Precise enumeration of living intracellular bacteria is the key step to estimate the invasion potential of pathogens and host immune responses to understand the mechanism and kinetics of bacterial pathogenesis. Therefore, quantitative assessment of host-pathogen interactions is essential for development of novel antibacterial therapeutics for infectious disease.
KEYWORDS: gentamicin protection assay, Staphylococcus aureus, bacteria, enzyme protection assay, host, lysostaphin
ABSTRACT
Precise enumeration of living intracellular bacteria is the key step to estimate the invasion potential of pathogens and host immune responses to understand the mechanism and kinetics of bacterial pathogenesis. Therefore, quantitative assessment of host-pathogen interactions is essential for development of novel antibacterial therapeutics for infectious disease. The gentamicin protection assay (GPA) is the most widely used method for these estimations by counting the CFU of intracellular living pathogens. Here, we assess the longstanding drawbacks of the GPA by employing an antistaphylococcal endopeptidase as a bactericidal agent to kill extracellular Staphylococcus aureus. We found that the difference between the two methods for the recovery of intracellular CFU of S. aureus was about 5 times. We prove that the accurate number of intracellular CFU could not be precisely determined by the GPA due to the internalization of gentamicin into host cells during extracellular bacterial killing. We further demonstrate that lysostaphin-mediated extracellular bacterial clearance has advantages for measuring the kinetics of bacterial internalization on a minute time scale due to the fast and tunable activity and the inability of protein to permeate the host cell membrane. From these results, we propose that accurate quantification of intracellular bacteria and measurement of internalization kinetics can be achieved by employing enzyme-mediated killing of extracellular bacteria (enzyme protection assay [EPA]) rather than the host-permeative drug gentamicin, which is known to alter host physiology.
INTRODUCTION
Host-pathogen interactions are steered by complicated defense and offense mechanisms between hosts and pathogens (1, 2). Professional phagocytes of the innate immune system, such as neutrophils and macrophages, play a key role in frontline defense against pathogens by engulfing them via phagocytosis and destroying them intracellularly (1, 3). Conversely, bacteria can also invade and/or induce their own internalization by a zipper or trigger mechanism for establishing a protected niche for survival (4). Intracellular pathogenic bacteria such as Mycobacterium tuberculosis (5), Salmonella enterica serovar Typhimurium (5), Listeria monocytogenes (4), and pathogenic Escherichia coli (6) enter and replicate within host cells. However, pathogens such as Helicobacter pylori (5) and Staphylococcus aureus (7) also invade and survive inside host cells, although they are known to be extracellular bacteria, which facilitate persistence and recurrence (8, 9). A small number of persistent intracellular bacteria can remain dormant as intracellular bacterial communities (IBCs), and thus, IBCs are difficult to treat with drugs. This very small population of IBCs is the primary cause of recurrent infections and chronic disease (5–7). However, the mechanisms of internalization, the persistence of pathogens, and the intracellular killing of pathogens by professional or nonprofessional phagocytes are not yet fully understood (9, 10). To investigate these processes, it is necessary to precisely quantify the internalized bacteria in infected host cells. The enumeration of intracellular living bacteria is further required for a systematic and comprehensive understanding of host-pathogen interactions during the innate immune response, for estimating bacterial virulence potential, and for evaluating the efficacy of new antibiotics.
There are several direct methods to measure the intracellular bacterial population, such as fluorescence-activated cell sorter (FACS) analysis and various microscopic techniques (11, 12). However, as there is a possibility that dead bacteria, or physiologically unfit and compromised bacteria, which hence are highly vulnerable to death, may also be counted. Therefore, direct counting methods, in many instances, are not considered reliable for the assessment of host-pathogen interactions or for the determination of the actual number of surviving intracellular bacteria. The most widely used enumeration method for intracellular living bacteria is a gentamicin protection assay (GPA) (13), in which CFU of bacteria infecting host cells are counted after killing extracellular bacteria with gentamicin (14, 15). The GPA relies on the ability of gentamicin to kill all extracellular and membrane-bound bacteria and is also based on the assumption of the inability of gentamicin to penetrate eukaryotic cells (16). However, many reports suspected that higher concentrations of gentamicin for long incubation times possibly cause the nonspecific killing of intracellular bacteria (13, 17–20), presumably by internalized gentamicin through pinocytosis (21). For this reason, the results of the GPA often exhibit significant variation (22).
To our knowledge, there are no reports that have quantitatively shown the internalization of gentamicin and the adverse effect on measuring the invasion potential and enumeration of surviving intracellular bacteria. Furthermore, precise kinetic measurement of bacterial internalization during bacterial invasion or host cell phagocytosis is hindered by the GPA, since the elimination of extracellular pathogens by gentamicin takes hours due to slow killing kinetics (20). Therefore, the widely used antibiotic protection assays, including GPA (23, 24), in basic host-pathogen interactions and clinical research need to be revisited. Alternatively, bacteriolytic enzymes, such as lysozyme and mutanolysin, have been introduced in enzyme-based protection assays (25). The moderate enzymatic killing activity under physiological conditions as well as the requirement for a specific pH and high temperature for optimal activity (26) have presumably limited their usage in protection assays. Among the bacteriolytic enzymes, lysostaphin has been successfully used in protection assays, mostly in combination with gentamicin, for the specific and efficient killing of extracellular and host cell surface-bound S. aureus (27–29). However, to the best of our knowledge, the exclusive use of lysostaphin for enumeration of intracellular S. aureus bacteria is rare, and comparative analyses of the GPA and EPA (enzyme protection assay) have not been performed.
