Staphylococcus aureus is a major human pathogen of the skin. The global burden of diabetes is high, with S. aureus being a major complication of diabetic wound infections.
KEYWORDS: Staphylococcus aureus, Clp protease, diabetes, host-pathogen interactions
ABSTRACT
Staphylococcus aureus is a major human pathogen of the skin. The global burden of diabetes is high, with S. aureus being a major complication of diabetic wound infections. We investigated how the diabetic environment influences S. aureus skin infection and observed an increased susceptibility to infection in mouse models of both type I and type II diabetes. A dual gene expression approach was taken to investigate transcriptional alterations in both the host and bacterium after infection. While analysis of the host response revealed only minor changes between infected control and diabetic mice, we observed that S. aureus isolated from diabetic mice had significant increases in the levels of genes associated with translation and posttranslational modification and chaperones and reductions in the levels of genes associated with amino acid transport and metabolism. One family of genes upregulated in S. aureus isolated from diabetic lesions encoded the Clp proteases, associated with the misfolded protein response. The Clp proteases were found to be partially glucose regulated as well as influencing the hemolytic activity of S. aureus. Strains lacking the Clp proteases ClpX, ClpC, and ClpP were significantly attenuated in our animal model of skin infection, with significant reductions observed in dermonecrosis and bacterial burden. In particular, mutations in clpP and clpX were significantly attenuated and remained attenuated in both normal and diabetic mice. Our data suggest that the diabetic environment also causes changes to occur in invading pathogens, and one of these virulence determinants is the Clp protease system.
INTRODUCTION
The number of patients with diabetes is expected to reach upwards of 440 million globally by 2030 (1). There are approximately 30 million patients with diabetes in the United States, where it is the 7th leading cause of death (2). Treatment of diabetic patients costs upwards of $245 billion to the U.S. economy, and a major comorbidity is nontraumatic limb amputations. Diabetic patients also have an increased risk of bacterial infection that is associated with increased morbidity and mortality as a result of slow-healing wounds, decreased blood flow to extremities, neuropathy, and a suppressed immune response (3–7). Bacterial infections also make slow-healing wounds fractious to repair. Diabetic patients have a much higher infection rate for wound, local, and cutaneous bacterial infections, with up to one-third of patients suffering from some form of skin disorder (7–9). The most common cause of skin and soft tissue infection is Staphylococcus aureus (10).
Staphylococcus aureus is carried by 30% of the general population (11) but at a much higher frequency within the diabetic population (12–14). Carriage of S. aureus is a significant risk factor for subsequent infection and death (15–17). Antibiotic resistance is also of increasing importance in the context of S. aureus infection, particularly methicillin-resistant S. aureus (MRSA) (18). A major strain in circulation is MRSA strain USA300, which is epidemic in the United States and is capable of causing infections in otherwise healthy individuals (19, 20).
We hypothesized that diabetes supports an environment for S. aureus infection, influencing cell function as well as causing changes in S. aureus that aid in pathogenesis. We show using a skin model of S. aureus infection that hyperglycemia in different models of diabetes leads to increased susceptibility to infection. Using a dual transcriptome sequencing (RNAseq) approach to investigate both mammalian and bacterial transcriptomes, we identified a multitude of transcriptional changes in response to the altered host environment. The response of S. aureus to the diabetic environment was associated with significant upregulation of the stress response and genes associated with translation. We observed that genes encoding the Clp proteases were significantly upregulated in diabetic mice, which, in our model of infection, played a significant role in S. aureus pathogenesis.
RESULTS
Hyperglycemic mice have increased susceptibility to S. aureus skin infection.
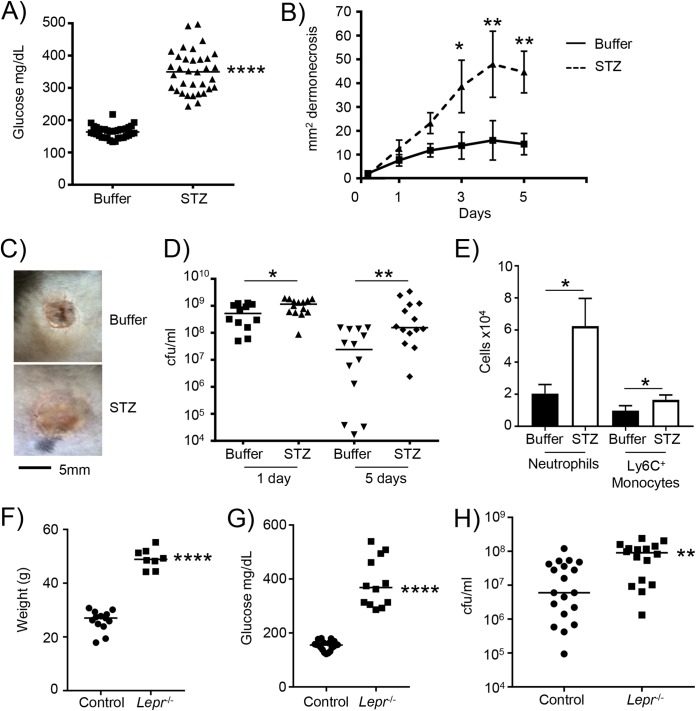
To better understand the interaction of S. aureus and the host in the context of diabetes, we utilized diabetic models of S. aureus skin infection. Our model of type I diabetes uses streptozotocin (STZ)-treated mice. Hyperglycemia was successfully shown by high blood glucose levels of 163 versus 350 mg/dl in control versus streptozotocin-treated mice (P < 0.0001) (Fig. 1A). Mice were subcutaneously infected and then monitored over a period of 5 days. Hyperglycemic mice had on average a 3-fold increase in the area of dermonecrosis compared to controls (P < 0.01) (Fig. 1B and C). Hyperglycemia also led to significant increases in bacterial burdens. One day after infection, we observed a 75% increase in bacterial numbers in hyperglycemic mice (P < 0.05) (Fig. 1D). By 5 days after infection, at which point control mice had started to reduce the bacterial burden, we observed 13-fold-higher bacterial numbers, on average, in hyperglycemic mice (P < 0.01) (Fig. 1D). Analysis of the cells recruited to the site of infection indicated that there were increased numbers of neutrophils and inflammatory monocytes (P < 0.05) (Fig. 1E), indicative of the increased bacterial burden evident in hyperglycemic mice. We quantified cytokine expression in skin biopsy samples from both 1 and 5 days after infection. We observed some reductions in monocyte chemoattractant protein 1 (MCP-1) (see Fig. S1 in the supplemental material) and tumor necrosis factor (TNF) (Fig. S2) in diabetic infected mice, but largely, no significant differences were observed, even though the diabetic mice had an increase in bacterial burden, indicating that the diabetic mice may have a reduced immune response.
FIG 1.
Type I and II diabetic mice are more susceptible to S. aureus skin infection. Diabetes was induced in mice using streptozotocin (STZ). (A) Glucose levels in control and STZ-treated mice prior to infection. (B) Control and STZ mice were subcutaneously infected with 2 × 106 CFU of S. aureus USA300, and the area of dermonecrosis was monitored over time (n = 14 for both genotypes). (C) Representative gross pathology of regions of dermonecrosis. (D) Bacterial quantification from punch biopsy specimens. (E) Flow cytometry analysis of cells from 5-day-infected punch biopsy homogenates (n = 3 for uninfected and 17 for infected groups). Data are from 4 independent experiments. (F and G) Weights (F) and glucose levels (G) of control (Lepr+/−) and obese (Lepr−/−) mice prior to infection. (H) Control and obese mice were infected with 2 × 106 CFU of S. aureus USA300 subcutaneously for 5 days. Data are from 3 independent experiments. Each point represents data for an individual mouse. ****, P < 0.0001; **, P < 0.01; *, P < 0.05.
To confirm that our observations were related to hyperglycemia and not due to other effects on the host, we repeated our experiments with an additional murine model of diabetic infection. The Leprdb mutation leads to obesity and hyperglycemia and is thus a model of type II diabetes (21, 22). Consistent with this, we observed that the weight of these mice was 49 g on average, compared to 26 g for their heterozygous controls (P < 0.0001) (Fig. 1F). The obese mice also had significant hyperglycemia, with glucose levels, on average, at 386 mg/dl, compared to 152 mg/dl in control mice (P < 0.0001) (Fig. 1G). The capacity to clear the infection in obese mice was reduced. Obese mice had on average a 3.8-fold increase in the bacterial burden compared to that in nonobese, euglycemic controls (P < 0.01) (Fig. 1H). These data show that high levels of glucose in mice perpetuate S. aureus skin infection.
Hyperglycemia inhibits macrophage killing.
Much has been reported about defects in innate immunity in diabetic patients. These observations have included reduced chemotaxis, phagocytosis, respiratory burst, and cytokine production (23–29). While it is clear that diabetic patients have increased susceptibility to infection, there is no consistent observation (30) regarding innate immune cell function between studies. Having observed a lack of a difference in immune responses in diabetic mice in lieu of an increased bacterial burden (Fig. S1 and S2), we sought to test the potential effect of glucose on macrophage function in response to S. aureus. We incubated immortalized bone marrow-derived murine macrophages with various levels of glucose for 2 days before washing the cells and weaning them to medium without glucose. Gentamicin protection assays indicated a residual glucose-dependent effect on cells that led to increased intracellular bacteria (10-fold increase with 30 versus 5 mM; P < 0.0001), showing evidence of impaired killing (Fig. S3A). The levels of glucose at which this effect became apparent (>10 mM) correspond to levels of glucose considered hyperglycemic in adult humans. This killing effect was not influenced by cell viability of macrophages (Fig. S3B). This observation was specific to glucose, as treatment with galactose did not alter numbers of intracellular bacteria (Fig. S3C). The reduction in bacterial killing was able to be titrated away using 2-deoxyglucose (2DG), a glucose analog that cannot be metabolized (Fig. S3D). This treatment did not affect cell viability (Fig. S3E). The increased numbers of bacteria in cells treated with 30 mM was not due to enhanced phagocytosis, as the uptake of labeled bacteria was reduced by 36% in 30 mM versus 5 mM glucose-treated cells (P < 0.0001) (Fig. S3F). Further support for the killing phenotype being unrelated to phagocytosis came from treatment with 2DG, which improved killing (Fig. S3D) and did not influence bacterial uptake in cells treated with 30 mM 2DG (Fig. S3G). This glucose effect, although increasing reactive oxygen species (ROS) activity in response to S. aureus (Fig. S4A and B), did not influence bacterial killing in a manner that would account for our phenotype (i.e., the enhanced ROS did not improve killing in glucose-treated cells) (Fig. S4C to E) and did not affect glycolytic activity (Fig. S5). The same effect of glucose on keratinocyte cell function (Fig. S6) did not appear to play a role, as a pyk (pyruvate kinase) mutant defective in bacterial glycolysis (31) did not display a phenotype. These data provide evidence that glucose can influence the host’s ability to deal with infection.
