Abstract
An effective framework for early warning and rapid response is a crucial element to prevent or mitigate the impact of biological invasions of plant pathogens, especially at ports of entry. Molecular detection of pathogens by using PCR-based methods usually requires a well-equipped laboratory. Rapid detection tools that can be applied as point-of-care diagnostics are highly desirable, especially to intercept quarantine plant pathogens such as Xylella fastidiosa, Ceratocystis platani and Phytophthora ramorum, three of the most devastating pathogens of trees and ornamental plants in Europe and North America. To this aim, in this study we developed three different loop mediated isothermal amplification (LAMP) assays able to detect each target pathogen both in DNA extracted from axenic culture and in infected plant tissues. By using the portable instrument Genie® II, the LAMP assay was able to recognize X. fastidiosa, C. platani and P. ramorum DNA within 30 min of isothermal amplification reaction, with high levels of specificity and sensitivity (up to 0.02 pg µL−1 of DNA). These new LAMP-based tools, allowing an on-site rapid detection of pathogens, are especially suited for being used at ports of entry, but they can be also profitably used to monitor and prevent the possible spread of invasive pathogens in natural ecosystems.
Keywords: Alien pathogens, Canker Stain Disease, Isothermal amplification, LAMP, Olive Quick Decline Syndrome, Portable diagnostics, Sudden Oak Death
Introduction
Invasive alien species represent a primary threat to biodiversity, economy and human health. International trade, tourism and other human activities break geographical barriers introducing non-native pathogenic organisms into new environments where they eventually find susceptible hosts and environments (Fisher et al. 2012; Migliorini et al. 2015; Santini et al. 2018). In Europe the accidental introduction of three quarantine pathogens, Xylella fastidiosa, Ceratocystis platani and Phytophthora ramorum with infected plants or wood material, has led to epidemics with heavy economic and ecological impacts.
Xylella fastidiosa is a bacterium reported on more than 350 different hosts (Denancè et al. 2017) and since 2013 is responsible for Olive Quick Decline Syndrome in Southern Italy (Apulia) (Saponari et al. 2013), more recently it has been found in Tuscany (Central Italy) (EPPO 2019); Ceratocystis platani is an ascomycetous fungus reported as the causal agent of Canker Stain Disease (CSD) of plane tree (Platanus) in urban and natural ecosystems (Lehtijärvi et al. 2018; Tsopelas et al. 2017). Phytophthora ramorum is an oomycete causing Sudden Oak Death (SOD) in the USA (Rizzo et al. 2002) but the pathogen has also been found in European ornamental nurseries (Werres et al. 2001) and in plantations of Japanese larch (Larix kaempferi) in Great Britain (Brasier and Webber 2010).
In the last decades alien plant pathogens are exponentially establishing in Europe (Santini et al. 2013). The European Union (EU) has an open-door phytosanitary system, which means that plants not specifically regulated can enter, therefore, inspections are concentrated on well-known pests and mostly limited to visual examination of aerial parts of plants. Traditional inspection methods are time consuming and labor-intensive, requiring specialized laboratories and expert operators. Furthermore, the first disease symptoms can occur after a long latent phase of the infection and they may be non-specific (e.g. X. fastidiosa), hampering detection efforts and, therefore, timely management of potential outbreaks. Serological and immunoassay-based methods are available, but their low sensitivity and specificity make them unreliable for phytosanitary inspections. For these reasons, sensitive and specific tools for effective phytosanitary inspection and interception are required to prevent new pathogen introductions. Nowadays, the high specificity and sensitivity of molecular DNA-based technologies allows detection of pathogens in the early stages of infection, when they are present at low DNA concentrations (Bilodeau et al. 2007; Chandelier et al. 2006; Harper et al. 2010; Luchi et al. 2013; Rollins et al. 2016). Although many of these methods have been used routinely in the laboratory, most of them are not transferable for field inspection, seriously limiting their adequacy for point-of-care application (Lau and Botella 2017). Point-of-care methods, besides being sensitive and specific, should also be simple and fast, producing results that are easy to interpret and demanding minimal equipment and facilities (Tomlinson et al. 2010a). For these purposes, an affordable LAMP (Loop mediated isothermal amplification) technique (Notomi et al. 2000), seems to be the most suitable. Recently several LAMP assays have been developed for both field and lab use especially for human and animal diseases and food safety control (Abdulmawjood et al. 2014; Lucchi et al. 2010). Up to now, even if many LAMP-based assays were developed for plant pathogens (Chen et al. 2013; Dai et al. 2012; Hansen et al. 2016; Harper et al. 2010; Moradi et al. 2014; Peng et al. 2013; Sillo et al. 2018; Tomlinson et al. 2007), only a few tests (Bühlmann et al. 2013; Franco Ortega et al. 2018; Harrison et al. 2017; Tomlinson et al. 2010b, 2013) were optimized and applied on portable instrument for on-site use. The use of portable detection instruments is a major driving force to achieve point-of-use, and real-time monitoring of analysed samples, allowing rapid detection.
The aim of this study was to optimize a reliable, fast and sensitive diagnostic assay using a LAMP portable instrument for early detection of X. fastidiosa, C. platani, and P. ramorum. These new protocols will be available to be used for research aims and for phytosanitary inspection, in order to prevent further introductions and spread of these pathogens.
Materials and methods
Microbial strains and DNA extraction
In addition to the targeted pathogens, fungal and bacterial species phylogenetically related to target pathogens, as well as out-group species and common host colonizers were used to optimize the molecular assay (Table 1).
Table 1.
List of isolates used in this study
| Species | Isolate code | Groupb | Host | Origin | Collectorc | Molecular assayd | ||
|---|---|---|---|---|---|---|---|---|
| LAMPe | qPCRf | |||||||
| tamp (min:s) | Ta (°C) | Detection | ||||||
| X. fastidiosa assay | ||||||||
| Xylella fastidiosa subsp. pauca | Co.Di.Roa | T | Olea europaea | Italy | M. Saponari | 7:15 | 88.98 | + |
| Xylella fastidiosa subsp. fastidiosa | Xffa | T | Prunus dulcis | USA | J. Chen | 14:20 | 88.78 | + |
| Xylella fastidiosa subsp. multiplex | Xfma | T | Liquidambar styraciflua | USA | S. Russell | 7:00 | 88.83 | + |
| Pseudomonas savastanoi pv. savastanoi | ITM05a | CHC | Olea europea | Italy | G. Marchi | – | – | − |
| Pantoea agglomerans | PaFL1a | CHC | Olea europea | Italy | G. Marchi | – | – | − |
| Pseudomonas fluorescens | KL218a | CHC | Actinidia deliciosa | Italy | G. Marchi | – | – | − |
| Xanthomonas arboricola pv. pruni | Xap | PR | Prunus laurocerasus | Italy | A. Raio | – | – | − |
| Pseudomonas savastanoi pv. nerii | Ps.sav | CHC | Nerium oleander | Italy | A. Raio | – | – | − |
| Pseudomonas koreensis | KL217a | NP, O | Actinidia deliciosa | Italy | G. Marchi | – | – | − |
| Pseudomonas syringae | KL34a | O | Actinidia deliciosa | Italy | G. Marchi | – | – | − |
| Pseudomonas syringae | KL32a | O | Actinidia deliciosa | Italy | G. Marchi | – | – | − |
| Pseudomonas viridiflava | KL24a | O | Actinidia deliciosa | Italy | G. Marchi | – | – | − |
| Pseudomonas mediterranea | C5P1rad1a | O | Chrysanthemum sp. | Italy | M. Fiori | – | – | − |
