The gut microbiome is an important factor in human health. It is affected by what we eat, what medicines we take, and what infections we acquire. In turn, it affects the way we absorb nutrients and whether we have excessive intestinal inflammation. Intestinal worms may have an important impact on the composition of the gut microbiome. Without a complete understanding of the impact of mass deworming programs on the microbiome, it is impossible to accurately calculate the cost-effectiveness of such public health interventions and to guard against any possible deleterious side effects. Our research examines this question in a “real-world” setting, using a longitudinal cohort, in which individuals with and without worm infections are treated with deworming medication and followed up at both three weeks and three months posttreatment. We quantify the impact of roundworms and hookworms on gut microbial composition, suggesting that the impact is small, but that treatment of hookworm infection results in significant changes. This work points to the need for follow-up studies to further examine the impact of hookworm on the gut microbiota and determine the health consequences of the observed changes.
KEYWORDS: 16S RNA, epidemiology, helminths, hookworm, microbial communities, microbial ecology, microbiota
ABSTRACT
Murine studies suggest that the presence of some species of intestinal helminths is associated with changes in host microbiota composition and diversity. However, studies in humans have produced varied conclusions, and the impact appears to vary widely depending on the helminth species present. To demonstrate how molecular approaches to the human gut microbiome can provide insights into the complex interplay among disparate organisms, DNA was extracted from cryopreserved stools collected from residents of 5 rural Kenyan villages prior to and 3 weeks and 3 months following albendazole (ALB) therapy. Samples were analyzed by quantitative PCR (qPCR) for the presence of 8 species of intestinal parasites and by MiSeq 16S rRNA gene sequencing. Based on pretreatment results, the presence of neither Ascaris lumbricoides nor Necator americanus infection significantly altered the overall diversity of the microbiota in comparison with age-matched controls. Following ALB therapy and clearance of soil-transmitted helminths (STH), there were significant increases in the proportion of the microbiota made up by Clostridiales (P = 0.0002; average fold change, 0.57) and reductions in the proportion made up by Enterobacteriales (P = 0.0004; average fold change, −0.58). There was a significant posttreatment decrease in Chao1 richness, even among individuals who were uninfected pretreatment, suggesting that antimicrobial effects must be considered in any posttreatment setting. Nevertheless, the helminth-associated changes in Clostridiales and Enterobacteriales suggest that clearance of STH, and of N. americanus in particular, alters the gut microbiota.
INTRODUCTION
Helminths and humans have coevolved for millennia. Some helminths are known to have far-reaching effects on the human immune system (1), and it is possible that some of these effects are mediated by changes in the gut microbiota (2). Changes in intestinal bacterial communities have been associated with both inflammatory diseases (3) and some helminth infections (4–6), all of which may interact (7, 8).
Studies in animal models and veterinary species provide evidence that the presence of intestinal helminths in the host can be associated with alterations in microbial communities, though the changes seen are not consistent across species of hosts and parasites (5, 6, 9–16). Changes in Bacteroidetes and Firmicutes (the two most common phyla in the human gut [17]) are frequently discussed in this body of literature. Changes (or lack thereof) in diversity vary by helminth species, but in general, the presence of helminth infections has been linked with increases in microbial diversity (18).
Some studies comparing humans with and without helminth infections have found differences in microbiota diversity and composition. One research consortium found that alpha diversity did not vary between comparison groups, but that beta diversity was higher among soil-transmitted helminths (STH)-infected individuals in Sri Lanka than in uninfected individuals (19). This group found that individuals in Italy who were infected with Strongyloides stercoralis had microbiota with higher alpha diversity and lower beta diversity than uninfected peers (20). Individuals in two Malaysian villages infected with Trichuris trichiura were found to have greater species richness and more Paraprevotellaceae than their uninfected neighbors (21). A study in a similar setting found that successful treatment of T. trichiura infections was followed by a reduction in Clostridiales and an increase in the levels of a proinflammatory Bacteroides species (8). A study in Indonesia and Liberia identified specific signatures in the microbiota that discriminated between individuals moderately/heavily infected with any STH and uninfected individuals (22). Other studies of individuals with Schistosoma haematobium and T. trichiura did not find large differences in the microbiota of individuals with and without STH or before or after anthelmintic treatment (23, 24). Recently, a placebo-controlled anthelmintic trial in Indonesia found that neither albendazole (ALB) nor helminth infection affected microbiota composition in the overall study population, but that ALB treatment was associated with lower levels of Bacteroidetes in a specific study population (25). These studies were based on naturally acquired infections (8, 21–25); experimental hookworm infections in humans were associated with an increase in microbial species richness but no significant change in microbial diversity or community structure (26, 27).
In the present study, our aim was to utilize stool samples collected longitudinally from an area where A. lumbricoides and N. americanus are endemic to examine the relationship between these STH and the human gut microbiota. Individuals in the study were infected with neither Trichuris trichiura nor Strongyloides stercoralis. Our data demonstrate that there were no obvious significant associations between STH infection and microbial diversity or richness, but successful anthelmintic treatment of N. americanus was followed by changes in microbial composition. Specifically, we found that Clostridiales increased and Enterobacteriales decreased following clearance of N. americanus infection. Depending on which metrics were used, smaller changes were also observed posttreatment in individuals who had been STH uninfected pretreatment, suggesting a direct ALB treatment effect as well.
RESULTS
16S rRNA gene sequencing results collected from individuals prior to treatment were compared with sequencing results from the same individuals 3 months posttreatment. Sequencing coverage was sufficiently higher in batch B (30% of the samples had fewer than 10,000 reads in batch A compared with 1% in batch B) to justify focusing our paired analyses on batch B alone, despite larger sample sizes being available using data from batch A. Additionally, since changes 3 months posttreatment could have been a result of helminth clearance, seasonal diet patterns, or some other time-dependent factor, samples were collected and sequenced from a subset (12 who had been infected with an STH pretreatment and 8 who had not) of participants 3 weeks following ALB treatment. At 3 weeks posttreatment, these individuals tested negative for STH infection, but the season had not yet changed. The cross-sectional component compared the stool microbiota of age-matched individuals who were infected and uninfected with STH pretreatment.
Alpha diversity and soil-transmitted helminths prior to ALB treatment.
Diversity is a common metric for the composition of the microbiota, though its significance is not definitively understood. Multiple metrics are available, and no definitive healthy range has been defined. Thus, the diversity in microbiota of uninfected individuals was compared to that of their peers who were infected with either A. lumbricoides or N. americanus, and the microbial diversity before and after treatment was compared for individuals with usable data at both time points.
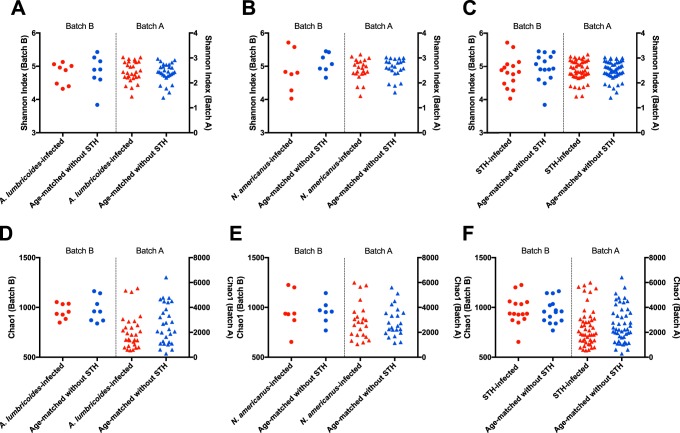
Shannon diversity index measurements for samples from STH-infected individuals were compared to samples from age-matched uninfected peers for A. lumbricoides (n = 8), N. americanus (n = 7), and for any STH (n = 15) using batch B sequencing (Fig. 1A to C, circles). There were no significant differences in the Shannon diversity index values between these comparison groups. The same comparisons are shown using data from batch A sequencing (Fig. 1, as triangles). Sample sizes are summarized in Fig. S1 in the supplemental material. No differences are seen between these comparisons using batch A data. Similarly, there were no differences in Chao1 index measurements between those who were uninfected and those infected with any STH at baseline (Fig. 1D to F), regardless of whether the analysis is based on batch A or batch B data. Many individuals who were uninfected with any STH pretreatment likely have an undocumented history of STH infection and treatment, since Kenya’s National School-Based Deworming Program began annual treatment in 2012, and treatment was less common prior to that year.