Host response to S. aureus in the skin under normal and hyperglycemic conditions.
To determine how the diabetic mice had impeded clearance of S. aureus, we undertook a dual RNAseq approach to investigate both the host and bacterial responses after 1 day of infection in the skin. We first examined the host response (see the data at www.daneparkerlab.com/data) of both control and diabetic mice to S. aureus infection by pathway analysis (Ingenuity; Qiagen). Irrespective of the diabetic status of the mice, we observed that infection led to increased expression of canonical pathways associated with neutrophil adhesion and migration, infection, inflammation, and immune cell trafficking. Downregulated genes included those associated with lipid, vitamin, and mineral metabolism; dermatological disease; and skeletal and muscular system development.
Direct comparison of infected mice revealed 231 differentially expressed genes in diabetic versus control mice (see the data at www.daneparkerlab.com/data). These genes (>1.5-fold changes, equal to log2 fold changes of >0.58; P < 0.05) were composed of 110 upregulated and 121 downregulated transcripts. Pathway analysis did not identify pathways significantly tied to skin infection with strong statistical significance. Investigation of the most differentially expressed genes (Table 1) showed that the levels of several transcripts increased above 2-fold, with highly significant P values. Several of the top 10 encoded proteins were related to cellular structure. Rho GTPase-activating protein 21 (encoded by Arhgap21) has GTPase-activating activity toward Cdc42 and RhoA and is important for α-catenin recruitment to the adherens junctions (32, 33). Sarcospan (encoded by Sspn) is a dystrophin-glycoprotein complex that links actin to the cytoskeleton and extracellular matrix (34), and filamin A interacting protein 1-like (encoded by Filip1l) binds to filamin A, an actin-binding protein (35). Pathway analysis of genes upregulated in diabetic infected compared to infected control mice revealed some changes. There were increases in genes associated with interleukin-10 (IL-10) signaling (16 versus 12 for infected mice treated with STZ versus control infected mice; P, 2.4E−12 versus 2.2E−8) and those associated with the role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis (27 versus 14 for infected mice treated with STZ versus control infected mice; P, 3.6E−9 versus 7.6E−3). Analysis of downregulated genes showed changes in EIF2 signaling (17 versus 39 genes downregulated for infected mice treated with STZ versus control infected mice; P, 3.3E−11 versus 7.4E−3), stearate signaling (11 versus 7 genes downregulated for infected mice treated with STZ versus control infected mice; P, 6.5E−6 versus 1.1E−3), and heme biosynthesis (0 versus 4 genes downregulated for infected mice treated with STZ versus control infected mice; P, 8E−4 versus 0). However, in many cases, these gene changes were due to only small changes in transcript levels, or they did not make the classification due to an insignificant P value. In summary, we observed similar changes upon infection in both control and diabetic animals, with only some changes noted in the canonical pathway analysis.
TABLE 1.
Top 10 up- and downregulated genes in mouse lesions in streptozotocin-treated versus untreated S. aureus-infected mice
| Target | Gene | Product | Log2 fold change (STZ/WT) | P value |
|---|---|---|---|---|
| Upregulated | ||||
| ENSMUSG00000036591 | Arhgap21 | Rho GTPase-activating protein 21 | 1.94 | 6.32E−14 |
| ENSMUSG00000022905 | Kpna1 | Karyopherin 1 | 1.92 | 5.68E−14 |
| ENSMUSG00000079184 | Mphosph8 | M-phase phosphoprotein 8 | 1.89 | 2.65E−13 |
| ENSMUSG00000030255 | Sspn | Sarcospan | 1.87 | 5.67E−13 |
| ENSMUSG00000038518 | Jarid2 | Jumonji | 1.68 | 8.57E−11 |
| ENSMUSG00000023806 | Rsph3b | Radial spoke 3B homolog | 1.64 | 1.55E−10 |
| ENSMUSG00000037965 | Zc3h7a | Zinc finger CCCH type containing 7A | 1.63 | 1.21E−10 |
| ENSMUSG00000043336 | Filip1l | Filamin A interacting protein 1-like | 1.58 | 9.54E−10 |
| ENSMUSG00000073471 | Rsph3a | Radial spoke 3A homolog | 1.45 | 1.56E−08 |
| ENSMUSG00000027104 | Atf2 | Activating transcriptional factor 2 | 1.45 | 7.09E−09 |
| Downregulated | ||||
| ENSMUSG00000029417 | Cxcl9 | Chemokine ligand 9 | −0.946 | 2.16E−04 |
| ENSMUSG00000027722 | Spata5 | Spermatogenesis associated 5 | −0.899 | 1.15E−07 |
| ENSMUSG00000091662 | Vmn1r69 | Vomeronasal 1 receptor 69 | −0.872 | 4.31E−04 |
| ENSMUSG00000078496 | Zfp982 | Zinc finger protein 982 | −0.862 | 5.76E−04 |
| ENSMUSG00000091259 | Vmn2r110 | Vomeronasal 2 | −0.839 | 1.12E−03 |
| ENSMUSG00000014905 | Dnajb9 | DnaJ heat shock family member B9 | −0.794 | 1.72E−03 |
| ENSMUSG00000038871 | Bpgm | Bisphosphoglycerate mutase | −0.781 | 1.99E−03 |
| ENSMUSG00000093528 | Nrg3os | Neuregulin 3 | −0.768 | 2.32E−03 |
| ENSMUSG00000020464 | Pnpt1 | Polyribonucleotide nucleotidyltransferase 1 | −0.762 | 1.68E−03 |
| ENSMUSG00000022146 | Osmr | Oncostatin M receptor | −0.756 | 1.17E−04 |
S. aureus adapts to the hyperglycemic environment in vivo.
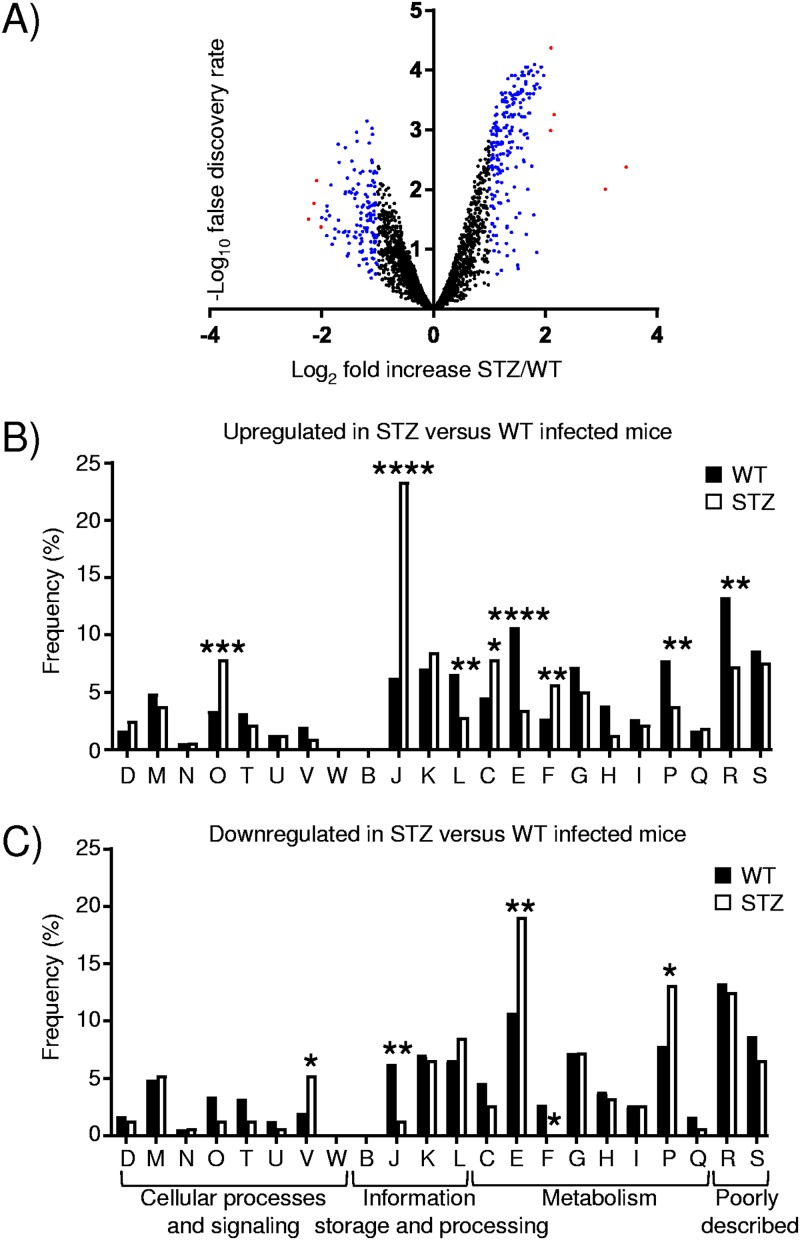
We next analyzed how S. aureus responds to the diabetic microenvironment between control and diabetic mice 1 day after infection. We observed a large number of S. aureus genes that were differentially expressed in diabetic compared to control mice (Fig. 2A). There were 513 differentially expressed genes (340 up- and 173 downregulated) (see the data at www.daneparkerlab.com/data) when a 1.5-fold cutoff (log2 fold change of >0.58) was used with a false discovery rate (FDR) of <0.05. This number went down to 205 when a 2-fold cutoff (log2 fold change of >1.00) and an FDR of <0.01 were used. Examination of the data (Table 2) shows a gene encoding a hypothetical protein as the most upregulated (10.9-fold; FDR of <0.01) (Table 2).
FIG 2.
Significant transcriptional changes to S. aureus occur in diabetic mice. RNAseq was conducted on RNA isolated from punch biopsy specimens from control and diabetic mice 1 day after subcutaneous infection with S. aureus USA300. (A) Volcano plot comparing diabetic (STZ-treated) and control mice infected with S. aureus. Red, >4-fold change; blue, >2-fold change (with a false discovery rate of <0.05). (B and C) S. aureus differentially expressed genes (513) were classified according to Clusters of Orthologous Groups (COG) designations, with changes in STZ-treated versus control infected mice analyzed among upregulated (B) and downregulated (C) genes. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (relative to the WT). D, cell cycle control, cell division, and chromosome partitioning; M, cell wall/membrane/envelope biogenesis; N, cell motility; O, posttranslational modification, protein turnover, and chaperones; T, signal transduction, intracellular trafficking, secretion, and vesicular transport; V, defense mechanisms; W, extracellular structures; B, chromatin structure and dynamics; J, translation, ribosomal structure, and biogenesis; K, transcription; L, replication, recombination, and repair; C, energy production and conversion; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q, secondary metabolite biosynthesis, transport, and catabolism; R, general function prediction; S, unknown function.
