Abstract
Although many studies have shown that species richness decreases from low to high latitudes (the Latitudinal Diversity Gradient), little is known about the relationship between latitude and phylogenetic diversity. Here we examine global latitudinal patterns of phylogenetic diversity using a dataset of 459 woody and 589 herbaceous plant communities. We analysed the relationships between community phylogenetic diversity, latitude, biogeographic realm and vegetation type. Using the most recent global megaphylogeny for seed plants and the standardised effect sizes of the phylogenetic diversity metrics ‘mean pairwise distance’ (SESmpd) and ‘mean nearest taxon distance’ (SESmntd), we found that species were more closely-related at low latitudes in woody communities. In herbaceous communities, species were more closely-related at high latitudes than at intermediate latitudes, and the strength of this effect depended on biogeographic realm and vegetation type. Possible causes of this difference are contrasting patterns of speciation and dispersal. Most woody lineages evolved in the tropics, with many gymnosperms but few angiosperms adapting to high latitudes. In contrast, the recent evolution of herbaceous lineages such as grasses in young habitat types may drive coexistence of closely-related species at high latitudes. Our results show that high species richness commonly observed at low latitudes is not associated with high phylogenetic diversity.
Subject terms: Community ecology, Evolutionary ecology, Macroecology, Phylogenetics, Speciation
Introduction
The latitudinal diversity gradient (LDG), which shows a pattern of decreasing species richness from the equator towards the poles1,2 is one of the earliest recorded and most striking global biogeographical patterns. Many hypotheses have been proposed to explain the LDG3–6. At the core of most of these hypotheses is the issue of the way in which evolution has shaped this pattern. For example, the time-integrated species-area hypothesis7 and the tropical conservatism hypothesis8 suggest that species diversity decreases with latitude because tropical biomes are larger and older than temperate biomes. Moreover, many currently extant clades originated in the tropics9,10, and therefore species in these clades have had more time, and a larger area in which to diversify, than would have been the case if they had originated at higher latitudes. It is widely believed that, following their evolution under tropical conditions, few of the species in these clades, or their descendants, subsequently migrated far from their region of origin8.
Using information about currently co-existing species to study the way in which evolution might have shaped the LDG makes the assumption that the species from which ecological communities are composed themselves provide valuable evolutionary information11. In particular, in addition to differing in species composition and richness, communities also display considerable differences in the evolutionary origin - i.e. the phylogenetic relatedness - of their constituent species. This aspect of biodiversity is reflected in community phylogenetic diversity. Two communities may have similar species diversity, but differ considerably in phylogenetic diversity because of differences in the evolutionary history of their constituent species. A community consisting of closely-related species has a low phylogenetic diversity compared to a community consisting of the same number of distantly-related species. Measuring and comparing phylogenetic diversity in different plant communities therefore offers the possibility of interpreting latitudinal diversity gradients not only on the basis of present-day ecological processes, but also by considering the role of evolutionary drivers including speciation and extinction.
Localized speciation causing low phylogenetic diversity has been observed repeatedly in a variety of plant communities. For example, neo-endemism, i.e. recent localized evolutionary radiation, is responsible for low phylogenetic diversity in Amazonian white-sand forests12. Similarly, localized radiations have resulted in low phylogenetic diversity in communities of the Qinghai-Tibetan Plateau, Central Asia13. The recently radiated lineages Restionaceae and Ericaceae have produced communities with low phylogenetic diversity in South African semi-arid shrublands14. Recent and rapid radiations in the genus Eucalyptus15 have resulted in low phylogenetic diversity in some Australian subtropical rainforest communities16. In contrast, low extinction rates, which result in long-term accumulation of distantly-related lineages, can result in communities with high phylogenetic diversity17,18. For example, coexistence of distantly-related plant lineages with resprouting ability in response to fire has promoted high phylogenetic diversity in low latitude Brazilian savannah communities throughout the last 8 million years19, and geographic isolation coupled with trait conservatism in distantly-related lineages has resulted in high phylogenetic diversity in the mountain forests of Malesia20.
Apart from speciation and extinction events, macroecological factors such as climate and regional stability can influence community phylogenetic diversity. Climate is a strong driver of phylogenetic diversity of angiosperm tree assemblages at regional scales21,22 and across the globe23,24 because of its tendency to select closely related species. Phylogenetically diverse tree assemblages are associated with long-term climatic stability25 and environmental heterogeneity26. Although various evolutionary and macroecological factors are major drivers of species diversity, little attention has been given to the way in which these factors combine to influence the LDG. To our knowledge, no comparative studies have been conducted on plant communities, and although studies have been carried out on specific taxonomic groups, including woody angiosperms24, amphibians27 and mammals28, no global-scale study has been undertaken to examine variation in phylogenetic diversity across a wide taxonomic range of plant communities.
In this study, we conduct a global meta-study to investigate patterns of phylogenetic diversity in contemporary plant communities along the latitudinal gradient from the tropics to polar regions (Fig. 1), in the context of evolutionary and macroecological processes. Specifically, we examine the relationships between the phylogenetic diversity of woody and herbaceous plant communities and latitude, biogeographic realms29, and vegetation type. We analysed woody and herbaceous communities independently because the presence of distinct growth forms (i.e. woody and herbaceous species) throughout the latitudinal gradient depends on the responses of many species in these categories to climatic variables that are correlated with latitude30. We calculated the standardised effect size of the mean pairwise distance (SESmpd), and standardised effect size of the mean nearest taxon distance (SESmntd) across the species recorded in each community. The first index measures phylogenetic distances across the whole phylogenetic tree by averaging all species pairwise distances, whereas the latter measures the phylogenetic distance at a shallower level, namely between closest relatives. Both indices have been shown to be independent of species richness when the number of species is held constant during randomizations31. Although these indices reflect “community phylogenetic structure” or “phylogenetic relatedness” of coexisting species, we use the term “phylogenetic diversity” of communities hereafter for simplicity.
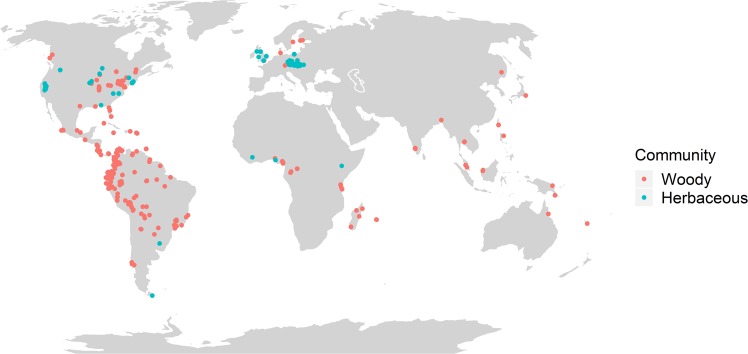
Figure 1.
