Advances in genomics and mass spectrometry have revealed several types of glycosylation systems in bacteria. However, why bacterial proteins are modified remains poorly defined. Here, we investigated the role of general N-linked glycosylation in a major food poisoning bacterium, Campylobacter jejuni. The aim of this study is to delineate the direct and indirect effects caused by disrupting this posttranslational modification. To achieve this, we employed a quantitative proteomic strategy to monitor alterations in the C. jejuni proteome. Our quantitative proteomic results linked general protein N-glycosylation to maintaining proteome stability. Functional analyses revealed novel roles for bacterial N-glycosylation in modulating multidrug efflux pump, enhancing nitrate reduction activity, and promoting host-microbe interaction. This work provides insights on the importance of general glycosylation in proteins in maintaining bacterial physiology, thus expanding our knowledge of the emergence of posttranslational modification in bacteria.
KEYWORDS: glycosylation, host-microbe interaction, microbial physiology, pathogenesis, proteomics
ABSTRACT
In eukaryotes, glycosylation plays a role in proteome stability, protein quality control, and modulating protein function; however, similar studies in bacteria are lacking. Here, we investigate the roles of general protein glycosylation systems in bacteria using the enteropathogen Campylobacter jejuni as a well-defined example. By using a quantitative proteomic strategy, we were able to monitor changes in the C. jejuni proteome when glycosylation is disrupted. We demonstrate that in C. jejuni, N-glycosylation is essential to maintain proteome stability and protein quality control. These findings guided us to investigate the role of N-glycosylation in modulating bacterial cellular activities. In glycosylation-deficient C. jejuni, the multidrug efflux pump and electron transport pathways were significantly impaired. We demonstrate that in vivo, fully glycosylation-deficient C. jejuni bacteria were unable to colonize its natural avian host. These results provide the first evidence of a link between proteome stability and complex functions via a bacterial general glycosylation system.
INTRODUCTION
Glycosylation is a prevalent protein posttranslational modification found in nature. In eukaryotes, the attachment of glycans to proteins has been shown to play a central role in modulating protein folding, stability, and signaling (1). Advances in genomics and mass spectrometry have revealed several types of glycosylation systems in bacteria. Among prokaryotes, general glycosylation systems have been reported in pathogenic bacteria, including Campylobacter jejuni (2), Neisseria gonorrhoeae (3), Burkholderia cepacia (4), as well as commensal bacteria such as Bacteroides fragilis (5), indicating their abundance across the kingdom. However, the reasons why bacterial proteins are specifically modified remains poorly defined, particularly among general protein glycosylation systems.
Genetic and biochemical studies have revealed that in C. jejuni, the N-oligosaccharyltransferase PglB is responsible for decorating at least 50 proteins in the C. jejuni cell with the heptasaccharide GalNAc-α1,4-GalNAc-[Glcβ1,3-]GalNAc-α1,4-GalNAc-α1,4-GalNAc-α1,3-diBacNAc-β1, where GalNAc is N-acetylgalactosamine, Glc is glucose, and diBacNAc is 2,4-diacetamido-2,4,6-trideoxyglucopyranose. This glycostructure is linked to the asparagine residue in the acceptor sequon D/E-X1-N-X2-S/T where X1 and X2 can be any amino acid except proline (6). Comparative genomic studies have revealed the conservation of general N-glycosylation gene clusters across the Campylobacter genus (7). Mutagenesis studies demonstrated that disruption of the N-glycosylation pathway led to pleiotropic effects in C. jejuni. These effects include a substantial decrease in bacterial adherence and invasion capability of INT 407 intestinal epithelial cells (8), a decrease in chicken colonization (9), and a reduction in competence (10). Similar findings have also been reported in other bacteria where inactivation of their corresponding general glycosylation systems resulted in a disruption in cellular physiology and a reduction in virulence (4, 5). These combined consequences were anticipated to be due to direct or indirect disruptions of several cellular processes in glycosylation-deficient strains. However, these studies did not address what perturbations may be occurring to the bacterial cell when glycosylation is no longer possible. To answer this question, we developed a platform to investigate the role of general N-linked glycosylation systems in prokaryotes, using C. jejuni as a model example.
We employed tandem mass tag (TMT) isobaric 6plex labeling quantitative proteomics on wild-type and glycosylation-deficient strains. Analysis of a glycosylation-deficient strain, revealed chaperones and stress-related proteins are more abundant, indicating a central alteration in bacterial cellular activities. Thus, contrary to current understanding, the loss of glycosylation predominantly has a mass effect on the C. jejuni proteome, rather than being restricted exclusively to proteins modified by glycosylation. Phenotypically this resulted in reduction in multidrug efflux pump activity as well as impairment in nitrate reductase activity. Furthermore, we demonstrate that general N-glycosylation plays an important role in host-microbe interactions in the chicken colonization model. This study provides deeper insights on the role of a general N-glycosylation system in bacteria, demonstrating that protein posttranslational modification is critical to a plethora of bacterial cellular activities.
RESULTS
N-glycosylation is essential for proteome stability.
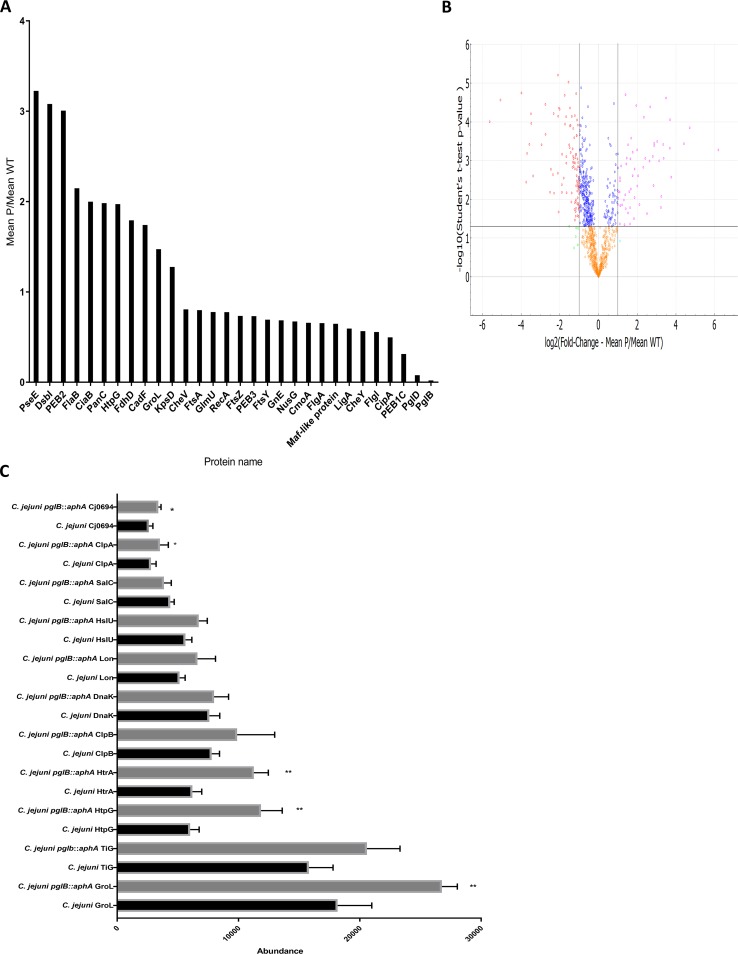
Quantitative proteomics serves as a tool to understand alterations in proteomes resulting from disruption of gene(s). To provide deeper insights into the role of the general glycosylation system in prokaryotes, we employed a mass spectrometry-based quantitative proteomic strategy to monitor differential protein abundance in glycosylation-deficient C. jejuni and compared this to wild-type bacteria. Proteins from both C. jejuni strains were reduced, alkylated, digested, and then labeled by tandem mass tag (TMT) 6plex isobaric labels to allow multiplex quantification for multiple samples. In total, 1,052 proteins out of 1,623 proteins were identified by matching peptides to the C. jejuni 11168 curated proteome. Proteins were quantitatively compared across the samples (6plex) and then filtered at 95% CI. After significance filtering, 502 proteins were found to be statistically different in the two strains; 120 proteins were more abundant and 382 proteins were less abundant in C. jejuni pglB::aphA lacking the oligosaccharyltransferase compared to the wild-type strain (Fig. 1A to C) (11, 12). Surprisingly, chaperones and proteases were found in significantly higher abundance in the glycosylation-deficient C. jejuni (Fig. 1C). Among these chaperones, we detected GroL, a periplasmic chaperone that binds to unfolded and partially folded proteins promoting folding (13), HtpG, a chaperone involved in binding to aggregated proteins and protecting the cell against environmental stresses (14), and Cj0694, a peptidyl-prolyl-cis/trans isomerase (PPIase) that interacts with the Sec translocon and promotes folding of polypeptides emerging from the translocon (15, 16). Protein degradation pathway proteins were also significantly highly abundant, notably HtrA, which is a multifunctional protein that plays a central role in proteolytically cleaving misfolded proteins as well as preventing aggregation of nonnative proteins (15, 17). This observation correlates with a significant reduction in the abundance of HspR in the same strain. HspR is a negative transcriptional regulator of chaperone-encoding genes such as clpB, dnaK, and groLS. Disruption of hspR enhanced thermotolerance in C. jejuni (18). Cytoplasmic chaperones and proteases such as ClpB, Lon, Trigger factor (Tig), and HslU that are reported to be involved in various arrays of protein quality mechanisms from assisting protein folding to protein degradation were also found to be differentially, but not statistically significantly, abundant in C. jejuni pglB::aphA (19) (Fig. 1C). These results indicate that disrupting N-linked glycosylation leads to a global alteration in the abundance of protein quality control machineries. The high abundance of chaperones and proteases suggests a potential misfolding of proteins and a rise in protein aggregation in glycosylation-deficient C. jejuni.