TABLE 2.
Top 10 up- and downregulated genes in S. aureus in streptozotocin-treated versus untreated mice
| Target | Gene | Product | Log2 fold change (STZ/WT) | FDR |
|---|---|---|---|---|
| Upregulated | ||||
| SAUSA300_0080 | Hypothetical protein | 3.45 | 4.18E−03 | |
| SAUSA300_0068 | Cadmium-exporting ATPase | 3.07 | 9.82E−03 | |
| SAUSA300_1511 | rpmG | 50S ribosomal protein L33 | 2.16 | 5.55E−04 |
| SAUSA300_1541 | grpE | Heat shock protein | 2.10 | 4.24E−05 |
| SAUSA300_1545 | rpsT | 30S ribosomal protein S20 | 2.09 | 1.02E−03 |
| SAUSA300_0781 | Hypothetical protein | 1.97 | 1.23E−04 | |
| SAUSA300_1542 | hrcA | Heat-inducible transcription repressor | 1.93 | 8.88E−05 |
| SAUSA300_2195 | rpsQ | 30S ribosomal protein S17 | 1.93 | 8.88E−05 |
| SAUSA300_1540 | dnaK | Molecular chaperone | 1.92 | 8.88E−05 |
| SAUSA300_0884 | Hypothetical protein | 1.90 | 1.96E−04 | |
| Downregulated | ||||
| SAUSA300_1218 | ABC transporter permease | −2.23 | 3.12E−02 | |
| SAUSA300_0811 | Hypothetical protein | −2.14 | 1.70E−02 | |
| SAUSA300_1971 | phi77 ORF017-like protein | −2.09 | 7.01E−03 | |
| SAUSA300_2598 | cap1A | Capsular polysaccharide biosynthesis protein | −2.01 | 4.24E−02 |
| SAUSA300_1955 | Putative endodeoxyribonuclease RusA | −1.99 | 2.93E−02 | |
| SAUSA300_1968 | Putative phage transcriptional regulator | −1.92 | 3.13E−02 | |
| SAUSA300_1741 | Putative lipoprotein | −1.90 | 1.88E−02 | |
| SAUSA300_1936 | Hypothetical protein | −1.90 | 2.27E−02 | |
| SAUSA300_0804 | Putative transcriptional regulator | −1.86 | 2.73E−02 | |
| SAUSA300_2305 | Transposase | −1.83 | 8.35E−03 | |
Organization of genes into orthologous groups (Fig. 2B and C) revealed significant changes across the information storage and processing and the metabolism categories. The largest difference within upregulated genes was for those associated with translation, ribosomal structure, and biogenesis (group J; P < 0.0001) (Fig. 2B). This group constitutes 6.2% of genes normally but represented 23.3% of those that were upregulated. There was also a significant enrichment of genes associated with posttranslational modification, turnover, and chaperones (group O) (7.9% versus 3.4% for diabetic versus control mice; P < 0.001) (Fig. 2B); nucleotide transport and modification (group F) (5.7% versus 2.7%; P < 0.01) (Fig. 2B); and energy production (group C) (7.9% versus 4.6%; P < 0.05) (Fig. 2B). We also observed concomitant decreases in these categories with genes that were downregulated in S. aureus in response to diabetic versus control mice (Fig. 2C). Categories decreased within upregulated genes were groups E (amino acid transport and metabolism) (3.5% versus 10.7% for diabetic versus control mice; P < 0.0001) (Fig. 2B), P (inorganic ion transport and metabolism) (3.8% versus 7.8%; P < 0.05) (Fig. 2B), L (replication recombination and repair) (2.8% versus 6.6%; P < 0.01) (Fig. 2B), and R (general function prediction) (7.3% versus 13.3%; P < 0.01) (Fig. 2B) We also observed that these categories were typically increased among the downregulated S. aureus genes (Fig. 2C).
Examination of genes differentially expressed by S. aureus revealed a number of significant changes in ribosomal, heat shock, and stress genes (Table 2; also see the data at www.daneparkerlab.com/data). The heat shock-related genes grpE, dnaK, and hrcA (Table 2) were all significantly upregulated, in addition to dnaJ (see the data at www.daneparkerlab.com/data). These heat shock-related genes are involved in combating a variety of cellular stresses and are located adjacent to one another in the genome (36). Several other heat shock-encoding genes were also upregulated. The levels of both groEL and groES were increased, 2.9- and 2.5-fold, respectively (Fig. S7; also see the data at www.daneparkerlab.com/data). Genes encoding the Clp proteases were also upregulated in S. aureus under diabetic conditions. The Clp proteases are ATPases that are conserved across species, perform multiple roles in degrading misfolded proteins, and are associated with oxidative and heat stress as well as biofilm formation and intracellular replication (37–39). The Clp proteins have a variety of regulatory targets; having been associated with pathogenesis, they have been targeted for antimicrobial development (38, 40, 41). The genes clpP (2.6-fold; FDR of <0.001), clpX (1.9-fold; FDR of <0.01), clpB (1.9-fold; FDR of <0.01), and clpC (1.8-fold; FDR of <0.01) were all upregulated in S. aureus isolated from infected diabetic mice compared to euglycemic controls (see the data at www.daneparkerlab.com/data). Additional stress-related genes encoding superoxide dismutase, a ferric uptake regulator, and a peroxide-responsive repressor were also upregulated (see the data at www.daneparkerlab.com/data). We observed that the genes agrA, hla, and sarA were associated with virulence factors and that their regulation was also upregulated (see the data at www.daneparkerlab.com/data). Virulence factor-encoding genes observed to be downregulated included lukD, sek, and seq (see the data at www.daneparkerlab.com/data).
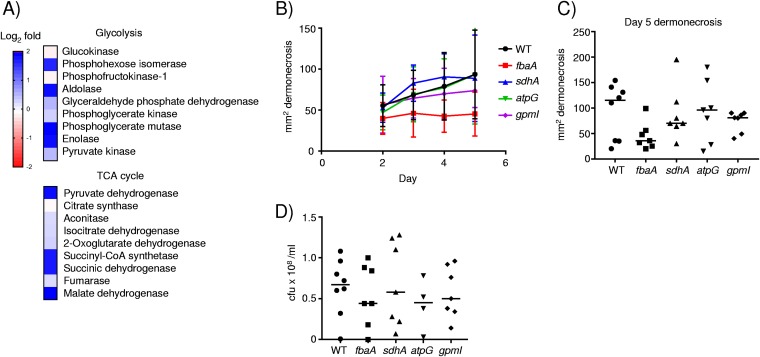
Analysis of KEGG pathways (42) also revealed significant enrichment for genes involved in glycolysis (P < 0.05), the tricarboxylic acid (TCA) cycle (P < 0.001), and oxidative phosphorylation (P < 0.0001). Examination of altered genes revealed changes in phosphohexose isomerase (pgi), aldolase (fbaA), phosphoglycerate mutase (gpmI), enolase, pyruvate dehydrogenase (pdhA and pdhB), succinyl-CoA synthetase (sucC and sucD), succinic dehydrogenase (sdhA), malate dehydrogenase, and the gamma subunit of ATP synthase (atpG) (see the data at www.daneparkerlab.com/data). The transcriptional analysis of S. aureus isolated from control and diabetic lesions indicates that in response to the diabetic environment, it alters its physiology to increase translation machinery, adopting a stress-like response and adapting to a glucose-rich environment.
Increased expression of glycolysis and TCA cycle genes does not contribute to S. aureus virulence.
As we observed increases in the expression of several S. aureus genes involved in glycolysis and the TCA cycle (Fig. 3A), we posited that this may aid the ability of S. aureus to survive in the skin environment. We examined mutants in genes that we observed to be upregulated in diabetic mice, in glycolysis (fbaA [fructose biphosphate aldolase] and gpmI [phosphoglyceromutase]), the TCA cycle (sdhA [succinate dehydrogenase]), as well as oxidative phosphorylation (atpG [ATP synthase γ-subunit]). We infected each of these strains along with our wild-type (WT) USA300 strain of S. aureus in our model of skin infection and monitored the mice for 5 days. We did not observe a significant change in the areas of dermonecrosis with any of the mutants (Fig. 3B and C). The mutation in the aldolase gene fbpA, involved in glycolysis, gave the greatest change. At day 5, the fbpA mutant had an average reduction in dermonecrosis of 48% (P = 0.052) (Fig. 3C). No changes in bacterial burdens were observed (Fig. 3D). Based on these data, it is likely that the increased expression of these genes is a direct consequence of increased glucose availability or that functional redundancy limits the observation of phenotypic differences with single mutations.
FIG 3.
Influence of the diabetic environment on glycolysis and the TCA cycle in S. aureus. (A) Heat map analysis of S. aureus genes involved in glycolysis or the TCA cycle. Transcript levels were compared between bacterial RNAs isolated from diabetic and control mouse lesion material (diabetic/control). (B) Wild-type mice were infected subcutaneously with 2 × 106 CFU of WT Je2 or mutant strains of S. aureus for 5 days, and dermonecrosis was monitored (n = 8 for the WT and 7 for each mutant strain). (C) Quantification of dermonecrosis at day 5. (D) Bacterial enumeration in punch biopsy specimens at day 5 of infection. Each point represents data for a mouse. Lines display medians.
Contribution of Clp proteases to the pathogenesis of S. aureus skin infection.