Locations of plant communities included in this study. Red circles = woody communities (N = 459); blue circles = herbaceous communities (N = 589). Multiple data points may be superimposed. The global map was plotted using the Robinson projection.
Results
Results of all tests are listed in Tables 1 and 2. Parameter estimates of the top-ranked linear mixed effects models are also shown in the Supplementary Information (Supplementary Tables S1, S2). In response to the wide range of values in latitude, there was considerable variation in phylogenetic diversity across the communities in our dataset, with SESmpd ranging from −10.50 to 9.54, and SESmntd ranging from −7.82 to 3.56. Model selection indicated the best models for each measure of phylogenetic diversity in each of the two types (i.e. woody, herbaceous) of communities (Table 1). The contribution of fixed effect variables (i.e. marginal R2) to the variability in phylogenetic diversity in the top-ranked models generally amounted to approximately half of the variability explained by both fixed and random effects variables for SESmpd in woody communities and SESmntd in herbaceous communities. It corresponds to 25% and 28% of the total variance explained by the models respectively. The fixed effect variables in the top-ranked models of SESmpd in herbaceous communities and SESmntd in woody communities explained 13% and 11% of the total variance respectively (Table 1). The inclusion of sampling unit size in the models did not affect the relationships in any community type (Table 2, Supplementary Tables S1, S2).
Table 1.
Ranking of the linear mixed effects models for linear and quadratic (latitude2) effects of latitude and other studied variables on community phylogenetic diversity (standardised effect size of the mean pairwise distance – SESmpd, and standardised effect size of the mean nearest taxon distance – SESmntd) in woody and herbaceous communities.
| Community type | Phylogenetic diversity | Model | logLik | AICc | ∆AICc | AICw | R2m | R2c |
|---|---|---|---|---|---|---|---|---|
| Woody | SESmpd | latitude + latitude2 + size + vegetation | −947.968 | 1916.4 | 0.00 | 0.400 | 0.25 | 0.46 |
| latitude + latitude2 + size + realm | −945.322 | 1917.5 | 1.03 | 0.238 | 0.28 | 0.47 | ||
| latitude + latitude2 + size | −950.605 | 1917.5 | 1.10 | 0.230 | 0.23 | 0.45 | ||
| latitude + latitude2 + size + realm + vegetation | −943.783 | 1918.6 | 2.22 | 0.132 | 0.29 | 0.48 | ||
| SESmntd | latitude + latitude2 + size | −843.544 | 1703.4 | 0.00 | 0.850 | 0.11 | 0.45 | |
| latitude + latitude2 + size + vegetation | −843.302 | 1707.1 | 3.69 | 0.135 | 0.11 | 0.45 | ||
| latitude + latitude2 + size + realm | −842.417 | 1711.7 | 8.24 | 0.014 | 0.12 | 0.44 | ||
| latitude + latitude2 + size + realm + vegetation | −842.286 | 1715.7 | 12.25 | 0.002 | 0.12 | 0.44 | ||
| Herbaceous | SESmpd | latitude + latitude2 + size + realm + vegetation | −997.599 | 2019.7 | 0.00 | 0.805 | 0.13 | 0.48 |
| latitude + latitude2 + size + vegetation | −1002.130 | 2022.6 | 2.83 | 0.195 | 0.13 | 0.51 | ||
| latitude + latitude2 + size + realm | −1008.466 | 2039.4 | 19.65 | 0.000 | 0.08 | 0.48 | ||
| latitude + latitude2 + size | −1012.197 | 2040.6 | 20.90 | 0.000 | 0.06 | 0.49 | ||
| SESmntd | latitude + latitude2 + size + realm + vegetation | −861.604 | 1747.7 | 0.00 | 0.762 | 0.28 | 0.38 | |
| latitude + latitude2 + size + vegetation | −865.881 | 1750.1 | 2.32 | 0.238 | 0.15 | 0.40 | ||
| latitude + latitude2 + size + realm | −882.025 | 1786.5 | 38.76 | 0.000 | 0.11 | 0.36 | ||
| latitude + latitude2 + size | −886.475 | 1789.2 | 41.45 | 0.000 | 0.01 | 0.41 |
The models are sorted by corrected Akaike information criterion value (AICc), with log likelihood (logLik), difference in AICc from the top-ranked model (∆AICc), model weight (AICw), marginal R2 (R2m), and conditional R2 (R2c). Size = sampling unit size; vegetation = vegetation type (closed, open, semi-open); realm = biogeographic realm (Afrotropical, Australasian, Indo-Malayan, Nearctic, Neotropical, Palearctic)29. Study identification was included as a random effect variable in all models.
Table 2.
Estimated effects of the variables in the top-ranked mixed effects models on community phylogenetic diversity (standardised effect size of the mean pairwise distance – SESmpd, and standardised effect size of the mean nearest taxon distance – SESmntd) in woody and herbaceous communities.