FIG 1.
Quantitative comparison of wild-type C. jejuni and C. jejuni pglB::aphA proteomes. (A) Selected proteins that are statistically differentially expressed in wild-type C. jejuni and C. jejuni pglB::aphA proteomes. Proteins were analyzed by Perseus and presented according to their fold change with 95% CI. (B) Volcano plot analysis of significantly abundant proteins. The −log10 (Student’s t test) is plotted against log2 mean fold change of C. jejuni pglB::aphA,P C. jejuni, WT. The nonaxial horizontal line denotes P = 0.05, while the nonaxial vertical lines denote 0.5- and 2-fold changes, respectively. The volcano plot was generated using PANDA-view (12). (C) Differential expression of chaperones and proteases in wild-type C. jejuni and C. jejuni pglB::aphA. Data are from three biological replicates. The error bars represent standard deviations. Values that are significantly different by Student’s t test are indicated by asterisks as follows: *, P < 0.05; **, P < 0.01.
Alteration in glycoprotein abundance in glycosylation-deficient C. jejuni.
Biochemical and biophysical studies of glycoproteins have revealed that N-linked glycans play a role in stabilizing proteins (20), promoting protein folding (21), and preventing protein aggregation (22). Enrichment analysis of the glycoproteome of C. jejuni has revealed 53 proteins to be decorated by the C. jejuni heptasaccharide GalNAc-α1,4-GalNAc-[Glcβ1,3-]GalNAc-α1,4-GalNAc-α1,4-GalNAc-α1,3-diBacNAc covalently linked to the asparagine residue in D/E-X1-N-X2-S/T where X1 and X2 can be any amino acid except proline (6, 23). Our quantitative proteomic approach managed to detect and correctly evaluate 35 glycoproteins in the samples. Out of the 35 glycoproteins detected, only 8 proteins (22.8%) were more abundant in glycosylation-deficient C. jejuni and 6 proteins (17.1%) were found to be unchanged. Interestingly, 21 proteins (60%) were found to be less abundant in C. jejuni pglB::aphA. It is unclear whether the low abundance of these proteins is due to misfolding/degradation of the proteins or is a consequence of global defects resulting from disruption of the N-linked glycosylation pathway. To gain more insights into the role of the C. jejuni heptasaccharide in promoting protein stability, we sought to study the functional activity of glycoproteins in C. jejuni.
N-glycosylation modulates multidrug efflux pump activity.
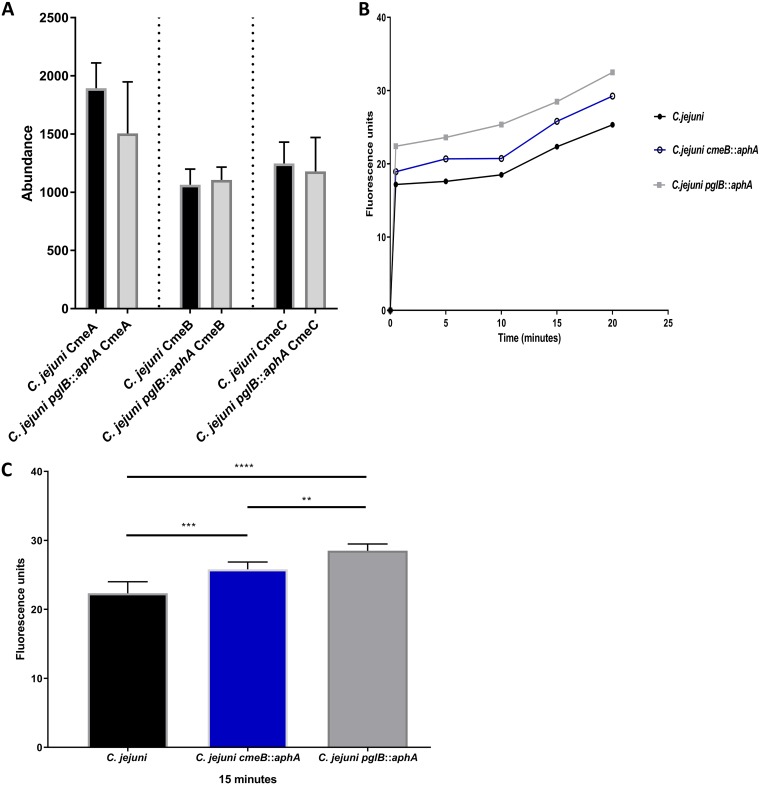
A difficulty in studying the role of general glycosylation systems in bacteria is that the modified proteins often have no known function. Using a genetically tractable bacterium that modifies proteins with predictable phenotypes could help to assign more definitively the role of glycosylation. Only two protein assemblies out of the 35 detected glycoproteins in our study on C. jejuni have known phenotypes (6, 24, 25). CmeABC shares amino acid sequence homology and similar molecular architecture with other tripartite resistance-nodulation-division (RND) type multidrug efflux pumps in other Gram-negative bacteria (26). Acting as the major multidrug efflux pump in C. jejuni, CmeABC plays an important role in antibiotic resistance and host colonization (27). The tripartite RND of C. jejuni is a molecular assembly of CmeB, an inner membrane multidrug transport protein, CmeA, a periplasmic fusion protein, and CmeC, an outer membrane-associated protein channel (24), where all three complex components were found to be glycosylated (6). Our bioinformatic analysis showed the presence of two glycosylation sequons at each protein (CmeA, CmeB, and CmeC) of the CmeABC assembly. In C. jejuni pglB::aphA, CmeA was found to be 0.8-fold less abundant, while no change was seen in the levels of CmeB and CmeC compared to the wild type (Fig. 2A). To investigate the role of the heptasaccharide in multidrug resistance, we sought to assess the antibiotic resistance profile of C. jejuni and glycosylation-deficient C. jejuni. A C. jejuni cmeB::aphA mutant was constructed and showed a substantial increase in antibiotic sensitivity. In glycosylation-deficient C. jejuni, a 100% increase in sensitivity against ampicillin, erythromycin, tetracycline, and ciprofloxacin was observed compared to the wild type (see Table S1 in the supplemental material). The results may be due to a defect in CmeABC multidrug efflux leading to an accumulation of antibiotics. In order to gain further insights on the effect of glycosylation on CmeABC activity, we used the fluorescent intercalating agent ethidium bromide in an accumulation assay that enables monitoring of efflux pump activity in real time. Ethidium bromide accumulation in C. jejuni cmeB::aphA was 15% higher (P = 0.0016) than in the wild-type strain. This result is in agreement with the antibiotic susceptibility tests. The ethidium bromide accumulation in C. jejuni pglB::aphA was 27.5% higher than in the wild type (P < 0.0001) (Fig. 2B and C). The results show that the effect of glycosylation on C. jejuni ethidium bromide accumulation kinetics is more detrimental than inactivation of the major multidrug efflux complex. Our results demonstrate the pivotal role of the heptasaccharide in efflux activity and subsequent multidrug resistance in C. jejuni.
FIG 2.
Differential expression and functional analysis of CmeABC in C. jejuni. (A) Differential expression of CmeABC in wild-type C. jejuni and C. jejuni pglB::aphA. Data are from three biological replicates, and error bars represent standard deviations. Data were analyzed by Student’s t test. (B) Ethidium bromide accumulation test in C. jejuni strains. Thirty milliliters of brucella broth was separately inoculated with overnight cultures of C. jejuni (black circles), C. jejuni cmeB::aphA (open circles), and C. jejuni pglB::aphA (gray squares) to an OD600 of 0.1. Cells were grown until an OD600 of 0.4 to 0.5 was reached, spun down, washed, and resuspended to an OD600 of 0.2 in 10 mM sodium phosphate buffer (pH 7). Cells were then incubated in a VAIN for 15 min at 37°C, and then ethidium bromide was added to a final concentration of 0.2 mg/ml. Fluorescence was read at excitation and emission for 20 min at 37°C. (B) Ethidium bromide accumulation in C. jejuni strains throughout 20 min. (C) Ethidium bromide accumulation in C. jejuni strains at 15 min. The data are means for three biological replicates, two technical replicates each, and the error bars represent standard deviations. Significance was calculated using one-way ANOVA test with multiple comparison and indicated by asterisks as follows: **, P < 0.01; ***, P < 0.001; ****, P < 0.0001.