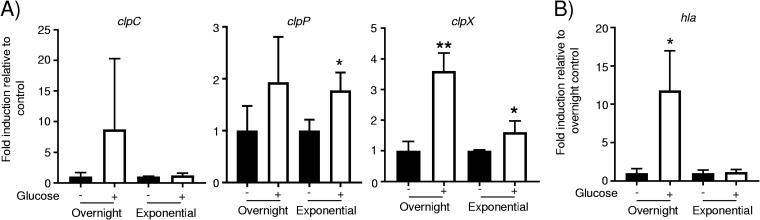
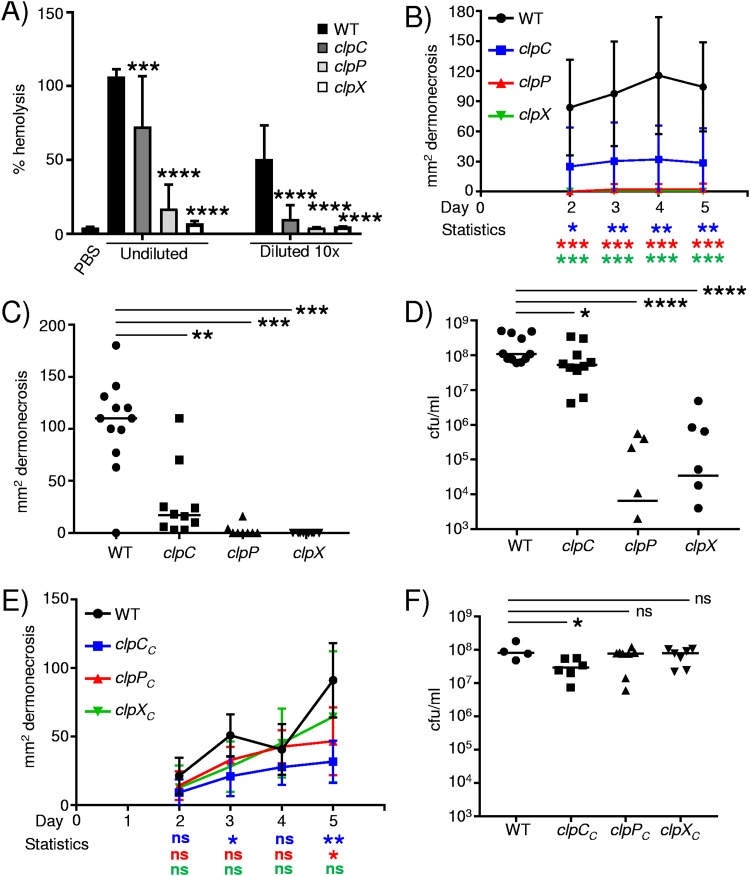
As the genes encoding the Clp proteases were upregulated in diabetic mice, we first tested whether their expression is influenced by glucose levels. While variable in cultures grown overnight, clpC showed no changes in expression in response to glucose (Fig. 4A). Likewise, cultures grown overnight did not alter clpP production in response to glucose but had an 80% increase (P < 0.05) (Fig. 4A) during exponential-phase growth. The expression of clpX was significantly influenced by glucose, with a 260% increase (P < 0.01) (Fig. 4A) in cultures grown overnight and a 60% increase (P < 0.05) (Fig. 4A) during exponential phase in response to glucose. Alpha-toxin expression has been reported to be repressed by glucose in strains of S. aureus (43); with our USA300 strain, we observed a significant increase in the expression of hla in response to glucose in cultures grown overnight (12-fold; P < 0.05) (Fig. 4B). These data suggest that some of the clp genes (clpX and to clpP to some degree) may be responsive to glucose levels.
FIG 4.
Influence of glucose on clp gene expression. RNA was extracted from overnight-grown and exponential-phase cultures of S. aureus, and transcripts for clp genes (A) and hla (B) were quantified (n = 3). **, P < 0.01; *, P < 0.05 relative to the no-glucose or PBS control.
We next investigated the pathogenic potential of each of the clp mutant strains. As previous work has shown that the expression of agrA is decreased in a clpP background (41, 44) and we observed increases in the expression of both agrA and hla in S. aureus isolated from diabetic mice, we first quantified the hemolytic activity of each of our clp strains. In the absence of either clpC, clpP, or clpX, we observed significant reductions in hemolytic activity (Fig. 5A). Spent cultures from the clpC mutant had 33% less hemolytic activity (P < 0.001) (Fig. 5A), while the activities of clpP and clpX cultures were reduced by 87% and 97%, respectively (P < 0.0001) (Fig. 5A). When the culture material was diluted, the clpP and clpX mutants had no activity above our phosphate-buffered saline (PBS) control, and the clpC material retained only 5% of the activity of the WT strain (P < 0.0001) (Fig. 5A). As alpha-toxin is an important virulence factor in S. aureus infection of the skin (45, 46), we hypothesized that each of these strains would be attenuated in our skin infection model. Infection of mice with the clp mutants led to significant reductions in dermonecrosis apparent over the course of infection (Fig. 5B). The majority of mice infected with the clpP or clpX mutant had no signs of dermonecrosis, with average sizes after 5 days of 2.5 mm2 for the clpP mutant and 0 mm2 for the clpX mutant (P < 0.001) (Fig. 5C). Infection with the clpC mutant was also significantly reduced, with an average area of dermonecrosis at day 5 of 25 mm2, compared to WT mice, with an average area of 85 mm2 (P < 0.01) (Fig. 5C). The attenuation of virulence with the clpP and clpX mutants was also evident with the bacterial burden: bacterial levels were reduced by 1,400-fold in clpP mutant-infected mice and 262-fold in clpX mutant-infected mice compared to those seen in WT-infected mice (P < 0.0001) (Fig. 5D). Mice infected with the clpC mutant also had 53% fewer bacteria than WT mice (P < 0.05) (Fig. 5D). The profound phenotype observed with mutations in clpP and clpX was reverted upon complementation (Fig. 5E and F and Fig. S8). Complemented strains of the clpP and clpX mutants gave rise to regions of dermonecrosis and bacterial burdens equivalent to those of the WT (Fig. 5E and F). The clpC mutation was successfully complemented and led to dermonecrosis; however, it showed a slight decrease in bacterial burden (Fig. 5F and Fig. S8). Since only partial complementation was observed for the clpC mutant in vivo, this suggests that the transposon insertion within clpC may have some polar effects on genes within its operon.
FIG 5.
The Clp proteases contribute to S. aureus subcutaneous skin infection. (A) Hemolytic activity of spent culture supernatants from WT S. aureus and clp mutants (n = 12 for PBS, 13 for the WT, 12 for the clpC mutant, 10 for the clpP mutant, and 6 for the clpX mutant). WT C57BL/6J mice were infected subcutaneously with WT S. aureus and clp mutants for 5 days. (B) Areas of dermonecrosis over time. (C) Areas of dermonecrosis quantified at day 5. (D) Bacterial counts from punch biopsy specimens at day 5. (E and F) Chromosomally complemented clp mutants were assessed in the skin infection model quantifying dermonecrosis (E) and bacterial burden (F). Each point represents data for a mouse. Lines display medians. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05; ns, not significant (relative to the WT control).
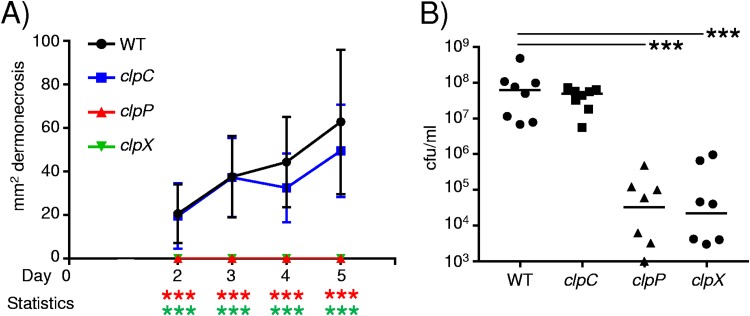
As we identified the expression of the clp genes in diabetic mice, we also tested their role in pathogenesis in the context of hyperglycemia. Similarly to what was observed previously, both the clpP and clpX mutants gave no indication of dermonecrosis (P < 0.001) (Fig. 6A) compared to WT diabetic mice. We note that although the clpC mutant showed some decrease in dermonecrosis at days 4 and 5, these differences were not statistically significant (Fig. 6A). Analysis of the bacterial burden at day 5 indicated that the clpP and clpX genes were important in skin pathogenesis in the context of diabetes. Bacterial levels in the absence of clpP were 1,084-fold lower than those of the WT strain (P < 0.001) (Fig. 6B), and those in the absence of clpX were 487-fold lower (P < 0.001) (Fig. 6B). Mice infected with the clpC mutant also had a 58% reduction in bacteria compared to the WT strain; however, this was not statistically significant. These data indicate the importance of the ClpXP protease under both normal and diabetic conditions.
FIG 6.
The ClpXP protease contributes to S. aureus skin infection under diabetic conditions. Diabetes was induced in mice using streptozotocin (STZ). (A) Mice were subcutaneously infected with 2 × 106 CFU of S. aureus USA300, and the area of dermonecrosis was monitored over time. (B) Bacterial quantification from punch biopsy specimens (n = 8). Data are from two independent experiments. ***, P < 0.001.
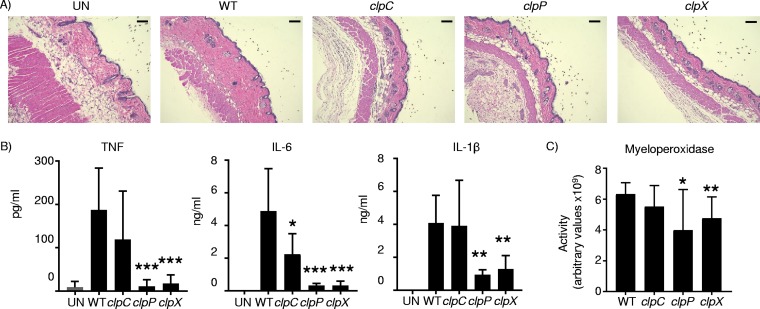
The immune response to the Clp mutants was also significantly attenuated. Histopathological examination of day 5 skin biopsy specimens showed that the dermis was thickened and inflamed in WT mice compared to uninfected mice, and this was significantly reduced in the clp mutant-infected mice (Fig. 7A). Consistent with the pathology, we observed that cytokine production was significantly reduced. TNF production was reduced by 94% and 90% in the clpP and clpX strains, respectively (P < 0.001), while that in the clpC strain was not statistically different (Fig. 7B). IL-6 production was reduced in all strains. Both the clpP and clpX strains had IL-6 levels 93% lower than those of the WT (P < 0.001), and that in the clpC mutant was reduced by 54% (P < 0.05) compared to WT levels (Fig. 7B). The IL-β levels were reduced 76% (P < 0.01) in the clpP strain and 68% (P < 0.01) in the clpX strain, with no difference observed in clpC mutant-infected skin samples (Fig. 7B). The level of myeloperoxidase (MPO), as a measure of neutrophil recruitment, was significantly reduced in both the clpP and clpX strain-infected mice (Fig. 7C). MPO activity was reduced by 37% in clpP mutant samples (P < 0.05) and by 25% in clpX mutant samples (P < 0.01), while MPO activity was not significantly reduced (12% decrease; P = 0.18) in the clpC mutant samples. These data indicate a significant role for the Clp proteases in pathogenesis, and their upregulation in S. aureus during hyperglycemia likely contributes to the increased disease observed in diabetic mice.
FIG 7.