| Community type | Phylogenetic diversity | Top-ranked model | Variables | Coefficient | F-value | p-value |
|---|---|---|---|---|---|---|
| Woody | SESmpd | latitude + latitude2 + size + vegetation | Intercept | 0.2437 | 2.9568 | 0.0868 |
| Latitude | — | 37.6293 | <0.0001 | |||
| Linear term | 1.0241*** | — | — | |||
| Quadratic term | 0.5636*** | — | — | |||
| Size | 0.2065 | 3.1324 | 0.0781 | |||
| Vegetation | — | 2.6106 | 0.0758 | |||
| Open | −3.3202* | — | — | |||
| Semi-open | –0.3985 | — | — | |||
| SESmntd | latitude + latitude2 + size | Intercept | −2.7331 | 538.3517 | <0.0001 | |
| Latitude | — | 13.9199 | <0.0001 | |||
| Linear term | 0.5869*** | — | — | |||
| Quadratic term | 0.0522 | — | — | |||
| Size | 0.1664 | 3.2111 | 0.0745 | |||
| Herbaceous | SESmpd | latitude + latitude2 + size + realm + vegetation | Intercept | 13.1456 | 4.3958 | 0.0365 |
| Latitude | — | 5.2857 | 0.0053 | |||
| Linear term | 0.5858 | — | — | |||
| Quadratic term | −1.4841** | — | — | |||
| Size | −0.0714 | 0.9973 | 0.3184 | |||
| Realm | — | 2.8835 | 0.0541 | |||
| Nearctic | −14.3492* | — | — | |||
| Neotropical | −12.4259* | — | — | |||
| Paleactic | −13.1274 | — | — | |||
| Vegetation | — | 21.4457 | <0.0001 | |||
| Open | −1.3713*** | — | — | |||
| SESmntd | latitude + latitude2 + size + realm + vegetation | Intercept | −1.3252 | 0.1169 | 0.7325 | |
| Latitude | — | 3.4684 | 0.0318 | |||
| Linear term | 5.9833 | — | — | |||
| Quadratic term | 11.0210 | — | — | |||
| Size | −0.0380 | 0.4748 | 0.4910 | |||
| Realm | — | 2.8352 | 0.0569 | |||
| Nearctic | 1.9078 | — | — | |||
| Neotropical | 0.8520 | — | — | |||
| Paleactic | 0.5757 | — | — | |||
| Vegetation | — | 41.9209 | <0.0001 | |||
| Open | −1.3868*** | — | — |
Variables were tested with ‘anova.lme’ function from nlme package72, using the marginal significance of each fixed effect variable coefficient when all other fixed effects variables are present in the model. Size = sampling unit size; vegetation = vegetation type (closed, open, semi-open); realm = biogeographic realm (Afrotropical, Australasian, Indo-Malayan, Nearctic, Neotropical, Palearctic)29. Significance of individual coefficients are flagged with stars: *p < 0.05, **p < 0.01, ***p < 0.001. Estimates for biogeographic realms refer to the deviation from Afrotropical, and for vegetation type to the deviation from closed. All parameter estimates of the top-ranked models are shown in Supplementary Tables S1 and S2. Study identification was included as a random effect variable in all models.
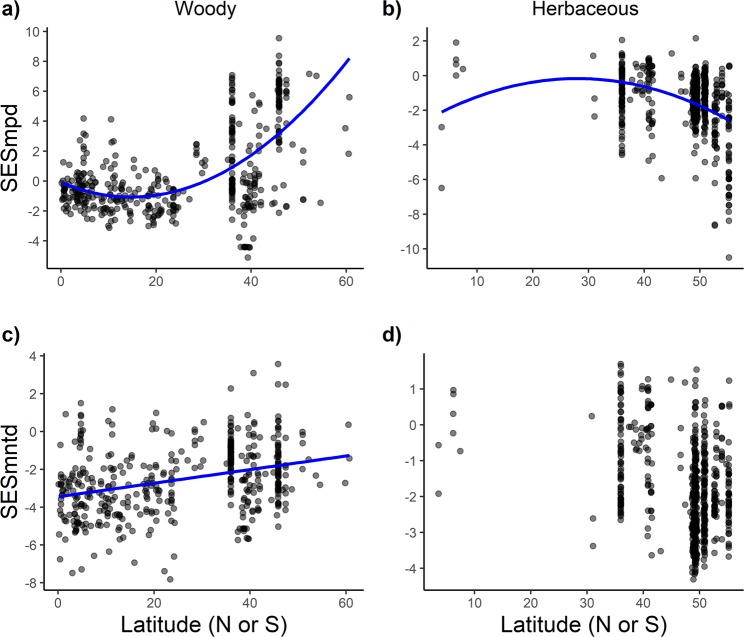
There were contrasting patterns of phylogenetic diversity in woody and herbaceous communities along the latitudinal gradient. In woody communities there were positive relationships between both measures of phylogenetic diversity and latitude (Fig. 2a,c). SESmpd showed a quadratic relationship, whereas SESmntd showed a positive linear relationship with latitude, with species being more closely-related at low latitudes in both cases. Vegetation type affected the relationship between SESmpd and latitude in woody communities (Supplementary Table S1 and Supplementary Fig. S1).
Figure 2.
Effect of latitude on phylogenetic diversity of plant communities. Standardised effect size of mean pairwise distance (SESmpd) in woody (a) and herbaceous (b) communities. Standardised effect size of mean nearest taxon distance (SESmntd) in woody (c) and herbaceous (d) communities. Multiple data points may be superimposed. Lines are based on the predicted values from the top-ranked models (see Table 2).
In contrast, the coefficient of the quadratic term for latitude in the top-ranked model of SESmpd had a negative value (Supplementary Table S2) which means that phylogenetic diversity was lower at high latitudes than at intermediate latitudes (Fig. 2b). SESmntd did not show a significant relationship with latitude (Fig. 2d, Supplementary Table S2). Vegetation type was the most important determining variable in the models for both measures of phylogenetic diversity: phylogenetic diversity was lower in open vegetation types than in closed vegetation types (Table 2, Supplementary Fig. S2b,d). Biogeographic realm was a significant determinant of SESmpd: phylogenetic diversity was higher in communities from the Afrotropical realm than in those from the Nearctic and Neotropical (Table 2, Supplementary Table S2, Supplementary Fig. S2a).
Excluding gymnosperms from woody communities resulted in a negative linear relationship between SESmpd and latitude (Supplementary Results, Supplementary Tables S3, S4, Supplementary Fig. S3a), and a positive linear relationship between SESmntd and latitude (Supplementary Results, Supplementary Tables S3, S4, Supplementary Fig. S3b).
Discussion
In accordance with several recent studies22,24,26–28,32–35, our results demonstrate that evolutionary and macroecological mechanisms contribute to explaining differences in the phylogenetic diversity of contemporary plant communities along the latitudinal gradient. In this study, data from a wide range of plant communities, distributed from the tropics to extreme polar regions, displayed significant effects of latitude on phylogenetic diversity. While local assembly processes acting at small scales may also influence community phylogenetic structure36–39, the inclusion of sampling unit size as a covariate in the models did not affect the latitudinal trends in phylogenetic diversity that we report (Table 2, Supplementary Tables S1, S2). This suggests strongly that the trends we report are unlikely to be altered by processes acting at very small scales.
Woody species were more closely-related in communities located at lower latitudes. The variance explained by fixed effect variables was 25% and 11% for SESmpd and SESmntd respectively. The higher level of explanation in the case of SESmpd is probably due to latitude being a major factor, coupled with vegetation type along the latitudinal gradient. Low phylogenetic diversity in communities at low latitudes probably arises because most woody lineages originated in warm climates such as those found in humid tropical regions10,40–42 in the Early Cretaceous (c. 146–100 myr43,44). In contrast to the results reported here, previous studies have demonstrated a pattern of lower phylogenetic diversity in woody communities located in colder and/or dryer regions21–24. This difference probably arises because the species lists used in our study involved complete lists of all woody species, including gymnosperms (110 gymnosperm species were included in the dataset in total). In contrast, earlier studies21–24 omitted gymnosperms because records of these species were rare in the data and consequently generated exceptionally long evolutionary branches in the phylogeny. We included gymnosperms because at the global scale they dominate many communities and occupy large areas of many regions around the globe (e.g. the Podocarpaceae and Araucariaceae in the Southern Hemisphere, and the Pinaceae in the Northern Hemisphere).
