Abstract
Transient receptor potential melastatin member 8 (TRPM8) is a calcium ion (Ca2+)-permeable cation channel that serves as the primary cold and menthol sensor in humans. Activation of TRPM8 by cooling compounds relies on allosteric actions of agonist and membrane lipid phosphatidylinositol 4,5-bisphosphate (PIP2), but lack of structural information has thus far precluded a mechanistic understanding of ligand and lipid sensing by TRPM8. Using cryo-electron microscopy, we determined the structures of TRPM8 in complex with the synthetic cooling compound icilin, PIP2, and Ca2+, as well as in complex with the menthol analog WS-12 and PIP2. Our structures reveal the binding sites for cooling agonists and PIP2 in TRPM8. Notably, PIP2 binds to TRPM8 in two different modes, which illustrate the mechanism of allosteric coupling between PIP2 and agonists. This study provides a platform for understanding the molecular mechanism of TRPM8 activation by cooling agents.
One Sentence Summary:
Structures reveal the molecular basis for cooling agonists sensing and unique PIP2 dependence in the TRPM8 channel.
Introduction
The transient receptor potential melastatin (TRPM) family, part of the TRP channel superfamily, is composed of eight members (TRPM1 to TRPM8) and is involved in various processes such as temperature and redox sensing (1–3). Both in vivo and in vitro studies have shown that TRPM8 acts as a cold and menthol receptor (4–8). In addition, TRPM8 plays crucial roles in cold-related pain and migraine (9, 10) and mediates menthol-induced analgesia of acute and inflammatory pain (11). Therefore, TRPM8 is a therapeutic target for treatments of cold allodynia, chronic pain, and migraine (12–15).
As a polymodal sensor, TRPM8 gating is shaped by multiple physicochemical stimuli. The channel is activated by exposure to cold temperatures or menthol and is allosterically modulated by phosphatidylinositol 4,5-bisphosphate (PIP2) and Ca2+. TRPM8 activation requires PIP2; at high concentrations, PIP2 alone appears to be sufficient for channel activation, whereas its depletion desensitizes the channel (16–19). Furthermore, binding of PIP2 and cooling compounds is allosterically coupled, as binding of one increases the potency of the other (17, 20). Although intracellular Ca2+ is required for activation of several TRPM channels (21, 22), it is not necessary for cold- or menthol-dependent TRPM8 activation (4, 23).
Because of the physiological and therapeutic importance of TRPM8, many studies have focused on dissecting the mechanisms by which cooling agents bind and gate the channel. (12, 13, 18, 24–28). Despite their apparent overlapping binding sites in TRPM8 (24), the natural cooling compound menthol and the synthetic cooling compound icilin activate the channel via distinct mechanisms (26). First, Ca2+ is required for TRPM8 activation by icilin but not menthol (23). Second, intracellular pH does not affect menthol-dependent TRPM8 gating but does modulate the effect of icilin on the channel (29). Third, although menthol is a common agonist for all known orthologs of TRPM8, icilin does not activate avian TRPM8 channels, which require a mutation of Ala to Gly (Ala805→Gly) on the transmembrane segment 3 (S3) to induce sensitivity to icilin (23).
The recent cryo-electron microscopy (cryo-EM) structure of the ligand-free TRPM8 offered the first opportunity to place residues previously identified as important for agonist binding in a structural context (30). However, this structure could not elucidate the specific mechanisms by which natural and synthetic cooling agents bind to and activate the channel or how PIP2 allosterically affects their potency. To answer these questions, we determined two cryo-EM structures of TRPM8: one in complex with icilin, PIP2, and Ca2+; the other in complex with the menthol analog WS-12 and PIP2 (Fig. 1, A to D). Our structural and functional studies identify a distinct binding site for PIP2 and unveil the structural basis for menthol, icilin, and Ca2+ recognition by TRPM8. Moreover, structural analyses provide insights into the allosteric coupling between lipid and agonists and illuminate the principles of ligand-dependent gating in TRPM8.
Figure 1. Structures and functions of TRPM8 in complex with PIP2 and cooling compounds.
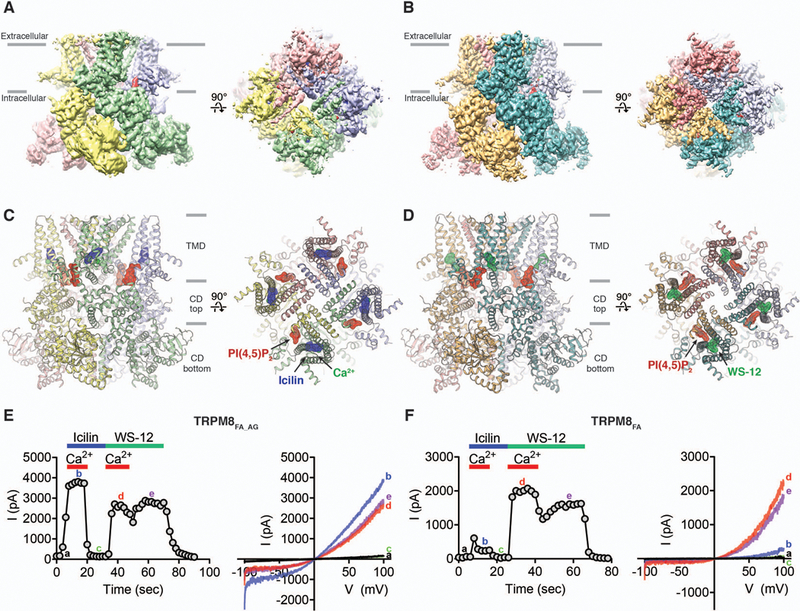
(A and B) Cryo-EM reconstructions and (C and D) structures of the class 1 icilin-PIP2-Ca2+ complex [(A) and (C)] and the WS-12-PIP2 complex [(B) and (D)], viewed from the membrane plane and from the extracellular side. Ligands are highlighted in surface representations. Gray bars indicate division of channel layers. (E and F) Functional characterization of the TRPM8FA_AG (E) and TRPM8FA (F) channels by icilin, WS-12 and Ca2+. Channel activation was elicited via application of 20 µM icilin or 40 µM WS-12 in the presence or absence of 12.5 µM Ca2+ to the cytoplasmic side of inside-out patches. Time course of the peak current (I) amplitudes at +100 mV from the voltage (V) ramp (−100 mV to +100 mV) is shown on the left and representative current traces are shown on the right. Quantifications of current amplitudes are shown in fig. S2B.