| Pseudomonas corrugata | C2P1 rada | O | Chrysanthemum sp. | Italy | M. Fiori | – | – | − |
| Pseudomonas syringae pv. photiniae | CFBP2899a | O | Photinia glabra | Japan | CFBP | – | – | − |
| Pectobacterium carotovorum | C24a | O | Zantedeschia aethiopica | Italy | G. Marchi | – | – | − |
| Pectobacterium carotovorum | C6a | O | Zantedeschia aethiopica | Italy | G. Marchi | – | – | − |
| Pantoea agglomerans pv. gypsophilae | Ehg824-1a | O | Gypsophila paniculata | Israel | G. Marchi | – | – | − |
| Pseudomonas syringae pv. actinidiae | KL103a | O | Actinidia deliciosa | Italy | G. Marchi | – | – | − |
| Pseudomonas syringae pv. tabaci | GSPB1209a | O | Nicotiana sp. | Italy | G. Marchi | – | – | − |
| Pseudomonas syringae pv. viburni | CFBP1702a | O | Viburnum sp. | USA | CFBP | – | – | − |
| Sphingomonas sp. | KVPT7FAa | NP, CHC | Actinidia deliciosa | Italy | G. Marchi | – | – | − |
| Stenotrophomonas malthopylia | St | NP, PR | Capsicum annum | Italy | A. Raio | – | – | − |
| Xanthomonas campestris pv. campestris | Xcc | PR | Brassica spp. | Italy | A. Raio | – | – | − |
| Xanthomonas arboricola pv. juglandis | Xaj | PR | Juglans regia | Italy | A. Raio | – | – | − |
| Erwinia amylovora | Ea12 | O | Pyrus communis | Italy | C. Cainelli | – | – | − |
| Agrobacterium tumefaciens | LMG37 | O | Prunus spp. | USA | BCCM/LMG | – | – | − |
| Agrobacterium vitis | CFBP5523 | O | Vitis vinifera | Australia | CFBP | – | – | − |
| C. platani assay | ||||||||
| Ceratocystis platani | CBS117355 | T | Platanus sp. | France | CBS-KNAW | 10:08 | 88.14 | + |
| Ceratocystis platani | Cp3 | T | Platanus × acerifolia | Italy | IPSP-CNR | 12:15 | 88.55 | + |
| Ceratocystis platani | Cp6 | T | Platanus × acerifolia | Italy | IPSP-CNR | 9:08 | 88.30 | + |
| Ceratocystis platani | G160 | T | Platanus × acerifolia | Turkey | T. Dogmus | 16:58 | 88.93 | + |
| Ceratocystis platani | DB203 | T | Platanus × acerifolia | Turkey | T. Dogmus | 14:08 | 88.78 | + |
| Ceratocystis platani | CBS115162 | T | Platanus occidentalis | USA | CBS-KNAW | 8:00 | 88.83 | + |
| Ceratocystis platani | Cp24 | T | Platanus × acerifolia | Italy | IPSP-CNR | 8:00 | 88.43 | + |
| Ceratocystis fimbriata | CBS 115167 | PR | Syngonium sp. | USA | CBS-KNAW | 10:05 | 88.45 | − |
| Ceratocystis fimbriata | CBS 118126 | PR | Syzgium aromaticum | Sulawesi | CBS-KNAW | 15:10 | 88.70 | − |
| Ceratocystis fimbriata | CBS 115175 | PR | Mangifera indica | Brazil | CBS-KNAW | 9:30 | 88.20 | − |
| Ceratocystis fimbriata | CBS 115174 | PR | Eucalyptus sp. | Brazil | CBS-KNAW | 14:00 | 88.50 | − |
| Ceratocystis fimbriata | CBS 115171 | PR | Colocasia esculenta | Brazil | CBS-KNAW | 10:13 | 88.60 | − |
| Ceratocystis fimbriata | CBS 74040 | PR | Crotolaria juncea | Brazil | CBS-KNAW | 13:38 | 88.00 | − |
| Sarcodontia pachyodon | Sp5 | O, CHC | Platanus × acerifolia | Italy | IPSP-CNR | – | – | − |
| P. ramorum assay | ||||||||
| Phytophthora ramorum | Prama | T | Rhododendron sp. | Greece | P. Tsopelas | 7:58 | 88.68 | + |
| P. ramorum | LSVM123a | T | Rhododendron sp. | France | R. Ioos-N. Schenck | 7:07 | 88.48 | + |
| P. ramorum | LSVM362a | T | Rhododendron sp. | France | R. Ioos-N. Schenck | 6:15 | 88.73 | + |
| P. ramorum | LSVM386a | T | Rhododendron sp. | France | R. Ioos-N. Schenck | 6:72 | 88.68 | + |
| P. ramorum | LSVM390a | T | Rhododendron sp. | France | R. Ioos-N. Schenck | 7:00 | 88.53 | + |
| P. ramorum | LSVM391a | T | Leucothoe sp. | France | R. Ioos-N. Schenck | 6:30 | 88.53 | + |
| P. ramorum | LSVM401a | T | Rhododendron sp. | France | R. Ioos-N. Schenck | 7:07 | 88.53 | + |
| P. ramorum | LSVM402a | T | Rhododendron sp. | France | R. Ioos-N. Schenck | 7:00 | 88.53 | + |
| P. ramorum | LSVM405a | T | Rhododendron sp. | France | R. Ioos-N. Schenck | 7:15 | 88.48 | + |
| P. lateralis | Plata | PR | Chamaecyparis lawsoniana | France | C. Robin | 9:00 | 88.43 | + |
| P. alni subsp. uniformis | Ph68 | PR | Alnus cordata | Italy | G. P. Barzanti | 23:27 | 90.47 | − |
| P. cactorum | PCA1a | PR | Aesculus hippocastanum | Germany | J. Schumacher | – | – | − |
| P. × cambivora | Ph21a | PR | Castanea sativa | Italy | A. Vettraino | 25:22 | 90.67 | − |
| P. cinnamomi | 28SA | PR | Laurus nobilis | Italy | IPSP-CNR | 16:37 | 89.62 | − |
| P. cinnamomi | Ncfca | PR | Unknown | Italy | IPSP-CNR | – | – | − |
| P. cryptogea | 13SA | PR | Prunus laurocerasus | Italy | IPSP-CNR | 22:07 | 89.78 | − |
| P. citricola | 51RC | PR | Viburnum lucidum | Italy | IPSP-CNR | – | – | − |
| P. citricola | Pcl1a | PR | Unknown | Germany | T. Jung | 27:27 | 89.82 | − |
| P. citrophthora | 33SB | PR | Euonymus spp. | Italy | IPSP-CNR | 20:05 | 89.23 | − |
| P. citrophthora | Ph9a | PR | Convolvolus sp. | Italy | S. O. Cacciola | 18:15 | 89.58 | − |
| P. europaea | PE1a | PR | Unknown | Germany | T. Jung | 15:12 | 90.42 | − |
| P. foliorum | 2015–1454a | PR, CHC | Rhododendron | UK | A. Pérez-Sierra | 11:15 | 89.08 | − |
| P. gonapodyides | PG7a | PR | Quercus robur | Germany | S. Leonhard | – | – | − |
| P. gonapodyides | IHTM | PR | Alnus cordata | Italy | IPSP-CNR | – | – | − |
| P. megasperma | Ph78 | PR | Prunus avium | Italy | G. P. Barzanti | – | – | − |
| P. megasperma | PMIa | PR | Quercus robur | Germany | S. Leonhard | – | – | − |
| P. nicotianae | 1RB | PR | Myrtus communis | Italy | IPSP-CNR | – | – | − |
| P. palmivora | 44RC | PR | Prunus laurocerasus | Italy | IPSP-CNR | – | – | − |
| P. quercina | PQ4a | PR | Quercus robur | Germany | S. Leonhard/J. Schumacher | – | – | − |
| P. syringae | Psy2a | PR | Unknown | Germany | J. Schumacher | 17:45 | 89.48 | − |
| Elongisporangium anandrum | PYAa | O | Quercus robur | Germany | S. Leonhard | – | – | − |
| Phytopythium litorale | 40SB | O | Prunus laurocerasus | Italy | IPSP-CNR | – | – | − |
| Elongisporangium undulatum | 76SB | O | Cupressus sempervirens | Italy | IPSP-CNR | – | – | − |
| Mortariella sp. | 26RA | O | Arbutus unedo | Italy | IPSP-CNR | – | – | − |
| Diplodia mutila | Dm | O | Quercus spp. | Italy | IPSP-CNR | – | – | − |
| D. pinea | 128 | O | Pinus resinosa | USA | M. A. Palmer | – | – | − |
| D. scrobiculata | 124 | O | Pinus resinosa | USA | M. A. Palmer | – | – | − |
| D. seriata | UCD 352 | O | Vitis vinifera | USA | J. R. Urbez-Torres | – | – | − |
| D. seriata | WP-J10 | O | Vitis vinifera | Australia | S. Savocchia | – | – | − |
| Geosmithia pallida | IVV7 | O | Ulmus spp. | Italy | IPSP-CNR | – | – | − |
| Ophiostoma novo ulmi subsp. americana | HI72 | O | Ulmus spp. | USA | IPSP-CNR | – | – | − |
aSamples provided as DNA
bFor each molecular assay developed in this study different groups of isolates were tested: target species (T); phylogenetically related species (PR), CHC = common host colonizers species (CHC); out-group species (O)
cCBS Centraalbureau voor Schimmelcultures (CBS) Fungal Biodiversity Centre—Royal Netherlands Academy of Arts and Sciences (KNAW), Utrecht, The Netherlands; CFBP French Collection of Plant Pathogenic Bacteria, INRA, France; IPSP-CNR Institute for Sustainable Plant Protection—National Research Council, Firenze, Italy; BCCM/LMG Bacteria Collection Laboratorium voor Microbiologie Universiteit Gent Bacteria Collection, Laboratorium voor Microbiologie, Universiteit Gent, Belgium
dMolecular assays are referred to LAMP and qPCR described in Table 2
etamp amplification time, Ta annealing temperature, – not detected
f+ positive, − negative
Mycelium of fungal and oomycete isolates (stored at 4 °C in the IPSP-CNR collection) was grown on 300PT cellophane discs (Celsa, Varese, Italy) on potato dextrose agar (PDA; Difco, Detroit, MI, USA) in 90 mm Petri dishes and maintained in the dark at 20–25 °C according to species requirements. After 7–10 days the mycelium was scraped from the cellophane surface and stored in 1.5 mL microfuge tubes at − 20 °C.