Antimicrobial resistance of C. jejuni strains. Etest strips were used to assess minimum inhibitory concentration (MIC) of C. jejuni strains. The data represent the means from three biological replicates, with two technical replicates each. Download Table S1, TIF file, 0.1 MB (103.2KB, tif) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Nitrite production impairment in glycosylation-deficient C. jejuni.
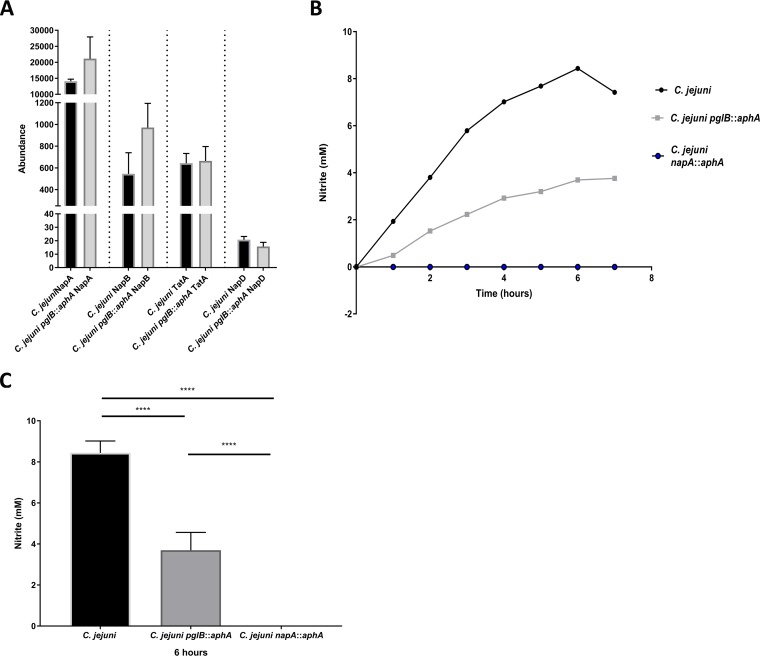
To analyze whether the loss of glycosylation affects other complex assemblies in C. jejuni, we interrogated the activity of NapAB for nitrite production. NapAB is a two-subunit periplasmic nitrate reductase enzyme that is responsible for the reduction of nitrate to nitrite. NapAB consists of a twin arginine translocation substrate; NapA is a 90-kDa catalytic bis-molybdenum guanosine dinucleoside (MGD) cofactor binding enzyme, while NapB is a 16-kDa diheme C-type cytochrome glycoprotein (6, 25). Inactivation of napA in C. jejuni results in reduction in chicken colonization (28). Mass spectrometric analysis of the C. jejuni glycoproteome demonstrated that NapB is glycosylated at 50N; however, there is no experimental evidence that NapA is a glycoprotein (6). Our quantitative proteomic analysis unexpectedly revealed a 1.7 ± 0.2-fold increase in both NapA and NapB levels in C. jejuni pglB::aphA compared to the wild type (Fig. 3A). Interestingly, there was no change in abundance in twin arginine translocase (TatA) or the NapA-accompanying chaperone NapD. To discern the role of N-glycosylation in modulating NapAB complex activity, we sought to investigate the ability to reduce nitrate in C. jejuni and C. jejuni pglB::aphA and a newly constructed C. jejuni napA::aphA mutant. The C. jejuni strains were grown in oxygen-limited conditions for 8 h. A biphasic pattern of nitrite production was observed whereby nitrite production increases until 6 h and then starts to decline (Fig. 3B). After 1 h, a difference in nitrite production between the wild type and its glycosylation-deficient counterpart was observed, while nitrite production in the C. jejuni napA::aphA mutant could not be detected (below the limit of detection). At 6 h, C. jejuni pglB::aphA produced 63.7% (P = 0.0018) less nitrite than the wild type did (Fig. 3C). Our results indicate a potential correlation between N-glycosylation and nitrate reductase activity in C. jejuni. It also suggests that despite the increase in NapAB abundance in the glycosylation-deficient strain, the activity of the NapAB complex was severely impaired compared to its glycosylated counterpart. These results indicate that N-glycosylation plays a role in enhancing NapAB complex efficiency in nitrite production.
FIG 3.
Differential expression and functional analysis of NapAB complex in C. jejuni. (A) Differential expression of NapAB, TatA, and NapD in C. jejuni and C. jejuni pglB::aphA. Data are from three biological replicates, and error bars represent standard deviations. Data were analyzed by Student’s t test. (B) Nitrate reduction in C. jejuni strains. Ninety milliliters of brucella broth supplemented with 200 mM nitrate was inoculated separately with wild-type C. jejuni (black circles), C. jejuni pglB::aphA (gray squares), and C. jejuni napA::aphA (blue circles) to an OD600 of 0.1 from an overnight culture. Cultures were incubated statically in a VAIN at 37°C. Samples were withdrawn every hour, C. jejuni cells were removed by centrifugation, and nitrate reduction was measured straight from the supernatant against nitrite standards. (B) Nitrite accumulation in the supernatant over 7 h. (C) Nitrite accumulation in the supernatant at 6 h. The data represents the mean of three biological replicates and two technical replicates, and error bars represent standard deviations. Significance was calculated using one-way ANOVA test with multiple comparison. ****, P < 0.0001.
Disruption of N-glycosylation causes alterations in the isoprenoid biosynthetic pathway and cell morphology abnormalities.
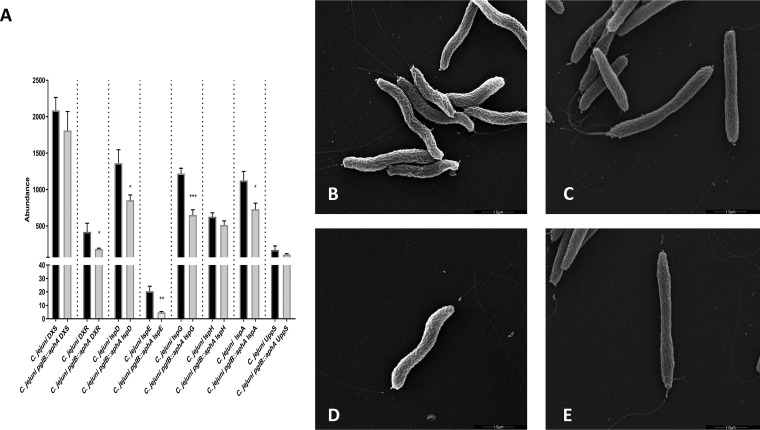
Isoprenoids are one of the largest naturally occurring organic compounds that are essential for maintaining bacterial cell physiology (29). They are synthesized from a five-carbon universal precursor, isopentyl diphosphate (IPP) or its isomer dimethylallyl diphosphate (DMAPP) (30). In C. jejuni, IPP is produced exclusively via 2C-methyl-D-erythriotol 4-phosphate (MEP) pathway that requires eight enzymatic steps. Our quantitative proteomic results show a significant reduction in MEP pathway enzymes in glycosylation-deficient C. jejuni compared to the wild-type strain (Fig. 4A). Interestingly, a 47% reduction in UppS abundance was observed in C. jejuni pglB::aphA compared to its wild-type counterpart. UppS catalyzes the sequential addition of IPP with farnesyl diphosphate to yield undecaprenyl diphosphate (Und-PP) (30). Und-P is an essential lipid carrier that plays a key role in the biosynthesis of lipooligosaccharide (LOS), capsular polysaccharide (CPS), N-linked glycan, and peptidoglycan in C. jejuni (31). The low abundance of UppS was accompanied by an alteration in the cell morphology of glycosylation-deficient C. jejuni, when examined via scanning electron microscopy (SEM).
FIG 4.
Abundance of undecaprenyl phosphate biosynthetic enzymes and cell morphology in C. jejuni strains. (A) Differential expression of MEP pathway enzymes and UppS. Data are from three biological replicates, and error bars represent standard deviations. Data were analyzed by Student’s t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B to E) Scanning electron micrographs of wild-type C. jejuni 11168H (B) and C. jejuni 11168H pglB::aphA (C) and of a single bacterial cell of wild-type C. jejuni 11168H (D) and a single bacterial cell of C. jejuni 11168H pglB::aphA (E). The bars represent 1.0 μm.