FIG 1.
Microbial diversity and richness in STH-infected individuals and age-matched uninfected individuals. (A) Shannon index comparisons for individuals infected with A. lumbricoides (red) and age-matched uninfected individuals (blue). For A-F in this figure, data from batch B are shown on the left of each graph (as circles), and data from batch A are shown on the right of each graph (as triangles). Each dot represents a single individual. (B) Shannon index comparisons for individuals infected with N. americanus (red) and age-matched individuals uninfected with any STH (blue). (C) Shannon index comparisons for individuals infected with either STH compared to STH-uninfected individuals. (D) Chao 1 comparisons for individuals infected with A. lumbricoides compared to STH-uninfected individuals. (E) Chao1 comparisons for individuals infected with N. americanus compared to STH-uninfected individuals. (F) Chao1 comparisons for individuals infected with either STH compared to STH-uninfected individuals.
Samples used in the longitudinal and cross-sectional (age-matched) sections of this study. The top row describes the total samples collected as part of the larger study, whereas information below shows the number of samples sequenced for the microbiome study described here. Download FIG S1, EPS file, 4.7 MB (4.7MB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Effect of ALB treatment on microbial diversity.
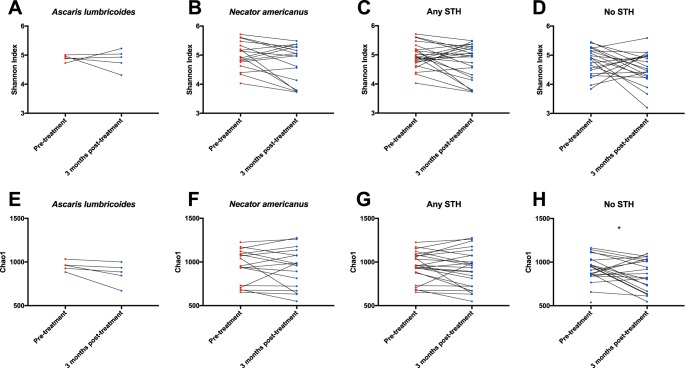
There were no significant changes in Shannon diversity index measurements 3 months following treatment in any group of individuals (Fig. 2A to D). As can be seen, the Shannon index changed slightly in many individuals who were infected with any STH pretreatment (Fig. 2C), but these changes were not significant (Wilcoxon matched-pairs signed-rank test, P = 0.12). Results based on the Chao1 richness index values were similar, except that there was a significant decrease in Chao1 at 3 months posttreatment in individuals who were uninfected at baseline (Wilcoxon test, P = 0.026), suggesting that the anthelmintic agent (ALB) by itself may alter the microbiota.
FIG 2.
Microbial diversity and richness pretreatment and 3 months posttreatment in longitudinal cohorts. Shannon diversity index measurements from individuals with results from both pretreatment and 3 months posttreatment are connected by lines in order to show changes in the diversity of their microbiota (A-D). Samples from individuals currently infected with any STH are shown in red, and samples from uninfected individuals are shown in blue. (A) Individuals infected with A. lumbricoides pretreatment, and STH-uninfected 3 months posttreatment. (B) Individuals infected with N. americanus pretreatment, and STH-uninfected 3 months posttreatment. (C) Individuals infected with either STH pretreatment, and STH-uninfected 3 months posttreatment. (D) STH-uninfected individuals pretreatment, who were treated with ALB and remained STH-uninfected 3 months posttreatment. Results are shown in panels E to H for the same individuals, using the Chao1 index to examined changes in microbial richness in these patients over time. The only significant change is a decrease in Chao1 among people uninfected with STH pretreatment (Wilcoxon matched-pairs signed rank test P value = 0.026).
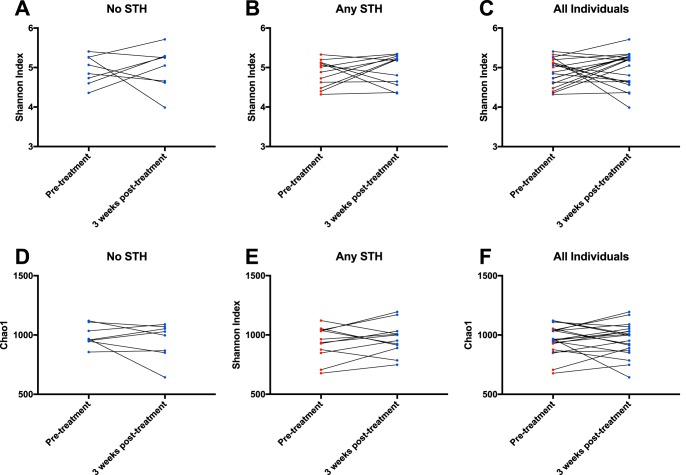
Finally, the same analyses were done comparing samples from individuals taken pretreatment and 3 weeks posttreatment. This sample size was more limited, so individuals were split into 3 groups, as follows: those with no STH infection at baseline, those with any STH infection at baseline, and all individuals with data from both time points together (to determine if there were any changes as a result of treatment with ALB). No changes were seen 3 weeks posttreatment in any of these comparison groups, whether the metric used was the Shannon index (Fig. 3A to C) or the Chao1 index (Fig. 3D to F). When the same analyses were performed as those shown in Fig. 2 and 3 but based on the limited data from samples in batch A, no changes were seen over time (Fig. S2 and S3).
FIG 3.
Microbial diversity and richness pretreatment and 3 weeks posttreatment in longitudinal cohorts. Shannon diversity index measurements from individuals with results from both pretreatment and 3 months posttreatment are connected by lines in order to show changes in the diversity of their microbiota (A to C). Samples from individuals currently infected with any STH are shown in red, and samples from uninfected individuals are shown in blue. (A) STH-uninfected individuals pretreatment, who were treated with ALB and remained STH-uninfected 3 weeks posttreatment. (B) Individuals infected with either STH pretreatment, and STH-uninfected 3 weeks posttreatment. A. lumbricoides-infected and N. americanus-infected individuals are combined due to small samples sizes available from 3 weeks posttreatment. (C) All individuals with sequencing results at both pretreatment and 3 weeks posttreatment, regardless of initial infection status. Pretreatment samples are color coded based on infection status: those infected with any STH are shown in red, whereas all others are shown in blue. Results are shown in panels D to F for the same individuals, using the Chao1 index. Wilcoxon matched-pairs signed rank test P values were insignificant for all comparisons.
Pretreatment versus 3-months-posttreatment diversity and richness comparisons based on batch A sequencing. In order to test whether the results based on batch A data diverged from those based on batch B (shown in Fig. 2), we performed the same comparisons. There is no overlap in the set of study participants in this figure and in Fig. 2. Download FIG S2, EPS file, 0.1 MB (143.8KB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Pretreatment versus 3-weeks-posttreatment diversity and richness comparisons based on batch A sequencing. In order to test whether the results based on batch A data diverged from those based on batch B (shown in Fig. 3), we performed the same comparisons. Download FIG S3, EPS file, 0.2 MB (161.5KB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Multivariate analysis of variance.
