Abstract
The mammalian olfactory system uses hundreds of specialized G-protein-coupled olfactory receptors (ORs) to discriminate a nearly unlimited number of odorants. Cognate agonists of most ORs have not yet been identified and potential non-olfactory processes mediated by ORs are unknown. Here, we used molecular modeling, fingerprint interaction analysis and molecular dynamics simulations to show that the binding pocket of the prototypical olfactory receptor Olfr73 is smaller, but more flexible, than binding pockets of typical non-olfactory G-protein-coupled receptors. We extended our modeling to virtual screening of a library of 1.6 million compounds against Olfr73. Our screen predicted 25 Olfr73 agonists beyond traditional odorants, of which 17 compounds, some with therapeutic potential, were validated in cell-based assays. Our modeling suggests a molecular basis for reduced interaction contacts between an odorant and its OR and thus the typical low potency of OR-activating compounds. These results provide a proof-of-principle for identifying novel therapeutic OR agonists.
Subject terms: Computational biophysics, Virtual drug screening, Drug screening, Protein function predictions
Shuguang Yuan et al. report molecular modeling of the prototypical olfactory receptor Olfr73 and find that the binding pocket is much smaller but more flexible than that of typical G-protein-coupled receptors. They virtually screen 1.6 million compounds and predict 25 Olfr73 agonists, 17 of which are validated by cell-based assays.
Introduction
The mammalian olfactory system has the intriguing capability of sensing and distinguishing a huge variety of chemical compounds, often with a considerable sensitivity and specificity1–3. The perception of smell of volatile molecules starts with the activation of olfactory receptors (ORs), translating the environmental chemical signals with the involvement of heterotrimeric G proteins into neuronal electrical responses. ORs are assumed to fold into a seven-transmembrane (7TM) helix structure establishing the largest class of G-protein-coupled receptors (GPCRs)2. More than 1000 ORs in mice and about 350 ORs in humans are dedicated to smell3, capable of detecting an enormous repertoire of chemical compounds by a combinatorial coding scheme. Typically, one OR recognizes multiple odorants and one odorant is recognized by multiple ORs, but different odorants are recognized by different combinations of ORs1,3,4. Although some progress in functional identification of ligands for ORs has been achieved in recent years, most ORs are orphan receptors, whose ligand repertoires remain to be determined5–8. This, together with the lack of high-resolution 3D structures of ORs, is the major reason why the fundamental mechanistic principles of odorant recognition and discrimination by ORs are largely unresolved at a molecular level. This is in contrast to non-olfactory GPCRs for which an increasing number of high-resolution 3D structures have fostered understanding of ligand/receptor interactions, of ligand-mediated transmembrane signaling, and of how they serve as critical templates for rational drug design9.
In the case of missing experimental structural and functional data, computer modeling can deliver reliable propositions on ligand structures, which may interact and activate a specific GPCR10–14. As ORs share similarities with the structure and function of class A GPCRs15, known structures of class A GPCRs have been used as templates for homology modelling of ORs. Combined with functional data on odorant-induced activation of wild-type and mutant ORs, computer modelling have generated reliable propositions for the structure of the ligand-binding sites and specific ligand-binding modes of particular ORs thereby gaining insight into odorant selectivity16–21.
The large number of ORs and their promiscuity in ligand-binding related to the combinatorial code opens a huge chemical range of OR specific agonists and antagonists7,22,23. Estimates under discussion range from thousands to billions of OR-activating compounds1,3,24–26. Decoding the large odorant library would be of interest for several reasons. First, to understand the fundamental principles of the combinatorial code of ligand binding to ORs, and how ligand binding eventually activates or suppresses signal transmission to the intracellular side of a particular olfactory receptor and finally induce neuronal responses. Second, to understand better olfaction related diseases and to find novel therapeutic strategies for their treatment. As an example, the correlation between the appearance of neurodegenerative diseases and dysfunction of the olfactory system is well known27. Interestingly, ORs are expressed in many non-olfactory tissues including, testis, tongue, heart, spleen, pancreas, lung, kidney and placenta28,29. This suggests that ORs influence non-olfactory processes and there are indications for that in sperm chemotaxis30, embryonic development and cell–cell recognition31, chemosensory functions in kidney32, proliferation of cancer cells33,34, and other metabolic processes intimately linked with the endocrine systems35 that regulate the bodies energy balance, revealing a connection between the appearance of diabetes and the dysfunction of the olfactory system36–38. To date mostly synthetic odorants have been identified to activate ectopically expressed ORs, for instance, in wound healing processes39 or by influencing gut motility through OR activation and the release of serotonin in human gastrointestinal enterochromaffin cells40. In pathologic processes synthetic odorant molecules were shown to influence different stages of cancer development41,42. More recent findings suggest that ORs might be targetable not only by classical odorant molecules but even by therapeutic compounds. The anesthetic drug ketamine, which elicits various neuropharmacological effects, including sedation, analgesia, and anti-depressant activity, has been shown to activate odorant receptors in olfactory sensory neurons and in the mouse brain43. In the present study we find that Olfr73 can also be a target of drug-like molecules. Such findings are important to unravel the poly-pharmacology of therapeutic agents interacting with multiple intended but also unintended targets44.
On the other hand, OR-activating compounds have been found to also activate non-olfactory receptors45. A better understanding of the odorant-OR structure–activity relationship will have impact beyond medical applications, including production of fragrances, perfumes, food and beverages, and offering new ways of rodent and non-insect pest control, to mention a few7,46.
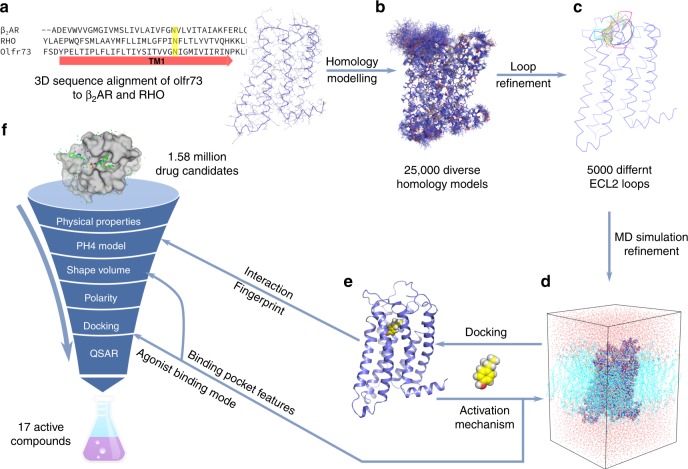
Here we report on a computer-directed structure-function study to discover fundamental principles of how odorants bind to a prototypical olfactory receptor and to find subsequently new receptor-activating compounds beyond traditional chemical odorant libraries. For our study, we have chosen the mouse eugenol olfactory receptor Olfr73, as this receptor has been functionally expressed in heterologous mammalian cells for compound screening16,17,47. Figure 1 outlines the approach of our study. First, we built a homology model of the structure of Olfr73 and refined it by molecular dynamics simulations. Second, we used ligand docking to identify interaction fingerprints of various agonists in the receptor’s binding pocket, which together with molecular dynamics simulations of the agonist-receptor complexes revealed structural details on the first steps of receptor activation following ligand binding. Finally, the structural models served as a reliable base to perform a virtual computer screening of the ZINC library of 1.6 million drug-like molecules. The in silico screen selected from the large compound library considerably reduced the number of potential OR-activating compounds out of which a high percentage showed functional activity in cell-based assays. This result underscores the capacity of our in silico approach for rapid identification of potentially activating compounds out of large chemical libraries. Combining the 25 already known agonists with the 17 newly identified compounds establishes one of the largest collections of active compounds for a specific OR from which substantially improved conclusions on the characteristics of ligand–OR interactions can be extracted.