Bacterial strains (stored at − 80 °C in the IPSP-CNR collection) were grown on Luria–Bertani (LB) agar for 24 h at 25 ± 2 °C. Single colonies were picked-up and transferred to tubes containing 5 mL of LB that were incubated in an orbital shaker at 25 ± 2 °C and 90 rpm overnight. One millilitre of each suspension was used for DNA extraction. Fungal and oomycete DNA suitable for molecular analysis was extracted from mycelium by using the EZNA Plant DNA Kit (Omega Bio-tek, Norcross, GA, USA), as described by Migliorini et al. (2015). DNA from bacteria was extracted by using EZNA Bacteria DNA Kit (Omega Bio-tek) according to the procedure described by the manufacturer. DNA from the quarantine pathogens X. fastidiosa, E. amylovora, P. ramorum and P. lateralis were kindly provided by different collectors (see Table 1). Concentration of extracted DNA was measured using a Nanodrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Plant DNA samples
Plant samples were analyzed from naturally infected hosts including: (i) Two symptomatic plants of each of the following Mediterranean maquis species were collected in March 2019: Rhamnus alaternus, Calicotome spinosa, Cistus incanus, Spartium junceum, Prunus dulcis, affected by X. fastidiosa subsp. multiplex (recently detected by Tuscany Regional Phytosanitary Service—EPPO 2019); (ii) 10 Platanus × acerifolia symptomatic trees infected by C. platani (Florence, Italy).
About 80 mg (fresh weight) of plant material, i.e. leaves of Mediterranean maquis species and wood of P. × acerifolia plants, were used for genomic DNA extraction by using two different extraction protocols: (i) on-site by using Plant Material DNA extraction kit (OptiGene), according to manufacturer’s instructions. Briefly, small pieces of plant material (c.a. 80 mg) were placed in a 5 mL bijou with ball bearing and 1 mL lysis buffer. Bijous were shaken vigorously for 1 min to ground the plant material. Plant material solution (10 μL) was transferred into a vial containing 2 mL dilution buffer and mixed. Finally, 3 μL of dilution buffer containing DNA has been used as template in a LAMP assay;
ii) in laboratory by using EZNA Plant DNA Kit (Omega Bio-tek). Plant material of all the collected samples for DNA extraction was transferred to 2 mL microfuge tubes with two tungsten beads (3 mm) (Qiagen) and 0.4 mL lysis buffer P1 EZNA Plant DNA Kit (Omega Bio-tek, Norcross, GA, USA) then ground with a TissueLyser (Qiagen) (30 oscillations/s for 1 min). DNA was extracted from all samples using the EZNA Plant DNA Kit (Omega Bio-tek) (Migliorini et al. 2015).
In addition to the above samples, the optimization of LAMP assay was conducted by using the following DNA samples stored at − 80 °C (IPSP-CNR DNA collection): (i) 10 DNA samples extracted from symptomatic Olea europaea leaves with X. fastidiosa subsp. pauca infections. DNA was kindly provided by M. Saponari (IPSP-CNR, Bari) and extracted in CTAB buffer (Loconsole et al. 2014); (ii) 10 DNA samples from symptomatic Viburnum tinus leaves affected by P. ramorum extracted by using EZNA Plant DNA Kit (Omega Bio-tek).
As negative control, fresh tissue collected from 10 healthy plant of each tested species (Olea europaea, Rhamnus alaternus, Calicotome spinosa, Cistus incanus, S. junceum, Prunus dulcis, Platanus × acerifolia and Viburnum tinus) were extracted by using both Plant Material DNA extraction kit (OptiGene) and EZNA Plant DNA Kit (Omega Bio-tek), as previously described.
LAMP primer design
The six LAMP primers included: two outer primers (forward primer, F3; backward primer, B3) two inner primers (forward inner primer, FIP; backward inner primer, BIP) and two loop primers (forward loop primer, FLP; backward loop primer, BLP), as required by LAMP reaction (Notomi et al. 2000). Primers were designed using LAMP Designer software (OptiGene Limited, Horsham, UK) (Table 2) on the basis of the consensus sequences of the ribosomal RNA gene (ITS1-5.8 S-ITS2) for P. ramorum (KC473522) and C. platani (EU426554.1), while for X. fastidiosa the ribosome maturation factor (RimM) gene belonging to Co.Di.Ro strain was chosen (JUJW01000001). All designed primers were synthesized by MWG Biotech (Ebersberg, Germany) and are reported in Table 2. The specificity of newly designed primers was further tested using nucleotide–nucleotide BLAST® (Basic Local Alignment Search Tool; http://www.ncbi.nlm.nih.gov/BLAST) (Altschul et al. 1990).
Table 2.
