Abstract
Biological soil crusts (biocrusts) occur within drylands throughout the world, covering ~12% of the global terrestrial soil surface. Their occurrence in the deserts of the Arabian Peninsula has rarely been reported and their spatial distribution, diversity, and microbial composition remained largely unexplored. We investigated biocrusts at six different locations in the coastal and central deserts of Oman. The biocrust types were characterized, and the bacterial and fungal community compositions of biocrusts and uncrusted soils were analysed by amplicon sequencing. The results were interpreted based on the environmental parameters of the different sites. Whereas at lowland sites, mainly cyanobacteria-dominated biocrusts were observed, both cyanobacteria- and lichen-dominated biocrusts occurred at mountain sites. The majority of bacterial sequences (32–83% of total sequences) belonged to Actinobacteria, Cyanobacteria, Alphaproteobacteria, and Bacteroidetes, whereas fungal sequences belonged to Ascomycota, Basidiomycota, and Chytridiomycota (>95%). With biocrust development, a notable increase in cyanobacterial and decrease in actinobacterial proportions was observed for cyanobacteria-dominated crusts. In coastal areas, where salinity is high, biocrusts were replaced by a unique marine mat-like microbial community, dominated by halotolerant taxa. Redundancy analysis revealed a significant contribution of soil texture, cover type, carbon content, and elevation to the variations in bacterial and fungal communities. Multivariate analysis placed microbial communities in significantly separated clusters based on their carbon content, elevation and electrical conductivity. We conclude that Oman hosts a variety of cyanobacteria- and lichen-dominated crusts with their bacterial and fungal communities being largely dictated by soil properties and environmental parameters.
Subject terms: Soil microbiology, Microbial ecology
Introduction
Biological soil crusts (biocrusts), composed of cyanobacteria, algae, lichen and mosses, are distributed worldwide in diverse environments ranging from hot to cold deserts and are estimated to cover 30–40% of the total arid and semi-arid landscapes1–4. The distribution patterns and heterogeneity of biocrusts can differ depending on the spatial scale, and they are often dictated by several biotic (e.g. plant cover and human disturbance) and abiotic (e.g. climatic and edaphic) factors5–12. These parameters can be distinct, but can also interrelate and depend on each other, and some vary spatially (from meters to kilometers) and temporally (days or seasons). Several studies have been devoted to gain a better understanding of the different parameters that determine the heterogeneity of biocrusts and their microbial communities10–15. Biocrust communities varied among different geographical locations within Africa, Australia, Asia and North America14,16–18 and even when comparison was carried out across continents and between hot and temperate deserts10,19,20. Patchiness in biocrust microenvironments has even been detected at the microscale level, using microsensors, indicating that different microhabitats supported the growth of different microorganisms2. Early studies from the arid and semi-arid Australia revealed a significant relationship between crusts’ species composition and soil physical and chemical properties11–14. Climatic changes in temperature and precipitation lead to replacement of certain biocrust species by others21,22. Soil properties including texture class, pH, salinity, nutrients and porosity strongly correlated with the distribution patterns of biocrust species23,24. Anthropogenic disturbance has also been shown to reduce diversity, abundance and activity of biocrust microorganisms25–28.
Most of the research related to distribution patterns of biocrusts was performed on samples originating from Australia, North America and Africa, while there is a clear gap in our knowledge on the biogeography of biocrust microorganisms on the Arabian Peninsula. Moreover, only recently modern molecular techniques have been used to correlate species compositions of biocrusts to their environments17,29. On the Arabian Peninsula, deserts are a prominent feature, covering more than half of the total area and characterized by limited plant cover. Deserts in this region (e.g. Oman) vary from semiarid in the central desert to arid in the coastal regions, with clear differences in thermal regimes and precipitation rates. While the central desert experiences freezing temperatures and an annual rainfall of ca. 300 mm during November to March, these values may reach 40 °C and <100 mm in the coastal deserts. Some biocrusts in coastal regions are exposed to frequent inundation by seawater, resulting in a clear transition from completely desiccated biocrusts to salt-influenced biocrusts, which are then considered as intertidal marine microbial mats. So far, the distribution of biocrusts and the parameters that drive the heterogeneity of their microbial communities have not been investigated for Oman. Such information provides the baseline knowledge that can be used to predict vegetation development and consequences of climate change (e.g. soil warming and altered precipitation) in the future.
In this study, we expand our understanding of the biogeography and species composition of different biocrust types from Oman and explore the determinants that contribute to the variation of their bacterial and fungal communities. We collected samples of bare soils, cyanobacteria- and lichen-dominated crusts from six localities (Fig. 1). Marine microbial mats from only one location (i.e. Shana) were included in our sampling for comparison of microbial communities with salt-influenced biocrusts. The sampling sites included the coastal and the central desert, covered an altitudinal gradient, and were chosen because of the clear variability in climatic and edaphic properties.
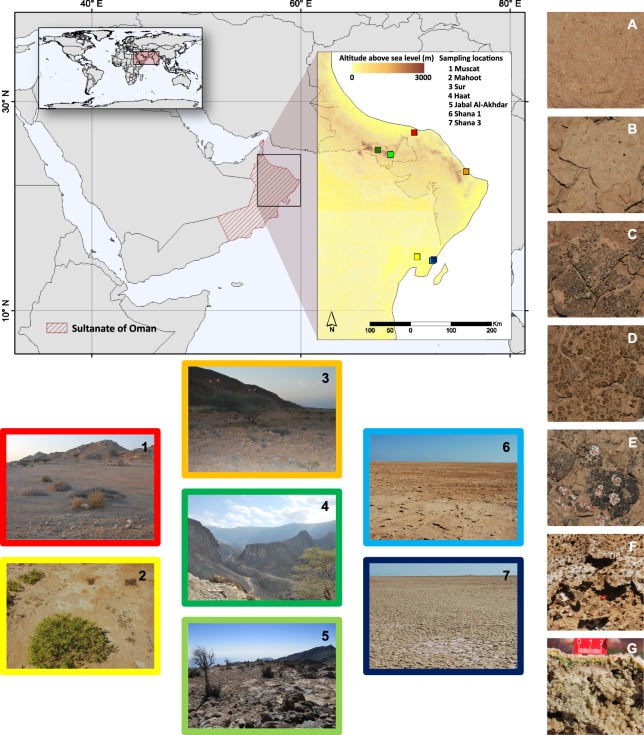
Figure 1.
Map composition presenting the sampling locations and sample types in the Sultanate of Oman. Framed and serially numbered pictures show the different sampling locations. On the right hand side close-up pictures of different biocrust types and bare soil samples are shown (A) bare soil; (B) cyanobacteria-dominated biocrust; (C) cyanobacteria-dominated biocrust with cyanolichen; (D) chlorolichen-dominated biocrust with Placidium sp.; (E) chlorolichen-dominated biocrust with Psora sp. and cyanolichens; (F) salt crust with microbial communities growing inside the crust; (G) soft, marine microbial mats (cross section)). Topographic information obtained from the NASA Shuttle Radar Topography Mission (SRTM30). All maps were created using ArcGIS Desktop 10.3 (Copyright(©) 1995–2015 ESRI; License acquired by the university of Almeria thought the agreement between University and Centro Informático Científico de Andalucía, CICA). Country boundaries were obtained from the FGGD coastal and country boundaries of the world 1.0 (2007; http://ref.data.fao.org/map?entryId = 18329470-472d-11db-88e0-000d939bc5d8&tab = metadata).
Materials and Methods
Study sites and sample collection
Sampling was carried out at six different locations (termed as Jabal Al-Akhdar, Muscat, Mahoot, Sur, Haat, and Shana) in the Northern and Eastern parts of the Sultanate of Oman in January 2016. The chosen sites represent variable environmental and ecological conditions, differing in soil texture, elevation, climatic conditions, human disturbance and distance to the sea (Table 1). A map depicting the sampling locations was created using ArcGIS Desktop 10.3 (Copyright(C) 1995–2015 ESRI). Topographic information were obtained from the NASA Shuttle Radar Topography Mission (SRTM30).
Table 1.
Sample locations and soil characteristics of bare soils, biocrusts and mats from the Sultanate of Oman.
