Abstract
The genetic regulation of nephron patterning during kidney organogenesis remains poorly understood. Nephron tubules in zebrafish are composed of segment populations that have unique absorptive and secretory roles, as well as multiciliated cells (MCCs) that govern fluid flow. Here, we report that the transcription factor iroquois 2a (irx2a) is requisite for zebrafish nephrogenesis. irx2a transcripts localized to the developing pronephros and maturing MCCs, and loss of function altered formation of two segment populations and reduced MCC number. Interestingly, irx2a deficient embryos had reduced expression of an essential MCC gene ets variant 5a (etv5a), and were rescued by etv5a overexpression, supporting the conclusion that etv5a acts downstream of irx2a to control MCC ontogeny. Finally, we found that retinoic acid (RA) signaling affects the irx2a expression domain in renal progenitors, positioning irx2a downstream of RA. In sum, this work reveals new roles for irx2a during nephrogenesis, identifying irx2a as a crucial connection between RA signaling, segmentation, and the control of etv5a mediated MCC formation. Further investigation of the genetic players involved in these events will enhance our understanding of the molecular pathways that govern renal development, which can be used help create therapeutics to treat congenital and acquired kidney diseases.
Subject terms: Pattern formation, Organogenesis
Introduction
The vertebrate kidney is an architecturally intricate organ composed of nephron functional units that cleanse the circulation of metabolic waste and maintain fluid homeostasis1. Nephrons contain a blood filter, a segmented epithelial tubule, and collecting duct2. The blood filter disperses fluid across sieve comprised of a fenestrated network of capillaries that are situated within a specialized basement membrane that is opposed by podocytes with interdigitating cellular extensions3. Once inside the tubule, the renal filtrate passes by a series of specialized proximal and distal epithelial segment populations that are responsible for step-wise nutrient reabsorption and metabolite secretion. Fine-tuning of salt balance and final excretion occurs through the distal segments and subsequent collecting duct.
While the composition and precise organization of nephron regions is essential for proper kidney physiology, the developmental processes that control formation of cell types are not yet fully understood4,5. In part, this is due to the complicated nature of kidney organogenesis, which involves the coordinated creation of nephrons that each contains a multitude of differentiated cell types in precise arrangements. For example, in its final postnatal form, a single human kidney can contain hundreds of thousands to upwards of 1.5 million nephrons comprised of over 20 differentiated cell types6. Further, multiple kidney forms are made during development, which involves the successive formation and degradation of several nephron-based organs. In mammals, the adult metanephros is preceded by a transitory structure termed the mesonephros, which develops after the embryonic pronephros arises from the intermediate mesoderm (IM)7. In fish, the mesonephros is the adult kidney and a simple pronephros functions during embryonic stages7. Despite these differences, the fundamental genetic and molecular composition of nephrons is conserved across vertebrate species based on the common expression of transcription factors and solute transporters that define the various differentiated cell compartments8,9.
In recent years, the zebrafish pronephros has emerged as a powerful model of vertebrate renal ontogeny10,11 and disease12–14 because of its rapid development and nephron conservation. During the initial stages of nephrogenesis, renal progenitors arise from bilateral stripes of IM precursors15 and have been characterized by dynamic spatiotemporal gene expression patterns that are modulated at the outset by retinoic acid (RA) signaling16–18. Following a mesenchymal-to-epithelial transition (MET) of the renal progenitors19,20, the zebrafish pronephros consists of two parallel nephrons that are connected caudally by a common collecting duct, and will later undergo morphogenesis events rostrally to form a shared blood filter apparatus21,22. The epithelial tubule of the nephron is subdivided into functional regions known as the proximal convoluted and straight tubule segments (PCT, PST) and the distal early and late (DE, DL) segments, the latter where the associated corpuscle of Stannius (CS) gland arises16. Each of these segments is comprised of unique differentiated transporter cells that possess a single primary cilium, which is a microtubule-based projection that extends from the apical surface into the extracellular space. Additionally, a population of multiciliated cells (MCCs) is dispersed in the intermediate region of the nephron corresponding to the PST and parts of the adjacent PCT and DE segments23,24. Furthermore, all cilia within the zebrafish kidney are motile and participate in the movement of filtrate through the nephron tubule25. Interestingly, while MCCs are a major renal epithelial cell type in zebrafish, analogous MCCs in humans have only been documented in the fetal kidney26,27 and in case reports of kidney diseases such as hypercalcemia, congenital nephrosis and glomerulonephritis27–34. These findings suggest that understanding the mechanisms that guide MCC formation may provide insights about kidney development and disease.
A growing list of genes and signaling pathways have been identified as participants in the networks that direct segmentation and MCC development during zebrafish renal organogenesis. RA both controls proximo-distal segment pattern and promotes MCC development by inhibiting expression of the mds1/evi1 complex (mecom) transcription factor35. mecom subsequently promotes Notch signaling which negatively modulates a binary transporter cell versus MCC fate choice23,24,35. In part, Notch signaling inhibits expression of the etv5a transcription factor, while RA promotes etv5a to stimulate MCC development36. Moreover, etv5a and etv4 were found to have redundant functions since their combined deficiency caused a significantly greater decrease in MCC number than the knockdown of either factor alone36. These studies indicate that the MCC developmental pathway must be highly regulated, as the loss of any single factor did not completely impair MCC formation35,36. Consequently, the exact mechanisms that direct the specification of MCC remains are not yet completely elucidated.
The iroquois (iro/Irx) genes encode transcription factors that belong to the TALE superclass of homeodomain proteins and regulate the patterning of tissue territories during embryogenesis in invertebrates and vertebrates, respectively37,38. Several Irx genes are expressed during kidney development in amphibians, mammals and zebrafish17,39–43. For example, irx1/2/3 transcripts are localized to the intermediate tubule region of the Xenopus pronephros41, and in the mouse metanephros are located in the intermediate region in the S-shaped body of developing nephrons and later to the middle section of the nephron, known as the Loop of Henle41,44,45. Knockdown of Irx2 did not alter pronephros tubule development in the frog embryo, however, leading to the hypothesis that it shares redundant activities with Irx1 and/or Irx341. Interestingly, the zebrafish homologs, irx2a and irx3b, are expressed in the central region of the pronephros tubule, in a domain which includes the PST and DE segments along with MCCs17,40. irx3b is required for DE specification, where the loss of this transcription factor results in the abrogation of slc12a1+ tubule cells, expanded proximal segments, and an expanded CS lineage17,46. In contrast, the function(s) of irx2a during pronephros development have remained undefined until now.
Here, we report novel roles for irx2a in PST and DL segment development as well as MCC formation in the zebrafish pronephros. We found that irx2a regulates expression of etv5a in part to control MCC fate choice, and that this regulation occurs downstream of RA signaling. These findings provide the first account that irx2a coordinates nephron segmentation and MCC development, which has implications for understanding kidney organogenesis across vertebrates.
Results
irx2a is expressed in the central renal progenitor field and subsequent pronephros
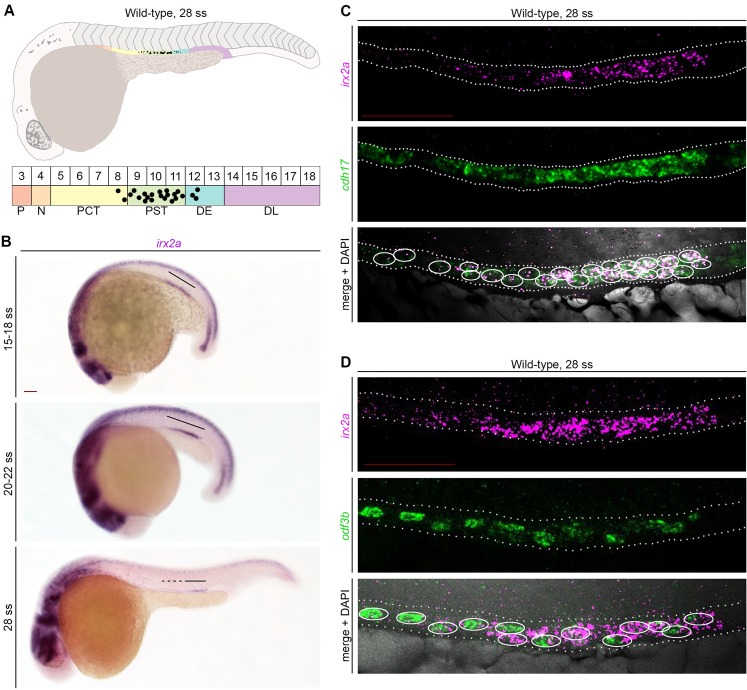
Nephron segmentation during zebrafish pronephros ontogeny is completed by the 28 somite stage (ss) and forms a series of cell types which will comprise the tubule, and others that later contribute to formation of the blood filter16 (Fig. 1A). The epithelial tubule populations are interspersed with MCCs, which occupy the PCT, PST, and DE segments in a “salt and pepper” like distribution23,24 (Fig. 1A). Further, the segment populations occupy an anatomical location in close proximity to the trunk somites, where they are situated adjacent to somites 3 through 18 (Fig. 1A). To further explore the association of irx2a with renal progenitor development, we performed whole mount in situ hybridization (WISH) on wild-type zebrafish embryos between the 5–28 ss to assess its spatiotemporal expression domain in the emerging kidney. irx2a transcripts were detected first at the 15 ss in a pattern consistent with the central region of the developing pronephros tubule, where irx2a transcripts continued to be expressed at the 20–22 and 28 ss (Fig. 1B). Next, we performed a series of double whole mount fluorescent in situ hybridization (FISH) experiments to further define the occupancy of irx2a transcripts within renal progenitors. Prior studies have shown that cadherin17 (cdh17) expression demarcates the nephron tubule at the 28 ss47, while outer dense fiber of sperm tails 3B (odf3b) demarcates MCCs23. We found that irx2a transcripts localized both to cdh17+ and odf3b+ nephron cells (Fig. 1C,D), confirming pronephros-specific expression of irx2a. There was substantial overlap between cells that expressed both irx2a and odf3b transcripts (Fig. 1D). Interestingly, some irx2a+ cells were not odf3b+, and visa versa; additionally, we noted that some cells showed very low co-expression of these markers (Fig. 1D). As the onset of odf3b expression in the pronephros occurs at approximately the 20 ss of embryogenesis36, and irx2a was detected as early as the 15 ss, this may suggest that irx2a marks MCC precursors and diminishes as MCC fate is selected and/or as MCCs differentiate. Therefore, we next explored whether irx2a has roles in nephrogenesis during events such as segmentation and MCC ontogeny.