List of primer set used in this study
| Target species | Molecular assay | Primer code | Sequence | References |
|---|---|---|---|---|
| Phytophthora ramorum | LAMP | Phy-r_F3 | 5′-ACGTTGTTGGTTGTGGAG-3′ | This study |
| Phy-r_B3 | 5′-CCAATTGAGATGCCAGCA-3′ | |||
| Phy-r_FLP | 5′-CGCATTGTTCAGCCGAAG-3′ | |||
| Phy-r_BLP | 5′-GAATCGACGGTGTTGTGC-3′ | |||
| Phy-r_FIP | 5′-AGTCATTACCGCCACAGCAGTGTTCGATTCGCGGTA-3′ | |||
| Phy-r_BIP | 5′-CGTAGCTGTGCAGGGCTTGAACCGCCACTCTACTTC-3′ | |||
| qPCR | PramF | 5′-GCAGGGCTTGGCTTTTGA-3′ | Migliorini et al. (2018) | |
| PramR | 5′-GCCGAACCGCCACTCTACT-3′ | |||
| Pram_PR | 5′-FAM-TCGACGGTGTTGTGCG-MGBNFQ-3′ | |||
| Xylella fastidiosa | LAMP | XF_F3 | 5′-TAGAGTCTTGGACTGAGCC-3′ | This study |
| XF_B3 | 5′-ATCGACCCAGTAATACTCGT-3′ | |||
| XF_FLP | 5′-AGGAGAACGTAATAACCACGG-3′ | |||
| XF_BLP | 5′-TCCTGGCATCAATGATCGTAAT-3′ | |||
| XF_FIP | 5′-CACCATTCAACATGGACTCGGTGCGATCTTCCGTTACCAG-3′ | |||
| XF_BIP | 5′-CTACGAGACTGGCAAGCGTTCGTACCACAGATCGCTTC-3′ | |||
| qPCR | Xf_Fw | 5′-CGGGTACCGAGTCCATGTTG-3′ | This study | |
| Xf_Rev | 5′-CAATCAAACGCTTGCCAGTCT-3′ | |||
| Xf_Pr | 5′-FAM-TGGTGCCCGTGGCTA-MGBNFQ-3′ | |||
| Ceratocystis platani | LAMP | CPL_F3 | 5′-CAGCGAAATGCGATAAGTAATG-3′ | This study |
| CPL_B3 | 5′-TTTATACTACACAGGGGAGTTG-3′ | |||
| CPL_FIP | 5′-AATGACGCTCGGACAGGCTCGAATCTTTGAACGCACA-3′ | |||
| CPL_BIP | 5′-TGTTCTTGGCGTTGGAGGTCGCAAGTATAACAGCCGATACA-3′ | |||
| CPL_FLP | 5′-TGCCTGGCAGAATACTGC-3′ | |||
| CPL_BLP | 5′-GTTCTCCCCTGAACAGGC-3′ | |||
| qPCR | CpITS-F | 5′-GCCTGTCCGAGCGTCATT-3′ | Luchi et al. (2013) | |
| CpITS-R | 5′-CCTCCAACGCCAAGAACAAA-3′ | |||
| CpITS-Pr | 5′-FAM-CACCACTCAAGGACTC-MGB-3′ | |||
| Cytochrome oxidase (COX) endogenous plant gene | LAMP | COX F3 | 5′-TATGGGAGCCGTTTTTGC-3′ | Tomlinson et al. (2010a, b) |
| COX B3 | 5′-AACTGCTAAGRGCATTCC-3′ | |||
| COX FLP | 5′-ATGTCCGACCAAAGATTTTACC-3′ | |||
| COX BLP | 5′-GTATGCCACGTCGCATTCC-3′ | |||
| COX FIP | 5′-ATGGATTTGRCCTAAAGTTTCAGGGCAGGATTTCACTATTGGGT-3′ | |||
| COX BIP | 5′-TGCATTTCTTAGGGCTTTCGGATCCRGCGTAAGCATCTG-3′ |
Real-time LAMP assay conditions
Real-time LAMP reactions were performed and optimised on the portable real-time fluorometer Genie® II (OptiGene Limited, Horsham, UK). DNA samples were amplified for 30 min in Genie® Strips (OptiGene Limited, Horsham, UK) with eight 0.2 mL isothermal reaction tubes with a locking cap providing a closed-tube system. Each isothermal amplification reaction was performed in duplicate, in a final volume of 25 μL. The reaction mixture contained 15 μL Isothermal Master Mix (ISO-001) (OptiGene Limited, Horsham, UK), 7 μL LAMP primer mixture (at final concentrations 0.2 μM of each F3 and B3, 0.4 μM of each FLP and BLP and 0.8 μM of each FIP and BIP) and 3 μL of template DNA. For each run two tubes including 3 μL dd-water were tested as No Template Control (NTC). LAMP amplification reactions were run at 65 °C for 30 min, followed by an annealing analysis from 98 to 80 °C with ramping at 0.05 °C per second that allow the generation of derivative melting curves (Abdulmawjood et al. 2014).
The main parameters used by Genie® II system to assess the positivity of a sample are: amplification time (tamp) and amplicon annealing temperature (Ta). The tamp is the time (expressed in min) where the fluorescence second derivative of the signal reaches its peak above the baseline value, while the Ta is the temperature (expressed in °C) at which double-stranded DNA product dissociates into single strands.
Specificity and sensitivity of real-time LAMP assays
For each target pathogen (X. fastidiosa, C. platani and P. ramorum) the specificity of the real-time LAMP assay was tested by using genomic DNA extracted from bacterial, fungal or oomycete strains (Table 1), at a final concentration of 10 ng μL−1. The limit of detection (LOD) of the LAMP assay was tested by using an 11-fold 1:5 serial dilution (ranging from 10 ng μL−1 to 0.001 pg μL−1) of each standard DNA template (X. fastidiosa - Co.Di.Ro strain; C. platani - isolate Cp24; P. ramorum - isolate Pram).
Real-time LAMP assay in naturally infected plants
To check the suitability of extracted plant DNA for downstream analysis the cytochrome oxidase (COX) gene was used as endogenous plant gene according to Tomlinson et al. (2010a) (Table 2).
The effectiveness of the real-time LAMP assay was then tested on DNA extracted from naturally infected hosts (Olea europaea, Rhamnus alaternus, Calicotome spinosa, Cistus incanus, S. junceum, Prunus dulcis, Platanus × acerifolia and Viburnum tinus) to detect each respective target pathogen (X. fastidiosa, C. platani and P. ramorum). For each plant species, additional healthy plants DNA were also included as negative control.
Real-time quantitative PCR assay
To validate the LAMP assay, for each target pathogen, DNA samples (from microbial and plant tissue) were also tested by real-time quantitative PCR (qPCR) based on TaqMan chemistry.
Primers and TaqMan® MGB probe for the DNA quantification of X. fastidiosa with the StepOnePlus™ Real-Time PCR System (Applied Biosystems, Forster City, CA, USA) were designed using Primer Express™ 3.0 software (Applied Biosystems). The DNA sequence of the ribosome maturation factor (RimM) gene (CoDiRO strain) was obtained from the ‘National Center for Biotechnology Information’ (NCBI) (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi) (accession number JUJW01000001). The TaqMan® MGB probe was labelled with 6-carboxy-fluorescein (FAM) at the end, and a non-fluorescent quencher (NFQ) with minor groove binder (MGB) ligands, at the 3′ end. Primers and probe are reported in Table 2. The length of the amplification product was 60 bp. The identity of the amplicon sequence was determined by comparing with other fungal species with the Standard nucleotide–nucleotide BLAST (blast n) of the NCBI.
DNA samples were assayed in MicroAmp Fast 96-well Reaction Plates (0.1 mL) closed with optical adhesive and using the StepOnePlus™ Real-Time PCR System (Applied Biosystems).
The real-time PCR reaction was performed in a final volume of 25 µL. Each tube contained: 300 nM forward primer (Eurofins MWG Operon, Ebersberg, Germany); 300 nM reverse primer (Eurofins MWG Operon); 200 nM fluorogenic probe (Applied Biosystems); 12.5 µL TaqMan™ Universal Master Mix (Applied Biosystems); 5 µL DNA template.
Each DNA sample was assayed in three replicates. Four wells containing 5 µL sterile water each were used for a No-Template Control (NTC) without any nucleic acid. The PCR protocol was 50 °C (2 min); 95 °C (10 min); 40 cycles of 95 °C (30 s), 60 °C (1 min).
For each replicate the Ct value, defined as the point at which the Reporter fluorescent signal first became statistically significant against the background, was utilised to quantify the sample. Measurements of X. fastidiosa DNA in unknown samples were achieved by interpolation from a standard curve generated with a DNA standard (Co.Di.Ro. strain), which was amplified in the same PCR run.
Real time PCR protocols for C. platani and P. ramorum were those described in Luchi et al. (2013) and Migliorini et al. (2018), respectively.
Statistical analysis
For each 1:5 serial dilution (ranging from 10 ng µL−1 to 0.128 pg µL−1) of each target pathogen, the correlation analysis was carried out between amplification time (tamp) for LAMP assay and threshold cycle (Ct) for qPCR.
Results
Specificity of real-time LAMP assay
For each target pathogen (X. fastidiosa, C. platani and P. ramorum) the nucleotide–nucleotide BLAST ® search showed a complete homology (100%) between the LAMP amplicon sequences designed in the current study and the sequences of the same pathogen available in GenBank database (NCBI).
BLAST ® search did not find sequence identity between the LAMP X. fastidiosa amplicon and the other species present in GenBank, while the P. ramorum LAMP amplicon showed 99% homology (due to only 2 bases of differences in the ITS region) with P. lateralis sequences. Similarly, the C. platani LAMP amplicon showed complete homology (100%) with C. fimbriata and 99% homology with C. neglecta, C. ecuadoriana and C. manginecans.