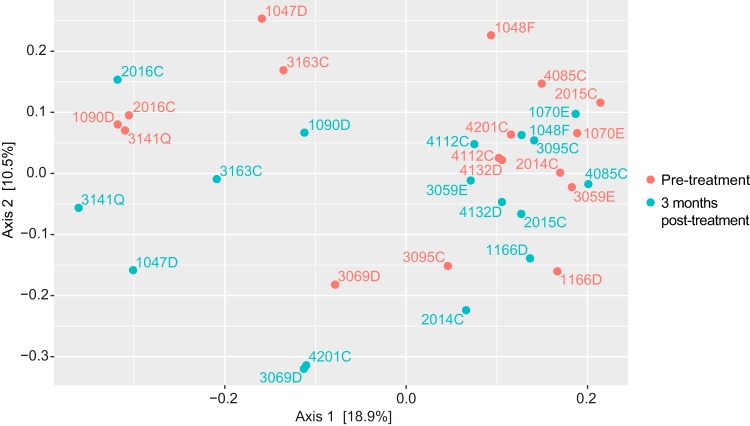
We performed principal-coordinate analysis (PCoA) to compare the overall community structures between microbiota from different individuals (an example is shown in Fig. 4). Clustering of similar microbiota within comparison groups was assessed using permutational analysis of variance. This analysis was based on unweighted UniFrac distances, though analysis based on Bray-Curtis distances is shown as well (Table 1). This analysis found that there were no major differences in microbial communities of those with and without A. lumbricoides or N. americanus at baseline, nor were there changes following treatment in any comparison group. One of these comparisons is shown in Fig. 4, where it can be seen that the pretreatment (red) samples and posttreatment (green) samples from the same people do not cluster separately. There appear to be two clusters in Fig. 4, but each cluster includes pre- and posttreatment samples, in accordance with the nonsignificant P value for separation of these two groups (P = 0.268, shown in Table 1). Many pretreatment samples are similar to the posttreatment sample from the same person (such as 2016D) whereas others are not (such as 3095C). This analysis suggests that whatever differences may be found in the microbiota of STH-infected and uninfected people, they are not markedly different in their overall composition. The most highly significant difference between comparison groups based on the Bray-Curtis distances was between individuals who were uninfected with any STH at baseline and the same individuals 3 weeks posttreatment (P = 0.001). Unweighted UniFrac distances do not consider the relative frequencies of taxa, which may explain why the differences seen in the representation of specific taxa do not translate into overall separation in terms of PCoA.
FIG 4.
Principal-coordinate analysis (PCoA) of samples from individuals infected with N. americanus pretreatment, but uninfected 3 months posttreatment. PCoA of unweighted UniFrac distances based on 16s rRNA gene sequencing results from 17 individuals infected with N. americanus pretreatment, and from the same people 3 months posttreatment. This is a visualization of one of the comparisons summarized in Table 1. Pretreatment samples are shown in red, and posttreatment samples from the same 17 people are shown in green. Samples are labeled with the study ID.
TABLE 1.
Permutational analysis of variation shows no overall separation between comparison groups
| Comparison group | n |
P value by: |
|
|---|---|---|---|
| Unweighted UniFrac | Bray-Curtis | ||
| Individuals with: | |||
| A. lumbricoides at baseline versus age-matched individuals with no STH at baseline | 8 | 0.365 | 0.453 |
| N. americanus at baseline versus age-matched individuals with no STH at baseline | 7 | 0.141 | 0.089 |
| Either STH at baseline (evenly split between A. lumbricoides and N. americanus) versus age-matched individuals with no STH at baseline | 15 | 0.066 | 0.075 |
| 3 mo posttreatment, individuals with: | |||
| A. lumbricoides at baseline | 5 | 0.984 | 0.774 |
| N. americanus at baseline | 17 | 0.268 | 0.065 |
| Either STH at baseline | 24 | 0.173 | 0.019 |
| No STH at baseline | 21 | 0.275 | 0.081 |
| 3 wk posttreatment, individuals: | |||
| With no STH at baseline | 8 | 0.063 | 0.001 |
| With either STH at baseline | 12 | 1 | 0.995 |
| In total | 20 | 0.717 | 0.037 |
Relative abundances of microbial phyla and soil-transmitted helminths.
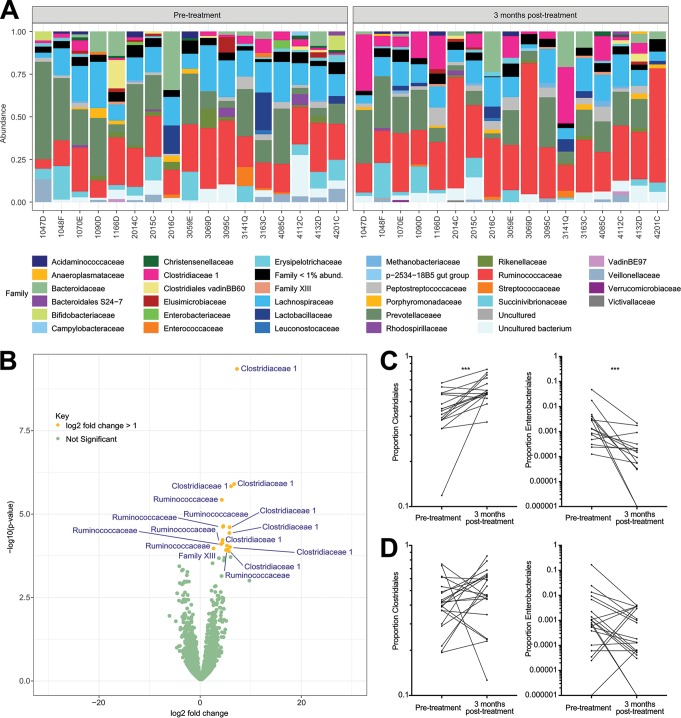
We next looked for specific taxa that were significantly more expressed in one comparison group than in another. Highly significant changes 3 months posttreatment (found at the 1% false-discovery rate [FDR]) were found in individuals with N. americanus at baseline or with any STH at baseline (which included samples with N. americanus at baseline). All changes significant at 1% FDR and 5% FDR are shown in Table 2. Clostridiales were less common at baseline in individuals infected with N. americanus than at 3 months following successful treatment, and Enterobacteriales were more common at baseline in individuals with any STH at baseline than in those same individuals posttreatment (Table 2). Similar results were found at the 5% false-discovery rate, though results were significant at a greater number of taxonomic levels (Table 2). Comparisons between the 17 individuals infected with N. americanus pretreatment and the same people 3 months posttreatment are displayed in Fig. 5. As can be seen in Fig. 5A, Clostridiaceae 1 and Ruminococcaceae appear to be expanded in the posttreatment samples (though the expansion of Ruminococcaceae at the family level is only significant at an FDR of 7%). The increase in members of these two families can also be seen clearly in a volcano plot (Fig. 5B), where several taxa in these two families have significant positive log2 fold changes greater than 1 posttreatment (Fig. 5B). The posttreatment increase in Clostridiales and decrease in Enterobacteriales among individuals infected with N. americanus pretreatment are examined in detail in Fig. 5C, showing the consistency across individuals of these changes within this group of participants. In Fig. 5D, these same comparisons are shown for individuals who were uninfected with any STH pretreatment. As expected (based on the fact that no taxa were significantly altered 3 months posttreatment in this group), no significant changes are seen, suggesting that the changes seen in other groups are not the result of seasonality.
TABLE 2.
Significant differences in the representation of family- to phylum-level taxonomic groups between comparison groups of individuals
| Measurement | Group | n | Taxa significant at FDR of: |
|
|---|---|---|---|---|
| 1% | 5% | |||
| Baseline | Individuals with A. lumbricoides at baseline | 8 | ||
| Individuals with N. americanus at baseline | 7 | |||
| Individuals with either STH at baseline (evenly split between species) | 15 | More common posttreatment, Actinobacteria/Bifidobacteriales | ||
| Longitudinal | 3 mo posttreatment | |||
| Individuals with A. lumbricoides at baseline | 5 | |||
| Individuals with N. americanus at baseline | 17 | More common posttreatment, Clostridia/Clostridiales; more common at baseline, Enterobacteriales/Enterobacteriaceae | More common posttreatment, Firmicutes/Clostridia/Clostridiales; more common at baseline, Enterobacteriales/Enterobacteriaceae | |
| Individuals with either STH at baseline | 24 | More common posttreatment, Firmicutes/Clostridia/Clostridiales; more common at baseline, Proteobacteria/Enterobacteriales/Enterobacteriaceae | More common posttreatment, Firmicutes/Clostridia/Clostridiales; more common at baseline, Proteobacteria/Gammaproteobacteria/Enterobacteriales/Enterobacteriaceae | |
| Individuals with no STH at baseline | 21 | |||
| 3 wk posttreatment | ||||
| Individuals with no STH at baseline | 8 | |||
| Individuals with either STH at baseline | 12 | |||
| All individuals | 20 | More common at baseline, Gammaproteobacteria/Aeromonadales | More common at baseline, Gammaproteobacteria/Aeromonadales | |
FIG 5.