C. jejuni has a helical cell morphology that along with the polar flagella, facilitates the corkscrew motility in viscous media, giving it an advantage over other bacteria (32). It has been postulated that the helical cell shape is critical for C. jejuni pathogenicity (33). Interestingly, glycosylation-deficient C. jejuni cells were shown to adapt a rod shape and are more flattened compared to the natural helical cell morphology of wild-type C. jejuni (Fig. 4D and E). We also noted a nonuniformity among the cell length in glycosylation-deficient C. jejuni that is not evident in the wild type (Fig. 4B and C). This might be due to the disruption of N-glycosylation in C. jejuni which also led to an alteration in Und-PP production, as shown by the reduction in isoprenoid biosynthesis and UppS, hence affecting PG biosynthesis in the cell. Our results show that N-glycosylation plays an important role in maintaining cell physiology as well as cellular morphology.
Role of N-glycosylation in host-pathogen interaction.
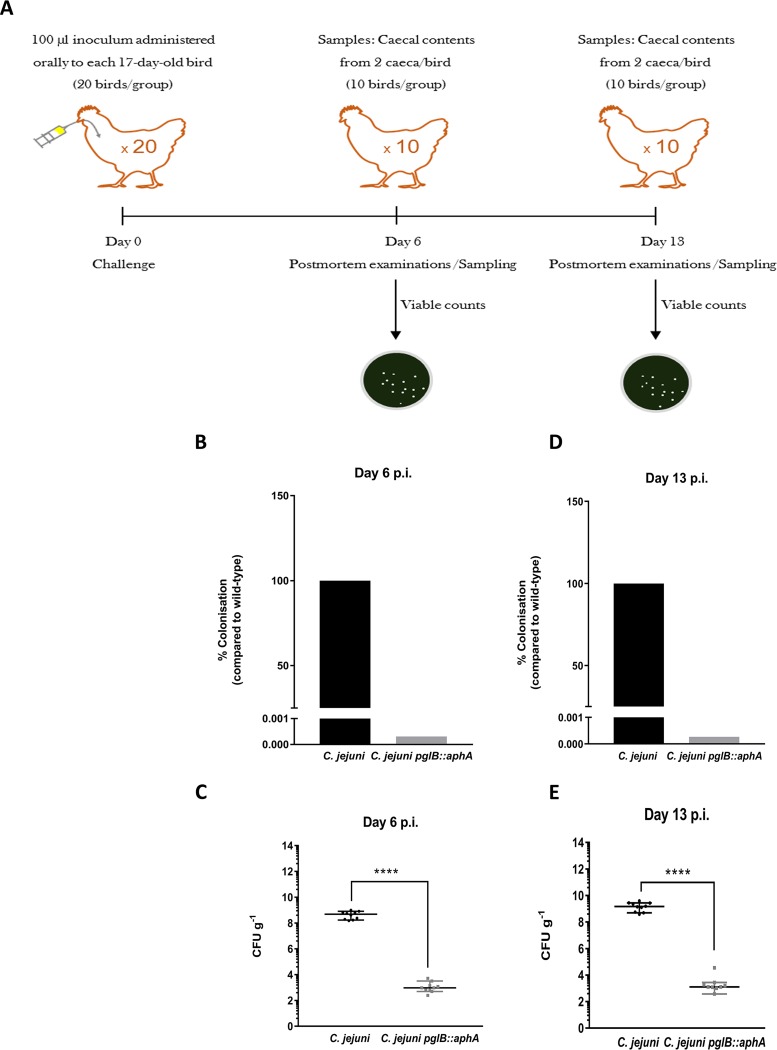
To determine the impact of protein glycosylation in host-pathogen interaction, we conducted chicken colonization experiments. Owing to its prevalence in chicken and other avian species, many reports have used chicken models of infection to test the effects of mutations in C. jejuni (9, 34). We have previously reported that disruption of pglH substantially reduced colonization in 2-week-old Light Sussex chickens (9). The glycosyltransferase PglH catalyzes the addition of three α1,4- GalNAc to GalNAc-α1,4-GalNAc-α1,3-diBacNAc-β1, an important step in the production of the heptasaccharide. Proteins from C. jejuni pglH::aphA can still be glycosylated by the truncated heptasaccharide (GalNAc-α1,4-GalNAc-α1,3-diBacNAc-β1). To understand the role of glycosylation in chicken colonization, we sought to investigate the impact of a C. jejuni pglB mutant devoid of the ability to fully glycosylate proteins and compare it to the wild type. We separately inoculated groups of twenty 2-week-old Campylobacter-free White Leghorn chickens with 106 CFU of wild-type strain of C. jejuni or the pglB::aph mutant (Fig. 5A). Ten chickens from each group were sacrificed at day 6 or 13 postinoculation, and viable C. jejuni in the ceca (a key site of persistence) were enumerated postmortem. Glycosylation-deficient C. jejuni pglB::aph showed a six-log10-unit reduction in cecal colonization compared to the wild type at day 6 (P < 0.0001) and day 13 (P < 0.0001) (Fig. 5B and C). This reduction in colonization is in agreement with our previous work on C. jejuni pglH::aphA. However, while glycan truncation in C. jejuni pglH::aphA reduced chicken colonization (9), the complete abolishment of glycosylation in C. jejuni pglB::aphA had a more severe impact on chicken colonization. The data highlight the importance of glycosylation in Campylobacter survival in the avian reservoir primarily responsible for zoonotic infections.
FIG 5.
Colonization of C. jejuni and C. jejuni pglB::aphA in 17-day-old White Leghorn chickens. (A) Schematic diagram of the chicken colonization experiment. Seventeen-day-old chicken were inoculated with 100 μl of 106 CFU of wild-type C. jejuni or C. jejuni pglB::aphA. Chickens were then sacrificed at day 6 and day 13, and postmortem examination (PM) was carried out by directly enumerating C. jejuni on CCDA plates. (B) CFU counts of C. jejuni strains on CCDA plates on day 6 postinoculation. (C) Percentage of C. jejuni strains colonization on day 6 postinoculation. (D) CFU counts of C. jejuni strains on CCDA plates on day 13 postinoculation. (E) Percentage of C. jejuni strain colonization on day 13 postinoculation. Statistical significance was calculated using Mann-Whitney test. ****, P < 0.0001.
DISCUSSION
Our results provide in-depth analyses on the alteration of the C. jejuni proteome as a consequence of disrupting the N-linked protein glycosylation pathway. Loss of glycosylation predominantly has a mass effect on the C. jejuni proteome, rather than being restricted exclusively to glyco-modified proteins. Previous studies have reported that loss of glycosylation affects bacterial motility and virulence (4, 35). However, none of these studies investigated the relative quantities of individual protein(s) and their function(s). Our TMT LC-MS/MS strategy shows that disruption of N-linked glycosylation correlates with a marked increase in the abundance of not only periplasmic but also cytoplasmic chaperones and proteases likely to be involved in folding, disaggregation, and degradation of proteins. These results suggest a critical defect in protein folding that potentially impairs protein cellular function. Differential abundance of protein quality control machineries was not translated into an obvious change in growth kinetics or cell dry weight. The cell count of the C. jejuni pglB::aphA strain at an OD600 of 0.4 to 0.5 showed a twofold reduction compared to wild-type C. jejuni; however, both strains had similar dry cell weights and growth kinetics (see Fig. S1 in the supplemental material). The reason(s) why the cell count was reduced at this OD600 in glycosylation-deficient C. jejuni strains remains unclear. One explanation may involve the observed reduction in the abundance of the cell division protein FtsZ (P = 0.006) (Fig. 1A), suggesting major cellular alterations upon abolishing N-linked glycosylation. Functional analysis demonstrates that molecular assemblies and protein functions were statistically impaired in glycosylation-deficient C. jejuni. We postulate that the difference in extrusion kinetics of ethidium bromide in C. jejuni strains were dependent on the C. jejuni heptasaccharide. While the abundance of CmeABC was similar in wild-type C. jejuni and C. jejuni pglB::aphA, we observed a significant impairment in ethidium bromide extrusion in the glycosylation-deficient strain, even when compared to the C. jejuni cmeB::aphA. This might be due to a central role of glycans in stabilizing not only CmeABC as the major multidrug efflux pump but also CmeDEF as a secondary pump (36). Indeed, CmeE, a periplasmic fusion glycoprotein in C. jejuni (6), was significantly reduced in C. jejuni pglB::aphA (P < 0.0001). The low abundance of CmeE suggests that CmeDEF is inefficiently assembled, and therefore functionally impaired. By testing other molecular assembly functions, we noticed a general role played by the C. jejuni heptasaccharide. Notably, the high abundance in the protein level of NapAB in C. jejuni pglB::aphA was not accompanied by an increase in the nitrate reduction function of the complex, indicating a severe impairment in the function of NapAB resulting from the loss of N-linked glycans. The critical effects caused by N-glycosylation disruption are extended beyond cellular activities and the assembly of protein complexes. In C. jejuni, various oligosaccharides such as LOS and CPS and N-linked glycan pathways compete for the limited pool of Und-P. Disrupting N-glycosylation in C. jejuni (and other bacteria with general glycosylation systems) may lead to a secondary effect such as sequestering of Und-P so that it will not be readily available for other pathways such as peptidoglycan biosynthesis, leading to the cell morphology alterations observed in C. jejuni pglB::aphA. In similar studies, interruption of the biosynthesis of the O antigen in Escherichiacoli led to morphological abnormalities in cell shape (37). This phenotype was rescued by overexpressing UppS to increase the pool of Und-P (37). Scrutiny of our proteomic data shows a reduction in the abundance of UppS as well as MEP pathway enzymes in glycosylation-deficient C. jejuni. This suggests either a feedback loop mechanism that prevents the accumulation of Und-P intermediates, which might result in cell toxicity, or perhaps a biophysical role played by N-linked glycans that affects cell shape.