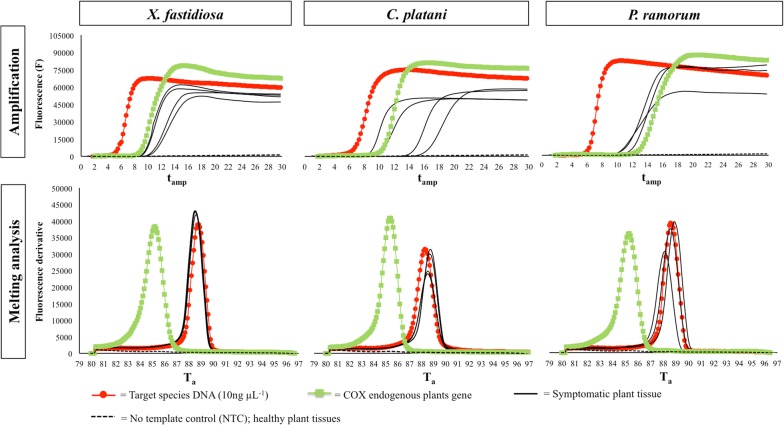
LAMP assay was able to detect DNA of each target pathogen (X. fastidiosa, C. platani and P. ramorum) with positive results in the first time of the isothermal amplification (tamp c.a. 7 min for P. ramorum and X. fastidiosa; c.a. 8 min for C. platani) (Fig. 1). All DNA samples of X. fastidiosa that include X. fastidiosa (Co.Di.Ro), X. fastidiosa subsp. fastidiosa (Xff) and X. fastidiosa subsp. multiplex (Xfm) were positively amplified by LAMP assay, and the melting curve showed a specific peak (Ta ranged between 88.78 and 88.98 °C) (Table 1). Bacterial DNA extracted from the other strains were not amplified by LAMP assay (Table 1). LAMP results were also confirmed by qPCR by using the designed primers (Xf_Fw and Xf_Rev) and probe (Xf_Pr) for X. fastidiosa (Tables 1, 2).
Fig. 1.
Selection of kinetics. Real time LAMP results reported as amplification and melting derivative plot for Xylella fastidiosa, Ceratocystis platani and Phytophthora ramorum including target species DNA (10 ng μL−1) in red, COX endogenous plant gene (green) and symptomatic plant tissues (black continuous line). No template control (NTC) and healthy plant tissue were also included (black dotted line). Ta annealing temperature, tamp amplification time
The real-time LAMP assay designed for C. platani was able to detect C. fimbriata strains belonging to different hosts and geographic origin (Table 1), whereas the qPCR assay gave negative results for these isolates. Similarly, the LAMP primers designed for P. ramorum were able to amplify P. lateralis DNA with melting temperatures very close to each other (Table 1). The other Phytophthora species included in this work either were not amplified or showed different amplification curves (with different tamp) or melting curves (with different Ta) (Table 1). For each designed LAMP assay DNA from outgroup species and common host colonizer species were not amplified, as confirmed by qPCR (Table 1).
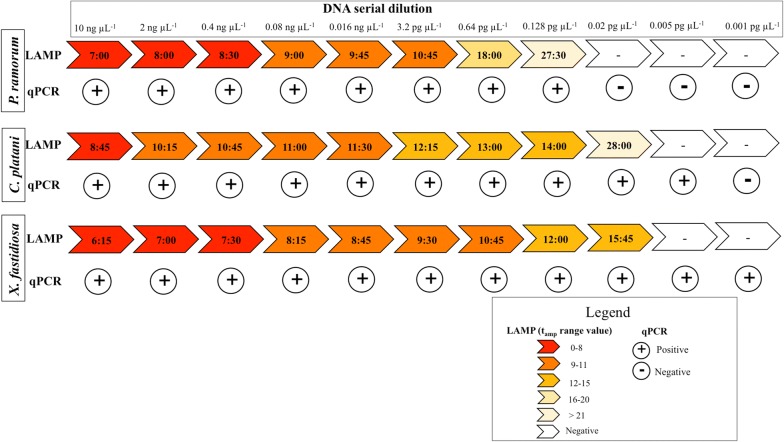
Sensitivity of real-time LAMP assays
The values of limit of detection of LAMP assays (LODLAMP) were always very low, ranging from 0.02 pg μL−1 for X. fastidiosa and C. platani and 0.128 pg μL−1 for P. ramorum, (Fig. 2; Table 3). P. ramorum qPCR assays had the same sensitivity as LAMP (LODqPCR = 0.128 pg μL−1). The qPCR assays for the other two pathogens were more sensitive than LAMP with lower detection limits (X. fastidiosa, LODqPCR = 0.001 pg μL−1; C. platani, LODqPCR = 0.005 pg μL−1) (Fig. 2).
Fig. 2.
Sensitivity results obtained by testing both on LAMP and qPCR 11-fold 1:5 serial dilution (ranged from 10 ng μL−1 to 0.001 pg μL−1) of each standard DNA template (X. fastidiosa—Co.Di.Ro strain; C. platani—isolate Cp24; P. ramorum- isolate Pram). LAMP results are inserted in a scale from positive (red) to negative (white) based on amplification time (tamp; min:s). Real-time qPCR results are reported as positive (+) ore negative (−)
Table 3.
Comparison of different DNA extraction and LAMP protocols for Xylella fastidiosa, Ceratocystis platani and Phytophthora ramorum detection
| Protocol | This study | Tomlinson et al. (2007) | Harper et al. (2010) | ||
|---|---|---|---|---|---|
| DNA extraction | |||||
| Target pathogen | Xylella fastidiosa, Ceratocystis platani, Phytophthora ramorum | Phytophthora ramorum | Xylella fastidiosa | ||
| Commercial kit | Plant Material Lysis Kit (OptiGene) | EZNA Plant DNA Kit (Omega Bio-tek) | QuickPick Plant DNA kit (Bio-Nobile) | Invimag Plant DNA Mini Kit (Invitek) | DNeasy Plant Minikit (Qiagen) |
| Use | Field | Laboratory | Field | Laboratory | Laboratory |
| Sample requirement | Fresh plant tissue (80–100 mg) | Fresh plant tissue (80–100 mg) | Fresh plant tissue (15–25 mg) | Lyophilized petiole (200 mg) | Fresh plant tissue (200 mg) |
| Advantages | Rapid and simple protocol with few reagents and steps; no laboratory instruments are required | Protocol kit with spin columns and buffer supplied | Processing up to 24 samples in parallel | Simplified sample processing | Grounding with beads; kit with spin column and buffer supplied |
| Disadvantage | Difficult for large number of samples | Required laboratories facilities for grinding and DNA extraction | Extremely basic equipment is needed | Required laboratories facilities for grinding and for DNA extraction | Required laboratories facilities for grinding and for DNA extraction |
| Time per sample | 5 min | 1 h | 40–50 min | > 30 min | 1 h |
| Isothermal DNA amplification | |||||
| Instrument | Genie II (OptiGene) | Smart Cycler (Cepheid) | ABI 9700 Thermocycler (Applied Biosystems) | ||
| Use | Field | Laboratory | Laboratory | ||
| Sensitivity (LOD) | P. ramorum (0.128 pg) | P. ramorum (10 pg) | – | ||
| X. fastidiosa (0.02 pg) | – | X. fastidiosa (1.4 pg) | |||
| C. platani (0.02 pg) | – | – | |||
| Specificity | P. ramorum (high specific; P. lateralis) | P. ramorum (high specific; P. lateralis) | – | ||
| X. fastidiosa (very high specific) | – | X. fastidiosa (very high specific) | |||
| C. platani (high specific; C. fimbriata) | – | – | |||
| Advantages | Rapid detection results; amplification and detection reaction is carried out in the same instrument (16 sample per run) | High number of samples to be processed | High number of samples to be processed | ||
| Disadvantage | Strip tubes with amplification mix need to be prepared before in laboratory | Additional steps to visualize amplified products (electrophoresis gel, colorimetric detection, fluorescent dye) | Electrophoresis gel to visualize amplified products | ||
| Time per sample | 30 min | > 1 h | > 1 h | ||
LOD limit of detection
We also observed a very strong correlation between the tamp of the LAMP assay and Ct value of the qPCR in the same set of DNA samples (X. fastidiosa: R2 = 0.97; C. platani R2 = 0.95; P. ramorum R2 = 0.98) (Fig. 3).
Fig. 3.