Clostridiales and Enterobacteriales proportions at baseline versus 3 months posttreatment in individuals initially infected with N. americanus. The posttreatment changes shown here are a visualization of the longitudinal changes identified in Table 2. A) Barplots of the 17 individuals infected with N. americanus pretreatment and STH-uninfected 3 months posttreatment. Individuals are ordered by study ID both pre- and posttreatment. B) A volcano plot showing posttreatment changes in OTUs among these 17 individuals. Those OTUs shown in yellow, and labeled by the family to which they belong, have a log2 fold change greater than 1 following treatment, and are significantly different (adjusted P < 0.05) posttreatment. (C) Pairwise comparison of the proportion of the microbiota assigned to the Clostridiales and Enterobacteriales orders pre- and 3 months posttreatment, among the 17 N. americanus-infected individuals pretreatment. (D) Pairwise comparison of the proportion of the microbiota assigned to the Clostridiales and Enterobacteriales orders pretreatment and 3 months posttreatment, among the 21 STH-uninfected individuals.
At 3 weeks posttreatment, the proportion of the microbiota across all sampled individuals made up by Aeromonadales (order) had decreased. At the 5% false-discovery rate, the only difference seen in baseline comparisons was that Bifidobacteriales (Actinobacteria) were more common in individuals with an STH than in those uninfected with any STH (Table 2).
DISCUSSION
There have been a number of recent studies that have examined the relationship between the human stool microbiota and infection with a variety of STH at study sites in Indonesia (22, 25), Liberia (22), Ecuador (24), and Malaysia (8). However, there is still no clear consensus of the impact of STH on microbiota diversity and composition. This could be in part because of differences in the STH species, prevalence, and intensity of infection in these different locations, as well as differences in the laboratory and analytical methodologies employed (28–31). In the present study, we used samples collected in Kenya as part of a larger study to examine differences in the stool microbiota of individuals with and without STH infections and before and after deworming treatment. Unlike previous studies where T. trichiura was a major intestinal pathogen (8, 24), the helminth infections in this population were primarily A. lumbricoides and N. americanus. Our data largely suggest that neither the presence of N. americanus nor A. lumbricoides infection was associated with marked dysbiosis. However, it is important to note that adult N. americanus and A. lumbricoides worms reside in the small intestine, whereas adult Trichuris spp. reside in the colon. It has been seen in animal models that the microbiota of the small intestine differs from that of the large intestine and feces (32, 33), which means that any local impact of N. americanus and A. lumbricoides may be attenuated by subsequent changes that occur in the large intestine.
In contrast with published literature showing that helminths are associated with increased microbial diversity (18, 21), people in the present study all had similar levels of microbial richness (34, 35) and diversity (36), regardless of whether they were infected with N. americanus or A. lumbricoides (Fig. 1 and Table 1). Since STH are prevalent in the study setting and likely have been for generations, helminth infection may have already imposed changes on the gut microbiota of all individuals in this region. STH infection could have influenced the microbiota of currently uninfected individuals during past STH infections or through transfer of microbiota from an individual’s mother (37, 38) or close contacts who had helminth infections. Additionally, research from other groups on STH and microbial diversity has led to a mixture of results, including some where helminths are associated with decreased diversity (39) and several that found find no impact of STH on diversity (15, 23, 26, 27, 40). The clearance of helminth infections was not associated with changes in microbial diversity, except for a slight decrease in Chao1 index 3 months posttreatment. It seems unlikely that this is a biologically meaningful trend, because if that were so, there would have to be a treatment effect decreasing microbial richness that was attenuated by past infection with helminths.
Similarly, the permutational analysis of variance (PERMANOVA) and associated PCoA did not find any striking differences between any of the comparison groups based on unweighted UniFrac distances (Table 1). The studies in Ecuador, Indonesia, and Liberia all also found that there was no clear separation of samples from the STH-infected and uninfected comparison groups (22, 24, 25). In contrast, the study in Malaysia found that samples from helminth-infected indigenous Malaysians clustered separately from those from urban study participants (8).
There were, however, some significant differences seen in the differential abundances of certain taxa. The most striking differences seen in terms of microbial composition between comparison groups were between individuals with N. americanus at baseline and those same individuals at 3 months posttreatment. In particular, the proportion of the microbiota made up by Clostridiales increased and that made up by Enterobacteriales decreased in these individuals following treatment. A recent study of STH following treatment with ALB in humans (8) found that Clostridiales was less abundant after treatment. A study in Indonesia (25) did not see any changes in Clostridiales or Enterobacteria and used the level of Firmicutes in each sample as a reference. A study in Indonesia and Liberia (22) identified taxa that discriminate between STH-infected and uninfected individuals. Seven out of the 12 taxa that were positively associated with infection were within Firmicutes, and 4 of these were from Clostridiales. In an analysis of the data originally collected in Ecuador (24), 7 taxa positively associated with STH were Firmicutes. The original study had concluded that infection with T. trichiura may have no effect on fecal microbiota but that A. lumbricoides might be associated with changes (24). Several Ruminococcaceae strains were also more common posttreatment in N. americanus-infected individuals (Fig. 5B). Similarly, Ruminococcaceae has been negatively associated with Hymenolepis spp. and Syphacia spp. infection in wild mice (18), Trichuris suis in pigs (14), and Trichostrongylus retortaeformis in rabbits (41). Ruminococcaceae are highly cellulolytic, and thus, reduction during helminth infection could possibly reduce a host’s ability to digest plant material.
Any microbial population affected acutely by STH infection might be expected to return to the uninfected state following clearance of the infection. In Malaysia, the Clostridiales/Bacteroidales ratio appeared to revert following successful treatment of T. trichiura infections (8). In Indonesia and Liberia, however, posttreatment microbial communities were more similar to pretreatment microbial communities than those of uninfected individuals (22). Since there were no pretreatment differences between STH-infected and uninfected individuals in the present study (nor in one study in Indonesia [25]), there was no possibility of the microbiota reverting to an uninfected state.
The known relationship between differential abundance of microbial taxa in the intestines and parasitic infections is more extensive when nonhuman hosts and non-STH parasites are considered. Firmicutes (phylum) and Clostridiales (order) have been positively associated with Heligmosomoides polygyrus bakeri (42) and Trichuris muris (8, 43) infections in mice. However, Clostridiaceae (family) were negatively associated with Nippostrongylus brasiliensis infections in mice (44), and Firmicutes were negatively associated with Trichostrongylus retortaeformis infection in rabbits (41) and Schistosoma haematobium infection in humans (23). Enterobacteriaceae were positively associated with Heligmosomoides polygyrus infection in mice (45, 46), and the broader phylum (Proteobacteria) was positively associated with T. muris in mice (43) and T. retortaeformis in rabbits (41). These and other differences associated with helminth infections are summarized in references 5 and 6.