Results
Structure determination of TRPM8 in complex with PIP2 and cooling agents
The previously developed TRPM8 construct from the collared flycatcher Ficedula albicollis (TRPM8FA) was used to determine the structure of TRPM8 in complex with the menthol analog WS-12 and PIP2 (fig. S1 and Methods) (30). WS-12 was chosen because of its higher potency and shared activation mechanism with menthol (31) (fig. S2A). To confer icilin sensitivity to TRPM8FA, we introduced an Ala805→Gly mutation (23) and used the construct (TRPM8FA_AG) for determining the structure of TRPM8 in complex with icilin, Ca2+, and PIP2. When applied to the inside-out patches of human embryonic kidney (HEK) 293 cells expressing TRPM8FA_AG, icilin elicits robust currents in the presence of Ca2+ (Fig. 1E and fig. S2B). TRPM8FA_AG shows a Ca2+-dependent desensitization at high concentrations of Ca2+ (fig. S3, A to C), similar to mammalian TRPM8 channels (4). TRPM8FA also exhibits icilin-evoked currents but to a much lesser extent than TRPM8FA_AG, consistent with previous data (23) (Fig. 1F and fig. S2B). In contrast to icilin, WS-12 elicits large outward-rectifying currents in both TRPM8FA and TRPM8FA_AG independent of intracellular Ca2+ (Fig. 1, E and F and fig. S2B). Icilin shows apparently higher efficacy and potency than WS-12 in TRPM8FA_AG activation (Fig. 1E and fig. S2B).
TRPM8FA_AG or TRPM8FA channels were purified in digitonin and incubated with short chain PIP2 (diC8-PIP2), icilin, and Ca2+, or with diC8-PIP2 and WS-12, respectively. The complex structures were determined by cryo-EM (see methods and figs. S4 and S5). The final three-dimensional (3D) reconstructions of the icilin-PIP2-Ca2+-TRPM8FA_AG complex (henceforth, the icilin-PIP2-Ca2+ complex) yielded two 3D classes: The major class (~100,000 particles), containing a strong density for icilin, PIP2, and Ca2+, was resolved to ~3.4 Å (referred to as the “class 1 reconstruction”), and the minor class (~9000 particles), containing weak or no density for the ligands, was resolved to ~4.3 Å (referred to as the “class 2 reconstruction”) (Fig. 1A and figs. S4 and S6). The final 3D reconstruction of the WS-12-PIP2-TRPM8FA complex (henceforth, the WS-12-PIP2 complex) was resolved to ~4 Å (Fig. 1B and fig. S5). The overall quality of the 3D reconstructions in the transmembrane region (fig. S7) was improved compared with that of the ligand-free TRPM8, but the transmembrane helical segment 6 (S6) and the pore helix (PH) are not well resolved in the class 1 reconstruction of the icilin-PIP2-Ca2+ complex. Therefore, these regions were built as poly-alanine chains (fig. S4 and Table S1).
The overall structure of the homotetrameric TRPM8 complex can be divided into three layers, in which the top layer comprises the transmembrane domain (TMD) and the bottom two layers form the cytoplasmic domain (CD) (Fig. 1, C and D). Each subunit contains an N-terminal region composed of melastatin homology regions 1 to 4 (MHR1 to MHR4), a transmembrane channel region, and a C-terminal region (fig. S2, E and F). The TMD is composed of the voltage-sensor-like domain (VSLD) (S1 to S4), with the pore domain (S5, S6, and PH) arranged in a domain-swapped manner. The CD is composed of the N-terminal and C-terminal regions assembled into the bottom two layers.
Icilin binding site within VSLD
In the class 1 reconstruction of the icilin-PIP2-Ca2+ complex, we observed a strong EM density located in the cavity between the VSLD and the TRP domain (the VSLD cavity). Because the elongated density matches the size and shape of an icilin molecule (Fig. 2A) and is present in the half-map reconstructions (fig. S6), we assigned this EM density to icilin. From the possible assignments of icilin (fig. S6E) we selected the one with the better fit to the density and fewer steric clashes. Within the VSLD cavity, the icilin molecule is surrounded by aromatic residues including Tyr745 in S1 and Tyr1004 in the TRP domain, both of which have previously been proposed to be important for menthol and icilin binding (24). In this structure, we observed two additional residues in the C-terminal part of S4 (termed S4b) that establish prominent side chain interactions with icilin. The side chains of Arg841, positioned underneath the central pyrimidine moiety of icilin, and His844, which points towards the center of the VSLD cavity, interact with icilin. Notably, S4b adopts a 310-helical conformation, in contrast to the α-helical conformation observed in the ligand-free TRPM8FA and the class 2 (low occupancy) icilin-PIP2-Ca2+ complex structures (fig. S8). The transition from α- to 310-helical conformation results in a register shift in S4b, which enables the interactions of Arg841 and His844 with icilin.
Figure 2. Structural basis of icilin recognition and its Ca2+ dependence in TRPM8.
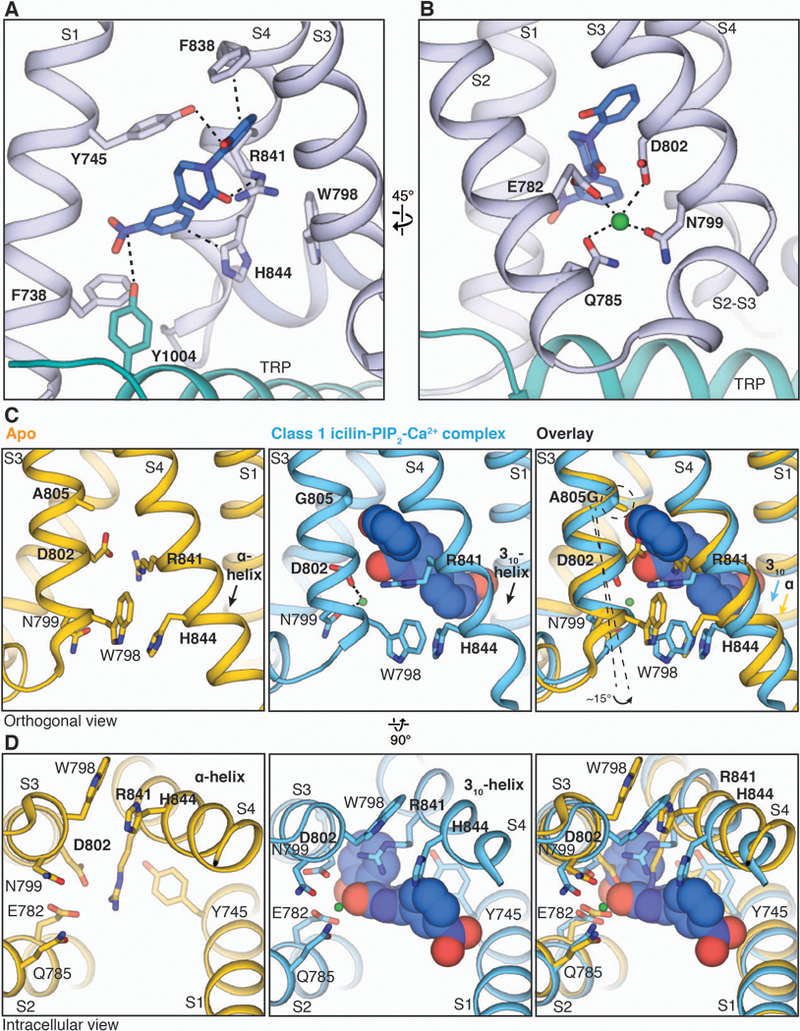
(A and B) Close-up views of the VSLD cavity showing key residues interacting with icilin [(A), blue sticks] and Ca2+ [(B), green sphere]. S2 and the S2-S3 linker are omitted in (A) for clarity. (C and D) Viewed orthogonally to the membrane (C) or from the intracellular side (D), comparison of the conformational changes in the VSLD cavity upon icilin and Ca2+ binding (blue) with the apo TRPM8 structure (yellow, PDB 6BPQ). The icilin molecule (blue) and Ca2+ ion (green) are shown as spheres.