Bacterial cell count in Campylobacter jejuni at an OD600 of 0.4 to 0.5. (A) Cell enumeration at log 106 of wild-type C. jejuni and C. jejuni pglB::aphA. (B) Dry cell weight of C. jejuni and C. jejuni pglB::aphA. Data show the means for three biological replicates with two technical replicates each, and error bars represent standard deviations. Significance was calculated using Student’s t test. ***, P < 0.0005 Download FIG S1, PDF file, 0.3 MB (350.8KB, pdf) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We performed reverse transcriptase PCR (RT-PCR) to rule out any polar effects resulting from disrupting pglB. Thus, transcription of the pglA and pglC genes downstream of pglB were similar in both wild-type and glycosylation-deficient C. jejuni (Fig. S2). Our results show a dramatic decrease in chicken colonization with C. jejuni pglB::aphA exhibiting a far greater decrease than that previously reported for C. jejuni pglH::aphA (9). This means that in contrast to the C. jejuni pglH::aphA mutant where a truncated glycan is produced, the full N-linked glycan is produced in the C. jejuni pglB::aphA strain but is not transferred to the acceptor proteins. This observation provides deeper insight into the essential role of the first two or three monosaccharides (GalNAc-α1,4-GalNAc-α1,3-diBacNAc) at the reducing end (which are absolutely conserved among strains in the Campylobacter genus) (7, 38) in host-microbe interactions. This study adds to understanding of the biological role of this general N-linked glycosylation system. We present a platform that enables the study of glycan-protein structural and functional relationship and could be applied to other prokaryotic protein glycosylation systems. Efforts to couple transcriptional and proteomic analysis to interrogate molecular changes resulting from altering or abolishing general glycosylation pathways in bacteria will enrich our understanding of the evolution and emergence of protein posttranslational modifications.
Organization of Campylobacter jejuni 11168H protein glycosylation locus. (A) Genes in the C.jejuni 11168H glycosylation locus indicated by arrows upstream and downstream of pglB. Arrow sizes are not to scale. Glycan biosynthesis genes (gray arrows), glycan translocation (light blue arrow), glycosyltransferases (dark blue arrows), oligosaccharyltransferases (black arrows), and the aphA gene from C. coli inserted to inactivate pglB in C. jejuni pglB::aphA strain (dark red rectangle) are indicated. (B) Transcription analysis of genes upstream and downstream to pglB in wild-type C. jejuni and C. jejuni pglB::aphA. Lanes 1, 5, and 9, RNA template from C. jejuni; lanes 3, 7, and 11, RNA template from C. jejuni pglB::aphA; lane 2, cDNA from C. jejuni probed by pglJ primers; lane 4, cDNA from C. jejuni pglB::aphA probed by pglJ primers; lane 6, cDNA from C. jejuni probed by pglA primers; lane 8, cDNA from C. jejuni pglB::aphA probed by pglA primers; lane 10, cDNA from C. jejuni pglB probed by pglC primers; lane 12, cDNA from C. jejuni pglB::aphA probed by pglC primers. Download FIG S2, TIF file, 0.2 MB (173.1KB, tif) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Campylobacter jejuni 11168H (39) and its derivatives, C. jejuni pglB::aphA, C. jejuni cmeB::aphA, and C. jejuni napA::aphA, were used in this study. C. jejuni 11168H was grown on Columbia-based agar or Mueller-Hinton-based agar supplemented with 5% horse blood according to the manufacturer’s instructions. Campylobacter strains were grown at 37°C in a variable atmospheric incubator (VAIN) cabinet (Don Whitley Scientific, UK) maintaining microaerophilic conditions of 85% nitrogen, 5% oxygen, and 10% carbon dioxide.
Reverse transcriptase PCR.
Overnight cultures of wild-type C. jejuni and C. jejuni pglB::aphA were diluted to an OD600 of 0.1 and then incubated in the VAIN at 37°C under shaking conditions. At an OD600 of 0.4 to 0.5, cells were harvested, and RNA was extracted by using a Monarch total RNA extraction kit (New England Biolabs) following the manufacturer’s protocol. The extracted RNA (2 μg) was treated with Ambion Turbo DNase (Invitrogen) according to the manufacturer’s instructions. cDNA was generated using Superscript III kit (Invitrogen) from DNase-treated RNA. cDNA and DNase-treated RNA were used as the templates in PCR using MyTaq mastermix (Bioline, UK) using the pglA, pglJ, and pglC oligonucleotide primers in Table 1.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotide primers used in this study
| Bacterial strain, plasmid, or primer |
Description or sequence | Source or reference |
|---|---|---|
| E. coli strains | ||
| DH10B | F−
mcrA Δ(mrr-hsdRMS-mcrBC) φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara leu)7697 araD139 galU galK nupG rpsL λ− |
New England Biolabs, UK |
| dam dcm mutant |
ara-14 leuB6 fhuA31 lacY1 tsx78 glnV44 galK2 galT22 mcrA dcm-6 hisG4 rfbD1 R(zgb210::Tn10) Tets endA1 rspL136 (Strr) dam13::Tn9 (Camr) xylA-5 mtl-1 thi-1 mcrB1 hsdR2 |
New England Biolabs, UK |
| C. jejuni strains | ||
| 11168H pglB::aphA | C. jejuni 11168H pglB is inactivated by aphA encoding kanamycin resistance cassette | This study |
| 11168H | Hypermotile variant of C. jejuni 11168 | 9 |
| 11168H cmeB::aphA | C. jejuni 11168H cmeB is inactivated by aphA encoding kanamycin resistance cassette | This study |
| 11168H napA::aphA | C. jejuni 11168H napA is inactivated by aphA encoding kanamycin resistance cassette | This study |
| Plasmids | ||
| pJMK30 | aphA gene cloned in BamHI restriction site | 41 |
| pRSA | cmeB amplicon cloned in pJET1.2 | This study |
| pRSF | aphA gene cloned in BamHI to inactivate cmeB in pRSA | This study |
| pATT3 | napA cloned in pJET1.2 | This study |
| pATT3F | aphA cloned in ClaI site inactivates napA in pATT3 | This study |
| Oligonucleotides | ||
| FWDcmeB | GACGTAATGAAGGAGAGCCA | |
| REVcmeB | CTGATCCACTCCAAGCTATG | |
| FWDnapA | ACCGCTATTGCAAGTGCTGCTAG | |
| REVnapA | GAAAGCGGACAAGTCGCATCC | |
| RTFWDPglA | ATG GGC AAA AAT TCC TTA TCG TTT | |
| RTREVPglA | GCT CGA CTA TAT CAC TTC TAG | |
| RTFWDPglC | CTC CGG TGA TTT TAA TCA CTG | |
| RTREVPglC | TTG CTT ACC CCA CTT CGT TT | |
| RTFWD PglJ | TTA GGA AGT GGT GGT GCT GA | |
| RTREV PglJ | AAA GCT ATG TCT TCA AGG GC | |
Protein preparation and digestion.
C. jejuni bacteria were grown overnight in brucella broth under microaerophilic conditions at 37°C under shaking conditions at 75 rpm. The following day, 3.5 ml was withdrawn to inoculate a 30-ml culture to a final OD600 of 0.1. The cultures were then incubated as described above until an OD600 of 0.4 to 0.5 was reached. Cells were harvested by centrifugation at 4,000 × g for 45 min. Supernatant was discarded, and cells were resuspended in lysis buffer (7 M urea, 2 M thiourea, 5 mM DTT, and 2% [wt/vol] CHAPS) and ultrasonicated following the manufacturer’s procedure for extraction of native protein samples from yeast cells (Covaris, US). Protein concentration was then measured using Bradford reagant (Sigma, UK) following the manufacturer’s protocol and then normalized to a total protein amount of 25 μg for enzymatic digestion. Prior to enzymatic digestion and labeling, samples were loaded into a stack gel for lysis buffer cleanup to eliminate any chemical interference at the labeling stage and to compress the whole proteome into a single band. Sample volumes were dried to half their original volume in a SpeedVac (Thermo Fisher Scientific, UK) with the volume replaced by Laemmli buffer (2×) and boiled for 10 min at 96°C. Reduced samples were loaded on to a 10% BisTris NuPAGE gel (Invitrogen, UK) and resolved for 10 min (100 V; 59 mA; 6 W) to “stack” the whole sample into a single band. Protein bands were visualized using Imperial protein stain (Thermo Fisher Scientific, UK). Gel bands were then excised, chopped, washed with acetonitrile (ACN), dried, and reduced by rehydration in 10 mM DTT for 30 min at 56°C. Gel slices were then washed with ACN, dried, and alkylated by incubation in 55 mM IAA at ambient temperature for 20 min in the dark, followed by washing with 100 mM TEAB, drying, and incubation in 40 μl trypsin (1.25 μg) at 37°C overnight. Peptides were extracted by collecting reaction supernatant.