Fig. 1.
Workflow of virtual screening for new agonists of Olfr73. a Homology modelling of Olfr73 based on 3D structure-sequence alignment of Olfr73 to β2AR and Rho. b Refinement of ECL2 loop. c Molecular dynamics (MD) simulations of agonist bound Olfr73. d Docking agonist molecule into the 3D structural model of Olfr73. e Interaction fingerprint analysis by docking 25 reported compounds. This information was subsequently used for PH4 model building in virtual screening. f Virtual screening for Olfr73 agonists in the ZINC compound library composed of 1.58 million drug candidates. Applying stepwise selection filtering based on shape volume, ionization penalty and polarity, downscaled the chemical library successively to 204 compounds. The shape volume features are deduced from the results of MD simulations. Finally, quantitative structure–activity relationship evaluations reduced the chemical library to 64 compounds with predicted potential to activate Olfr73. Out of this final list, agonist binding modes were verified manually based on the activation mechanism deduced from MD simulations, and 25 available compounds were tested by cellular functional assays yielding 17 active compounds
Results
Homology models of Olfr73
To obtain a reliable 3D model of Olfr73 through the initial homology modelling, we first compared the sequence of Olfr73 with sequences of other class A GPCRs in the PDB database. We found that it shared in the best case 19% sequence identity with beta-2-adrenergic receptor (β2AR, pdb code: 4LDE)48 and 16% sequence identity with rhodopsin (RHO, pdb code: 4BEY)49. Since multiple modelling templates can considerably improve the reliability of homology models50–52, we used the crystal structures of both receptors as templates for model building. 3D sequence alignment (Supplementary Fig. 1) indicates that most highly conserved residues/motifs in class A GPCRs9 including N1.50, D2.50, DRY motif, W4.50, Y5.58, F6.44 and NPxxY motif are also present in Olfr7353. However, residue P5.50 and motif CWxP that are typically found in non-OR class A GPCRs are missing in Olfr73. Moreover, one gap in TM3 is observed between the sequence of C3.25 and DRY motif of Olfr73 (Supplementary Fig. 1).
Interaction fingerprints (IFP) between agonists and Olfr73
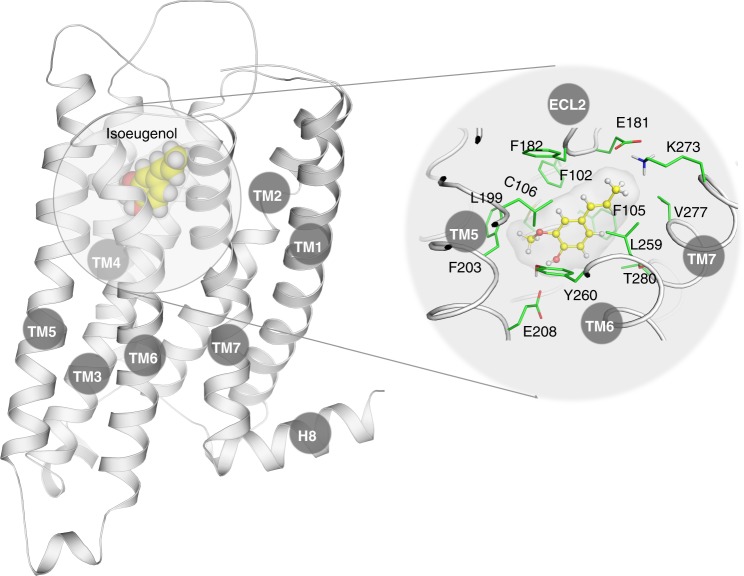
The final refined homology model of Olfr73 (Fig. 2) shared many of the common features of non-OR class A GPCRs outlined elsewhere9,54. We then docked isoeugenol, a potent agonist for Olfr7347, into the predicted extracellular ligand-binding pocket to explore atomistic details of the interaction between the ligand and its receptor. As depicted in Fig. 2, the hydrophobic moiety of isoeugenol is surrounded by several aromatic residues including F1023.30, F1053.33, F182ECL2, F2035.42, Y2606.52 which were also found by functional analysis of mutant receptors to play a crucial role in agonist binding47. Furthermore, the three non-aromatic hydrophobic residues L1995.38, L2596.51 and V2777.39 are contacting the agonist molecule. The hydroxyl group in isoeugenol forms an H-bond with Y2606.52, which in turn forms an H-bond with E2085.47 and water mediated hydrogen bonding to S1133.41. Both Y2606.52, E2085.47 and S1133.41 had been shown elsewhere to be important for Olfr73 activation47.
Fig. 2.
The 3D structural model of Olfr73 (left) and enlarged view of the binding mode of the agonist isoeugenol (right). Amino acid side chains in contact with bound isoeugenol are shown in green
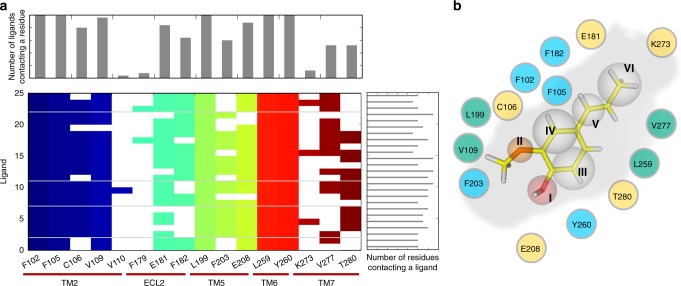
To further validate these observations, we performed an IFP analysis (Fig. 3a), which encodes specific interactions between a particular ligand and specific amino acids in the binding pocket. IFP analyses have been used for computational drug discovery for non-olfactory GPCRs55. Here we docked 25 previously reported Olfr73 agonist molecules16,45 (Fig. 4) into the binding pocket of Olfr73 and obtained the interaction fingerprints of the different agonists with residues in the binding pocket (Fig. 3a). The IFP analysis showed that all docked agonists could interact with five residues in the receptor’s binding pocket including F1023.30, F1053.33, L1995.38, L2596.51 and Y2606.52. Furthermore, C1063.34(80%), V1093.37(96%), E181ECL2(80%), F182ECL2(64%), F2035.42(60%), E2085.47(88%), V2777.39(52%) and T2807.42(52%) are also found frequently (percentage in parentheses) contacting the agonists (Fig. 3a, top histogram). In addition, the three residues V1103.38(5%), F179ECL2(8%) and K2737.35(12%) were found sometimes in contact with the agonists. Each particular ligand was found interacting with at least 80% of all the residues in the binding pocket (Fig. 3a, right histogram). Most of these mentioned residues were found by functional analysis of mutant receptors to play a crucial role in agonist binding47.
