Abstract
Shoot meristems, which harbor a small population of stem cells, are responsible for generating new above-ground organs in plants. The proliferation and differentiation of these stem cells is regulated by a genetic pathway involving two key meristematic genes: CLAVATA3 (CLV3) and WUSCHEL (WUS). However, it is not well understood how CLV3 and WUS expression domains in the shoot meristems are specified and maintained during post-embryogenic development. Here, we show that a tomato mutant with fasciated stems, flowers and fruits, due to impaired stem cell activity, is defective in a LITTLE ZIPPER gene denoted as DEFECTIVE TOMATO MERISTEM (DTM). DTM forms a negative feedback loop with class III homeodomain-leucine zipper (HD-ZIP III) transcription factors to confine CLV3 and WUS expression to specific domains of the shoot meristems. Our findings reveal a new layer of complexity in the regulation of plant stem cell homeostasis.
Subject terms: Shoot apical meristem, Plant signalling
Qian Xu et al. show that shoot apical meristem activity in tomato is maintained by a LITTLE ZIPPER protein known as DTM. This protein negatively regulates a class of HD-ZIP III transcription factors, ultimately maintaining stem cell homeostasis.
Introduction
Shoot apical meristems (SAMs) provide meristematic cells for the formation of plant aerial organs, and maintaining SAM activity is crucial for plants to complete their life cycles and to adapt to changing environments1. Within the SAM, stem cell niches are maintained by the CLAVATA-WUSHEL (CLV-WUS) feedback loop2–5. In this regulatory module, WUS controls stem cell fate by preventing cell differentiation, while CLV3 regulates cell division to restrict SAM size6,7. At the transcription level, CLV3 restricts WUS expression to a limited number of cells, while low WUS activity promotes CLV3 expression8,9. In addition, SHOOTMERISTEMLESS (STM) prevents the entrance of stem cell differentiation in the SAM10–13. It has been well documented that the coordinated actions of these three meristematic genes in maintaining the SAM are highly dependent on their layer-specific and overlapping expression2,4,14.
Class III homeodomain-leucine zipper (HD-ZIP III) genes regulate SAM activity in addition to the identities of the adaxial tissues of lateral organs in Arabidopsis thaliana15,16. The A. thaliana loss-of-function HD-ZIP III triple mutant, revoluta phabulosa phavoluta (rev phb phv), only forms pin-like shoots, whereas high expression of HD-ZIP III genes causes enlarged SAMs17–19. Recently, it was shown that HD-ZIP III proteins regulate WUS expression through interaction with A-type ARABIDOPSIS RESPONSE REGULATOR proteins during de novo shoot regeneration20. Furthermore, REV directly activates STM expression in the meristematic cells at the leaf axil for axillary meristem formation21. However, regulation of SAM activity by HD-ZIP III transcription factors can be WUS- dependent or independent19,22, suggesting the involvement of additional modifiers.
HD-ZIP III transcription factors are regulated at the transcriptional, post-transcriptional, and post-translational levels23–27. For example, LITTLE ZIPPER proteins (ZPRs) inhibit HD-ZIP III activities by forming presumably nonfunctional heterodimers with them25,27. Thus, ZPRs control SAM development through suppression of HD-ZIP III activities. The A. thaliana double mutant of ZPR3 and ZPR4, zpr3–2 zpr4–2, produces ectopic shoot meristems, while overexpression of ZPR3 causes early meristem termination25,27.
The genetic program governing the SAM stem cell system is generally conserved across plant species2. Changes in meristem size have also been observed in the tomato (Solanum lycopersicum) locule number and fasciated (fas) mutants, which have mutations in the WUS and CLV3 orthologs SlWUS and SlCLV3, respectively28,29. However, unlike A. thaliana, tomato is a typical sympodial plant: its shoot development is indeterminate since after primary shoot meristems terminate into inflorescences, sympodial meristems (SYM) are formed at the leaf axil immediately beneath the inflorescence meristems to sustain its growth14. The difference in SAM activity between the two species is likely due to diversified regulatory mechanisms, as exemplified by SAM doming being regulated by LATE TERMINATION, which occurs in tomato but is absent in A. thaliana30. To identify new genetic components that regulate SAM activity in tomato, we screened for defective meristem mutants in a large ethyl methanesulfonate mutagenized tomato population. Characterization of one such mutant revealed that a LITTLE ZIPPER protein DTM regulates SAM activity through its negative post-translational regulation of SlREV, the tomato ortholog of REV. Mutations in DTM and SlREV cause mutually exclusive changes in expression patterns of the meristematic genes SlCLV3 and SlWUS in the SAM. We propose that a DTM-SlREV feedback loop defines the domain-specific action of CLV-WUS signaling, thereby maintaining stem cell homeostasis during post-embryogenic development.
Results
The DTM gene encodes a LITTLE ZIPPER protein
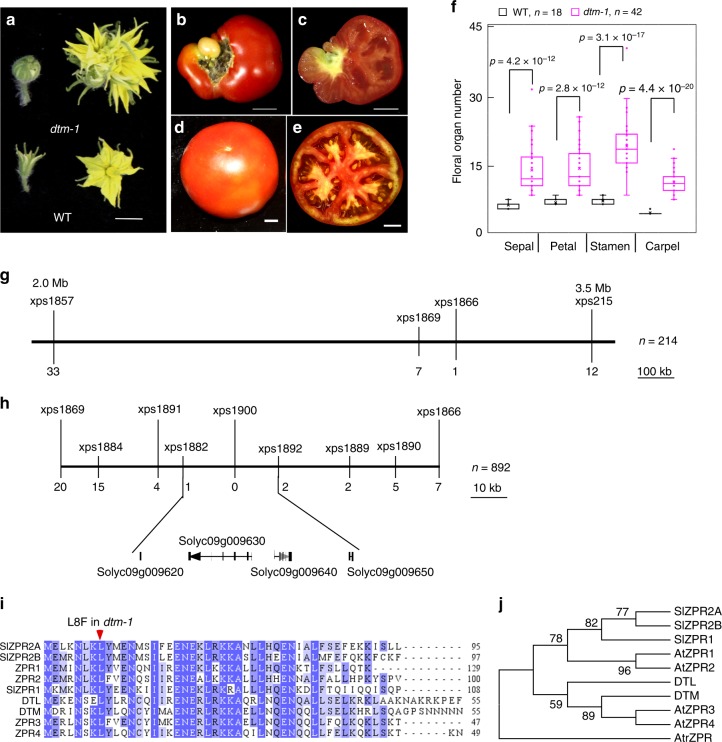
The SAMs of plants provide stem cells for the formation of aerial organs, such as flowers and fruits, which are often associated with the target traits of crop breeding. Through screening of an ethyl methanesulfonate mutagenized population of the cultivated tomato LA2397 for mutations affecting flower and fruit development, we identified a meristem defect mutant, which we named defective tomato meristems-1 (dtm-1). This mutant was first noted for its enlarged fasciated flowers and increased numbers of floral organ (Fig. 1a–f). dtm-1 has seedless fruits, which we determined was due to female sterility, since the mutation was transmitted to wild type plants by pollinating with samples of dtm-1 pollens. To identify the causal mutation underlying the mutant phenotype, dtm-1 was crossed as the pollen donor to Solanum pimpinellifolium LA1781, an accession of a wild relative of cultivated tomato, to generate a F2 mapping population. Using 214 dtm-1 plants, DTM was mapped to a 1.5 Mb interval (SL2.50ch09:2.0–3.5 Mb) between markers xps1857 and xps215 (Fig. 1g). Fine mapping, using an additional 678 dtm-1 plants and newly developed markers, further narrowed down the DTM locus to a 25.3 kb region between the xps1882 and xps1892 markers (Fig. 1h). Based on the ITAG2.4 annotation from the sol genomics network database (SGN, https://solgenomics.net/), this region was determined to contain four protein encoding sequences, but no mutation was found in any of them in dtm-1. We then performed ab initio gene prediction with the 25.3 kb genomic sequences using FGENESH (www.softberry.com), and found that Solyc09g009620 was annotated incorrectly, missing a noncoding exon and 45 nucleotides (15 amino acids) at its 5′-end. This was confirmed by rapid amplification of cDNA ends analysis (Supplementary Fig. 1). After resequencing the corrected Solyc09g009620 coding sequence, we found that dtm-1 has a single point mutation near the beginning of the second exon: a C to T transversion (C22T) that causes the conserved leucine at position 8 to be changed to phenylalanine (L8F) (Fig. 1i).
Fig. 1.