Figure 1.
irx2a expression localizes to a region of the zebrafish pronephros that corresponds to the PST, DE and MCC domains. (A) Schematic of a zebrafish embryo (lateral view) at 24 hpf, which is equivalent to the 28 ss. Schematic below depicts color coded segments, corresponding somite numbers, and the expression pattern of MCCs in black within the nephron. MCC number is not to scale. (B) WISH in wild-type zebrafish embryos at the 15–18 ss, 20–22 ss, and 28 ss demonstrates irx2a transcript expression (purple) in the middle of the developing pronephros. Black lines highlight the irx2a expression domain. Scale bar is 50 μm. (C) FISH in wild-type embryos at the 28 ss demonstrates that irx2a expression (magenta) colocalizes with cdh17+ epithelial cells and (D) odf3b+ MCCs of the nephron tubule (green), where DAPI labels the nuclei (grey). Scale bar is 50 μm. White circles indicate nuclei with co-expression of the respective markers. P - podocytes, N - neck, PCT - proximal convoluted tubule, PST - proximal straight tubule, DE - distal early, DL - distal late, MCC - multiciliated cell, ss - somite stage, WISH – whole mount in situ hybridization, FISH – fluorescent in situ hybridization.
irx2a deficiency alters PST and DL segment development in the pronephros
To explore the functional role of irx2a during kidney development, we conducted knockdown studies by utilizing a morpholino (MO) that specifically targeted the splicing boundary between intron 1–2 and exon 2 of the irx2a pre-mRNA sequence48 (Fig. S1A). Through RT-PCR and sequencing analysis, we confirmed that the majority of irx2a transcripts were misspliced in irx2a morphants compared to wild-type embryos (Fig. S1B,C). The aberrantly spliced irx2a transcript is predicted to produce a truncated peptide that lacks the homeobox domain due to inclusion of intron 1, which leads to a premature stop codon shortly after exon 1 (Fig. S1D).
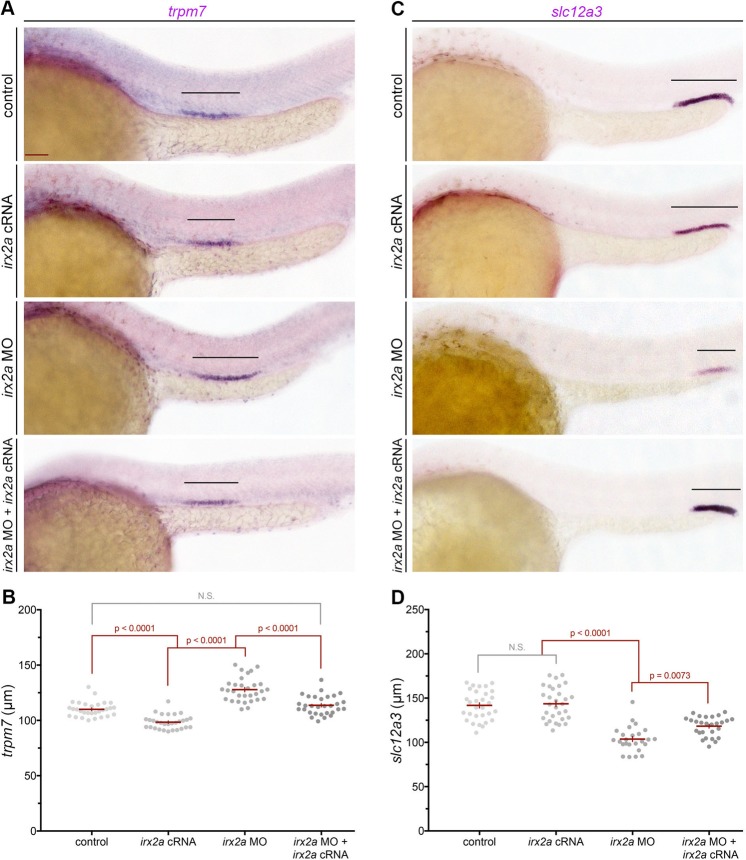
Next, we performed WISH to characterize the effect of irx2a deficiency on pronephros development. Using the somites, marked by smyhc1, as a point of reference to demarcate distinct segment boundaries, we analyzed each segment based on the specific expression of solute transporter genes: slc20a1a (PCT), trpm7 (PST), slc12a1 (DE), and slc12a3 (DL)16. irx2a deficient embryos formed PCT and DE segments that were indistinguishable in absolute length from wild-type controls (Fig. S2). However, the position of the DE was shifted posteriorly with respect to the trunk, where it was situated adjacent to somites 13–14 compared to the wild-type location adjacent to somites 12–13 (Fig. S2). Further, irx2a deficient embryos exhibited an expanded PST segment and a reduced DL segment (Figs 2, S2). Measurement and statistical analysis of the showed that these segment alterations in irx2a deficient embryos were significant (Fig. 2B,D). To assess whether morpholino toxicity could affect pronephros development, we examined segment development by WISH following microinjection of a standard control morpholino, and observed no alteration to segment pattern formation (Fig. S3).
Figure 2.
irx2a restricts the PST and promotes the DL during pronephros segmentation. (A) Lateral view of WISH analysis with the PST marker trpm7 at the 28 ss. Black bars highlight the trpm7 expression domain in the pronephros. Scale bar is 50 μm. (B) Quantification of trpm7 length (μm). Each dot represents one nephron and data is presented +/− SEM. Statistical significance was determined with ANOVA. (C) WISH in 28 ss embryos with the DL marker slc12a3 shown in a lateral view. Black bars denote slc12a3 expression. (D) Quantification of slc12a3 domain length (μm), where each dot represents one nephron. Data is presented +/− SEM and statistical significance was determined by ANOVA. WISH – whole mount in situ hybridization, ss – somite stage, PST – proximal straight tubule, DL – distal late tubule.
To explore the irx2a morphant phenotype further, and next test the MO specificity for these renal phenotypes, rescue studies were conducted in irx2a deficient embryos by provision of irx2a capped mRNA (cRNA). Expression of irx2a was sufficient to rescue both PST and DL development in irx2a deficient embryos, which was found to be statistically significant based on measurement of absolute segment lengths (Fig. 2). Additionally, irx2a overexpression studies were performed in wild-type embryos to assess whether irx2a cRNA was sufficient to induce alterations in the PST or DL lineages. However, at irx2a cRNA dosages that did not grossly affect embryogenesis, there was no significant difference observed in the populace of either the PST or DL segments (Fig. 2). Taken together, these results indicate that irx2a is necessary but not sufficient for PST and DL segment fate during pronephros development.
irx2a deficiency alters MCC development in the pronephros
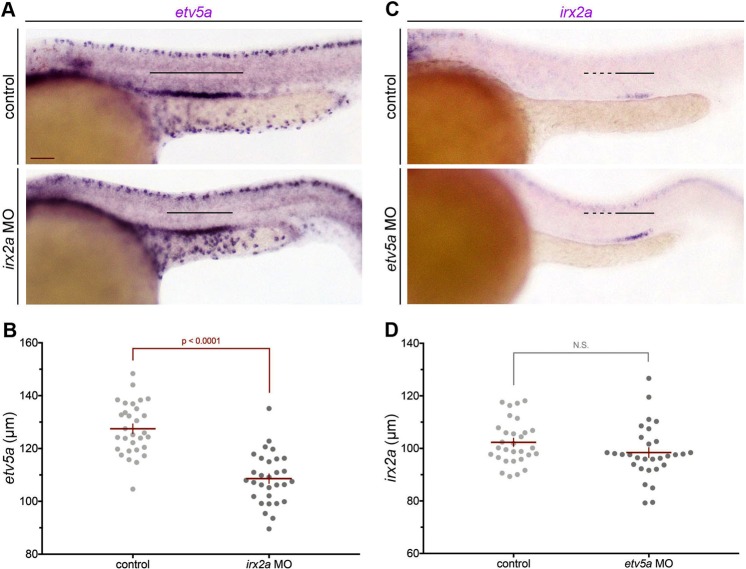
Given the expression domain of irx2a in maturing MCCs, we next explored whether MCC ontogeny in the pronephros is influenced by irx2a activity. MCC development is known to be reliant on expression of the transcription factor etv5a, which establishes the domain of MCC progenitors through interplay with Notch signaling36. Upon using WISH to examine etv5a expression in irx2a deficient embryos, we found that the pronephros domain of etv5a was significantly reduced in absolute length compared to wild-type controls (Fig. 3A,B). Next, irx2a expression was assessed in etv5a deficient embryos using WISH. We found that the pronephros expression of irx2a was comparable between wild-type and etv5a deficient embryos, where measurement of the irx2a expression domain within the nephron tubules confirmed that there was no statistically significant difference in length compared to wild-type embryo controls (Fig. 3C,D). Taken together, these data suggested that irx2a acts upstream of etv5a during nephrogenesis, and led us to hypothesize that irx2a was likely requisite for MCC fate choice.