Fig. 3.
Interaction fingerprints of 25 known Olfr73 agonists grouped in classes 1–5 according to Fig. 4. a In the interaction histogram, each contact of a particular residue with the ligand is indicated by a color. The color code distinguishes the residue location in a particular TM helix. Each class of compounds is separated by a horizontal gray line. b The pharmacophore model based on 25 known Olfr73 agonists. As a prototypical example, the position of isoeugenol in the Olfr73 binding pocket showing the interaction fingerprint. Assignments: H-bond donor (I), H-bond acceptor (II), hydrophobic moiety (III, IV, V, VI); polar residues (yellow), aromatic residues (cyan), hydrophobic residues (green)
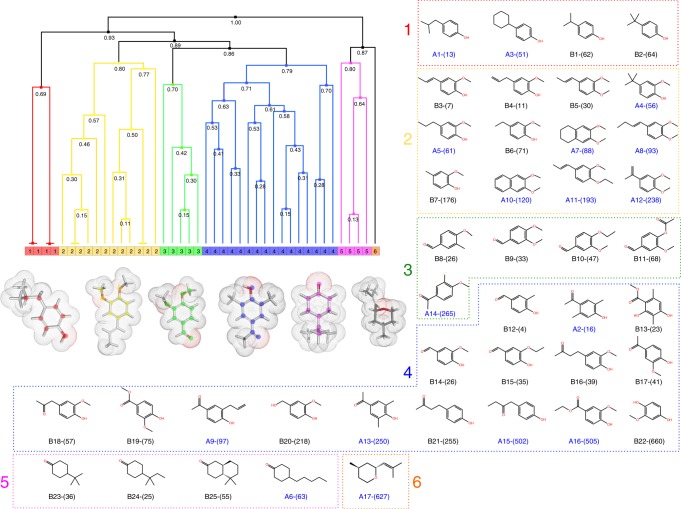
Fig. 4.
Hierarchical clustering of Olfr73 agonist molecules. Six different classes of agonists are identified (distinguished by a color code) according to their PH4 features. In the Hierarchical diagram, the links between the chemical compounds are represented as branched vertical lines. The height of the lines, coupled with merging distance (numbers showed in each node), indicate the normalized dissimilarity distance between the adjacent compounds. A higher line or a larger merging distance denotes a larger dissimilarity. A typical representative molecular structure of each class is shown below the dendrogram together with their molecular surfaces indicating hydrophobic moieties in grey and polar moieties in red. The commonly shared atoms within a certain class of molecules are labeled with colored dots accordingly. The molecular structures of the six classes of agonists are grouped in boxes. The 17 newly found agonists are represented as A1-A17 in blue. The 25 previously reported agonists are represented as B1-B25 in black. The agonist isoeugenol is B3 and p-isobutylphenol is A1. In all cases the corresponding micromolar EC50 values are indicated in brackets. Names of A- and B-compounds are listed in Supplementary Tables 2 and 3
Structural characteristics of Olfr73 from molecular dynamics simulations
We modelled the Olfr73 with crystal structures of activated GPCRs. To explore reliable atomic details, we performed 2 × 500 ns all-atom molecular dynamics simulations for both the apo form of the receptor (apo-Olfr73) and the receptor with agonist isoeugenol (iEG-Olfr73) (Fig. 2). The molecular dynamics simulations showed that the volumes of the binding pocket for apo-Olfr73 and iEG-Olfr73 were 190 ± 3 Å3 and 220 ± 3 Å3, respectively. This is probably because of the induced fit effect (IFD), which has been widely observed in GPCR system and many others56,57.
Since the Olfr73 was modelled with low sequence identity templates, it was necessary to restrain the backbone of the modeled OR structure during molecular dynamics simulations to keep correct secondary structure50,52. Thus, we added a small force constrain during all our molecular dynamics simulations (see methods section). Transmembrane (TM) movements are a hallmark of GPCR activation. Since the templates used for the molecular dynamics simulations are based on receptors in activated states, the cytoplasmic TM regions of Olfr73 have been kept in the active open conformation by the end of molecular dynamics simulations (Supplementary Fig. 2).
Virtual agonist screening
We established a refined 3D structural homology model of Olfr73, the agonist-receptor interaction fingerprint and the structural framework explaining the mechanism of receptor activation. To validate these findings, we performed a virtual screen on a large chemical compound library to find new candidates of agonists for Olfr73 (Fig. 1) beyond classical odorant compound libraries, which finally will be tested by cellular functional assays.
First, we evaluated the physicochemical properties of all reported compounds (see Methods section on physical properties filtering) and used them for setting the conditions for an initial filter according to which 312,800 compounds were selected from the initial 1.58 million drug-like compounds of the ZINC library (Supplementary Table 1). For details about this procedure, please see the Methods section on virtual screening).
We applied the next round of selection criteria to our downsized chemical compound library using pharmacophore search (PH4)58, a screening method selecting compounds according to their chemical shape (Fig. 3b). The PH4 screen heavily relies on our results obtained from the IFP analysis and the molecular dynamics simulations. According to the molecular dynamics simulations, the oxygen at site I is crucial for agonist binding, forming distinct H-bonds with Y2606.52 and E2085.47. IFP analysis further confirmed that the interaction with Y2606.52 in this position is highly conserved (Fig. 3a). Since the -OH group could be either H-bond donor or H-bond acceptor, it was featured allocated preferentially the PH4 selection filter further downsized the library to 266,000 compounds.
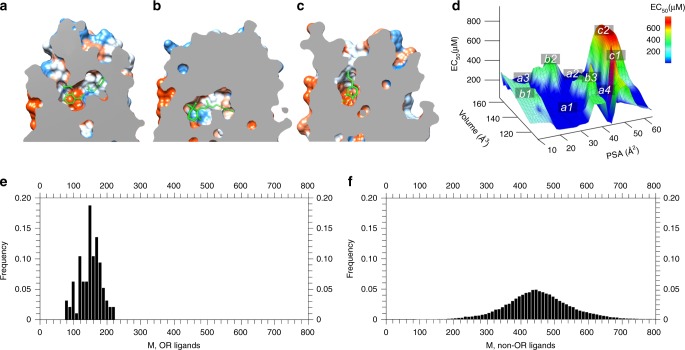
Interestingly, the empty ligand-binding pocket of Olfr73 has a volume of 190 Å3 and is noticeably smaller than that of other GPCRs including A2AR (270 Å3)59, rhodopsin (260 Å3)11 (Fig. 5), the 5-HT1A60 receptor (360 Å3), or the μ-opioid receptor57 (510 Å3), and therefore acts as a size-selection filter for potential binders. This explains why all currently reported agonists of Olfr73 are small (MW = 130–220) and the corresponding EC50 values are relatively high, due to the limited interactions in such small binding pocket. On this basis we created a volume counter along the 3D space of the sixteen superimposed ligands further reducing the library of potential Olfr73 binders to 493 compounds. We then continued selection filtering applying first an ionization penalty and then a molecular polarity counter, which narrowed the library further down first to 371 and then to 204 compounds.
Fig. 5.