Isolation of the DTM gene. a–e images of flower buds and flowers at anthesis (a) and fruits (b–e) from dtm-1 (a–c) and wild type (a, d, and e) plants in the LA2397 background. Images of longitudinally sectioned dtm-1 fruits (c) showed that no seeds formed inside, compared with seeded wild type fruits (e). Scale bars, 1 cm. f floral organ number of dtm-1 and wild type. Data were collected from flowers at anthesis from 10 plants per genotype. The boxplots show the first quartile and third quartiles, split by the median. Means are indicated by an X in each box. A Welch’s t-test was applied to compare the differences in means between dtm-1 and wild type. g low resolution mapping of the DTM locus. DTM was mapped to a 1.5 Mb interval using 214 dtm-1 plants from a S. pimpinellifolium LA1781 × dtm-1 F2 population. h fine mapping of DTM. The DTM locus was further narrowed down to a 25.3 kb region between the xps1882 and xps1892 markers using a total of 892 mutants. The numbers under the lines marking the genetic markers are the numbers of recombinants. i protein sequence alignment of DTM and its homologs in tomato and Arabidopsis thaliana. The missense mutation identified in dtm-1 is indicated by a red arrowhead. j maximum parsimony tree of DTM and its homologs in tomato and A. thaliana. The consensus phylogenetic tree was constructed using MEGA7.0 with 1000 bootstrap replications and the tree was rooted by an outgroup ZPR protein, AtrZPR (NCBI accession: XP 006851800.1), from Amborella trichopoda. The numbers next to branches show the percentages of the replicate trees (only 50% or higher reported) in the bootstrap test
DTM shares high amino acid sequence similarity with the A. thaliana ZPR4 protein (77% identity, 35/47), a member of a small gene family that is widely present in land plants25,27. The tomato genome encodes four additional ZPR proteins, named DTM-like (DTL, Solyc11g007100), SlZPR1 (Solyc01g091490), SlZPR2A (Solyc08g007570), and SlZPR2B (Solyc08g079690) based on their homology with A. thaliana ZPR proteins (Fig. 1j). In a phylogenetic analysis, DTM and its closest homolog DTL (69% identity) grouped with A. thaliana ZPR3 and ZPR4, while the other three tomato ZPR proteins were more similar to A. thaliana ZPR1 and ZPR2 (Fig. 1j).
dtm-1 exhibits multiple meristem defects
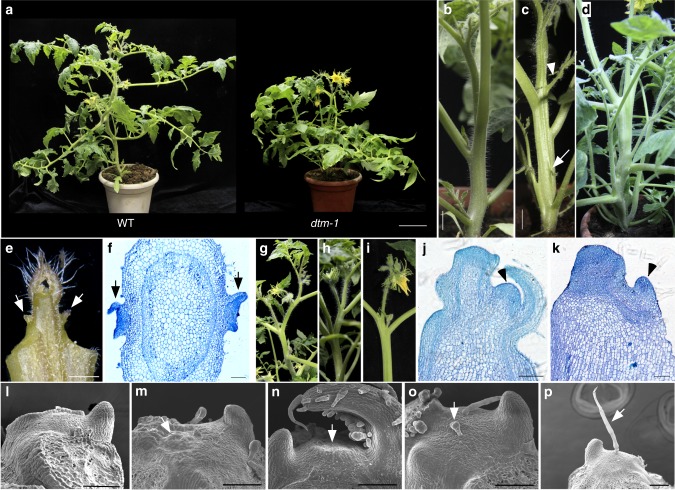
Although dtm-1 plants looked similar to wild type at the seedling stage, close examination revealed that the mutant had defects in axillary shoot formation. Specifically, axillary buds were formed above the leaf axils, rather than at the leaf axils, as in wild type (Fig. 2a–d). Occasionally, leafy organs, instead of axillary buds, were formed on the stems between two consecutive leaves (Fig. 2c). Additionally, two lateral shoots were often observed on opposite sides of the stems, and anatomical analysis of cross-sections of the upper shoots revealed that two axillary meristems formed simultaneously (Fig. 2e, f). These axillary buds were able to develop into functional shoots with fasciated flowers like those on the primary shoots (Fig. 2g–i). However, all branches terminated in single inflorescences, indicating that SYM development on the branches was defective (Fig. 2i). Compared with wild type, SYM formation on primary shoots was slightly delayed in the dtm-1 mutant (Fig. 2j, k). These observations suggest that SAM activity is not properly maintained in the dtm-1 mutant.
Fig. 2.
Phenotypes of the dtm-1 mutant and wild type. a images of representative adult dtm-1 and wild type plants. Scale bar, 10 cm. b–d images of main stems showing axillary bud development in wild type (LA2397, b) and dtm-1 (c, d). The arrowhead and arrow in (c) indicate a side shoot and a leafy structure ectopically formed between two consecutive leaves rather than at the leaf axils. Scale bars, 1 cm. e a dissected shoot apex from dtm-1 plants showing two symmetrical buds (indicated by arrows) formed on the stem. Scale bar, 100 μm. f cross section of a dtm-1 primary stem showing two axillary meristems (indicated by arrows) formed almost simultaneously at opposite positions. Scale bar, 100 μm. g–i images of primary (g, h) and side (i) shoots of wild type (g) and dtm-1 plants (h, i). j–k longitudinal sections of a wild type (j) and a dtm-1 (k) shoot apex at 15 days after germination (DAG). Arrowheads indicate sympodial meristems (SYMs). l–p scanning electron microscopy (SEM) images of shoot apical meristems (SAMs) from wild type (l) and dtm-1 (m–p) seedlings. The dtm-1 SAMs had a rough surface (m), doming defect (n), and produced precocious leaf primordia (o) and ectopic trichomes (o, p) indicated by arrows. Scale bars, 100 μm
By dissecting the shoot apices, we found that dtm-1 SAMs were flatter and wider than those of wild type, as revealed by scanning electron microscopy (SEM) (Fig. 2l, m). In addition, the dome surface of dtm-1 SAMs appeared wrinkled, and ectopic trichomes often formed on the epidermis (Fig. 2n–p). As in many other plant species, the SAM gradually develops into a dome structure in tomato30. To better understand the timing of DTM action on SAM development, we analyzed the doming of dtm-1 SAMs. At 3 days after germination (DAG), we observed no difference in SAM morphology between dtm-1 and wild type, except that the SAMs of the mutant were wider (Supplementary Fig. 2). However, doming was limited in dtm-1 and its SAMs remained flat or slightly bulged until the developmental transition to inflorescence meristems.
Interestingly, in the F2 population derived from a cross between dtm-1 and LA1781, we observed extremely elongated flower stalks and a wide range of variations in fruit fasciation (Supplementary Fig. 3). This suggests that the dtm-1 mutation has pleiotropic effects on flower and fruit development due to undetermined genetic modifiers in the LA1781 background, or to recombination events. To test whether the mutation has different extents of phenotypic penetration in different genetic backgrounds, we also introduced the dtm-1 allele into S. lycopersicum cv. Moneymaker, a cultivar that has been widely used for genetic and molecular studies31, and into the wild species accession LA1781. In the three genetic backgrounds, the dtm-1 mutation had very similar, if not identical, effects on SAM development. All mutant phenotypes observed in the LA2397 background were observed in both the LA1781 and Moneymaker backgrounds (Supplementary Fig. 4).
Phenotypes of null dtm alleles created by CRISPR-Cas9
Since the phenotypic abnormality observed in the dtm-1 mutant is caused by a missense mutation, to gain insight into its loss-of-function effects on SAM development, we generated several null alleles of the DTM gene by clustered regularly interspaced short palindromic repeats (CRISPR)-Cas9 genome editing, using two single guide sequences located 169 bp apart on the second exon (Fig. 3a). In total, we obtained four lines with 2–176 bp deletions in the protein coding region that likely disrupted DTM functionality, from more than ten primary transformants (Fig. 3b). The four loss-of-function alleles had the same phenotypes and we chose line dtm-cr5, which has a 176 bp deletion (nucleotide 64–239), for further analysis.
Fig. 3.
Phenotypes of the loss-of-function dtm-cr5 alleles created by CRISPR-Cas9. a schematic illustration of the gRNA target design aiming for large deletions in the coding region of DTM. The two single gRNA target sites are indicated by two red arrowheads. b alignment of translated protein sequences from the four dtm-cr alleles containing putative loss-of-function mutations. Sequences identical to wild type DTM are shaded and the first 15 amino acids not changed by mutations in the four alleles are highlighted in red. The nature of the mutations in each of the four alleles is indicated on the right. c–f representative images of dtm-cr5 (d, f) and wild type (Moneymaker, c, e) seedlings at 3 and 9 days after germination (DAG). The four mutant alleles had identical phenotypes including tricot formation (d) and precocious leaf formation (f). Only dtm-cr5 images are shown. g–i dissected dtm-cr5 apices showing shoot apical meristem (SAM)developmental abnormality as revealed by stereomicroscopy (g), paraffin sectioning (h), and scanning electron microscopy (SEM) (i). The dtm-cr5 apex was flat with a very rough surface (g) and two SAM-like structures (indicated by arrows) were observed in longitudinal sections (h). The SEM image (i) shows dense trichomes formed on a dtm-cr5 SAM. j images of dtm-cr5 and wild type adult plants. k–l two close-up images showing an extremely fasciated stem (k) and flower (l) from the dtm-cr5 plant shown in (j). Scale bars, 5 cm (c, d), 1 cm (e, f, j) and 100 μm (g–i)
Unlike the dtm-1 allele, dtm-cr5 seedlings had abnormal cotyledon numbers, with almost all of the mutants (104 out of 107) having more than two (Fig. 3c–f). Examination of the dtm-cr5 apices revealed that its SAM development was more severely perturbed than in dtm-1. Domed SAM structures were rarely present in the dtm-cr5 seedlings at 3–12 DAG, although multiple SAM-like structures were identified in longitudinal sections of their shoot apices (Fig. 3g, h; Supplementary Fig. 5). Moreover, dtm-cr5 apices were densely covered by trichomes (Fig. 3i). Later in development, the adult dtm-cr5 plants were dwarfed, with extremely fasciated and twisted stems, clustered leaves (Fig. 3j, k), and occasionally flowers formed without petals (Fig. 3l).
Since SlCLV3 is also involved in SAM doming, we tested for genetic interactions between SlCLV3 and DTM by crossing dtm-1 with S. lycopersicum cv. Super Beefsteak, which has a chromosomal conversion that downregulates SlCLV3 expression29. The double mutant, dtm-1 fas, was indistinguishable from dtm-1 in SAM morphology: both produced flat SAMs with ectopic trichomes on the epidermis (Supplementary Fig. 6). However, SYM development was abolished in dtm-1 fas mutants and side shoots were replaced by inflorescence-like structures (Supplementary Fig. 6), indicating that the two genes may have additive effects on lateral meristem development.