| Sampling sites | Type/ | Latitude | Longitude | Elevation (m) | Disturbance | pH | Conductivity | N content | C content | Soil texture | FAO* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Replicate | (°) | (°) | (µS/m) | (%) | (%) | Major class | |||||
| Muscat | Bare1 | 23.580318 | 58.201758 | 38 | + | 8.8 | 114 | 0.00 | 9.73 | Sand | S (sand) |
| Bare2 | 23.580318 | 58.201758 | 38 | + | 8.6 | 210 | 0.01 | 10.00 | Sand | S (sand) | |
| Bare3 | 23.580318 | 58.201758 | 38 | + | 8.7 | 189 | 0.01 | 8.49 | Sand | S (sand) | |
| Bare4 | 23.580318 | 58.201758 | 38 | + | 8.8 | 120 | 0.01 | 7.20 | Sand | S (sand) | |
| Cyano1 | 23.580318 | 58.201758 | 38 | + | 7.8 | 1930 | 0.08 | 6.01 | Clay | SiC (silt clay) | |
| Cyano2 | 23.580318 | 58.201758 | 38 | + | 7.9 | 301 | 0.16 | 6.03 | Loam | SCL (sandy clay loam) | |
| Cyano3 | 23.580318 | 58.201758 | 38 | + | 8.1 | 175 | 0.11 | 6.16 | Loam | CL (clay loam) | |
| Sur | Bare1 | 22.714222 | 59.353199 | 20 | + | 8.5 | 128 | 0.04 | 7.51 | Sand | LS (loamy sand) |
| Bare2 | 22.714222 | 59.353199 | 20 | + | 8.6 | 256 | 0.03 | 9.14 | Sand | LS (loamy sand) | |
| Bare3 | 22.714222 | 59.353199 | 20 | + | 8.7 | 148 | 0.02 | 6.95 | Sand | LS (loamy sand) | |
| Bare4 | 22.714222 | 59.353199 | 20 | + | 8.5 | 130 | 0.03 | 7.93 | Sand | LS (loamy sand) | |
| Cyano1 | 22.714222 | 59.353199 | 20 | + | 8.6 | 148 | 0.04 | 6.37 | Clay | SC (sandy clay) | |
| Cyano2 | 22.714222 | 59.353199 | 20 | + | 8.4 | 245 | 0.03 | 6.47 | Clay | SiC (silt clay) | |
| Cyano3 | 22.714222 | 59.353199 | 20 | + | 8.7 | 107 | 0.03 | 6.41 | Clay | SiC (silt clay) | |
| Mahoot | Bare1 | 20.829529 | 58.266045 | 37 | + | 8.6 | 166 | 0.01 | 3.18 | Loam | SL (sandy loam) |
| Bare2 | 20.829529 | 58.266045 | 37 | + | 8.7 | 370 | 0.01 | 4.00 | Loam | SL (sandy loam) | |
| Bare3 | 20.829529 | 58.266045 | 37 | + | 8.7 | 1228 | 0.02 | 3.41 | Loam | SL (sandy loam) | |
| Bare4 | 20.829529 | 58.266045 | 37 | + | 8.6 | 1134 | 0.01 | 2.42 | Loam | SL (sandy loam) | |
| Cyano1 | 20.829529 | 58.266045 | 37 | + | 8.3 | 228 | 0.03 | 3.65 | Loam | SiL (silt loam) | |
| Cyano2 | 20.829529 | 58.266045 | 37 | + | 8.5 | 232 | 0.01 | 3.88 | Loam | SCL (sandy clay loam) | |
| Cyano3 | 20.829529 | 58.266045 | 37 | + | 8.2 | 584 | 0.01 | 3.49 | Loam | SCL (sandy clay loam) | |
| Haat | Bare1 | 23.193069 | 57.396991 | 989 | − | 8.3 | 97 | 0.15 | 5.19 | Sand | LS (loamy sand) |
| Bare2 | 23.193069 | 57.396991 | 989 | − | 8.4 | 160 | 0.15 | 5.03 | Sand | LS (loamy sand) | |
| Bare3 | 23.193069 | 57.396991 | 989 | − | 8.3 | 121 | 0.16 | 4.16 | Sand | LS (loamy sand) | |
| Bare4 | 23.193069 | 57.396991 | 989 | − | 8.3 | 110 | 0.22 | 5.28 | Sand | LS (loamy sand) | |
| Cyano1 | 23.193069 | 57.396991 | 989 | − | 8.2 | 160 | 0.14 | 6.25 | Loam | SL (sandy loam) | |
| Cyano2 | 23.193069 | 57.396991 | 989 | − | 8.3 | 115 | 0.16 | 5.14 | Loam | SL (sandy loam) | |
| Cyano3 | 23.193069 | 57.396991 | 989 | − | 8.4 | 91 | 0.18 | 4.56 | Clay | SiC (silt clay) | |
| Lichen1 | 23.193069 | 57.396991 | 989 | − | 8.5 | 95 | 0.21 | 4.41 | Sand | LS (loamy sand) | |
| Lichen2 | 23.193069 | 57.396991 | 989 | − | 8.1 | 171.2 | 0.14 | 7.33 | Clay | SiC (silt clay) | |
| Lichen3 | 23.193069 | 57.396991 | 989 | − | 8.5 | 103 | 0.17 | 5.40 | Clay | SiC (silt clay) | |
| Jabal Al-Akhdar | Cyano1 | 23.091781 | 57.680719 | 2022 | − | 8.3 | 341 | 0.17 | 7.88 | Loam | CL (clay loam) |
| Cyano2 | 23.091781 | 57.680719 | 2022 | − | 8.4 | 111 | 0.08 | 4.91 | Clay | SC (sandy clay) | |
| Cyano3 | 23.091781 | 57.680719 | 2022 | − | 8.3 | 122 | 0.12 | 5.27 | Loam | SCL (sandy clay loam) | |
| Lichen1A | 23.091781 | 57.680719 | 2022 | − | 8.2 | 184.3 | 0.13 | 5.91 | Loam | SiL (silt loam) | |
| Lichen2A | 23.091781 | 57.680719 | 2022 | − | 8.0 | 255 | 0.22 | 6.24 | Loam | CL (clay loam) | |
| Lichen3A | 23.091781 | 57.680719 | 2022 | − | 8.3 | 133 | 0.12 | 4.43 | Clay | SiC (silt clay) | |
| lichen1B | 23.091781 | 57.680719 | 2022 | − | 8.4 | 133 | 0.13 | 5.14 | Clay | SiC (silt clay) | |
| Lichen2B | 23.091781 | 57.680719 | 2022 | − | 8.3 | 100 | 0.12 | 5.39 | Clay | SiC (silt clay) | |
| Lichen3B | 23.091781 | 57.680719 | 2022 | − | 8.2 | 140 | 0.18 | 4.58 | Loam | CL (clay loam) | |
| Shana1 | Cyano1 | 20.743597 | 58.603713 | 7 | − | 8.3 | 65800 | 0.01 | 2.30 | Sand | S (sand) |
| Cyano2 | 20.743597 | 58.603713 | 7 | − | 8.3 | 45900 | 0.00 | 2.40 | Sand | S (sand) | |
| Cyano3 | 20.743597 | 58.603713 | 7 | − | 8.3 | 108800 | 0.01 | 1.93 | Sand | S (sand) | |
| Shana3 | Mat1 | 20.769648 | 58.639686 | 12 | − | 8.9 | 34700 | 0.06 | 0.67 | Sand | S (sand) |
| Mat2 | 20.769648 | 58.639686 | 12 | − | 9.1 | 34800 | 0.05 | 0.60 | Sand | S (sand) | |
| Mat3 | 20.769648 | 58.639686 | 12 | − | 9.5 | 35400 | 0.04 | 0.54 | Sand | S (sand) |
*According to FAO (2006): Guidelines for soil description http://www.fao.org/3/a-a0542e.pdf.
Haat and Jabal Al-Akhdar are both situated within the Al-Hajar Mountain ranges and sampling sites were located at an elevation of approx. 989 and 2022 m a.s.l., respectively. Haat and Jabal Al-Akhdar are fairly pristine locations, characterized by a stony and rocky ground with small areas or sinks in between, where biocrusts and bare soil can be found. All other samples were collected at lowland sites ranging between 7 to 38 m a.s.l. (Table 1). The locations Mahoot, Muscat and Sur are anthropogenically influenced areas. The Mahoot site, about 25 km away from the sea, was situated next to a big road, where tyre and hoof prints indicated anthropogenic influences as well as an impact by grazing. The sampling site Sur was located between a highway road and the sea, whereas the Muscat site was a ruderal site located at the edge of the city of Muscat. While the non-elevated sites experience temperatures between 40–50 °C in summer and as low as 12 °C in winter, with a mean annual precipitation of ca. <100 mm (from November to March), the temperature at the elevated sites reaches a maximum of 25–35 °C in summer but can freeze in winter, with a mean annual precipitation of about 300 mm31. The Shana samples were collected close to the sea under marine influence. Shana1 was a salt pan area presenting hard and desiccated salt crusts with interspersed microbial communities, whereas Shana3 was inundated by seawater during high tides, forming soft marine mats with a typical striped appearance.
At each site, samples of the different types of biocrusts as well as bare soils were collected in triplicates. The topmost 1 cm of cyanobacteria- and lichen-dominated crusts (when found) as well as bare soils (for comparison) were collected in small petri dishes (5.5 cm diameter) using aseptic tools. Biocrusts from Shana1 and microbial mats from Shana3 sites were also sampled in triplicate in a similar way. Samples for molecular work were transported on dry ice to guarantee a continuous cooling chain and finally stored in a freezer at −80 °C until analysis.
Environmental and soil quality parameters
The textural classes were estimated by a simple field test based on feeling the constituents of the soil32. For this, soil samples have to be in a moist to slightly wet state and gravel and other constituents >2 mm have to be removed. The constituents feel as follows, depending on the dominating compound: (1) Clay, such as clayey loam, feels sticky. Soil between fingers, is cohesive (sticky), is formable, has a high plasticity and a shiny surface when squeezed between fingers; (2) Silt, such as silty loam or silty clay, feels smooth. Soil between fingers, is non-sticky, only weakly formable, has a rough and rippled surface after squeezing between fingers and feels very floury (like talcum powder), and (3) Sand, such as sandy loam or sandy clay, has a gritty texture. It cannot be formed, does not stick to fingers and feels very grainy.
For determination of soil pH and electrical conductivity, 1 g of soil were thoroughly mixed with 2.5 ml of distilled water and allowed to stand for 45 min. The filtrate was collected and the pH and electrical conductivity were measured from the filtrate using glass electrodes (Minitrode, Hamilton Messetechnik GmbH, Höchst, Germany). Total nitrogen and carbon were analysed in the soil samples using a C/N analyser (cube EL, Elementar Analysensysteme GmbH, Hanau, Germany), after air drying the soil, followed by grinding and homogenization. The results are expressed as oven-dry soil (105 °C).
DNA sequencing
For DNA extraction, we combined many small subsamples from one petridish to obtain one DNA sample per petridish. The subsamples of the biocrusts and mats were taken from the photoautotrophic layer and the heterotrophic layer directly below. For bare soil, many subsamples taken from the uppermost millimetres were combined in one sample. DNA was extracted from biocrusts, bare soils and microbial mats (total 45 samples) using PowerSoil DNA extraction kit (MOBIO laboratories, Inc., Carlsbad, CA) according to the manufacturer’s instructions. Purified DNA extracts were then submitted to Molecular Research MR DNA laboratory (www.mrdnalab.com, Shallowater, TX, USA) for paired-end Illumina amplicon sequencing of the bacterial 16S rRNA genes V4 variable region using the primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) with barcode on the forward primer33. A single-step 30 cycle PCR using HotStarTaq Plus Master Mix kit (Qiagen, USA) was performed at 94 °C for 30 seconds, 53 °C for 40 seconds and 72 °C for 1 minute. The fungal internal transcribed spacer (ITS) region was also amplified from the same DNA extracts using the primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′)34. After amplification, PCR products were checked in 2% agarose gel to determine the success of amplification and the relative intensity of bands. Multiple samples were pooled together in equal proportions based on their molecular weight and DNA concentrations. Pooled samples were purified using calibrated AMPure XP beads according to the manufacturer’s instructions (BioLabs, NewEngland). The pooled and purified PCR products were used to prepare a DNA library by following Illumina DNA library. Details for library preparation protocol and quality control can be found in TrueSeq Nano DNA Library Prep, Reference Guide, Illumina. Sequencing was performed on a MiSeq following the manufacturer’s guidelines.
Sequence data were demultiplexed and adapters and primers were removed using the software FASTqProcessor version 1.1.4.19846 (http://www.mrdnafreesoftware.com/; date accessed: 23.02.2019) provided by MR DNA (MR DNA, Shallowater, TX, USA). For the fungal data set, primers were further removed from the 3′ end of each read using cutadapt version 1.9.135. In compliance with the Minimal Information about any (X) Sequence (MIxS) standard36, the demultiplexed and primer-clipped sequences were deposited at the European Nucleotide Archive37 using the data brokerage service of the German Federation for Biological Data38. They are accessible under the project accession number PRJEB31699. Further sequence processing steps were conducted in R version 3.5.239 using the package dada2 version 1.10.140. Briefly, sequences were quality filtered at a maximum expected error rate of 3 after trimming both forward and reverse reads to 230 bp (bacterial data set) or removing reads shorter than 50 bp (fungal data set). Error learning, dereplication, and denoising were conducted with default parameters. Forward and reverse reads were merged with a minimum overlap of 10 bp. For chimera detection and taxonomic classification default settings were selected. Bacterial sequences were classified using the SILVA reference database version 13241 and fungal sequences were classified using the fungal release of the UNITE reference database42. Only non-singleton and non-doubleton sequence variants, hereafter referred to as operational taxonomic units (OTUs), that were classified on phylum level were retained for further analysis. Chloroplast and mitochondrial sequences were further removed from the bacterial data set. The code for the sequence analysis has been submitted to PANGAEA alongside the environmental and soil parameters (10.1594/PANGAEA.899529).