Cross-section through several GPCRs along the membrane normal showing the vertical part of the ligand-binding pocket of (a) A2AR in complex with ZMA, (b) rhodopsin in complex with retinal, and (c) Olfr73 in complex with isobutylphenol. d Plot of EC50 values versus agonist volumes and agonist polar surface areas (PSA) for Olfr73 based on all reported agonists. Highly potent agonists are located in regions a1, a2, a3 and a4; agonists with medium potency are in the regions of b1, b2 and b3; agonists with lower potency are found in regions c1 and c2. e Molecular mass distribution of OR ligands. f Molecular mass distribution of non-olfactory GPCR ligands
Finally, we selected potential agonists from the remaining 204 compounds using quantitative structure–activity relationships (QSAR) based on comparative molecular field analysis (CoMFA) methods61. We docked the top 100 ranked compounds by QSAR into the MD refined homology model and found that 64 compounds fitted into the ligand-binding pocket of Olfr73, close to the activation trigger F1053.33. However, only 25 out of the 64 selected compounds have been commercially available for testing biological activity.
Cell-based functional tests
Next, we used the SEAP reporter assay, monitoring changes in cyclic adenosine monophosphate (cAMP) second messenger signalling as a read-out for cellular responses of odorant-induced receptor activation and found ligands capable of activating Olfr73 in Hana3A cells47 We tested 25 compounds of the molecules predicted from virtual screening and identified 17 (Fig. 4, blue labelled compounds; Supplementary Table 1 and Supplementary Fig. 3) inducing a noticeable SEAP signal in a concentration-dependent activation of Olfr73. It would be interesting to test by additional experiments whether of the eight compounds, which did not show agonist activity, there are antagonists for Olfr73.
The diversity of OR agonists
In the following, we used a hierarchical agglomerative clustering method62 to classify both the newly found and the previously known Olfr73 agonists based on their PH4 characteristics. As shown in Fig. 4, the 42 compounds can be grouped into 6 different classes. The four agonists of class-1 comprise a common phenol group with bulky hydrophobic groups (cyclohexyl or branched methyl containing alkyl chains) in para position. The EC50 values of class-1 agonists range from 13 to 64 μM. The agonists of class-2 share a central modified pyrocatechol structure (primarily in form of monometoxy-phenol or dimetoxy-phenon) with an additional linear, branched or cyclic hydrophobic group attached. The EC50 values of these agonists range from 7 to 240 μM. The agonists of class-3 contain a central benzaldehyde structure. The para positions carry primarily a metoxy- or ethoxy-substitute; the meta positions are substituted mostly by methoxy groups or for one case by a methyl group. The EC50 of class-3 agonists range from 26 to 270 μM. The 16 agonists of class-4, share a central phenol structure with oxygen carrying groups in the para and sometimes also in the ortho position. The class-4 agonists are the most polar ones in our collection; ten of them show EC50 values in the range of 4–100 μM, the remaining six have EC50 values between 200 and 660 μM. The class-5 agonists are quite different from the initial four classes; they do not contain an aromatic ring but instead carry a central cyclohexanone structure preferentially with a linear or branched alkyl substituent at the para position. The four class-5 agonists show EC50 values from 36 to 63 μM. Only one agonist is listed in class-6. It is composed of a tetrahydro-2H-pyran structure carrying two hydrophobic substitutions in the ring and has an EC50 value of 630 μM.
Therapeutic potential of the newly discovered agonists
We found p-isobutylphenol (4-isobutylphenol) as the most potent ligand activating Olfr73 in our functional assay (Fig. 4). It is a known degradation product of Ibuprofen which is widely used as analgesic anti-inflammatory drug but p-isobutylphenol has also been shown to exhibit antibiotic activity63. The estrogenic activity of the compound 4-cyclohexylphenol has been documented by in-vitro assays64. The Olfr73 activating compound 4′-hydroxy-3′,5′-dimethoxyacetophenone (Acetosyringone) has anti-asthmatic and anti-inflammatory properties65. And finally, 4′-hydroxypropiophenone is a predicted inhibitor of metalloproteinase 10, which has an active role in lung cancer development (Kiresee et al., 2016). Thus, our results have revealed some insights regarding the potential poly-pharmacological profile of these drugs acting not only on a defined medicinal target but also activating an OR. Similar observations of unintended interactions and activation by medicinal drugs have also been documented for the bitter taste receptor TAS2R1466. The complete list of newly discovered compounds can be found in Supplementary Table 2. The full list of reported compounds is in Supplementary Table 3.
Limited volume of OR binding pocket
ORs in general and Olfr73 in particular show some interesting structural and functional differences to their class A GPCR relatives. Most of the known OR-agonists are smaller in size than the typical agonists of non-olfactory class A GPCRs. Considering a large panel of reported OR ligands67 together with the new ligands (105 compounds in total) from this work shows that the molecular mass (M) of OR ligands distribute between 80 and 220 Da (maximum around 150 Da) (Fig. 5e). In contrast, non-olfactory GPCR ligands68 (161,083 compounds in total) distribute primarily between 300 and 600 Da (maximum around 450 Da) (Fig. 5f). Additionally, EC50 values for OR-agonists are usually much higher than those of the agonists of the non-olfactory class A GPCRs69. In our present study, this can be explained by the volume of the ligand-binding pocket of the Olfr73 which in the apo form is considerably smaller than comparable regions of the non-olfactory class A GPCRs reducing the number of interaction points between ligand and receptor (Fig. 5). Obviously, the ligand-binding pocket of a particular receptor acts as a size exclusion filter for potential ligands. To test this hypothesis and to probe the flexibility of the ligand-binding pocket for the Olfr73-agonists, we submitted Olfr73 with bound compounds A1, A2 and A3 (Fig. 4) to additional 2 × 500 ns all-atom molecular dynamics simulations. Similar to the agonist isoeugenol, the volume of the binding pocket of Olfr73 increased from 190 ± 3 Å3 in the empty state to 220 ± 3 Å3 for A1 (MW = 149) to 225 ± 5 Å3 for A2 (MW = 166), and 240 ± 2 Å3 for A3 (MW = 171), respectively. In general, larger ligands induce a larger volume increase in the occupied binding pocket56. Obviously, the binding pocket of Olfr73 is within a certain range quite flexible and adjusts perfectly to the size of the bound ligand with volume changes between 15 and 25%. We previously made similar observations for non-olfactory class A GPCRs such as the P2Y1 receptor70 changing the volume of the binding pocket from 230 ± 4 to 280 ± 5 Å3 (22% change), the 5-HT1A60 receptor from 360 ± 5 to 425 ± 3 Å3 (18%), the A2AR59 from 270 ± 2 to 315 ± 4 Å3 (17%), and the μ-opioid receptor57 from 510 ± 3 to 575 ± 5 Å3 (13%). In the next step, we performed an interaction fingerprint analysis for our newly found 17 agonists and compared the outcome with that in Fig. 3 of the 25 known agonists (Supplementary Fig. 4). The IFPs of both sets of agonists are quite similar. Moreover, IFPs indicated that there are only two hydrogen bond interactions between Olfr73 and its agonists, whereas there are much more polar interactions in other GPCRs71,72. These results further confirm our conclusions that the volume of Olfr73 is limited which is responsible for the small size of its agonists and weak EC50 values.
