Abstract
Nonalcoholic fatty liver disease (NAFLD) is a complex disorder that has evolved in recent years as the leading global cause of chronic liver damage. The main obstacle to better disease management pertains to the lack of approved pharmacological interventions for the treatment of nonalcoholic steatohepatitis (NASH) and NASH-fibrosis-the severe histological forms. Over the past decade, tremendous advances have been made in NAFLD research, resulting in the discovery of disease mechanisms and novel therapeutic targets. Hence, a large number of pharmacological agents are currently being tested for safety and efficacy. These drugs are in the initial pharmacological phases (phase 1 and 2), which involve testing tolerability, therapeutic action, and pharmacological issues. It is thus reasonable to assume that the next generation of NASH drugs will not be available for clinical use for foreseeable future. The expected delay can be mitigated by drug repurposing or repositioning, which essentially relies on identifying and developing new uses for existing drugs. Here, we propose a drug candidate selection method based on the integration of molecular pathways of disease pathogenesis into network analysis tools that use OMICs data as well as multiples sources, including text mining from the medical literature.
Keywords: Drug discovery, Drug repositioning, Fibrosis, Genetics, Treatment, Systems biology
Core tip: As a proof-of-concept of the advantages that can be yielded by applying multi-omics systems-based approaches to the analysis of potential candidates to the treatment of nonalcoholic steatohepatitis (NASH) we selected the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway map of nonalcoholic fatty liver disease (NAFLD), which illustrates a stage-dependent progression of the disease. After generating a protein−chemical interaction network, we predicted remarkable examples of potential drug repurposing for the treatment of NASH based on the NAFLD-KEGG connectivity map.
INTRODUCTION
Nonalcoholic fatty liver disease (NAFLD) is a complex disorder that has emerged as the leading global cause of chronic liver damage in recent years[1]. The disease course progresses through highly dynamic histological stages, ranging from simple steatosis or nonalcoholic fatty liver (NAFL) to nonalcoholic steatohepatitis (NASH), NASH-fibrosis and cirrhosis[1,2]. NASH-fibrosis and its complications, including cirrhosis and hepatocellular carcinoma, not only significantly reduce life expectancy by increasing liver-related mortality[3] but also represent a challenge for the healthcare system because much of the affected population is also affected by NAFLD-associated comorbidities, including obesity, type 2 diabetes (T2D), and cardiovascular disease[1,4-6]. Absence of reliable noninvasive biomarkers that allow identification of patients at a high risk of fibrosis and /or disease progression is one of the obstacles facing disease management[7,8]. Similarly, while a large number of drugs against NASH are currently being tested for efficacy and safety, no pharmacological interventions are presently approved for treating NASH[2,5,9,10].
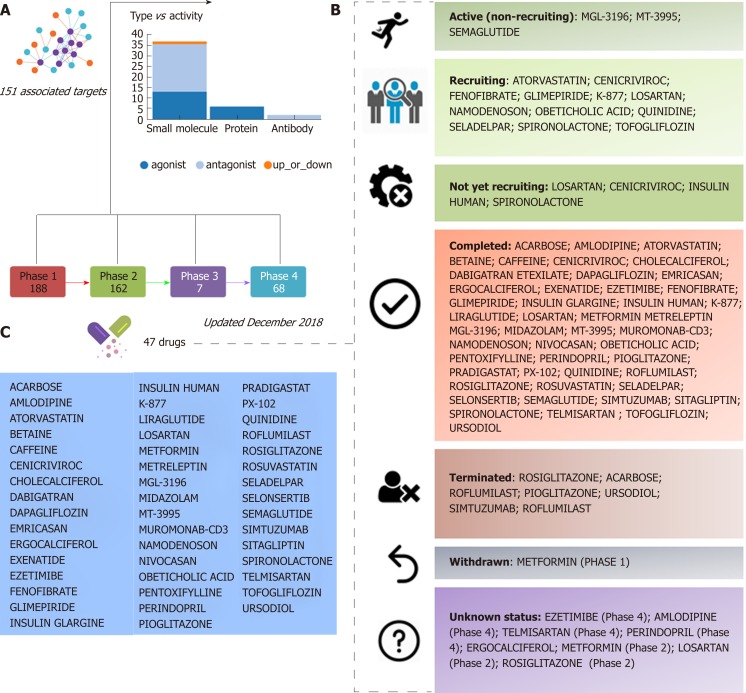
Information retrieved from public domain data sources and clinical ClinicalTrials.gov (updated December 2018), a resource provided by the U.S. National Library of Medicine, indicates that approximately 47 different drugs that target NASH and NASH-fibrosis are currently being tested in different pharmacological stages, including 188 drugs in phase 1 and 162 in phase 2 studies (Figure 1). A significant proportion of these drugs are small molecules or proteins that either antagonize or act as exogenous agonists of one or more targets of interest; the 47 aforementioned NASH drugs are in fact predicted to be linked to 151 molecular targets (Figure 1). Considering that a large majority of these drugs are in the earliest pharmacological phases that involve testing tolerability, therapeutic action, and pharmacological issues, it is reasonable to conclude that there will be a significant time lag before the next generation of NASH drugs is available for clinical use.
Figure 1.
Clinical trials for the treatment of nonalcoholic steatohepatitis. A and B: Figure highlights 47 drugs that are currently under investigation for the treatment of nonalcoholic steatohepatitis in different pharmacological phases (from phase 1 to phase 4): Information on clinical trial status (recruitment status) as well as prediction of potential associated targets were retrieved from the Target Validation Platform available at https://www.targetvalidation.org; C: Drugs listed in the most advanced pharmacological phase updated December 2018 concerning to privately and publicly funded clinical studies. Not yet recruiting: The study has not started recruiting participants; Recruiting: The study is currently recruiting participants; Active, not recruiting: The study is ongoing, and participants are receiving an intervention or being examined, but potential participants are not currently being recruited or enrolled; Terminated: The study has stopped early and will not start again; participants are no longer being examined or treated; Completed: The study has ended normally, and participants are no longer being examined or treated (that is, the last participant's last visit has occurred); Withdrawn: The study stopped early, before enrolling its first participant; Unknown: A study on ClinicalTrials.gov whose last known status was recruiting; not yet recruiting; or active, not recruiting but that has passed its completion date, and the status has not been last verified within the past 2 years).