Statistical analysis
A principle component analysis (PCA) was conducted to discern patterns among samples based on their environmental characteristics. The following parameters were included in the PCA calculation using the R package PCAmixdata (version 3.1)43: elevation, pH, electrical conductivity, carbon and nitrogen content, human disturbance, soil texture class, and soil cover type (i.e. bare, cyano and lichen). For simplicity, textural classes were divided into sand, loam, and clay according to the dominant particle size.
Alpha diversity of the bacterial and fungal communities was assessed based on richness (number of OTUs), the Shannon-Wiener Index, and the Inverse Simpson Index after randomly rarefying the data set repeatedly to the minimum library size (bacteria: 16,338 sequences; fungi: 15,827 sequences). Samples with less than 10,000 sequences were excluded from the alpha diversity analysis. Cluster analysis and non-metric multidimensional scaling (NMDS) were performed to visualize patterns in bacterial and fungal community composition. While for the cluster analysis Bray-Curtis dissimilarities were calculated based on relative sequence abundances of OTUs without further transformation, for NMDS, the Wisconsin double standardization was applied to the relative sequence abundances prior to the calculation of Bray Curtis dissimilarities. Bacterial communities collected at Shana were excluded from the NMDS since their Bray-Curtis dissimilarity to any of the other samples was by far the highest between any two samples, which would have reduced the resolution of the dissimilarities between the remaining samples. Analysis of similarity (ANOSIM) was used to assess the separation of bacterial and fungal communities between groups of samples based on similar environmental parameters.
Redundancy analyses (RDA) were run to evaluate the ability of environmental parameters (cover type, soil texture, human disturbance, elevation, electrical conductivity, pH, carbon and nitrogen content) to explain the variation in bacterial and fungal community composition. Nitrogen content was excluded as an explanatory variable in the RDA models due to a high collinearity with elevation. Prior to RDA, OTUs that did not occur in at least 2 replicates per location and sample type were removed from the dataset. Furthermore, sequence counts were centered log ratio (clr)-transformed. Best-fitting RDA models were selected based on maximum adjusted R² and minimum AIC (Akaike Information Criterion, an estimator of the relative quality of statistical models for a given dataset). Variance inflation factors of the explanatory variables in the best-fitting models were below 10. All statistical analyses were conducted using the R software package, version 3.5.139 and R-Studio, version 1.0.15344. Additional packages used within R were ‘PCAmixdata’43 and ‘vegan’45.
Results
Variation among sampling sites and soil samples
Field observations showed that cyanobacteria-dominated crusts were encountered at all sampling sites, whereas lichen-dominated crusts were mainly restricted to the elevated sites (i.e. Haat and Jabal Al-Akhdar). Bare soils (except from Mahoot) and Shana biocrusts and mats had coarse sandy texture, all Mahoot soils had loamy texture and the remaining biocrusts had either loamy or clayey texture (Table 1). A PCA analysis captured 54.7% of the variation among sampling sites on the first two principle components using the measured environmental parameters and soil characteristics (Fig. 2). Elevation of the sampling site co-varied with nitrogen (N) content in soils and both were the major contributors to PC1, which accounted for 33.4% of the variation. PC2 explained 21.3% of total variance and this was mainly attributed to electrical conductivity and carbon content. Based on the PCA patterns, samples collected from elevated sites (i.e. Jabal Al-Akhdar and Haat) were clearly separated from the rest (Green hull in Fig. 2). These soils had the highest N levels with values between 0.08 and 0.22% (Table 1). The sampling sites Mahoot, Sur and Muscat, characterized by their exposure to human and/or animal disturbance, clustered together (Red hull in Fig. 2). Shana biocrust and mat samples formed a distinct cluster along the second PC axis, significantly different from all other samples (Blue hull Fig. 2). This separation was largely driven by the high electrical conductivity values, which reached as high as ca. 109 mS/m (Table 1).
Figure 2.
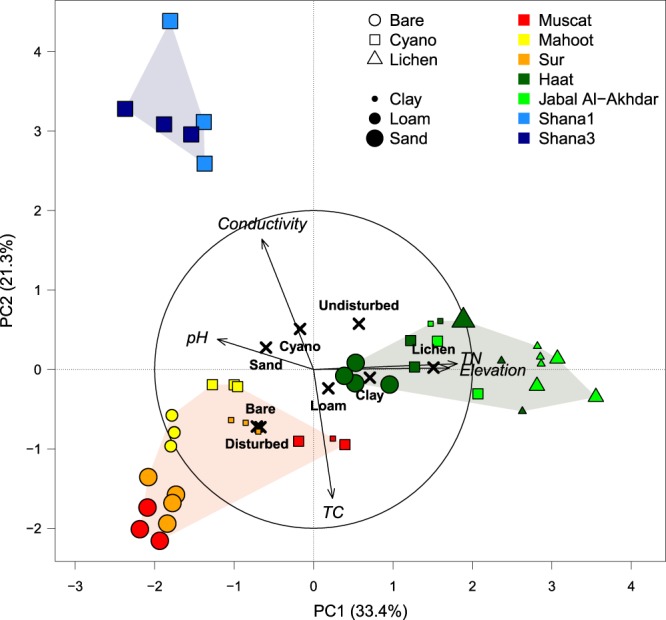
PCA plot of environmental parameters. Color, shape, and size of points indicate sampling location, surface cover type, and soil texture, respectively. Based on their environmental characteristics, samples group into 3 distinct categories, shown as shaded hulls: disturbed (red), marine (blue), and elevated (green) sites. TC and TN refer to total carbon and total nitrogen, respectively.
Redundancy analysis revealed that soil cover type, soil texture, carbon content, elevation, and human disturbance explained 32% of the variation in bacterial communities but only 19% in fungal communities (Table 2). Individually, all tested parameters contributed significantly to the variations in bacterial communities while accounting for the effect of the other parameters (pure effects), although the total effects were often considerably larger. Among the observed environmental parameters, the most important determinants of bacterial communities were cover type (pure: 6.5%, total: 10.6%; Table 2), carbon content (pure: 5.7%, total: 6.3%), and elevation (pure: 4.3%, total: 8.0%) followed by soil texture (pure: 3.9%, total: 7.1%) and human disturbance (pure: 3.8%, total: 7.0%; Table 2). A similar pattern was observed for fungal communities. Soil texture, elevation, human disturbance, and cover type were the most important factors explaining 3.5% (pure: 2.0%), 4.9% (pure: 2.1%), 6.2% (pure: 2.2%), and 3.8% (pure: 4.3%) of the variation in fungal communities, respectively (Table 2). Electrical conductivity and pH were not selected in the best fitting model, presumably because they did not substantially contribute to explaining community composition within the observed range, or because they were redundant (collinear) with parameters already included in the model.
Table 2.
Effects of contextual parameters on variation in bacterial and fungal communities of studied soils.
| Explanatory factors | Bacteria | Fungi | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Effect | R² | F | df | p | R² | F | df | p | |
| Soil texture | Pure | 3.9 | 2.1 | 2, 37 | 0.001*** | 2.0 | 1.5 | 2, 36 | 0.004** |
| Total | 7.1 | 2.7 | 2, 42 | 0.001*** | 3.5 | 1.8 | 2, 41 | 0.001*** | |
| Cover type | Pure | 6.5 | 2.8 | 2, 37 | 0.001*** | 4.3 | 2.0 | 2, 36 | 0.001** |
| Total | 10.6 | 3.6 | 2, 42 | 0.001*** | 3.8 | 1.9 | 2, 41 | 0.001*** | |
| Carbon content | Pure | 5.7 | 4.1 | 1, 37 | 0.001*** | 3.2 | 2.5 | 1, 36 | 0.001*** |
| Total | 6.3 | 3.9 | 1, 43 | 0.001*** | 3.5 | 2.6 | 1, 42 | 0.001*** | |
| Elevation | Pure | 4.3 | 3.4 | 1, 37 | 0.001*** | 2.1 | 2.0 | 1, 36 | 0.001*** |
| Total | 8.0 | 4.8 | 1, 43 | 0.001*** | 4.9 | 3.2 | 1, 42 | 0.001*** | |
| Human disturbance | Pure | 3.8 | 3.1 | 1, 37 | 0.001*** | 2.2 | 2.0 | 1, 36 | 0.002** |
| Total | 7.0 | 4.3 | 1, 43 | 0.001*** | 6.2 | 3.8 | 1, 42 | 0.001*** | |
| Nitrogen content | Total | 5.1 | 3.4 | 1, 43 | 0.001*** | 3.9 | 2.7 | 1, 42 | 0.001*** |
| Conductivity | Total | 1.3 | 1.6 | 1, 43 | 0.035* | 0.1 | 1.1 | 1, 42 | ns |
| pH | Total | 1.3 | 1.6 | 1, 43 | 0.027* | 0.6 | 1.2 | 1, 42 | ns |
| All | Pure | 31.5 | 3.9 | 7, 37 | 0.001*** | 19.1 | 2.5 | 7, 36 | 0.001*** |
| Total | 32.1 | 3.1 | 10, 34 | 0.001*** | 19.5 | 2.0 | 10, 33 | 0.001*** | |
Total and the pure effects (i.e. when controlling for all other factors of the analysis) of explanatory factors were calculated by using canonical redundancy analysis (RDA) models. The proportion of explained community variation is expressed as R2 values. Significances of the respective F-ratios were tested by performing 1000 Monte Carlo permutation tests and are indicated by *significant (P ≤ 0.05), **very significant (P ≤ 0.01), ***highly significant (P ≤ 0.001), and ns when not significant (P > 0.05). df: degrees of freedom (numerator, denominator).
We were not always able to isolate the effect of a particular parameter on bacterial community composition, while accounting for all of the other parameters in the model as evidenced by the differences between pure and total effects (Table 2). For instance, since samples with the highest electrical conductivity were all sandy soils collected at Shana from cyanobacteria-dominated crusts/mats, it was not possible to reliably attribute the differences observed in the bacterial communities at Shana to any of these parameters. Besides, bare soils were mostly collected at disturbed non-elevated locations, whereas lichen-crusts were mainly found at elevated undisturbed sites, resulting in a non-orthogonal sampling design and making a distinction between the effects of the different factors very difficult. Therefore, the pure effects of the parameters included in the RDA were considerably lower than the total effects (Table 2).