Figure 3.
irx2a acts upstream of etv5a. WISH at the 28 ss revealed decreased etv5a expression in the pronephros of irx2a morphants. Black bars denote the etv5a expression domain. Scale bar is 50 μm. (B) Quantification of etv5a length in μm at the 28 ss. Each dot represents one nephron and data is presented +/− SEM. Significance was evaluated with an unpaired student’s T-test. (C) The irx2a domain is unchanged in etv5a morphants at the 28 ss. Solid black bars denote strong irx2a expression and the dashed black lines denote faint expression in the pronephros. (D) Quantification of irx2a length (μm) at the 28 ss. Each dot represents one nephron and data is presented +/− SEM. An unpaired student’s T-test was used to determine significance. WISH – whole mount in situ hybridization, ss – somite stage.
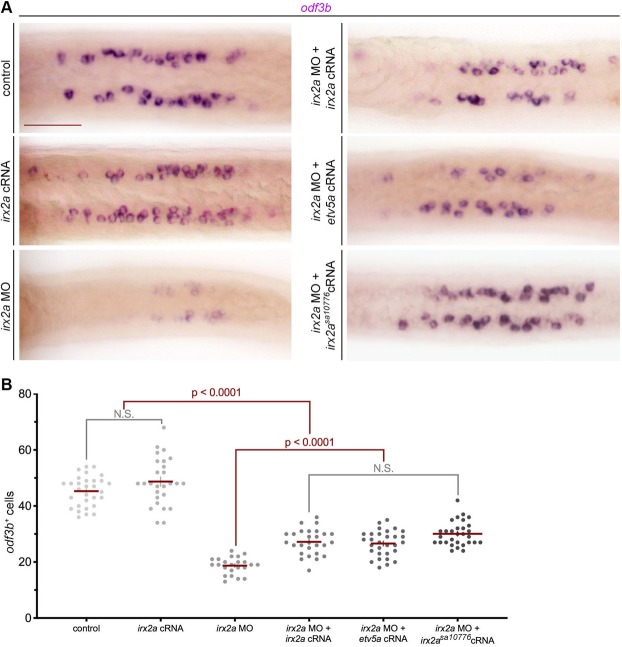
To explore this notion, MCC development in the pronephros was assessed in irx2a gain and loss of function studies (Fig. 4). WISH was performed to evaluate odf3b+ cell number as this marker specifically labels maturing MCCs23. Overexpression of irx2a was not associated with a significant increase in absolute MCC number compared to wild-type controls (Fig. 4A,B). However, irx2a deficient embryos developed significantly fewer numbers of nephron MCCs compared to wild-type controls or irx2a overexpressing cohorts at the 28 ss (Fig. 4A,B). Similarly, reduced MCC numbers were observed at 36 hpf in irx2a deficient embryos, likely latter ruling out a phenotype attributable to developmental delay (Fig. S4). Next, we found that the reduction of nephron MCC number in irx2a deficient embryos was partially rescued by provision of irx2a cRNA (Fig. 4A,B).
Figure 4.
irx2a is required upstream of etv5a for MCC genesis. (A) Dorsal view of odf3b in the pronephros by WISH at the 28 ss shows a decrease in maturing MCCs in irx2a deficient embryos, which was rescued with irx2a cRNA, etv5a cRNA, and irx2asa10776 cRNA. Scale bar is 50 μm. (B) Quantification of odf3b+ MCCs at the 28 ss. Each dot represents one pronephros. Data is presented +/− SEM and statistical significance was determined with ANOVA. WISH – whole mount in situ hybridization, ss – somite stage, MCC – multiciliated cell.
In light of the observation that irx2a deficiency led to reduced etv5a expression, and prior knowledge that the latter is essential for MCC fate choice36, we subsequently tested if etv5a could rescue MCC ontogeny in irx2a morphants. Provision of etv5a cRNA in irx2a deficient embryos partially rescued MCC number, which was statistically significant compared to irx2a knockdown alone, and statistically equivalent to MCC rescue following irx2a cRNA expression (Fig. 4A,B). Based on these results, we conclude that irx2a is essential for MCC fate where it acts upstream of etv5a in a shared pathway that promotes MCC development in renal progenitors.
Phenotypic analysis of an irx2a genetic mutant
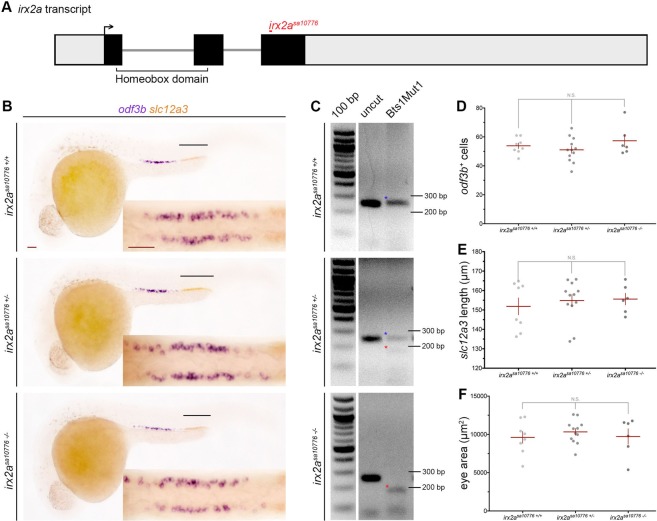
To perform further studies of the roles of irx2a in development, we obtained the irx2asa10776 line from the Zebrafish International Resource Center, which was generated by the Zebrafish Mutation Project49. The irx2asa10776 line was reported to harbor a C- > T mutation that encodes a nonsense mutation, which is predicted to produce a truncated protein that contains only 260 of the 432 amino acids that are present in the wild-type protein (Fig. 5A). We developed a restriction fragment length polymorphism genotyping assay that utilized digestion of a PCR product to confirm and identify the mutant allele. Adult irx2asa10776 heterozygotes were identified and incrossed to collect clutches for analysis of renal progenitor development using WISH followed by genotyping. Interestingly, irx2asa10776 homozygote and heterozygote embryos displayed both normal nephron segmentation and MCC development (Fig. 5B–D). Assessment of MCC number through quantification of odf3b+ cells showed that there were no significant differences between wild-type, irx2asa10776 heterozygote and homozygote embryos (Fig. 5B–D). Assessment of DL segment development based on the absolute domain length of slc12a3+ cells also showed no significant differences between these groups (Fig. 5B–D). Eye development was normal as well based on gross morphology and size, in contrast to a role for irx2a in this organ48 (Fig. 5E). Other studies have found inconsistencies between knockdown and mutant phenotypes due to genetic compensation in the latter50. Thus, irx2asa10776 mutants may have normal renal progenitor development due to mechanisms that otherwise compensate for the loss of irx2a within the genome. Alternatively, it is also possible that the location of this nonsense mutation is not sufficient to interrupt Irx2a function.
Figure 5.
irx2a genetic mutants do not have a renal or eye phenotype. (A) Schematic of the irx2a transcript and location of the irx2asa10716 mutation, where grey is non-coding and black represents coding sequence. The forward arrow demarcates the ATG start site. (B) WISH analysis at the 28 ss shows odf3b and slc12a3 expression in irx2asa10716+/+, irx2asa10716+/−, and irx2asa10716−/− embryos. The black lines highlight the DL marker slc12a3. An enlarged dorsal view of the odf3b+ MCCs for each embryo is shown in the inset. Scale bars are 50 μm. (C) Gel image of the restriction digest product for each genotype. Wild-type bands are marked with a blue star, and mutant bands with a red star. (D) Quantification of odf3b+ cells at the 28 ss. Each dot represents one pronephros. (E) Quantification of the slc12a3 length (μm) at the 28 ss. Each dot represents one nephron. (F) Quantification of eye area (μm2). Each dot represents one eye. For all graphs, data is presented +/− SEM and statistical significance was determined by ANOVA. WISH – whole mount in situ hybridization, ss – somite stage, DL – distal late, MCC – multiciliated cell.
To explore the latter, we performed site-directed mutagenesis on our irx2a expression construct to modify the sequence so that it would encode the irx2asa10776 allele. Following synthesis of irx2asa10776 cRNA and microinjection into irx2a morphants, MCC development was assessed by WISH for expression of odf3b transcripts. Interestingly, we found that irx2asa10776 was sufficient to rescue MCC number comparable to irx2a cRNA (Fig. 4A,B). Given this result, and as the nonsense mutation in irx2asa10776 is situated after the homeobox domain, we concluded that the mutant protein in fact retains the key functional attribute(s) necessary to support pronephros development. Thus, the normal appearance of renal MCCs and segments in irx2asa10776 mutants is not likely due to genetic compensation by other factors.
RA signaling is an upstream regulator of irx2a during nephrogenesis
We next sought to determine the relationship between irx2a and RA signaling to explore how irx2a further fits within the genetic cascades that regulate pronephros segment and MCC development. RA is a diffusible morphogen that is necessary to establish rostrocaudal patterning among pronephros progenitors during the earliest stages of nephrogenesis, and subsequently modulates the expression of many transcription factors to direct segment formation16,17. Furthermore, treatment with exogenous all-trans RA results in an expansion of proximal segment identities including MCCs at the expense of the distal segment regions16,35. Conversely, the inhibition of RA signaling by blocking its biosynthesis with the inhibitor N,N-diethylaminobenzaldehyde (DEAB) causes distal fates to be favored, while decreasing proximal lineages and abrogating MCCs16,35.