Icilin-mediated activation of TRPM8 has been shown to depend on Ca2+ (23). In the class 1 reconstruction, we identified a strong EM density peak between S2 and S3 and assigned it as Ca2+ on the basis of its presence in both half-maps (fig. S6) and the analogous Ca2+ location identified in other TRPM channels (32, 33). Previous studies have identified Asn799, Asp802, and Gly805 in TRPM8 as important for icilin-mediated activation (23). Instead of directly participating in icilin binding, Asn799 and Asp802 in S3, along with Glu782 and Gln785 in S2, participate in Ca2+ coordination (Fig. 2B). Despite its proximity to icilin in the VSLD cavity, Ca2+ does not directly interact with the agonist, suggesting that binding of Ca2+ may induce a conformational change that primes the site for icilin binding. In the ligand-free TRPM8 structure, Asp802 in S3 and Arg841 in S4 apparently form a salt bridge and occupy the position where the central pyrimidine ring of icilin is located in the icilin-PIP2-Ca2+ complex structure. Binding of Ca2+ is associated with a slight rotation of the N-terminal part of S3, allowing Asn799 and Asp802 in S3 to coordinate Ca2+. This moves the Asp802 side chain away from Arg841, breaking the salt bridge; as a result, the VSLD cavity widens to accommodate icilin, and Arg841 is positioned below icilin’s central pyrimidine moiety (Fig. 2, C and D). The residue at position 805, which accounts for the differential sensitivity to icilin in mammalian and avian orthologs (23), is located one helical turn above Asp802 in S3, facing the hydroxyphenyl moiety of icilin (Fig. 2, C and D). We propose that the Ala805→Gly substitution confers icilin sensitivity by (i) providing flexibility for rotation of the N-terminal part of S3, which contains Asp802 and Asn799, thus leading to the assembly of the Ca2+ binding site, and (ii) enlarging the VSLD cavity to accommodate the hydroxyphenyl moiety of icilin (Fig. 2C).
The binding site for the menthol analog WS-12
We identified a strong, asymmetric dumbbell-shaped EM density within the VSLD cavity of the WS-12-PIP2 complex 3D reconstruction that is present in both half-maps (Fig. 3A and fig. S9). We assigned this density to WS-12 because its size and shape are consistent with those of a WS-12 molecule. The density is sandwiched between Tyr745 in S1 and Tyr1004 in the TRP domain, with the menthol-like moiety and the methoxyphenyl ring near Tyr745 and Tyr1004, respectively (Fig. 3A). Side chains of Arg841 in S4 and Tyr1004 and Arg1007 in the TRP domain are within the interacting distances of the central amide bond in WS-12. This structure is consistent with previous functional studies; Tyr745, Tyr1005 (Tyr1004 in TRPM8FA), and Arg842 (Arg841 in TRPM8FA) have all been identified as important for menthol sensing (18, 24). Because WS-12 is composed of a menthol-like moiety linked to a methoxyphenyl group via a central amide bond, we anticipate that menthol binding to TRPM8 will be similar to that of the menthol-like moiety in WS-12 in our structure.
Figure 3. The binding site for menthol analog WS-12 in TRPM8.
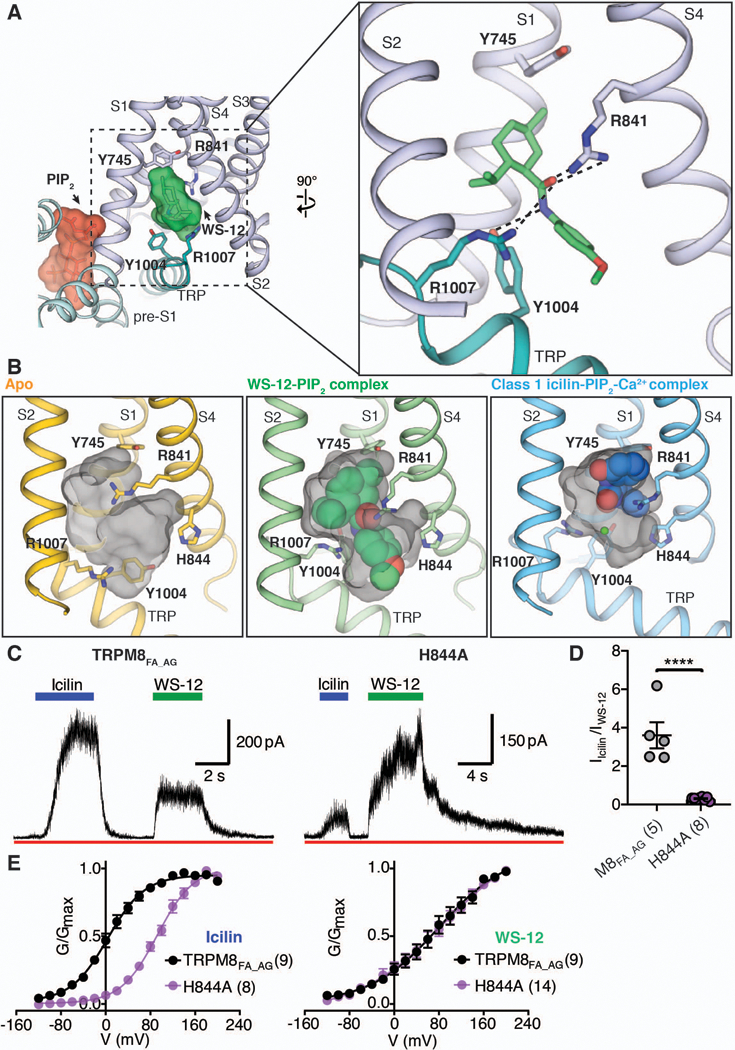
(A) WS-12 binding in the VSLD cavity (left) and close-up view of the binding site (right). PIP2 (red) and WS-12 (green) molecules are shown as sticks and highlighted with surfaces in the left panel. The S2-S3 linker and S3 are omitted in right panel. (B) Comparison of the VSLD cavity in the apo TRPM8 (yellow), WS-12-PIP2-complex (green), and class 1 icilin-PIP2-Ca2+ complex (blue) structures. Agonists are shown as spheres. The green sphere in the right panel represents Ca2+. Grey surfaces represent the binding pocket shaped by residues lining the VSLD cavity. The S2-S3 linker and S3 are omitted. (C) Representative recordings showing differential channel activation by icilin and WS-12 in TRPM8FA_AG and TRPM8FA_AG with the His844→Ala (H844A) mutation in response to 20 µM icilin or 40 µM WS-12 in the presence of 12.5 µM Ca2+. The membrane was held at +60 mV. (D) Icilin- and WS-12-evoked current ratio from recordings in (C). A two-tailed unpaired Student’s t-test was used for the comparison; ****P < 0.0001. (E) Conductance-voltage (G-V) relationship of TRPM8FA_AG and TRPM8FA_AG H844A in response to 20 µM icilin (left) or 40 µM WS-12 (right) as measured by their normalized peak tail currents (see Methods and fig. S10 for details). Numbers of individual recordings are noted in parentheses. Error bars indicate SEM.