Alpha and beta diversity analysis
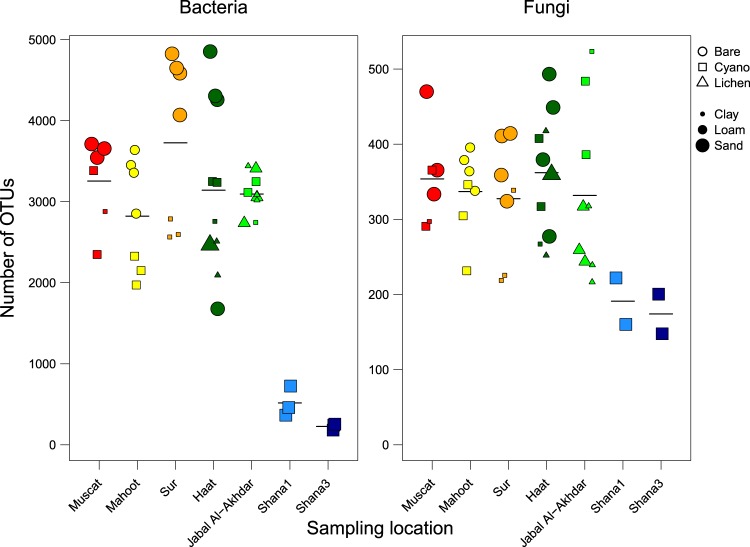
Bacterial and fungal diversity estimators were lower in the samples from Shana compared to all other samples (Fig. 3; Supplementary Table S1). For instance, bacterial and fungal species richness, as determined by number of OTUs, reached an average of 3196 ± 796 and 343 ± 80 in Muscat, Mahoot, Sur, Jabal Al-Akhdar and Haat soils but only 371 ± 200 and 183 ± 35 in Shana soils, respectively. Among the locations, where both bare and crusted soils were sampled, the average numbers of OTUs and Shannon index in bare soils at Mahoot, Sur and Haat were considerably higher than in crusted soils at these sites, whereas at Muscat similarly high numbers were reached by cyanobacteria-dominated crust (Fig. 3, Supplementary Table S1). The averages of OTU richness of crusted soils from all sites (except from Shana) were comparable.
Figure 3.
Bacterial and fungal richness (number of OTUs). Color, shape, and size of points indicate sampling location, surface cover type, and soil texture, respectively. The sample color codes are as in Fig. 2.
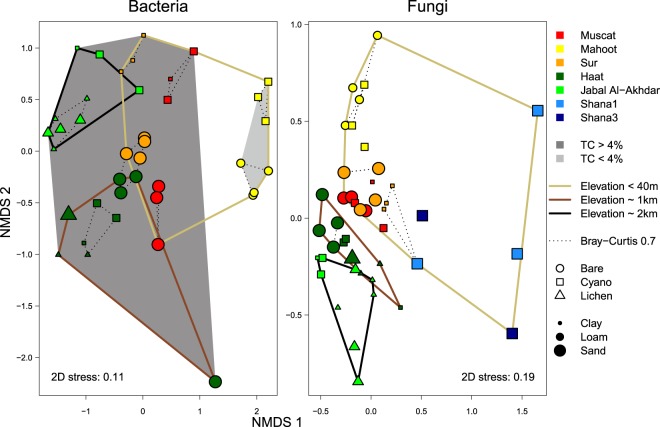
When variations in bacterial and fungal communities among bare soils and biocrusts were visualized in a two-dimensional space using multidimentional scaling (NMDS), both communities showed distinct clusters according to the significant parameters determined by the RDA analysis (Fig. 4). Soils of the same cover type and from the same site clustered together, although there was a clear heterogeneity among replicates (Fig. 4). Bacterial communities of soils containing more than 4% carbon content were clearly different from those found in less than 4% carbon-containing soils from similar elevation and disturbance conditions (Fig. 4). The NMDS ordination further suggested a clustering of bacterial as well as fungal communities based on the elevation of the sampling site, which was, however, only moderately supported by ANOSIM (R = 0.31–0.57, P < 0.05), due to a high intra-group dissimilarity. Bacterial communities of Shana samples, exposed to marine influence and characterized by high electrical conductivity, were very different from the other locations and formed a well-separated, although very heterogeneous, cluster (Fig. 5, ANOSIM R = 0.61–0.89, P < 0.002). Fungal communities at Shana were also distinct from the other sites (Fig. 4, ANOSIM R = 0.57–0.81, P < 0.002), although given the generally much higher heterogeneity of fungal communities (Fig. 6) this separation was not as pronounced as for the bacterial communities (Fig. 5).
Figure 4.
Non-metric multidimensional scaling (NMDS) plot based on Bray-Curtis dissimilarity of bacterial and fungal communities. Color, shape, and size of points indicate sampling location, surface cover type, and soil texture, respectively. Dotted lines show clusters with a maximum intra-group dissimilarity of 0.7. Solid lines group samples with similar elevation. Shaded hulls indicate organic carbon content (TC in %).
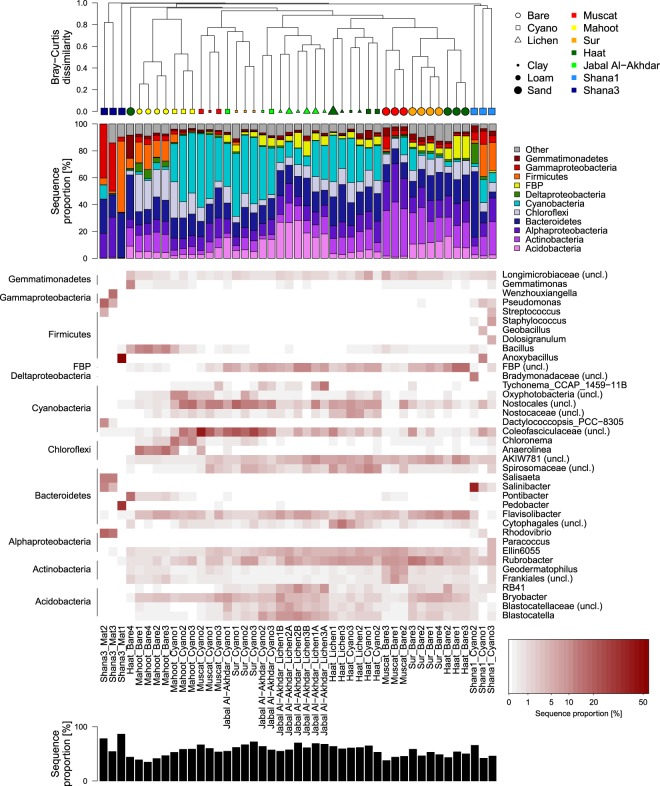
Figure 5.
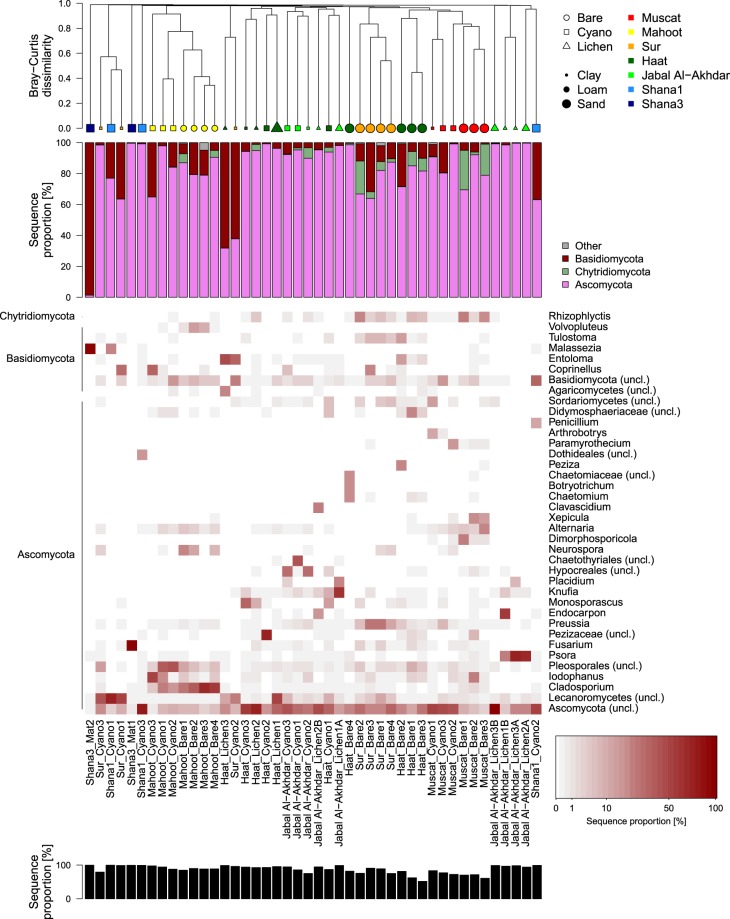
Bacterial community composition on phylum and genus level. Order of plots from top to bottom: hierarchical cluster diagram based on Bray-Curtis dissimilarity, bar plot of relative sequence proportions of dominant bacterial phyla (class-level taxonomy for Proteobacteria), heatmap of dominant genera (white: no sequences), total sequence proportion of genera displayed in heatmap.
Figure 6.
Fungal community composition on phylum and genus level. Order of plots from top to bottom: hierarchical cluster diagram based on Bray-Curtis dissimilarity, bar plot of relative sequence proportions of dominant bacterial phyla, heatmap of dominant genera (white: no sequences), total sequence proportion of genera displayed in heatmap.
Cluster analysis based on Bray-Curtis dissimilarities provided more details about the pattern in community dissimilarity than NMDS. It confirmed that bacterial and fungal communities clustered together based on cover type (i.e. bare, cyanobacteria- or lichen-dominated crust), as well as their sampling site (Figs 5 and 6). Bare soils were found in two clusters: the first cluster comprising Muscat, Sur and Haat samples, while the second cluster was formed by bare soil samples from Mahoot (Figs 5 and 6). In general, bacterial and fungal communities displayed a surprising congruency in diversity patterns.
Variation in bacterial communities
In all bare and crusted soils, the majority of sequences belonged to Actinobacteria (average 13% across all samples), Alphaproteobacteria (12%), Bacteroidetes (16%) and Cyanobacteria (21%), encompassing between 32 to 83% of the total number of sequences per sample (Fig. 5). Notably a considerable proportion of all sequences (45%) could not be confidently classified on genus level. Actinobacteria exhibited their highest relative abundance in bare soils (21 ± 9% of total sequences), followed by lichen-dominated (11 ± 3%) and cyanobacteria-dominated crusts (7 ± 6%). In bare soils, the relative abundance of Actinobacteria was highest in Muscat soils, but lowest in Mahoot soils, whereas in cyanobacteria-dominated crusts, they were higher in Haat than in Jabal Al-Akhdar samples (Fig. 5). The relative abundance of Actinobacteria in Haat soils was comparable, regardless of the surface cover type, with an average of 16 ± 2% in the bare soil, 11 ± 4% in the cyanobacteria- and 13 ± 2% in the lichen-dominated crusts. In contrast, their relative abundance in lichen-dominated crusts from Jabal Al-Akhdar was considerably higher than in the cyanobacteria-dominated crusts (6–15% vs. 3–4% of total sequences, respectively). At Shana3, Actinobacteria were completely absent. At the genus level, most of the actinobacterial sequences across all samples belonged to Rubrobacter.