One of the strengths of this study is that participants were examined at 2 time points posttreatment. The 3-week time point allowed us to look at the acute impact of ALB treatment on the microbiota, before seasonal changes in diet and environment could impact the microbiota of participants. The 3-month time point allowed us to examine the longer-term impact of helminth clearance. The uninfected comparison group, which was also evaluated pretreatment and 3 months posttreatment, allowed us to identify changes that were likely due to seasonality or to ALB. The study in Indonesia (25) accomplished this by having a control group that was uninfected with STH and not treated with ALB. In our study, as in a study by Ramanan et al. (8), treatment was implemented as it would be in a mass deworming program, meaning that there was no untreated group. At our 3-weeks-posttreatment time point, there were no changes in diversity or by PCoA (except in individuals who were uninfected at baseline, according to Bray-Curtis distances). This result is not likely to be very meaningful, unless this is a change that was prevented by some kind of stabilizing effect of recent STH infection and thus not seen in STH-infected individuals posttreatment. There were no significant changes in the frequencies of particular microbial taxa at 3 weeks posttreatment, except that Aeromonadales (Gammaproteobacteria) was less common at the 3-week time point. This could mean that Aeromonadales are transiently reduced by ALB treatment. One study did not detect a direct ALB treatment effect on the gut microbiota (47), whereas another found differences in the gut microbiota between individuals given ALB versus a placebo, but only among individuals who remained helminth-infected posttreatment (25). A study of ALB treatment of pregnant women suggested a direct drug effect on the risk of allergy in infancy (48). In conjunction with prior work, our study suggests that the direct effects of ALB treatment should be considered in the design of future studies. They should also be further examined to determine whether changes mediated by ALB have any effect on a host’s susceptibility to opportunistic pathogens. Potential direct drug effects on the microbiome, in combination with concern that frequent single-drug therapy could select for resistance (49–51) in helminths and future improvements in low-cost diagnostic techniques, may influence the debate over when mass deworming is preferable to test-and-treat strategies (52).
This study contributes to the ongoing discussion over whether there are measurable impacts of STH infection on the human stool microbiota. In particular, we are able to identify changes in the microbiota associated with clearance of N. americanus infection, which are not seen posttreatment in individuals who were uninfected pretreatment. By sampling at two different time points posttreatment, we are able to identify some changes in microbial community structure that may be transient and possibly are the result of treatment with ALB or of worm expulsion, as well as other changes that are probably longer-term consequences of the microbiota readjusting to the lack of helminth presence. We welcome the purpose-built helminth/microbiota studies that are under way and believe that microbiota sequencing from samples collected as part of deworming program monitoring and evaluation studies will contribute to an improved understanding of which changes are consistent across geographies for each STH species. We also welcome efforts to standardize the methodology for comparing microbiota sequencing between groups and suggest that further research is needed to understand whether stool microbiota sequencing is sufficiently representative of the human gut microbiota in the small intestine (in which N. americanus and A. lumbricoides reside). This study contributes to our understanding of how microbial communities differ between STH-infected and uninfected individuals; the next step will be to understand the impact of the identified differences on human health.
MATERIALS AND METHODS
Sample collection and study design.
Stool samples were collected from 5 villages in rural western Kenya, as described elsewhere (53). Stools were collected and frozen (−20°C) within 10 h of production. During the delay between production in the morning, time in the field lab where parasitological examination and aliquoting took place, and transport to the hospital where they were stored, samples were kept in cool shady areas as much as possible. Baseline (pretreatment) stool collection occurred in January through March 2014. All study participants (both STH infected and uninfected) were subsequently treated under observation with 400 mg ALB. Stool samples were collected again 3 weeks following treatment from all STH-infected individuals and from a subset of STH-uninfected individuals. All posttreatment samples sequenced for this study were STH negative by quantitative PCR (qPCR). Three months following treatment, stool samples were collected from the entire study population again prior to everyone being treated again with ALB. Samples were shipped on dry ice to the NIH. DNA was extracted from cryopreserved stools and examined by qPCR for 8 intestinal parasites, as previously described (53).
Study populations.
This study was nested within a broader (previously described) study of 796 people (53). Samples from individuals infected with STH pretreatment were compared with samples from the same individuals posttreatment. Seventeen individuals were infected with N. americanus pretreatment and uninfected 3 months posttreatment (age 3 to 84 years; median age, 51 years). Five A. lumbricoides were infected with A. lumbricoides pretreatment but uninfected at 3 months posttreatment (age 3 to 12 years; median age, 7 years). A combined comparison group was constructed to include these 22 individuals, plus two A. lumbricoides/N. americanus-coinfected individuals (age 3 to 84 years; median age, 27.5 years). In order to examine acute changes posttreatment, comparisons were made between pretreatment and 3-weeks-posttreatment samples. Pretreatment samples from individuals with any STH (including 7 with A. lumbricoides, 4 with N. americanus, and one with both; age 3 to 75 years; median age, 11 years), or no STH infection (n = 8; age 6 to 13 years; median age, 9 years) were compared with paired samples from the same individuals at 3 weeks posttreatment.
Additionally, pretreatment age-matched comparison groups were constructed to compare individuals with and without STH. Individuals infected with N. americanus were 3 to 71 years of age (median age, 11 years; n = 7), and individuals with A. lumbricoides were 2 to 15 years of age (median age, 9.5 years; n = 8). A combined comparison group was created from 7 individuals with N. americanus, 7 individuals with A. lumbricoides, and one coinfected individual. This group included individuals 3 to 71 years of age (median age, 10 years). These samples are summarized in Fig. S1. Infection with Giardia lamblia was not controlled for, as most individuals had a transient asymptomatic G. lamblia infection at one point in this study. We were unable to control for recent antibiotic and antiparasitic use, as we determined during the pilot phase of this study that the majority of participants were unsure whether they had taken any medicines in these categories. Additional individual-level metadata are available for each sample at https://www.ncbi.nlm.nih.gov/bioproject/510835.
16S rRNA gene sequencing.
16S rRNA gene sequencing was performed in 2 broad batches, referred to here as batch A and batch B. Methodological differences and batch effects limited our analysis to comparisons of individuals within the same batch. Technical limitations in batch A prevented the acquisition of a broad distribution of read coverage, and a majority of samples exhibited low read counts, necessitating rarefaction at a low depth. Furthermore, batch A was split across 3 different sequencing runs, and batch effects limited the number of possible comparisons. This spurred a single resequencing run, focusing on sequencing longitudinal samples within the same batch, which had improved breadth of coverage (batch B). For this paper, only data from batch B are used for longitudinal comparisons; however, pretreatment comparisons of microbial richness and diversity between individuals with and without STH infections are shown using both batch A and batch B data. In these comparisons, 23 individuals with and without N. americanus were compared, 29 individuals with and without A. lumbricoides were compared, and 48 individuals with and without any STH (evenly split between N. americanus and A. lumbricoides) were compared using these batch A data. These results are shown alongside the comparable samples from batch B, described in the previous paragraph. There was no overlap in the set of individuals compared using batch A and batch B data sets.
For 16S rRNA amplicon Illumina MiSeq sequencing, the DNA from each sample was amplified using the AccuPrime high-fidelity Taq polymerase (Invitrogen Life Technologies) with universal primers flanking the V4 hypervariable region of the 16S rRNA gene (primers 515F, GTGYCAGCMGCCGCGGTAA, and 806R, GGACTACNVGGGTWTCTAAT). For each sample, the universal primers were tagged with unique sequence barcodes to allow for multiplexing/demultiplexing. PCR products were then purified using the Agencourt AMPure XP kit (Beckman Counter Genomics) and quantitated using the Quant-iT double-stranded DNA (dsDNA) high-sensitivity assay kit (Invitrogen Life Technologies). Approximately equivalent amounts of each PCR product were then pooled before sequencing on an Illumina MiSeq instrument.
Sequence processing and analysis.
Samples were demultiplexed using BaseSpace (Illumina). Further read processing was done using the QIIME 1.9 pipeline available through Nephele (28). The preprocessing steps included read pair joining using default parameters (perc_max_diff = 25, min_overlap = 10), removal of reads with average Phred score of ≤20, and removal of chimeras using vsearch (https://github.com/torognes/vsearch). Taxonomy assignments were done against SILVA version 128, with a default sequence similarity threshold of 0.97.
Statistical analysis.