Interactions between DTM and HD-ZIP III proteins
LITTLE ZIPPER proteins function as post-translational suppressors of class III HD-ZIP transcription factors by inhibiting their homodimerization25,27. In tomato, there are six HD-ZIP III members, of which Solyc11g069470 has been named SlREV, based on its sequence similarity to A. thaliana class III HD-ZIP transcription factors32. We assigned the following names to the remaining tomato HD-ZIP III proteins based on sequence similarity and phylogenetic analysis: SlPHB (Solyc02g024070), SlPHV (Solyc02g069830), SlHB15A (Solyc03g120910), SlHB15B (Solyc12g044410), and SlHB8 (Solyc08g066500) (Supplementary Fig. 7). We tested the interactions between DTM and the tomato HD-ZIP III proteins by yeast two hybrid and pulldown assays. In yeast, DTM interacted with full-length SlREV, SlPHV and SlHB8 (Fig. 4a), and with the individual N-terminal regions, which each contains a homeodomain and a leucine zipper domain, of all the tomato HD-ZIP III members except SlHB15A (Supplementary Fig. 8a). Using Escherichia coli expressed proteins, cmyc-DTM was immunoprecipitated with an HA antibody when incubated with protein extractions containing HA-tagged SlPHV, SlREV, SlHB8, SlHB15A or SlHB15B, but not HA-SlPHB (Fig. 4c). This suggests that DTM may interact with several members of the tomato HD-ZIP III transcription factor family.
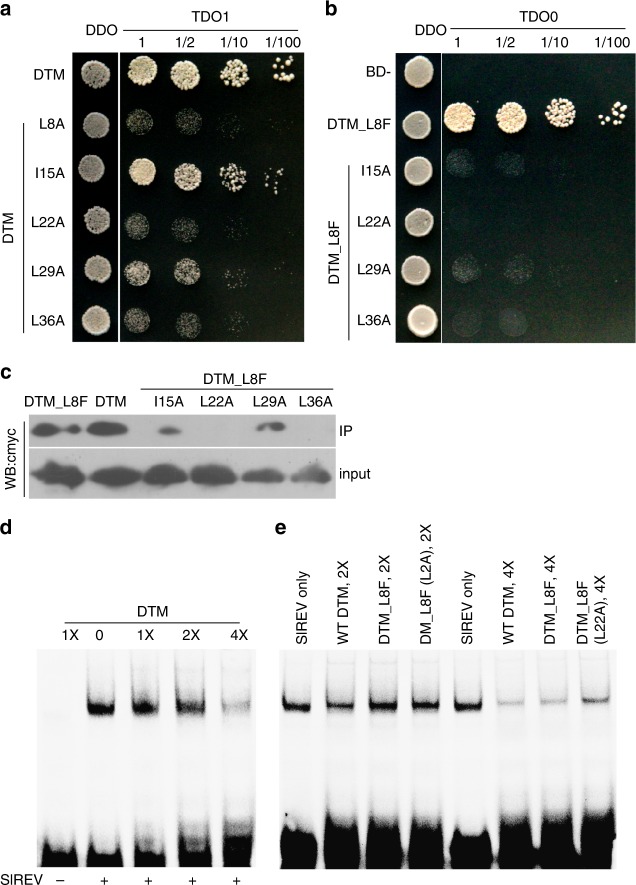
Fig. 4.
The L8F mutation in DTM weakens the interactions between DTM and tomato HD-ZIP III proteins. a interactions between DTM and tomato HD-ZIP III proteins SlREV, SlPHB, SlPHV, SlHB8, SlHB15A, and SlHB15B in yeast cells. DTM cDNA was fused in frame to the coding sequence of the GAL4 DNA binding domain (BD), and the full-length cDNA of the six HD-ZIP III genes was fused to the coding sequence of the GAL4 activation domain (AD). The numbers above the images represent cell densities obtained by 0, 1-, 10-, and 100-times dilutions from yeast cell suspensions at an OD600 of 1.0. b interactions between DTM mutants and the six tomato HD-ZIP III proteins in yeast cells. The cDNA encoding DTM_L8F, dtmcr3, dtmcr5, and dtmcr7 indicated by the numbers above the images were PCR amplified from dtm-1, dtm-cr3, dtm-cr5 and dtm-cr7 DNA. c interactions between DTM and the six tomato HD-ZIP III proteins tested by a pulldown assay. d comparison of binding ability between DTM and DTM_L8F to the HD-ZIP III proteins SlREV, SlPHV, SlHB15A, and SlHB15B. Pulldown assays were conducted using Escherichia coli expressed proteins either fused to the cmyc (DTM and DTM_L8F) or the HA (HD-ZIP IIIs) tag. Protein complexes were immunoprecipitated using an anti-HA antibody (c, d). e–o interactions between DTM and the mutant form DTM_L8F and HD-ZIP III proteins in tobacco leaves. DTM (or DTM_L8F), and the six tomato HD-ZIP III proteins were fused to the N- and C-terminal portion of LUC, respectively. Constructs expressing two terminal halves of LUC only, or one terminal half of LUC not fused with a target protein in combination with another half fused with the target to be tested, served as negative controls. The white field image (j) depicts the experiment setup, in which ‘X’ represents a specific HD-ZIP III member to be tested as shown under each leaf in (k–o). DDO, SD/-Leu-Trp; TDO0 and TDO1, SD/-Leu-Trp-His with 0 and 1 mM 3-amino-1,2,4- triazole (3-AT), respectively; QDO, SD/-Leu-Trp-His-Ade. IP, immunoprecipitation; WB, Western blot
The dtm-1 mutant has a missense mutation (L8F) at the interhelical interface in the second heptad of the leucine zipper domain, which presumably affects its interaction with the HD-ZIP III proteins25. Indeed, compared to wild type DTM, the mutant form DTM_L8F and the three truncated proteins created by CRISPR-Cas9 showed weaker and no interaction, respectively, with HD-ZIP III members in yeast (Fig. 4b; Supplementary Fig. 8a). The weakened interaction between DTM_L8F and full-length or the N-terminal regions of HD-ZIP III proteins was further confirmed by pulldown assays (Fig. 4d; Supplementary Fig. 8b, c). To investigate the interactions between DTM and the HD-ZIP III proteins in vivo, we conducted a biomolecular fluorescence complementation assay in tobacco leaves33. DTM or DTM_L8F were separately heterologously expressed in tobacco leaves as translational fusions with the N-terminal half of the firefly luciferase (LUC) reporter protein, together with the C-terminus of LUC fused with different HD-ZIP III proteins. We observed that wild type DTM interacted with all six tomato HD-ZIP III proteins, while its mutant form, DTM_L8F, showed much weaker interactions (Fig. 4e–o).
When the conserved Leu or Ile residues in the five heptads (2nd–6th) of the leucine zipper domain in DTM were individually mutated to Ala, its interaction with SlREV in yeast was weakened (Fig. 5a). However, mutation of the Ile in the third heptad caused only a slightly attenuation of the interaction between DTM_I15A and SlREV, suggesting that it is less important for the DTM-SlREV interaction. Mutagenesis of DTM_L8F further confirmed the weak effect of the I15A mutation as we still observed an interaction between DTM_L8F(I15A) and SlREV using both the yeast two hybrid and pulldown assays (Fig. 5b, c). DTM_L8F(L29A) interacted with SlREV in pulldown assays, but not in yeast two hybrid assays. No interaction was found between DTM_L8F(L22A), DTM_L8F(L36A) and SlREV. The mutagenesis analysis suggests that the leucine residues in heptad two, four, and six are critical for the interaction between DTM and HD-ZIP III transcription factors.
Fig. 5.
Mutagenesis of the DTM protein. a interactions between SlREV (fused to the activation domain, AD) and five DTM mutants with single mutations in the conserved Leu or Ile residues in each heptad (fused to the binding domain, BD) in yeast. The five conserved Leu or Ile residues were individually mutated to Ala. b interactions between SlREV and four DTM double mutants derived from DTM_L8F in yeast. c binding activities of DTM and its mutant forms to its partner, SlREV. DTM and its mutant forms were fused to cmyc and their binding affinities to SlREV (fused to HA) were assayed by pulldown using an anti-HA antibody. d electrophoretic mobility shift assay showing dosage-dependent inhibition of DTM on SlREV binding ability to its target sequence. e inhibitive effect of single and double mutations in the conserved residues of DTM on SlREV binding ability to its target sequence. Both SlREV (1–264 aa) and DTM were synthesized using wheat germ extract, and different concentrations of DTM (d) and/or its mutated forms (e) were tested for their inhibitive effects on SlREV binding to a Cy5-labeled HB9 DNA duplex. 3-AT, 3-amino-1,2,4- triazole; IP, immunoprecipitation; WB, Western blot
Previous studies have shown that LITTLE ZIPPER proteins inhibit DNA binding of HD-ZIP III transcription factors25,27. We confirmed that DTM inhibited SlREV binding to the HB9 duplex containing consensus sequence for REV using an electrophoretic mobility shift assay, and further showed that the inhibition by DTM was dosage-dependent (Fig. 5d, e). Moreover, consistent with the weakened interactions between the DTM mutants and SlREV in vitro and in vivo, DTM_L8F and DTM_L8F(L22A) had a weaker inhibition effect on SlREV-DNA binding than did wild type DTM.