One potential solution to this expected delay is drug repurposing or repositioning, which relies on identifying and developing new uses for existing drugs[11]. The advantage of drug repurposing is not limited to the fact that drugs selected for a novel indication have already passed the time-consuming pharmacokinetics, pharmacodynamics, and toxicity profiling evaluation, but are also already approved by major regulatory agencies, including the United States Food and Drug Administration and/or the European Medicines Agency.
Drug repurposing can be addressed by different approaches. Most common ones involve the selection of drug candidate/s based on known targets involved in the pathogenesis of the disease of interest. More recently, system biology strategies based on a broad search into genomic resources, as well as large-scale gene expression libraries, have been proposed as an attractive and innovative solution, particularly for the treatment of complex diseases like NAFLD that shares disease mechanisms with diseases of the metabolic syndrome[12-14]. Hence, we propose a drug candidate selection method based on the integration of molecular pathways of disease pathogenesis into network analysis tools that use OMICs data as well as multiples sources, including text mining from pertinent medical literature.
including text mining from pertinent medical literature
including text mining from pertinent medical literature
DRUG REPURPOSING FOR THE TREATMENT OF NASH BASED ON THE NAFLD-KEGG CONNECTIVITY MAP
As a proof-of-concept of the advantages of using multi-omics systems-based approaches for the analysis of potential NASH treatment candidates, we selected the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway map of NAFLD (pathway ID: hsa04932), which illustrates a stage-dependent progression of the disease (Figure 2). This pathway is composed of 149 genes/proteins involved not only in the progression of NAFL to NASH and to cirrhosis, but also genes/proteins shared with obesity and T2D (Table 1). Significant disease-related pathogenic processes, including de novo fatty acid biosynthesis, lipid peroxidation, endoplasmic reticulum stress and mitochondrial dysfunction[15-17], as well as apoptosis and cell death related mechanisms are represented in the NAFLD-KEGG pathway (Figure 2). Thus, we generated a protein−chemical interaction network by mapping the significant genes/proteins that are represented in the pathway to chemicals/drugs that are annotated in the Comparative Toxicogenomics Database. The 149 genes (seeds) yielded by our analysis were then mapped to the corresponding molecular interaction database; this procedure produced an extensive network comprising of approximately 2000 nodes. One of the largest subnetworks included 3212 smaller nodes (that represent the number of gene/protein–chemical interactions in this subnetwork), with 13314 interactions among node members. For simplicity, we manually curated some chemical−drug interactions focusing specifically on certain genes/proteins of potential interest, including members of the caspase family (CASP3 and CASP7), interleukins (IL1A, IL1B, and IL6), tumor necrosis factor α (TNFα), nuclear factor kappa B subunit 1 (NFKB1) and inhibitor of nuclear factor kappa B kinase subunit beta, Jun proto-oncogene (JUN), transcription factor subunit, and AKT serine/threonine kinase 1 (Figure 3). Remarkably, several drugs were predicted to have a significant interaction with the highlighted targets. For example, minocycline that is a broad spectrum long-acting derivative of the antibiotic tetracycline was mapped in the pathway of caspases, whereas IL1B (Figure 3) or pomalidomide that is a derivative of thalidomide with immuno-modulating, antiangiogenic and antineoplastic activities was mapped in the network of TNF, NFKB1, and interleukins (Figure 3).
Figure 2.
Nonalcoholic fatty liver disease-Kyoto Encyclopedia of Genes and Genomes pathway and mechanisms of disease pathogenesis. Pathway was retrieved from https://www.genome.jp/dbget-bin/www_bget?pathway+hsa04932; figure was modified to highlight key molecular processes. This map shows a stage-dependent progression of nonalcoholic fatty liver disease (NAFLD). In the first stage of NAFLD, pathway highlights excess lipid accumulation associated with the induction of insulin resistance, which leads to a defect in insulin suppression of free fatty acids (FAAs) disposal. In addition, two transcription factors, SREBP-1c and PPARα, activate key enzymes of lipogenesis and increase the synthesis of FAAs in liver. In the second stage, pathway is presented as a consequence of the progression to nonalcoholic steatohepatitis (NASH); the production of reactive oxygen species is enhanced due to oxidation stress through mitochondrial beta-oxidation of fatty acids and endoplasmic reticulum (ER) stress, leading to lipid peroxidation. The lipid peroxidation can further cause the production of cytokines [Fas ligand, tumor necrosis factor α (TNF-α), IL-8 and transforming growth factor], promoting cell death, inflammation and fibrosis. The activation of JNK, which is induced by ER stress, TNF-α and FAAs, is also associated with NAFLD progression. Increased JNK promotes cytokine production and initiation of hepatocellular carcinoma. Major organelles involved in the pathogenesis of NASH are also highlighted in the NAFLD-pathway, including mitochondria and mitochondrial dysfunction. In the figure, molecular targets that were further selected to explore protein-chemical interactions are highlighted by red squares. NAFLD: Nonalcoholic fatty liver disease; NASH: Nonalcoholic steatohepatitis; ER: Endoplasmic reticulum; HCC: Hepatocellular carcinoma; NAFL: Nonalcoholic fatty liver; FAAs: Free fatty acids; TNFα: tumor necrosis factor α.
Table 1.