Microbial alpha diversity within each sample was calculated using the Shannon diversity index as implemented in QIIME (54, 55). Species richness was calculated using the Chao1 index (36). Chao1 considers rare sequence variants but not the abundance of each sequence, whereas the Shannon index also considers the abundance. Between-group and before/after comparisons were done using Wilcoxon matched-pairs signed rank tests (Prism; GraphPad Software, San Diego, CA, USA).
Differences in community structure were examined broadly by permutational multivariate analysis using unweighted UniFrac distances (‘Adonis’ function within the ‘vegan’ R package). Specifically, we examined whether there were greater differences between comparison groups than within comparison groups. Unweighted UniFrac distances were chosen because they incorporate the relatedness of different taxa and are frequently used in microbial ecology. Bray-Curtis distances were also calculated and analyzed but were not the primary focus here.
Finally, we delved into specific differences in the abundances of different types of bacteria within the microbiota of individuals studied here. We performed Wilcoxon exact paired tests (in R) comparing the representation of operational taxonomic units (OTUs). This was done at different levels from phylum to genus. All phylogenetic levels were inspected separately in order to identify trends occurring at any level. The Benjamini-Hochberg false-discovery rate calculation was used to correct for multiple comparisons. False-discovery rates of 1 and 5% were considered. Further exploratory analysis was performed using the negative binomial distribution method in DESeq2 to identify OTUs that made up a significantly different proportion of the microbiota in one comparison group versus another.
Ethics approval.
This study was approved by the ethics review committee of the Kenya Medical Research Institute (Scientific Steering Committee protocol number 2688) and the Imperial College Research Ethics Committee (ICREC_ 13_1_15). Informed written consent was obtained from all adults and parents or guardians of each child. Minor assent was obtained from all children age 12 to 17 years. Anyone found to be infected with any STH was treated with 400 mg ALB during each phase of the study, and all previously untreated village residents were offered ALB at the end of each study phase.
Consent for publication.
Individuals consented to the publication of their results, without any patient identifying information.
Availability of materials.
All data sets and scripts used during the current study are available from the corresponding author on reasonable request.
Data availability.
The data sets supporting the conclusions of this article are available in the Sequence Read Archive repository (accession number PRJNA510835; https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP173959).
Clostridiales and Enterobacteriales proportional representation relative to N. americanus egg burden. (A) Clostridiales percent makeup of the microbiome versus N. americanus DNA as measured by qPCR at baseline (r = 0.14, P = 0.54). (B) Enterobacteriales percent makeup of the microbiome versus N. americanus DNA as measured by qPCR at baseline (r = 0.43, P = 0.04). (C) Percent change in proportional representation of Clostridiales posttreatment relative to the N. americanus infection burden in those people (r = 0.20, P = 0.43). (D) Percent change in proportional representation of Enterobacteriales posttreatment relative to the N. americanus infection burden in those people (r = −0.30, P = 0.23). Download FIG S4, EPS file, 0.2 MB (181.1KB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
ACKNOWLEDGMENTS
We thank the school children, schoolteachers, and Bungoma administrators for their support. We would like to extend special thanks to all the members of the study team: Bungoma County Hospital, Siangwe, Siaka, Sang’alo, Nasimbo, and Ranje village administrators and Community Health Workers. Particular thanks go to Charles S. Mwandawiro, Sammy Njenga, and Jimmy H. Kihara (KEMRI) and Simon J. Brooker (BMGF) for making the fieldwork possible in Kenya, and for their invaluable scientific and logistical advice. We are grateful to Joseph Kubofcik, Elise O’Connell, and Sasisekhar Bennuru for assistance in the laboratory.
This work was supported in part by the Division of Intramural Research (DIR) of the National Institute of Allergy and Infectious Diseases, NIH. The 16S rRNA gene sequencing was supported by the NIAID Microbiome Program within the National Institute of Allergy and Infectious Diseases. Fieldwork was supported by a grant from the Bill and Melinda Gates Foundation to the London Centre for Neglected Tropical Disease Research. A.V.E. was supported by a Ph.D. training fellowship from the Marshall Commission (Foreign and Commonwealth Office, UK) and the NIH Oxford-Cambridge Scholars Program.
A.V.E., Y.B., and T.B.N. designed the study. A.V.E., R.G.O., S.K., and M.R.O. designed and facilitated data collection and logistics in Kenya. A.V.E., M.Q., and I.V.-C. undertook the statistical analyses. A.V.E. wrote the manuscript. A.V.E. and R.M.A. designed the larger study from which these samples were drawn. All authors read, edited, and approved the final manuscript.
We declare no competing interests. R.M.A. was a Non-Executive Director of GlaxoSmithKline (GSK). GSK played no role in the funding of this research or this publication. The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Citation Easton AV, Quiñones M, Vujkovic-Cvijin I, Oliveira RG, Kepha S, Odiere MR, Anderson RM, Belkaid Y, Nutman TB. 2019. The impact of anthelmintic treatment on human gut microbiota based on cross-sectional and pre- and postdeworming comparisons in western Kenya. mBio 10:e00519-19. https://doi.org/10.1128/mBio.00519-19.
Contributor Information
Martin J. Blaser, New York University.
Philip Cooper, St George's University of London.
Rojelio Mejia, Baylor College of Medicine.
REFERENCES
- 1.Zaiss MM, Harris NL. 2016. Interactions between the intestinal microbiome and helminth parasites. Parasite Immunol 38:5–11. doi: 10.1111/pim.12274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung JM, Loke P. 2013. A role for IL-22 in the relationship between intestinal helminths, gut microbiota and mucosal immunity. Int J Parasitol 43:253–257. doi: 10.1016/j.ijpara.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strober W. 2013. Impact of the gut microbiome on mucosal inflammation. Trends Immunol 34:423–430. doi: 10.1016/j.it.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glendinning L, Nausch N, Free A, Taylor DW, Mutapi F. 2014. The microbiota and helminths: sharing the same niche in the human host. Parasitology 141:1255–1271. doi: 10.1017/S0031182014000699. [DOI] [PubMed] [Google Scholar]
- 5.Bhattacharjee S, Kalbfuss N, Prazeres da Costa C. 2017. Parasites, microbiota and metabolic disease. Parasite Immunol 39:e12390. doi: 10.1111/pim.12390. [DOI] [PubMed] [Google Scholar]
- 6.Peachey LE, Jenkins TP, Cantacessi C. 2017. This gut ain’t big enough for both of us. Or is it? Helminth-microbiota interactions in veterinary species. Trends Parasitol 33:619–632. doi: 10.1016/j.pt.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Mishra PK, Palma M, Bleich D, Loke P, Gause WC. 2014. Systemic impact of intestinal helminth infections. Mucosal Immunol 7:753–762. doi: 10.1038/mi.2014.23. [DOI] [PubMed] [Google Scholar]