Therefore, we tested whether alterations in RA levels affect irx2a expression within the pronephros. Wild-type embryos were treated with RA or DEAB at 60% epiboly and fixed at the 28 ss for WISH analysis. We found that treatment of wild-type embryos with exogenous all-trans RA resulted in a domain expansion and a distal shift of irx2a expression (Fig. 6). Alternately, treatment with DEAB caused a reduction and concomitant proximal shift of the irx2a expression domain in the pronephros (Fig. 6). These results indicate that RA controls the expression of irx2a either directly or indirectly during renal ontogeny, which suggests irx2a acts downstream of RA to pattern the PST, DE, and MCCs.
Figure 6.
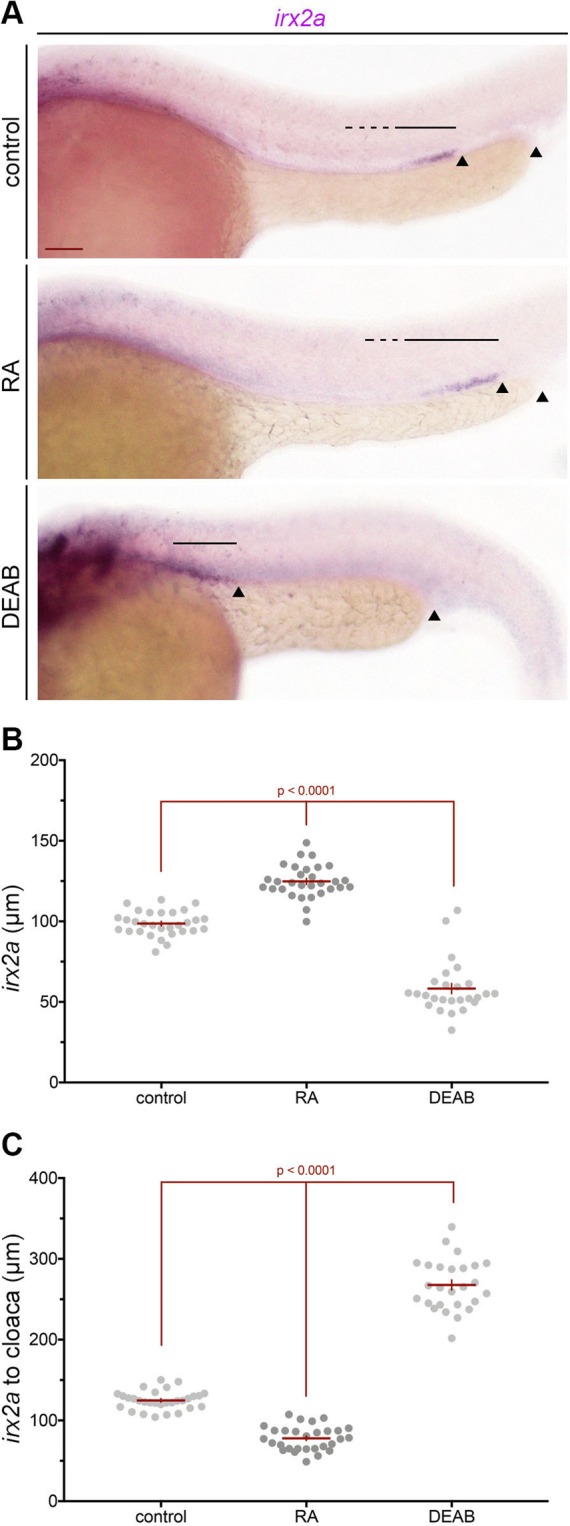
RA signaling regulates irx2a expression during nephrogenesis. (A) WISH analysis at the 28 ss demonstrates alterations in the irx2a pronephros domain after either treatment of RA or DEAB. The black arrowheads show the distance from distal irx2a expression to the cloaca, and the black lines highlight irx2a expression. (B) Quantification of irx2a length (μm) at the 28 ss. Each dot represents one pronephros. (C) Quantification of the length from irx2a expression to the cloaca (μm), where each dot represents one pronephros. For each graph, data is represented +/− SEM and significance was determined with an ANOVA. WISH – whole mount in situ hybridization, ss- somite stage, RA – retinoic acid, DEAB - N,N-diethylaminobenzaldehyde.
Discussion
There is an increasing appreciation of the genetic factors that control nephrogenesis processes across vertebrate species9,51–57. However, the molecular mechanisms that direct the specification of renal progenitors into the distinct cell populations that comprise each nephron segment is still not completely understood. Further defining the genetic mechanisms that govern renal organogenesis has direct relevance to deepening our knowledge about the mechanisms of kidney disease and regeneration58,59. The present work has enhanced our understanding of the renal transcription factor code that currently defines the zebrafish pronephros by defining several essential roles of irx2a within the regulatory networks that guide renal progenitor fate decisions (Fig. 7). Previous research has established that RA acts upstream of mecom and Notch signaling to influence MCC renal development in the zebrafish embryo, where etv5a is necessary for MCC fate downstream of RA35,36. In the present report, our results indicate that RA signaling regulates irx2a expression, either directly or indirectly, in the developing nephron, where irx2a is necessary for a normal balance of PST, DL and MCC development (Fig. 7). Given that irx2a transcripts are not expressed in the DL, it is intriguing that the knockdown of irx2a expanded the PST at the expense of the DL. Thus, it is possible that the effect on the DL may be an indirect consequence of altered proximal segment development in irx2a deficient embryos. With regard to MCC genesis, our results here are consistent with the conclusion that irx2a regulates etv5a during the specification of MCCs (Fig. 7). Again, the present study does not resolve whether these interactions are direct or indirect (Fig. 7). However, it is likely that irx2a influences additional targets during MCC genesis, as etv5a was only partially able to rescue MCC numbers in irx2a deficient embryos.
Figure 7.
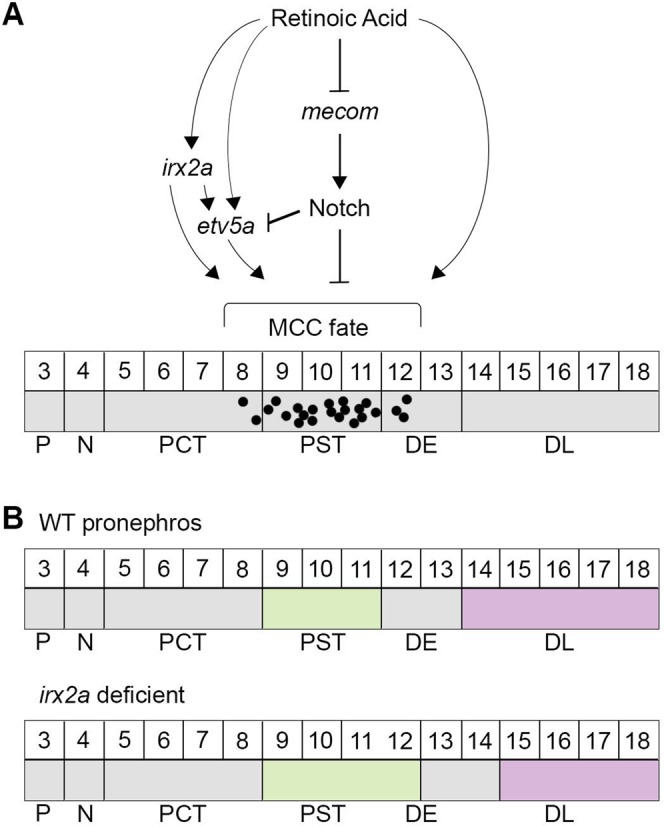
Model of irx2a function during segmentation and MCC specification during zebrafish nephrogenesis. (A) Renal MCC ontogeny is known to be reliant on RA signaling which acts upstream of the transcription factors mecom and etv5a, where negative regulation by Notch signaling restricts MCC fate choice. The present work adds to this model by demonstrating for the first time that irx2a functions downstream of RA signaling to regulate the expression of the etv5a transcription factor, thereby directing MCC fate. irx2a may act directly or indirectly on etv5a. irx2a likely acts on other targets as well to control MCC fate. (B) Diagram summarizing wild-type and irx2a morphant nephron segments with respect to their somite boundaries. MCC - multiciliated cell, P - podocytes, N - neck, PCT - proximal convoluted tububle, PST - proximal straight tubule, DE - distal early, DL - distal late, WT – wild-type.
Three ancestral Irx genes—araucan, caupolican, and mirror—were originally identified in Drosophila melanogaster where they comprise the ‘Iro-C complex’ that controls segmentation of the body and regulates later formation of sense organs, the thorax, and wing vasculature60,61. Irx gene functions have been ascribed to many aspects of vertebrate organogenesis as well37,38, including the kidney17,41–45,62. During Xenopus embryonic kidney development, Irx1 and Irx3 have an early role in maintaining the pronephric territory42, and subsequently Irx3 is essential for intermediate nephron tubule fate, in part by controlling expression of Irx1 and Irx2 in renal progenitors41. Interestingly, knockdown of Irx1 or Irx2 did not perturb nephron development, suggesting redundant functions for these genes during nephrogenesis41. In zebrafish, irx3b knockdown led to the abrogation of the DE with compensatory expansions of the PCT and PST, resulting in an overall increase in the proximal pronephros17. This data highlights a clear requirement for irx3b in DE segment development, and suggests that irx3b may also negatively modulate the PCT and PST segment domains, perhaps by regulating the position of the PCT-PST boundary17. As the current work indicates that irx2a negatively regulates PST size, assessment of irx2a/3b doubly deficient zebrafish embryos may provide further insights into segmentation mechanisms.