Non-alcoholic fatty liver disease-Kyoto Encyclopedia of Genes and Genomes pathway (hsa04932)
| Gene symbol; description |
| IL6; interleukin 6 |
| IL6R; interleukin 6 receptor |
| SOCS3; suppressor of cytokine signaling 3 |
| TNF; tumor necrosis factor |
| TNFRSF1A; TNF receptor superfamily member 1A |
| NFKB1; nuclear factor kappa B subunit 1 |
| RELA; RELA proto-oncogene, NF-kB subunit |
| INS; insulin |
| INSR; insulin receptor |
| IRS1; insulin receptor substrate 1 |
| IRS2; insulin receptor substrate 2 |
| PIK3CA; phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| PIK3CD; phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| PIK3CB; phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit beta |
| PIK3R1; phosphoinositide-3-kinase regulatory subunit 1 |
| PIK3R2; phosphoinositide-3-kinase regulatory subunit 2 |
| PIK3R3; phosphoinositide-3-kinase regulatory subunit 3 |
| AKT1; AKT serine/threonine kinase 1 |
| AKT2; AKT serine/threonine kinase 2 |
| AKT3; AKT serine/threonine kinase 3 |
| GSK3A; glycogen synthase kinase 3 alpha |
| GSK3B; glycogen synthase kinase 3 beta |
| NR1H3; nuclear receptor subfamily 1 group H member 3 |
| RXRA; retinoid X receptor alpha |
| SREBF1; sterol regulatory element binding transcription factor 1 |
| MLX; MLX, MAX dimerization protein |
| MLXIP; MLX interacting protein |
| MLXIPL; MLX interacting protein like |
| PKLR; pyruvate kinase L/R |
| LEP; leptin |
| LEPR; leptin receptor |
| ADIPOQ; adiponectin, C1Q and collagen domain containing |
| ADIPOR1; adiponectin receptor 1 |
| ADIPOR2; adiponectin receptor 2 |
| PRKAA1; protein kinase AMP-activated catalytic subunit alpha 1 |
| PRKAA2; protein kinase AMP-activated catalytic subunit alpha 2 |
| PRKAB1; protein kinase AMP-activated non-catalytic subunit beta 1 |
| PRKAB2; protein kinase AMP-activated non-catalytic subunit beta 2 |
| PRKAG1; protein kinase AMP-activated non-catalytic subunit gamma 1 |
| PRKAG3; protein kinase AMP-activated non-catalytic subunit gamma 3 |
| PRKAG2; protein kinase AMP-activated non-catalytic subunit gamma 2 |
| PPARA; peroxisome proliferator activated receptor alpha |
| CDC42; cell division cycle 42 |
| RAC1; Rac family small GTPase 1 |
| MAP3K11; mitogen-activated protein kinase kinase kinase 11 |
| MAPK8; mitogen-activated protein kinase 8 |
| MAPK10; mitogen-activated protein kinase 10 |
| MAPK9; mitogen-activated protein kinase 9 |
| ITCH; itchy E3 ubiquitin protein ligase |
| ERN1; endoplasmic reticulum to nucleus signaling 1 |
| TRAF2; TNF receptor associated factor 2 |
| MAP3K5; mitogen-activated protein kinase kinase kinase 5 |
| JUN; Jun proto-oncogene, AP-1 transcription factor subunit |
| IL1A; interleukin 1 alpha |
| IL1B; interleukin 1 beta |
| IKBKB; inhibitor of nuclear factor kappa B kinase subunit beta |
| XBP1; X-box binding protein 1 |
| CEBPA; CCAAT enhancer binding protein alpha |
| CYP2E1; cytochrome P450 family 2 subfamily E member 1 |
| FASLG; Fas ligand |
| CXCL8; C-X-C motif chemokine ligand 8 |
| TGFB1; transforming growth factor beta 1 |
| EIF2AK3; eukaryotic translation initiation factor 2 alpha kinase 3 |
| EIF2S1; eukaryotic translation initiation factor 2 subunit alpha |
| ATF4; activating transcription factor 4 |
| DDIT3; DNA damage inducible transcript 3 |
| BCL2L11; BCL2 like 11 |
| BAX; BCL2 associated X, apoptosis regulator |
| FAS; Fas cell surface death receptor |
| CASP8; caspase 8 |
| BID; BH3 interacting domain death agonist |
| CYCS; cytochrome c, somatic |
| CASP3; caspase 3 |
| CASP7; caspase 7 |
| NDUFV1-3; NADH:ubiquinone oxidoreductase core subunit V1 –V3 |
| NDUFA1-3; NADH:ubiquinone oxidoreductase subunit A1-3 |
| NDUFA4; NDUFA4, mitochondrial complex associated |
| NDUFA4L2; NDUFA4, mitochondrial complex associated like 2 |
| NDUFA5-13; NADH:ubiquinone oxidoreductase subunit A5-A13 |
| NDUFAB1; NADH:ubiquinone oxidoreductase subunit AB1 |
| NDUFB1-11; NADH:ubiquinone oxidoreductase subunit B1-B11 |
| NDUFS1-S8; NADH:ubiquinone oxidoreductase core subunit S1 –S8 |
| NDUFC1; NADH:ubiquinone oxidoreductase subunit C1 |
| NDUFC2; NADH:ubiquinone oxidoreductase subunit C2 |
| NDUFC2-KCTD14; NDUFC2-KCTD14 readthrough |
| SDHA; succinate dehydrogenase complex flavoprotein subunit A |
| SDHB; succinate dehydrogenase complex iron sulfur subunit B |
| SDHC; succinate dehydrogenase complex subunit C |
| SDHD; succinate dehydrogenase complex subunit D |
| UQCRFS1; ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| CYTB; cytochrome b |
| CYC1; cytochrome c1 |
| UQCRC1; ubiquinol-cytochrome c reductase core protein 1 |
| UQCRC2; ubiquinol-cytochrome c reductase core protein 2 |
| UQCRH; ubiquinol-cytochrome c reductase hinge protein |
| UQCRHL; ubiquinol-cytochrome c reductase hinge protein like |
| UQCRB; ubiquinol-cytochrome c reductase binding protein |
| UQCRQ; ubiquinol-cytochrome c reductase complex III subunit VII |
| UQCR10; ubiquinol-cytochrome c reductase, complex III subunit X |
| UQCR11; ubiquinol-cytochrome c reductase, complex III subunit XI |
| COX3; cytochrome c oxidase III |
| COX1; cytochrome c oxidase subunit I |
| COX2; cytochrome c oxidase subunit II |
| COX4I2; cytochrome c oxidase subunit 4I2 |
| COX4I1; cytochrome c oxidase subunit 4I1 |
| COX5A; cytochrome c oxidase subunit 5A |
| COX5B; cytochrome c oxidase subunit 5B |
| COX6A1; cytochrome c oxidase subunit 6A1 |
| COX6A2; cytochrome c oxidase subunit 6A2 |
| COX6B1; cytochrome c oxidase subunit 6B1 |
| COX6B2; cytochrome c oxidase subunit 6B2 |
| COX6C; cytochrome c oxidase subunit 6C |
| COX7A1; cytochrome c oxidase subunit 7A1 |
| COX7A2; cytochrome c oxidase subunit 7A2 |
| COX7A2L; cytochrome c oxidase subunit 7A2 like |
| COX7B; cytochrome c oxidase subunit 7B |
| COX7B2; cytochrome c oxidase subunit 7B2 |
| COX7C; cytochrome c oxidase subunit 7C |
| COX8C; cytochrome c oxidase subunit 8C |
| COX8A; cytochrome c oxidase subunit 8A |
https://www.genome.jp/kegg-bin/show_pathway? hsa04932.