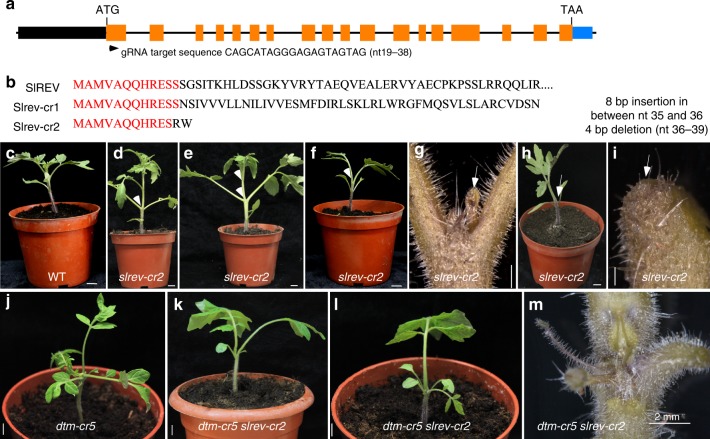
Since dtm-1 seedlings produce ectopic axillary meristems, as observed in plants overexpressing SlREV32, we generated putative loss-of-function mutations in the SlREV gene using CRISPR-Cas9 (Fig. 6a, b). Two mutant alleles, slrev-cr1 and slrev-cr2, exhibited SAM arrest and leaf phyllotaxis defects of differing severities, ranging from relatively normal SAM development, with an opposite leaf arrangement or two leaves (sometimes fused) with slowly initiated leaf primordia, to single leaves without shoot meristems (Fig. 6c–i). These developmental defects are indicative of premature stem cell consumption in the tomato rev mutants, contrasting with the overactive SAM activity in the dtm mutants. To further verify that DTM regulates SAM development through SlREV, we analyzed their genetic interaction by crossing dtm-cr5 with slrev-cr2 (both in the Moneymaker background). The resulting dtm-cr5 slrev-cr2 seedlings were tricots, but looked more similar to slrev-cr2 seedlings (Fig. 6j–m). This suggests that DTM regulates post-embryogenic SAM development in a SlREV-dependent manner, whereas its roles in embryogenesis is SlREV-independent.
Fig. 6.
SlREV regulates shoot apical meristem (SAM) maintenance in tomato. a design of CRISPR-Cas9 editing for the tomato REV gene. The start codon ATG and the gRNA target sequence are indicated. The arrowhead indicates the gRNA target site. b deduced protein sequences of two slrev alleles created by CRISPR-Cas9. c–i representative images showing meristem defects of different severity observed in slrev mutants. slrev-cr mutants displayed weak (d) or strong (e) defects in leaf phyllotaxis (indicated by arrowheads). slrev seedlings often produced only one or two true leaves (f, h) with few small leaf-like tissues (g) or barren SAMs (i) (indicated by arrows). Since no allele-specific phenotypic abnormality was observed between slrev-cr1 and slrev-cr2, only those of slrev-cr2 are shown here. Scale bars represent 1 cm (c–f, h) and 100 μm (g, i), respectively. j–l seedling phenotypes of the dtm slrev double mutant. Scale bars, 1 cm. m dissected dtm-cr5 slrev-cr2 apices showing SAM defects as revealed by stereomicroscopy. Scale bar, 2 mm
Although the closest homolog of DTM, DTL, also interacts with tomato HD-ZIP III proteins, it has a different binding affinity, as revealed by yeast two hybrid and in vitro pulldown assays (Supplementary Fig. 9). Two dtl knockout mutants, created by CRISPR-Cas9 editing, had no obvious phenotypes (Supplementary Fig. 10a–g). Consistent with the phenotypic observation, in situ hybridization analysis suggested that DTL is likely not expressed in the SAM (Supplementary Fig. 10h–k). To test the possibility that DTL affects SAM development by interacting with DTM, we analyzed the phenotype of the double dtm-cr5 dtl-cr1 mutant, and observed that it was indistinguishable from dtm-cr5 (Supplementary Fig. 10l–n). These results suggest that DTL is not essential for post-embryogenic meristem development in tomato.
DTM-SlREV defines expression domains of meristematic genes
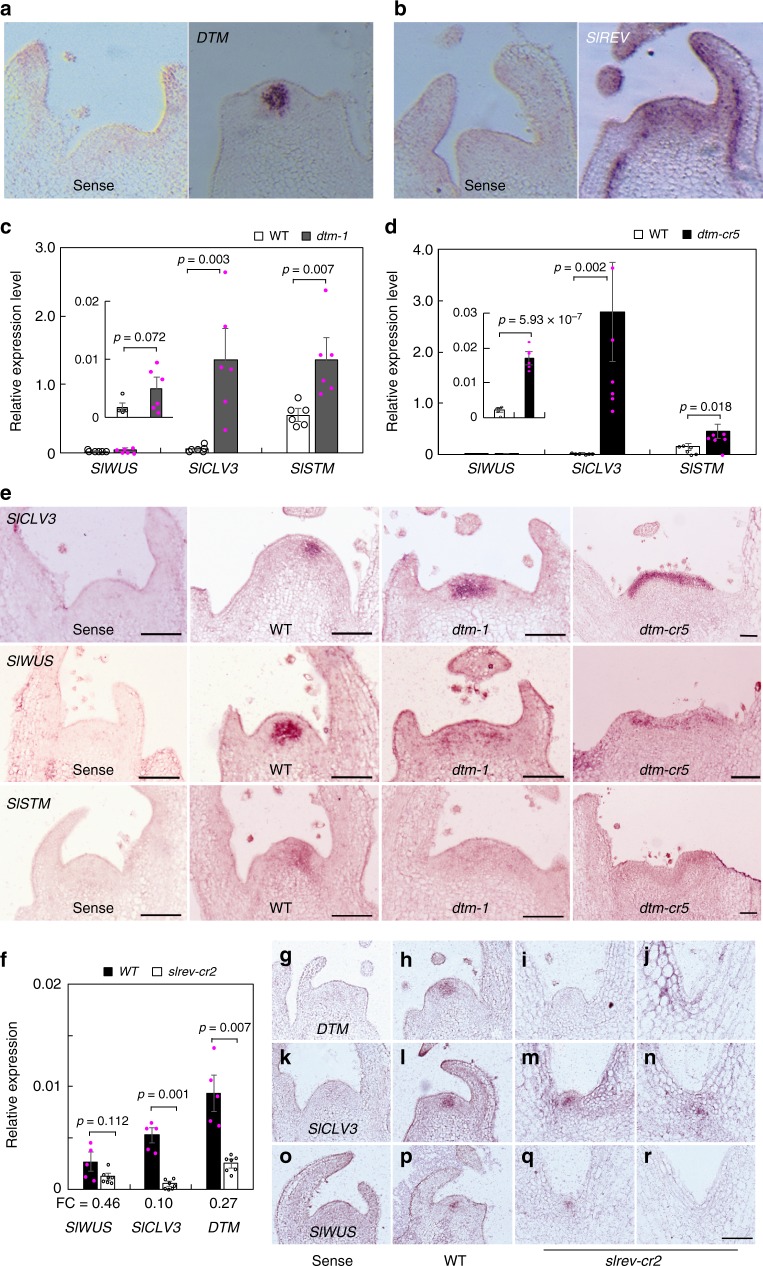
Since DTM expression was detected mainly in the central zone of the SAM that marks the expression domains of SlWUS and SlCLV3 (Fig. 7a)29, the observed SAM defects in the dtm mutants were likely caused by altered expression of these meristematic genes. We then monitored the expression patterns of the three meristem marker genes SlCLV3, SlWUS, and SlSTM in the shoot apices of dtm-1, dtm-cr5, and their corresponding wild types, at 3, 6 and 9 DAG. Expression analysis by quantitative reverse transcription PCR (qRT-PCR) revealed that SlCLV3 transcript levels were substantially elevated in both the dtm-1 and dtm-cr5 shoot apices at all three time points, except for a marginal difference in expression level, which was detected between dtm-cr5 and its wild type at 3 DAG (Fig. 7c–d; Supplementary Fig. 11a–d). SlWUS expression was also much higher in dtm-cr5 shoot apices at all three time points and in dtm-1 at 6 DAG, while SlSTM expression was apparently only upregulated in the two dtm alleles at 6 DAG.
Fig. 7.
DTM defines the SlCLV3 and SlWUS expression domains. a, b expression of DTM and SlREV in wild type (Moneymaker) shoot apices as revealed by RNA in situ hybridization. c, d expression patterns of the meristematic SlCLV3, SlWUS and SlSTM genes in the shoot apices of dtm-1 (c), dtm-cr5 (d), and their corresponding wild types (LA2397 and Moneymaker) at 6 days after germination (DAG) determined by quantitative reverse transcription (qRT)-PCR. SleIF4α6 was used as the reference gene to normalize gene expression. Data represent means ± sem, n = 6 biologically independent samples. A two-tailed t-test was applied to compare the differences in means between dtm-1 and wild type. e expression patterns of the SlCLV3, SlWUS, and SlSTM genes in the shoot apices of dtm-1, dtm-cr5 and wild type (Moneymaker) at 6 DAG revealed by RNA in situ hybridization. Scale bars, 100 μm. f expression levels of DTM and the meristematic SlCLV3, SlWUS, and SlSTM genes in slrev-cr2 shoot apices and wild type (Moneymaker) at 6 DAG determined by qRT-PCR. Data represent means ± sem, n = 5 (wild type) or 7 (slrev-cr2) biologically independent samples. Fold changes (FC, mutant vs wild type (WT)) of gene expression levels are given under the graph. A Welch’s t-test was applied to compare the differences in means between slrev-cr2 and wild type. g–r expression of DTM (g–j), SlCLV3 (k–n), and SlWUS (o–r) in slrev-cr2 (i, j, m, n, q and r) and wild type (Moneymaker, h, l and p) shoot apices revealed by in situ hybridization. Hybridization using sense probes served as negative controls (g, k and o). Scale bar, 100 μm. The experiments were repeated at least twice using different batches of plants with similar results. Expression data collected from the dtm-1 and dtm-cr5 seedlings at 3 and 9 DAG are provided in Supplementary Fig. 10c-d. SlSTM expression in slrev-cr2 and wild type apices is available in Supplementary Fig. 10e
Similarly, in situ hybridization analysis revealed that SlCLV3 expression was higher and spatially expanded in the dtm-1 and dtm-cr5 shoot apices at 3, 6, and 9 DAG (Fig. 7e; Supplementary Fig. 12). In wild type, SlCLV3 was expressed in the central zone of the SAM, apart from in the L1 layer. SlCLV3 expression in the weak dtm-1 allele was expanded but still detected in the central zone, whereas in the dtm-cr5 null allele it was only detected in the outermost two layers of the whole apex, indicating that DTM restricts SlCLV3 expression in the central zone. SlWUS, which is mainly expressed in the organizer zone, is likely also required for defining the size of the stem cell population in tomato29,34. In the dtm-1 and dtm-cr5 apices, SlWUS expression appeared patchy and expanded. Similarly, SlSTM expression was also affected. Unlike in wild type, the SlSTM expression in dtm-1 and dtm-cr5 apices was diffuse and no apparent expression domain could be defined.