Comparison of the VSLD cavities from ligand-free, WS-12-bound, and icilin-bound TRPM8 structures shows that residues lining the binding pocket within the cavity, including Glu782, Arg841, His844, Tyr1004, and Arg1007, can adopt multiple conformations, enabling the cavity to fit structurally distinct agonist molecules (Fig. 3B). Notably, His844 does not interact with WS-12 but does with icilin. We found that introduction of the His844→Ala mutation to TRPM8FA_AG strongly suppresses icilin-dependent activation (Fig. 3, C and D). Whereas the His844→Ala mutation profoundly right-shifts the conductance-voltage (G-V) curve in response to icilin, it exerts no effect on TRPM8 activation by WS-12 (Fig. 3E and fig. S10). Previous studies have shown that menthol-dependent TRPM8 activation is not sensitive to intracellular pH, but intracellular pH does affect icilin-dependent activation (29). On the basis of our studies, we posit that the differential contribution of His844 to icilin and WS-12 binding may be the cause of the differences in pH sensitivity between menthol and icilin activation.
Distinct location of the PIP2 binding site in TRPM8
TRPM8FA and TRPM8FA_AG possess a pronounced PIP2 dependence, as depletion by poly-L-lysine (34) strongly induces channel desensitization, which is rapidly reversed by exogenous application of diC8-PIP2 (Fig. 4A and fig. S2, C and D), similar to mammalian TRPM8 channels (16, 17, 19). In our class 1 reconstruction of the icilin-PIP2-Ca2+ complex, we observed a strong non-protein density located at the interface between the TMD and the top layer of the CD ring (Fig. 4B). We assigned this density to PIP2, which is present in both half-maps (fig. S6), because the shape of the density is consistent with PIP2 and is absent in the reconstruction of the ligand-free TRPM8FA (30). The inositol 1,4,5-trisphosphate head group of the PIP2 molecule is positioned in the interfacial region (the interfacial cavity) formed by the pre-S1 domain, the junction between S4 and S5, the TRP domain, and the MHR4 from the adjacent subunit (Fig. 4B), with the acyl chains extended upward into the putative membrane region. Our structure reveals that basic amino acid residues from different subdomains—Lys605 from the neighboring MHR4 domain (MHR4’), Arg688 from the pre-S1 domain, Arg850 from the junction between S4 and S5, and Arg997 from the TRP domain—cluster at the interfacial cavity and interact with PIP2. Specifically, Lys605 and Arg850 interact with both the C-4 and the C-5 phosphate groups, whereas Arg688 and Arg997 form electrostatic interactions with the C4 phosphate moiety of PIP2 (Fig. 4C). Among these residues, only Arg997 has previously been implicated in PIP2 binding (17). Consistent with our structure, neutralizing mutations of residues Lys605, Arg850, and Arg997 via Gln substitutions impaired channel activation, as evidenced by the rightward shift in the G-V curves (Fig. 4D and fig. S11, A and B). As PIP2 binding and membrane depolarization have been demonstrated to be coupled for channel gating (17), the large increase in the voltage required for channel opening in these PIP2 binding mutations is consistent with reduced binding of PIP2. Therefore, our structural data in combination with the functional studies identify the PIP2 binding site in TRPM8.
Figure 4. PIP2 binding in TRPM8.
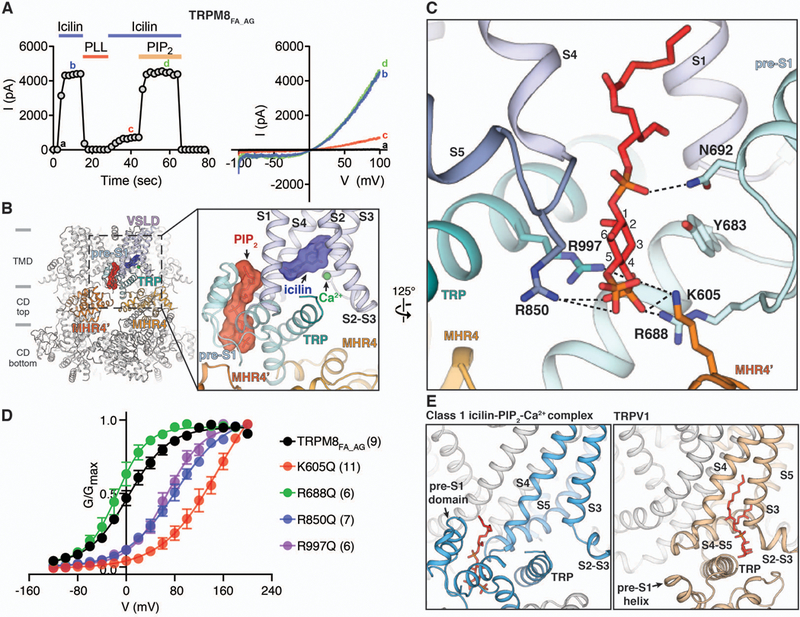
A) Functional characterization of TRPM8FA_AG by icilin and PIP2. 20 µM icilin in the presence of 12.5 µM Ca2+ was used to activate TRPM8FA_AG channels using inside-out patches. Membrane PIP2 was depleted by poly-L-lysine (PLL, 50 µg/ml) in the absence of icilin and Ca2+. 40 µM diC8-PIP2 was applied together with icilin and Ca2+ to recover channel activity. Time course of the peak current amplitudes at +100 mV from the voltage ramp (−100 mV to +100 mV) is shown on the left, and representative current traces are shown on the right. Quantifications of current amplitudes are in fig. S2D. (B) Global and close-up views (inset) of the PIP2 binding site in the class 1 icilin-PIP2-Ca2+ complex structure. PIP2 and icilin molecules are shown as sticks and surfaces. Gray bars indicate division of channel layers. (C) Key residues in the interfacial cavity that interact with PIP2. PIP2 and amino acid side chains are shown as sticks. (D) Conductance-voltage (G-V) relationship of K605Q, R688Q, R850Q, and R997Q mutant TRPM8FA_AG channels in response to 20 µM icilin and 12.5 µM Ca2+, as measured by their normalized peak tail currents (see Methods and fig. S11A for details). Numbers of individual recordings are noted in parentheses. Error bars indicate SEM. (E) Comparison of the PIP2 binding site in TRPM8 (left) and the phosphatidylinositol binding site in TRPV1 (right, PDB 5IRZ). Lipid molecules are shown as red sticks.