Discussion
Olfactory receptors represent almost 50% of the human GPCRs and may have additional physiological and pathological functions in the human body beyond their role in olfaction. A critical step allowing studies of the different functional roles of ORs relies on the discovery of their activating ligands. By employing a combination of homology modelling, interaction fingerprint analysis and molecular dynamics simulations to finally find novel agonists beyond classical odorant compounds by in silico screening a large drug compound library. We find that even drug-like molecules can target Olfr73, and compared their OR-activation mechanism to that mediated by classical odorant molecules.
In summary, our study revealed a structural framework for ligand–receptor interactions. We found that the limited volume of the binding pocket is responsible for the small size and weak EC50 values of Olfr73 agonists, which is typical for many ORs. Molecular fingerprint analysis discovered the principal interactions between the agonists and the binding pocket comprising many aromatic and hydrophobic residues. Interestingly, when plotting the EC50 values vs. ligand volume and ligand polar surface area (PSA), we identified four favorable regions for the most potent agonists with the following characteristics (Fig. 5d): (a1) small volume (120–140 Å3) and small PSA (20–35 Å2); (a2) large volume (140–160 Å3) and medium PSA (35–40 Å2); (a3) large volume (>160 Å3) and small PSA (10–20 Å2); (a4) small volume (120–130 Å3) and medium PSA (42–45 Å2). Three regions with moderate potencies were also identified: (b1) small or medium volume (120–150 Å3) and small PSA (10–20 Å2); (b2) large volume (150–160 Å3) and small PSA (20–30 Å2); (b3) medium volume (130–150 Å3) and medium PSA (35–40 Å2). Two regions with poor potencies were also found: (c1) small volume (<130 Å3) and large PSA (45–55 Å2); (c2) large volume (>145 Å3) and large PSA (>50 Å2).
Comparing with previous work on olfactory receptors15,43,73, we found several new aspects in this area including the structural principles leading to higher EC50 values for OR activation and the molecular diversity of ligands and interactions with their OR. In conclusion, molecular dynamics simulations in combination with structure based in silico screening offer a promising way to deorphanize the mammalian OR repertoire and thereby contribute to a better understanding of the molecular basis of ligand–OR interactions in olfactory and non-olfactory processes.
Methods
Homology modelling of Olfr73
The initial homology models of Olfr73 were obtained by Modeller 9.1074 using the crystal structure of two GPCRs in the active state as templates, β2AR (pdb code: 4LDE)48 and rhodopsin (pdb: 4BEY)49; β2AR shares 19% and rhodopsin 16% sequence identity with Olfr73. They share the highest sequence identity with Olfr73. 3D multiple sequence alignments were performed by Promals3D75 with default settings and were adjusted manually for properly aligning conserved motifs and disulfide bridges. 25,000 structural models (5 × 5000 with different random seeds) were created from the two template structures for Olfr73 in Modeller with fully annealing protocol, and the optimal model was chosen for further study based on Discrete Optimized Protein Energy (DOPE) score. Models from Modeller were submitted to Rosetta for kinematic loop modelling refinement76. Over 20,000 structures for loop region were generated. The loop refinement was done in Rosetta76.
Refinement of structural model and protein-ligand docking
The initial Olfr73 structure models generated from Modeller were optimally aligned with the structure of β2AR (pdb code: 4LDE) using OPM (Orientations of Proteins in Membranes) database77. The pre-aligned Olfr73 structure models were imported into the Maestro v9.3 program78. Hydrogens were added to the structures corresponding to physiological pH 7.0. For details please see our previous work11.
Molecular dynamics simulations
We performed restrained molecular dynamics simulations to achieve local improvement of the homology models52,79. Using the g_membed80 tool in Gromacs81,82, the well-prepared Olfr73 structure model was embedded into a pre-equilibrated lipid bilayer of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) solvated in 0.15 M NaCl. All molecular dynamics simulations were performed in Gromacs81,82. For details please see our previous work11,83.
Interaction fingerprint analysis and compound clustering
Both the interaction fingerprint (IFP) analysis and compound clustering analysis were performed in Schrodinger software78. Each compound was first docked into the binding pocket of Olfr73 in Schrodinger, then the interaction fingerprint analysis was performed using the Canvas module84. Interaction fingerprints are calculated as a set of bits for the presence or absence of particular types of interactions between a set of ligands and the residues in the active site of a receptor. The IFP divides protein/ligand interactions into four different types: hydrophobic interaction, ion-lock interaction, and H-bond interactions. The frequency of each interaction was calculated by the sum of contacts over the whole frames during molecular dynamics simulations.
The 3D coordinates of each docked compound were imported into Canvas for compound clustering analysis. Canvas first calculates the pharmacophore features (such as H-bond interaction and hydrophobic contact) of imported compounds, then performs hierarchical agglomerative clustering on a set of structures using a similarity matrix. The Hierarchical clustering was generated in Canvas. Hierarchical clustering is a method of cluster analysis which seeks to build a hierarchy of clusters. The merges and splits are determined in a greedy manner85. The results of hierarchical clustering are usually presented in a dendrogram. In order to decide which clusters should be combined or split, a measure of dissimilarity between sets of observations is required. In the hierarchical clustering, this is achieved by use of “Average distance between all inter-cluster pairs” method in Schrodinger software85, and the Kelley criterion86 was used for the linkage which specifies the dissimilarity of sets as a function of the pairwise distances of observations in the sets.
Virtual screening
We first considered the physicochemical properties of 25 known agonists of Olfr7317,47 such as molecular mass (M), calculated logP (clogP), number of rotatable bonds (nRot) and bond valence. However, we extended the scale of each criterium. For instance, as the molecular mass M of the reported compounds are within 134–218 Da, we used a filter for M of 110–320 Da.
The pharmacophore (PH4) search was performed using the Phase58 module in the Schrodinger software. PH4 is a fast and efficient tool for shape-based superposition and similarity searching of pharmacophores. A scoring function rank-orders potential pharmacophores by their performance in virtual screening and ligand alignment. The PH4 models were built according to the results obtained from both IFP analysis and molecular dynamics simulations. A H-bond donor/acceptor featured sphere (site I) was created next to the -OH group of Y2606.52. A H-bond acceptor descriptor (site II) was placed next to site I. Additional four hydrophobic featured spheres (site III, IV, V and VI) were created according to the IFP analysis.
On the basis of the outcome of the PH4 search, we created a volume counter (230 Å3) along the 3D space of the 25 docked ligands to further reduce the compound library. Then we continued the selection filtering applying first an ionization penalty and then a molecular polarity counter. All these steps were performed in the Schrodinger software package.
We selected potential agonists from the remaining compound library using quantitative structure–activity relationships (QSAR) based on comparative molecular field analysis (CoMFA) methods in SYBYL-X 1.387 software. CoMFA considers steric and electrostatic interactions, which would block ligand–receptor interations88. As a result, each molecule is located within a three-dimensional grid of defined dimensions. A probe calculates the energy within the bound molecule and neighboring residues of the receptor, in all directions over the entire grid yielding thousands of interactions88. Here we used the 25 previously reported47 active compounds as training sets. Good correlations (R2 = 0.73) were obtained between reported experimental results from functional assays and QSAR predicted ligands.