Since DTM acts on SAM development in a SlREV-dependent manner, and the two genes have overlapping expression domains in the SAM (Fig. 7a, b), we examined the expression of SlCLV3 and SlWUS in the SAMs of slrev seedling apices at 6 DAG. SlCLV3 expression, measured by qRT-PCR, was substantially downregulated in slrev-cr2, and while there were indications that the expression of SlWUS was also slightly lower (Fig. 7f). We saw no evidence that SlSTM expression was affected by the slrev mutation (Supplementary Fig. 11e). Further, in situ hybridization analyses revealed that SlCLV3 was expressed in a much deeper region of the SAM, likely in the organizer zone (Fig. 7k–n). The SlWUS expression level correlated with the severity of SAM defects; expression was not detected in the shoot apices without SAMs, but was maintained in the organizer zone with SAM-like structures (Fig. 7o–r). Consistent with previous results identifying ZPR genes as REV targets in A. thaliana35, DTM expression was downregulated in slrev-cr2 shoot apices at 6 DAG (Fig. 7f–j). These results suggest that DTM and SlREV form a feedback loop to confine the expression of meristematic genes to specific cell layers in the SAM.
Mutations in DTM impair leaf phyllotaxis
In addition to the defects in stem and axillary bud development, the leaf formation rate was also affected by dtm mutations, since five or more leaves were observed in dtm-cr5 seedlings when the wild type had only three visible leaves (Fig. 3e, f). This likely reflects faster leaf initiation and the less serrated leaves in the dtm-cr5 seedlings appeared to be clustered on a short stem region (Supplementary Fig. 13a, b). After removal of visible leaves, four small leaves or primordia forming on the dtm-cr5 apices were observed by stereomicroscopy and SEM (Supplementary Fig. 12c, d). Although the leaf formation rate was less affected in dtm-1, longitudinal sectioning revealed that two precocious leaves formed almost simultaneously on the flank of the SAMs at 6 DAG (Supplementary Fig. 13e–g).
LEAFLESS (LFS) is required for leaf formation and its expression predicts leaf initiation36. Since dtm mutations affected leaf phyllotaxis, we used in situ hybridization to investigate whether LFS expression was altered in dtm mutants during leaf formation. Consistent with previous reports36, we observed that LFS was expressed in the incipient regions of leaf primordia. Although LFS expression remained unchanged in both the dtm-1 and dtm-cr5 apices at 3 DAG, its expression was barely detectable after this time point (Supplementary Fig. 13h-p). This effect on LFS expression is consistent with the observation that leaf formation was accelerated in dtm-1 and dtm-cr5 after germination.
Discussion
It is well established that maintenance of SAM activity is primarily governed by the conserved CLV-WUS pathway2, but it is not well understood how this pathway is modified to allow diversified growth patterns in different plant species. In this study, we characterized a tomato LITTLE ZIPPER protein, DTM, and elucidated its role in SAM maintenance by defining WUS and CLV3 expression domains.
Our data indicate that DTM is crucial for maintaining SAM activity in tomato, a model plant with a sympodial growth habit. Loss of, or weakened, DTM activity affects primary and secondary shoot meristem development, resulting in enlarged flattened SAMs, ectopic formation of axillary buds and leaves, and termination of SYMs on side shoots, as well as enlarged fasciated flowers and fruits. Such flower and fruit fasciation are often associated with failures in SAM maintenance, as observed in the tomato fasciated inflorescence, fasciated and branched and fas mutants29. Furthermore, the defects in SAM development caused by dtm mutations are characteristic features of A. thaliana mutants containing mutations in CLV3 or genes involved in the transmission of the CLV3 peptide signal, such as clavata1 and the quadruple mutant of the CLAVATA3 INSENSITIVE RECEPTOR KINASE genes37–40. Given that dtm mutants produce tricots, or fused cotyledons, DTM is also involved in embryogenic meristem formation. However, its primary role is to maintain post-embryogenic meristem development. The presence of multiple slightly domed SAM-like structures in the dtm seedlings indicates that DTM limits stem cell activity. Thus, impairing DTM activity causes overproduction of stem cells in the SAM. Under field growth conditions, the excessive number of stem cells ensures the development of extremely long inflorescences and large flowers.
Like tomato dtm mutants, the A. thaliana double mutant, zpr3–2 zpr4–2, shows a similar phenotypic abnormality resulting from SAM defects, including a high frequency of tricots, early leaf initiation and flower fasciation. This suggests that DTM and its A. thaliana homologs have conserved roles in SAM maintenance. However, DTM has an additional role in the regulation of cell differentiation in tomato because dtm mutations cause ectopic trichomes to be formed on the SAM epidermis, as also observed in the petunia (Petunia hybrida) and pepper (Capsicum annuum) hairy meristem mutants that have mutations in GRAS family genes, which encode proteins that interact with WUS homologs41,42. The regulation of cell differentiation by DTM is likely CLV3-independent, since ectopic trichomes are still formed on the SAM epidermis of the double dtm-1 fas mutants.
Consistent with a function in the regulation of SAM activity, DTM is mainly expressed in the central zone of the SAM that marks the SlCLV3 and SlWUS expression domains28,29. Based on phenotypic observations and expression changes of these meristematic genes caused by the dtm mutations, we conclude that DTM regulates SAM maintenance through defining the expression domains of the meristem genes SlCLV3 and SlWUS. Disrupting DTM activity leads to SlCLV3 expression spreading to the L1 layer in the SAM, where it is not expressed in wild type29. Furthermore, high SlCLV3 activity likely causes the patchy SlWUS expression observed in dtm SAMs, a phenomenon that has been observed in CLV3 overexpressing A. thaliana9.
Although previous studies have shown that CLV3 restricts WUS expression to a limited set of cells in the central zone of the SAM in A. thaliana9, high SlCLV3 expression caused by dtm mutations does not repress overall SlWUS transcription, consistent with a failure in CLV3 peptide signal transmission in the dtm mutants. Thus, the expanded SlCLV3 expression caused by dtm mutations did not reduce SAM size, but instead led to enlarged SAMs. Such SAM enlargement, coupled with high CLV3 expression, is well recognized in A. thaliana mutants such as clavata1, receptor-like protein kinase2 2 and the quadruple mutant of the CLAVATA3 INSENSITIVE RECEPTOR KINASE genes, in which mutations disrupt the transmission of the CLV3 peptide signal37–40. Expanded CLV3 and WUS expression has also been observed in the enlarged shoot meristems of an A. thaliana HD-ZIP III triple mutant22.
Our data also demonstrated that DTM can interact with HD-ZIP III transcription factors in vitro and in vivo, in agreement with previous studies in A. thaliana25,27. Both dtm and zpr3–2 zpr4–2 have SAM defects similar to the A. thaliana phv-1d mutant25,27. Thus, DTM regulates SAM activity in tomato through a similar mechanism to A. thaliana ZPR proteins, by acting as a competitor to suppress HD-ZIP III transcription factor activity. Ectopic AM formation in dtm mutants is also observed in tomato plants overexpressing miR166-resistant SlREV32. Furthermore, loss-of-function mutations in the tomato SlREV gene result in opposite meristem phenotypes, including SAM arrest and disordered leaf phyllotaxis, suggesting that DTM controls SAM development mainly through suppression of SlREV activity. This conclusion is further supported by attenuated and domain-shifted expression of the meristematic genes SlCLV3 and SlWUS in the SAMs of slrev seedlings. Moreover, the characteristic SAM defect phenotypes of the dtm-cr5 slrev-cr2 mutants suggest that DTM acts in a SlREV-dependent manner. We further observed that DTM competes for HD-ZIP III binding in a dosage-dependent manner, supporting the hypothesis that ZPR proteins form heterotetramers, rather than heterodimers, with HD-ZIP III proteins43.
Given that HD-ZIP III transcription factors directly activate WUS and STM expression during de novo shoot regeneration and AM formation in A. thaliana20,21, we propose that the elevated expression of SlWUS and SlSTM in dtm apices is due to weak, or lost, inhibition of SlREV and other HD-ZIP III transcription factors that have yet to be identified. We conclude that a DTM-SlREV feedback loop plays an essential role in regulating the CLV-WUS signaling pathway by defining its expression domains.
Methods
Plant growth conditions and phenotypic analysis
dtm-1 was identified by screening for defective meristem mutants in an ethyl methanesulfonate mutagenesis population in the cultivated tomato LA2397 background. Since dtm-1 is female sterile, the mutant is maintained in a heterozygous state by backcrossing to wild type LA2397. Unless specified, all phenotypic observations and gene expression analysis were conducted with phytotron-grown seedlings or plants derived from selfed BC2 or BC3 heterozygotes. The phytotron growth conditions were maintained at 20–25 °C, a relative humidity of 70–80%, and 16 h daily illumination by 150 μmol m−2 s−1 light from metal halide and high-pressure sodium lamps. F2 mapping populations were grown under natural radiation in plastic greenhouses located in Songjiang, Shanghai.