Our TRPM8 complex structure reveals an unforeseen binding site for PIP2 previously not observed in other TRP channels (Fig. 4E and fig. S12). Phosphatidylinositol lipids have been shown to bind to a cleft formed by S3, S4, and the S4-S5 linker in TRPV1 (Fig. 4E) and TRPV5 (35, 36). In marked contrast, PIP2 binds to the TRPM8 channel on the opposite side of S4 and S5, facing the interfacial cavity formed by multiple subdomains, including the pre-S1 and MHR4 domains, both of which are absent in TRPV channels. Our comparison of TRPM8 with the recently published TRPM4 and TRPM2 structures (33, 37) suggests that the observed PIP2 site may not be conserved throughout TRPM channels. In TRPM8 the pre-S1 domain preceding the TMD makes extensive interactions with the MHR4 of the CD at the membrane interface, which forms a large part of the PIP2 binding site (fig. S12A). By contrast, these interactions are absent in TRPM4 and TRPM2, and the amino acids involved in PIP2 binding are not conserved in these channels (fig. S12, B and C). Consistent with this structural observation, mutation of the PIP2-interacting Lys605 in MHR4 to Gln severely impairs the TRPM8 channel opening by icilin and WS-12 (Fig. 4D and fig. S11). Notably, among mutations of the PIP2 binding residues, Lys605→Gln exhibits the largest rightward shifts in the G-V curves, further indicating the importance of the PIP2-mediated interactions between CD and TMD for TRPM8 channel gating.
Two conformations of the interfacial cavity capture distinct PIP2 binding modes
Notably, structures of the ligand-free TRPM8FA, the WS-12-PIP2 complex, and the class 2 low-occupancy icilin-PIP2-Ca2+ complex possess similar interfacial cavity conformations, which are wider than that of the class 1 icilin-PIP2-Ca2+ complex structure (Fig. 5, A to E). The PIP2 density is also present in the 3D reconstruction of the WS-12-PIP2 complex (fig. S9). However, owing to the wider interfacial cavity in the WS-12-PIP2 complex structure, PIP2 fits less snugly in its binding site than it does in the class 1 icilin-PIP2-Ca2+ complex structure (Fig. 5, F and G). Structural analyses show that the difference in the size of the interfacial cavity stems from secondary structure rearrangements in S4b. In the structures of ligand-free TRPM8FA, the WS-12-PIP2 complex, and the class 2 icilin-PIP2-Ca2+ complex, the S4 is entirely α-helical, which positions Arg850 away from the PIP2 binding cavity. By contrast, in the class 1 high-occupancy icilin-PIP2-Ca2+ complex, S4b adopts a 310-helical conformation, which leads to a register change and repositions Arg850 towards the PIP2 binding cavity. This secondary structure change is accompanied by an upward tilt of the TRP domain and movement of S5 toward the interfacial cavity. Therefore, two distinct arrangements of the interfacial cavity provide different PIP2 binding modes (Fig. 5). We suggest that the class 1 icilin-PIP2-Ca2+ complex represents a conformation of TRPM8 with fully engaged PIP2 because (i) Arg850 forms additional interactions with PIP2, (ii) PIP2 fits more snugly in the binding pocket, and (iii) the ligand-free and the class 2 low-occupancy icilin-PIP2-Ca2+ structures adopt a distinct, wider interfacial cavity conformation.
Figure 5. Comparison of PIP2 binding in TRPM8 complex structures.
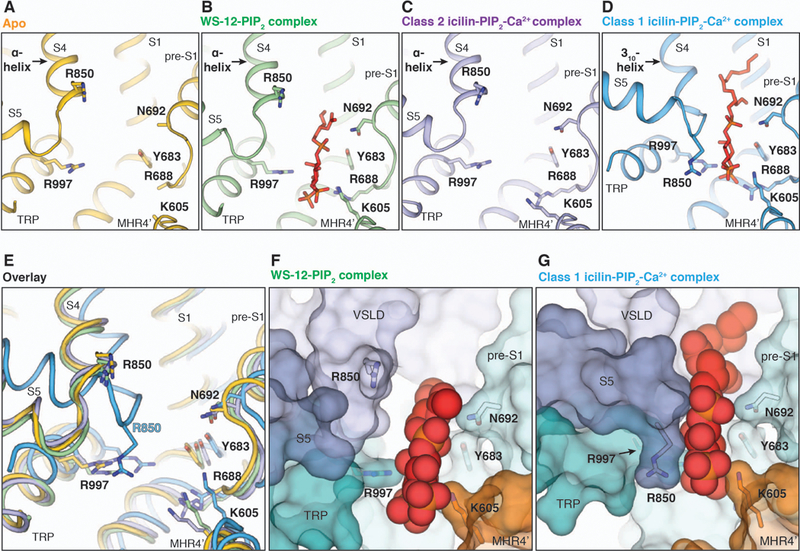
(A to D) Side-by-side comparison of the PIP2 binding site in the ligand-free TRPM8 [(A), yellow], WS-12-PIP2 complex [(B), green], class 2 icilin-PIP2-Ca2+ complex [(C), purple], and class 1 icilin-PIP2-Ca2+ complex structures [(D), blue]. PIP2 was not modeled in (C) owing to the weak EM density. (E) Overlay of the PIP2 binding site shown in (A) to (D). PIP2 molecules are omitted for clarity. (F and G) Surface representations comparing PIP2 binding in the WS-12-PIP2 complex (F) and in the class 1 icilin-PIP2-Ca2+ complex (G) structures. PIP2 molecules are shown as spheres.
Allosteric coupling between agonists and PIP2
Our studies illustrate a structural basis of coupling between PIP2 and cooling agonists, especially icilin (Fig. 6). In the class 1 icilin-PIP2-Ca2+ complex structure, PIP2 binds more optimally to the interfacial cavity because S4b adopts a 310-helical configuration, S5 is bent, and the TRP domain moves closer to PIP2 (Figs. 2A and6A). Likewise, icilin binding prefers a 310 configuration of S4b in which Arg841 and His844 rotate toward the center of the VSLD cavity. Therefore, by binding to a different region of S4b, PIP2 enhances the conformational change to accommodate icilin binding; conversely, icilin binding to S4b and the TRP domain induces the structural rearrangements favorable for PIP2 binding. By contrast, PIP2 is not fully engaged in the WS-12-PIP2 complex structure (Fig. 5F). We postulate that allosteric coupling of WS-12 with PIP2 likely proceeds via a similar mechanism, indicating that the WS-12-PIP2 complex structure represents a state in which the allosteric coupling between PIP2 and agonist is not yet established. The difference in the conformations of the interfacial cavity between the WS-12-PIP2 complex and the class 1 icilin-PIP2-Ca2+ complex likely originates from the higher potency and efficacy of icilin compared with WS-12 (Figs. 1E and4D and fig. S11G).