Figure 3.
Protein-chemical interactions and potential repurposing drugs to target nonalcoholic steatohepatitis. We generated a protein-chemical interaction network by mapping the significant genes/proteins that are represented in the nonalcoholic fatty liver disease-Kyoto Encyclopedia of Genes and Genomes pathway to chemicals/drugs that are annotated in the Comparative Toxicogenomics Database. The 149 genes (seeds) from our analysis were mapped to the corresponding molecular interaction database; full list of seed genes is listed in Table 1. This analysis generated a huge network composed of approximately 2000 nodes. Current figure shows chemical-drug-interactions specifically focused on selected genes/proteins of potential interest, including members of the caspase family (CASP3 and CASP7), interleukins (IL1B, IL1A, and IL6), tumor necrosis factor α (TNF-α), NFKB1 (Nuclear factor kappa B subunit 1) and IKBKB (inhibitor of nuclear factor kappa B kinase subunit beta), JUN (Jun proto-oncogene, AP-1 transcription factor subunit), AKT1 (AKT serine/threonine kinase 1). In green charts we summarized information on current use and known action of selected drugs. Interaction network was predicted by the Networkanalyst resource available at https://www.networkanalyst.ca/faces/home.xhtml. The network is shown as a Cytoscape graph.
Additional targets predicted in the minoclycline interaction network are arachidonate 5-lipoxygenase (which is involved in the synthesis of leukotrienes from arachidonic acid), cytochrome C (a central component of the electron transport chain in mitochondria), matrix metallopeptidase 9 (involved in the breakdown of extracellular matrix), vascular endothelial growth factor A (which induces proliferation and migration of vascular endothelial cells, particularly during pathological angiogenesis) and Poly(ADP-ribose) polymerase 1 (which is involved in the regulation of a myriad of cellular processes, such as differentiation, proliferation, and tumor transformation, as well as in the regulation of the molecular events implicit in the cell recovery from DNA damage). Further two candidate targets predicted in the network of pomalidomide are prostaglandin-endoperoxide synthase 2 (also known as cyclooxygenase, which is the key enzyme in prostaglandin biosynthesis) and CRBN (a calcium channel membrane protein, thought to play a role in brain development).
Additional examples of drugs that could be potentially tested for the treatment of NASH based on the concept of drug repositioning are illustrated in Figure 3. Drugs in the category of angiotensin II receptor type 1 (AGTR1) antagonists that were predicted in the network of JUN, for instance irbersartan-a nonpeptide AGTR1 antagonist with antihypertensive activity-might indeed be regarded as an indication expansion rather than drug repositioning because, as mentioned above, NAFLD and components of the Metabolic Syndrome, including arterial hypertension, present shared disease mechanisms (12-14). Therefore, given the pleiotropic effects of AGTR1 blockers[18] it is plausible to suggest that drugs in this pharmacological group-sartans-would synergize or potentiate the benefits of blocking the renin angiotensin system in the liver[19-22]. Remarkably, the pharmacological properties and toxicity profiles of some of the drugs presently undergoing NASH clinical trials are already known, such as atorvastatin, ezetimibe, fenofribrate, losartan, and pioglotazone, just to mention a few (Figure 1).
PLEIOTROPY: CHALLENGES AND OPPORTUNITIES FOR THE TREATMENT OF NASH
It is also important to acknowledge the possibility that some of the novel pharmacotherapy options for the treatment of NASH might eventually present pleiotropic effect/s. This point represents the paradox of a drug covering multiple pathways and cell types, which could be either harmful or beneficial for patients. Remarkable examples of the advantages of pleiotropic effects of pharmacological targets for the treatment of complex traits are, as already mentioned, agents that modulate or interfere with the rennin−angiotensin system, which not only reduce cardiovascular risk but also improve systemic inflammation, oxidative stress, and even present anti-fibrogenic properties in the liver. Similar effects have also been demonstrated for statins[23,24].