Further, the relationship between Irx2a and the transcription factor genes sim1a and ppargc1a will be necessary to define, as the proper balance of Sim1a/Ppargc1a is necessary to form the PST segment in the zebrafish pronephros63,64. With regard to the role of irx2a in regulating DL development: as noted above, this effect could be a secondary consequence of the PST phenotype, or could involve direct effects on distal renal progenitors. As the emx1 transcription factor was recently found to promote the DL and to be a negative regulator of irx3b and irx1a in the DE, it will be of interest to test if and how irx2a interfaces with this network65. irx2a deficiency also phenocopies the segment changes (PST & DL) observed in prostaglandin signaling gain of function studies66, and mecom loss of function studies35. Moreover, the tbx2a/2b transcription factors are requisite for DL development67. In the zebrafish kidney, expression of four other irx genes has been documented as well (irx1a in the DE; irx2a, irx4a, and irx5b in both the PST and DE), denoting these genes as potential regulators of these segments40,68. Determining the relationship of irx2a with these various irx family members, and exploring the hypothesis that functional redundancy may exist between these related genes in renal progenitors, will further our understanding of the networks that underlie pronephros segmentation.
In addition to the contrast between our findings and the Xenopus pronephros where Irx2 knockdown does not cause renal anomalies41,42, homozygous Irx2 mutant mice are healthy and lack obvious organ deficiencies69. Here, lacZ expression faithfully recapitulated the endogenous Irx2 domain in various tissues including the central nervous system, kidneys, pancreas, and lungs in Irx2LacZ/+ mice where the null allele was generated by targeted introduction of an IRES-lacZ reporter and neomycin resistance gene69. The intercross of these Irx2LacZ/+ mice produced progeny according to normal Mendelian ratios, and analysis of homozygous Irx2 mutants showed that the Irx2 transcript level was markedly reduced but did not result in protein production, signifying loss of function69. One explanation for the normal renal formation and function in Irx2 knockout mice may be functional redundancy with other Irx family members. Indeed, Irx1/2 were placed downstream of HNF1B during intermediate tubule development in the murine metanephros44,45. It is possible that evolutionary differences exist between Irx gene functions in zebrafish and other vertebrates including mammals70. Most vertebrates have six Irx genes that are organized into two clusters, IrxA and IrxB, arising from a duplication event that occurred early in their evolution71. Following teleost divergence from the tetrapod lineage, zebrafish underwent another whole genome duplication, which gave rise to four irx clusters and a single isolated gene locus. Of these, irx2b was deleted from the zebrafish genome72–77. Consequently, it is possible that neofunctionalization occurred where irx2a accrued adaptive mutations allowing it to perform unique functions during cell specification rather than dependently operating under the control of irx3b as the findings in the Xenopus studies predict41.
The current work implicates for the first time that an Irx transcription factor has essential functions in MCC development. MCCs have motile cilia and are responsible for stimulating fluid propulsion across tissues in a synchronized and directional manner78,79. In these MCCs, each cilium is characterized by a bundle of microtubules organized in a 9 + 2 configuration that is surrounded by a plasma membrane and anchored to the surface of the cell by a basal body78,79. The motility of a cilium is then stimulated by dynein arms located in between the nine microtubule doublets, resulting in a bending movement due to the action of these motor proteins78,79. In contrast, most cells in vertebrates contain a single, non-motile primary cilium that predominantly functions in signal transduction78,79. Despite the known significance of MCC function during development and homeostasis, the anatomical location of these intriguing cells in mammals throughout the brain ventricles and airways, for instance, has historically precluded them from comprehensive genetic examinations. In recent years, however, the application of functional genomics has led to numerous advancements due to the use of various animal and cell culture models79,80. Efforts to better define the transcriptional programs and ancillary modulating factors of MCC differentiation have established Notch signaling as an essential regulator of multiciliated fate choice23,24,81,82. This mechanism was found to be conserved between MCCs located in the zebrafish embryonic kidney, murine respiratory tract, and Xenopus epidermis, where the latter has also served as an excellent model for the live imaging of MCCs82–85. Furthermore, the microRNA, miR-499, was recently discovered to control the expression levels of Notch and Delta-like1, thereby contributing another layer of control to the pathways that influence MCC differentiation86. Additional studies in Xenopus and mice lead to the identification of a Notch target, Multicilin, which is a small coiled-coil protein that acts in a complex with the E2f4/5 transcription factors to regulate multiciliogenesis through the activation of key centriole replication genes87,88. Other research in these model organisms have identified that members of the Rfx family89, including Rfx2/3, C-Myb, and FoxJ1, operate in evolutionarily conserved functions downstream of Multicilin to promote the genesis of motile cilia80,81,90,91.
As we have now determined newfound roles for irx2a in MCC development (Fig. 7), irx2a is a promising candidate for further exploration in terms of conserved ciliary transcriptional programs. Epistasis experiments between irx2a and Notch signaling are necessary to understand their relationship during MCC emergence. Additionally, the expression of rfx2, foxj1a/b, gmnc, and mcidasl has been documented in the zebrafish pronephros and/or MCCs23,24,92. Studies should be conducted to examine possible relationships between these regulators of ciliogenesis and irx2a. Further, it will be valuable to ascertain whether Irx2a affects etv5a expression directly or indirectly, as etv5a was recently linked to the control of MCC differentiation by its regulation of prostaglandin signaling93. These genetic analyses will hold two-fold significance where they can help refine our understanding of the MCC molecular mechanisms of programming from precursor cells in the zebrafish kidney, and provide data for additional comparison of ciliary regulatory networks between species and tissues.
The importance of MCCs can be appreciated by their diverse roles in multiple human tissues without which dysfunction and disease would ensue78,79. With respect to the respiratory tract, MCCs promote the clearance of mucus, an important protective mechanism against pathogenic agents. Moreover, disruption of fluid depth between the mucosal layer and airway epithelium can lead to faulty mucus clearance by MCCs, and complicate respiratory diseases such as cystic fibrosis78,79. In the ventricles of the brain, MCCs enable the circulation of cerebrospinal fluid, while in the female reproductive system they facilitate the movement of the egg through the Fallopian tubes78,79. Failure of MCCs to perform these tasks increases the risk of hydrocephalus and infertility, respectively78,79.
In contrast, only a few isolated reports have documented the presence of MCCs in the human fetal kidney26,27 and in kidney disease states27–34—a stark contrast to the zebrafish kidney, where MCCs and the motile primary cilia on transportor cells are required for fluid propulsion33. It has been hypothesized that renal MCCs were evolutionarily lost due to the elevation of blood pressure in higher vertebrates, such as mammals, which negated the need for MCCs and motile primary cilia to regulate fluid flow33,34,94,95. Although MCCs are absent in human post-natal, healthy kidneys, their presence in the fetal kidney may suggest roles during early development. The manifestation of MCCs in human kidney disease, like nephrotic syndrome and renal sarcoidosis associated with hypercalcemia, also incites curiosity to the function of this cell type in these conditions. One hypothesis is that the kidney is reverting to a more primitive state in response to damage34. For example, the formation of cysts in the kidney could promote the reappearance of MCCs as a means to combat fluid accumulation. Consistent with this notion, the Foxj1 transcription network is upregluated after kidney injury in the zebrafish pronephros and in both mammalian cystic kidneys and following acute reperfusion injury96. The appearance of MCCs may thus constitute a regenerative response. Future study of MCC dynamics during renal regeneration in zebrafish could be used to assess their biological functions in this context as well97. While MCCs are differentially expressed between zebrafish and mammalian kidneys, it is likely that some degree of conservation exists between the transcriptional cascades that guide MCC formation among vertebrates. Therefore, the zebrafish pronephros is a useful setting for high-throughput analyses such as chemical screening98 to discover MCC relevant factors. Such future studies can advance our knowledge about ciliogenesis mechanisms, and provide insight into the etiology of ciliopathies while aiding the identification of potential therapeutic agents.
Materials and Methods
Zebrafish husbandry and ethics statement
Zebrafish were housed and cared for in the Center for Zebrafish Research at the University of Notre Dame Freimann Life Science Center, where the Institutional Animal Care and Use Committee approved the experiments documented here under protocol 16-025. All methods were carried out in accordance with relevant guidelines and regulations. Tübingen strain wild-type zebrafish were used, and staged as described99,100.
Expression analysis and image acquisition
Whole mount in situ hybridization (WISH) was conducted as described101,102. Anti-sense RNA probes were digoxigenin-labeled (etv5a, odf3b, cetn4, scl, irx2a, cdh17, slc20a1a, trmp7, slc12a1, slc12a3, jag2b) or fluorescein-labeled (smyhc1, cdh17, pax2a), and generated by in vitro transcription using plasmid or PCR templates as described16,17,36,63,103. Embryos were mounted in glycerol and images were taken using a Nikon Eclipse Ni with a DS-Fi2 camera. Whole mount fluorescent in situ hybridization (FISH) was performed as described103 with the digoxigenin and fluorescein-labeled RNA probes used in WISH. Stains were developed with the TSA plus Cy3 and TSA plus Fluorescein kits (Perkin Elmer). Embryos were mounted in Poly Aqua-mount as described104 and imaged on a Nikon C2 confocal microscope. Z-stacks were processed with FIJI into max image projections, and all figures were assembled using Adobe Photoshop CS5.
