Competition experiments were performed in microcosms (6 mL LB medium in 30 mL glass vials, 6 replicas per treatment), containing 60 μL of every culture used in the experiment. Competition experiments were incubated at 37°C at 100 rpm and a daily transfer of 120 μL of the competition experiments into fresh microcosms was carried out. After 2 days cells were plated on LB agar containing kanamycin (50 mg/L) and X-gal (50 mg/L). Colony counting of blue and white colonies was used to determine relative fitness of CRISPR-immune (white colonies) and CRISPR-sensitive strains (blue colonies). Experiments were performed in presence or absence of pOX38-Cm to measure the effect of plasmid presence on the fitness of the bacterial strains. Statistical analysis was performed using JMP10 Software.
RESULTS
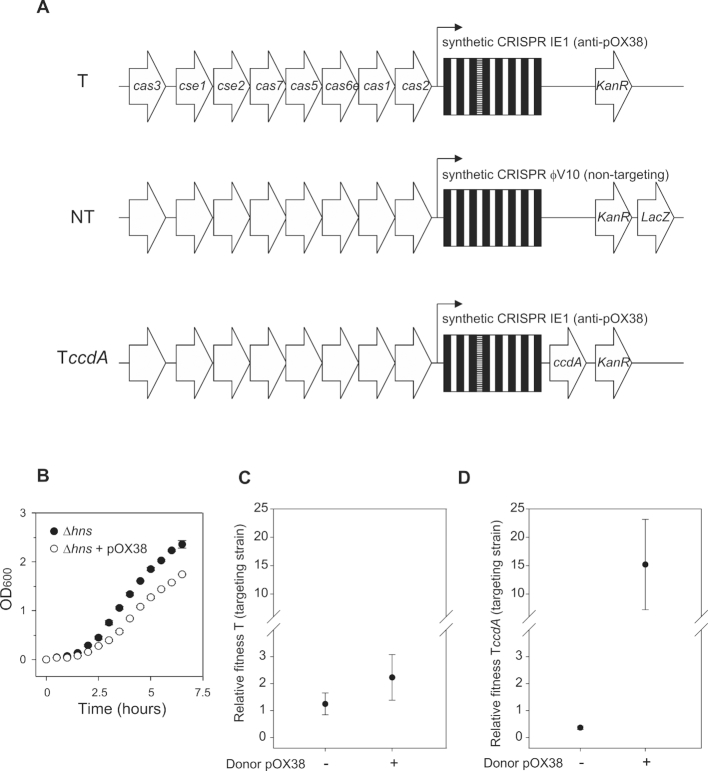
It has previously been shown that the Type I-E CRISPR-Cas adaptive immune system of E. coli (Fig. 1A) can effectively protect against conjugative transfer of plasmid pOX38-Cm (Westra et al. 2013), which is a derivative of the well-studied plasmid F and encodes chloramphenicol (Cm) resistance. Measurements of the specific growth rates of E. coli Δhns reveal that carrying plasmid pOX38-Cm reduces growth rates with 27% when antibiotics are absent (Fig. 1B and Table 1; F1,7 = 50.67, P = 0.0004). Based on the difference in growth rate between plasmid-containing and plasmid-free cells in monoculture, CRISPR-mediated immunity against the plasmid would be expected to result in a large fitness benefit. To measure the relative fitness associated with CRISPR immunity, competition experiments were performed between CRISPR-immune and susceptible E. coli K12 derived strains. To this end, the genome of E. coli K12 Δhns was engineered to replace the CRISPR 2.1 locus flanking the cas genes with synthetic CRISPR arrays that either target (strain T) plasmid pOX38-Cm or that do not target (non-targeting; strain NT) the plasmid (Fig. 1A). After competing strain T and strain NT for two days the resulting relative fitness is approximately one, indicating that the two strains have comparable fitness (Fig. 1C; 1-sample t-test, T5 = 1.19, P = 0.29). Surprisingly, the presence of a donor strain that carries conjugative plasmid pOX38-Cm did not cause a significant fitness increase of the T strain compared to when the plasmid was absent (Fig. 1C; F1,11 = 4.23, P = 0.067). These data therefore suggest that the cost of immunity is of the same order of magnitude as the cost of carrying the plasmid.
Figure 1.