Excluding gymnosperms from woody communities revealed a negative relationship between SESmpd and latitude, which is in accordance with results from previous studies, and a positive relationship between SESmntd and latitude. A probable explanation for the negative relationship when gymnosperms are excluded, is that harsh conditions at high latitudes exclude angiosperm taxa that have evolved in warmer climates and favour taxa adapted to colder temperatures. The positive relationship between SESmntd and latitude probably arises from the fact that the division between angiosperms and gymnosperms deep in the phylogeny does not affect the relationships at the tips of the phylogeny. Gymnosperms were the most abundant plant species until the emergence of the angiosperms in the Late Cretaceous (c. 94–66 myr45), and global cooling at the end of the Eocene would have led to polewards expansion of temperate forests46. This might have created new regeneration niches for gymnosperms. Thus, the presence of phylogenetically distant gymnosperm species may have made a significant contribution to the high phylogenetic diversity we observed in woody communities located at high latitudes. Given that ecosystems at low latitudes are a major source of woody species, and that most gymnosperm species have physiological advantages over the angiosperms in cold, nutrient-poor environments47,48, the tendency for higher phylogenetic diversity at higher latitudes could result both from the combination of in situ speciation of woody angiosperms at low latitudes, and from niche conservatism of woody gymnosperms at high latitudes.
In contrast to the results for woody communities, we found that phylogenetic diversity in herbaceous communities decreased towards extreme latitudes, and that this effect was influenced mainly by vegetation type. Low phylogenetic diversity at high latitudes accords with the relatively recent origin of temperate and boreal climates and habitats7 compared to the tropics, and with the young age of some lineages present at high latitudes, such as the grasses. Herbaceous lineages may be able to cope with low availability of resources at high latitudes through niche evolution49. Additionally, most herbaceous species at high latitudes are well-adapted to freezing conditions because of their ability to produce and replace short-lived aboveground tissue at low cost50.
Our results indicate that phylogenetic diversity has a weaker relationship with latitude in herbaceous communities than in woody communities. The weaker relationships in herbaceous communities could result from the concentration of numerous studies exhibiting a high level of variability in phylogenetic diversity in Central Europe (Fig. 1), even though the spatial aggregation of those studies did not result in any significant spatial autocorrelation. Species tended to be more closely-related in herbaceous communities at high latitudes relative to moderate latitudes, but the latitudinal trend is more pronounced when calculated from the whole phylogenetic tree (SESmpd) than when calculated among closest relatives (SESmntd) (Fig. 2b, Supplementary Table S2). Low SESmpd values at high latitudes are probably a result of recent rapid diversification in several herbaceous lineages following the rise of angiosperm-dominated herbaceous flora in open habitats51 during global cooling in the Late Cretaceous (c. 83–66 myr52) and Eocene-Oligocene (c. 51–33 myr53). For example, the Ranunculaceae, which originated as a forest clade, rapidly diversified c. 83 myr into 11 herbaceous lineages in mid- to high latitudes51. Likewise, the family Valerianaceae originated in the Himalayas of Asia in the early Cenozoic (66–40 myr54,55) and subsequently dispersed into temperate latitudes and higher elevation habitats, eventually reaching the Andes55, where strong diversification occurred56. Despite the large number of herbaceous communities analysed in our study, the results need to be interpreted with caution because of spatial limitation in the distribution of the original studies. Herbaceous communities were restricted both in latitude (98% were from latitudes above 30°N) and in the climate space occupied (mostly temperate climate, Supplementary Fig. S5). Our results also showed that vegetation type might be a major determinant of phylogenetic diversity in herbaceous communities, as shown in Table S2, and by the difference in the marginal R2 values between the top-ranked models for SESmpd and SESmntd (13% and 28% respectively). As in the case of SESmntd, latitude is not a significant predictor.
Our results show that open communities were generally less phylogenetically diverse than closed communities. This agrees with results of Lososova et al.57, which showed that the phylogenetic diversity of species pools in grasslands and other open habitats was lower than that in forests in Central Europe. Lososova et al.57 attributed this to the historical age of the vegetation types: grasslands were formed during the late Tertiary, with a limited number of lineages having enough time to adapt to the open conditions, whereas high phylogenetic diversity in forests results from continuous accumulation of lineages since the Mesozoic. Proches et al.14 also found lower phylogenetic diversity in vegetation types that have undergone recent radiation, such as grassland, fynbos and Nama-Karoo, compared to evolutionarily older subtropical scrub vegetation in South Africa.
We only found a significant effect of biogeographical realm on SESmpd in herbaceous communities. This is surprising because different biogeographic realms are known to support the development of distinct patterns of biodiversity as a result of historical factors58. The failure to observe effects of biogeographic realm in several cases may have been caused by communities from some realms, such as the Afrotropics, being relatively poorly represented in our dataset. Alternatively, our results indicate that the latitudinal gradient in phylogenetic diversity may be a general macroecological trend across biogeographical realms.
Although most lineages exhibit higher species richness at lower latitudes1,6, our results demonstrate that if phylogenetic diversity is measured using indices that are independent of species richness, it cannot be uncritically assumed that the same pattern will be observed28,59. Instead, the results of this study suggest that the impacts of the mechanisms proposed to explain latitudinal diversity gradients may differ between woody and herbaceous growth forms. Several speciation and extinction events have affected lineages during evolutionary time, causing a variety of patterns of phylogenetic diversity to develop in plant communities, with the important consequence that lineages with different growth forms have responded idiosyncratically to changing environmental circumstances60.
This study adds further insight into the cradle/museum debate about the causes of regional diversity18, although the data do not allow us to test these hypotheses directly. Nevertheless, our results support the proposal that the latitudinal diversity gradient may arise from different regions acting as either cradles or museums for lineages, with communities containing closely-related species (i.e. with low phylogenetic diversity) arising in regions acting as evolutionary cradles, and communities consisting of more distantly-related species (i.e. with high phylogenetic diversity) being found in regions acting as museums, in which conditions promote the preservation of a high degree of evolutionary diversity. Testing these hypotheses would require data on speciation and extinction rates in lineages of both woody and herbaceous growth forms.