Genetic tools
An antisense morpholino (MO) was used to block irx2a splicingwith the following sequence48: 5′-ACGGAGAGCCCTTCAAAAATAAC-3′ (ZFIN Annotation MO2-irx2a) at 133 µM. The antisense standard control MO was used at was: 5′-CCTCTTACCTCAGTTACAATTTATA-3′ at 133 µM. MOs were synthesized and purified by Gene Tools, LLC (Philomath, OR), and resolubilized with DNase/RNase free water for storage at −20 °C. irx2asa10716 were obtained from the Zebrafish International Resource Center49. For genotyping irx2asa10716, we developed a restriction fragment length polymorphism assay. Genomic DNA was isolated from individual adult fins or embryos as described105. PCR amplification was performed with primers flanking the mutation site: 5′-GACCTTGTTTGCGATTCTGGAGCTGAAATC-3′ and 5′- GGGGTCCGAAGTGGCGATCTCTGCTAATGA-3′. The cycling conditions were as follows: initial 4 minute denaturation step at 94 °C followed by 45 cycles of 94 °C for 30 seconds, annealing for 30 seconds at 68 °C, 72 °C for 1 minute and a final extension step at 72 °C for 10 minutes. PCR products were purified using a Qiagen PCR Purification Kit and sent to the University of Notre Dame Genomics Core for sequencing. Restriction digest of the purified PCR product was performed using the enzyme Bts1Mut1 (NEB). The digest reaction (10 μL PCR product, 7 μL molecular dH2O, 2 μL CutSmart Buffer, 1 μL Bts1Mut1 enzyme) was incubated at 55 °C overnight. 10 μL of the digest product was then run on a 2% agarose gel.
RT-PCR verification of splice-blocking morpholinos
20–30 uninjected and injected embryos at the 28 ss were homogenized in 500 μL TRIZOL (Ambion) and RNA was isolated according to manufacturer insructions. PCR amplification was performed using the SuperScript® IV First-Strand Synthesis System (Invitrogen) according to manufacturer instructions with primers to amplify the region between irx2a exons 2 and 3: forward 5′-GACGAAGACGAAGATGACGGAGATG-3′, reverse 5′-CTCGCATTTGTCCTGGATTTCAGCT-3′. Products were isolated by agarose gel extraction (Qiagen QIAquick Gel Extraction) and sequenced.
cRNA synthesis and rescue experiments
The zebrafish irx2a open reading frame (ORF) was PCR amplified using high fidelity TAQ polymerase from the Expand PCR kit (Roche) in combination with a PCR Mix solution (100 mM dNTPs, 1 M MgCl2, 1 M Tris-HCl (pH 8.4), 4 M KCl, 1% Gelatin, 100 mg/mL BSA, and sterile H2O) and primers specific for irx2a: forward, 5′-ATTCGAATTCGCCGCCACC atgtcctatcctcagggttacctctaccagcccccgggctc-3′ and reverse, 5′-CGACCTCGAG ttaactcgacaggtaagattgggatcttactgtgaaaacctcgctggggt-3′. The forward primers were designed with a 4 bp anchor (bold print) followed by the 6 bp EcoR1 sequence (italicized), the Kozak consensus (underlined), and finally a sequence beginning at the start site (lowercase). Alternatively, the reverse primer contains the stop sequence (lowercase), 6 bp Xho1 sequence (italicized), and a 4 bp anchor (bold print). Amplified ORF was ligated into the pCS2 vector and transformed into DH5α competent cells (Invitrogen). Mutagenesis was performed on the irx2a.pCS2 construct to create irx2asa10776.pCS2 using the QuikChange Site Directed Mutagenesis Kit (Agilent Technologies, 200518-12) as previously described106. The mutagenesis primers were forward, 5′-GGTGACGCGCCTCACTGCCTTTCCTCC-3′ and reverse, 5′-GGAGGAAAGGCAGTGAGGCGCGTCACC-3′. etv5a was synthesized from a previously reported rescue construct template33. All capped RNA (cRNA) was synthesized in vitro using the mMESSAGE mMACHINE SP6 Transcription kit (Ambion) and stored at −80 °C. Overexpression experiments were performed by injecting 20 pg of irx2a cRNA into wild-type embryos. For irx2a and irx2asa10776 rescue experiments, 20 pg of cRNA was co-injected with 133 µM irx2a MO. For etv5a rescue experiments, a combination of 220 pg etv5a cRNA with 133 µM irx2a MO was injected into wild-type embryos. For all rescue studies, replicate group sizes were a minimum of 30 embryos, and typically ranged between 40–60 embryos for each cohort.
Quantification of phenotypes and statistical analysis
To measure domain length in the pronephros, embryos were mounted laterally in glycerol on a bridge slide. Either the polyline tool on the Nikon imaging software was used to trace the expression domain, or the segment line tool in FIJI. MCC cell number was assessed by viewing embryos dorsally at the highest magnification on a Nikon SMZ1000 stereomicroscope, and counting the individual odf3b+ cells in both nephrons93. Each experiments was completed in biological triplicate, with group sizes of at least n = 10 embryos, and confocal imaging was performed with group sizes of a least n = 3 representative embryos. ANOVA and student t-tests were used to assess statistical significance. Graphs were created using GraphPad Prism software.
Chemical treatments
all-trans RA and DEAB (Sigma-Aldrich) were dissolved in 100% DMSO to make 1 M stock solutions, as previously described16. For all chemical treatments, wild-type embryos were incubated and protected from light in vehicle control, 1 × 10−7 M RA/DMSO or 1.6 × 10−5 M DEAB/DMSO made with E3 from 60% epiboly to 24 hpf. The chemical was then washed off and the embryos were fixed in 4% PFA. These chemical treatments were fully penetrant and produced consistent results over three replicates with n = 40–60 embryos per replicate analyzed for these studies.
Supplementary information
Acknowledgements
NIH Grant R01DK100237 to RAW, and a NSF-GRFP to ANM supported this work. We are grateful to Elizabeth and Michael Gallagher for a generous gift to the University of Notre Dame on behalf of their family for the support of stem cell research. The funders had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation. We thank the staffs of the Department of Biological Sciences and the Center for Zebrafish Research at the University of Notre Dame for their dedication and care of our aquarium. Finally, we thank the members of our lab for support, discussions, and insights about this work.
Author Contributions
A.N.M., C.N.C., B.A. and R.A.W. designed the experiments. A.N.M., C.N.C., B.A., A.A. and R.A.W. performed experiments, collected and analyzed the data. H.M.W., B.E.C. and J.M.C. analyzed data and prepared figures. A.N.M., C.N.C. and R.A.W. prepared figures and wrote the manuscript.
Data Availability
The data associated with this report are provided in the figures and supplemental figures.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Amanda N. Marra and Christina N. Cheng contributed equally.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42943-y.
References
- 1.Saxen, L. Organogenesis of the kidney. (Cambridge University Press, 1987).
- 2.Romagnani P, Lasagni L, Remuzzi G. Renal progenitors: an evolutionary conserved strategy for kidney regeneration. Nat. Rev. Nephrol. 2013;9:137–146. doi: 10.1038/nrneph.2012.290. [DOI] [PubMed] [Google Scholar]
- 3.Kroeger PT, Jr., Wingert RA. Using zebrafish to study podocyte genesis during kidney development and regeneration. Genesis. 2014;52:771–792. doi: 10.1002/dvg.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McMahon AP. Development of the mammalian kidney. Curr. Top. Dev. Biol. 2016;117:31–64. doi: 10.1016/bs.ctdb.2015.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Little, M. H., Kumar, S. V. & Forbes, T. Recapitulating kidney development: Progress and challenges. Semin. Cell. Dev. Biol, 10.1016/j.semcdb.2018.08.015 (2018). [DOI] [PMC free article] [PubMed]
- 6.McCampbell KK, Wingert RA. Renal stem cells: fact or science fiction? Biochem. J. 2012;444:153–168. doi: 10.1042/BJ20120176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dressler GR. The cellular basis of kidney development. Annu. Rev. Cell Dev. Biol. 2006;22:509–529. doi: 10.1146/annurev.cellbio.22.010305.104340. [DOI] [PubMed] [Google Scholar]
- 8.Wingert RA, Davidson AJ. The zebrafish pronephros: a model to study nephron segmentation. Kidney Int. 2008;73:1120–1127. doi: 10.1038/ki.2008.37. [DOI] [PubMed] [Google Scholar]
- 9.Desgrange A, Cereghini S. Nephron patterning: lessons from Xenopus, zebrafish and mouse studies. Cells. 2015;4:483–499. doi: 10.3390/cells4030483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerlach GF, Wingert RA. Kidney organogenesis in the zebrafish: insights into vertebrate nephrogenesis and regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2013;2:559–85. doi: 10.1002/wdev.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Naylor RW, Qubisi SS, Davidson AJ. Zebrafish pronephros development. Results Probl. Cell Differ. 2017;60:27–53. doi: 10.1007/978-3-319-51436-9_2,. [DOI] [PubMed] [Google Scholar]