Among the proteobacterial classes, Alphaproteobacteria were most frequently encountered in all samples, with few sequences from Gammaproteobacteria (Fig. 5). The relative abundance of Gammaproteobacteria did not exceed 10% in any of the soils, except in Shana3 mats, where they made up between 3–40% of the total number of sequences, respectively (Fig. 5). Gammaproteobacterial sequences belonged mainly to the genera Wenzhouxiangella, Pseudomonas, Delftia and the Burkholderia-Caballeronia-Paraburkholderia complex. The contribution of Alphaproteobacteria to the total number of sequences did not vary much among the different surface cover types, with an average proportion between 11 and 14% across all samples. The highest relative abundance of Alphaproteobacteria was detected in Muscat bare soils and Shana3 mats, with up to 28% and 30% of total sequences, respectively. While in Muscat soils these sequences belonged mainly to Sphingomonas (Ellin6055), they belonged to Rhodovibrio in Shana3 mats (Fig. 5). In Muscat soils, the relative abundance of Alphaproteobacteria in bare soils was comparatively higher than in cyanobacteria-dominated crusts (24 ± 4% vs. 13 ± 2%).
Bacteroidetes constituted between 5 to 49% of the total number of sequences in each sample (average 17 ± 7%). Both of these extremes were recorded in samples from Shana1 (Fig. 5). Samples from Shana3 and Haat generally displayed higher proportions of Bacteroidetes (16–33%) compared to the remaining locations with similar proportions between 10 and 22%. At each sampling location, the relative abundance of Bacteroidetes among bare and crusted soils was comparable (Fig. 5). In Shana soils, most Bacteroidetes sequences belonged to Salisaeta, Salinibacter and Pedobacter, although these genera were barely detectable in any other soil (<0.2% of total sequences; Fig. 5). Conversely, sequences affiliated to Flavisolibacter were found in all bare and crusted soils from the other sites (Fig. 5).
Cyanobacteria exhibited the most remarkable differences between the different types of soils and biocrusts in all locations (Fig. 5). While cyanobacteria were barely detectable in most bare soil samples (average of 4 ± 5%), their average percentage across the cyanobacteria- and lichen-dominated crusts reached 29 ± 18% and 20 ± 9%, respectively (Fig. 5). In bare soils, cyanobacteria exhibited their highest relative abundance in one of the Muscat and Sur samples (13% and 16% of total sequences, respectively) but made up only less than 5% in each of the other samples (Fig. 5). The relative abundance of cyanobacteria in cyanobacteria-dominated crusts was notably lower at Shana1 compared to all other sites and they were almost absent in the Shana3 mats, with the exception of Dactylococcopsis, which constituted 10% of the sequences in one sample (Fig. 5). At Jabal Al-Akhdar, cyanobacteria had higher relative abundance in cyanobacteria- than in lichen-dominated crusts with an average of 35 ± 4 and 16 ± 8%, respectively, but this difference was not observed in samples from Haat (25 ± 8 vs. 28 ± 6%). Notably, the most frequent cyanobacterial OTUs could not be classified on genus level, but were affiliated with uncultured representatives of the Nostocales. Closest relatives were Coleofasciculus (sequence identity 89–95%), Loriellopsis (88–89%), Planktothricoides (89%), Brasilonema (92–96%, Supplementary Dataset 1). These so far not well characterized OTUs dominated the cyanobacterial communities of most crusts, except those from Shana, but displayed variable distribution among different samples. For instance, while most presumptive Coleofasciculus OTUs were prevalent in cyanobacteria-dominated crusts of Muscat and Sur samples, other presumptive Coleofasciculus as well as potential Brasilonema OTUs were frequently found in cyanobacteria- and lichen-dominated crusts of Haat, and an OTU most closely related to Loriellopsis characterized the cyanobacteria-dominated crusts of Mahoot.
Sequences belonging to Acidobacteria were encountered in all soils at a relative abundance of less than 8% of the total number of sequences in each sample, except in bare soils of Sur and Haat (8–18% of total sequences in each sample) and crusted soils of Jabal Al-Akhdar (14–15% in the cyanobacteria-dominated crusts and 16–28% in lichen-dominated crusts). Most of these sequences belonged to the genus Bryobacter and representatives of the Blastocatellaceae (Fig. 5). While of low overall sequence abundance, Firmicutes contributed substantially to samples from bare soils of Mahoot (12–18%), and cyanobacteria-dominated crusts of Shana1 (2–24%) and Shana3 (2–53%). Similarly, Chloroflexi were only found in high proportions in both bare soils and cyanobacteria-dominated crusts of Mahoot (12–28%; Fig. 5).
Variation in fungal communities
Fungal sequences belonged to three main phyla (i.e. Ascomycota, Chytridiomycota, and Basidiomycota), with an average of 83 ± 20% of the total number of sequences belonging to Ascomycota across all samples (Fig. 6). Similar to the bacterial dataset, a large proportion of fungal sequences (49%) remained unclassified on genus level. Ascomycota were predominantly encountered in most bare and crusted soils (≥50% of total sequences), except three samples from Shana3, Haat, and Sur (Fig. 6). This group constituted 99% of the total sequences in one microbial mat from Shana3 (Shana3_Mat1) but was almost absent in another mat sample (Shana3_Mat2; Fig. 6). This high heterogeneity between sampling locations and cover type, but also between replicate samples from similar conditions, was even more pronounced at higher taxonomic resolution, where each sample was often dominated by a (different) single genus. Patterns of Ascomycota genera among samples were further obscured by a large proportion of Ascomycota OTUs, which all had less than 85% sequence identity to known fungal species (Supplementary Dataset 2), but which showed highly variable sequence proportions among samples. For instance, unclassified OTUs of the Lecanoromycetes characterized cyanobacterial-dominated crusts in Sur and Shana1. Among classified representatives of the Ascomycota, Cladosporium was found predominantly in both bare soils (28–66%) and cyanobacteria-dominated crusts (4–23%) of Mahoot, while Neurospora was mostly restricted to bare soils from this location (Fig. 6). OTUs of the genera Psora, Endocarpon, Knufia, and Placidium dominated most of the lichen-crusts of Jabal Al-Akhdar comprising up to 69%, 58%, 60%, and 21% of the fungal community, respectively (Fig. 6). The highest proportions of Fusarium and Penicillium were encountered at Shana3 and Shana1, respectively, however, only in one out of three replicate samples. The genera Dimorphosporicola, Xepicula, and Alternaria were predominantly found in bare soils from Muscat with up to 24%, 17%, and 21%, respectively, whereas they comprised mostly less than 1% in all other sample (Fig. 6). In bare soils of Sur, Preussia was consistently enriched (5–27%), while Monosporascus was found in high proportions in several crusts from Haat (Fig. 6).
The relative abundance of Chytridiomycota was considerably higher in bare soils with up to 26% compared to cyanobacteria- and lichen-dominated crusts, where they accounted for less than 7% and 4% of the total number of sequences in each sample, respectively (Fig. 6). Nevertheless, there were clear differences among replicates from the same site (Fig. 6), with percentages ranging between 2–26%, 0.3–9%, and 2–21% in bare soils from Muscat, Haat and Sur. Most Chytridiomycota sequences belonged to the genus Rhizophlyctis, which in some replicates accounted for 8–25% of the sequences in bare soils from Muscat, Sur and Haat sites (Fig. 6).
Sequences belonging to Basidiomycota were also detected in bare soils, mat samples, cyanobacteria- and lichen-dominated crusts from all sites (Fig. 6). Their relative abundance in a mat sample from Shana3 and a cyanobacteria- and a lichen-dominated crusts from Sur and Haat was highest and reached 62 to 99% of the total number of sequences (Fig. 6). The main genera determined for this phylum were Malassezia, Coprinellus, Volvopluteus, Entoloma and Tulstoma, of which Malassezia was found mainly in Shana samples, while Volvopluteus was characteristic for bare soils from Mahoot, and Tulstoma for bare soils from Sur (Fig. 6). Sequences belonging to Coprinellus accounted for 33% of the total number of sequences in one cyanobacteria-dominated crust from Mahoot and Sur (Fig. 6).
Discussion
Field observations have shown that lichen-dominated biocrusts become more prevalent as aridity decreases and precipitation rates increase20,23. This is congruent with our observations at the elevated sites of Haat and Jabal Al Akhdar and with the previously reported lichen vegetation of the Arabian Peninsula, Colorado Plateau and Gurbantunggut Desert46,47. At the Jabal Al Akhdar site, the higher annual precipitation and the mountain slopes supported greater cover of lichens and the samples were characterized by fine-textured soils.
While the relative abundance of Cyanobacteria was much higher in crusted than in bare soils, the relative abundance of Actinobacteria showed an opposite trend. Shifts in microbial communities at different successional stages of crust development have been previously reported18,48–50. This is ascribed to the fact that growth of Cyanobacteria results in higher levels of organic matter and nutrients in crusted soils, relative to bare soils, thus promoting the growth of different microbial communities51,52. The improvement in soil physiochemical properties under crusts also promotes hyphal proliferation and fungal growth53,54. However, the competition for organics and nutrients in crusts could result in the growth of highly specific and competitive microbial populations and this could explain our lower OTU richness of bacteria and fungi in crusted than in bare soils.
The detected bacterial phyla including Cyanobacteria, Actinobacteria and Proteobacteria, were also numerically abundant in other crusts from the Colorado Plateau and Sonoran Desert55,56. The Cyanobacteria Coleofasciculus (formerly Microcoleus), Brasilonema (a genus placed within Scytonema in the silva reference database41 and Nostocales have been previously encountered in crusts from Oman and elsewhere30,31,56–58. The prevalence of Coleofasciculus spp. in most crusts from all locations is consistent with previous observations, and highlights their crucial role in the consolidation of soil particles and crust formation55,56,59. Actinobacteria, exhibiting their highest relative abundance in bare soils, are common in soil environments and play a vital role in the decomposition of organic matter and recycling of nutrients60. Actinobacteria was the most dominant phylum in biocrusts from the Colorado Plateau, Sonoran Desert and Tengger Desert, China17,48,55,56, but were detected at a very low dominance in biocrusts from the Gurbantunggut Desert and Cape Code Seashore18,61,62. Bacteroidetes and Proteobacteria, which are known to produce exopolysaccharides, may contribute to soil stabilization and biocrust formation56. Proteobacteria and Bacteroidetes were shown to increase with biocrust formation in the Gurbantunggut Desert18. Also in biocrusts of the Succulent Karoo in South Africa, the relative abundance of some bacterial phyla (i.e., Acidobacteria, Chloroflexi, Verrucomicrobia) was higher, whereas that of Thermi was lower compared to uncrusted soils50. The detection of some similar taxa in bare soils and cyanobacteria- and lichen-dominated crusts suggests that some bacterial populations are retained during the crust development process.