We have demonstrated that plant communities consisting of species that have arisen through different evolutionary lineages, and are dominated by species with different growth forms, display contrasting patterns of phylogenetic diversity across the latitudinal gradient. These findings reinforce the view that both evolutionary and ecological processes should be taken into consideration in future efforts to explain latitudinal diversity gradients.
Methods
Plant community data
We compiled a global data set using the libraries of ISI Web of Knowledge for papers published between 1945 and August 2016, and JSTOR for papers published between 1700 and 1945. We searched for simultaneous occurrence of the key words “community”, “vegetation”, “species list” and “plant”, to find papers with full species lists for plant communities (Supplementary Dataset S1). We considered all sampling methods used by the authors of the selected original studies. We use the term “community” to refer to locally coexisting species that were recorded in sampling units (e.g. plots, transects, regional surveys) ranging in size from <1 m2 in herbaceous communities, to >1 km2 in woody communities. Number of sampling units ranged from 1 to 367 in woody communities and from 1 to 160 in herbaceous communities. We separated the dataset into woody communities composed only of woody species (shrubs and trees), and herbaceous communities composed only of herbaceous species, based on the sampling information provided by the authors of the original studies. Communities composed of both woody and herbaceous species were excluded from the dataset. We also excluded studies with only partial species lists (e.g. lists only for dominant species, or only for some taxonomic groups). We excluded aquatic and urban communities, and communities with fewer than three species to avoid including anthropogenic communities, e.g. arable fields, plantations. The latitudinal range of the locations of the studied communities ran from 0° to a maximum of 60.67°N of the equator and a maximum of 54.79°S of the equator (Fig. 1).
We also included 223 woody communities from Alwyn Gentry’s dataset61, available from http://salvias.net/pages/database_info.php. In total, this resulted in 459 woody communities in which a total of 8,753 species were recorded, and 589 herbaceous communities in which a total of 1,847 species were recorded (Supplementary Dataset S1 and Supplementary References S1). Woody communities included forests, scrublands and savannas, whereas herbaceous communities included the herbaceous layer of forests, grasslands, meadows, salt marshes, outcrops and dunes (Supplementary Dataset S1).
We extracted data from the database WorldClim62 for mean annual temperature (hereafter temperature) and annual precipitation (hereafter precipitation) at the locations of every community. These data were extracted at a spatial resolution of 30 arc seconds.
Phylogenetic data
A phylogeny of species was obtained based on the most up to date megaphylogeny for seed plants63, which comprises 79,881 taxa. We standardised the species names in our dataset according to The Plant List using the R package ‘Taxonstand’64. For those taxa identified to genus level or with misspelled names, we standardised their names manually according to The Plant List. We then used the R function S.PhyloMaker65 to link the species names in our dataset with those in the megaphylogeny, and the scenario 3 approach65 to add species to the phylogeny. We used this same approach for taxa identified to genus level. Scenario 3 adds missing taxa (e.g. genera or species) to the phylogeny within the taxa with known branch lengths, in a similar way to the approach implemented in Phylomatic and BLADJ66. We pruned our complete phylogeny to create two trees which included either (i) only those species recorded in the woody communities or (ii) only those species recorded in the herbaceous communities in our dataset. These two trees were then used as reference lists from which phylogenetic diversity could be calculated for every community in the dataset.
Phylogenetic diversity, which often depends on species richness, can be quantified in several ways31. We used the index ‘mpd’, which calculates phylogenetic distances across the whole phylogenetic tree by averaging all species pairwise distances, and the index ‘mntd’, which calculates phylogenetic distances at a shallower level, between the most closely-related pairs of species. These indices are opposite in sign to the net relatedness index (NRI) and nearest taxon index (NTI), respectively36,66, and they measure phylogenetic diversity between species at different depths in the phylogenetic tree. To produce indices of phylogenetic diversity that were independent of species richness, we calculated the standardised effect sizes of both indices (SESmpd and SESmntd), by comparing the observed community phylogenetic diversity to the null distribution of randomly assembled communities with equal richness. Negative values of SESmpd and SESmntd indicate lower phylogenetic diversity than expected under the assumption of the null model, whereas values greater than zero indicate higher phylogenetic diversity than predicted by the null model. We calculated phylogenetic diversity using the R software program67 and the package ‘picante’68.
Statistical analysis
Latitude is widely recognized as a surrogate for environmental variables including temperature6 and precipitation69. To determine whether to include both temperature6 and precipitation in further analyses, we first analysed the correlation between latitude and both mean annual temperature and annual precipitation, using Pearson’s product moment correlation. We found strong correlations in both cases (Supplementary Table S5), and therefore used only latitude in further analyses, in combination with other non-correlated variables (see below). The distribution of woody and herbaceous communities in the climate space is shown through a Whittaker biomes plot70, produced using the function ‘whittaker_base_plot’ in the package ‘plotbiomes’71 (Supplementary Fig. S5).
We analysed the patterns of community phylogenetic diversity (SESmpd and SESmntd) along the latitudinal gradient in woody and herbaceous communities using linear mixed-effect models (function ‘lme’) in the ‘nlme’ package72. For each of the two categories of community (woody and herbaceous), we built alternative mixed-effect models with different combinations of the following fixed effects variables: latitude, biogeographical realm29 (Afrotropical, Australasian, Indo-Malayan, Nearctic, Neotropical, Palearctic) and vegetation type. As we expected latitude to have an effect on phylogenetic diversity, but were unable to predict whether the relationship would be linear, we included both linear and quadratic terms in the models. The effect of vegetation type on community phylogenetic structure was examined by extracting information about vegetation type from each of the published studies (e.g. forests, dunes, scrublands) and reclassifying them as either closed (forest communities), open (grassland, meadow, salt marsh, outcrop and dune communities), and semi-open (savanna and scrubland communities). To account for the influence of differences in sampling unit size, we included this as a covariate in the fixed effects variables of the models. All continuous fixed effects variables were centered and standardised to have zero mean and unit variance before parameter estimation in order to make them comparable within models. We fitted models with all combinations of fixed effects variables, excluding interactions between them using maximum likelihood (“ML”) to allow comparison between models with different predictors. We then ranked all models based on the Akaike information criterion corrected for small sample size (AICc). The most parsimonious models were refitted using restricted maximum likelihood (“REML”). The fixed effects variables in the most parsimonious models were then compared with the ‘anova.lme’ function in ‘nlme’72, using the marginal significance of each fixed effect variable when all other fixed effects variables were present in the model. Additionally, we used two types of R2 (marginal and conditional R2) proposed by Nakagawa & Schielzeth73 for mixed-effect models, using the ‘MuMIn’ package74. Marginal R2 represents the variance explained by the fixed effects variables, whereas the conditional R2 represents the variance explained by the whole model, including both fixed and random effects variables73.