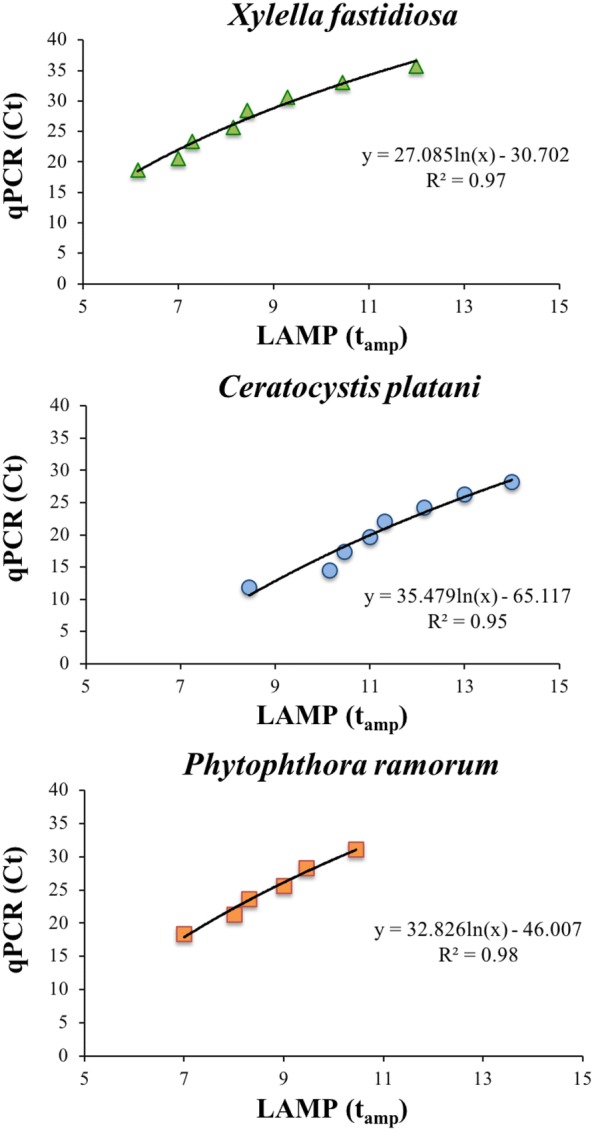
Statistical correlation between LAMP amplification time (tamp) and qPCR threshold cycle (Ct) obtained by testing with both methods each 1:5 serial dilution (ranged from 10 ng μL−1 to 0.128 pg μL−1) of each standard DNA template (X. fastidiosa—Co.Di.Ro strain; C. platani—isolate Cp24; P. ramorum-isolate Pram)
Real-time LAMP detection in plant samples
LAMP analyses carried out on plant host DNA were further validated by COX gene amplification, showing a specific melting peak at Ta = 85 °C for each analysed plant sample (both healthy and infected tissues) (Fig. 1). COX gene amplification was a reliable internal positive control, confirming host DNA extractions were successful by using both on-site DNA extraction kit (OptiGene) and laboratory commercial kit (Omega Bio-tek).
All symptomatic host plant samples (Olea europaea, Rhamnus alaternus, Calicotome spinosa, Cistus incanus, S. junceum, Prunus dulcis, Platanus × acerifolia and Viburnum tinus) were amplified successfully with the LAMP assay designed for each target pathogen (X. fastidiosa, C. platani and P. ramorum, respectively).
Symptomatic plant tissue showed similar Ta obtained from DNA of axenic cultures of each target pathogen (Table 1; Fig. 1), confirming the specificity of each LAMP assay to detect pathogens in infected plant tissues.
No amplification nor melting curve was obtained by applying the LAMP primers to healthy samples confirming the specificity of the LAMP optimized assay.
Discussion
In this work LAMP assays for detecting X. fastidiosa, P. ramorum and C. platani, optimized for a portable instrument in real time were developed. LAMP-based assays optimized in this study allow a complete analysis (amplification and annealing) in only 30 min, starting to have positive amplification from ca. 7 min (Table 1). To our best knowledge no previous LAMP assay has been developed for C. platani. qPCR showed higher sensitivity with respect to LAMP in X. fastidiosa and C. platani detection, while for P. ramorum LOD was the same as that of LAMP.
The opportunity to have an accurate and rapid detection of the three quarantine pathogens considered in this study directly in the field by a portable instrument, represents a great advantage to preventing introductions and for applying control measures. Most of the LAMP-based assays recently developed for plant pathogens, including the one developed for P. ramorum by Tomlinson et al. (2007) and for X. fastidiosa by Harper et al. (2010), are based on laborious and time-consuming isothermal amplification reactions (Table 3). As an example, the LAMP protocol adopted by EPPO for X. fastidiosa detection and developed by Harper et al. (2010), requires ca. 60 min to amplify all the isolates tested by the author and to consistently amplify ca. 250 copies of template for reaction (corresponding to 1.4 pg μL−1 pathogen DNA) in host (Vitis vinifera) DNA. In comparison, the assay developed in the current study requires only ca. 15 min to amplify 0.02 pg μL−1 of X. fastidiosa DNA in dd-water. The use of a simple colour change method to assess the positive result of LAMP-tested samples (e.g. Hydroxynaphtal blue dye used in Harper et al. 2010), could be particularly suited for use in the field, but opening the tube to add the colorimetric dye makes the method extremely vulnerable to carryover contamination due to the very large amount of product generated by LAMP reaction (Tomlinson et al. 2007). Furthermore, some colorimetric dyes reagents can completely inhibit the LAMP reaction at the concentration needed to produce a colour change visible with the naked eye (Tomlinson et al. 2007) and even though they may be possible to observe in a laboratory environment, they are difficult to detect in the field due to the different light conditions at different times of the day (Lau and Botella 2017), leading to false negative results or to losses in detection sensitivity. In addition, the interpretation of positive results from colour changes in colorimetric dyes is very subjective, requiring experienced staff. On the contrary, the main parameters used to assess the positivity of a sample in a LAMP real-time assay, as the one developed in the present work, are amplification time (tamp) and annealing temperature (Ta) resulting by fluorescence analysis results provided by the instrument.
The EPPO diagnostic protocol (PM 7/24) for X. fastidiosa describes a field LAMP assay based on the paper by Yaseen et al. (2015). In this paper authors optimized the Harper et al. (2010) assay for a portable instrument, but they do not report the sensitivity of the assay, strongly limiting its application due to the risk of false negatives.
LAMP assays developed in this study are specific and able to detect the target species, both from pure DNA and from DNA obtained from plant infected tissues. Some cross reactions have been observed in species genetically closely related to target species (for C. platani/C. fimbriata and P. ramorum/P. lateralis); however, their Ta is one-two degrees higher than that of the target organisms (89–90 °C vs. 88 °C), allowing a correct detection (Table 1).
A positive amplification sharing the same Ta of that of P. ramorum and C. platani (88 °C) was obtained only with P. lateralis and C. fimbriata, respectively. These species are almost morphologically indistinguishable and phylogenetically very close (De Beer et al. 2014; Kroon et al. 2012; Martin et al. 2014), but they were reported on very different hosts: P. lateralis attacks Chamaecyparis spp. and other Cupressaceae (Hansen et al. 2000; Robin et al. 2011), and C. fimbriata is the agent of sweet potato black rot (Okada et al. 2017).
The results of LAMP assays were also validated by those obtained from qPCR assays. The new TaqMan qPCR assay developed in this study for targeting X. fastidiosa is able to amplify all the X. fastidiosa tested subspecies with high efficiency excluding other tested bacteria species (Table 1). Furthermore, its sensitivity (0.001 pg μL−1) is much higher than that of the qPCR TaqMan assays developed by Harper et al. (2010) and by Francis et al. (2006) (both EPPO official diagnostic qPCR for X. fastidiosa) which has a detection limit of 0.05 pg μL−1, corresponding to 20 copies of template for reaction.
The use of rapid, specific and sensitive point-of-care methods like the LAMP assays developed in this study could enable phytosanitary services to make immediate management decisions, helping in containing environmental and economic losses. The application of such a portable diagnostic tool, requiring minimum equipment and a few, if any, specific scientific skills could be profitably used to check the health status of live plants or plant parts at the points of entry or in field, thus reducing time of analyses, thus allowing a prompt reaction. In conclusion, the results presented in this study show how an advance in technology can provide efficient tools to prevent the introduction or limit the spread of diseases that can have severe economic, ecological and sociological consequences.