Peptide TMT labeling.
Each sample was treated individually with labels added at a 1:1 ratio. TMT6plex labels (Thermo Fisher Scientific, UK) were resuspended in 41 μl ACN, vortexed, and centrifuged for 1 min at 14,000 rpm. Each vial of TMT reagent was added to the appropriate sample, vortexed, and briefly centrifuged at 14,000 rpm before incubation at room temperature for 1 h. The reaction was stopped by adding 8 μl of 5% hydroxylamine (Sigma, UK) to each sample and incubated for a further 15 min at room temperature. Samples were then combined for each TMT6plex and incubated at room temperature for 15 min, followed by freezing at –80°C and drying in a SpeedVac.
LC-MS/MS tandem mass spectrometry.
The combined TMT-labeled peptide sample was resuspended in a solution containing water-acetonitrile-trifluoroacetic acid (98%:2%:0.05%) and analyzed by LC-MS/MS. Chromatographic separations were performed using an Ultimate 3000 UHPLC system (Thermo Fisher Scientific, UK). A 3-μl injection of peptides (equivalent to 15 μg peptides) was resolved by reversed phase chromatography on a 75-μm C18 column (50 cm) using a three-step linear gradient of acetonitrile in 0.1% formic acid. The gradient was designed to elute the peptides at a flow rate of 250 nl/min over 120 min. The eluate was ionized by electrospray ionization using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific, UK) and Xcalibur v4.1 software. The instrument was programmed to acquire data in the automated data-dependent switching mode, selecting precursor ions based on their intensity for sequencing by higher-energy C-trap dissociation (HCD) for peptide identification and reporter ion fragmentation. Selection of precursor ions was based on their intensity for sequencing by HCD in a TopN method. The MS/MS analyses were conducted using higher than normal collision energy profiles that were chosen based on the mass-to-charge ratio (m/z) and the charge state of the peptide. To increase fragmented peptide coverage and reporter ion intensities, a further synchronous precursor scan (SPS) of the top five most intense peaks using MS3 was performed.
Raw mass spectrometry data were processed into peak list files using Proteome Discoverer (Thermo Scientific; v2.2). The raw data file was processed and searched using the Mascot search algorithm (v2.6.0) (Matrix Science, Boston, MA, USA) and the Sequest search algorithm against the current Campylobacter jejuni 11168 proteome curated within Uniprot. Following processing with Proteome Discoverer, the resulting file was exported into Perseus (v1.6.1) (http://www.perseus-framework.org) for qualitative and quantitative data analysis.
Construction of C. jejuni pglB::aphA, C. jejuni cmeB::aphA, and C. jejuni napA::aphA.
Construction of C. jejuni pglB::aphA was achieved by nonpolar insertion of the aphA gene into a unique restriction site present in the pglB gene of the pUC18 plasmid generated from a random genomic library used in sequencing C. jejuni 11168 (40). Briefly, pUC18 was digested using PsiI in the pglB gene, and linearized plasmid was then blunt ended using T4 DNA polymerase (Promega) according to the manufacturer’s protocol. The antibiotic resistance gene aphA was amplified and cut with StuI and blunt end ligated to the linearized plasmid. The construct was then electroporated in C. jejuni 11168H, and colonies were selected on CBA supplemented with kanamycin (30 μg/ml). Inactivation of cmeB in C. jejuni 11168H was achieved using the method described by Pumbwe and Piddock (26). Briefly, 1 kbp of cmeB was amplified with Phusion polymerase (New England Biolabs, UK) by primers FWDcmeB and REVcmeB using C. jejuni 11168H genomic DNA as the template. The amplicon was cloned into pJET1.2 to give pRSA. In order to disrupt the gene, the kanamycin resistance gene aphA was cloned in the middle of the cmeB fragment to give pRSF. Construction of C. jejuni 11168H napA::aphA was achieved by amplifying the napA gene with Phusion polymerase (New England Biolabs, UK) using primers FWDnapA and REVnapA using C. jejuni 11168H genomic DNA as the template. The amplicon was then ligated into pGEM T-easy to give pATT3, which was transformed to E. coli dam dcm mutant. To disrupt napA, aphA was cut with BamHI from pJWK30, blunt ended by CloneJET PCR cloning kit blunting enzyme (Thermo Fisher Scientific, UK), and then ligated to pATT3 that was previously cut by ClaI and blunt ended to give pATT3F. pRSF and pATT3F were electroporated in C. jejuni 11168H, and colonies were selected on CBA supplemented with kanamycin (30 μg/ml). All insertional inactivation were checked by gene-specific and aphA-specific primers.
Antibiotic sensitivity.
C. jejuni 11168H was grown in suspension in Mueller-Hinton broth equivalent to a 1.0 McFarland standard, and 100-μl aliquots were spread plated on dry Mueller-Hinton agar plates supplemented with 5% sheep blood (Oxoid, UK). The plates were left for 5 to 10 min to dry before the antibiotic strip (Oxoid, UK) was added. The plates were incubated at 37°C overnight. The MIC was read directly from the strip at the point where the zone of inhibition of bacterial growth intersected with the antibiotic concentration on the strip.
Ethidium bromide accumulation assay.
Bacterial cells were grown to mid-log phase (OD600 of 0.4 to 0.5). Cells were harvested, washed, and resuspended in 0.1 M sodium phosphate buffer (pH 7) (previously incubated in the VAIN) to an OD600 of 0.2. Cells were then incubated in the VAIN for 15 min at 37°C before a 100-μl aliquot was withdrawn to indicate time zero. Ethidium bromide (Sigma, UK) was added to a final concentration of 2 μg/ml, and fluorescence was measured at 530 nm excitation and 600 nm emission using a plate reader (Molecular Devices M3 plate reader, USA).
Determination of the nitrite concentrations in the culture supernatants.
Determining the nitrite concentrations in the culture supernatants was done according to the method of Pittman et al. (25) with slight modifications. Briefly, an overnight culture of C. jejuni grown in brucella broth was diluted to an OD600 of 0.1 in oxygen-limited condition. Samples were drawn every hour and centrifuged at 12,000 × g for 1 min. Supernatant was then diluted 1:5 with deionized water. Fifty microliters of diluted culture supernatant was mixed with 850 μl of 1% (wt/vol) sulfanilamide dissolved in 1 M HCl and 100 μl of 0.02% (wt/vol) naphthylethylenediamine. After 15 min, the absorbance at 540 nm was measured using a plate reader (Molecular Devices M3 plate reader, USA), and nitrite concentrations were determined by reference to a standard curve.
Scanning electron microscopy.
C. jejuni cells were resuspended in PBS after being scraped from an overnight culture grown on Columbia-based agar plate supplemented with blood and Skirrow supplement. The bacterial suspension was then mixed with fixing buffer (0.1 M sodium cacodylate buffer containing 4% paraformaldehyde [pH 7.4]) and fixed on polylysine-coated coverslips overnight at 4°C. Samples were then washed twice for 10 min in 0.15 M sodium cacodylate buffer (pH 7.3) and then treated for 1 h at room temperature with 1% osmium tetroxide in 0.15 M cacodylate phosphate buffer. Samples were dehydrated in an ethanol series consisting of the following steps: 10 min in 10% ethanol, 30 min in 70% ethanol, and three times in 100% ethanol (20 min each). The coverslips were then transferring to 100% ethanol in critical point drying holders and then critical point dried using carbon dioxide in a Polaron E3000 critical point dryer with >3 flushes over a 45-min soaking period. The coverslips were mounted on stubs (TAAB, UK) with conductive carbon adhesive discs (TAAB), and samples were sputter coated with gold in an Emitech K550 sputter coater (45 mA current, 2 min). The cells were viewed, and images were recorded using a FEI Quanta 200 field emission scanning electron microscope (FEI UK Ltd.) operated at 10 to 20 kV in high vacuum mode.
Chicken colonization.