Spatial autocorrelation is commonly found in ecological data observed across geographical space75. To account for heterogeneous and spatial aspects of the dataset, we used ‘study’ as a random factor in all models. In addition, as our dataset involved a clustered spatial arrangement of communities (in particular those in Central Europe and Northwestern South America), we fitted all models with an additional term describing the within-group correlation structure using the ‘corExp’ function in ‘nlme’ package72. We also plotted the normalised residuals of the top-ranked models on the global map (Supplementary Fig. S6).
In order to examine how the inclusion of gymnosperm species affected the pattern of phylogenetic diversity in woody communities along the latitudinal gradient, we ran mixed-effects models with gymnosperm species excluded (Supplementary results, Supplementary Tables S3, S4, Supplementary Figs S3, S4). All figures were produced in the package ggplot276. Statistical analyses were performed in R software program version 3.4.367.
Supplementary information
Acknowledgements
This study was supported by the Estonian Research Council (grant PUT 1006 to J.C.M. and P.G., grant PUT 1409 to L.L., grant IUT 21–1 to K.T., and grant IUT20-33 to A.K.). We are grateful to Alwyn H. Gentry, and his co-workers for their plot data, the Missouri Botanical Garden, and all authors of the papers used in this study. We thank Meelis Pärtel, James (JC) Cahill, and Andreas Prinzing for insightful discussions. We also thank the Estonian Government for continuously keeping up our hopes about raising research funding to 1% of GDP.
Author Contributions
P.G. conceived and designed the study, K.T. and T.H. collected the data, J.C.M. analysed the data with support of L.G., L.L. and A.K. J.C.M. and P.G. wrote the first draft of the manuscript. M.J.H. contributed to revising the manuscript. All authors contributed to the final version.
Data Availability
All data analysed during this study are included in this article and in its Supplementary Dataset and Supplementary Information files. R scripts including species name standardisation, calculation of phylogenetic diversity indexes, and all statistical analyses are also available as Supplementary Information.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
5/22/2019
A correction to this article has been published and is linked from the HTML and PDF versions of this paper. The error has been fixed in the paper.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42827-1.
References
- 1.Hillebrand H. On the generality of the latitudinal diversity gradient. Am. Nat. 2004;163:192–211. doi: 10.1086/381004. [DOI] [PubMed] [Google Scholar]
- 2.Mittelbach GG, et al. Evolution and the latitudinal diversity gradient: Speciation, extinction and biogeography. Ecol. Lett. 2007;10:315–331. doi: 10.1111/j.1461-0248.2007.01020.x. [DOI] [PubMed] [Google Scholar]
- 3.Fine PVA. Ecological and evolutionary drivers of geographic variation in species diversity. Annu. Rev. Ecol. Evol. Syst. 2015;46:369–392. doi: 10.1146/annurev-ecolsys-112414-054102. [DOI] [Google Scholar]
- 4.Pianka ER. Latitudinal gradients in species diversity: a review of concepts. Am. Nat. 1966;100:33–46. doi: 10.1086/282398. [DOI] [Google Scholar]
- 5.Rohde K. Latitudinal gradients in species diversity: the search for the primary cause. Oikos. 1992;65:514–527. doi: 10.2307/3545569. [DOI] [Google Scholar]
- 6.Willig MR, Kaufman DM, Stevens RD. Latitudinal gradients of biodiversity: pattern, process, scale, and synthesis. Annu. Rev. Ecol. Evol. Syst. 2003;34:273–309. doi: 10.1146/annurev.ecolsys.34.012103.144032. [DOI] [Google Scholar]
- 7.Fine PVA, Ree RH. Evidence for a time‐integrated species‐area effect on the latitudinal gradient in tree diversity. Am. Nat. 2006;168:796–804. doi: 10.1086/508635. [DOI] [PubMed] [Google Scholar]
- 8.Wiens JJ, Donoghue MJ. Historical biogeography, ecology and species richness. Trends Ecol. Evol. 2004;19:639–644. doi: 10.1016/j.tree.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 9.Fischer AG. Latitudinal Variations in Organic Diversity. Evolution. 1960;14:64–81. doi: 10.1111/j.1558-5646.1960.tb03057.x. [DOI] [Google Scholar]
- 10.Ricklefs RE. Historical and ecological dimensions of global patterns in plant diversity. Biol. Skr. 2005;55:583–603. [Google Scholar]
- 11.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 2002;33:475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448. [DOI] [Google Scholar]
- 12.Guevara JE, et al. Low phylogenetic beta diversity and geographic neo-endemism in Amazonian white-sand forests. Biotropica. 2016;48:34–46. doi: 10.1111/btp.12298. [DOI] [Google Scholar]
- 13.Yan Y, Yang X, Tang Z. Patterns of species diversity and phylogenetic structure of vascular plants on the Qinghai-Tibetan plateau. Ecol. Evol. 2013;3:4584–4595. doi: 10.1002/ece3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Proches S, Wilson JR, Cowling RM. How much evolutionary history in a 10 × 10 m plot? Proc. R. Soc. B Biol. Sci. 2006;273:1143–1148. doi: 10.1098/rspb.2005.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crisp M, Cook L, Steane D. Radiation of the Australian flora: what can comparisons of molecular phylogenies across multiple taxa tell us about the evolution of diversity in present-day communities? Philos. Trans. R. Soc. B Biol. Sci. 2004;359:1551–1571. doi: 10.1098/rstb.2004.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kooyman R, Rossetto M, Cornwell W, Westoby M. Phylogenetic tests of community assembly across regional to continental scales in tropical and subtropical rain forests. Glob. Ecol. Biogeogr. 2011;20:707–716. doi: 10.1111/j.1466-8238.2010.00641.x. [DOI] [Google Scholar]
- 17.Chown SL, Gaston KJ. Areas cradles and museums: The latitudinal gradient in species richness. Trends Ecol. Evol. 2000;15:311–315. doi: 10.1016/S0169-5347(00)01910-8. [DOI] [PubMed] [Google Scholar]
- 18.Stebbins, G. L. Flowering Plants: Evolution Above The Species Level. (Mass: Belknap Press of Harvard University Press, 1974).