Reduced immune response to Clp mutants. WT C57BL/6J mice were infected subcutaneously with WT S. aureus and clp mutants for 5 days before punch biopsies were performed and samples were homogenized. (A) H&E-stained sections. Bars = 100 μm. UN, uninfected. (B and C) Clarified homogenized samples were used to quantify cytokine levels (B) and myeloperoxidase activity (C). Graphs show means with standard deviations (n = 12 for the WT and 8 for clp mutants, except for TNF [n = 7 for the clpP mutant]). ***, P < 0.001; **, P < 0.01; *, P < 0.05 (relative to the WT control).
DISCUSSION
Diabetes is a major cause of morbidity and mortality in the community, affecting a significant proportion of the population. Studies on the immunological effects of diabetes are focused on changes to the host immune system, while in this study, we examined both the host and the influence that it has on a major pathogen, S. aureus. We observed an increased susceptibility to skin infection in mouse models of diabetes. The host responses to infection at the cytokine level were similar in vivo between control and diabetic mice. In vitro, we observed significant effects on immune cell function. By analyzing the global transcriptional response of the host-pathogen interaction, we observed several interesting changes to the pathogen. S. aureus from diabetic mice adapted to the environment by increasing its glycolytic machinery, stress- and heat shock-responsive genes, as well as the Clp proteases, which were demonstrated to be important in a model of skin infection. These data indicate that in the diabetic environment, in addition to defects in the host response, S. aureus also adapts, leading to a worse host outcome.
We observed that diabetes significantly influences host outcome. With S. aureus infection being the most common infection of diabetics, we sought to better understand why this is the case (47). In both models of diabetic skin infection, we observed a significant increase in susceptibility, which is consistent with data from other studies that have examined foot infection, orthopedic implantation, and wound healing models, all seeing an impediment to clearance (23, 48–50). These studies and others have observed a variety of host immune defects. Much has been reported about defects in innate immunity in diabetic patients. These observations have included reduced chemotaxis, phagocytosis, and respiratory burst of immune cells, as well as cytokine release, but not in the context of bacteria (10, 51). Infection studies have shown evidence for deficiencies in neutrophil phagocytosis and killing (23, 24, 49) as well as killing of bacteria by blood. While we observed that glucose influences macrophage function in vitro, our expression analysis in vivo showed only subtle changes to host pathways. We observed that cytokine production was largely unaltered. This indicates that a blunted immune response was occurring given that the diabetic mice had significantly increased bacterial burdens.
In contrast to the host response, the transcriptional responses of S. aureus varied significantly between control and diabetic mice. We observed a significant increase in gene induction associated with translation, while we observed decreases in amino acid transport genes as well as genes of unknown function. This contrasts slightly with a study that examined S. aureus expression in diabetic rats in an endocarditis model, whereby amino acid transport genes were upregulated (52); however, this may just be a reflection of the different infection models. Many of the upregulated genes of S. aureus under diabetic conditions were associated with heat shock and cellular stress, an interesting observation given our host expression data, suggesting that the infection site was not a more hostile environment for growth. Glucose is known to alter the transcription of S. aureus significantly. Thus, some of these changes may be a reflection of that. A group of genes associated with this stress response was the clp protease genes.
A major observation was the increased expression of the Clp protease genes in S. aureus USA300 under diabetic conditions and their involvement in the pathogenesis of skin infection. Each of the mutants analyzed, clpP, clpX, and clpC, displayed significant attenuation. The degree of dermonecrosis and bacterial burden were also largely correlated with their capacity to produce alpha-toxin, which was significantly reduced in all the mutants but was reduced almost entirely in the clpP and clpX backgrounds, which had the most significant in vivo phenotypes. The loss of alpha-toxin and decreases in agr expression have been noted previously for a different strain of S. aureus, while a mechanism for this is still to be determined (40, 41, 44, 53). Various mutations in the Clp proteases have been examined in models of S. aureus systemic infection, abscess formation, and foreign-body infection, with similar observations that inactivation led to attenuation of virulence (39, 40, 54, 55). The roles of the other Clp proteases, ClpB and ClpL, have not been examined yet in S. aureus USA300. Given that the Clp proteases are involved in various functions within the cell (37, 40, 53, 56, 57), it is likely that these proteins will also play a significant role in pathogenesis.
The Clp proteins function like heat shock proteins (58), and we noted several other significant changes that likely contributed to the overall persistence of S. aureus in the diabetic mice. The increased expression of ribosomal proteins along with the change in glycolytic machinery might indicate an adaption to the environment and facilitate improved growth. Several other stress response genes associated with heat shock were also induced. Heat shock proteins such as DnaK, DnaJ, and GrpE have various roles, such as influencing lipopeptides in Pseudomonas aeruginosa (59). In S. aureus, DnaK has been shown to be important in systemic infection as well as having roles in adhesion and biofilm formation (36, 60). The heat shock protein GroEL has also shown promise as a target for antimicrobials (61). The respective roles of these various heat shock proteins have not been investigated in the context of skin infection; however, given their various functions and increased expression in susceptible mice, they are likely to also play a role in the pathogenesis of S. aureus in the skin.
Our results concur with those observed in patients, showing an increase in susceptibility to infection. Our data suggest that the diabetic environment not only causes an effect on host immunity and immune cell function but also causes physiological adaptations to occur in S. aureus. These changes increase the virulence of S. aureus by increasing the expression of translational and stress- and virulence-related genes. We show that one set of these genes encoding the Clp proteases are important in the pathogenesis of S. aureus skin infection with MRSA strain USA300.
MATERIALS AND METHODS
Bacterial culture.
Staphylococcus aureus USA300 LAC as well as the JE2, pyk, sodA, katE, fbaA, sdhA, atpG, gpmI, clpP, and clpC strains from the S. aureus transposon library (62) and the clpX strain (63) and its complement (44) were grown in Luria-Bertani broth. Bacteria were diluted 1:100 from cultures grown overnight and grown to mid-exponential phase before resuspension in PBS for experiments. For complementation, Escherichia coli ElectroTen-Blue was grown in LB medium or LB agar supplemented with 100 μg/ml ampicillin, and S. aureus strains were grown in tryptic soy broth supplemented with 0.1 mM cadmium chloride when required. Hemolysis assays were performed on spent cultures from suspensions of bacteria grown overnight. Clarified medium was filter sterilized and incubated with 1% sheep red blood cells in PBS for 30 min at 37°C in 5% CO2. Red cells were removed at 2,000 rpm in V-bottom plates before measurement at 415 nm in a Tecan plate reader. Triton X-100 at 1% was used as a positive control. Protease assays were performed as described previously (64). Briefly, concentrated bacteria were applied to gelatin-containing agar plates and incubated for 18 h at 37°C before precipitation with 15% (wt/vol) HgCl2 dissolved in 20% HCl at room temperature until zones of proteolysis were visible.
Chromosomal complementation.
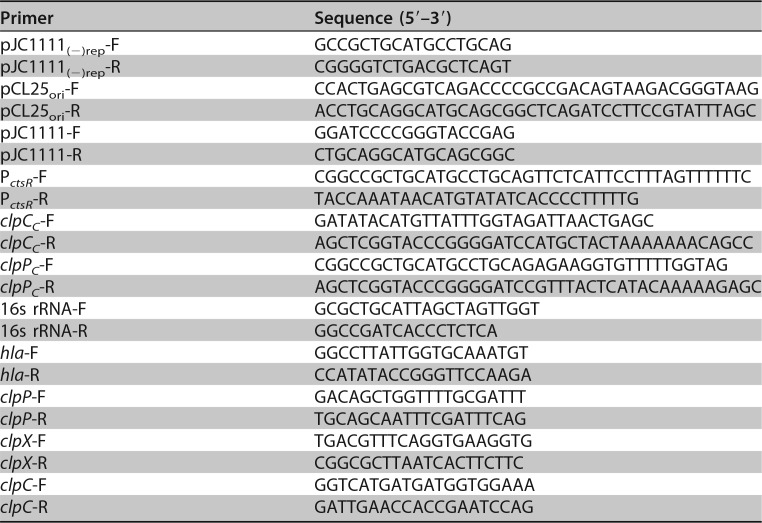
The clpP mutant was chromosomally complemented using pJC1111, a previously described SapI site integration vector (65). Essentially, clpP was amplified along with its native promoter using primers clpPC-F and clpPC-R and assembled into pJC1111 (PCR amplified by primers pJC1111-F and pJC1111-R) to generate pAQ57 (Table 3). The clpC mutant was complemented using pAQ56, a low-copy-number derivative of pJC1111. For the construction of plasmid pAQ56, the pSC101ori region of pCL25 (66) was initially amplified using primers pCL25ori-F and pCL25ori-R and assembled into pJC1111 devoid of its colE1 replication origin [amplified using primers pJC1111(−)rep-F and pJC1111(−)rep-R] to generate pAQ59. Finally, clpC (PCR amplified using clpCC-F and clpCC-R) and its native promoter (PCR amplified using PctsR-F and PctsR-R) were independently amplified and assembled into pAQ59 (PCR amplified using pJC1111_fwd and pJC1111_rev) to generate pAQ56. All DNA assembly reactions were performed using the NEBuilder high-fidelity DNA assembly cloning kit. The resulting plasmids were maintained in E. coli ElectroTen-Blue (Stratagene) prior to SapI site integration in S. aureus RN9011. The chromosomally integrated plasmids (pAQ57 and pAQ59) were subsequently moved from S. aureus RN9011 to the clpP and clpC mutants, respectively, by Φ11 phage-mediated transduction.
TABLE 3.
Primers used in this study
Mouse studies.
C57BL/6J, Lepr+/−, and Lepr−/− mice were bred at Columbia University. Obese Lepr−/− mice were compared with littermate Lepr+/− controls, and both sexes were used. Hyperglycemic C57BL/6J mice were generated through daily injections over 5 days with 50 mg/kg of body weight of streptozotocin (STZ) (67). STZ experiments were done using 8-week-old male mice, with glucose levels measured 10 days after the last injection. Glucose measurements were obtained from tail vein blood and quantified using an Accu-Chek Aviva Plus monitoring kit (Roche). Mice with glucose levels over 250 mg/dl were included in hyperglycemic groups for study.
Mice were infected with 2 × 106 CFU of S. aureus USA300 subcutaneously while under anesthesia. Streptozotocin-treated and control mice were anesthetized using ketamine and xylazine, and Lepr−/− and Lepr+/− mice were anesthetized with isoflurane prior to infection. Analysis of areas of dermonecrosis was performed using homogenized material from 5-mm punch biopsy specimens. Bacterial counts were quantified after growth on CHROMagar S. aureus bacteriological medium (Becton Dickinson). Flow cytometry on cells from skin homogenates was performed as described previously (68), using an LSRII instrument (BD Biosciences). Clarified supernatants from skin homogenates were used to quantify cytokine levels by an enzyme-linked immunosorbent assay (ELISA) (BioLegend) (according to the manufacturer’s instructions) and myeloperoxidase (MPO) activity using the EnzChek MPO activity assay kit (Life Technologies). Skin biopsy specimens were fixed in 4% paraformaldehyde overnight to prepare paraffin blocks for hematoxylin and eosin (H&E)-stained slides.