Genetic mapping of the DTM gene and sequence analysis
dtm-1 was crossed with the wild relative S. pimpinellifolium accession LA1781 to produce a F2 mapping population. Based on the flower phenotypes of the mutant, 892 dtm-1 plants identified from a total of 3,964 F2 progenies were used to map the DTM locus. Genomic DNA was extracted from 1–2 cm young leaves using a high throughput miniprep method44. Leaves were ground in 350 μL DNA extraction buffer [100 mM Tris, 5 mM EDTA, 150 mM sorbitol, 30 mM sodium bisulfite, 0.1% cetyltrimethylammonium ammonium bromide, 2% sarkosyl, pH 8.0] on a mixer mill MM 400 (Retsch). After extraction with 350 μL chloroform:isoamyl alcohol (24:1), DNA was precipitated with an equal volume of isopropanol from cleared supernatant by centrifugation and dissolved in 100 μL TE (10 mM Tris-Cl, 1 mM EDTA, pH 8.0). Molecular markers, comprising PCR and cleaved amplified polymorphic sequence markers, were developed based on single nucleotide polymorphisms and insertions/deletions identified comparison of genome sequence of cv. Heinz 1706 and S. pimpinellifolium LA1589, which were downloaded from SGN (https://solgenomics.net/). Marker information is provided in Supplementary Table 1.
After fine mapping, gene prediction was conducted in the 25.3 kb interval of the Heinz 1706 reference genome containing the DTM locus, using the FGENESH program in Softberry (http://www.softberry.com/). To identify the causal dtm-1 mutation, genomic and complementary DNA (cDNA) sequences of the DTM genes from the mutant and the LA2397 wild type were determined by Sanger sequencing. Sequence assembly and comparisons were performed using the Sequencher® (Gene Codes Inc.) DNA analysis software.
To identify the tomato LITTLE ZIPPER gene family members, protein sequences of DTM and A. thaliana ZPR proteins were used as queries in a BLAST search of the ITAG2.5 genome database downloaded from SGN. Similarly, tomato HD-ZIP III proteins were identified using protein sequences of the five A. thaliana HD-ZIP III proteins as queries. Maximum parsimony trees of tomato and A. thaliana DTM/ZPR and HD-ZIP III proteins were constructed using MEGA7 (ver. 7.0.26)45. Phylogenetic relationships were tested with 1000 bootstrap replications and the trees were inferred using the Subtree-Pruning-Regrafting (SPR) method embedded in the program.
Mutagenesis by CRISPR-Cas9
To create null dtm and dtl alleles using CRISPR-Cas9 genome editing technology, two single guide RNAs were designed using the CRISPR-P (http://cbi.hzau.edu.cn/crispr/) online tool46. After annealing, oligos were subsequently cloned into the psgR-Cas9-At vector47. Sequence-validated psgR-Cas9-At fragments containing guide sequences of the target genes were digested with the EcoRI and HindIII (New England BioLabs) restriction enzymes, and then cloned into the pCAMBIA1300 binary vector. The CRISPR-Cas9 vector used to create the slrev-cr mutants was constructed similarly, using a single guide RNA. These constructs were transformed into cv. Moneymaker (LA2706) by Agrobacterium-mediated transformation48,49. Briefly, cotyledons harvested from 7–10 days old seedlings were co-cultured for two days on Murashige and Skoog medium containing 100 μM acetosyringone with Agrobacterium strain GV3101 (OD600 = 0.5–0.6) harboring individual CRISPR-Cas9 constructs in the dark under standard tissue culture conditions. Then, the transformants were selected on Murashige and Skoog medium supplemented with 15 mg L−1 hygromycin B (H370, PhytoTechnology Laboratories®). Plants of the T0 generation were genotyped by PCR using Cas9-specific primers and the mutations created by Cas9 were identified by sequencing. Cas9-free T1 plants harboring gene-edited mutations were backcrossed to Moneymaker, and their selfed F2 plants were used for phenotypic observations and gene expression analysis. Oligo and primer information used for vector construction and genotyping described here and hereafter is available in Supplementary Table 2.
Microscopy of SAM morphology
dtm-1, dtm-cr5, and their corresponding wild type SAMs were dissected from seedlings at different developmental stages by removing any visible leaves and examined with a stereomicroscope (M125/DFC420, Leica). For SEM and histological analysis, the dissected meristems were fixed in formalin-acetic acid-alcohol (10% formaldehyde, 5% acetic acid, and 50% alcohol by volume). The meristems were gradually dehydrated by incubation in an ethanol series (30–100%). Half of these meristem samples were used for SEM analysis and so further dried using a critical-point dryer (Tousimis, USA), before being sputter coated in gold particles. SEM images were collected using a high-resolution field emission SEM (Zeiss Merlin Compact, Zeiss or JSM-6360LV, JEOL). The other half were embedded in Paraplast (Sigma-Aldrich) for histological analysis and 8 μm sections made using a Leica microtome (Leica). The sections were briefly stained with 0.05% toluidine blue.
Gene expression analysis
Total dtm-1, dtm-cr5, and wild type RNA was extracted from shoot apices with visible leaves removed at specific stages using Trizol® reagent (ThermoFisher scientific)48. Residual genomic DNA was removed with DNAase I (NEB). To validate DTM coding sequences, 5′ and 3′ rapid amplification of cDNA ends was carried out using the SMARTer® RACE Kit (Clontech) following the supplier’s instructions. For qRT-PCR, 1 μg RNA was used to synthesize cDNA in a 20 μL reaction volume with the First Strand cDNA Synthesis kit (NEB). Two microliter 10fold-diluted cDNA was subjected to qRT-PCR with a 0.2 μM primer concentration. The PCR reaction containing 1× AceQ qPCR SYBR Green Master Mix (Vazyme Biotech, Nanjing) and 0.4 μL Rox Reference Dye 2, was performed using a QuantStudio3 (Applied Biosystems, Thermo) with the following PCR conditions: 45 cycles of 10 s at 95 °C and 30 s at 60oC. The SleIF4α6 gene was used as a reference to normalize the expression levels of target genes50.
For in situ hybridization, dissected shoot apices were fixed in formalin-acetic acid-alcohol overnight at 4 °C. The fixed tissues were then embedded in Paraplast (Sigma-Aldrich) and 8 μm sections made using a microtome (Leica). After dewaxing and rehydration, in situ hybridization was performed as previously described using digoxigenin-labeled gene-specific probes48. Probes were prepared by in vitro transcription in the presence of 0.1 mM DIG-UTP (Roche) using 0.5–1.0 μg PCR amplified gene coding regions as DNA templates with the T7 promoter sequence incorporated. Sections were probed at 50 °C overnight in hybridization buffer [50% deionized formamide, 300 mM Tris pH 8.0, 100 mM sodium phosphate pH 6.8, 300 mM NaCl, 5 mM EDTA, 5× Denhardt’s solution, 5% dextran sulphate, 1 mg L−1 tRNA] containing gene-specific riboprobes of about 100 bp fragmented by carbonate hydrolysis. After post hybridization washes, gene expression was detected using anti-digoxigenin-AP Fab fragments (1:3000, # 11093274910, Roche) with nitro-blue tetrazolium and 5-bromo-4-chloro-3′-indolyphosphate (NBT/BCIP, Roche) as substrates for color detection. The primers used for qRT-PCR analysis and for preparing probe templates are listed in Supplementary Table 2.
Bimolecular fluorescence complementation assay
The coding regions of DTM, the DTM_L8F mutant and the six tomato HD-ZIP III proteins (SlREV, SlPHB, SlPHV, SlHB8, SlHB15A, and SlHB15B) were amplified by PCR. The DTM sequences were then cloned into pJW771 (Pro35S:nLUC) and the HD-ZIP III sequences into pJW772 (Pro35S:cLUC), allowing expression of fusion proteins containing the N- and C-terminal halves of firefly luciferase, respectively33. For each interaction to be tested, equal amounts of Agrobacterium cells harboring the N- and C-terminal fusion expression cassettes were mixed. The cell mixture at an OD600 of 0.8 in infiltration buffer (10 mM MES, 10 mM MgCl2, 150 mM acetosyringone, pH 5.6) was infiltrated into Nicotiana benthamiana leaves using 1 mL needleless syringes51. Three days after infiltration, 1 mM Luciferin (Promega, WI) was infiltrated into the tobacco leaves. After 10 min incubation in the dark, luminescence was detected using an automatic chemiluminescence image analysis system (Tanon Science & Technology Co., Ltd., Shanghai) with an exposure time of 3 min.
Yeast two hybrid assay
The DTL and DTM coding regions and those of its mutant forms DTM_L8F, dtm-cr3, dtm-cr5, and dtm-cr7 were amplified by PCR and cloned into the pBD-GAL4 Cam bait vector (Stratagene). Similarly, the coding sequences (full-length or the N-terminal) of the six tomato HD-ZIP III genes were cloned into the pAD-GAL4–2.1 prey vector (Stratagene). The bait and prey plasmids were co-transformed into yeast strain AH109 (Clontech) using the LiCl-PEG method according to the manufacturer’s instructions. After transformants were selected on synthetic dropout nutrient medium (SD/-Leu-Trp) plates, protein interactions were tested by cell growth on SD/-Trp-Leu-His and SD/-Trp-Leu-His-Ade plates with or without 1 mM 3-amino-1,2,4-triazole added to the media.