- 19.Silva IA, Batalha MA. Phylogenetic structure of Brazilian savannas under different fire regimes. J. Veg. Sci. 2010;21:1003–1013. doi: 10.1111/j.1654-1103.2010.01208.x. [DOI] [Google Scholar]
- 20.Culmsee H, Leuschner C. Consistent patterns of elevational change in tree taxonomic and phylogenetic diversity across Malesian mountain forests. J. Biogeogr. 2013;40:1997–2010. [Google Scholar]
- 21.Qian H, Zhang Y, Zhang J, Wang X. Latitudinal gradients in phylogenetic relatedness of angiosperm trees in North America. Glob. Ecol. Biogeogr. 2013;22:1183–1191. doi: 10.1111/geb.12069. [DOI] [Google Scholar]
- 22.Kerkhoff AJ, Moriarty PE, Weiser MD. The latitudinal species richness gradient in New World woody angiosperms is consistent with the tropical conservatism hypothesis. Proc. Natl. Acad. Sci. 2014;111:8125–8130. doi: 10.1073/pnas.1308932111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian H, Jin Y, Ricklefs RE. Patterns of phylogenetic relatedness of angiosperm woody plants across biomes and life-history stages. J. Biogeogr. 2017;44:1383–1392. doi: 10.1111/jbi.12936. [DOI] [Google Scholar]
- 24.Kubota Y, Kusumoto B, Shiono T, Ulrich W. Environmental filters shaping angiosperm tree assembly along climatic and geographic gradients. J. Veg. Sci. 2018;29:607–618. doi: 10.1111/jvs.12648. [DOI] [Google Scholar]
- 25.Feng G, et al. Phylogenetic age differences in tree assemblages across the Northern Hemisphere increase with long-term climate stability in unstable regions. Glob. Ecol. Biogeogr. 2017;26:1035–1042. doi: 10.1111/geb.12613. [DOI] [Google Scholar]
- 26.Carlucci MB, et al. Phylogenetic composition and structure of tree communities shed light on historical processes influencing tropical rainforest diversity. Ecography. 2017;40:521–530. doi: 10.1111/ecog.02104. [DOI] [Google Scholar]
- 27.Fritz SA, Rahbek C. Global patterns of amphibian phylogenetic diversity. J. Biogeogr. 2012;39:1373–1382. doi: 10.1111/j.1365-2699.2012.02757.x. [DOI] [Google Scholar]
- 28.Davies TJ, Buckley LB. Phylogenetic diversity as a window into the evolutionary and biogeographic histories of present-day richness gradients for mammals. Philos. Trans. R. Soc. B Biol. Sci. 2011;366:2414–2425. doi: 10.1098/rstb.2011.0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olson DM, et al. Terrestrial ecoregions of the world: a new map of life on earth; a new global map of terrestrial ecoregions provides an innovative tool for conserving biodiversity. Bioscience. 2001;51:933–938. doi: 10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2. [DOI] [Google Scholar]
- 30.Engemann K, et al. Patterns and drivers of plant functional group dominance across the Western Hemisphere: a macroecological re-assessment based on a massive botanical dataset. Bot. J. Linn. Soc. 2016;180:141–160. doi: 10.1111/boj.12362. [DOI] [Google Scholar]
- 31.Miller ET, Farine DR, Trisos CH. Phylogenetic community structure metrics and null models: a review with new methods and software. Ecography. 2017;40:461–477. doi: 10.1111/ecog.02070. [DOI] [Google Scholar]
- 32.Gerhold P, Cahill JF, Winter M, Bartish IV, Prinzing A. Phylogenetic patterns are not proxies of community assembly mechanisms (they are far better) Funct. Ecol. 2015;29:600–614. doi: 10.1111/1365-2435.12425. [DOI] [Google Scholar]
- 33.Pigot AL, Etienne RS. A new dynamic null model for phylogenetic community structure. Ecol. Lett. 2015;18:153–163. doi: 10.1111/ele.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weber MG, Wagner CE, Best RJ, Harmon LJ, Matthews B. Evolution in a community context: on integrating ecological interactions and macroevolution. Trends Ecol. Evol. 2017;32:291–304. doi: 10.1016/j.tree.2017.01.003. [DOI] [PubMed] [Google Scholar]
- 35.Gerhold P, Carlucci MB, Proches S, Prinzing A. The deep past controls the phylogenetic structure of present, local communities. Annu. Rev. Ecol. Evol. Syst. 2018;49:477–497. doi: 10.1146/annurev-ecolsys-110617-062348. [DOI] [Google Scholar]
- 36.Webb CO. Exploring the phylogenetic struture of ecological communities: an example for rain forest trees. Am. Nat. 2000;156:145–155. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 37.Cavender-bares J, Keen A, Miles B. Phylogenetic structure of Floridian plant communities depends on taxonomic and spatial scale. Ecology. 2015;87:S109–S122. doi: 10.1890/0012-9658(2006)87[109:PSOFPC]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 38.Swenson NG, et al. The problem and promise of scale dependency in community phylogenetics. Ecology. 2006;87:2418–2424. doi: 10.1890/0012-9658(2006)87[2418:TPAPOS]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 39.Vamosi SM, Heard SB, Vamosi JC, Webb CO. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 2009;18:572–592. doi: 10.1111/j.1365-294X.2008.04001.x. [DOI] [PubMed] [Google Scholar]
- 40.Axelrod DI. Origin of deciduous and evergreen habits in temperate forests. Evolution. 1966;20:1–15. doi: 10.1111/j.1558-5646.1966.tb03339.x. [DOI] [PubMed] [Google Scholar]
- 41.Ricklefs RE. Evolutionary diversification and the origin of the relationship of diversity-environment. Ecology. 2006;87:3–13. doi: 10.1890/0012-9658(2006)87[3:EDATOO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 42.Richards, P. W. The Tropical Rain Forest: An Ecological Study. (Cambridge University Press, 1996).