Authors’ contributions
NL, AS conceived and designed the experiments. CA, NL, ALP, PB, FP, AS performed the field work and the experiments. AR provided bacterial strains. CA, NL, AS analyzed the data. CA, NL, AS wrote the paper. AR, PC made contribution to the revision of the manuscript. All authors read and approved the final manuscript.
Acknowledgements
The authors wish to thank colleagues who kindly provided DNA and isolates of fungi and bacteria species used in this work: R. Ioos and N. Schenck (ANSES, France), G.P. Barzanti (CREA, Italy), S.O. Cacciola (Università di Catania, Italy), T. Dogmus (Süleyman Demirel University, Isparta, Turkey), T. Jung (University of Algarve, Portugal), S. Leonhard, J. Schumacher (BBA, Germany), A. Pérez-Sierra (Forest Research, UK), C. Robin (INRA, France), P. Tsopelas, (NAGREF, Greece), A. Vettraino (Università della Tuscia, Italy), M. Saponari, D. Boscia (IPSP-CNR, Bari, Italy), G. Marchi (University of Florence, Italy). Authors are also grateful to Tuscany Regional Phytosanitary Service for helping to the sampling in the field. Authors wish to warmly thank Dr. Trudy Paap (FABI, University of Pretoria, South Africa) for the thorough critical review of the text and for English language editing of the manuscript. The authors would like to thank the editor and the anonymous referees for their comments and suggestions that greatly improved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
The data supporting the conclusions of this article are included within the article. Data and materials can also be requested from the corresponding author.
Consent for publication
All authors gave their consent for publication.
Ethical approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Funding
This study was funded by European Union’s Horizon 2020 Research and Innovation Programme (grant No 771271). Part of this work has been funded by the project “PATINVIVA—Invasive pathogens in nurseries: new tools for the certification of pathogen exemption of material for export”—Fondazione Cassa di Risparmio di Pistoia e Pescia (grant No 255).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Abbreviations
- X. fastidiosa
Xylella fastidiosa
- C. platani
Ceratocystis platani
- P. ramorum
Phytophthora ramorum
- LAMP
loop mediated isothermal amplification
- qPCR
real-time quantitative polymerase chain reaction
- Ct
threshold cycle
- tamp
amplification time
- Ta
amplicon annealing temperature
- LOD
limit of detection
Contributor Information
Chiara Aglietti, Email: chiara.aglietti@unifi.it.
Nicola Luchi, Phone: +390555225531, Email: nicola.luchi@ipsp.cnr.it.
Alessia Lucia Pepori, Email: alessia.pepori@ipsp.cnr.it.
Paola Bartolini, Email: paola.bartolini@ipsp.cnr.it.
Francesco Pecori, Email: francesco.pecori@ipsp.cnr.it.
Aida Raio, Email: aida.raio@ipsp.cnr.it.
Paolo Capretti, Email: paolo.capretti@unifi.it.
Alberto Santini, Email: alberto.santini@cnr.it.
References
- Abdulmawjood A, Grabowski N, Fohler S, Kittler S, Nagengast H, Klein G. Development of loop-mediated isothermal amplification (LAMP) assay for rapid and sensitive identification of ostrich meat. PLoS ONE. 2014;9:e100717. doi: 10.1371/journal.pone.0100717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bilodeau GJ, Lévesque CA, De Cock AW, Duchaine C, Brière S, Uribe P, Martin FN, Hamelin RC. Molecular detection of Phytophthora ramorum by real-time polymerase chain reaction using TaqMan, SYBR Green, and molecular beacons. Phytopathology. 2007;97:632–642. doi: 10.1094/PHYTO-97-5-0632. [DOI] [PubMed] [Google Scholar]
- Brasier C, Webber J. Plant pathology: sudden larch death. Nature. 2010;466:824–825. doi: 10.1038/466824a. [DOI] [PubMed] [Google Scholar]
- Bühlmann A, Pothier JF, Rezzonico F, Smits TH, Andreou M, Boonham N, Duffy B, Frey EJ. Erwinia amylovora loop-mediated isothermal amplification (LAMP) assay for rapid pathogen detection and on-site diagnosis of fire blight. J Microbiol Meth. 2013;92:332–339. doi: 10.1016/j.mimet.2012.12.017. [DOI] [PubMed] [Google Scholar]
- Chandelier A, Ivors K, Garbelotto M, Zini J, Laurent F, Cavelier M. Validation of a real-time PCR method for the detection of Phytophthora ramorum. EPPO Bull. 2006;36:409–414. doi: 10.1111/j.1365-2338.2006.01020.x. [DOI] [Google Scholar]
- Chen Q, Li B, Liu P, Lan C, Zhan Z, Weng Q. Development and evaluation of specific PCR and LAMP assays for the rapid detection of Phytophthora melonis. Eur J Plant Pathol. 2013;137:597–607. doi: 10.1007/s10658-013-0273-9. [DOI] [Google Scholar]
- Dai TT, Lu CC, Lu J, Dong S, Ye W, Wang Y, Zheng X. Development of a loop-mediated isothermal amplification assay for detection of Phytophthora sojae. FEMS Microbiol Lett. 2012;334:27–34. doi: 10.1111/j.1574-6968.2012.02619.x. [DOI] [PubMed] [Google Scholar]
- De Beer ZW, Duong TA, Barnes I, Wingfield BD, Wingfield MJ. Redefining Ceratocystis and allied genera. Stud Mycol. 2014;79:187–219. doi: 10.1016/j.simyco.2014.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denancé N, Legendre B, Briand M, Olivier V, De Boisseson C, Poliakoff F, Jacques MA. Several subspecies and sequence types are associated with the emergence of Xylella fastidiosa in natural settings in France. Plant Pathol. 2017;66:1054–1064. doi: 10.1111/ppa.12695. [DOI] [Google Scholar]
- EPPO 2019- Xylella fastidiosa subsp. multiplex detected in Toscana region, Italy. N. 2019/16. EPPO Reporting Service n.01-2019
- Fisher MC, Henk DA, Briggs CJ, Brownstein JS, Madoff LC, McCraw SL, Gurr SJ. Emerging fungal threats to animal, plant and ecosystem health. Nature. 2012;484:186194. doi: 10.1038/nature10947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis M, Lin H, Cabrera-La Rosa J, Doddapaneni H, Civerolo EL. Genome-based PCR primers for specific and sensitive detection and quantification of Xylella fastidiosa. Eur J Plant Pathol. 2006;115:203–213. doi: 10.1007/s10658-006-9009-4. [DOI] [Google Scholar]
- Franco Ortega S, Tomlinson J, Hodgetts J, Spadaro D, Gullino LM, Boonham N. Development of loop-mediated isothermal amplification assays for the detection of seedborne fungal pathogens, Fusarium fujikuroi and Magnaporthe oryzae, in rice seed. Plant Dis. 2018;102:1549–1558. doi: 10.1094/PDIS-08-17-1307-RE. [DOI] [PubMed] [Google Scholar]
- Hansen EM, Goheen DJ, Jules ES, Ullian B. Managing Port-Orford-cedar and the introduced pathogen Phytophthora lateralis. Plant Dis. 2000;84:4–14. doi: 10.1094/PDIS.2000.84.1.4. [DOI] [PubMed] [Google Scholar]
- Hansen ZR, Knaus BJ, Tabima JF, Press CM, Judelson HS, Grünwald NJ, Smart CD. Loop-mediated isothermal amplification for detection of the tomato and potato late blight pathogen, Phytophthora infestans. J Appl Microbiol. 2016;120:1010–1020. doi: 10.1111/jam.13079. [DOI] [PubMed] [Google Scholar]
- Harper SJ, Ward LI, Clover GRG. Development of LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology. 2010;100:1282–1288. doi: 10.1094/PHYTO-06-10-0168. [DOI] [PubMed] [Google Scholar]
- Harrison C, Tomlinson J, Ostoja-Starzewska S, Boonham N. Evaluation and validation of a loop-mediated isothermal amplification test kit for detection of Hymenoscyphus fraxineus. Eur J Plant Pathol. 2017;149:253–259. doi: 10.1007/s10658-017-1179-8. [DOI] [Google Scholar]