(A), Overview of the engineered CRISPR locus of the T strain (targeting pOX38-Cm), the NT strain (not targeting pOX38-Cm, encoding LacZ) and the TccdA strain (targeting pOX38-Cm, encoding CcdA). Genes are indicated by arrows. The CRISPR locus consists of repeats (black) and spacers (white). The spacer targeting pOX38-Cm is indicated by horizontal stripes. (B), Optical densities at 600 nm (OD600) of plasmid-free (Δhns) cells and plasmid-containing (Δhns + pOX38-Cm) cells at different time points after inoculation. Measurements of dry weight were used to determine specific growth rates (g new cells·g cells−1·hr−1, Table 1). (C), Relative fitness (mean ± 95% CI) of T strain in the absence or presence of the pOX38-Cm donor strain after 2 days of competition with NT strain. (D), Relative fitness (mean ± 95% CI) of TccdA strain in the absence or presence of the pOX38-Cm donor strain after 2 days of competition with NT strain.
Table 1.
Specific growth rates of bacteria with and without plasmid pOX38-Cm.
| Strain | Specific growth rate (g new cells*g cells−1*hr−1) |
| Δhns | 0.90 ± 0.06 |
| Δhns + pOX38-Cm | 0.66 ± 0.04 |
Given the T and NT strains did not differ in fitness in the absence of the plasmid, we hypothesized that the cost of immunity associated with CRISPR-Cas could be due to gene expression from the invading plasmid prior to detection by the immune system, analogous to the expression of anti-CRISPR genes from phage genomes prior to CRISPR-mediated cleavage of the phage genomes (Bondy-Denomy et al. 2013; Borges et al. 2018; Landsberger et al. 2018). Although any of the plasmid genes could contribute to this cost, it is well documented that expression of plasmid-encoded addiction systems would be particularly harmful. Addiction systems prevent plasmid curing, since removal of the plasmid results in rapid depletion of the anti-toxin whereas the toxin will persist for longer periods of time to eventually cause cell death (Gerdes and Maisonneuve 2012; Cook et al. 2013). The toxin–anti-toxin (TA) system of plasmid pOX38-Cm is encoded by the ccdAB genes; the CcdB toxin is neutralized by the CcdA anti-toxin. In the absence of the short-lived CcdA anti-toxin the CcdB toxin inhibits DNA gyrase, which eventually leads to cell death (Cook et al. 2013).
To test the hypothesis that this TA system contributes to the cost of resistance we engineered an E. coli strain to express the CcdA anti-toxin from the genome in addition to carrying the T CRISPR (TccdA strain; Fig. 1A). This strain is immune to plasmid pOX38-Cm and to the detrimental effect of toxin CcdB since the toxin is neutralized by CcdA. Competition between the TccdA and the NT strain in the absence of the conjugative plasmid reveals that encoding CcdA on the genome is associated with a fitness cost (relative fitness = 0.37) (Fig. 1D; T5 = −19.2, P < 0.0001) after two days of competition. By contrast, when competing TccdA and NT for two days in the presence of a donor strain that carries the pOX38-Cm plasmid, the TccdA strain has a large fitness benefit (relative fitness = 15.2) (Fig. 1D; T5 = 3.5, P = 0.017). Hence, these data demonstrate that expression of the anti-toxin from the bacterial chromosome alleviates the cost of CRISPR immunity, suggesting that TA expression from the plasmid prior to its degradation by CRISPR-Cas immune systems may be an important contributor to the observed cost of immunity.
DISCUSSION
Costs of resistance potentially have profound effects on co-evolutionary dynamics (Agrawal and Lively 2003; Lopez-Pascua and Buckling 2008;Gomez and Buckling 2011; Buckling and Brockhurst 2012) and are directly responsible for the existence of trade-offs between immunity and other life-history traits (Boots and Begon 1993; Boots and Bowers 2004; Little and Killick 2007; Kempel et al. 2011). Our data suggest that TA systems encoded by plasmids may cause CRISPR immunity against an invading plasmid to be costly, due to the time-lag between infection and clearance of the infection during which the TA system may already be expressed. As a result of this cost the CRISPR system may have little net benefit against costly plasmids if they encode TA-systems. This could explain the limited spread and degeneration of CRISPR-Cas systems, and may also help to explain observations of high degrees of susceptibility to costly plasmids in E. coli (Touchon et al. 2012). Furthermore, many antibiotic resistance genes are carried on conjugative plasmids containing TA systems (Maiden 1998, Dionisio et al. 2002; Svara and Rankin 2011; Carattoli 2013). As such, there will be relatively weak selection to resist these plasmids via CRISPR-Cas, even in the absence of antibiotic selection. Our findings also indicate that using CRISPR-Cas as a tool to resensitize bacteria to antibiotics by selectively removing antibiotic resistance-carrying conjugative plasmids (Pursey et al. 2018) may be challenging when these plasmids encode TA systems.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Søren Høgh for experimental contributions and Arjan de Visser for stimulating discussions. This work was financially supported by grants from the Natural Environment Research Council and from the Royal Society of Biological Sciences to AB, from the Netherlands Organization of Scientific Research (NWO) to JVDO (NWO-TOP, 854.10.003), and to SJJB (NWO Vidi, 864.11.005). ERW received funding from the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme (FP7/2007–2013) under REA grant agreement n⁰[327606], as well as funding from the ERC (ERC-STG-2016–714478–EVOIMMECH), the BBSRC (BB/N017412/1), the Wellcome Trust (109776/Z/15/Z) and the NERC (NE/M018350/1).