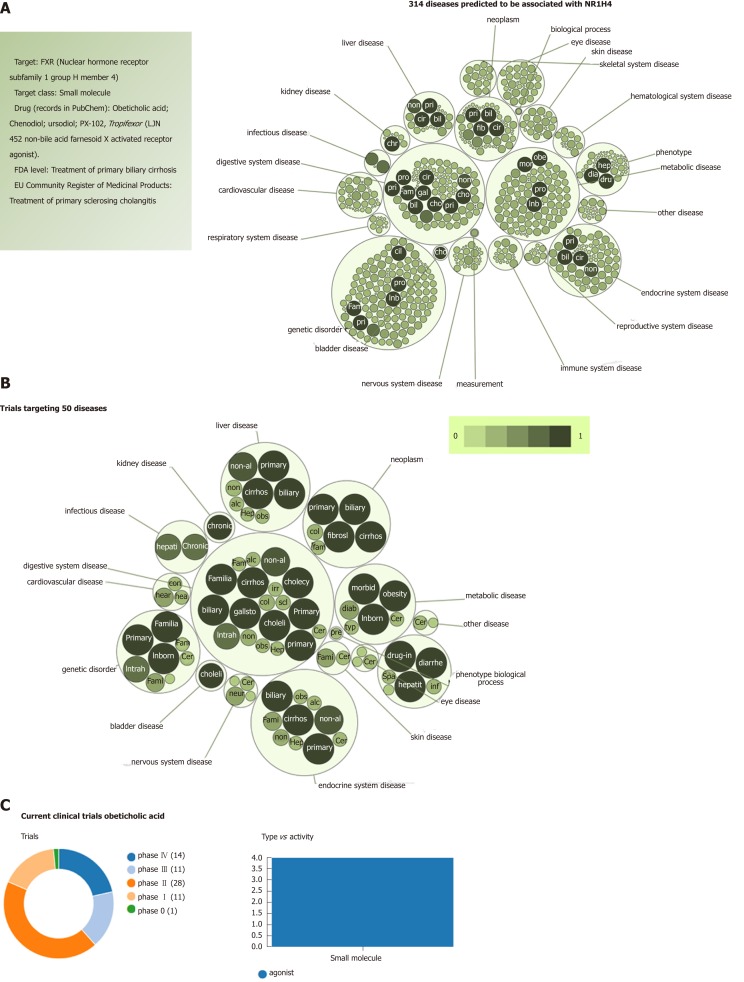
When focusing on the new generation of NASH targets, obeticholic acid (OCA), a synthetically-modified bile acid (a dihydroxy-5beta-cholanic acid), is a remarkable example of the potential systemic effects of a drug targeting nuclear receptors. OCA exhibits a potent agonist effect on the farnesoid X nuclear receptor (FXR). More importantly, its target-FXR (formally Nuclear hormone receptor subfamily 1 group H member 4, NR1H4, also known as BAR) is predicted to be involved in the pathogenesis of multiple phenotypes that practically cover the full range of human diseases and traits (Figure 4). It is well known that OCA is currently used to treat not only NASH but other chronic liver diseases as well, including primary biliary cholangitis[25]. However, there are at least 65 registered clinical trials in various pharmacological phases for ~50 different diseases (Figure 4).
Figure 4.
Farnesoid X nuclear receptor (nuclear hormone receptor subfamily 1 group H member 4): Analysis of pleiotropy. A: Graph shows all predicted diseases associated with farnesoid X nuclear receptor; B: Clinical trials of drugs that target farnesoid X nuclear receptor. Predictions were explored in The Open Targets Platform that allows prioritisation of drug targets based on the strength of their association with a disease (https://www.targetvalidation.org/); C: Evidence curated from ClinicalTrials.gov, a database of privately and publicly funded clinical studies conducted around the world. https://clinicaltrials.gov/. Diseases are presented as bubbles grouped into therapeutic areas using their Experimental Factor Ontology relationships. The size and shade of the color of each bubble is proportional to the strength of association between the disease and farnesoid X nuclear receptor. The concept of a target-disease association is based on the analysis of several resources, including genetic associations (GWAS Catalog, UniProt, European Variation Archive, Gene2Phenotype), somatic mutations (Cancer Gene Census, European Variation Archive somatic, IntOGen), RNA expression (expression atlas), drugs (ChEMBL), affected pathways (Reactome), animal models (PhenoDigm) and text mining (Europe PMC). The platform is available at https://www.targetvalidation.org. Data last updated December 2018.
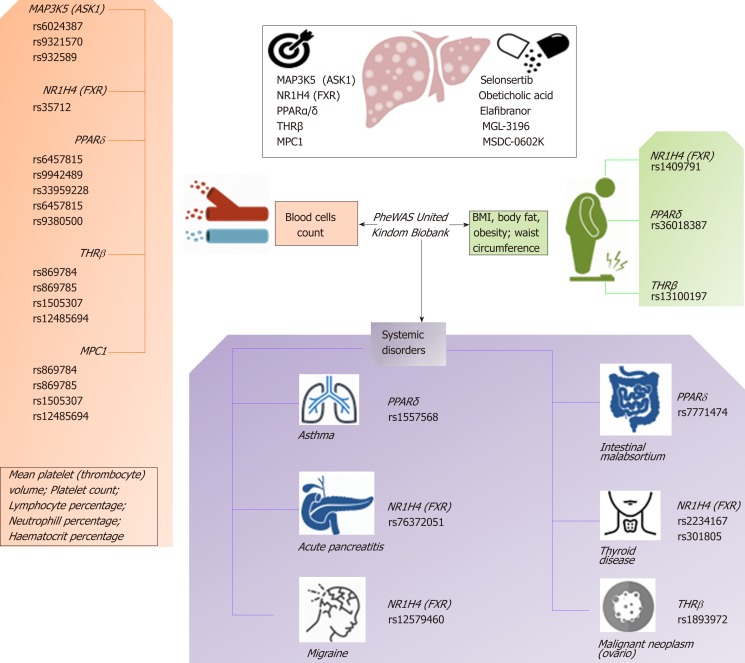
Based on this evidence, one may presume that the pleiotropic effects, and thus the clinical consequences, of the novel NASH drugs that are predicted to concurrently modulate a broad range of molecular pathways could be surprisingly extensive and therefore largely beneficial for treating multiple phenotypes. However, potential pleiotropic effects of the novel anti-NASH drugs could produce undesirable effects that we need to understand in order to anticipate their management. Some of these potential pleiotropic effects are indeed related to the primary biological and molecular network associated with the drug target itself. To illustrate the importance of this issue, we randomly selected five molecular targets (MAP3K5 or ASK1, FXR, PPARα/δ, THRβ, and MPC1) against which five drugs are currently being tested in patients with NASH (selonsertib[26], OCA[27], elafibranor[28], MGL-3196 (https://clinicaltrials.gov/ct2/show/NCT02912260), and MSDC-0602K[29] https://clinicaltrials.gov/ct2/show/NCT02784444). Next, we explored the potential pleiotropic effect/s of modulating these targets in humans by searching for associations of genetic variants in the aforementioned targets with different phenotypes and traits, known as PheWAS (Phenome-wide association studies). We specifically retrieved publically available information from the United Kindom Biobank that explored genetic variations in 452264 United Kindom Biobank White British individuals (http://geneatlas.roslin.ed.ac.uk/)[30].