Cell Culture and Transfection
As described in detail elsewhere16,45,47 HEK293T-derived Hana3A cells (provided by Prof. Matsunami, Duke University, USA) were grown in DMEM/F12 medium (Invitrogen, Netherlands) supplemented with 10% fetal calf serum (FCS) (Invitrogen, Netherlands), maintained under selective conditions with 1 μg/ml of puromycin (Sigma, Switzerland) and kept in the incubator at 37 °C and 5% CO2. Cells were transfected with plasmid DNA using Lipofectamine 2000 (Invitrogen, Netherlands).
Quantification of OR responses by SEAP reporter assay
The assays were performed as described in detail elsewhere16,45,47 twenty hours before transfection Hana3A cells were seeded into 96 wells (Greiner, Germany) at a concentration of 3.5 × 106 cells per ml of medium. 75 ng of pRTP1S, 150 ng of the cAMP response element fused to the secreted alkaline phosphatase (pCRE-SEAP)89 and either 75 ng of pOlfr7347 or 75 ng of calf thymus DNA, used as a control, were co-transfected. Compounds to be tested (Sigma, Switzerland) were diluted in DMEM/F12 without FCS and added to the cells 7 h after transfection. Cells were incubated for 16 h at 37 °C in the incubator. For the SEAP reporter assay, the culture medium was mixed with an equal volume of 1 M diethanolamine-bicarbonate, pH 9.8, containing 20 mM para-nitrophenyl-phosphate (pNPP) (Sigma, Switzerland) and 1 mM MgCl2 (Sigma, Switzerland). Absorbance was measured at 410 nm using a multiwell absorbance plate reader (Molecular Devices, USA) at 1- to 4-min intervals for a period of 5 min to determine SEAP expression dependent pNPP hydrolysis rates (A410 /min). EC50 values were determined from dose-response curves fitting experimental data with the Hill equation using IGOR Pro software (WaveMetrics):
f(x) is the background-corrected response signal at concentration x of the investigated ligand, f(max) the maximal, background-corrected amplitude of the response signal, EC50 the half maximal effective ligand concentration, and n the Hill coefficient. Experiments were performed in triplicate.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
Most computational work has been done at the Interdisciplinary Centre for Mathematical and Computational Modelling in Warsaw (grant no. GB71–3 and GB70-3). S.F. was funded by the National Center of Science, Poland (grant no. 2013/08/M/ST6/00788). H.V. was supported by the European Community (project SynSignal, grant no. FP7-KBBE-2013-613879) and funds of the EPFL.
Author contributions
S.Y. and H.V. initiated and designed the project. S.Y. performed the modelling and molecular dynamics simulations. S.Y. and S.F. analyzed the results of modelling. T.D., M.C.B. and H.P. performed biological testing. All authors wrote the manuscript.
Data Availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Shuguang Yuan, Email: shuguang.yuan@gmail.com.
Horst Vogel, Email: horst.vogel@epfl.ch.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s42003-019-0384-8.
References
- 1.Firestein S. How the olfactory system makes sense of scents. Nature. 2001;413:211–218. doi: 10.1038/35093026. [DOI] [PubMed] [Google Scholar]
- 2.Buck L, Axel R. A novel multigene family may encode odorant receptors: a molecular basis for odor recognition. Cell. 1991;65:175–187. doi: 10.1016/0092-8674(91)90418-X. [DOI] [PubMed] [Google Scholar]
- 3.Mombaerts P. Genes and ligands for odorant, vomeronasal and taste receptors. Nat. Rev. Neurosci. 2004;5:263–278. doi: 10.1038/nrn1365. [DOI] [PubMed] [Google Scholar]
- 4.Malnic B, Hirono J, Sato T, Buck LB. Combinatorial receptor codes for odors. Cell. 1999;96:713–723. doi: 10.1016/S0092-8674(00)80581-4. [DOI] [PubMed] [Google Scholar]
- 5.von der Weid B, et al. Large-scale transcriptional profiling of chemosensory neurons identifies receptor-ligand pairs in vivo. Nat. Neurosci. 2015;18:1455–1463. doi: 10.1038/nn.4100. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Y, et al. Molecular profiling of activated olfactory neurons identifies odorant receptors for odors in vivo. Nat. Neurosci. 2015;18:1446–1454. doi: 10.1038/nn.4104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunkel A, et al. Nature’s chemical signatures in human olfaction: a foodborne perspective for future biotechnology. Angew. Chem. 2014;53:7124–7143. doi: 10.1002/anie.201309508. [DOI] [PubMed] [Google Scholar]
- 8.Peterlin Z, Firestein S, Rogers ME. The state of the art of odorant receptor deorphanization: a report from the orphanage. J. Gen. Physiol. 2014;143:527–542. doi: 10.1085/jgp.201311151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Venkatakrishnan AJ, et al. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 10.Nygaard R, et al. The dynamic process of beta(2)-adrenergic receptor activation. Cell. 2013;152:532–542. doi: 10.1016/j.cell.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan S, Filipek S, Palczewski K, Vogel H. Activation of G-protein-coupled receptors correlates with the formation of a continuous internal water pathway. Nat. Commun. 2014;5:4733. doi: 10.1038/ncomms5733. [DOI] [PubMed] [Google Scholar]
- 12.Rose AS, et al. Position of transmembrane helix 6 determines receptor G protein coupling specificity. J. Am. Chem. Soc. 2014;136:11244–11247. doi: 10.1021/ja5055109. [DOI] [PubMed] [Google Scholar]
- 13.Kohlhoff KJ, et al. Cloud-based simulations on Google Exacycle reveal ligand modulation of GPCR activation pathways. Nat. Chem. 2014;6:15–21. doi: 10.1038/nchem.1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lakkaraju SK, Lemkul JA, Huang J, MacKerell AD., Jr. DIRECT-ID: an automated method to identify and quantify conformational variations-application to beta2 -adrenergic GPCR. J. Comput. Chem. 2016;37:416–425. doi: 10.1002/jcc.24231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park JH, et al. Opsin, a structural model for olfactory receptors? Angew. Chem. 2013;52:11021–11024. doi: 10.1002/anie.201302374. [DOI] [PubMed] [Google Scholar]
- 16.Baud O, et al. Exchanging ligand-binding specificity between a pair of mouse olfactory receptor paralogs reveals odorant recognition principles. Sci. Rep. 2015;5:14948. doi: 10.1038/srep14948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Katada S, Hirokawa T, Oka Y, Suwa M, Touhara K. Structural basis for a broad but selective ligand spectrum of a mouse olfactory receptor: mapping the odorant-binding site. J. Neurosci. 2005;25:1806–1815. doi: 10.1523/JNEUROSCI.4723-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Di Pizio A, Niv MY. Computational Studies of Smell and Taste Receptors. Isr. J. Chem. 2014;54:1205–1218. doi: 10.1002/ijch.201400027. [DOI] [Google Scholar]
- 19.Doszczak L, et al. Prediction of perception: probing the hOR17-4 olfactory receptor model with silicon analogues of bourgeonal and lilial. Angew. Chem. 2007;46:3367–3371. doi: 10.1002/anie.200605002. [DOI] [PubMed] [Google Scholar]
- 20.Gelis L, Wolf S, Hatt H, Neuhaus EM, Gerwert K. Prediction of a ligand-binding niche within a human olfactory receptor by combining site-directed mutagenesis with dynamic homology modeling. Angew. Chem. 2012;51:1274–1278. doi: 10.1002/anie.201103980. [DOI] [PubMed] [Google Scholar]
- 21.Bavan S, Sherman B, Luetje CW, Abaffy T. Discovery of novel ligands for mouse olfactory receptor MOR42-3 using an in silico screening approach and in vitro validation. PloS One. 2014;9:e92064. doi: 10.1371/journal.pone.0092064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sell, C. S. Chemistry and the Sense of Smell (John Wiley & Sons, Inc., Hoboken, 2014).