All procedures were conducted under Home Office project license PPL 60/4420, according to the requirements of the Animal (Scientific Procedures) Act 1986, with the approval of the Ethical Review Committee of the Moredun Research Institute. A total of 40 White Leghorn chickens, obtained on the day of hatch from a Home Office licensed breeding establishment were used. Chickens were housed in groups of 20 in colony cages under specific-pathogen-free conditions. Groups contained female and male chickens, and the wings of the chickens were tagged for individual identification. Water and sterile irradiated feed based on vegetable protein (DBM Ltd., UK) was provided ad libitum. Fresh fecal samples collected from the cages were plated onto mCCDA plates to confirm the absence of C. jejuni in the chickens. When the chickens were 2 weeks old, they were challenged with 106 CFU of C. jejuni 11168H or 11168H pglB::aphA in a volume of 100 μl. At 1 and 2 weeks after challenge, postmortem examinations of 10 birds from each group were performed. The contents of each cecum from each bird were weighed and serially diluted in phosphate-buffered saline (PBS) and then plated on mCCDA plates to enumerate viable Campylobacter per gram of cecal contents. Statistical analysis was performed using the Mann-Whitney test in Graphpad Prism 7.
Data availability.
The raw quantitative proteomic data have been deposited to the ProteomeXchange consortium (http://www.proteomexchange.org/) via the PRIDE partner repository with identifier PXD011452.
Full list of proteins identified and compared in the wild-type C. jejuni and C. jejuni pglB::aphA strains in this study. Download Table S2, CSV file, 0.1 MB (140.2KB, csv) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of proteins found to show statistically different abundances in wild-type C. jejuni and C. jejuni pglB::aphA. Download Table S3, XLS file, 0.3 MB (276.5KB, xls) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
ACKNOWLEDGMENTS
We acknowledge Wellcome Trust grant 102978/Z/13/Z and Biotechnology & Biological Sciences Research Council grant BB/N001591/1 for funding.
We thank Steven Lynham (King’s College, London, UK) for his help with isobaric labeling mass spectrometry and Gema Vizcay-Barrena (King’s College, London, UK) for assistance with scanning electron microscopy.
S.A. designed the experiments, performed most of the experiments and data analysis, and wrote the manuscript. P.H. and S.N. assisted with the quantitative proteomics. P.V. and C.C.-U. conducted the chicken colonization experiment and data analysis. M.S., A.D., J.C., and B.W.W. revised the manuscript. B.W.W. directed the research.
We declare that we have no conflicts of interest.
ADDENDUM
While this article was in process, an article on a similar topic was published in Molecular & Cellular Proteomics: Cain JA, et al. 2019. Proteomics reveals multiple phenotypes associated with N-linked glycosylation in Campylobacter jejuni. Mol Cell Proteomics 18:715–734. https://doi.org/10.1074/mcp.ra118.001199.
Footnotes
Citation Abouelhadid S, North SJ, Hitchen P, Vohra P, Chintoan-Uta C, Stevens M, Dell A, Cuccui J, Wren BW. 2019. Quantitative analyses reveal novel roles for N-glycosylation in a major enteric bacterial pathogen. mBio 10:e00297-19. https://doi.org/10.1128/mBio.00297-19.
REFERENCES
- 1.Varki A. 2017. Biological roles of glycans. Glycobiology 27:3–49. doi: 10.1093/glycob/cww086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Linton D, Allan E, Karlyshev AV, Cronshaw AD, Wren BW. 2002. Identification of N-acetylgalactosamine-containing glycoproteins PEB3 and CgpA in Campylobacter jejuni. Mol Microbiol 43:497–508. doi: 10.1046/j.1365-2958.2002.02762.x. [DOI] [PubMed] [Google Scholar]
- 3.Vik A, Aas FE, Anonsen JH, Bilsborough S, Schneider A, Egge-Jacobsen W, Koomey M. 2009. Broad spectrum O-linked protein glycosylation in the human pathogen Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 106:4447–4452. doi: 10.1073/pnas.0809504106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lithgow KV, Scott NE, Iwashkiw JA, Thomson ELS, Foster LJ, Feldman MF, Dennis JJ. 2014. A general protein O-glycosylation system within the Burkholderia cepacia complex is involved in motility and virulence. Mol Microbiol 92:116–137. doi: 10.1111/mmi.12540. [DOI] [PubMed] [Google Scholar]
- 5.Fletcher CM, Coyne MJ, Villa OF, Chatzidaki-Livanis M, Comstock LE. 2009. A general O-glycosylation system important to the physiology of a major human intestinal symbiont. Cell 137:321–331. doi: 10.1016/j.cell.2009.02.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scott NE, Parker BL, Connolly AM, Paulech J, Edwards AVG, Crossett B, Falconer L, Kolarich D, Djordjevic SP, Højrup P, Packer NH, Larsen MR, Cordwell SJ. 2011. Simultaneous glycan-peptide characterization using hydrophilic interaction chromatography and parallel fragmentation by CID, higher energy collisional dissociation, and electron transfer dissociation MS applied to the N-linked glycoproteome of Campylobacter jejuni. Mol Cell Proteomics 10:M000031-MCP201. doi: 10.1074/mcp.M000031-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nothaft H, Scott NE, Vinogradov E, Liu X, Hu R, Beadle B, Fodor C, Miller WG, Li J, Cordwell SJ, Szymanski CM. 2012. Diversity in the protein N-glycosylation pathways within the Campylobacter genus. Mol Cell Proteomics 11:1203–1219. doi: 10.1074/mcp.M112.021519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymanski CM, Burr DH, Guerry P. 2002. Campylobacter protein glycosylation affects host cell interactions. Infect Immun 70:2242–2244. doi: 10.1128/IAI.70.4.2242-2244.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones MA, Marston KL, Woodall CA, Maskell DJ, Linton D, Karlyshev AV, Dorrell N, Wren BW, Barrow PA. 2004. Adaptation of Campylobacter jejuni NCTC11168 to high-level colonization of the avian gastrointestinal tract. Infect Immun 72:3769–3776. doi: 10.1128/IAI.72.7.3769-3776.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Larsen JC, Szymanski C, Guerry P. 2004. N-linked protein glycosylation is required for full competence in Campylobacter jejuni 81-176. J Bacteriol 186:6508–6514. doi: 10.1128/JB.186.19.6508-6514.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Binns D, Dimmer E, Huntley R, Barrell D, O’Donovan C, Apweiler R. 2009. QuickGO: a web-based tool for Gene Ontology searching. Bioinformatics 25:3045–3046. doi: 10.1093/bioinformatics/btp536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C, Xu K, Guo C, Wang J, Yan Q, Zhang J, He F, Zhu Y. 2018. PANDA-view: an easy-to-use tool for statistical analysis and visualization of quantitative proteomics data. Bioinformatics 34:3594–3596. doi: 10.1093/bioinformatics/bty408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenton WA, Horwich AL. 2003. Chaperonin-mediated protein folding: fate of substrate polypeptide. Q Rev Biophys 36:229–256. doi: 10.1017/S0033583503003883. [DOI] [PubMed] [Google Scholar]
- 14.Nakamoto H, Fujita K, Ohtaki A, Watanabe S, Narumi S, Maruyama T, Suenaga E, Misono TS, Kumar PKR, Goloubinoff P, Yoshikawa H. 2014. Physical interaction between bacterial heat shock protein (Hsp) 90 and Hsp70 chaperones mediates their cooperative action to refold denatured proteins. J Biol Chem 289:6110–6119. doi: 10.1074/jbc.M113.524801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merdanovic M, Clausen T, Kaiser M, Huber R, Ehrmann M. 2011. Protein quality control in the bacterial periplasm. Annu Rev Microbiol 65:149–168. doi: 10.1146/annurev-micro-090110-102925. [DOI] [PubMed] [Google Scholar]
- 16.Taylor AJ, Zakai SAI, Kelly DJ. 2017. The periplasmic chaperone network of Campylobacter jejuni: evidence that SalC (Cj1289) and PpiD (Cj0694) are involved in maintaining outer membrane integrity. Front Microbiol 8:531. doi: 10.3389/fmicb.2017.00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehm M, Lind J, Backert S, Tegtmeyer N. 2015. Campylobacter jejuni serine protease HtrA plays an important role in heat tolerance, oxygen resistance, host cell adhesion, invasion, and transmigration. Eur J Microbiol Immunol (Bp) 5:68–80. doi: 10.1556/EUJMI-D-15-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersen MT, Brøndsted L, Pearson BM, Mulholland F, Parker M, Pin C, Wells JM, Ingmer H. 2005. Diverse roles of HspR in Campylobacter jejuni revealed by the proteome, transcriptome and phenotypic characterization of an hspR mutant. Microbiology 151:905–915. doi: 10.1099/mic.0.27513-0. [DOI] [PubMed] [Google Scholar]
- 19.Butler SM, Festa RA, Pearce MJ, Darwin KH. 2006. Self-compartmentalized bacterial proteases and pathogenesis. Mol Microbiol 60:553–562. doi: 10.1111/j.1365-2958.2006.05128.x. [DOI] [PubMed] [Google Scholar]