The dominance of Ascomycota in our biocrusts has also been shown for biocrusts from the Negev Desert, Wyoming and Utah, Colorado Plateau, and the Chihuahuan Desert63–67. While Ascomycota showed a similar relative abundance across all bare and crusted soils, Chytridiomycota were more prevalent in bare than in crusted soils. Members of Chytridiomycota have not been detected in any biocrusts studied up to date, except at a low abundance in the Oman and the Chihuahuan deserts65,68. This suggests that Chytridiomycota belong to an early stage of crust development, indicating their tolerance to stress environments. The dominance of different fungi in bare soils compared with biocrusts indicates successional changes of fungal communities during crust formation from bare to lichen-dominated crusts, which have been previously demonstrated69. The dominance of a single fungus in every soil supports the notion that fungal succession occurs through the selection of the best-adapted species to a certain ecological niche69,70. The fungi belonging to the detected genera in our biocrusts are often very common components of crusts and aridland soils worldwide63,64,71. Interestingly, the lichen-forming Endocarpon was described for the first time for crusts from this region, whereas Psora has been known to be widespread68,72.
Crust microorganisms and soil characteristics
Microbial communities in our crusts were strongly affected by soil texture and carbon (and nitrogen) contents as could be inferred from RDA analysis. This is expected since clear differences in soil texture were observed, ranging from fine loamy texture only in Mahoot samples to coarse sandy texture in most bare soils and Shana crusts and mats. Soil texture affects crust development by influencing soil stability, aeration and water retention ability, as coarser soils are unstable and lose more water relative to fine soils73. Previously, it was shown that cyanobacteria develop on sandy soils whereas lichens preferably grow on fine texture soils74. However, biocrusts have also been shown to influence soil texture, as soil mean particle size decreased with succession of biocrusts of the Gurbantunggut Desert of northwestern China75. Lichens were also found associated with high soil N and C content76. This is in agreement with the greater cover of lichens at the elevated Jabal Al-Akhdar and Haat sites, characterized by high levels of carbon and nitrogen and a fine soil texture (Table 1, Fig. 2). Also for N and C contents, it is difficult to distinguish between the preference and impact of biocrust types on these factors, as well-developed biocrusts have been shown to also contribute to increased N and C contents in soils77,78.
Among the bacterial genera encountered in this study, the relative abundance of presumptive Coleofasciculus, and Blastocatella, was high in biocrusts with loam and clay textures in Jabal Al Akhdar, Haat, Sur and Muscat sites, whereas Bacillus exhibited their maximum relative abundance in biocrusts with loamy texture at the Mahoot site. The bacterial phyla Chloroflexi and Firmicutes, detected in Mahoot soils, are known to include species that are either strictly or facultatively anaerobic79,80. It is plausible that after heavy rainfalls, the fine-textured soil in Mahoot may cause limited oxygen diffusion, thus resulting in the development of anaerobic niches. Sequences belonging to Firmicutes and Chloroflexi have been previously reported from different bare desert soils and crusts from Oman, Atacama, Gurbantunggut, Tabernas, South African and Sonoran deserts18,50,80–84. Similar to bacterial communities, the distribution of fugal genera was also driven by soil texture. For instance, the fungal genera Cladosporium and Neospora were associated with loamy texture at Mahoot, the lichen-forming Psora, Endocarpon and Monosporascus were detected in loamy and clayey soils of Jabal Al-Akhdar and Haat, and Rhizophlyctis and Malassezia were mainly detected in coarse soils of Muscat and Shana, respectively.
Crustal microorganisms and elevation
RDA analysis indicated elevation to be a major contributor to variations in crust bacterial and fungal communities, and this was further supported by NMDS plots. Sites at different elevations clearly vary in climate, precipitation amounts and patterns, temperature regimes (daily and seasonal), UV and solar radiation, moisture and soil texture and chemistry. All these parameters have been shown to directly affect crustal microbial communities10,15,64. For instance, microbial communities in crusts from the Negev and Mojave Deserts have been shown to vary depending on altitude and annual precipitation64. Crust abundance and development was shown to increase with increasing moisture and precipitation levels, whereas with increasing aridity, biocrusts were restricted to cyanobacteria-dominated types10,85. Presumptive Brasilonema, Blastocatella, and some Nostocales OTUs showed the strongest differences along the elevation gradient. The sequence proportion of these bacterial genera increased with elevation, with the exception of Nostocales OTUs, which exhibited divergent trends. Cyanobacteria could play an active role in nitrogen fixation, contributing to the measured higher levels of N at the elevated compared to the non-elevated sites (Fig. 2). Indeed, previous research has demonstrated an increase in nitrogen fixation rates with increasing temperature, precipitation, and altitude86.
While the relative abundance of the lichen genera Psora and Endocarpon increased with elevation, showing their maximum percentage at Jabal Al- Akhdar, the relative abundance of Cladosporium and several unclassified Ascomycota decreased. These genera accounted for most of the differences in fungal communities between elevated and non-elevated biocrusts. The gelatinous fungal material surrounded by Endocarpon can absorb and retain water during precipitation events87. Therefore, elevation could promote the growth of fungi that are adapted to sporadic precipitation events.
Marine influence on crustal and mat microorganisms
The remarkable difference, especially in bacterial, but also in fungal communities, of the Shana samples, as shown by cluster and NMDS analyses, suggests that salinity may be a major determinant shaping these biocrust communities. This marine influence lead to significantly higher electrical conductivities and lower nutrient concentrations in Shana crusts (Table 1) and shifted the microbial communities. The infrequent exposure to seawater during high tides, followed by desiccation due to very high evaporation rates, resulted in salt precipitation in the Shana1 crusts. The Shana3 mat samples were regularly inundated, mostly soft, and had a layered structure with greenish and pink strata. Thus, the samples of Shana1 and Shana3 reflect a transition from salt-influenced biocrusts to intertidal mats. While the Shana1 crusts comprised Cyanobacteria, Actinobacteria, and Acidobacteria, these phyla were almost completely absent in Shana3 mats, where in some samples Rhodovibrio, Pedobacter and Anoxybacillus occurred in large numbers. This transition from microbial mats to crusts in coastal deserts has been previously described for the Sebkhas of Tunisia15. Earlier studies on various ecosystems demonstrated a decrease in microbial diversity with increasing salinity88–91. This is consistent with our observation, where OTU richness of both bacterial and fungal communities in Shana samples was indeed significantly lower than in other crusts. At Shana1, cyanobacteria were made up by halotolerant types such as Dactylococcopsis and presumptive Coleofasciculus, which are typically found in hypersaline mats92,93. The increased proportion of Bacteroidetes, known for their ability to degrade polymeric substances, in some samples could be linked to the increased production of EPS by cyanobacteria under salt stress94,95. The detection of many halophilic genera such as Salisaeta and Salinibacter is also consistent with the high salinities in these samples.
Elevated soil salinities are known to be less favourable for the development of lichen-dominated crusts19. Although high salinities do not support the growth of fungi96, several types of fungi were detected in Shana samples. The most prevalent fungi in Shana biocrusts were Malassezia, Fusarium, Penicillium, as well as many so far unclassified OTUs. Fungi belonging to some of these genera have been reported for other crusts from Oman, Colorado Plateau, Gurbantunggut Desert, Utah and Wyoming63,67–69. Nevertheless, their detection predominantly in Shana biocrusts suggests the ability of these genera to tolerate elevated salinities. Indeed, various taxa of Penicillium and Fusarium have been detected in salterns and hypersaline microbial mats and played a role in the degradation of EPS and growth on cyanobacterial exudates97,98.
We conclude that spatial heterogeneity in bacterial and fungal communities in the cyanobacteria- and lichen-dominated crusts from the central and coastal deserts of Oman is highly correlated with soil and site characteristics. Striking shifts in microbial communities were detected in crusts along an elevation gradient and in crusts/mats exposed to marine influence. Our results confirm previously reported effects of the studied parameters on other crusts. This strengthens the notion that crust microbial communities are highly selective to fit specific habitat conditions. Furthermore, the large proportion of unclassified OTUs in our data set emphasizes that biological soil crusts in Oman are a severely understudied environment.
Supplementary information
Acknowledgements
We would like to thank Mohammed Gomaa, Wadih Rassy and Derek Roberts for their help during the field trip. This research was funded by a research grant from Sultan Qaboos University (grant No. CL/SCI/BIOL/17/1). RA would like to thank the Hanse-Wissenschaftskolleg (HWK), Institute for Advanced Study, Germany for supporting his study group. BW, AT and RA would like to thank Ulrich Pöschl for his overall support and the provision of lab space and the Max-Planck Society for financial support.
Author Contributions
A.T., B.W., R.A. and A.A. collected soil samples; A.T. and B.W. took photographs of the sampling sites; A.T. and S.M. performed the DNA extraction and the molecular work; R.A. and C.H. did the bioinformatics and statistical analysis; E.R. prepared the map; S.F. analysed the soil physical and chemical properties; R.A. wrote the main manuscript and R.A. and C.H. prepared the figures and tables. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-42911-6.
References
- 1.Whitford, W. Ecology of desert systems (Academic Press, 2002).
- 2.Garcia-Pichel, F. & Belnap, J. In Biological soil crusts: Structure, function, and management (eds Belnap, J. & Lange, O. L.) 193–201 (Springer Berlin Heidelberg, 2003).
- 3.Elbert W, et al. Contribution of cryptogamic covers to the global cycles of carbon and nitrogen. Nature Geoscience. 2012;5:459–462. doi: 10.1038/ngeo1486. [DOI] [Google Scholar]
- 4.Rodriguez-Caballero E, et al. Dryland photoautotrophic soil surface communities endangered by global change. Nature Geoscience. 2018;11:185–189. doi: 10.1038/s41561-018-0072-1. [DOI] [Google Scholar]
- 5.Ferrenberg S, Reed SC, Belnap J. Climate change and physical disturbance cause similar community shifts in biological soil crusts. Proceedings of the National Academy of Sciences. 2015;112:12116–12121. doi: 10.1073/pnas.1509150112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maestre FT, et al. Do biotic interactions modulate ecosystem functioning along stress gradients? Insights from semi-arid plant and biological soil crust communities. Philosophical Transactions of the Royal Society B: Biological Sciences. 2010;365:2057–2070. doi: 10.1098/rstb.2010.0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tamm A, et al. Ecophysiological properties of three biological soil crust types and their photoautotrophs from the Succulent Karoo, South Africa. Plant and Soil. 2018;429:127–146. doi: 10.1007/s11104-018-3635-4. [DOI] [Google Scholar]
- 8.Zhang, Y., Aradottir, A. L., Serpe, M. & Boeken, B. Interactions of Biological Soil Crusts with Vascular Plants in Biological soil crusts: An Organizing Principle in Drylands, Ecological Studies 226 (eds Weber, B., Büdel, B. & Belnap, J.) 385–406 (Springer International Publishing, 2016).
- 9.Zhao HL, Guo YR, Zhou RL, Drake S. Biological soil crust and surface soil properties in different vegetation types of Horqin Sand Land, China. Catena. 2010;82:70–76. doi: 10.1016/j.catena.2010.05.002. [DOI] [Google Scholar]
- 10.Bowker, M. A. et al. In Biological soil crusts: An organizing principle in drylands (eds Weber, B., Büdel, B. & Belnap, J.) 173–197 (Springer International Publishing, 2016).