- 23.Ruddigkeit L, Awale M, Reymond JL. Expanding the fragrance chemical space for virtual screening. J. Chemin-. 2014;6:27. doi: 10.1186/1758-2946-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerkin, R. C. & Castro, J. B. The number of olfactory stimuli that humans can discriminate is still unknown. eLife4, e08127 (2015). [DOI] [PMC free article] [PubMed]
- 25.Meister M. On the dimensionality of odor space. eLife. 2015;4:e07865. doi: 10.7554/eLife.07865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bushdid C, Magnasco MO, Vosshall LB, Keller A. Humans can discriminate more than 1 trillion olfactory stimuli. Science. 2014;343:1370–1372. doi: 10.1126/science.1249168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doty RL. Olfactory dysfunction in Parkinson disease. Nat. Rev. Neurol. 2012;8:329–339. doi: 10.1038/nrneurol.2012.80. [DOI] [PubMed] [Google Scholar]
- 28.Flegel C, Manteniotis S, Osthold S, Hatt H, Gisselmann G. Expression profile of ectopic olfactory receptors determined by deep sequencing. PLoS. One. 2013;8:e55368. doi: 10.1371/journal.pone.0055368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang N, Koo J. Olfactory receptors in non-chemosensory tissues. BMB Rep. 2012;45:612–622. doi: 10.5483/BMBRep.2012.45.11.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spehr M, et al. Identification of a testicular odorant receptor mediating human sperm chemotaxis. Science. 2003;299:2054–2058. doi: 10.1126/science.1080376. [DOI] [PubMed] [Google Scholar]
- 31.Feldmesser E, et al. Widespread ectopic expression of olfactory receptor genes. BMC Genom. 2006;7:121. doi: 10.1186/1471-2164-7-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pluznick JL, et al. Functional expression of the olfactory signaling system in the kidney. Proc. Natl Acad. Sci. USA. 2009;106:2059–2064. doi: 10.1073/pnas.0812859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiese H, et al. Quantitative phosphoproteomics reveals the protein tyrosine kinase Pyk2 as a central effector of olfactory receptor signaling in prostate cancer cells. Biochim. Biophys. Acta. 2015;1854:632–640. doi: 10.1016/j.bbapap.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 34.Neuhaus EM, et al. Activation of an olfactory receptor inhibits proliferation of prostate cancer cells. J. Biol. Chem. 2009;284:16218–16225. doi: 10.1074/jbc.M109.012096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fleischer J, Bumbalo R, Bautze V, Strotmann J, Breer H. Expression of odorant receptor Olfr78 in enteroendocrine cells of the colon. Cell Tissue Res. 2015;361:697–710. doi: 10.1007/s00441-015-2165-0. [DOI] [PubMed] [Google Scholar]
- 36.Kang N, et al. Olfactory receptor Olfr544 responding to azelaic acid regulates glucagon secretion in alpha-cells of mouse pancreatic islets. Biochem. Biophys. Res. Commun. 2015;460:616–621. doi: 10.1016/j.bbrc.2015.03.078. [DOI] [PubMed] [Google Scholar]
- 37.Varkonyi T, Korei A, Putz Z, Kempler P. Olfactory dysfunction in diabetes: a further step in exploring central manifestations of neuropathy? Angiology. 2014;65:857–860. doi: 10.1177/0003319714526971. [DOI] [PubMed] [Google Scholar]
- 38.Palouzier-Paulignan B, et al. Olfaction under metabolic influences. Chem. Senses. 2012;37:769–797. doi: 10.1093/chemse/bjs059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Busse D, et al. A synthetic sandalwood odorant induces wound-healing processes in human keratinocytes via the olfactory receptor OR2AT4. J. Invest. Dermatol. 2014;134:2823–2832. doi: 10.1038/jid.2014.273. [DOI] [PubMed] [Google Scholar]
- 40.Braun T, Voland P, Kunz L, Prinz C, Gratzl M. Enterochromaffin cells of the human gut: sensors for spices and odorants. Gastroenterology. 2007;132:1890–1901. doi: 10.1053/j.gastro.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 41.Massberg D, et al. Monoterpene (−)-citronellal affects hepatocarcinoma cell signaling via an olfactory receptor. Arch. Biochem. Biophys. 2015;566:100–109. doi: 10.1016/j.abb.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 42.Weber L, et al. Activation of odorant receptor in colorectal cancer cells leads to inhibition of cell proliferation and apoptosis. PLoS ONE. 2017;12:e0172491. doi: 10.1371/journal.pone.0172491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ho J, et al. Molecular recognition of ketamine by a subset of olfactory G protein-coupled receptors. Sci. Signal. 2015;8:ra33. doi: 10.1126/scisignal.2005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keiser MJ, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pick H, et al. Dual activities of odorants on olfactory and nuclear hormone receptors. J. Biol. Chem. 2009;284:30547–30555. doi: 10.1074/jbc.M109.040964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert AN, Firestein S. Dollars and scents: commercial opportunities in olfaction and taste. Nat. Neurosci. 2002;5:1043–1045. doi: 10.1038/nn937. [DOI] [PubMed] [Google Scholar]
- 47.Baud O, et al. The mouse eugenol odorant receptor: structural and functional plasticity of a broadly tuned odorant binding pocket. Biochemistry. 2011;50:843–853. doi: 10.1021/bi1017396. [DOI] [PubMed] [Google Scholar]
- 48.Ring AM, et al. Adrenaline-activated structure of beta2-adrenoceptor stabilized by an engineered nanobody. Nature. 2013;502:575–579. doi: 10.1038/nature12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Singhal A, et al. Insights into congenital stationary night blindness based on the structure of G90D rhodopsin. EMBO Rep. 2013;14:520–526. doi: 10.1038/embor.2013.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flohil JA, Vriend G, Berendsen HJ. Completion and refinement of 3-D homology models with restricted molecular dynamics: application to targets 47, 58, and 111 in the CASP modeling competition and posterior analysis. Proteins. 2002;48:593–604. doi: 10.1002/prot.10105. [DOI] [PubMed] [Google Scholar]
- 51.Larsson P, Wallner B, Lindahl E, Elofsson A. Using multiple templates to improve quality of homology models in automated homology modeling. Protein Sci. 2008;17:990–1002. doi: 10.1110/ps.073344908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan H, Mark AE. Refinement of homology-based protein structures by molecular dynamics simulation techniques. Protein Sci. 2004;13:211–220. doi: 10.1110/ps.03381404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Isberg V, et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Granier S, Kobilka B. A new era of GPCR structural and chemical biology. Nat. Chem. Biol. 2012;8:670–673. doi: 10.1038/nchembio.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vass M, et al. Molecular interaction fingerprint approaches for GPCR drug discovery. Curr. Opin. Pharmacol. 2016;30:59–68. doi: 10.1016/j.coph.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J. Med. Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- 57.Yuan S, et al. The mechanism of ligand-induced activation or inhibition of mu- and kappa-opioid receptors. Angew. Chem. 2015;54:7560–7563. doi: 10.1002/anie.201501742. [DOI] [PubMed] [Google Scholar]