Figure 6. Allosteric coupling between agonists and PIP2.
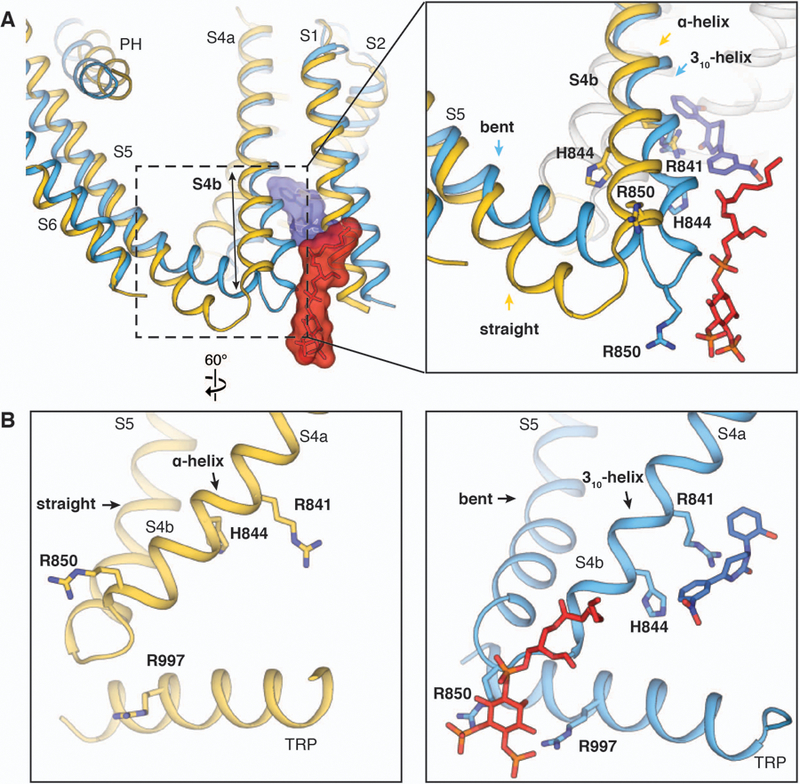
(A) Aligned at TMD, comparison between the ligand-free TRPM8 (yellow) and the class 1 icilin-PIP2-Ca2+ complex (blue) structures shows that the binding of PIP2 and icilin (in sticks and transparent surfaces colored red and blue, respectively) induces secondary structure rearrangements in S4b and the S4-S5 junction. (B) Side-by-side comparison illustrating the allosteric coupling between PIP2 and icilin.
Structural rearrangement in the pore upon ligand binding
In TRPM8, the binding of PIP2 along with icilin and Ca2+ or WS-12 triggers global conformational changes in the TMD that are propagated to the CD (fig. S13A). In contrast to the VSLD of TRPV1, which remains stationary during channel activation (35), the binding of cooling agonists and PIP2 to TRPM8 results in a rigid body rotation of the VSLD away from the pore domain as well as conformational changes in PH and S6 (Fig. 7, A and B and fig. S13, B and C). Compared to the apo structure, the ligand-induced conformational changes appeared to be most pronounced in the class 1 icilin-PIP2-Ca2+ complex, so we focus our analysis on the structural rearrangements induced by icilin, Ca2+, and PIP2. In the ligand-free TRPM8 structure, all transmembrane helices are straight and α-helical (30). S6 in the pore domain is restrained by its apparently tight interactions with VSLD (Trp798 in S3); thus, the pore is locked in a closed conformation (Fig. 7, C and D). In contrast, icilin, Ca2+, and PIP2 binding in the VSLD domain triggers an α- to 310-helical transition in S4b (Fig. 2C), bending of S5 [analogous to the S4-S5 linker bend in other TRP channels (35, 38)] (Fig. 7B), and movement of the TRP domain. These rearrangements generate more-extensive interactions between the TRP domain and S5 (fig. S13E) and, more importantly, disrupt the interactions between VSLD (Trp798 in S3) and S6, thereby enabling the rearrangements of S6 and the PH for channel gating (Fig. 7, C and D). As a result, substantial tilt and bending occur in the pore helix and S6 in the class 1 icilin-PIP2-Ca2+ complex structure; both S6 and PH in the icilin- and Ca2+-bound TRPM8 display curved conformations compared with the ligand-free TRPM8 structure, suggesting a potential presence of π-helical turns in these helices (Fig. 7E). The curved S6 in the icilin-PIP2-Ca2+-bound structure is reminiscent of the π helix containing S6 of the sensitized but non-conductive conformation of the human TRPV3 (39), and similar PH arrangements due to bending in S5 have been observed upon resiniferatoxin binding in TRPV2 (38). It is noteworthy that both PH and S6 in the class 1 reconstruction are poorly resolved whereas robust density is present for S5, indicating the mobile nature of these regions in the icilin-PIP2-Ca2+ complex. By contrast, in both the class 2 icilin-PIP2-Ca2+ complex and WS-12-PIP2 complex structures, slight rotation of the VSLD and slight widening of the S6 gate were observed but S5 maintains a straight conformation (Fig. 7E and fig. S13, B and C).
Figure 7. Structural rearrangements in response to the binding of PIP2 and agonists.
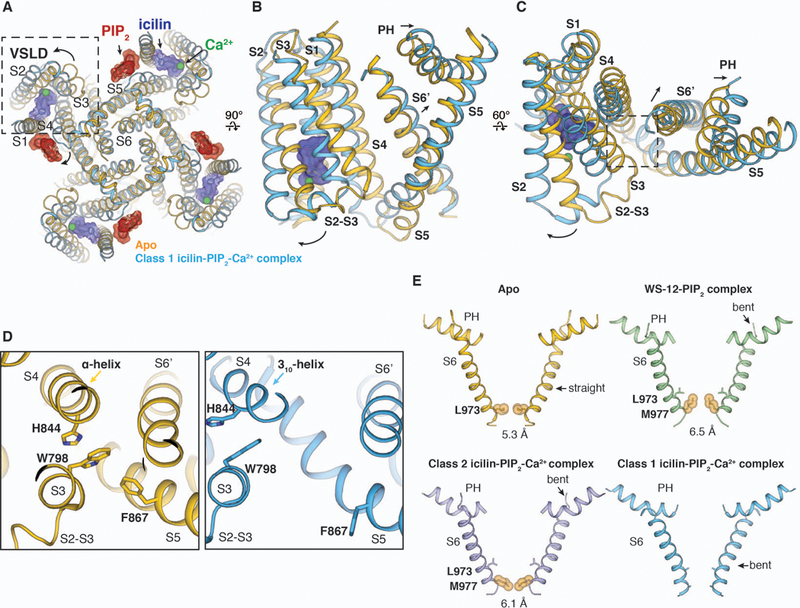
(A) As viewed from the intracellular side, comparison of the ligand-free TRPM8 (yellow) and the class 1 icilin-PIP2-Ca2+ complex (blue) structures reveals a large rotation of VSLD. (B and C) As viewed from the membrane (B) and from the extracellular side (C), overlay of the TMD in the ligand-free TRPM8 (yellow) and the class 1 icilin-PIP2-Ca2+ complex structure (blue). Arrows indicate the movement of VSLD, PH, and the neighboring S6 (S6’). (D) Close-up views of the region highlighted by the dashed square in (C), comparing differences in the interaction network between VSLD (Trp798 in S3) and the pore (S6’) in the two structures. (E) Comparison of S6 helices and PH in TRPM8 apo (yellow) and complex structures (WS-12, green; class 2 icilin-PIP2-Ca2+ complex, purple; class 1 icilin-PIP2-Ca2+ complex structure, blue). Residues at the narrowest point at the S6 gate are shown in orange spheres with diagonal distances indicated in angstroms. S6 and PH in the class 1 icilin-PIP2-Ca2+ complex structure were modeled as poly-alanine.