Animal work in this study was carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (69), the Animal Welfare Act, and U.S. federal law. Protocols were approved by the Animal Care and Use Committees of Columbia University and Rutgers New Jersey Medical School.
Tissue culture.
Immortalized bone marrow-derived macrophages (BMM) were grown as described previously (70). The human keratinocyte line HaCaT was grown in RPMI 1640 medium with 10% fetal calf serum (FCS), penicillin, and streptomycin. Primary human keratinocytes (HKn) were grown in Dermalife (Lifeline Cell Technologies) with 1% FCS, penicillin, and streptomycin. Fibronectin (Corning) was added at 10 μg/ml 1 h prior to infection. To analyze the effect of glucose on cell function, BMM were grown in normal (5 mM) and high (30 mM) glucose. Likewise, skin cells were grown in 6 mM for normal levels and in 50 mM for high levels of glucose. Cells in normal and high glucose were grown for 2 days prior to experiments, with cells being removed to no glucose 2 h prior to bacterial stimulation. 2-Deoxyglucose (2DG) was used to inhibit glucose utilization at 1 mM during growth under normal- and high-glucose conditions. The effect of reactive oxygen species (ROS) was tested by growing cells in the presence of 5 μM hydrogen peroxide. Viability of cells was determined with a Countess cell counter (Life Technologies).
Gentamicin protection assays were performed at an S. aureus multiplicity of infection (MOI) of 20 for 2 h, before cells were washed and treated with 500 μg/ml of gentamicin for 2 h. Cells were then washed, detached with TrypleExpress (Life Technologies), and serially diluted onto LB agar plates for quantitation. Extracellular killing assays were conducted 2 h after bacterial stimulation.
Bacterial uptake assays were conducted using Alexa Fluor 647 (AF647) (Life Technologies)-labeled S. aureus. Mammalian cells were incubated for 20 min with labeled bacteria, prior to washing, detachment, and analysis via flow cytometry.
ROS were measured using the CellROX (cellular ROS) and MitoSOX (mitochondrial ROS) kits using flow cytometry according to the manufacturer’s instructions (Life Technologies).
Glycolytic stress.
BMM cells were seeded at 20,000 cells/well in a Seahorse XF24 well plate (Agilent) in the presence of 5 mM or 30 mM glucose and incubated at 37°C with 5% CO2 for 2 days. A sensor cartridge was calibrated according to the manufacturer’s instructions overnight at 37°C without CO2. On the day of infection, BMM were washed once in XF base medium supplemented with 2 mM glutamine. The cells were infected at an MOI of 10 and incubated at 37°C without CO2 for 3 h. The extracellular acidification rate (ECAR) was measured using an XF24 analyzer (Seahorse Bioscience). Each measurement cycle consisted of a mixing time of 3 min and a data acquisition period of 3 min (12 data points). Glucose was injected at a final concentration of 10 mM to stimulate glycolysis, followed by the addition of oligomycin at 1 μM to suppress oxidative phosphorylation and 2-deoxyglucose at 50 mM to inhibit glycolysis. The metabolic activity of the bacteria was determined by adding the same amount of bacteria used during tissue culture infection to XF24 well plates containing XF base medium only.
RNA extraction and cDNA synthesis.
RNA was isolated from punch biopsy homogenates of 1-day-old infection sites using the Direct-zol RNA extraction kit (Zymo), after treatment for 5 min with 50 μg/ml of lysostaphin. Isolated RNA was further treated with DNase according to the manufacturer’s instructions (Ambion). Quantitative reverse transcriptase PCR (qRT-PCR) was performed using SYBR green on cDNA generated using a high-capacity cDNA synthesis kit (Applied Biosystems). Transcripts were normalized to 16S rRNA levels.
For dual RNAseq analysis, RNA was treated with a Ribo-Zero gold epidemiology kit (Illumina) according to the manufacturer’s instructions. Enrichment for bacterial transcripts was performed as described previously (71). The RNA was fragmented using the NEBNext magnesium RNA fragmentation module (72) with a 3-min incubation at 94°C, followed by a cleanup step using RNA Clean & Concentrator-5 (Zymo Research). Double-stranded, tagged cDNA was generated from 50 ng of fragmented RNA as previously described (71, 73). Noncaptured samples were prepared by diluting the double-stranded cDNA 100-fold and then adding sequencing adaptors and 6-mer ScriptSeq (Epicentre) barcodes with 12 cycles of PCR. Biotinylated probes specific to S. aureus were generated from S. aureus genomic DNA using the BioPrime DNA labeling system (Life Technologies) according to the manufacturer’s instructions. The hybridization reaction was set up with 20 ng of double-stranded tagged cDNA mixed with 2 μg of probes and then dehydrated using a vacuum centrifuge and resuspended in 10 μl of hybridization buffer (NimbleGen). The hybridization mixture was denatured using a thermocycler at 95°C for 5 min, followed by incubation overnight (16 to 18 h) at 60°C. Monomeric avidin agarose (Pierce)-packed Micro-Spin columns (Pierce) were made by adding 60 μl of the mixture and centrifuging the mixture for 1 min at 600 × g. All subsequent steps were carried out in a 60°C incubator, with all buffers and equipment prewarmed in the incubator for at least 2 h. The packed column was washed twice with 200 μl of 0.1 M PBS, and the hybridized sample was then applied directly to the packed resin. After 5 min of incubation, the sample was washed 15 times with 200 μl 2× stringent wash buffer (NimbleGen). The probe-bound sequences were then eluted off the column with two additions of 25 μl of biotin elution buffer (Pierce). The eluted fraction was cleaned and concentrated to a final volume of 13 μl using 1.8 volumes of AMPure XP beads (Beckman Coulter).
Library preparation and sequencing.
A quantitative PCR (qPCR) assay was performed on the captured cDNA samples to determine the correct number of cycles required to generate the final sequencing library as well as to assess the efficacy of normalization and enrichment (73). Typically, higher threshold cycle (CT) values indicate greater depletion of host transcripts. The final library was created by PCR amplifying the samples with the full-length sequencing adaptors and 6-mer ScriptSeq (Epicentre) barcodes. Following PCR, the samples were cleaned and size selected in a two-step cleanup using 0.75 volumes of AMPure XP beads followed by 0.15 volumes to achieve a final library with an average size of ∼330 bp. Libraries were combined with 12 samples each, in equal amounts, and concentrated using DNA Clean & Concentrator-5 (Zymo Research). Prior to sequencing, the final libraries were quantified using qPCR. Combined libraries were sequenced on an Illumina NextSeq instrument using 75-cycle kits. Samples were loaded at 1.8 pM, and 75-base single-end reads were obtained.
Transcriptome analysis.
Host gene analysis was performed as previously reported, using the in-house-developed YAnTra software pipeline (71). Low-complexity and low-quality sequences from FASTQ files were filtered out as previously described (74, 75). Filtered reads were then aligned to the mouse reference genome using TopHat2 (76) and Bowtie2 (77) to produce SAM/BAM files that were then used for alignment maps. Cufflinks (78) was used to analyze SAM/BAM files to obtain normalized expression values for genes as fragments per kilobase of exon per million fragments mapped (FPKM) values. Data were analyzed through the use of IPA (Qiagen Inc.). For bacterial sequence information, read abundance was quantified using the S. aureus USA300 FPR3757 reference genome (GenBank accession number NC_007793) and Kallisto (79), a kmer-based pseudoalignment tool, with analysis and visualization using Degust (https://github.com/drpowell/degust). Degust uses Voom/Limma (80) and generates an interactive website to analyze and explore the data. Differential expression was considered statistically significant by applying threshold cutoffs of a >1.5-fold change and a P value of <0.05 (adjusted for false discovery). Bacterial genes were categorized into orthologous groups (81), and pathways were interpreted using Genome2D (42).
Statistics.
Animal data were assessed using a nonparametric Mann-Whitney test. Graphs display means with standard deviations. All experiments were conducted on at least two separate occasions. In vitro experiments were assessed using parametric Student’s t test. Multiple comparisons were conducted using one-way analysis of variance (ANOVA) with Bonferroni’s multiple-comparison test. Clusters of Orthologous Groups (COG) were compared with Fisher’s exact test. Statistics were performed with Prism software (GraphPad, La Jolla, CA, USA).
Supplementary Material
ACKNOWLEDGMENTS
A.E.L., K.P., and R.J.M. were supported by Sandia National Laboratories Laboratory Directed Research and Development (LDRD) Program project 173021 (R.J.M.). Sandia National Laboratories is a multimission laboratory managed and operated by National Technology and Engineering Solutions of Sandia LLC, a wholly owned subsidiary of Honeywell International Inc., for the U.S. Department of Energy’s National Nuclear Security Administration under contract DE-NA0003525. V.C.T. and A.A.A. were funded by NIH/NIAID R01AI125588. S.P. was supported by the NIDDK Medical Student Research Program. Research reported in this publication was performed in the CCTI Flow Cytometry Core, supported in part by the Office of the Director, National Institutes of Health, under award S10RR027050. This work was supported by a Pilot Feasibility Grant from the Columbia University Diabetes Research Center (to D.P.).
This paper describes objective technical results and analysis. Any subjective views or opinions that might be expressed in the paper do not necessarily represent the views of the U.S. Department of Energy or the U.S. Government.
R.J. and L.A. were involved in diabetic models of infection in vivo and in vitro. S.P. investigated effects of glucose in vitro. T.W.F.L. conducted the Seahorse analyzer experiments. A.E.L. constructed libraries from RNA for sequencing. K.P. and T.P.S. analyzed transcriptomic data. A.A.A. and V.C.T. complemented the mutants. R.J., S.P., A.E.L., T.W.F.L., K.P., T.P.S., V.C.T., R.J.M., and D.P. edited the manuscript. D.P. conducted experiments, analyzed the data, and wrote the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00163-19.