- 58.Sastry GM, Dixon SL, Sherman W. Rapid shape-based ligand alignment and virtual screening method based on atom/feature-pair similarities and volume overlap scoring. J. Chem. Inf. Model. 2011;51:2455–2466. doi: 10.1021/ci2002704. [DOI] [PubMed] [Google Scholar]
- 59.Yuan S, Hu Z, Filipek S, Vogel H. W246 opens a gate for a continuous intrinsic water pathway during activation of the adenosine a receptor. Angew. Chem. 2014;54:556–559. doi: 10.1002/anie.201409679. [DOI] [PubMed] [Google Scholar]
- 60.Yuan S, Peng Q, Palczewski K, Vogel H, Filipek S. Mechanistic studies on the stereoselectivity of the serotonin 5-HT1A receptor. Angew. Chem. 2016;55:8661–8665. doi: 10.1002/anie.201603766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Parra-Delgado H, et al. Synthesis and comparative molecular field analysis (CoMFA) of argentatin B derivatives as growth inhibitors of human cancer cell lines. Bioorg. Med Chem. 2006;14:1889–1901. doi: 10.1016/j.bmc.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 62.Murtagh F, Legendre P. Ward’s hierarchical agglomerative clustering method: which algorithms implement ward’s criterion? J. Classif. 2014;31:274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 63.Caviglioli G, et al. Identification of degradation products of ibuprofen arising from oxidative and thermal treatments. J. Pharm. Biomed. Anal. 2002;30:499–509. doi: 10.1016/S0731-7085(02)00400-4. [DOI] [PubMed] [Google Scholar]
- 64.Ogawa Y, et al. Estrogenic activities of chemicals related to food contact plastics and rubbers tested by the yeast two-hybrid assay. Food Addit. Contam. 2006;23:422–430. doi: 10.1080/02652030500482371. [DOI] [PubMed] [Google Scholar]
- 65.Muller AA, Reiter SA, Heider KG, Wagner H. Plant-derived acetophenones with antiasthmatic and anti-inflammatory properties: inhibitory effects on chemotaxis, right angle light scatter and actin polymerization of polymorphonuclear granulocytes. Planta Med. 1999;65:590–594. doi: 10.1055/s-1999-14029. [DOI] [PubMed] [Google Scholar]
- 66.Levit A, et al. The bitter pill: clinical drugs that activate the human bitter taste receptor TAS2R14. FASEB J. 2014;28:1181–1197. doi: 10.1096/fj.13-242594. [DOI] [PubMed] [Google Scholar]
- 67.Modena D, Trentini M, Corsini M, Bombaci A, Giorgetti A. OlfactionDB: a database of olfactory receptors and their ligands. Adv. Life Sci. 2011;1:5. [Google Scholar]
- 68.Gaulton A, et al. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Saito H, Chi Q, Zhuang H, Matsunami H, Mainland JD. Odor coding by a Mammalian receptor repertoire. Sci. Signal. 2009;2:ra9. doi: 10.1126/scisignal.2000016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan S, et al. The molecular mechanism of P2Y1 receptor activation. Angew. Chem. Int. Ed. 2016;55:10331–10335. doi: 10.1002/anie.201605147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang W, et al. Structural insights into mu-opioid receptor activation. Nature. 2015;524:315–321. doi: 10.1038/nature14886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chung KY, et al. Conformational changes in the G protein Gs induced by the beta2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Poivet E, et al. Applying medicinal chemistry strategies to understand odorant discrimination. Nat. Commun. 2016;7:11157. doi: 10.1038/ncomms11157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eswar, N., et al. Comparative protein structure modeling using Modeller. Curr. Protoc. Bioinformatics 15, 5.6.1–5.6.30 (2006). [DOI] [PMC free article] [PubMed]
- 75.Pei J, Grishin NV. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics. 2007;23:802–808. doi: 10.1093/bioinformatics/btm017. [DOI] [PubMed] [Google Scholar]
- 76.Mandell DJ, Coutsias EA, Kortemme T. Sub-angstrom accuracy in protein loop reconstruction by robotics-inspired conformational sampling. Nat. Methods. 2009;6:551–552. doi: 10.1038/nmeth0809-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 78.Schrödinger Release 2018-2: Maestro, Schrödinger, LLC, New York, NY, USA (2018).
- 79.Thompson, J. D., Gibson, T. J. & Higgins, D. G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics Chapter 2, (2002). [DOI] [PubMed]
- 80.Wolf MG, Hoefling M, Aponte-Santamaria C, Grubmuller H, Groenhof G. g_membed: efficient insertion of a membrane protein into an equilibrated lipid bilayer with minimal perturbation. J. Comput. Chem. 2010;31:2169–2174. doi: 10.1002/jcc.21507. [DOI] [PubMed] [Google Scholar]
- 81.Pronk S, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hess Berk, Kutzner Carsten, van der Spoel David, Lindahl Erik. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. Journal of Chemical Theory and Computation. 2008;4(3):435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 83.Chan HCS, et al. Exploring a new ligand binding site of G protein-coupled receptors. Chem. Sci. 2018;9:6480–6489. doi: 10.1039/C8SC01680A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Singh J, Deng Z, Narale G, Chuaqui C. Structural interaction fingerprints: a new approach to organizing, mining, analyzing, and designing protein-small molecule complexes. Chem. Biol. Drug. Des. 2006;67:5–12. doi: 10.1111/j.1747-0285.2005.00323.x. [DOI] [PubMed] [Google Scholar]
- 85.Schrödinger Suite 2012: Maestro, version 9.3.5. Schrödinger, LLC; New York, NY, USA.
- 86.Kelley LA, Gardner SP, Sutcliffe MJ. An automated approach for clustering an ensemble of NMR-derived protein structures into conformationally related subfamilies. Protein Eng. 1996;9:1063–1065. doi: 10.1093/protein/9.11.1063. [DOI] [PubMed] [Google Scholar]
- 87.Sybyl-X Molecular Modeling Software Packages, Version 1.3. TRIPOS Associates, Inc; St. Louis, MO, USA (2012).
- 88.Ghasemi JB, Tavakoli H. Improvement of the prediction power of the CoMFA and CoMSIA models on histamine h3 antagonists by different variable selection methods. Sci. Pharm. 2012;80:547–566. doi: 10.3797/scipharm.1204-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Durocher Y, et al. A reporter gene assay for high-throughput screening of G-protein-coupled receptors stably or transiently expressed in HEK293 EBNA cells grown in suspension culture. Anal. Biochem. 2000;284:316–326. doi: 10.1006/abio.2000.4698. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.