Pulldown assay
Pulldown assays were conducted with E. coli expressed fusion proteins containing either cmyc (DTM and its mutant forms, DTL) or human influenza virus hemagglutinin (HA) tags (the six tomato HD-ZIP III proteins) and the Novagen® pET28 vector (Merck Millipore). Protein expression in E. coli BL21 star (DE3, Invitrogen) cells grown in liquid Luria-Bertani broth at an OD600 of 0.5–0.6 was induced with 1 mM isopropylthio-β-galactoside for 2 h at 28 °C. The cell pellets were collected by centrifugation at 12,000 × g for 2 min at 4 °C and resuspended in BugBuster Master Mix (Novagen) (200 μL per 1 mL cell culture). For soluble cmyc-tagged wild type DTL, DTM and its mutant forms as well as HA-tagged N-terminal portions of the HD-ZIP IIIs, the lysate was cleared by centrifugation at 14,000 × g for 15 min at 4 °C and directly used for immunoprecipitation. For HA-tagged full-length HD-ZIP III proteins that predominantly accumulated in protein inclusion bodies, the protein pellets collected by centrifugation at 14,000 × g for 15 min at 4 °C were solubilized in buffer A containing 10 mM Tris-HCl, pH 7.8, 50 mM NaCl, 1 mM EDTA, 6 M urea and 1 mM phenylmethanesulfonyl fluoride52, and then cleared by centrifugation at 12,000 × g for 15 min at 4 °C.
Pulldown assays were performed essentially as described in immunoprecipitation technical guide and protocols (Thermo Scientific) with some modifications. Before pulldown, 20 μL anti-HA Agarose resin (Sigma-Aldrich) (for a single reaction) was washed three times with 500 μL lysis buffer (10 m M HEPES, pH 7.5, 1.5 mM MgCl2, 10 mM KCl and 1 mM phenylmethanesulfonyl fluoride, 0.5% Triton X-100) containing 300 mM NaCl and once with 500 μL lysis buffer. The resin was suspended in 500 μL lysis buffer after the last wash and incubated with an aliquot of 50 μL freshly prepared lysate containing HA-HD-ZIP III proteins for 4 h at 4 °C. Anti-HA Agarose resin conjugated with HD-ZIP III fusion proteins was collected by a short centrifugation at 5000 × g at 4 °C. The resin was then washed three times with 500 μL lysis buffer containing 300 mM NaCl at 4 °C, followed by three washes with 500 μL lysis buffer containing 0.5% Triton X-100. After washing, the resin was suspended in 500 μL lysis buffer and incubated with 50 μL cmyc-DTM or cmyc-DTL lysate overnight at 4 °C. The resin containing the protein complex was then pelleted and washed as described above. The immunoprecipitation protein complex was released from the resin with 50 μL of 0.1 M glycine (pH2.5) for 30 min at 4 °C.
For Western blot analysis, IP prepared proteins and aliquots of input controls (2% of the amount used for each IP reaction) were separated in 10% (cmyc-DTL, -DTM and its mutants) or 12% (HA-HD-ZIP III proteins) SDS-PAGE gels and transferred to Amersham Hybond P 0.45 PVDF membranes (GE Healthcare Life Science). The membranes were subsequently blotted with primary anti-HA (1:1,000, # 3724S, CST) or anti-cmyc (1: 2,000, # A00172–40, Genscript) antibodies and then with secondary anti-rabbit HRP antibodies (1:5,000, # Ab6721, Abcam) as previously described53. To detect the antibody signals, the membranes were incubated in the buffer provided with the Super Signal West Pico Chemiluminescent substrate kit (Pierce). All full-length blots and gels are shown in Supplementary Fig. 14–17.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay was performed essentially according to the procedures48 we established previously with some modifications. Briefly, 2 μg purified SlREV (nucleotide 1–795) plasmid (pTNT™ vector, Promega) and PCR fragments of DTM, DTM_L8F, and DTM_L8F(L22A) coding sequences with the SP6 and T7 polymerase promoter sequences incorporated were used as transcription templates in the Wheat Germ Protein Expression System (Promega). Individual proteins were then synthesized in 10 μL reaction volumes using the TnT™ SP6 High-Yield Wheat Germ Protein Expression System (# L3261, Promega) following the supplier’s instructions. For the DNA binding assay, 1 μL aliquots of the synthesized SlREV (1–265aa) protein was first mixed with specified amounts (0, 1, 2,and 4 μL) of wild type DTM protein and its mutant forms DTM_L8F and DTM_L8F(L22A) in gel-shift binding buffer from the LightShift EMSA Optimization and Control Kit (Thermo). After 20 min incubation on ice, the Cy5-labeled HB-9 DNA duplexes (5’-CAGATCTGTAATGATTACGAGAAT-3′) recognized by REV25 were added and the reactions were incubated at room temperature for another 20 min. The mixtures were then subjected to electrophoresis in a 6% native PAGE gel.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Description of additional supplementary items
Acknowledgements
We would like to thank the Tomato Genetics Resource Center (TGRC) at University of California, Davis for kindly providing the tomato seeds and JW Wang for the pJW771 and pJW772 vectors. The work was supported by grants from the Ministry of Science and Technology of the People’s Republic of China (2016YFD0100506 and 2012CB113900), the National Natural Science Foundation of China (31672164 to H.X., 31501773 to Q.X.), and the China Postdoctoral Science Foundation (2015M580363 to Q.X.).
Author contributions
Q.X., R.L., L.W., S.Y., M.L performed the experiments and analyzed the data; H.X. conceived the study; Q.X., R.L. and H.X. wrote the paper.
Data availability
Tomato sequence data for ZPR, HD-ZIP III, and other genes used in this study can be found at the Sol Genomics Network (SGN, https://solgenomics.net/) under the following accession numbers: DTM (Solyc09g009620), DTL (Solyc11g007100), SlZPR1 (Solyc01g091490), SlZPR2A (Solyc08g007570), SlZPR2B (Solyc08g079690), SlREV (Solyc11g069470), SlPHB (Solyc02g024070), SlPHV (Solyc02g069830), SlHB8 (Solyc08g066500), SlHB15A (Solyc03g120910), SlHB15B (Solyc12g044410), SlWUS/LC (Solyc02g083950), SlCLV3/FAS (Solyc11g071380), SlSTM (Solyc02g081120), LFS (Solyc05g013540). A. thaliana ZPR and HD-ZIP III gene sequences can be found at the Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/index.jsp): ZPR1 (At2g45450), ZPR2 (At3g60890), ZPR3 (At3g52770), ZPR4 (At2g36307), ATHB15 (At1g52150), ATHB8 (At4g32880), REV (At5g60690), PHB (At2g34710) and PHV (At1g30490). Source data of floral organ number measurements used for making the boxplot in Fig. 1f is shown in Supplementary data 1, and source data for expression analysis by qRT-PCR in Fig. 7 and Supplementary Fig. 10 is available in Supplementary data 2 and 3, respectively. All other data generated in this study are available from the authors upon request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0368-8.
References
- 1.Pfeiffer A, Wenzl C, Lohmann JU. Beyond flexibility: controlling stem cells in an ever changing environment. Curr. Opin. Plant. Biol. 2017;35:117–123. doi: 10.1016/j.pbi.2016.11.014. [DOI] [PubMed] [Google Scholar]
- 2.Somssich M, Je BI, Simon R, Jackson D. CLAVATA-WUSCHEL signaling in the shoot meristem. Development. 2016;143:3238–3248. doi: 10.1242/dev.133645. [DOI] [PubMed] [Google Scholar]
- 3.Pautler M, Tanaka W, Hirano HY, Jackson D. Grass meristems I: shoot apical meristem maintenance, axillary meristem determinacy and the floral transition. Plant Cell Physiol. 2013;54:302–312. doi: 10.1093/pcp/pct025. [DOI] [PubMed] [Google Scholar]
- 4.Aichinger E, Kornet N, Friedrich T, Laux T. Plant stem cell niches. Annu. Rev. Plant. Biol. 2012;63:615–636. doi: 10.1146/annurev-arplant-042811-105555. [DOI] [PubMed] [Google Scholar]
- 5.Doerner P. Plant meristems: a merry-go-round of signals. Curr. Biol. 2003;13:R368–R374. doi: 10.1016/S0960-9822(03)00280-X. [DOI] [PubMed] [Google Scholar]
- 6.Brand U, Fletcher JC, Hobe M, Meyerowitz EM, Simon R. Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science. 2000;289:617–619. doi: 10.1126/science.289.5479.617. [DOI] [PubMed] [Google Scholar]
- 7.Schoof H, et al. The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell. 2000;100:635–644. doi: 10.1016/S0092-8674(00)80700-X. [DOI] [PubMed] [Google Scholar]
- 8.Perales M, et al. Threshold-dependent transcriptional discrimination underlies stem cell homeostasis. Proc. . Natl Acad. Sci. USA. 2016;113:E6298–E6306. doi: 10.1073/pnas.1607669113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reddy GV, Meyerowitz EM. Stem-cell homeostasis and growth dynamics can be uncoupled in the Arabidopsis shoot apex. Science. 2005;310:663–667. doi: 10.1126/science.1116261. [DOI] [PubMed] [Google Scholar]
- 10.Clark SE, Jacobsen SE, Levin JZ, Meyerowitz EM. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development. 1996;122:1567–1575. doi: 10.1242/dev.122.5.1567. [DOI] [PubMed] [Google Scholar]
- 11.Endrizzi K, Moussian B, Haecker A, Levin JZ, Laux T. The SHOOT MERISTEMLESS gene is required for maintenance of undifferentiated cells in Arabidopsis shoot and floral meristems and acts at a different regulatory level than the meristem genes WUSCHEL and ZWILLE. Plant J. 1996;10:967–979. doi: 10.1046/j.1365-313X.1996.10060967.x. [DOI] [PubMed] [Google Scholar]
- 12.Gallois JL, Woodward C, Reddy GV, Sablowski R. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development. 2002;129:3207–3217. doi: 10.1242/dev.129.13.3207. [DOI] [PubMed] [Google Scholar]
- 13.Lenhard M, Jurgens G, Laux T. The WUSCHEL and SHOOTMERISTEMLESS genes fulfil complementary roles in Arabidopsis shoot meristem regulation. Development. 2002;129:3195–3206. doi: 10.1242/dev.129.13.3195. [DOI] [PubMed] [Google Scholar]