REFERENCES
- 1.Wild S, Roglic G, Green A, Sicree R, King H. 2004. Global prevalence of diabetes: estimates for the year 2000 and projections for 2030. Diabetes Care 27:1047–1053. doi: 10.2337/diacare.27.5.1047. [DOI] [PubMed] [Google Scholar]
- 2.CDC. 2014. National diabetes statistics report. CDC, Atlanta, GA. [Google Scholar]
- 3.Joshi MB, Lad A, Bharath Prasad AS, Balakrishnan A, Ramachandra L, Satyamoorthy K. 2013. High glucose modulates IL-6 mediated immune homeostasis through impeding neutrophil extracellular trap formation. FEBS Lett 587:2241–2246. doi: 10.1016/j.febslet.2013.05.053. [DOI] [PubMed] [Google Scholar]
- 4.Lepper PM, Ott S, Nuesch E, von Eynatten M, Schumann C, Pletz MW, Mealing NM, Welte T, Bauer TT, Suttorp N, Juni P, Bals R, Rohde G, German Community Acquired Pneumonia Competence Network . 2012. Serum glucose levels for predicting death in patients admitted to hospital for community acquired pneumonia: prospective cohort study. BMJ 344:e3397. doi: 10.1136/bmj.e3397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rao Kondapally Seshasai S, Kaptoge S, Thompson A, Di Angelantonio E, Gao P, Sarwar N, Whincup PH, Mukamal KJ, Gillum RF, Holme I, Njolstad I, Fletcher A, Nilsson P, Lewington S, Collins R, Gudnason V, Thompson SG, Sattar N, Selvin E, Hu FB, Danesh J, Emerging Risk Factors Collaboration . 2011. Diabetes mellitus, fasting glucose, and risk of cause-specific death. N Engl J Med 364:829–841. doi: 10.1056/NEJMoa1008862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mantey I, Hill RL, Foster AV, Wilson S, Wade JJ, Edmonds ME. 2000. Infection of foot ulcers with Staphylococcus aureus associated with increased mortality in diabetic patients. Commun Dis Public Health 3:288–290. [PubMed] [Google Scholar]
- 7.Trivedi U, Parameswaran S, Armstrong A, Burgueno-Vega D, Griswold J, Dissanaike S, Rumbaugh KP. 2014. Prevalence of multiple antibiotic resistant infections in diabetic versus nondiabetic wounds. J Pathog 2014:173053. doi: 10.1155/2014/173053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lima AL, Illing T, Schliemann S, Elsner P. 2017. Cutaneous manifestations of diabetes mellitus: a review. Am J Clin Dermatol 18:541–553. doi: 10.1007/s40257-017-0275-z. [DOI] [PubMed] [Google Scholar]
- 9.Sayampanathan AA. 2016. Systematic review of complications and outcomes of diabetic patients with burn trauma. Burns 42:1644–1651. doi: 10.1016/j.burns.2016.06.023. [DOI] [PubMed] [Google Scholar]
- 10.Casqueiro J, Casqueiro J, Alves C. 2012. Infections in patients with diabetes mellitus: a review of pathogenesis. Indian J Endocrinol Metab 16(Suppl 1):S27–S36. doi: 10.4103/2230-8210.94253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. 2008. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J Infect Dis 197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- 12.McKinnell JA, Miller LG, Eells SJ, Cui E, Huang SS. 2013. A systematic literature review and meta-analysis of factors associated with methicillin-resistant Staphylococcus aureus colonization at time of hospital or intensive care unit admission. Infect Control Hosp Epidemiol 34:1077–1086. doi: 10.1086/673157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luzar MA, Coles GA, Faller B, Slingeneyer A, Dah GD, Briat C, Wone C, Knefati Y, Kessler M, Peluso F. 1990. Staphylococcus aureus nasal carriage and infection in patients on continuous ambulatory peritoneal dialysis. N Engl J Med 322:505–509. doi: 10.1056/NEJM199002223220804. [DOI] [PubMed] [Google Scholar]
- 14.Smith JA, O’Connor JJ. 1966. Nasal carriage of Staphylococcus aureus in diabetes mellitus. Lancet ii:776–777. [DOI] [PubMed] [Google Scholar]
- 15.Stanaway S, Johnson D, Moulik P, Gill G. 2007. Methicillin-resistant Staphylococcus aureus (MRSA) isolation from diabetic foot ulcers correlates with nasal MRSA carriage. Diabetes Res Clin Pract 75:47–50. doi: 10.1016/j.diabres.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Gupta K, Martinello RA, Young M, Strymish J, Cho K, Lawler E. 2013. MRSA nasal carriage patterns and the subsequent risk of conversion between patterns, infection, and death. PLoS One 8:e53674. doi: 10.1371/journal.pone.0053674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Muller LM, Gorter KJ, Hak E, Goudzwaard WL, Schellevis FG, Hoepelman AI, Rutten GE. 2005. Increased risk of common infections in patients with type 1 and type 2 diabetes mellitus. Clin Infect Dis 41:281–288. doi: 10.1086/431587. [DOI] [PubMed] [Google Scholar]
- 18.Ambrosch A, Haefner S, Jude E, Lobmann R. 2011. Diabetic foot infections: microbiological aspects, current and future antibiotic therapy focusing on methicillin-resistant Staphylococcus aureus. Int Wound J 8:567–577. doi: 10.1111/j.1742-481X.2011.00849.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klevens RM, Morrison MA, Nadle J, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Zell ER, Fosheim GE, McDougal LK, Carey RB, Fridkin SK, Active Bacterial Core Surveillance MRSA Investigators . 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- 20.Klevens RM, Morrison MA, Fridkin SK, Reingold A, Petit S, Gershman K, Ray S, Harrison LH, Lynfield R, Dumyati G, Townes JM, Craig AS, Fosheim G, McDougal LK, Tenover FC, Active Bacterial Core Surveillance of the Emerging Infections Program Network . 2006. Community-associated methicillin-resistant Staphylococcus aureus and healthcare risk factors. Emerg Infect Dis 12:1991–1993. doi: 10.3201/eid1212.060505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen H, Charlat O, Tartaglia LA, Woolf EA, Weng X, Ellis SJ, Lakey ND, Culpepper J, Moore KJ, Breitbart RE, Duyk GM, Tepper RI, Morgenstern JP. 1996. Evidence that the diabetes gene encodes the leptin receptor: identification of a mutation in the leptin receptor gene in db/db mice. Cell 84:491–495. doi: 10.1016/S0092-8674(00)81294-5. [DOI] [PubMed] [Google Scholar]
- 22.Coleman DL. 1978. Obese and diabetes: two mutant genes causing diabetes-obesity syndromes in mice. Diabetologia 14:141–148. doi: 10.1007/BF00429772. [DOI] [PubMed] [Google Scholar]
- 23.Park S, Rich J, Hanses F, Lee JC. 2009. Defects in innate immunity predispose C57BL/6J-Leprdb/Leprdb mice to infection by Staphylococcus aureus. Infect Immun 77:1008–1014. doi: 10.1128/IAI.00976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yano H, Kinoshita M, Fujino K, Nakashima M, Yamamoto Y, Miyazaki H, Hamada K, Ono S, Iwaya K, Saitoh D, Seki S, Tanaka Y. 2012. Insulin treatment directly restores neutrophil phagocytosis and bactericidal activity in diabetic mice and thereby improves surgical site Staphylococcus aureus infection. Infect Immun 80:4409–4416. doi: 10.1128/IAI.00787-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bagdade JD, Root RK, Bulger RJ. 1974. Impaired leukocyte function in patients with poorly controlled diabetes. Diabetes 23:9–15. doi: 10.2337/diab.23.1.9. [DOI] [PubMed] [Google Scholar]
- 26.Hanses F, Park S, Rich J, Lee JC. 2011. Reduced neutrophil apoptosis in diabetic mice during staphylococcal infection leads to prolonged Tnfalpha production and reduced neutrophil clearance. PLoS One 6:e23633. doi: 10.1371/journal.pone.0023633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hill HR, Augustine NH, Rallison ML, Santos JI. 1983. Defective monocyte chemotactic responses in diabetes mellitus. J Clin Immunol 3:70–77. doi: 10.1007/BF00919141. [DOI] [PubMed] [Google Scholar]
- 28.Pettersson US, Christoffersson G, Massena S, Ahl D, Jansson L, Henriksnas J, Phillipson M. 2011. Increased recruitment but impaired function of leukocytes during inflammation in mouse models of type 1 and type 2 diabetes. PLoS One 6:e22480. doi: 10.1371/journal.pone.0022480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Repine JE, Clawson CC, Goetz FC. 1980. Bactericidal function of neutrophils from patients with acute bacterial infections and from diabetics. J Infect Dis 142:869–875. doi: 10.1093/infdis/142.6.869. [DOI] [PubMed] [Google Scholar]
- 30.Grishman EK, White PC, Savani RC. 2012. Toll-like receptors, the NLRP3 inflammasome, and interleukin-1beta in the development and progression of type 1 diabetes. Pediatr Res 71:626–632. doi: 10.1038/pr.2012.24. [DOI] [PubMed] [Google Scholar]
- 31.Wickersham M, Wachtel S, Wong Fok Lung T, Soong G, Jacquet R, Richardson A, Parker D, Prince A. 2017. Metabolic stress drives keratinocyte defenses against Staphylococcus aureus infection. Cell Rep 18:2742–2751. doi: 10.1016/j.celrep.2017.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sousa S, Cabanes D, Archambaud C, Colland F, Lemichez E, Popoff M, Boisson-Dupuis S, Gouin E, Lecuit M, Legrain P, Cossart P. 2005. ARHGAP10 is necessary for alpha-catenin recruitment at adherens junctions and for Listeria invasion. Nat Cell Biol 7:954–960. doi: 10.1038/ncb1308. [DOI] [PubMed] [Google Scholar]
- 33.Basseres DS, Tizzei EV, Duarte AA, Costa FF, Saad ST. 2002. ARHGAP10, a novel human gene coding for a potentially cytoskeletal Rho-GTPase activating protein. Biochem Biophys Res Commun 294:579–585. doi: 10.1016/S0006-291X(02)00514-4. [DOI] [PubMed] [Google Scholar]
- 34.Crosbie RH, Heighway J, Venzke DP, Lee JC, Campbell KP. 1997. Sarcospan, the 25-kDa transmembrane component of the dystrophin-glycoprotein complex. J Biol Chem 272:31221–31224. doi: 10.1074/jbc.272.50.31221. [DOI] [PubMed] [Google Scholar]
- 35.Kwon M, Hanna E, Lorang D, He M, Quick JS, Adem A, Stevenson C, Chung JY, Hewitt SM, Zudaire E, Esposito D, Cuttitta F, Libutti SK. 2008. Functional characterization of filamin a interacting protein 1-like, a novel candidate for antivascular cancer therapy. Cancer Res 68:7332–7341. doi: 10.1158/0008-5472.CAN-08-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, Chamberlain NR. 2007. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology 153:3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- 37.Chatterjee I, Becker P, Grundmeier M, Bischoff M, Somerville GA, Peters G, Sinha B, Harraghy N, Proctor RA, Herrmann M. 2005. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J Bacteriol 187:4488–4496. doi: 10.1128/JB.187.13.4488-4496.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Frees D, Gerth U, Ingmer H. 2014. Clp chaperones and proteases are central in stress survival, virulence and antibiotic resistance of Staphylococcus aureus. Int J Med Microbiol 304:142–149. doi: 10.1016/j.ijmm.2013.11.009. [DOI] [PubMed] [Google Scholar]