- 12.Poureetezadi SJ, Wingert RA. Little fish, big catch: zebrafish as a model for kidney disease. Kidney Int. 2016;89:1204–1210. doi: 10.1016/j.kint.2016.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morales EE, Wingert RA. Zebrafish as a model of kidney disease. Results Probl. Cell Differ. 2017;60:55–75. doi: 10.1007/978-3-319-51436-9_3. [DOI] [PubMed] [Google Scholar]
- 14.Elmonem MA, et al. Genetic renal diseases: the emerging role of zebrafish models. Cells. 2018;7:E130. doi: 10.3390/cells7090130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drummond IA, Davidson AJ. Zebrafish kidney development. Methods Cell Biol. 2016;134:391–429. doi: 10.1016/bs.mcb.2016.03.041. [DOI] [PubMed] [Google Scholar]
- 16.Wingert RA, et al. The cdx genes and retinoic acid control the positioning and segmentation of the zebrafish pronephros. PLoS Genet. 2007;3:1922–1938. doi: 10.1371/journal.pgen.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wingert RA, Davidson AJ. Zebrafish nephrogenesis involves dynamic spatiotemporal expression changes in renal progenitors and essential signals from retinoic acid and irx3b. Dev. Dyn. 2011;240:2011–2027. doi: 10.1002/dvdy.22691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naylor RW, et al. BMP and retinoic acid regulate anterior-posterior patterning of the non-axial mesoderm across the dorsal-ventral axis. Nat. Commun. 2016;7:12197. doi: 10.1038/ncomms12197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerlach GF, Wingert RA. Zebrafish pronephros tubulogenesis and epithelial identity maintenance are reliant on the polarity proteins Prkc iota and zeta. Dev. Biol. 2014;396:183–200. doi: 10.1016/j.ydbio.2014.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McKee R, Gerlach GF, Jou J, Cheng CN, Wingert RA. Temporal and spatial expression of tight junction genes during zebrafish pronephros development. Gene Expr. Patterns. 2014;16:104–113. doi: 10.1016/j.gep.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drummond IA, et al. Early development of the zebrafish pronephros and analysis of mutations affecting pronephric function. Development. 1998;125:4655–4667. doi: 10.1242/dev.125.23.4655. [DOI] [PubMed] [Google Scholar]
- 22.Kramer-Zucker AG, et al. Organization of the pronephric filtration apparatus in zebrafish requires Nephrin, Podocin and the FERM domain protein Mosaic eyes. Dev. Biol. 2005;285:316–329. doi: 10.1016/j.ydbio.2005.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Y, Narendra P, Kramer-Zucker A, Drummond IA. Notch signaling controls the differentiation of transporting epithelia and multiciliated cells in the zebrafish pronephros. Development. 2007;134:1111–1122. doi: 10.1242/dev.02806. [DOI] [PubMed] [Google Scholar]
- 24.Ma M, Jiang YJ. Jagged2a-Notch signaling mediates cell fate choice in the zebrafish pronephric duct. PLoS. Genet. 2007;3:e18. doi: 10.1371/journal.pgen.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kramer-Zucker AG, et al. Cilia-driven fluid flow in the zebrafish pronephros, brain and Kupffer’s vesicle is required for normal organogenesis. Development. 2005;132:1907–1921. doi: 10.1242/dev.01772. [DOI] [PubMed] [Google Scholar]
- 26.Zimmerman DH. Cilia in the fetal kidney of man. Beitr. Pathol. 1971;143:227–240. [PubMed] [Google Scholar]
- 27.Katz SM, Morgan JJ. Cilia in the human kidney. Ultrastruct. Pathol. 1984;6:285–294. doi: 10.3109/01913128409018587. [DOI] [PubMed] [Google Scholar]
- 28.Duffy JL, Suzuki Y. Ciliated human renal proximal tubular cells. Observations in three cases of hypercalcemia. Am. J. Pathol. 1968;53:609–616. [PMC free article] [PubMed] [Google Scholar]
- 29.Datsis SA, Boman IA. Ciliated renal tubular epithelium in congenital nephrosis. Beitr. Pathol. 1974;151:297–303. doi: 10.1016/S0005-8165(74)80006-5. [DOI] [PubMed] [Google Scholar]
- 30.Larsen TE, Ghadially FN. Cilia in lupus nephritis. J. Pathol. 1974;114:69–73. doi: 10.1002/path.1711140203. [DOI] [PubMed] [Google Scholar]
- 31.Lungarella G, de Santi MM, Tosi P. Ultrastructural study of the ciliated cells from renal tubular epithelium in acute progressive glomerulonephritis. Ultrastruct. Pathol. 1984;6:1–7. doi: 10.3109/01913128409016659. [DOI] [PubMed] [Google Scholar]
- 32.Ong AC, Wagner B. Detection of proximal tubular motile cilia in a patient with renal sarcoidosis associated with hypercalcemia. Am. J. Kidney Dis. 2005;45:1096–1099. doi: 10.1053/j.ajkd.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 33.Marra AN, Li Y, Wingert RA. Antennas of organ morphogenesis: the roles of cilia in vertebrate kidney development. Genesis. 2016;54:457–469. doi: 10.1002/dvg.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spassky N, Meunier A. The development and functions of multiciliated epithelia. Nat. Rev. Mol. Cell Biol. 2017;18:423–436. doi: 10.1038/nrm.2017.21. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, Cheng CN, Verdun VA, Wingert RA. Zebrafish nephrogenesis is regulated by interactions between retinoic acid, mecom, and Notch signaling. Dev. Biol. 2014;386:111–122. doi: 10.1016/j.ydbio.2013.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marra AN, Wingert RA. Epithelial cell fate in the nephron tubule is mediated by the ETS transcription factors etv5a and etv4 during zebrafish kidney development. Dev. Biol. 2016;411:231–245. doi: 10.1016/j.ydbio.2016.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavodeassi F, Modolell J, Gómez-Skarmeta JL. The Iroquois family of genes: from body building to neural patterning. Development. 2001;128:2847–2855. doi: 10.1242/dev.128.15.2847. [DOI] [PubMed] [Google Scholar]
- 38.Gómez-Skarmeta JL, Modolell J. Iroquois genes: genomic organization and function in vertebrate neural development. Curr. Opin. Genet. Dev. 2002;12:403–408. doi: 10.1016/S0959-437X(02)00317-9. [DOI] [PubMed] [Google Scholar]
- 39.Cheng CW, Hui C, Strähle U, Cheng SH. Identification and expression of zebrafish Iroquois homeobox gene irx1. Dev. Genes. Evol. 2001;211:442–444. doi: 10.1007/s004270100168. [DOI] [PubMed] [Google Scholar]
- 40.Lecauday V, Anselme I, Dildrop R, Rüther U, Schneider-Maunoury S. Expression of the Iroquois genes during early nervous system formation and patterning. J. Comp. Neurol. 2005;492:289–302. doi: 10.1002/cne.20765. [DOI] [PubMed] [Google Scholar]
- 41.Reggiani L, Raciti D, Airik R, Kispert A, Brändli AW. The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev. 2007;21:2358–2370. doi: 10.1101/gad.450707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alarcon P, et al. A dual requirement for Iroquois genes during Xenopus kidney development. Development. 2008;135:3197–3207. doi: 10.1242/dev.02397,. [DOI] [PubMed] [Google Scholar]
- 43.Marra AN, Wingert RA. Roles of Iroquois transcription factors in kidney development. Cell Dev. Biol. 2014;3:1000131. doi: 10.4172/2168-9296.1000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heliot C, et al. HNF1B controls proximal-intermediate nephron segment identity in vertebrates by regulating Notch signalling components and Irx1/2. Development. 2013;140:873–885. doi: 10.1242/dev.086538. [DOI] [PubMed] [Google Scholar]
- 45.Massa F, et al. Hepatocyte nuclear factor 1β controls nephron tubular development. Development. 2013;140:886–896. doi: 10.1242/dev.086546. [DOI] [PubMed] [Google Scholar]
- 46.Naylor RW, Chang HG, Qubisi S, Davidson AJ. A novel mechanism of gland formation in zebrafish involving transdifferentiation of renal epithelial cells and live cell extrusion. Elife. 2018;7:e38911. doi: 10.7554/eLife.38911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Horsfield J, et al. Cadherin-17 is required to maintain pronephric duct integrity during zebrafish development. Mech. Dev. 2002;115:15–26. doi: 10.1016/S0925-4773(02)00094-1. [DOI] [PubMed] [Google Scholar]
- 48.Choy SW, et al. A cascade of irx1a and irx2a controls shh expression during retinogenesis. Dev. Dyn. 2010;239:3204–3214. doi: 10.1002/dvdy.22462. [DOI] [PubMed] [Google Scholar]
- 49.Busch-Nentwich, E. et al. Sanger Institute Zebrafish Mutation Project mutant data submission. ZFIN Direct Data Submission, http://zfin.org (2013).