As shown in Figure 5 and Table 2, MAP3K5/ASK1, FXR, PPARα/δ, THRβ, and MPC1 variants are involved in multiple pleiotropic effects, including modulation of blood cell count, body mass index, and general body adiposity, along with complex systemic disorders, such as asthma, acute pancreatitis, migraine, intestinal malabsortium, thyroid disease, and malignant neoplasm. Hence, understanding the pleiotropic effects of the novel NASH drugs is the key to optimizing their use as well as preventing emergent-yet poorly understood-undesirable systemic complications that could potentially jeopardize their short- or long-term use.
Figure 5.
The complexity of molecular targets and novel nonalcoholic steatohepatitis drugs: Pleiotropy assessed in the PheWAS United Kindom Biobank. Figure shows associations between gene variants in five nonalcoholic steatohepatitis-related molecular targets (MAP3K5/ASK1, FXR, PPARα/δ, THRβ, and MPC1) with different traits and phenotypes in the UK-PheWAS (Phenome-wide association study). Information regarding single nucleotide polymorphisms and associations were retrieved from the United Kindom Biobank (http://geneatlas.roslin.ed.ac.uk/).
Table 2.
Associations between variants in locus that are targets of novel drugs for the treatment of nonalcoholic steatohepatitis and multiple traits from individuals of the United Kindom Biobank
| Trait | Variant | Position | -log10(p-value) |
| NR1H4 (FXR) Farnesoid X-Activated Receptor | |||
| K85 Acute pancreatitis | rs76372051 | 100945711 | 6.963890333 |
| Immature reticulocyte fraction | rs35712 | 100971355 | 5.607954097 |
| Impedance of arm (right) | rs1409791 | 100851307 | 5.152661824 |
| Impedance of whole body | rs1409791 | 100851307 | 4.772216099 |
| migraine | rs12579460 | 100966714 | 4.639293011 |
| high cholesterol | rs7967468 | 100853792 | 4.543497322 |
| N30-N39 Other diseases of urinary system | rs79306023 | 100938470 | 4.420628035 |
| H81 Disorders of vestibular function | rs140644635 | 100923359 | 4.069764347 |
| PPARδ (Peroxisome Proliferator Activated Receptor Delta) | |||
| Whole body fat-free mass | rs36018387 | 35386872 | 59.74853212 |
| Hip circumference | rs36018387 | 35386872 | 49.20670564 |
| Whole body fat mass | rs36018387 | 35386872 | 37.00113934 |
| Body fat percentage | rs36018387 | 35386872 | 20.45328464 |
| Monocyte percentage | rs9469982 | 35267548 | 45.86340625 |
| Platelet crit | rs33959228 | 35259397 | 21.6726615 |
| White blood cell (leukocyte) count | rs9380500 | 35266231 | 21.54556677 |
| Platelet count | rs9658111 | 35364534 | 17.88276186 |
| Neutrophill count | rs9380500 | 35266231 | 17.11253462 |
| Eosinophill percentage | rs2395625 | 35405461 | 15.34904201 |
| Lymphocyte percentage | rs9658079 | 35327577 | 9.741626151 |
| asthma | rs1557568 | 35260530 | 9.184130164 |
| K90 Intestinal malabsorption | rs7771474 | 35320447 | 11.86097145 |
| MPC1 (Mitochondrial Pyruvate Carrier 1) | |||
| Mean platelet (thrombocyte) volume | rs10946160 | 166757818 | 7.378512135 |
| Platelet count | rs3728 | 166778679 | 5.285527735 |
| Red blood cell (erythrocyte) count | rs6916128 | 166759313 | 4.825911105 |
| M31 Other necrotising vasculopathies | rs7449594 | 166774429 | 4.699926505 |
| dyspepsia / indigestion | rs6909951 | 166758198 | 4.594790286 |
| MAP3K5 (ASK-1) (Mitogen-Activated Protein Kinase Kinase Kinase 5) | |||
| Mean platelet (thrombocyte) volume | rs6924387 | 137082948 | 14.48853109 |
| Eosinophill count | rs932589 | 137083138 | 13.39556873 |
| Lymphocyte percentage | rs6924387 | 137082948 | 10.84396601 |
| Neutrophill count | rs6924387 | 137082948 | 10.59715422 |
| Platelet count | rs9321570 | 137095679 | 9.792150289 |
| White blood cell (leukocyte) count | rs6924387 | 137082948 | 9.574319083 |
| Eosinophill percentage | rs932589 | 137083138 | 9.344890391 |
| Monocyte count | rs9385775 | 137144920 | 9.1157769 |
| Mean reticulocyte volume | rs9385775 | 137144920 | 8.817927896 |
| Platelet distribution width | rs6924387 | 137082948 | 8.001963098 |
| THRβ (Thyroid Hormone Receptor Beta) | |||
| Mean corpuscular volume | rs869785 | 24347800 | 152.2743497 |
| Mean corpuscular haemoglobin | rs869784 | 24348008 | 143.9371173 |
| Red blood cell (erythrocyte) count | rs869785 | 24347800 | 61.9076303 |
| Mean reticulocyte volume | rs869784 | 24348008 | 43.97976306 |
| Reticulocyte count | rs1505307 | 24343330 | 16.57632823 |
| Immature reticulocyte fraction | rs869784 | 24348008 | 15.67096843 |
| Monocyte count | rs12485694 | 24346109 | 11.11788547 |
| Lymphocyte count | rs13096529 | 24232035 | 10.58643203 |
| Red blood cell (erythrocyte) distribution width | rs2167115 | 24339734 | 10.44361306 |
| C56 Malignant neoplasm of ovary | rs189397255 | 24389732 | 12.2277003 |
| Trunk fat-free mass | rs13100197 | 24491484 | 8.731024419 |
| Trunk predicted mass | rs13100197 | 24491484 | 8.614769205 |
| Leg fat percentage (left) | rs1349265 | 24159387 | 8.323233252 |
The associations have been computed using 452264 United Kindom Biobank White British individuals. http://geneatlas.roslin.ed.ac.uk/.