- 14.Galli M, Gallavotti A. Expanding the regulatory network for meristem size in plants. Trends Genet. 2016;32:372–383. doi: 10.1016/j.tig.2016.04.001. [DOI] [PubMed] [Google Scholar]
- 15.Byrne ME. Shoot meristem function and leaf polarity: the role of class III HD-ZIP genes. PLoS. Genet. 2006;2:e89. doi: 10.1371/journal.pgen.0020089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prigge MJ, et al. Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic, and distinct roles in Arabidopsis development. Plant Cell. 2005;17:61–76. doi: 10.1105/tpc.104.026161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emery JF, et al. Radial patterning of Arabidopsis shoots by class III HD-ZIP and KANADI genes. Curr. Biol. 2003;13:1768–1774. doi: 10.1016/j.cub.2003.09.035. [DOI] [PubMed] [Google Scholar]
- 18.Mandel T, et al. Differential regulation of meristem size, morphology and organization by the ERECTA, CLAVATA and class III HD-ZIP pathways. Development. 2016;143:1612–1622. doi: 10.1242/dev.129973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Williams L, Grigg SP, Xie M, Christensen S, Fletcher JC. Regulation of Arabidopsis shoot apical meristem and lateral organ formation by microRNA miR166g and its AtHD-ZIP target genes. Development. 2005;132:3657–3668. doi: 10.1242/dev.01942. [DOI] [PubMed] [Google Scholar]
- 20.Zhang TQ, et al. A two-step model for de Novo activation of WUSCHEL during plant shoot regeneration. Plant Cell. 2017;29:1073–1087. doi: 10.1105/tpc.16.00863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shi B, et al. Two-step regulation of a meristematic cell population acting in shoot branching in Arabidopsis. PLoS. Genet. 2016;12:e1006168. doi: 10.1371/journal.pgen.1006168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee C, Clark SE. A WUSCHEL-independent stem cell specification pathway is repressed by PHB, PHV and CNA in Arabidopsis. PLoS. One. 2015;10:e0126006. doi: 10.1371/journal.pone.0126006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu H, et al. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011;145:242–256. doi: 10.1016/j.cell.2011.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou GK, Kubo M, Zhong R, Demura T, Ye ZH. Overexpression of miR165 affects apical meristem formation, organ polarity establishment and vascular development in Arabidopsis. Plant Cell Physiol. 2007;48:391–404. doi: 10.1093/pcp/pcm008. [DOI] [PubMed] [Google Scholar]
- 25.Wenkel S, Emery J, Hou BH, Evans MM, Barton MK. A feedback regulatory module formed by LITTLE ZIPPER and HD-ZIPIII genes. Plant Cell. 2007;19:3379–3390. doi: 10.1105/tpc.107.055772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith ZR, Long JA. Control of Arabidopsis apical-basal embryo polarity by antagonistic transcription factors. Nature. 2010;464:423–426. doi: 10.1038/nature08843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim YS, et al. HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. Plant Cell. 2008;20:920–933. doi: 10.1105/tpc.107.057448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Münos S, et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 2011;156:2244–2254. doi: 10.1104/pp.111.173997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu C, et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015;47:784–792. doi: 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- 30.Tal L, et al. Coordination of meristem doming and the floral transition by late termination, a kelch repeat protein. Plant Cell. 2017;29:681–696. doi: 10.1105/tpc.17.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biais B, et al. Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiol. 2014;164:1204–1221. doi: 10.1104/pp.113.231241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu G, et al. Overexpression of SlREV alters the development of the flower pedicel abscission zone and fruit formation in tomato. Plant Sci. 2014;229:86–95. doi: 10.1016/j.plantsci.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Gou JY, Felippes FF, Liu CJ, Weigel D, Wang JW. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell. 2011;23:1512–1522. doi: 10.1105/tpc.111.084525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reinhardt D, Frenz M, Mandel T, Kuhlemeier C. Microsurgical and laser ablation analysis of interactions between the zones and layers of the tomato shoot apical meristem. Development. 2003;130:4073–4083. doi: 10.1242/dev.00596. [DOI] [PubMed] [Google Scholar]
- 35.Brandt R, et al. Genome-wide binding-site analysis of REVOLUTA reveals a link between leaf patterning and light-mediated growth responses. Plant J. 2012;72:31–42. doi: 10.1111/j.1365-313X.2012.05049.x. [DOI] [PubMed] [Google Scholar]
- 36.Capua Y, Eshed Y. Coordination of auxin-triggered leaf initiation by tomato LEAFLESS. Proc. . Natl Acad. Sci. USA. 2017;114:3246–3251. doi: 10.1073/pnas.1617146114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu C, et al. A group of receptor kinases are essential for CLAVATA signalling to maintain stem cell homeostasis. Nat. Plants. 2018;4:205–211. doi: 10.1038/s41477-018-0123-z. [DOI] [PubMed] [Google Scholar]
- 38.Kinoshita A, et al. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- 39.Fletcher JC, Brand U, Running MP, Simon R, Meyerowitz EM. Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science. 1999;283:1911–1914. doi: 10.1126/science.283.5409.1911. [DOI] [PubMed] [Google Scholar]
- 40.Clark SE, Williams RW, Meyerowitz EM. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell. 1997;89:575–585. doi: 10.1016/S0092-8674(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 41.David-Schwartz R, Borovsky Y, Zemach H, Paran I. CaHAM is autoregulated and regulates CaSTM expression and is required for shoot apical meristem organization in pepper. Plant Sci. 2013;203-204:8–16. doi: 10.1016/j.plantsci.2012.12.011. [DOI] [PubMed] [Google Scholar]
- 42.Stuurman J, Jäggi F, Kuhlemeier C. Shoot meristem maintenance is controlled by a GRAS-gene mediated signal from differentiating cells. Genes Dev. 2002;16:2213–2218. doi: 10.1101/gad.230702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Husbands AY, Aggarwal V, Ha T, Timmermans MC. In Planta single-molecule pull-down reveals tetrameric stoichiometry of HD-ZIPIII:LITTLE ZIPPER Complexes. Plant Cell. 2016;28:1783–1794. doi: 10.1105/tpc.16.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fulton TM, Chunwongse J, Tanksley SD. Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant. Mol. Biol. Rep. 1995;13:207–209. doi: 10.1007/BF02670897. [DOI] [Google Scholar]
- 45.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lei Y, et al. CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol. Plant. 2014;7:1494–1496. doi: 10.1093/mp/ssu044. [DOI] [PubMed] [Google Scholar]
- 47.Liu WS, et al. A detailed procedure for CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana. Sci. Bull. 2015;60:1332–1347. doi: 10.1007/s11434-015-0848-2. [DOI] [Google Scholar]
- 48.Weng L, et al. The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiol. 2015;167:931–949. doi: 10.1104/pp.114.255174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McCormick S, et al. Leaf disc transformation of cultivated tomato (L. esculentum) using Agrobacterium tumefaciens. Plant Cell Rep. 1986;5:81–84. doi: 10.1007/BF00269239. [DOI] [PubMed] [Google Scholar]
- 50.Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 51.Lindbo JA. TRBO: a high-efficiency tobacco mosaic virus RNA-based overexpression vector. Plant Physiol. 2007;145:1232–1240. doi: 10.1104/pp.107.106377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang H, Mizukami Y, Hu Y, Ma H. Isolation and characterization of the binding sequences for the product of the Arabidopsis floral homeotic gene AGAMOUS. Nucleic Acids Res. 1993;21:4769–4776. doi: 10.1093/nar/21.20.4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schulman S, Wang B, Li W, Rapoport TA. Vitamin K epoxide reductase prefers ER membrane-anchored thioredoxin-like redox partners. Proc. Natl Acad. Sci. USA. 2010;107:15027–15032. doi: 10.1073/pnas.1009972107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of additional supplementary items
Data Availability Statement
Tomato sequence data for ZPR, HD-ZIP III, and other genes used in this study can be found at the Sol Genomics Network (SGN, https://solgenomics.net/) under the following accession numbers: DTM (Solyc09g009620), DTL (Solyc11g007100), SlZPR1 (Solyc01g091490), SlZPR2A (Solyc08g007570), SlZPR2B (Solyc08g079690), SlREV (Solyc11g069470), SlPHB (Solyc02g024070), SlPHV (Solyc02g069830), SlHB8 (Solyc08g066500), SlHB15A (Solyc03g120910), SlHB15B (Solyc12g044410), SlWUS/LC (Solyc02g083950), SlCLV3/FAS (Solyc11g071380), SlSTM (Solyc02g081120), LFS (Solyc05g013540). A. thaliana ZPR and HD-ZIP III gene sequences can be found at the Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/index.jsp): ZPR1 (At2g45450), ZPR2 (At3g60890), ZPR3 (At3g52770), ZPR4 (At2g36307), ATHB15 (At1g52150), ATHB8 (At4g32880), REV (At5g60690), PHB (At2g34710) and PHV (At1g30490). Source data of floral organ number measurements used for making the boxplot in Fig. 1f is shown in Supplementary data 1, and source data for expression analysis by qRT-PCR in Fig. 7 and Supplementary Fig. 10 is available in Supplementary data 2 and 3, respectively. All other data generated in this study are available from the authors upon request.