Because the open probability of the channel at 0 mV is low (~0.2 to 0.5) even in the saturating concentrations of ligands, and is further lowered by high concentrations of Ca2+ (Fig. 4D and figs. S3, A to C, and S11G), multiple conformational states exist in the cryo-EM samples. In spite of the considerable conformational differences with respect to the published ligand-free TRPM8FA structure, both the WS-12-PIP2-bound and icilin-PIP2-Ca2+-bound complex structures apparently adopt nonconducting states, suggesting that our cryo-EM reconstructions have captured conformations that either precede or follow the open state (Fig. 7E). We speculate that the class 1 icilin-PIP2-Ca2+ complex structure reflects a sensitized state of the channel because PIP2 is fully engaged in the interfacial cavity, S5 forms more extensive interactions with the TRP domain (fig. S13E), and the bent S6 conformation is analogous to that of TRPV3 in its sensitized-but-closed state (39). However, it is also possible that our class 1 icilin-PIP2-Ca2+ complex represents a desensitized state that could be populated by a high concentration of Ca2+ introduced in the cryo-EM sample. By contrast, we speculate that the WS-12-PIP2 complex structure adopts a presensitized state because PIP2 is not fully engaged in the interfacial cavity and no substantial conformational changes in S5 are observed when compared with the ligand-free TRPM8FA. Although WS-12 and icilin do not bind to TRPM8 in the same manner, we postulate that they might utilize use similar downstream mechanisms of channel activation once PIP2 is fully engaged, and thus the channel is sensitized. However, we cannot rule out the possibility that WS-12 and icilin have distinct mechanisms of activation.
Discussion
Our structural analyses revealed that TRPM8 adopts a sophisticated design principle and its mechanisms of coupled agonist and PIP2 sensing stand in stark contrast to those of TRPV1. First, unlike TRPV1 and other TRP channels (33, 35), the natural ligand-binding site is located within the VSLD cavity in TRPM8 channel. Ligand binding in the VSLD cavity offers an opportunity to directly engage and reposition the TRP domain to open the S6 gate. Moreover, the VSLD cavity in TRPM8 can adjust its shape to accommodate cooling compounds that are chemically distinct. This conformational flexibility of the VSLD cavity holds potential for the development of analgesic compounds to target TRPM8. Second, the PIP2 binding site in TRPM8 is strategically positioned in the interfacial cavity at the nexus of key subdomains including the VSLD. Therefore, PIP2 can effectively control conformational transitions associated with gating and enhance agonist binding. By contrast, in TRPV1, the location of phosphatidylinositol lipids overlaps the agonist-binding pocket, enabling phosphatidylinositol lipids to serve as competitive vanilloid antagonists (35). Therefore, we propose that the design of distinct but nearby binding sites for agonist and PIP2 in TRPM8 functions to exploit their structural allostery.
Members of the TRPM family have shown different levels of PIP2 dependence on channel activation (40). Our structural analyses indicate that the PIP2 site in TRPM8 is likely distinct from that in TRPM2 and TRPM4, as the latter two channels do not have the same quaternary structure arrangement at the interface between TMD and CD as TRPM8. It is tempting to speculate that this quaternary structure arrangement in TRPM8, which enables PIP2 binding and facilitates the conformational change required for menthol binding, is key to understanding the lack of menthol sensitivity in other TRPM channels (41).
Materials and methods summary
TRPM8FA and TRPM8FA_AG channels were expressed in HEK293S GnTI- cells and purified as described previously (30), with slight modifications. For cryo-EM study, purified TRPM8FA was incubated with WS-12 and PIP2, whereas TRPM8FA_AG was incubated with icilin, PIP2, and Ca2+ before vitrification. Data were collected on a Titan Krios electron microscope and processed with RELION 3.0 using standard procedures. Point mutations were introduced at the binding site for PIP2, icilin, and WS-12, and the effects of the mutations were examined by inside-out patch electrophysiology.
Supplementary Material
Acknowledgments
Cryo-EM data were collected at the Shared Materials Instrumentation Facility at Duke University as part of the Molecular Microscopy Consortium. We thank G. Lander and M. Wu at TSRI for help with preliminary studies and cryo-EM training for Y.Y. Cryo-EM image quality was monitored on the fly during data collection using routines developed by A. Bartesaghi. We thank L. Zubcevic and W. Borschel for critical manuscript reading and K. Yokoyama for help with icilin modeling.
Funding: This work was supported by the National Institutes of Health (R35NS097241 to S.-Y.L. and DP2GM126898 to H.Y.) and by the National Institute of Health Intramural Research Program, U.S. National Institutes of Environmental Health Sciences (ZIC ES103326 to M.J.B.).
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability: For the class 1 and class 2 icilin-PIP2-Ca2+ complex, and the WS-12-PIP2 complex, the coordinates have been deposited in the Protein Data Bank with IDs 6NR3, 6NR4, and 6NR2, respectively; and the cryo-EM density maps have been deposited in the Electron Microscopy Data Bank with IDs EMD-0488, EMD-0489, and EMD-0487, respectively.