In this study, we assessed the internalization of S. aureus USA300 strain FPR3757 (here referred to as S. aureus) in mouse macrophage cells (RAW264.7) and human embryonic kidney cells (HEK293) by applying the GPA and EPA to evaluate the drawbacks of the GPA and advantages associated with the EPA. S. aureus, a human pathogen that causes mild to life-threatening infections and possesses intracellular persistence properties (3, 30, 31), was chosen since it is one of the most important and widely studied pathogens due to its multiple-drug-resistance patterns and relevance to many human diseases (32, 33). The present study reveals that a lysostaphin-based EPA has advantages; this method has fast killing kinetics, tunability by quenching of lysostaphin activity, and negligible diffusion into live host cells. Thus, the application repertoire of the EPA can be expanded to the precise measurement of invasion potential, internalization kinetics, and intracellular killing kinetics of S. aureus on a minute time scale, which cannot be achieved when the antibiotic protection assay is applied.
RESULTS AND DISCUSSION
S. aureus infection of mammalian cells using gentamicin and enzyme protection assays.
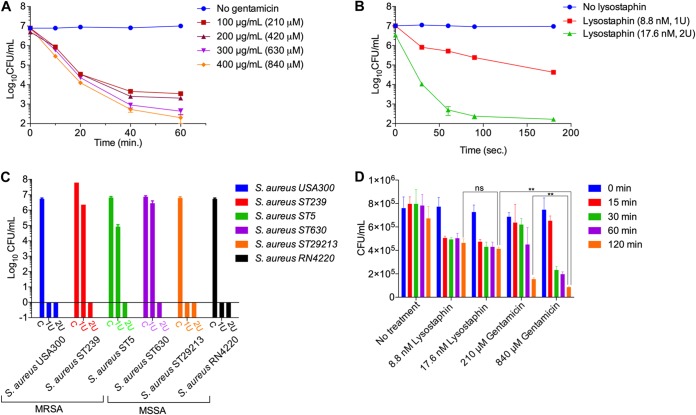
Lysostaphin kills S. aureus by hydrolyzing the polyglycine bridges that cross-link glycopeptide chains in the peptidoglycan of the S. aureus cell wall (34). By microscopic observations and measuring the CFU of S. aureus after treatment with gentamicin or lysostaphin at various concentrations and incubation times, we observed that 99.999% (5 log10 CFU difference) killing of S. aureus was achieved via treatment with gentamicin at 400 μg/ml (840 μM) for 60 min of incubation (Fig. 1A; see also Fig. S1A in the supplemental material). In contrast, 17.6 nM (2 U) lysostaphin was required to kill the same number of S. aureus cells in only 180 s (Fig. 1B and Fig. S1B). Considering that high-cell-density conditions are more likely to occur when in vitro infection experiments are conducted at a high multiplicity of infection (MOI), we investigated the killing efficiency of lysostaphin for high-cell-density S. aureus (optical density at 600 nm [OD600] of 1, or ∼1.0 × 109 cells per ml) by treatment with various concentrations of lysostaphin with a fixed incubation time of 180 s. The result showed that bacterial cells were eradicated by 4 U of lysostaphin (Fig. S2). These results suggest that lysostaphin has a much higher killing efficiency and faster killing kinetics than gentamicin.
FIG 1.
Comparison between the gentamicin protection assay (GPA) and the lysostaphin-mediated enzyme protection assay (EPA). (A) Quantitative assessment of gentamicin concentration and time-dependent killing efficiency for S. aureus cells at various concentrations from 100 μg/ml (210 μM) to 400 μg/ml (840 μM). Gentamicin at a concentration of 400 μg/ml (840 μM) could kill 99.999% of S. aureus cells within 60 min. (B) Concentration and time-dependent killing of S. aureus cells using lysostaphin. One unit (8.8 nM) and 2 U (17.6 nM) of lysostaphin were incubated with 1 × 107 S. aureus cells in 1 ml DMEM containing 10% FBS in a 5% CO2 environment at 37°C. Nearly all (99.999%) the S. aureus cells were killed within 180 s with 2 U lysostaphin. (C) Killing efficiency of lysostaphin against various methicillin-sensitive S. aureus (MSSA) (ST630, RN4220, and MSSA29213) and methicillin-resistant S. aureus (MRSA) (ST5, ST239, and USA300) strains. Two units of lysostaphin eradicated all the tested clinical methicillin-sensitive and methicillin-resistant strains of S. aureus. C, control. (D) Concentration and time-dependent killing of S. aureus in host cells. After infection of RAW264.7 cells (1 × 106) by S. aureus cells (1 × 107) for 30 min, gentamicin (210 μM and 840 μM) or lysostaphin (1 and 2 U) was applied to the host-pathogen mixture for 15, 30, 60, and 120 min. After rigorous washing using PBS to remove gentamicin or EDTA-quenched lysostaphin in the infection medium, the host cells were lysed to release internalized bacteria, followed by serial dilution and plating to count CFU of intracellular S. aureus. The GPA killed a significant number of intracellular S. aureus bacteria compared to the EPA. The intracellular killing of S. aureus during the GPA versus the EPA was analyzed using Student’s t tests. ns, nonsignificant; **, P < 0.01.