- 11.Rogers R. Soil surface lichens in arid and subarid south-eastern Australia. II. Phytosociology and geographic zonation. Australian Journal of Botany. 1972;20:215–227. doi: 10.1071/BT9720215. [DOI] [Google Scholar]
- 12.Rogers R. Soil surface lichens in arid and subarid South-Eastern Australia. III. The relationship between distribution and environment. Australian Journal of Botany. 1972;20:301–316. doi: 10.1071/BT9720301. [DOI] [Google Scholar]
- 13.Eldridge D. Distribution and floristics of terricolous lichens in soil crusts in arid and semi-arid New South Wales, Australia. Australian Journal of Botany. 1996;44:581–599. doi: 10.1071/BT9960581. [DOI] [Google Scholar]
- 14.Eldridge DJ, Tozer ME. Environmental factors relating to the distribution of terricolous bryophytes and lichens in semi-arid Eastern Australia. The Bryologist. 1997;100:28–39. doi: 10.2307/3244384. [DOI] [Google Scholar]
- 15.Ullmann, I. & Büdel, B. In Biological soil crusts: Structure, function, and management (eds Belnap, J. & Lange, O. L.) 107–118 (Springer Berlin Heidelberg, 2003).
- 16.Rivera-Aguilar V, Montejano G, Rodríguez-Zaragoza S, Durán-Díaz A. Distribution and composition of cyanobacteria, mosses and lichens of the biological soil crusts of the Tehuacán Valley, Puebla, México. Journal of Arid Environments. 2006;67:208–225. doi: 10.1016/j.jaridenv.2006.02.013. [DOI] [Google Scholar]
- 17.Steven B, Gallegos-Graves LV, Belnap J, Kuske CR. Dryland soil microbial communities display spatial biogeographic patterns associated with soil depth and soil parent material. FEMS Microbiology Ecology. 2013;86:101–113. doi: 10.1111/1574-6941.12143. [DOI] [PubMed] [Google Scholar]
- 18.Zhang B, Kong W, Wu N, Zhang Y. Bacterial diversity and community along the succession of biological soil crusts in the Gurbantunggut Desert, Northern China. J Basic Microbiol. 2016;56:670–679. doi: 10.1002/jobm.201500751. [DOI] [PubMed] [Google Scholar]
- 19.Büdel, B. In Biological soil crusts: Structure, function, and management (eds Belnap, J. & Lange, O. L.) 141–152 (Springer Berlin Heidelberg, 2003).
- 20.Büdel B, et al. Southern african biological soil crusts are ubiquitous and highly diverse in drylands, being restricted by rainfall frequency. Microbial Ecology. 2009;57:229–247. doi: 10.1007/s00248-008-9449-9. [DOI] [PubMed] [Google Scholar]
- 21.Lange, O. L. In Biological soil crusts: Structure, function, and management (eds Belnap, J. & Lange, O. L.) 217–240 (Springer Berlin Heidelberg, 2003).
- 22.Garcia-Pichel F, Loza V, Marusenko Y, Mateo P, Potrafka RM. Temperature drives the continental-scale distribution of key microbes in topsoil communities. Science. 2013;340:1574–1577. doi: 10.1126/science.1236404. [DOI] [PubMed] [Google Scholar]
- 23.Bowker MA, Belnap J, Davidson DW, Goldstein H. Correlates of biological soil crust abundance across a continuum of spatial scales: support for a hierarchical conceptual model. Journal of Applied Ecology. 2006;43:152–163. doi: 10.1111/j.1365-2664.2006.01122.x. [DOI] [Google Scholar]
- 24.George, D. B., Davidson, D., Schliep, K. C. & Patrell-Kim, L. J. Microtopography of microbiotic crusts on the Colorado Plateau, and distribution of component organisms. Vol. 60 (2000).
- 25.Belnap J, Harper KT, Warren SD. Surface disturbance of cryptobiotic soil crusts: Nitrogenase activity, chlorophyll content, and chlorophyll degradation. Arid Soil Research and Rehabilitation. 1994;8:1–8. doi: 10.1080/15324989309381373. [DOI] [Google Scholar]
- 26.Eldridge DJ, Koen TB. Cover and floristics of microphytic soil crusts in relation to indices of landscape health. Plant Ecology. 1998;137:101–114. doi: 10.1023/A:1008036214140. [DOI] [Google Scholar]
- 27.Kemmling A, Pfeiffer B, Daniel R, Hoppert M. Bacterial diversity in biological soil crusts from extrazonal mountain dry steppes in northern Mongolia Erforsch. biol. Ress. Mongolei (Halle/Saale) 2012;12:437–449. [Google Scholar]
- 28.Thomas AD. Impact of grazing intensity on seasonal variations in soil organic carbon and soil CO2 efflux in two semiarid grasslands in southern Botswana. Philosophical Transactions of the Royal Society B: Biological Sciences. 2012;367:3076–3086. doi: 10.1098/rstb.2012.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mogul, R. et al. Microbial community and biochemical dynamics of biological soil crusts across a gradient of surface coverage in the Central Mojave Desert. Frontiers in Microbiology8, 10.3389/fmicb.2017.01974 (2017). [DOI] [PMC free article] [PubMed]
- 30.Farr TG, et al. The Shuttle Radar Topography Mission. Rev. Geophys. 2007;45:RG2004. doi: 10.1029/2005RG000183. [DOI] [Google Scholar]
- 31.Abed RMM, Al Kharusi S, Schramm A, Robinson MD. Bacterial diversity, pigments and nitrogen fixation of biological desert crusts from the Sultanate of Oman. FEMS Microbiol Ecol. 2010;72:418–428. doi: 10.1111/j.1574-6941.2010.00854.x. [DOI] [PubMed] [Google Scholar]
- 32.FAO Guidelines for soil description http://www.fao.org/3/a-a0542e.pdf (2006).
- 33.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research. 2013;41:e1–e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.White, T. J. et al. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. Vol. 31 (1990).
- 35.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 36.Yilmaz P, et al. Minimum information about a marker gene sequence (MIMARKS) and minimum information about any (x) sequence (MIxS) specifications. Nature Biotechnology. 2011;29:415–420. doi: 10.1038/nbt.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrison PW, et al. The European Nucleotide Archive in 2018. Nucleic Acids Research. 2019;47:D84–D88. doi: 10.1093/nar/gky1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Diepenbroek, M. et al. Towards an Integrated Biodiversity and Ecological Research Data Management and Archiving Platform : The German Federation for the Curation of Biological Data (GFBio). Informatik 2014 – Big Data Komplexität meistern. GI-Edition Lect. Notes Informatics - Proc. 1711–1724 (2014).
- 39.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical computing, Vienna, Austria, https://www.R-project.org/ (2018).
- 40.Callahan BJ, et al. “DADA2: High-resolution sample inference from Illumina amplicon data.”. Nature Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.UNITE Community. UNITE general FASTA release for Fungi. Version 18.11.2018, 10.15156/BIO/786343 (2019).
- 43.Chavent, M., Kuentz-Simonet, V., Labenne, A. & Saracco, J. Multivariate analysis of mixed data: The R Package PCAmixdata arXiv preprint arXiv, https://arxiv.org/abs/1411.4911 (2017).
- 44.RStudio Team. RStudio: Integrated Development for R. RStudio, Inc., Boston, MA, http://www.rstudio.com/ (2016).
- 45.Oksanen, J. et al. Vegan: community ecology package. R package vegan, vers. 2.2-1, (2015).
- 46.Schultz M. Studies on lichens from southern Yemen (Arabian Peninsula) Lichenologist. 1998;30:293–297. doi: 10.1017/S0024282992000264. [DOI] [Google Scholar]
- 47.Galun, M. & Garty, J. In Biological soil crusts: Structure, function, and management (eds Belnap, J. & Lange, O. L.) 95–106 (Springer Berlin Heidelberg, 2003).
- 48.Liu L, et al. Development of bacterial communities in biological soil crusts along a revegetation chronosequence in the Tengger Desert, northwest China. Biogeosciences. 2017;14:3801–3814. doi: 10.5194/bg-14-3801-2017. [DOI] [Google Scholar]
- 49.Navarro-Noya YE, et al. Bacterial communities in soil under moss and lichen-moss crusts. Geomicrobiology Journal. 2014;31:152–160. doi: 10.1080/01490451.2013.820236. [DOI] [Google Scholar]
- 50.Maier S, et al. Photoautotrophic organisms control microbial abundance, diversity, and physiology in different types of biological soil crusts. Isme Journal. 2018;12:1032–1046. doi: 10.1038/s41396-018-0062-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DeFalco LA, Detling JK, Tracy CR, Warren SD. Physiological variation among native and exotic winter annual plants associated with microbiotic crusts in the Mojave Desert. Plant and Soil. 2001;234:1–14. doi: 10.1023/A:1010323001006. [DOI] [Google Scholar]
- 52.Pendleton RL, Pendleton BK, Howard GL, Warren SD. Growth and nutrient content of herbaceous seedlings associated with biological soil crusts. Arid Land Research and Management. 2003;17:271–281. doi: 10.1080/15324980301598. [DOI] [Google Scholar]
- 53.Gryndler M, et al. Hyphal growth and mycorrhiza formation by the arbuscular mycorrhizal fungus Glomus claroideum BEG 23 is stimulated by humic substances. Mycorrhiza. 2005;15:483–488. doi: 10.1007/s00572-005-0352-7. [DOI] [PubMed] [Google Scholar]
- 54.Joner EJ, Jakobsen I. Growth and extracellular phosphatase activity of arbuscular mycorrhizal hyphae as influenced by soil organic matter. Soil Biology and Biochemistry. 1995;27:1153–1159. doi: 10.1016/0038-0717(95)00047-I. [DOI] [Google Scholar]
- 55.Nagy ML, Pérez A, Garcia-Pichel F. The prokaryotic diversity of biological soil crusts in the Sonoran Desert (Organ Pipe Cactus National Monument, AZ) FEMS Microbiology Ecology. 2005;54:233–245. doi: 10.1016/j.femsec.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 56.Gundlapally SR, Garcia-Pichel F. The community and phylogenetic diversity of biological soil crusts in the Colorado Plateau studied by molecular fingerprinting and intensive cultivation. Microbial Ecology. 2006;52:345–357. doi: 10.1007/s00248-006-9011-6. [DOI] [PubMed] [Google Scholar]
- 57.Garcia-Pichel F, Lopez-Cortes A, Nubel U. Phylogenetic and morphological diversity of cyanobacteria in soil desert crusts from the Colorado Plateau. Appl Environ Microb. 2001;67:1902–1910. doi: 10.1128/Aem.67.4.1902-1910.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.De Winder, B., Stal, L. J. & Mur, L. R. Crinalium epipsammum sp. nov.: A filamentous cyanobacterium with trichomes composed of elliptical cells and containing poly-β-(1,4) glucan (cellulose). Vol. 136 (1990).