CONCLUSION
We provide new strategies and approaches by which known drugs can be repurposed for the treatment of NASH. Although we explored and mapped NAFLD-chemical interaction networks, it will be necessary to perform clinical trials not only to assess therapeutic response and optimize dosage and delivery routes, but also to explore the possibility that new uses of existing (old) drugs could act on novel or unanticipated targets. The presence of potential “off target”-pleiotropic-effects raises the mandatory necessity of pharmacological optimization, including the assessment of drug interactions and adjustment according to liver function tests.
Footnotes
Supported by grants PICT 2014-0432, PICT 2014-1816 and PICT 2015-0551, PICT 2016-0135 (Agencia Nacional de Promoción Científica y Tecnológica, FONCyT).
Conflict-of-interest statement: No potential conflicts of interest.
Manuscript source: Invited manuscript
Peer-review started: January 18, 2019
First decision: March 20, 2019
Article in press: March 30, 2019
Specialty type: Gastroenterology and hepatology
Country of origin: Argentina
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Kim DJ S-Editor: Yan JP L-Editor: A E-Editor: Ma YJ
Contributor Information
Silvia Sookoian, Department of Clinical and Molecular Hepatology, National Scientific and Technical Research Council (CONICET), University of Buenos Aires, Institute of Medical Research (IDIM), Ciudad Autónoma de Buenos Aires 1427, Argentina. ssookoian@intramed.net.
Carlos J Pirola, Department of Molecular Genetics and Biology of Complex Diseases, National Scientific and Technical Research Council (CONICET), University of Buenos Aires, Institute of Medical Research (IDIM), Ciudad Autonoma de Buenos Aires 1427, Argentina.
References
- 1.Brunt EM, Wong VW, Nobili V, Day CP, Sookoian S, Maher JJ, Bugianesi E, Sirlin CB, Neuschwander-Tetri BA, Rinella ME. Nonalcoholic fatty liver disease. Nat Rev Dis Primers. 2015;1:15080. doi: 10.1038/nrdp.2015.80. [DOI] [PubMed] [Google Scholar]
- 2.Chalasani N, Younossi Z, Lavine JE, Charlton M, Cusi K, Rinella M, Harrison SA, Brunt EM, Sanyal AJ. The diagnosis and management of nonalcoholic fatty liver disease: Practice guidance from the American Association for the Study of Liver Diseases. Hepatology. 2018;67:328–357. doi: 10.1002/hep.29367. [DOI] [PubMed] [Google Scholar]
- 3.Dulai PS, Singh S, Patel J, Soni M, Prokop LJ, Younossi Z, Sebastiani G, Ekstedt M, Hagstrom H, Nasr P, Stal P, Wong VW, Kechagias S, Hultcrantz R, Loomba R. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology. 2017;65:1557–1565. doi: 10.1002/hep.29085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sookoian S, Pirola CJ. Non-alcoholic fatty liver disease is strongly associated with carotid atherosclerosis: A systematic review. J Hepatol. 2008;49:600–607. doi: 10.1016/j.jhep.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 5.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med. 2018;24:908–922. doi: 10.1038/s41591-018-0104-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lonardo A, Sookoian S, Pirola CJ, Targher G. Non-alcoholic fatty liver disease and risk of cardiovascular disease. Metabolism. 2016;65:1136–1150. doi: 10.1016/j.metabol.2015.09.017. [DOI] [PubMed] [Google Scholar]
- 7.Pirola CJ, Sookoian S. Multiomics biomarkers for the prediction of nonalcoholic fatty liver disease severity. World J Gastroenterol. 2018;24:1601–1615. doi: 10.3748/wjg.v24.i15.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong VW, Adams LA, de Lédinghen V, Wong GL, Sookoian S. Noninvasive biomarkers in NAFLD and NASH - current progress and future promise. Nat Rev Gastroenterol Hepatol. 2018;15:461–478. doi: 10.1038/s41575-018-0014-9. [DOI] [PubMed] [Google Scholar]
- 9.Rotman Y, Sanyal AJ. Current and upcoming pharmacotherapy for non-alcoholic fatty liver disease. Gut. 2017;66:180–190. doi: 10.1136/gutjnl-2016-312431. [DOI] [PubMed] [Google Scholar]
- 10.Konerman MA, Jones JC, Harrison SA. Pharmacotherapy for NASH: Current and emerging. J Hepatol. 2018;68:362–375. doi: 10.1016/j.jhep.2017.10.015. [DOI] [PubMed] [Google Scholar]
- 11.Ashburn TT, Thor KB. Drug repositioning: Identifying and developing new uses for existing drugs. Nat Rev Drug Discov. 2004;3:673–683. doi: 10.1038/nrd1468. [DOI] [PubMed] [Google Scholar]
- 12.Sookoian S, Pirola CJ. Nonalcoholic fatty liver disease and metabolic syndrome: Shared genetic basis of pathogenesis. Hepatology. 2016;64:1417–1420. doi: 10.1002/hep.28746. [DOI] [PubMed] [Google Scholar]
- 13.Sookoian S, Pirola CJ. Nonalcoholic fatty liver disease: Biomarkers support decisions around pharmacological intervention. Hepatology. 2017;65:1417–1419. doi: 10.1002/hep.28866. [DOI] [PubMed] [Google Scholar]
- 14.Sookoian S, Pirola CJ. Review article: Shared disease mechanisms between non-alcoholic fatty liver disease and metabolic syndrome - translating knowledge from systems biology to the bedside. Aliment Pharmacol Ther. 2019;49:516–527. doi: 10.1111/apt.15163. [DOI] [PubMed] [Google Scholar]