- 8.Ramanan D, Bowcutt R, Lee SC, Tang MS, Kurtz ZD, Ding Y, Honda K, Gause WC, Blaser MJ, Bonneau RA, Lim YAL, Loke P, Cadwell K. 2016. Helminth infection promotes colonization resistance via type 2 immunity. Science 352:608–612. doi: 10.1126/science.aaf3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plieskatt JL, Deenonpoe R, Mulvenna JP, Krause L, Sripa B, Bethony JM, Brindley PJ. 2013. Infection with the carcinogenic liver fluke Opisthorchis viverrini modifies intestinal and biliary microbiome. FASEB J 27:4572–4584. doi: 10.1096/fj.13-232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walk ST, Blum AM, Ewing S-S, Weinstock JV, Young VB. 2010. Alteration of the murine gut microbiota during infection with the parasitic helminth Heligmosomoides polygyrus. Inflamm Bowel Dis 16:1841–1849. doi: 10.1002/ibd.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williamson LL, McKenney EA, Holzknecht ZE, Belliveau C, Rawls JF, Poulton S, Parker W, Bilbo SD. 2016. Got worms? Perinatal exposure to helminths prevents persistent immune sensitization and cognitive dysfunction induced by early-life infection. Brain Behav Immun 51:14–28. doi: 10.1016/j.bbi.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 12.McKenney EA, Williamson L, Yoder AD, Rawls JF, Bilbo SD, Parker W. 2015. Alteration of the rat cecal microbiome during colonization with the helminth Hymenolepis diminuta. Gut Microbes 6:182–193. doi: 10.1080/19490976.2015.1047128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broadhurst MJ, Ardeshir A, Kanwar B, Mirpuri J, Gundra UM, Leung JM, Wiens KE, Vujkovic-Cvijin I, Kim CC, Yarovinsky F, Lerche NW, McCune JM, Loke P. 2012. Therapeutic helminth infection of macaques with idiopathic chronic diarrhea alters the inflammatory signature and mucosal microbiota of the colon. PLoS Pathog 8:e1003000. doi: 10.1371/journal.ppat.1003000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li RW, Wu S, Li W, Navarro K, Couch RD, Hill D, Urban JF Jr.. 2012. Alterations in the porcine colon microbiota induced by the gastrointestinal nematode Trichuris suis. Infect Immun 80:2150–2157. doi: 10.1128/IAI.00141-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li RW, Li W, Sun J, Yu P, Baldwin RL, Urban JF. 2016. The effect of helminth infection on the microbial composition and structure of the caprine abomasal microbiome. Sci Rep 6:20606. doi: 10.1038/srep20606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu S, Li RW, Li W, Beshah E, Dawson HD, Urban JF. 2012. Worm burden-dependent disruption of the porcine colon microbiota by Trichuris suis infection. PLoS One 7:e35470. doi: 10.1371/journal.pone.0035470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marchesi JR. 2010. Prokaryotic and eukaryotic diversity of the human gut. Adv Appl Microbiol 72:43–62. doi: 10.1016/S0065-2164(10)72002-5. [DOI] [PubMed] [Google Scholar]
- 18.Kreisinger J, Bastien G, Hauffe HC, Marchesi J, Perkins SE. 2015. Interactions between multiple helminths and the gut microbiota in wild rodents. Philos Trans R Soc Lond B Biol Sci 370:20140295. doi: 10.1098/rstb.2014.0295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jenkins TP, Rathnayaka Y, Perera PK, Peachey LE, Nolan MJ, Krause L, Rajakaruna RS, Cantacessi C. 2017. Infections by human gastrointestinal helminths are associated with changes in faecal microbiota diversity and composition. PLoS One 12:e0184719. doi: 10.1371/journal.pone.0184719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenkins TP, Formenti F, Castro C, Piubelli C, Perandin F, Buonfrate D, Otranto D, Griffin JL, Krause L, Bisoffi Z, Cantacessi C. 2018. A comprehensive analysis of the faecal microbiome and metabolome of Strongyloides stercoralis infected volunteers from a non-endemic area. Sci Rep 8:15651. doi: 10.1038/s41598-018-33937-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee SC, Tang MS, Lim YAL, Choy SH, Kurtz ZD, Cox LM, Gundra UM, Cho I, Bonneau R, Blaser MJ, Chua KH, Loke P. 2014. Helminth colonization is associated with increased diversity of the gut microbiota. PLoS Negl Trop Dis 8:e2880. doi: 10.1371/journal.pntd.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosa BA, Supali T, Gankpala L, Djuardi Y, Sartono E, Zhou Y, Fischer K, Martin J, Tyagi R, Bolay FK, Fischer PU, Yazdanbakhsh M, Mitreva M. 2018. Differential human gut microbiome assemblages during soil-transmitted helminth infections in Indonesia and Liberia. Microbiome 6:33. doi: 10.1186/s40168-018-0416-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kay GL, Millard A, Sergeant MJ, Midzi N, Gwisai R, Mduluza T, Ivens A, Nausch N, Mutapi F, Pallen M. 2015. Differences in the faecal microbiome in Schistosoma haematobium infected children vs. uninfected children. PLoS Negl Trop Dis 9:e0003861. doi: 10.1371/journal.pntd.0003861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper P, Walker AW, Reyes J, Chico M, Salter SJ, Vaca M, Parkhill J. 2013. Patent human infections with the whipworm, Trichuris trichiura, are not associated with alterations in the faecal microbiota. PLoS One 8:e76573. doi: 10.1371/journal.pone.0076573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin I, Djuardi Y, Sartono E, Rosa BA, Supali T, Mitreva M, Houwing-Duistermaat JJ, Yazdanbakhsh M. 2018. Dynamic changes in human-gut microbiome in relation to a placebo-controlled anthelminthic trial in Indonesia. PLoS Negl Trop Dis 12:e0006620. doi: 10.1371/journal.pntd.0006620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Giacomin P, Zakrzewski M, Croese J, Su X, Sotillo J, McCann L, Navarro S, Mitreva M, Krause L, Loukas A, Cantacessi C. 2015. Experimental hookworm infection and escalating gluten challenges are associated with increased microbial richness in celiac subjects. Sci Rep 5:13797. doi: 10.1038/srep13797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantacessi C, Giacomin P, Croese J, Zakrzewski M, Sotillo J, McCann L, Nolan MJ, Mitreva M, Krause L, Loukas A. 2014. Impact of experimental hookworm infection on the human gut microbiota. J Infect Dis 210:1431–1434. doi: 10.1093/infdis/jiu256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weber N, Liou D, Dommer J, MacMenamin P, Quiñones M, Misner I, Oler AJ, Wan J, Kim L, Coakley McCarthy M, Ezeji S, Noble K, Hurt DE. 2018. Nephele: a cloud platform for simplified, standardized and reproducible microbiome data analysis. Bioinformatics 34:1411–1413. doi: 10.1093/bioinformatics/btx617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight R, Vrbanac A, Taylor BC, Aksenov A, Callewaert C, Debelius J, Gonzalez A, Kosciolek T, McCall L-I, McDonald D, Melnik AV, Morton JT, Navas J, Quinn RA, Sanders JG, Swafford AD, Thompson LR, Tripathi A, Xu ZZ, Zaneveld JR, Zhu Q, Caporaso JG, Dorrestein PC. 2018. Best practices for analysing microbiomes. Nat Rev Microbiol 16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 30.Human Microbiome Project Consortium. 2012. A framework for human microbiome research. Nature 486:215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goodrich JK, Di Rienzi SC, Poole AC, Koren O, Walters WA, Caporaso JG, Knight R, Ley RE. 2014. Conducting a microbiome study. Cell 158:250–262. doi: 10.1016/j.cell.2014.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Crespo-Piazuelo D, Estellé J, Revilla M, Criado-Mesas L, Ramayo-Caldas Y, Óvilo C, Fernández AI, Ballester M, Folch JM. 2018. Characterization of bacterial microbiota compositions along the intestinal tract in pigs and their interactions and functions. Sci Rep 8:12727. doi: 10.1038/s41598-018-30932-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu S, Chen D, Zhang J-N, Lv X, Wang K, Duan L-P, Nie Y, Wu X-L. 2013. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One 8:e74957. doi: 10.1371/journal.pone.0074957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hill TCJ, Walsh KA, Harris JA, Moffett BF. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol Ecol 43:1–11. doi: 10.1111/j.1574-6941.2003.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 35.Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J, Weitz JS. 2013. Robust estimation of microbial diversity in theory and in practice. ISME J 7:1092–1101. doi: 10.1038/ismej.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chao A. 1984. Nonparametric estimation of the number of classes in a population. Scandinavian J Stat 11:265–270. [Google Scholar]