Conflicts of interest. None declared.
REFERENCES
- Agrawal AF, Lively CM. Modelling infection as a two-step process combining gene-for-gene and matching-allele genetics. Proc Biol Sci. 2003;270:323–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl MI, Hansen LH, Sorensen SJ. Persistence mechanisms of conjugative plasmids. Methods Mol Biol. 2009;532:73–102. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H et al.. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. [DOI] [PubMed] [Google Scholar]
- Bikard D, Hatoum-Aslan A, Mucida D et al.. CRISPR interference can prevent natural transformation and virulence acquisition during in vivo bacterial infection. Cell Host Microbe. 2012;12:177–86. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL et al.. Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature. 2013;493:429–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boots M, Begon M. Trade-offs with resistance to a granulosis virus in the Indian meal moth, examined by a laboratory evolution experiment. Functional Ecology. 1993;7:528–34. [Google Scholar]
- Boots M, Bowers RG. The evolution of resistance through costly acquired immunity. Proc Biol Sci. 2004;271:715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges AL, Zhang JY, Rollins MF et al.. Bacteriophage cooperation suppresses CRISPR-Cas3 and Cas9 immunity. Cell. 2018;174:917–925 e910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouns SJ, Jore MM, Lundgren M et al.. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckling A, Brockhurst M. Bacteria-virus coevolution. Adv Exp Med Biol. 2012;751:347–70. [DOI] [PubMed] [Google Scholar]
- Carattoli A. Plasmids and the spread of resistance. Int J Med Microbiol. 2013;303:298–304. [DOI] [PubMed] [Google Scholar]
- Cook GM, Robson JR, Frampton RA et al.. Ribonucleases in bacterial toxin-antitoxin systems. Biochim Biophys Acta. 2013;1829:523–31. [DOI] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A. 2000;97:6640–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Pougach K, Tikhonov A et al.. Molecular memory of prior infections activates the CRISPR/Cas adaptive bacterial immunity system. Nat Commun. 2012;3:945. [DOI] [PubMed] [Google Scholar]
- Dionisio F, Matic I, Radman M et al.. Plasmids spread very fast in heterogeneous bacterial communities. Genetics. 2002;162:1525–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doron S, Melamed S, Ofir G et al.. Systematic discovery of antiphage defense systems in the microbial pangenome. Science. 2018;359:eaar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes K, Maisonneuve E. Bacterial persistence and toxin-antitoxin loci. Annu Rev Microbiol. 2012;66:103–23. [DOI] [PubMed] [Google Scholar]
- Gomez P, Buckling A. Bacteria-phage antagonistic coevolution in soil. Science. 2011;332:106–9. [DOI] [PubMed] [Google Scholar]
- Gophna U, Kristensen DM, Wolf YI et al.. No evidence of inhibition of horizontal gene transfer by CRISPR-Cas on evolutionary timescales. ISME J. 2015;9:2021–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes F. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science. 2003;301:1496–9. [DOI] [PubMed] [Google Scholar]
- Hullahalli K, Rodrigues M, Nguyen UT et al.. An attenuated CRISPR-Cas system in Enterococcus faecalis permits DNA acquisition. MBio. 2018;9:e00414-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iranzo J, Lobkovsky AE, Wolf YI et al.. Evolutionary dynamics of the prokaryotic adaptive immunity system CRISPR-Cas in an explicit ecological context. J Bacteriol. 2013; 195:3834–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A, Ogura T, Hiraga S. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 1985;163:841–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Maniv I, Arain F et al.. Dealing with the evolutionary downside of CRISPR immunity: bacteria and beneficial plasmids. PLoS Genet. 2013;9:e1003844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempel A, Schadler M, Chrobock T et al.. Tradeoffs associated with constitutive and induced plant resistance against herbivory. Proc Natl Acad Sci U S A. 2011;108:5685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8:317–27. [DOI] [PubMed] [Google Scholar]
- Landsberger M, Gandon S, Meaden S et al.. Anti-CRISPR phages cooperate to overcome CRISPR-Cas immunity. Cell. 2018;174:908–916 e912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin BR. Nasty viruses, costly plasmids, population dynamics, and the conditions for establishing and maintaining CRISPR-mediated adaptive immunity in bacteria. PLoS Genet. 2010;6:e1001171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little TJ, Killick SC. Evidence for a cost of immunity when the crustacean Daphnia magna is exposed to the bacterial pathogen Pasteuria ramosa. J Anim Ecol. 2007;76:1202–7. [DOI] [PubMed] [Google Scholar]