- 15.Pirola CJ, Gianotti TF, Burgueño AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castaño GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut. 2013;62:1356–1363. doi: 10.1136/gutjnl-2012-302962. [DOI] [PubMed] [Google Scholar]
- 16.Sookoian S, Rosselli MS, Gemma C, Burgueño AL, Fernández Gianotti T, Castaño GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: Impact of liver methylation of the peroxisome proliferator-activated receptor γ coactivator 1α promoter. Hepatology. 2010;52:1992–2000. doi: 10.1002/hep.23927. [DOI] [PubMed] [Google Scholar]
- 17.Sookoian S, Flichman D, Scian R, Rohr C, Dopazo H, Gianotti TF, Martino JS, Castaño GO, Pirola CJ. Mitochondrial genome architecture in non-alcoholic fatty liver disease. J Pathol. 2016;240:437–449. doi: 10.1002/path.4803. [DOI] [PubMed] [Google Scholar]
- 18.Chrysant SG, Chrysant GS. The pleiotropic effects of angiotensin receptor blockers. J Clin Hypertens (Greenwich) 2006;8:261–268. doi: 10.1111/j.1524-6175.2005.05264.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rosselli MS, Burgueño AL, Carabelli J, Schuman M, Pirola CJ, Sookoian S. Losartan reduces liver expression of plasminogen activator inhibitor-1 (PAI-1) in a high fat-induced rat nonalcoholic fatty liver disease model. Atherosclerosis. 2009;206:119–126. doi: 10.1016/j.atherosclerosis.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 20.Sookoian S, Fernández MA, Castaño G. Effects of six months losartan administration on liver fibrosis in chronic hepatitis C patients: A pilot study. World J Gastroenterol. 2005;11:7560–7563. doi: 10.3748/wjg.v11.i48.7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sookoian S, Castaño G, García SI, Viudez P, González C, Pirola CJ. A1166C angiotensin II type 1 receptor gene polymorphism may predict hemodynamic response to losartan in patients with cirrhosis and portal hypertension. Am J Gastroenterol. 2005;100:636–642. doi: 10.1111/j.1572-0241.2005.41168.x. [DOI] [PubMed] [Google Scholar]
- 22.Sookoian S, Gianotti TF, Rosselli MS, Burgueño AL, Castaño GO, Pirola CJ. Liver transcriptional profile of atherosclerosis-related genes in human nonalcoholic fatty liver disease. Atherosclerosis. 2011;218:378–385. doi: 10.1016/j.atherosclerosis.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 23.Oesterle A, Laufs U, Liao JK. Pleiotropic Effects of Statins on the Cardiovascular System. Circ Res. 2017;120:229–243. doi: 10.1161/CIRCRESAHA.116.308537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pose E, Trebicka J, Mookerjee RP, Angeli P, Ginès P. Statins: Old drugs as new therapy for liver diseases? J Hepatol. 2019;70:194–202. doi: 10.1016/j.jhep.2018.07.019. [DOI] [PubMed] [Google Scholar]
- 25.Hirschfield GM, Mason A, Luketic V, Lindor K, Gordon SC, Mayo M, Kowdley KV, Vincent C, Bodhenheimer HC, Jr, Parés A, Trauner M, Marschall HU, Adorini L, Sciacca C, Beecher-Jones T, Castelloe E, Böhm O, Shapiro D. Efficacy of obeticholic acid in patients with primary biliary cirrhosis and inadequate response to ursodeoxycholic acid. Gastroenterology. 2015;148:751–61.e8. doi: 10.1053/j.gastro.2014.12.005. [DOI] [PubMed] [Google Scholar]
- 26.Jayakumar S, Middleton MS, Lawitz EJ, Mantry PS, Caldwell SH, Arnold H, Mae Diehl A, Ghalib R, Elkhashab M, Abdelmalek MF, Kowdley KV, Stephen Djedjos C, Xu R, Han L, Mani Subramanian G, Myers RP, Goodman ZD, Afdhal NH, Charlton MR, Sirlin CB, Loomba R. Longitudinal correlations between MRE, MRI-PDFF, and liver histology in patients with non-alcoholic steatohepatitis: Analysis of data from a phase II trial of selonsertib. J Hepatol. 2019;70:133–141. doi: 10.1016/j.jhep.2018.09.024. [DOI] [PubMed] [Google Scholar]
- 27.Neuschwander-Tetri BA, Loomba R, Sanyal AJ, Lavine JE, Van Natta ML, Abdelmalek MF, Chalasani N, Dasarathy S, Diehl AM, Hameed B, Kowdley KV, McCullough A, Terrault N, Clark JM, Tonascia J, Brunt EM, Kleiner DE, Doo E NASH Clinical Research Network. Farnesoid X nuclear receptor ligand obeticholic acid for non-cirrhotic, non-alcoholic steatohepatitis (FLINT): a multicentre, randomised, placebo-controlled trial. Lancet. 2015;385:956–965. doi: 10.1016/S0140-6736(14)61933-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ratziu V, Harrison SA, Francque S, Bedossa P, Lehert P, Serfaty L, Romero-Gomez M, Boursier J, Abdelmalek M, Caldwell S, Drenth J, Anstee QM, Hum D, Hanf R, Roudot A, Megnien S, Staels B, Sanyal A GOLDEN-505 Investigator Study Group. Elafibranor, an Agonist of the Peroxisome Proliferator-Activated Receptor-α and -δ, Induces Resolution of Nonalcoholic Steatohepatitis Without Fibrosis Worsening. Gastroenterology. 2016;150:1147–1159.e5. doi: 10.1053/j.gastro.2016.01.038. [DOI] [PubMed] [Google Scholar]
- 29.Colca JR, McDonald WG, Adams WJ. MSDC-0602K, a metabolic modulator directed at the core pathology of non-alcoholic steatohepatitis. Expert Opin Investig Drugs. 2018;27:631–636. doi: 10.1080/13543784.2018.1494153. [DOI] [PubMed] [Google Scholar]
- 30.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]