- 59.Belnap J, Gardner JS. Soil microstructure in soils of the Colorado Plateau: the role of the cyanobacterium Microcoleus vaginatus. Great Basin Naturalist. 1993;53:40–47. [Google Scholar]
- 60.Prince, R. C., Gramain, A. & McGenity, T. J. In Handbook of hydrocarbon and lipid microbiology (ed Timmis, K. N.) 1669–1692 (Springer Berlin Heidelberg, 2010).
- 61.Smith SM, Abed RMM, Garcia-Pichel F. Biological soil crusts of sand dunes in Cape Cod National Seashore, Massachusetts, USA. Microbial Ecology. 2004;48:200–208. doi: 10.1007/s00248-004-0254-9. [DOI] [PubMed] [Google Scholar]
- 62.Moquin SA, Garcia JR, Brantley SL, Takacs-Vesbach CD, Shepherd UL. Bacterial diversity of bryophyte-dominant biological soil crusts and associated mites. Journal of Arid Environments. 2012;87:110–117. doi: 10.1016/j.jaridenv.2012.05.004. [DOI] [Google Scholar]
- 63.States JS, Christensen M. Fungi Associated with Biological Soil Crusts in Desert Grasslands of Utah and Wyoming. Mycologia. 2001;93:432–439. doi: 10.2307/3761728. [DOI] [Google Scholar]
- 64.Grishkan I, Zaady E, Nevo E. Soil crust microfungi along a southward rainfall gradient in desert ecosystems. European Journal of Soil Biology. 2006;42:33–42. doi: 10.1016/j.ejsobi.2005.09.014. [DOI] [Google Scholar]
- 65.Green LE, Porras-Alfaro A, Sinsabaugh RL. Tanslocation of nitrogen and carbon intergrates biotic crust and grass production in desert grassland. Journal of Ecology. 2008;96:1076–1085. doi: 10.1111/j.1365-2745.2008.01388.x. [DOI] [Google Scholar]
- 66.Bates ST, Garcia-Pichel F. A culture-independent study of free-living fungi in biological soil crusts of the Colorado Plateau: their diversity and relative contribution to microbial biomass. Environmental Microbiology. 2009;11:56–67. doi: 10.1111/j.1462-2920.2008.01738.x. [DOI] [PubMed] [Google Scholar]
- 67.Bates ST, III, Sweat THN, Garcia-Pichel KG. F. Fungal communities of lichen-dominated biological soil crusts: diversity, relative microbial biomas and their relationship to disturbance and crust cover. Journal of Arid Environments. 2010;74:1192–1199. doi: 10.1016/j.jaridenv.2010.05.033. [DOI] [Google Scholar]
- 68.Abed RMM, Al-Sadi AM, Al-Shehi M, Al-Hinai S, Robinson MD. Diversity of free-living and lichenized fungal communities in biological soil crusts of the Sultanate of Oman and their role in improving soil properties. Soil Biology and Biochemistry. 2013;57:695–705. doi: 10.1016/j.soilbio.2012.07.023. [DOI] [Google Scholar]
- 69.Zhang B, Zhang Y, Li X, Zhang Y. Successional changes of fungal communities along the biocrust development stages. Biology and Fertility of Soils. 2018;54:285–294. doi: 10.1007/s00374-017-1259-0. [DOI] [Google Scholar]
- 70.Yao M, et al. Rate-specific responses of prokaryotic diversity and structure to nitrogen deposition in the Leymus chinensis steppe. Soil Biology and Biochemistry. 2014;79:81–90. doi: 10.1016/j.soilbio.2014.09.009. [DOI] [Google Scholar]
- 71.Maier, S., Muggia, L., Kuske, C. R. & Grube, M. In Biological soil crusts: An organizing principle in drylands (eds Weber, B., Büdel, B. & Belnap, J.) 81–100 (Springer International Publishing, 2016).
- 72.Galun, M. & Garty, J. In Biological Soil Crusts: Structure, Function, and Management (eds Belnap, J. & Lange, O.) 95–106 (Springer-Verlag, 2001).
- 73.Belnap J, Gillette DA. Vulnerability of desert biological soil crusts to wind erosion: the influences of crust development, soil texture, and disturbance. Journal of Arid Environments. 1998;39:133–142. doi: 10.1006/jare.1998.0388. [DOI] [Google Scholar]
- 74.Root HT, McCune B. Surveying for biotic soil crust lichens of shrub steppe habitats in the Columbia Basin. 2012. 2012;7:21. doi: 10.2509/naf2012.007.007. [DOI] [Google Scholar]
- 75.Chen RY, et al. The variation of morphological features and mineralogical components of biological soil crusts in the Gurbantunggut Desert of Northwestern China. Environ Geol. 2009;57:1135–1143. doi: 10.1007/s00254-008-1410-1. [DOI] [Google Scholar]
- 76.Reed SC, et al. Changes to dryland rainfall result in rapid moss mortality and altered soil fertility. Nature Climate Change. 2012;2:752–755. doi: 10.1038/nclimate1596. [DOI] [Google Scholar]
- 77.Barger, N. N., Weber, B., Garcia-Pichel, F., Zaady, E. & Belnap, J. In Biological soil crusts: An organizing principle in drylands (eds Weber, B., Büdel, B. & Belnap, J.) 257–285 (Springer International Publishing, 2016).
- 78.Sancho, L. G., Belnap, J., Colesie, C., Raggio, J. & Weber, B. In Biological soil crusts: An organizing principle in drylands (eds Weber, B., Büdel, B. & Belnap, J.) 287–304 (Springer International Publishing, 2016).
- 79.Yamada T, et al. Bellilinea caldifistulae gen. nov., sp. nov. and Longilinea arvoryzae gen. nov., sp. nov., strictly anaerobic, filamentous bacteria of the phylum Chloroflexi isolated from methanogenic propionate-degrading consortia. Int J Syst Evol Microbiol. 2007;57:2299–2306. doi: 10.1099/ijs.0.65098-0. [DOI] [PubMed] [Google Scholar]
- 80.Lacap DC, Warren-Rhodes KA, McKay CP, Pointing SB. Cyanobacteria and chloroflexi-dominated hypolithic colonization of quartz at the hyper-arid core of the Atacama Desert, Chile. Extremophiles. 2011;15:31–38. doi: 10.1007/s00792-010-0334-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abed RMM, Al-Kindi S, Al-Kharusi S. Diversity of bacterial communities along a petroleum contamination gradient in desert soils. Microbial Ecology. 2015;69:95–105. doi: 10.1007/s00248-014-0475-5. [DOI] [PubMed] [Google Scholar]
- 82.Connon, S. A., Lester, E. D., Shafaat, H. S., Obenhuber, D. C. & Ponce, A. Bacterial diversity in hyperarid Atacama Desert soils. Journal of Geophysical Research: Biogeosciences112, 10.1029/2006JG000311 (2007).
- 83.Li K, Liu R, Zhang H, Yun J. The diversity and abundance of bacteria and oxygenic phototrophs in saline biological desert crusts in Xinjiang, Northwest China. Microbial Ecology. 2013;66:40–48. doi: 10.1007/s00248-012-0164-1. [DOI] [PubMed] [Google Scholar]
- 84.Maier S, et al. Analyses of dryland biological soil crusts highlight lichens as an important regulator of microbial communities. Biodiversity and Conservation. 2014;23:1735–1755. doi: 10.1007/s10531-014-0719-1. [DOI] [Google Scholar]
- 85.Kidron GJ, Vonshak A, Dor I, Barinova S, Abeliovich A. Properties and spatial distribution of microbiotic crusts in the Negev Desert, Israel. CATENA. 2010;82:92–101. doi: 10.1016/j.catena.2010.05.006. [DOI] [Google Scholar]
- 86.Blumthaler M, Ambach W, Ellinger R. Increase in solar UV radiation with altitude. Journal of Photochemistry and Photobiology B: Biology. 1997;39:130–134. doi: 10.1016/S1011-1344(96)00018-8. [DOI] [Google Scholar]
- 87.Chamizo, S., Belnap, J., Eldridge, D. J., Cantón, Y. & Malam Issa, O. In Biological soil crusts: An organizing principle in drylands (eds Weber, B., Büdel, B. & Belnap, J.) 321–346 (Springer International Publishing, 2016).
- 88.Benlloch S, et al. Prokaryotic genetic diversity throughout the salinity gradient of a coastal solar saltern. Environmental Microbiology. 2002;4:349–360. doi: 10.1046/j.1462-2920.2002.00306.x. [DOI] [PubMed] [Google Scholar]
- 89.Casamayor EO, et al. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environmental Microbiology. 2002;4:338–348. doi: 10.1046/j.1462-2920.2002.00297.x. [DOI] [PubMed] [Google Scholar]
- 90.Jungblut AD, et al. Diversity within cyanobacterial mat communities in variable salinity meltwater ponds of McMurdo Ice Shelf, Antarctica. Environmental Microbiology. 2005;7:519–529. doi: 10.1111/j.1462-2920.2005.00717.x. [DOI] [PubMed] [Google Scholar]
- 91.Rothrock MJ, Garcia-Pichel F. Microbial diversity of benthic mats along a tidal desiccation gradient. Environmental Microbiology. 2005;7:593–601. doi: 10.1111/j.1462-2920.2005.00728.x. [DOI] [PubMed] [Google Scholar]
- 92.Des Marais, D. J. In Advances in Microbial Ecology (ed Jones, G. J.) 251-274 (Springer US, 1995).
- 93.Abed RMM, Kohls K, de Beer D. Effect of salinity changes on the bacterial diversity, photosynthesis and oxygen consumption of cyanobacterial mats from an intertidal flat of the Arabian Gulf. Environ Microbiol. 2007;9:1384–1392. doi: 10.1111/j.1462-2920.2007.01254.x. [DOI] [PubMed] [Google Scholar]
- 94.Liu H, Buskey EJ. Hypersalinity enhances the production of extracellular polymeric substance (eps) in the texas brown tide alga, aureoumbra lagunensis (PELAGOPHYCEAE) Journal of Phycology. 2000;36:71–77. doi: 10.1046/j.1529-8817.2000.99076.x. [DOI] [Google Scholar]
- 95.Abdullahi AS, Underwood GJC, Gretz MR. Extracellular matrix assembly in diatoms (Bacillariophyceae). V. Environmental effects on polysaccharide synthesis in the model diatom. Phaeodactylum Tricornutum1. Journal of Phycology. 2006;42:363–378. doi: 10.1111/j.1529-8817.2006.00193.x. [DOI] [Google Scholar]
- 96.Gunde-Cimerman, N. et al. In Halophilic Microorganisms (ed Ventosa, A.) 103–113 (Springer Berlin Heidelberg, 2004).
- 97.Cantrell SA, Baez-Félix C. Fungal molecular diversity of a Puerto Rican subtropical hypersaline microbial mat. Fungal Ecology. 2010;3:402–405. doi: 10.1016/j.funeco.2010.04.001. [DOI] [Google Scholar]
- 98.Cantrell, S. & Duval-Pérez, L. Microbial mats: an ecological niche for fungi. Frontiers in Microbiology3, 10.3389/fmicb.2012.00424 (2013). [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.