References and Notes:
- 1.Farooqi AA et al. , TRPM channels: Same ballpark, different players, and different rules in immunogenetics. Immunogenetics 63, 773–787 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Fleig A, Penner R, The TRPM ion channel subfamily: molecular, biophysical and functional features. Trends Pharmacol Sci 25, 633–639 (2004). [DOI] [PubMed] [Google Scholar]
- 3.McKemy DD, TRPM8: The cold and menthol receptor in TRP Ion Channel Function in Sensory Transduction and Cellular Signaling Cascades, Liedtke WB, Heller S, Eds. (CRC Press/Taylor & Francis, 2007), pp. 177–188. [PubMed] [Google Scholar]
- 4.McKemy DD, Neuhausser WM, Julius D, Identification of a cold receptor reveals a general role for TRP channels in thermosensation. Nature 416, 52–58 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Peier AM et al. , A TRP channel that senses cold stimuli and menthol. Cell 108, 705–715 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Bautista DM et al. , The menthol receptor TRPM8 is the principal detector of environmental cold. Nature 448, 204–208 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Dhaka A et al. , TRPM8 is required for cold sensation in mice. Neuron 54, 371–378 (2007). [DOI] [PubMed] [Google Scholar]
- 8.Colburn RW et al. , Attenuated cold sensitivity in TRPM8 null mice. Neuron 54, 379–386 (2007). [DOI] [PubMed] [Google Scholar]
- 9.Knowlton WM et al. , A sensory-labeled line for cold: TRPM8-expressing sensory neurons define the cellular basis for cold, cold pain, and cooling-mediated analgesia. J Neurosci 33, 2837–2848 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chasman DI et al. , Genome-wide association study reveals three susceptibility loci for common migraine in the general population. Nat Genet 43, 695–698 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu B et al. , TRPM8 is the principal mediator of menthol-induced analgesia of acute and inflammatory pain. Pain 154, 2169–2177 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andrews MD et al. , Discovery of a Selective TRPM8 Antagonist with Clinical Efficacy in Cold-Related Pain. ACS Med Chem Lett 6, 419–424 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weyer AD, Lehto SG, Development of TRPM8 Antagonists to Treat Chronic Pain and Migraine. Pharmaceuticals (Basel) 10, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Almaraz L, Manenschijn JA, de la Pena E, Viana F, Trpm8. Handb Exp Pharmacol 222, 547–579 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Horne DB et al. , Discovery of TRPM8 Antagonist ( S)-6-(((3-Fluoro-4-(trifluoromethoxy)phenyl)(3-fluoropyridin-2-yl)methyl)carbamoy l)nicotinic Acid (AMG 333), a Clinical Candidate for the Treatment of Migraine. J Med Chem 61, 8186–8201 (2018). [DOI] [PubMed] [Google Scholar]
- 16.Liu B, Qin F, Functional control of cold- and menthol-sensitive TRPM8 ion channels by phosphatidylinositol 4,5-bisphosphate. J Neurosci 25, 1674–1681 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rohacs T, Lopes CM, Michailidis I, Logothetis DE, PI(4,5)P2 regulates the activation and desensitization of TRPM8 channels through the TRP domain. Nat Neurosci 8, 626–634 (2005). [DOI] [PubMed] [Google Scholar]
- 18.Voets T, Owsianik G, Janssens A, Talavera K, Nilius B, TRPM8 voltage sensor mutants reveal a mechanism for integrating thermal and chemical stimuli. Nat Chem Biol 3, 174–182 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Zakharian E, Cao C, Rohacs T, Gating of transient receptor potential melastatin 8 (TRPM8) channels activated by cold and chemical agonists in planar lipid bilayers. J Neurosci 30, 12526–12534 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voets T, Owsianik G, Nilius B, Trpm8. Handb Exp Pharmacol, 329–344 (2007). [DOI] [PubMed]
- 21.Launay P et al. , TRPM4 is a Ca2+-activated nonselective cation channel mediating cell membrane depolarization. Cell 109, 397–407 (2002). [DOI] [PubMed] [Google Scholar]
- 22.McHugh D, Flemming R, Xu SZ, Perraud AL, Beech DJ, Critical intracellular Ca2+ dependence of transient receptor potential melastatin 2 (TRPM2) cation channel activation. J Biol Chem 278, 11002–11006 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Chuang HH, Neuhausser WM, Julius D, The super-cooling agent icilin reveals a mechanism of coincidence detection by a temperature-sensitive TRP channel. Neuron 43, 859–869 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Bandell M et al. , High-throughput random mutagenesis screen reveals TRPM8 residues specifically required for activation by menthol. Nat Neurosci 9, 493–500 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Ma S, G G, Ak VE, Jf D, H H, Menthol derivative WS-12 selectively activates transient receptor potential melastatin-8 (TRPM8) ion channels. Pak J Pharm Sci 21, 370–378 (2008). [PubMed] [Google Scholar]
- 26.Kuhn FJ, Kuhn C, Luckhoff A, Inhibition of TRPM8 by icilin distinct from desensitization induced by menthol and menthol derivatives. J Biol Chem 284, 4102–4111 (2009). [DOI] [PubMed] [Google Scholar]
- 27.Malkia A, Pertusa M, Fernandez-Ballester G, Ferrer-Montiel A, Viana F, Differential role of the menthol-binding residue Y745 in the antagonism of thermally gated TRPM8 channels. Mol Pain 5, 62 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Janssens A, Voets T, Ligand stoichiometry of the cold- and menthol-activated channel TRPM8. J Physiol 589, 4827–4835 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Andersson DA, Chase HW, Bevan S, TRPM8 activation by menthol, icilin, and cold is differentially modulated by intracellular pH. J Neurosci 24, 5364–5369 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin Y et al. , Structure of the cold- and menthol-sensing ion channel TRPM8. Science 359, 237–241 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bodding M, Wissenbach U, Flockerzi V, Characterisation of TRPM8 as a pharmacophore receptor. Cell Calcium 42, 618–628 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Autzen HE et al. , Structure of the human TRPM4 ion channel in a lipid nanodisc. Science 359, 228–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang Y, Winkler PA, Sun W, Lu W, Du J, Architecture of the TRPM2 channel and its activation mechanism by ADP-ribose and calcium. Nature 562, 145–149 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Hille B, Dickson EJ, Kruse M, Vivas O, Suh BC, Phosphoinositides regulate ion channels. Biochim Biophys Acta 1851, 844–856 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gao Y, Cao E, Julius D, Cheng Y, TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature 534, 347–351 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hughes TET et al. , Structural insights on TRPV5 gating by endogenous modulators. Nat Commun 9, 4198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guo J et al. , Structures of the calcium-activated, non-selective cation channel TRPM4. Nature 552, 205–209 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zubcevic L, Le S, Yang H, Lee SY, Conformational Plasticity in the Selectivity Filter of the TRPV2 Ion Channel. Nat Struct Mol Biol, in press (2018). [DOI] [PMC free article] [PubMed]
- 39.Zubcevic L et al. , Conformational ensemble of the human TRPV3 ion channel. Nat Commun 9, 4773 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohacs T, Phosphoinositide regulation of TRP channels. Handb Exp Pharmacol 223, 1143–1176 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bae C, Jara-Oseguera A, Swartz KJ, TRPM channels come into focus. Science 359, 160–161 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Goehring A et al. , Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat Protoc 9, 2574–2585 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zheng SQ et al. , MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods 14, 331–332 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang K, Gctf: Real-time CTF determination and correction. J Struct Biol 193, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW, New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zivanov J, Nakane T, Scheres SHW, A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K, Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Adams PD et al. , PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen VB et al. , MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