- 37.Collado MC, Rautava S, Aakko J, Isolauri E, Salminen S. 2016. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci Rep 6:23129. doi: 10.1038/srep23129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Neu J. 2016. The microbiome during pregnancy and early postnatal life. Semin Fetal Neonatal Med 21:373–379. doi: 10.1016/j.siny.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Houlden A, Hayes KS, Bancroft AJ, Worthington JJ, Wang P, Grencis RK, Roberts IS. 2015. Chronic Trichuris muris infection in C57BL/6 mice causes significant changes in host microbiota and metabolome: effects reversed by pathogen clearance. PLoS One 10:e0125945. doi: 10.1371/journal.pone.0125945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krauth SJ, Coulibaly JT, Knopp S, Traoré M, N’Goran EK, Utzinger J. 2012. An in-depth analysis of a piece of shit: distribution of Schistosoma mansoni and hookworm eggs in human stool. PLoS Negl Trop Dis 6:e1969. doi: 10.1371/journal.pntd.0001969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cattadori IM, Sebastian A, Hao H, Katani R, Albert I, Eilertson KE, Kapur V, Pathak A, Mitchell S. 2016. Impact of helminth infections and nutritional constraints on the small intestine microbiota. PLoS One 11:e0159770. doi: 10.1371/journal.pone.0159770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zaiss MM, Rapin A, Lebon L, Dubey LK, Mosconi I, Sarter K, Piersigilli A, Menin L, Walker AW, Rougemont J, Paerewijck O, Geldhof P, McCoy KD, Macpherson AJ, Croese J, Giacomin PR, Loukas A, Junt T, Marsland BJ, Harris NL. 2015. The intestinal microbiota contributes to the ability of helminths to modulate allergic inflammation. Immunity 43:998–1010. doi: 10.1016/j.immuni.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Holm JB, Sorobetea D, Kiilerich P, Ramayo-Caldas Y, Estellé J, Ma T, Madsen L, Kristiansen K, Svensson-Frej M. 2015. Chronic Trichuris muris infection decreases diversity of the intestinal microbiota and concomitantly increases the abundance of lactobacilli. PLoS One 10:e0125495. doi: 10.1371/journal.pone.0125495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fricke WF, Song Y, Wang A-J, Smith A, Grinchuk V, Mongodin E, Pei C, Ma B, Lu N, Urban JF, Shea-Donohue T, Zhao A. 2015. Type 2 immunity-dependent reduction of segmented filamentous bacteria in mice infected with the helminthic parasite Nippostrongylus brasiliensis. Microbiome 3:40. doi: 10.1186/s40168-015-0103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rausch S, Held J, Fischer A, Heimesaat MM, Kühl AA, Bereswill S, Hartmann S. 2013. Small intestinal nematode infection of mice is associated with increased enterobacterial loads alongside the intestinal tract. PLoS One 8:e74026. doi: 10.1371/journal.pone.0074026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reynolds LA, Smith KA, Filbey KJ, Harcus Y, Hewitson JP, Redpath SA, Valdez Y, Yebra MJ, Finlay BB, Maizels RM. 2014. Commensal-pathogen interactions in the intestinal tract: lactobacilli promote infection with, and are promoted by, helminth parasites. Gut Microbes 5:522–532. doi: 10.4161/gmic.32155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneeberger PHH, Coulibaly JT, Gueuning M, Moser W, Coburn B, Frey JE, Keiser J. 2018. Off-target effects of tribendimidine, tribendimidine plus ivermectin, tribendimidine plus oxantel-pamoate, and albendazole plus oxantel-pamoate on the human gut microbiota. Int J Parasitol Drugs Drug Resist 8:372–378. doi: 10.1016/j.ijpddr.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mpairwe H, Webb EL, Muhangi L, Ndibazza J, Akishule D, Nampijja M, Ngom-Wegi S, Tumusime J, Jones FM, Fitzsimmons C, Dunne DW, Muwanga M, Rodrigues LC, Elliott AM. 2011. Anthelminthic treatment during pregnancy is associated with increased risk of infantile eczema: randomised-controlled trial results. Pediatr Allergy Immunol 22:305–312. doi: 10.1111/j.1399-3038.2010.01122.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brooker SJ. 2018. Soil-transmitted helminth treatment: multiple-drug regimens. Lancet Infect Dis 18:698–699. doi: 10.1016/S1473-3099(18)30268-8. [DOI] [PubMed] [Google Scholar]
- 50.Clarke NE, Doi SAR, Wangdi K, Chen Y, Clements ACA, Nery SV. 2019. Efficacy of anthelminthic drugs and drug combinations against soil-transmitted helminths: a systematic review and network meta-analysis. Clin Infect Dis 68:96–105. doi: 10.1093/cid/ciy423. [DOI] [PubMed] [Google Scholar]
- 51.Vercruysse J, Albonico M, Behnke JM, Kotze AC, Prichard RK, McCarthy JS, Montresor A, Levecke B. 2011. Is anthelmintic resistance a concern for the control of human soil-transmitted helminths? Int J Parasitol Drugs Drug Resist 1:14–27. doi: 10.1016/j.ijpddr.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hicks JH, Kremer M, Miguel E. 2015. The case for mass treatment of intestinal helminths in endemic areas. PLoS Negl Trop Dis 9:e0004214. doi: 10.1371/journal.pntd.0004214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Easton AV, Oliveira RG, O’Connell EM, Kepha S, Mwandawiro CS, Njenga SM, Kihara JH, Mwatele C, Odiere MR, Brooker SJ, Webster JP, Anderson RM, Nutman TB. 2016. Multi-parallel qPCR provides increased sensitivity and diagnostic breadth for gastrointestinal parasites of humans: field-based inferences on the impact of mass deworming. Parasit Vectors 9:38. doi: 10.1186/s13071-016-1314-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shannon CE. 1948. A mathematical theory of communication. Bell Syst Tech J 27:379–423. doi: 10.1002/j.1538-7305.1948.tb01338.x. [DOI] [Google Scholar]
- 55.Lande R. 1996. Statistics and partitioning of species diversity, and similarity among multiple communities. Oikos 76:5–13. doi: 10.2307/3545743. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Samples used in the longitudinal and cross-sectional (age-matched) sections of this study. The top row describes the total samples collected as part of the larger study, whereas information below shows the number of samples sequenced for the microbiome study described here. Download FIG S1, EPS file, 4.7 MB (4.7MB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Pretreatment versus 3-months-posttreatment diversity and richness comparisons based on batch A sequencing. In order to test whether the results based on batch A data diverged from those based on batch B (shown in Fig. 2), we performed the same comparisons. There is no overlap in the set of study participants in this figure and in Fig. 2. Download FIG S2, EPS file, 0.1 MB (143.8KB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Pretreatment versus 3-weeks-posttreatment diversity and richness comparisons based on batch A sequencing. In order to test whether the results based on batch A data diverged from those based on batch B (shown in Fig. 3), we performed the same comparisons. Download FIG S3, EPS file, 0.2 MB (161.5KB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Clostridiales and Enterobacteriales proportional representation relative to N. americanus egg burden. (A) Clostridiales percent makeup of the microbiome versus N. americanus DNA as measured by qPCR at baseline (r = 0.14, P = 0.54). (B) Enterobacteriales percent makeup of the microbiome versus N. americanus DNA as measured by qPCR at baseline (r = 0.43, P = 0.04). (C) Percent change in proportional representation of Clostridiales posttreatment relative to the N. americanus infection burden in those people (r = 0.20, P = 0.43). (D) Percent change in proportional representation of Enterobacteriales posttreatment relative to the N. americanus infection burden in those people (r = −0.30, P = 0.23). Download FIG S4, EPS file, 0.2 MB (181.1KB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Data Availability Statement
The data sets supporting the conclusions of this article are available in the Sequence Read Archive repository (accession number PRJNA510835; https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP173959).
Clostridiales and Enterobacteriales proportional representation relative to N. americanus egg burden. (A) Clostridiales percent makeup of the microbiome versus N. americanus DNA as measured by qPCR at baseline (r = 0.14, P = 0.54). (B) Enterobacteriales percent makeup of the microbiome versus N. americanus DNA as measured by qPCR at baseline (r = 0.43, P = 0.04). (C) Percent change in proportional representation of Clostridiales posttreatment relative to the N. americanus infection burden in those people (r = 0.20, P = 0.43). (D) Percent change in proportional representation of Enterobacteriales posttreatment relative to the N. americanus infection burden in those people (r = −0.30, P = 0.23). Download FIG S4, EPS file, 0.2 MB (181.1KB, eps) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.