- 43.Friis EM, Pedersen KR, Crane PR. Diversity in obscurity: fossil flowers and the early history of angiosperms. Philos. Trans. R. Soc. B Biol. Sci. 2010;365:369–382. doi: 10.1098/rstb.2009.0227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wing SL, Boucher LD. Ecological aspects of the cretaceous flowering plant radiation. Annu. Rev. Earth Planet. Sci. 1998;26:379–421. doi: 10.1146/annurev.earth.26.1.379. [DOI] [Google Scholar]
- 45.Augusto L, Davies TJ, Delzon S, de Schrijver A. The enigma of the rise of angiosperms: Can we untie the knot? Ecol. Lett. 2014;17:1326–1338. doi: 10.1111/ele.12323. [DOI] [PubMed] [Google Scholar]
- 46.Graham A. The age and diversification of terrestrial new world ecosystems through cretaceous and Cenozoic time. Am. J. Bot. 2011;98:336–351. doi: 10.3732/ajb.1000353. [DOI] [PubMed] [Google Scholar]
- 47.Berendse F, Scheffer M. The angiosperm radiation revisited, an ecological explanation for Darwin’s ‘abominable mystery’. Ecol. Lett. 2009;12:865–872. doi: 10.1111/j.1461-0248.2009.01342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bond WJ. The tortoise and the hare: ecology of angiosperm dominance and gymnosperm persistence. Biol. J. Linn. Soc. 1989;36:227–249. doi: 10.1111/j.1095-8312.1989.tb00492.x. [DOI] [Google Scholar]
- 49.Smith SA, Beaulieu JM. Life history influences rates of climatic niche evolution in flowering plants. Proc. R. Soc. B Biol. Sci. 2009;276:4345–4352. doi: 10.1098/rspb.2009.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zanne AE, et al. Three keys to the radiation of angiosperms into freezing environments. Nature. 2014;506:89–92. doi: 10.1038/nature12872. [DOI] [PubMed] [Google Scholar]
- 51.Wang W, et al. The rise of angiosperm-dominated herbaceous floras: Insights from Ranunculaceae. Sci. Rep. 2016;6:27259. doi: 10.1038/srep27259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linnert C, et al. Evidence for global cooling in the Late Cretaceous. Nat. Commun. 2014;5:4194. doi: 10.1038/ncomms5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Couvreur TLPP, et al. Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae) Mol. Biol. Evol. 2010;27:55–71. doi: 10.1093/molbev/msp202. [DOI] [PubMed] [Google Scholar]
- 54.Bell C, Edwards E, Kim S, Donoghue M. Dipsacales phylogeny based on chloroplast DNA sequences. Harvard Pap. Bot. 2001;6:481–499. [Google Scholar]
- 55.Bell CD, Donoghue MJ. Phylogeny and biogeography of Valerianaceae (Dipsacales) with special reference to the South American valerians. Org. Divers. Evol. 2005;5:147–159. doi: 10.1016/j.ode.2004.10.014. [DOI] [Google Scholar]
- 56.Moore BR, Donoghue MJ. Correlates of diversification in the plant clade dipsacales: geographic movement and evolutionary innovations. Am. Nat. 2007;170:S28–S55. doi: 10.1086/519460. [DOI] [PubMed] [Google Scholar]
- 57.Lososová Z, et al. Phylogenetic structure of plant species pools reflects habitat age on the geological time scale. J. Veg. Sci. 2015;26:1080–1089. doi: 10.1111/jvs.12308. [DOI] [Google Scholar]
- 58.Flynn JJ, Wyss AR. Recent advances in South American mammalian paleontology. Trends Ecol. Evol. 1998;13:449–54. doi: 10.1016/S0169-5347(98)01457-8. [DOI] [PubMed] [Google Scholar]
- 59.Mishler BD, et al. Phylogenetic measures of biodiversity and neo-and paleo-endemism in Australian acacia. Nat. Commun. 2014;5:4473. doi: 10.1038/ncomms5473. [DOI] [PubMed] [Google Scholar]
- 60.Willis KJ, Niklas KJ. The role of Quaternary environmental change in plant macroevolution: the exception or the rule? Philos. Trans. R. Soc. B Biol. Sci. 2004;359:159–172. doi: 10.1098/rstb.2003.1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Phillips, O. & Miller, J. S. Global Patterns Of Plant Diversity: Alwyn H. Gentry’s Forest Transect Data Set. (Missouri Botanical Press, 2002).
- 62.Hijmans RJ, Cameron SE, Parra JL, Jones PG, Jarvis A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. 2005;25:1965–1978. doi: 10.1002/joc.1276. [DOI] [Google Scholar]
- 63.Smith SA, Brown JW. Constructing a broadly inclusive seed plant phylogeny. Am. J. Bot. 2018;105:302–314. doi: 10.1002/ajb2.1019. [DOI] [PubMed] [Google Scholar]
- 64.Cayuela L, Granzow-de la Cerda Í, Albuquerque FS, Golicher DJ. Taxonstand: An r package for species names standardisation in vegetation databases. Methods Ecol. Evol. 2012;3:1078–1083. doi: 10.1111/j.2041-210X.2012.00232.x. [DOI] [Google Scholar]
- 65.Qian H, Jin Y. An updated megaphylogeny of plants, a tool for generating plant phylogenies and an analysis of phylogenetic community structure. J. Plant Ecol. 2016;9:233–239. doi: 10.1093/jpe/rtv047. [DOI] [Google Scholar]
- 66.Webb CO, Ackerly DD, Kembel SW. Phylocom: software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics. 2008;24:2098–2100. doi: 10.1093/bioinformatics/btn358. [DOI] [PubMed] [Google Scholar]
- 67.R Core Team R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org/ (2016).
- 68.Kembel SW, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–1464. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 69.Hartmann DL, et al. Observations: Atmosphere and surface. Clim. Chang. 2013 Phys. Sci. Basis Work. Gr. I Contrib. to Fifth Assess. Rep. Intergov. Panel Clim. Chang. 2013;9781107057:159–254. [Google Scholar]
- 70.Ricklefs, R. E. The Economy Of Nature. (W. H. Freeman and Company, 2008).
- 71.Stefan, V. & Levin, S. Plotbiomes: Plot Whittaker biomes with ggplot2. R package version 0.0.0.9001. https://rawgit.com/valentinitnelav/plotbiomes/master/html/Whittaker_biomes_examples.html (2018).
- 72.Pinheiro, J., Bates, D., DebRoy, S., D., S. & Team, R. C. {nlme}: Linear and Nonlinear Mixed Effects Models. R package version 3.1-131. http://CRAN.R-project.org/package=nlme (2017).
- 73.Nakagawa S, Schielzeth H. A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol. Evol. 2013;4:133–142. doi: 10.1111/j.2041-210x.2012.00261.x. [DOI] [Google Scholar]
- 74.Barton, K. MuMIn: multi-model inference. R package version 1.42.1 (2018).
- 75.Legendre P. Spatial autocorrelation: trouble or new paradigm? Ecology. 1993;74:1659–1673. doi: 10.2307/1939924. [DOI] [Google Scholar]
- 76.Wickham, H. Ggplot2: Elegant Graphics For Data Analysis. (Springer, 2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data analysed during this study are included in this article and in its Supplementary Dataset and Supplementary Information files. R scripts including species name standardisation, calculation of phylogenetic diversity indexes, and all statistical analyses are also available as Supplementary Information.