- Kroon LP, Brouwer H, De Cock AW, Govers F. The genus Phytophthora anno 2012. Phytopathology. 2012;102:348–364. doi: 10.1094/PHYTO-01-11-0025. [DOI] [PubMed] [Google Scholar]
- Lau HY, Botella JR. Advanced DNA-based point-of-care diagnostic methods for plant diseases detection. Front Plant Sci. 2017;8:2016. doi: 10.3389/fpls.2017.02016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtijärvi A, Oskay F, Doğmuş Lehtijärvi HT, Aday Kaya AG, Pecori F, Santini A, Woodward S. Ceratocystis platani is killing plane trees in Istanbul (Turkey) For Pathol. 2018;48:e12375. doi: 10.1111/efp.12375. [DOI] [Google Scholar]
- Loconsole G, Potere O, Boscia D, Altamura G, Djelouah K, Elbeaino T, Frasheri D, Lorusso D, Palmisano F, Pollastro P, Silletti MR, Trisciuzzi N, Valentini F, Savino V, Saponari M. Detection of Xylella fastidiosa in olive trees by molecular and serological methods. J Plant Pathol. 2014;96:7–14. doi: 10.4454/JPP.V96I1.041. [DOI] [Google Scholar]
- Lucchi NW, Demas A, Narayanan J, Sumari D, Kabanywanyi A, Kachur SP, Barnewell JW, Udhayakumar V. Real-time fluorescence loop mediated isothermal amplification for the diagnosis of malaria. PLoS ONE. 2010;5:e13733. doi: 10.1371/journal.pone.0013733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luchi N, Ghelardini L, Belbahri L, Quartier M, Santini A. Rapid detection of Ceratocystis platani inoculum by quantitative real-time PCR assay. Appl Environ Microbiol. 2013;79:5394–5404. doi: 10.1128/AEM.01484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FN, Blair JE, Coffey MD. A combined mitochondrial and nuclear multilocus phylogeny of the genus Phytophthora. Fungal Genet Biol. 2014;66:19–32. doi: 10.1016/j.fgb.2014.02.006. [DOI] [PubMed] [Google Scholar]
- Migliorini D, Ghelardini L, Tondini E, Luchi N, Santini A. The potential of symptomless potted plants for carrying invasive soilborne plant pathogens. Div Distrib. 2015;21:1218–1229. doi: 10.1111/ddi.12347. [DOI] [Google Scholar]
- Migliorini D, Ghelardini L, Luchi N, Capretti P, Onorari M, Santini A. Temporal patterns of airborne Phytophthora spp. in a woody plant nursery area detected using Real time PCR. Aerobiologia. 2018 doi: 10.1007/s10453-018-09551-1. [DOI] [Google Scholar]
- Moradi A, Almasi MA, Jafary H, Mercado-Blanco J. A novel and rapid loop-mediated isothermal amplification assay for the specific detection of Verticillium dahliae. J Appl Microbiol. 2014;116:942–954. doi: 10.1111/jam.12407. [DOI] [PubMed] [Google Scholar]
- Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28:e63. doi: 10.1093/nar/28.12.e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y, Kobayashi A, Tabuchi H, Kuranouchi T. Review of major sweetpotato pests in Japan, with information on resistance breeding programs. Breed Sci. 2017;67:73–82. doi: 10.1270/jsbbs.16145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J, Zhan Y, Zeng F, Long H, Pei Y, Guo J. Development of a real-time fluorescence loop-mediated isothermal amplification assay for rapid and quantitative detection of Fusarium oxysporum f. sp. niveum in soil. FEMS Microbiol Lett. 2013;349:127–134. doi: 10.1111/1574-6968.12305. [DOI] [PubMed] [Google Scholar]
- Rizzo DM, Garbelotto M, Davidson M, Slaughter JM, Koike GW. Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis. 2002;86:205–214. doi: 10.1094/PDIS.2002.86.3.205. [DOI] [PubMed] [Google Scholar]
- Robin C, Piou D, Feau N, Douzon G, Schenck N, Hansen EM. Root and aerial infections of Chamaecyparis lawsoniana by Phytophthora lateralis: a new threat for European countries. Forest Pathol. 2011;41:417–424. doi: 10.1111/j.1439-0329.2010.00688.x. [DOI] [Google Scholar]
- Rollins L, Coats K, Elliott M, Chastagner G. Comparison of five detection and quantification methods for Phytophthora ramorum in stream and irrigation water. Plant Dis. 2016;100:1202–1211. doi: 10.1094/PDIS-11-15-1380-RE. [DOI] [PubMed] [Google Scholar]
- Santini A, Ghelardini L, De Pace C, Desprez-Loustau ML, Capretti P, Chandelier A, Cech T, Chira D, Diamandis S, Gaitniekis T, Hantula J, Holdenrieder O, Jankovsky L, Jung T, Jurc D, Kirisits T, Kunca A, Lygis V, Malecka M, Marcais B, Schmitz S, Schumacher J, Solheim H, Solla A, Szabò I, Tsopelas P, Vannini A, Vettraino AM, Webber J, Woodward S, Stenlid J. Biogeographical patterns and determinants of invasion by forest pathogens in Europe. New Phytol. 2013;197:238–250. doi: 10.1111/j.1469-8137.2012.04364.x. [DOI] [PubMed] [Google Scholar]
- Santini A, Liebhold A, Migliorini D, Woodward S. Tracing the role of human civilization in the globalization of plant pathogens. ISME J. 2018;12:647–652. doi: 10.1038/s41396-017-0013-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saponari M, Boscia D, Nigro F, Martelli GP. Identification of DNA sequences related to Xylella fastidiosa in oleander, almond and olive trees exhibiting leaf scorch symptoms in Apulia (Southern Italy) J Plant Pathol. 2013;95:3. doi: 10.4454/JPP.V95I3.035. [DOI] [Google Scholar]
- Sillo F, Giordano L, Gonthier P. Fast and specific detection of the invasive forest pathogen Heterobasidion irregulare through a loop-mediated isothermal AMPlification (LAMP) assay. For Pathol. 2018;48:e12396. doi: 10.1111/efp.12396. [DOI] [Google Scholar]
- Tomlinson JA, Barker I, Boonham N. Faster, simpler, more-specific methods for improved molecular detection of Phytophthora ramorum in the field. Appl Environm Microbiol. 2007;73:4040–4047. doi: 10.1128/AEM.01389-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson JA, Dickinson MJ, Boonham N. Rapid detection of Phytophthora ramorum and P. kernoviae by two-minute DNA extraction followed by isothermal amplification and amplicon detection by generic lateral flow device. Phytopathol. 2010;100:143–149. doi: 10.1094/PHYTO-100-2-0143. [DOI] [PubMed] [Google Scholar]
- Tomlinson JA, Dickinson MJ, Boonham N. Detection of Botrytis cinerea by loop-mediated isothermal amplification. Lett Appl Microbiol. 2010;51:650–657. doi: 10.1111/j.1472-765X.2010.02949.x. [DOI] [PubMed] [Google Scholar]
- Tomlinson JA, Ostoja-Starzewska S, Webb K, Cole J, Barnes A, Dickinson M, Boonham N. A loop-mediated isothermal amplification-based method for confirmation of Guignardia citricarpa in citrus black spot lesions. Eur J Plant Pathol. 2013;136:217–224. doi: 10.1007/s10658-013-0168-9. [DOI] [Google Scholar]
- Tsopelas P, Santini A, Wingfield MJ, Wilhelm de Beer Z. Canker stain: a lethal disease destroying iconic plane trees. Plant Dis. 2017;101:645–658. doi: 10.1094/PDIS-09-16-1235-FE. [DOI] [PubMed] [Google Scholar]
- Werres S, Marwitz R, Man in ‘t Veld WA, De Cock WAM, Bonants PJM, De Weerdt M, Themann K, Ilieva E, Baayen RP. Phytophthora ramorum sp. nov., a new pathogen on Rhododendron and Viburnum. Mycol Res. 2001;105:1155–1165. doi: 10.1016/S0953-7562(08)61986-3. [DOI] [Google Scholar]
- Yaseen T, Drago S, Valentini F, Elbeaino T, Stampone G, Digiaro M, D’onghia AM. On-site detection of Xylella fastidiosa in host plants and in “spy insects” using the real-time loop-mediated isothermal amplification method. Phytopathol Medit. 2015;54:488–496. doi: 10.14601/Phytopathol_Mediterr-15250. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the conclusions of this article are included within the article. Data and materials can also be requested from the corresponding author.