- 39.Liu Q, Wang X, Qin J, Cheng S, Yeo WS, He L, Ma X, Liu X, Li M, Bae T. 2017. The ATP-dependent protease ClpP inhibits biofilm formation by regulating Agr and cell wall hydrolase Sle1 in Staphylococcus aureus. Front Cell Infect Microbiol 7:181. doi: 10.3389/fcimb.2017.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Frees D, Qazi SN, Hill PJ, Ingmer H. 2003. Alternative roles of ClpX and ClpP in Staphylococcus aureus stress tolerance and virulence. Mol Microbiol 48:1565–1578. doi: 10.1046/j.1365-2958.2003.03524.x. [DOI] [PubMed] [Google Scholar]
- 41.Michel A, Agerer F, Hauck CR, Herrmann M, Ullrich J, Hacker J, Ohlsen K. 2006. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J Bacteriol 188:5783–5796. doi: 10.1128/JB.00074-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baerends RJ, Smits WK, de Jong A, Hamoen LW, Kok J, Kuipers OP. 2004. Genome2D: a visualization tool for the rapid analysis of bacterial transcriptome data. Genome Biol 5:R37. doi: 10.1186/gb-2004-5-5-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duncan JL, Cho GJ. 1972. Production of staphylococcal alpha toxin. II. Glucose repression of toxin formation. Infect Immun 6:689–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stahlhut SG, Alqarzaee AA, Jensen C, Fisker NS, Pereira AR, Pinho MG, Thomas VC, Frees D. 2017. The ClpXP protease is dispensable for degradation of unfolded proteins in Staphylococcus aureus. Sci Rep 7:11739. doi: 10.1038/s41598-017-12122-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kennedy AD, Bubeck Wardenburg J, Gardner DJ, Long D, Whitney AR, Braughton KR, Schneewind O, DeLeo FR. 2010. Targeting of alpha-hemolysin by active or passive immunization decreases severity of USA300 skin infection in a mouse model. J Infect Dis 202:1050–1058. doi: 10.1086/656043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel AH, Nowlan P, Weavers ED, Foster T. 1987. Virulence of protein A-deficient and alpha-toxin-deficient mutants of Staphylococcus aureus isolated by allele replacement. Infect Immun 55:3103–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Eleftheriadou I, Tentolouris N, Argiana V, Jude E, Boulton AJ. 2010. Methicillin-resistant Staphylococcus aureus in diabetic foot infections. Drugs 70:1785–1797. doi: 10.2165/11538070-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 48.Hirsch T, Spielmann M, Zuhaili B, Koehler T, Fossum M, Steinau HU, Yao F, Steinstraesser L, Onderdonk AB, Eriksson E. 2008. Enhanced susceptibility to infections in a diabetic wound healing model. BMC Surg 8:5. doi: 10.1186/1471-2482-8-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rich J, Lee JC. 2005. The pathogenesis of Staphylococcus aureus infection in the diabetic NOD mouse. Diabetes 54:2904–2910. doi: 10.2337/diabetes.54.10.2904. [DOI] [PubMed] [Google Scholar]
- 50.Lovati AB, Drago L, Monti L, De Vecchi E, Previdi S, Banfi G, Romanò CL. 2013. Diabetic mouse model of orthopaedic implant-related Staphylococcus aureus infection. PLoS One 8:e67628. doi: 10.1371/journal.pone.0067628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pongcharoen S, Chansantor W, Supalap K, Jienmongkol P, Niumsup PR. 2011. Impaired interleukin-1beta expression by monocytes stimulated with Staphylococcus aureus in diabetes. Southeast Asian J Trop Med Public Health 42:1197–1203. [PubMed] [Google Scholar]
- 52.Hanses F, Roux C, Dunman PM, Salzberger B, Lee JC. 2014. Staphylococcus aureus gene expression in a rat model of infective endocarditis. Genome Med 6:93. doi: 10.1186/s13073-014-0093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frees D, Chastanet A, Qazi S, Sorensen K, Hill P, Msadek T, Ingmer H. 2004. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol Microbiol 54:1445–1462. doi: 10.1111/j.1365-2958.2004.04368.x. [DOI] [PubMed] [Google Scholar]
- 54.Mei JM, Nourbakhsh F, Ford CW, Holden DW. 1997. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol 26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 55.Farrand AJ, Reniere ML, Ingmer H, Frees D, Skaar EP. 2013. Regulation of host hemoglobin binding by the Staphylococcus aureus Clp proteolytic system. J Bacteriol 195:5041–5050. doi: 10.1128/JB.00505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Graham JW, Lei MG, Lee CY. 2013. Trapping and identification of cellular substrates of the Staphylococcus aureus ClpC chaperone. J Bacteriol 195:4506–4516. doi: 10.1128/JB.00758-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Frees D, Andersen JH, Hemmingsen L, Koskenniemi K, Bæk KT, Muhammed MK, Gudeta DD, Nyman TA, Sukura A, Varmanen P, Savijoki K. 2012. New insights into Staphylococcus aureus stress tolerance and virulence regulation from an analysis of the role of the ClpP protease in the strains Newman, COL, and SA564. J Proteome Res 11:95–108. doi: 10.1021/pr200956s. [DOI] [PubMed] [Google Scholar]
- 58.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi MR. 1994. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc Natl Acad Sci U S A 91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubern JF, Lagendijk EL, Lugtenberg BJ, Bloemberg GV. 2005. The heat shock genes dnaK, dnaJ, and grpE are involved in regulation of putisolvin biosynthesis in Pseudomonas putida PCL1445. J Bacteriol 187:5967–5976. doi: 10.1128/JB.187.17.5967-5976.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singh VK, Syring M, Singh A, Singhal K, Dalecki A, Johansson T. 2012. An insight into the significance of the DnaK heat shock system in Staphylococcus aureus. Int J Med Microbiol 302:242–252. doi: 10.1016/j.ijmm.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Abdeen S, Salim N, Mammadova N, Summers CM, Frankson R, Ambrose AJ, Anderson GG, Schultz PG, Horwich AL, Chapman E, Johnson SM. 2016. GroEL/ES inhibitors as potential antibiotics. Bioorg Med Chem Lett 26:3127–3134. doi: 10.1016/j.bmcl.2016.04.089. [DOI] [PubMed] [Google Scholar]
- 62.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bæk KT, Gründling A, Mogensen RG, Thøgersen L, Petersen A, Paulander W, Frees D. 2014. β-Lactam resistance in methicillin-resistant Staphylococcus aureus USA300 is increased by inactivation of the ClpXP protease. Antimicrob Agents Chemother 58:4593–4603. doi: 10.1128/AAC.02802-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Frazier WC. 1926. A method for the detection of changes in gelatin due to bacteria. J Infect Dis 39:302–309. doi: 10.1093/infdis/39.4.302. [DOI] [Google Scholar]
- 65.Chen J, Yoong P, Ram G, Torres VJ, Novick RP. 2014. Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid 76:1–7. doi: 10.1016/j.plasmid.2014.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohen SN, Chang AC. 1977. Revised interpretation of the origin of the pSC101 plasmid. J Bacteriol 132:734–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Motyl K, McCabe LR. 2009. Streptozotocin, type I diabetes severity and bone. Biol Proced Online 11:296–315. doi: 10.1007/s12575-009-9000-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parker D. 2018. CD80/CD86 signaling contributes to the proinflammatory response of Staphylococcus aureus in the airway. Cytokine 107:130–136. doi: 10.1016/j.cyto.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.National Research Council. 2011. Guide for the care and use of laboratory animals, 8th ed National Academies Press, Washington, DC. [Google Scholar]
- 70.Parker D, Martin FJ, Soong G, Harfenist BS, Aguilar JL, Ratner AJ, Fitzgerald KA, Schindler C, Prince A. 2011. Streptococcus pneumoniae DNA initiates type I interferon signaling in the respiratory tract. mBio 2:e00016-11. doi: 10.1128/mBio.00016-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bent ZW, Poorey K, LaBauve AE, Hamblin R, Williams KP, Meagher RJ. 2016. A rapid spin column-based method to enrich pathogen transcripts from eukaryotic host cells prior to sequencing. PLoS One 11:e0168788. doi: 10.1371/journal.pone.0168788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Minato Y, Ghosh A, Faulkner WJ, Lind EJ, Schesser Bartra S, Plano GV, Jarrett CO, Hinnebusch BJ, Winogrodzki J, Dibrov P, Häse CC. 2013. Na+/H+ antiport is essential for Yersinia pestis virulence. Infect Immun 81:3163–3172. doi: 10.1128/IAI.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Langevin SA, Bent ZW, Solberg OD, Curtis DJ, Lane PD, Williams KP, Schoeniger JS, Sinha A, Lane TW, Branda SS. 2013. Peregrine: a rapid and unbiased method to produce strand-specific RNA-Seq libraries from small quantities of starting material. RNA Biol 10:502–515. doi: 10.4161/rna.24284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bent ZW, Poorey K, Brazel DM, LaBauve AE, Sinha A, Curtis DJ, House SE, Tew KE, Hamblin RY, Williams KP, Branda SS, Young GM, Meagher RJ. 2015. Transcriptomic analysis of Yersinia enterocolitica biovar 1B infecting murine macrophages reveals new mechanisms of extracellular and intracellular survival. Infect Immun 83:2672–2685. doi: 10.1128/IAI.02922-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vandernoot VA, Langevin SA, Solberg OD, Lane PD, Curtis DJ, Bent ZW, Williams KP, Patel KD, Schoeniger JS, Branda SS, Lane TW. 2012. cDNA normalization by hydroxyapatite chromatography to enrich transcriptome diversity in RNA-seq applications. Biotechniques 53:373–380. doi: 10.2144/000113937. [DOI] [PubMed] [Google Scholar]
- 76.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. 2011. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol 12:R22. doi: 10.1186/gb-2011-12-3-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 80.Law CW, Alhamdoosh M, Su S, Smyth GK, Ritchie ME. 2016. RNA-seq analysis is easy as 1-2-3 with limma, Glimma and edgeR. F1000Res 5:1408. doi: 10.12688/f1000research.9005.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.