- 20.Jayaprakash NG, Surolia A. 2017. Role of glycosylation in nucleating protein folding and stability. Biochem J 474:2333–2347. doi: 10.1042/BCJ20170111. [DOI] [PubMed] [Google Scholar]
- 21.Hanson SR, Culyba EK, Hsu T-L, Wong C-H, Kelly JW, Powers ET. 2009. The core trisaccharide of an N-linked glycoprotein intrinsically accelerates folding and enhances stability. Proc Natl Acad Sci U S A 106:3131–3136. doi: 10.1073/pnas.0810318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biswas H, Chattopadhyaya R. 2016. Stability of Curcuma longa rhizome lectin: role of N-linked glycosylation. Glycobiology 26:410–426. doi: 10.1093/glycob/cwv110. [DOI] [PubMed] [Google Scholar]
- 23.Linton D, Dorrell N, Hitchen PG, Amber S, Karlyshev AV, Morris HR, Dell A, Valvano MA, Aebi M, Wren BW. 2005. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol Microbiol 55:1695–1703. doi: 10.1111/j.1365-2958.2005.04519.x. [DOI] [PubMed] [Google Scholar]
- 24.Lin J, Michel LO, Zhang Q. 2002. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother 46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pittman MS, Elvers KT, Lee L, Jones MA, Poole RK, Park SF, Kelly DJ. 2007. Growth of Campylobacter jejuni on nitrate and nitrite: electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol Microbiol 63:575–590. doi: 10.1111/j.1365-2958.2006.05532.x. [DOI] [PubMed] [Google Scholar]
- 26.Pumbwe L, Piddock LJ. 2002. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol Lett 206:185–189. doi: 10.1111/j.1574-6968.2002.tb11007.x. [DOI] [PubMed] [Google Scholar]
- 27.Lin J, Sahin O, Michel LO, Zhang Q. 2003. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun 71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weingarten RA, Grimes JL, Olson JW. 2008. Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl Environ Microbiol 74:1367–1375. doi: 10.1128/AEM.02261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gabrielsen M, Rohdich F, Eisenreich W, Gräwert T, Hecht S, Bacher A, Hunter WN. 2004. Biosynthesis of isoprenoids: a bifunctional IspDF enzyme from Campylobacter jejuni. Eur J Biochem 271:3028–3035. doi: 10.1111/j.1432-1033.2004.04234.x. [DOI] [PubMed] [Google Scholar]
- 30.MacCain WJ, Kannan S, Jameel DZ, Troutman JM, Young KD. 2018. A defective undecaprenyl pyrophosphate synthase induces growth and morphological defects that are suppressed by mutations in the isoprenoid pathway of Escherichia coli. J Bacteriol 200:e00255-18. doi: 10.1128/JB.00255-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouhss A, Trunkfield AE, Bugg TDH, Mengin-Lecreulx D. 2008. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol Rev 32:208–233. doi: 10.1111/j.1574-6976.2007.00089.x. [DOI] [PubMed] [Google Scholar]
- 32.Lertsethtakarn P, Ottemann KM, Hendrixson DR. 2011. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol 65:389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frirdich E, Biboy J, Adams C, Lee J, Ellermeier J, Gielda LD, DiRita VJ, Girardin SE, Vollmer W, Gaynor EC. 2012. Peptidoglycan-modifying enzyme Pgp1 is required for helical cell shape and pathogenicity traits in Campylobacter jejuni. PLoS Pathog 8:e1002602. doi: 10.1371/journal.ppat.1002602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Vries SP, Gupta S, Baig A, Wright E, Wedley A, Jensen AN, Lora LL, Humphrey S, Skovgård H, Macleod K, Pont E, Wolanska DP, L’Heureux J, Mobegi FM, Smith DGE, Everest P, Zomer A, Williams N, Wigley P, Humphrey T, Maskell DJ, Grant AJ. 2017. Genome-wide fitness analyses of the foodborne pathogen Campylobacter jejuni in in vitro and in vivo models. Sci Rep 7:1251. doi: 10.1038/s41598-017-01133-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iwashkiw JA, Seper A, Weber BS, Scott NE, Vinogradov E, Stratilo C, Reiz B, Cordwell SJ, Whittal R, Schild S, Feldman MF. 2012. Identification of a general O-linked protein glycosylation system in Acinetobacter baumannii and its role in virulence and biofilm formation. PLoS Pathog 8:e1002758. doi: 10.1371/journal.ppat.1002758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akiba M, Lin J, Barton YW, Zhang Q. 2006. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother 57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
- 37.Jorgenson MA, Young KD. 2016. Interrupting biosynthesis of O antigen or the lipopolysaccharide core produces morphological defects in Escherichia coli by sequestering undecaprenyl phosphate. J Bacteriol 198:3070–3079. doi: 10.1128/JB.00550-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jervis AJ, Butler JA, Lawson AJ, Langdon R, Wren BW, Linton D. 2012. Characterization of the structurally diverse N-linked glycans of Campylobacter species. J Bacteriol 194:2355–2362. doi: 10.1128/JB.00042-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karlyshev AV, Linton D, Gregson NA, Wren BW. 2002. A novel paralogous gene family involved in phase-variable flagella-mediated motility in Campylobacter jejuni. Microbiology 148:473–480. doi: 10.1099/00221287-148-2-473. [DOI] [PubMed] [Google Scholar]
- 40.Parkhill J, Wren BW, Mungall K, Ketley JM, Churcher C, Basham D, Chillingworth T, Davies RM, Feltwell T, Holroyd S, Jagels K, Karlyshev AV, Moule S, Pallen MJ, Penn CW, Quail MA, Rajandream MA, Rutherford KM, van Vliet AH, Whitehead S, Barrell BG. 2000. The genome sequence of the food-borne pathogen Campylobacter jejuni reveals hypervariable sequences. Nature 403:665–668. doi: 10.1038/35001088. [DOI] [PubMed] [Google Scholar]
- 41.Van Vliet AHM, Wooldridge KG, Ketley JM. 1998. Iron-responsive gene regulation in a Campylobacter jejuni fur mutant. J Bacteriol 180:5291–5298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Antimicrobial resistance of C. jejuni strains. Etest strips were used to assess minimum inhibitory concentration (MIC) of C. jejuni strains. The data represent the means from three biological replicates, with two technical replicates each. Download Table S1, TIF file, 0.1 MB (103.2KB, tif) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Bacterial cell count in Campylobacter jejuni at an OD600 of 0.4 to 0.5. (A) Cell enumeration at log 106 of wild-type C. jejuni and C. jejuni pglB::aphA. (B) Dry cell weight of C. jejuni and C. jejuni pglB::aphA. Data show the means for three biological replicates with two technical replicates each, and error bars represent standard deviations. Significance was calculated using Student’s t test. ***, P < 0.0005 Download FIG S1, PDF file, 0.3 MB (350.8KB, pdf) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Organization of Campylobacter jejuni 11168H protein glycosylation locus. (A) Genes in the C.jejuni 11168H glycosylation locus indicated by arrows upstream and downstream of pglB. Arrow sizes are not to scale. Glycan biosynthesis genes (gray arrows), glycan translocation (light blue arrow), glycosyltransferases (dark blue arrows), oligosaccharyltransferases (black arrows), and the aphA gene from C. coli inserted to inactivate pglB in C. jejuni pglB::aphA strain (dark red rectangle) are indicated. (B) Transcription analysis of genes upstream and downstream to pglB in wild-type C. jejuni and C. jejuni pglB::aphA. Lanes 1, 5, and 9, RNA template from C. jejuni; lanes 3, 7, and 11, RNA template from C. jejuni pglB::aphA; lane 2, cDNA from C. jejuni probed by pglJ primers; lane 4, cDNA from C. jejuni pglB::aphA probed by pglJ primers; lane 6, cDNA from C. jejuni probed by pglA primers; lane 8, cDNA from C. jejuni pglB::aphA probed by pglA primers; lane 10, cDNA from C. jejuni pglB probed by pglC primers; lane 12, cDNA from C. jejuni pglB::aphA probed by pglC primers. Download FIG S2, TIF file, 0.2 MB (173.1KB, tif) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Full list of proteins identified and compared in the wild-type C. jejuni and C. jejuni pglB::aphA strains in this study. Download Table S2, CSV file, 0.1 MB (140.2KB, csv) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of proteins found to show statistically different abundances in wild-type C. jejuni and C. jejuni pglB::aphA. Download Table S3, XLS file, 0.3 MB (276.5KB, xls) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
The raw quantitative proteomic data have been deposited to the ProteomeXchange consortium (http://www.proteomexchange.org/) via the PRIDE partner repository with identifier PXD011452.
Full list of proteins identified and compared in the wild-type C. jejuni and C. jejuni pglB::aphA strains in this study. Download Table S2, CSV file, 0.1 MB (140.2KB, csv) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
List of proteins found to show statistically different abundances in wild-type C. jejuni and C. jejuni pglB::aphA. Download Table S3, XLS file, 0.3 MB (276.5KB, xls) .
Copyright © 2019 Abouelhadid et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.