- 50.Kok FO, et al. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev. Cell. 2015;32:97–108. doi: 10.1016/j.devcel.2014.11.018.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cheng CN, Verdun V, Wingert RA. Recent advances in elucidating the genetic mechanisms of nephrogenesis using zebrafish. Cells. 2015;4:218–233. doi: 10.3390/cells4020218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grinstein M, Yelin R, Herzlinger D, Schultheiss TM. Generation of the podocyte and tubular components of an amniote kidney: timing of specification and a role for Wnt signaling. Development. 2013;140:4565–4573. doi: 10.1242/dev.097063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schneider J, Arraf AA, Grinstein M, Yelin R, Schultheiss TM. Wnt signaling orients the proximal-distal axis of chick kidney nephrons. Development. 2015;142:2686–2695. doi: 10.1242/dev.123968. [DOI] [PubMed] [Google Scholar]
- 54.Lindström NO, et al. Integrated Β-Catenin, BMP, PTEN, and Notch signalling patterns the nephron. Elife. 2015;3:e04000. doi: 10.7554/eLife.04000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Basta JM, Robbins L, Denner DR, Kolar GR, Rauchman MA. Sall1-NuRD interaction regulates multipotent nephron progenitors and is required for loop of Henle formation. Development. 2017;144:3080–3094. doi: 10.1242/dev.148692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chung E, Deacon P, Park JS. Notch is required for the formation of all nephron segments and primes nephron progenitors for differentiation. Development. 2017;144:4530–4539. doi: 10.1242/dev.156661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lindström NO, et al. Conserved and divergent features of human and mouse kidney organogenesis. J. Am. Soc. Nephrol. 2018;29:785–805. doi: 10.1681/ASN.2017080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schedl A. Renal abnormalities and their developmental origin. Nat. Rev. Genet. 2007;8:791–802. doi: 10.1038/nrg2205. [DOI] [PubMed] [Google Scholar]
- 59.Chambers BE, Wingert RA. Renal progenitors: roles in kidney disease and regeneration. World J. Stem Cells. 2016;8:367–375. doi: 10.4252/wjsc.v8.i11.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Leyns L, Gómez-Skarmeta JL, Dambly-Chaudiere C. Iroquois: a prepattern gene that controls the formation of bristles on the thorax of Drosophila. Mech. Dev. 1996;59:63–71. doi: 10.1016/0925-4773(96)00577-1. [DOI] [PubMed] [Google Scholar]
- 61.Gómez-Skarmeta JL, Modolell J. araucan and caupolican provide a link between compartment divisions and patterning of sensory organs and veins in the Drosophila wing. Genes Dev. 1996;10:2935–2946. doi: 10.1101/gad.10.22.2935. [DOI] [PubMed] [Google Scholar]
- 62.Holmquist Mengelbier Linda, Lindell-Munther Simon, Yasui Hiroaki, Jansson Caroline, Esfandyari Javanshir, Karlsson Jenny, Lau Kimberly, Hui Chi-chung, Bexell Daniel, Hopyan Sevan, Gisselsson David. The Iroquois homeobox proteins IRX3 and IRX5 have distinct roles in Wilms tumour development and human nephrogenesis. The Journal of Pathology. 2018;247(1):86–98. doi: 10.1002/path.5171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cheng CN, Wingert RA. Nephron proximal tubule patterning and corpuscles of Stannius formation are regulated by the sim1a transcription factor and retinoic acid in zebrafish. Dev. Biol. 2015;399:100–116. doi: 10.1016/j.ydbio.2014.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chambers JM, Poureetezadi SJ, Addiego A, Lahne M, Wingert RA. ppargc1a controls nephron segmentation during zebrafish embryonic kidney ontogeny. Elife. 2018;7:e40266. doi: 10.7554/eLife.40266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morales EE, et al. Homeogene emx1 is required for nephron distal segment development in zebrafish. Sci. Rep. 2018;8:18038. doi: 10.1038/s41598-018-36061-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Poureetezadi SJ, et al. Prostaglandin signaling regulates nephron segment patterning of renal progenitors during zebrafish kidney development. Elife. 2016;5:e17551. doi: 10.7554/eLife.17551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Drummond BE, Li Y, Marra AN, Cheng CN, Wingert RA. The tbx2a/b transcription factors direct pronephros segmentation and corpuscle of Stannius formation in zebrafish. Dev. Biol. 2017;421:52–66. doi: 10.1016/j.ydbio.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Thisse, B. & Thisse, C. Fast release clones: a high throughput expression analysis. ZFIN Direct Data Submission, http://zfin.org (2004).
- 69.Lebel M, et al. The Iroquois homeobox gene Irx2 is not essential for normal development of the heart and midbrain-hindbrain boundary in mice. Mol. Cell Biol. 2003;23:8216–8225. doi: 10.1128/MCB.23.22.8216-8225.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dildrop R, Ruther U. Organization of Iroquois in fish. Dev. Genes Evol. 2004;214:267–276. doi: 10.1007/s00427-004-0402-8. [DOI] [PubMed] [Google Scholar]
- 71.Houweling AC, et al. Gene and cluster-specific expression of the Iroquois family members during mouse development. Mech. Dev. 2001;107:169–174. doi: 10.1016/S0925-4773(01)00451-8. [DOI] [PubMed] [Google Scholar]
- 72.Amores A, et al. Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- 73.Postlethwait JH, et al. Vertebrate genome evolution and the zebrafish gene map. Nat. Genet. 1998;18:345–349. doi: 10.1038/ng0498-345. [DOI] [PubMed] [Google Scholar]
- 74.Meyer A, Schartl M. Gene and genome duplications in vertebrates: the one-to-four (to-eight in fish) rule and the evolution of novel gene functions. Curr. Opin. Cell Biol. 1999;11:699–704. doi: 10.1016/S0955-0674(99)00039-3. [DOI] [PubMed] [Google Scholar]
- 75.Aparicio S. Vertebrate evolution: recent perspectives from fish. Trends Genet. 2000;16:54–56. doi: 10.1016/S0168-9525(99)01934-4. [DOI] [PubMed] [Google Scholar]
- 76.Taylor JS, de Peer V, Braasch Y, Meyer I. A. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2001;356:1661–1679. doi: 10.1098/rstb.2001.0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 78.Brooks ER, Wallingford JB. Multiciliated cells. Curr. Biol. 2014;24:R973–R982. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meunier A, Azimzadeh J. Multiciliated cells in animals. Cold Spring Harb. Perspect. Biol. 2016;8:a028233. doi: 10.1101/cshperspect.a028233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Choksi SP, Lauter G, Swoboda P, Roy S. Switching on cilia: transcriptional networks regulating ciliogenesis. Development. 2014;141:1427–1441. doi: 10.1242/dev.074666. [DOI] [PubMed] [Google Scholar]
- 81.Chung MI, et al. Coordinated genomic control of ciliogenesis and cell movement by RFX2. Elife. 2014;3:e01439. doi: 10.7554/eLife.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Deblandre GA, Wettstein DA, Koyano-Nakagawa N, Kintner C. A two-step mechanism generates the spacing pattern of the ciliated cells in the skin of Xenopus embryos. Development. 1999;126:4715–4728. doi: 10.1242/dev.126.21.4715. [DOI] [PubMed] [Google Scholar]
- 83.Tsao P, et al. Notch signaling controls the balance of ciliated and secretory cell fates in developing airways. Development. 2009;136:2297–2307. doi: 10.1242/dev.034884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stubbs JL, Davidson L, Keller R, Kintner C. Radial intercalation of ciliated cells during Xenopus skin development. Development. 2006;133:2507–2515. doi: 10.1242/dev.02417. [DOI] [PubMed] [Google Scholar]
- 85.Morimoto M, et al. Canonical Notch signaling in the developing lung is required for determination of arterial smooth muscle cells and selection of Clara versus ciliated cell fate. J. Cell. Sci. 2010;123:213–224. doi: 10.1242/jcs.058669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marcet B, et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat. Genet. 2011;13:693–699. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 87.Stubbs JL, Vladar EK, Axelrod JD, Kintner C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 2012;14:140–147. doi: 10.1038/ncb2406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ma L, Quigley I, Omran H, Kintner C. Multicilin drives centriole biogenesis via E2f proteins. Genes Dev. 2014;28:1461–1471. doi: 10.1101/gad.243832.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chung MI, et al. RFX2 is broadly required for ciliogenesis during vertebrate development. Dev. Biol. 2012;363:155–165. doi: 10.1016/j.ydbio.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Baas D, et al. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. Eur. J. Neurosci. 2006;24:1020–1030. doi: 10.1111/j.1460-9568.2006.05002.x. [DOI] [PubMed] [Google Scholar]
- 91.Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiro SD. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am. J. Respir. Cell Mol. Biol. 2000;23:45–51. doi: 10.1165/ajrcmb.23.1.4070. [DOI] [PubMed] [Google Scholar]
- 92.Zhou F, et al. Gmnc is a master regulator of the multiciliated cell differentiation program. Curr. Biol. 2015;25:3267–3273. doi: 10.1016/j.cub.2015.10.062. [DOI] [PubMed] [Google Scholar]
- 93.Marra, A. M. et al. Prostaglandin signaling regulates renal multiciliated cell specification and maturation. Proc. Natl. Acad. Sci. USA10.1073/pnas.1813492116 (2019). [DOI] [PMC free article] [PubMed]
- 94.Marshall EK., Jr. The comparative physiology of the kidney in relation to theories of renal secretion. Physiol. Rev. 1934;14:133–159. doi: 10.1152/physrev.1934.14.1.133. [DOI] [Google Scholar]
- 95.Vu HTK, et al. Stem cells and fluid flow drive cyst formation in an invertebrate excretory organ. ELife. 2015;4:e07405. doi: 10.7554/eLife.07405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hellman NE, et al. The zebrafish foxj1a transcription factor regulates cilia function in response to injury and epithelial stretch. Proc. Natl. Acad. Sci. USA. 2010;107:18499–18504. doi: 10.1073/pnas.1005998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.McCampbell KK, Springer KN, Wingert RA. Atlas of cellular dynamics during zebrafish adult kidney regeneration. Stem Cells Int. 2015;2015:547636. doi: 10.1155/2015/547636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Poureetezadi SJ, Donahue EK, Wingert RA. A manual small molecule screen approaching high-throughput using zebrafish embryos. J. Vis. Exp. 2014;93:e52063. doi: 10.3791/52063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Westerfield, M. The Zebrafish Book. (University of Oregon Press, Eugene, 1993).
- 100.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 101.Galloway JL, et al. Combinatorial regulation of novel erythroid gene expression in zebrafish. Exp. Hematol. 2008;36:424–432. doi: 10.1016/j.exphem.2007.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng CN, et al. Flat mount preparation for observation and analysis of zebrafish embryo specimens stained by whole mount in situ hybridization. J. Vis. Exp. 2014;89:51604. doi: 10.3791/51604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lengerke C, et al. Interactions between Cdx genes and retinoic acid modulate early cardiogenesis. Dev. Biol. 2011;163:134–142. doi: 10.1016/j.ydbio.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Marra AN, et al. Visualizing multiciliated cells in the zebrafish through a combined protocol of whole mount fluorescent in situ hybridization and immunofluorescence. J. Vis. Exp. 2017;129:e56261. doi: 10.3791/56261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kroeger PT, Jr., et al. The zebrafish kidney mutant zeppelin reveals that brca2/fancd1 is essential for pronephros development. Dev. Biol. 2017;428:148–163. doi: 10.1016/j.ydbio.2017.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wingert RA, et al. Deficiency of glutaredoxin 5 reveals Fe-S clusters are required for vertebrate haem synthesis. Nature. 2005;436:1035–39. doi: 10.1038/nature03887. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data associated with this report are provided in the figures and supplemental figures.