- Lopez-Pascua L, Buckling A. Increasing productivity accelerates host-parasite coevolution. J Evol Biol. 2008;21:853–60. [DOI] [PubMed] [Google Scholar]
- Lopez-Sanchez MJ, Sauvage E, Da Cunha V et al.. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol. 2012;85:1057–71. [DOI] [PubMed] [Google Scholar]
- Maiden MC. Horizontal genetic exchange, evolution, and spread of antibiotic resistance in bacteria. Clin Infect Dis. 1998;27:Suppl 1:S12–20. [DOI] [PubMed] [Google Scholar]
- Marraffini LA. CRISPR-Cas immunity in prokaryotes. Nature. 2015;526:55–61. [DOI] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. Science. 2008;322:1843–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Hiraga S. Mini-F plasmid genes that couple host cell division to plasmid proliferation. Proc Natl Acad Sci U S A. 1983;80:4784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer KL, Gilmore MS. Multidrug-resistant Enterococci lack CRISPR-cas. MBio. 2010;1:e00227-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pul U, Wurm R, Arslan Z et al.. Identification and characterization of E. coli CRISPR-cas promoters and their silencing by H-NS. Mol Microbiol. 2010;75:1495–512. [DOI] [PubMed] [Google Scholar]
- Pursey E, Sunderhauf D, Gaze WH et al.. CRISPR-Cas antimicrobials: challenges and future prospects. PLoS Pathog. 2018;14:e1006990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern A, Keren L, Wurtzel O et al.. Self-targeting by CRISPR: gene regulation or autoimmunity? Trends Genet. 2010;26:335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svara F, Rankin DJ. The evolution of plasmid-carried antibiotic resistance. BMC Evol Biol. 2011;11:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Mosterd C, van Passel MW et al.. CRISPR interference directs strand specific spacer acquisition. PLoS One. 2012;7:e35888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Charpentier S, Pognard D et al.. Antibiotic resistance plasmids spread among natural isolates of Escherichia coli in spite of CRISPR elements. Microbiology. 2012;158:2997–3004. [DOI] [PubMed] [Google Scholar]
- Tyson GW, Banfield JF. Rapidly evolving CRISPRs implicated in acquired resistance of microorganisms to viruses. Environ Microbiol. 2008;10:200–7. [DOI] [PubMed] [Google Scholar]
- Vale PF, Lafforgue G, Gatchitch F et al.. Costs of CRISPR-Cas-mediated resistance in Streptococcus thermophilus. Proc Biol Sci. 2015;282:20151270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte S, Buckling A, Westra ER. Evolutionary Ecology of Prokaryotic Immune Mechanisms. Microbiol Mol Biol Rev. 2016a;80:745–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Houte S, Ekroth AK, Broniewski JM et al.. The diversity-generating benefits of a prokaryotic adaptive immune system. Nature. 2016b;532:385–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vercoe RB, Chang JT, Dy RL et al.. Cytotoxic chromosomal targeting by CRISPR/Cas systems can reshape bacterial genomes and expel or remodel pathogenicity islands. PLoS Genet. 2013;9:e1003454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger AD, Wolf YI, Lobkovsky AE, Gilmore MS & Koonin EV viral diversity threshold for adaptive immunity in prokaryotes. MBio. 2012;3:e00456–00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, Dowling AJ, Broniewski JM et al.. Evolution and ecology of CRISPR. Annu Rev Ecol Evol Syst. 2016;47:307–31. [Google Scholar]
- Westra ER, Semenova E, Datsenko KA et al.. Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLoS Genet. 2013;9:e1003742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westra ER, van Houte S, Oyesiku-Blakemore S et al.. Parasite exposure drives selective evolution of constitutive versus inducible defense. Curr Biol. 2015;25:1043–9. [DOI] [PubMed] [Google Scholar]
- Westra ER, Pul U, Heidrich N et al.. H-NS-mediated repression of CRISPR-based immunity in Escherichia coli K12 can be relieved by the transcription activator LeuO. Mol Microbiol. 2010;77:1380–93. [DOI] [PubMed] [Google Scholar]
- Yosef I, Goren MG, Qimron U. Proteins and DNA elements essential for the CRISPR adaptation process in Escherichia coli. Nucleic Acids Res. 2012;40:5569–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.