To test whether lysostaphin can be applied to enumerate internalized S. aureus bacteria by the EPA, we examined the killing efficiency of lysostaphin against various methicillin-sensitive S. aureus strains (ST630, RN4220, and MSSA29213) and methicillin-resistant S. aureus strains (ST5, ST239, and USA300) (35–37). All the tested strains were found to be susceptible to lysostaphin, and CFU could not be recovered when strains were incubated for 5 min with 2 U of lysostaphin (Fig. 1C). However, the possibility of the presence or occurrence of lysostaphin-resistant S. aureus cannot be ruled out entirely based on our present experiments, as a lysostaphin-resistant S. aureus mutant with increased lysostaphin resistance has been reported (38).
We next compared the numbers of bacteria internalized into host cells using the EPA and the GPA (Fig. 1D). In each assay, RAW264.7 cells infected with S. aureus for 30 min were treated with bactericidal reagents at optimized concentrations for various time periods. Lysostaphin was promptly deactivated, removed, and washed using cold phosphate-buffered saline (PBS) (pH 7.4), while gentamicin was removed by aspiration and repeated washing using cold PBS. After removal of gentamicin and quenching of lysostaphin, CFU of S. aureus were measured using lysates of infected RAW264.7 cells. From this analysis, we found that the CFU count reached a near-constant value within 15 min for the EPA, while the CFU value continued to decrease with increasing incubation times of gentamicin treatment during the GPA. Moreover, the CFU count for the EPA was about 5 times higher than that for the GPA. The discrepancies in CFU recoveries between the two methods were within a range of 1 log10 value, which primarily depends upon the removal of bacteria, subsequent washing after infection, and the duration and concentration of gentamicin used for killing extracellular bacteria (Fig. 1D).
Visualization of intracellular killing of bacteria during the GPA.
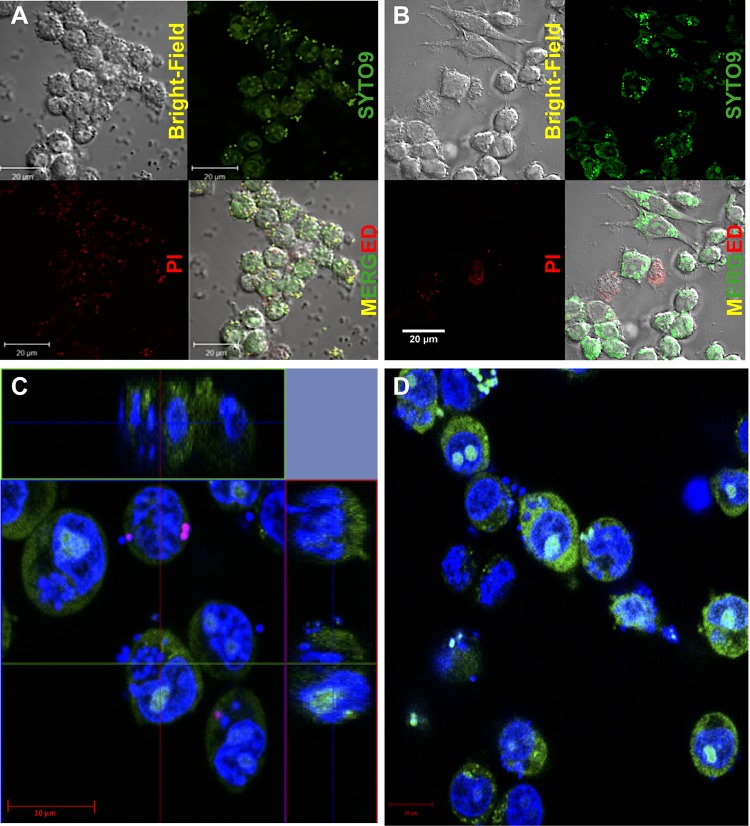
To investigate what caused the discrepancy in CFU recovery between the EPA and GPA, we monitored the internalization of S. aureus into RAW264.7 cells using confocal microscopy after gentamicin (Fig. 2A) and lysostaphin (Fig. 2B) treatments. To visualize the bacteria, we introduced the BacLight cell viability staining assay, in which total bacteria were stained by green fluorescence emitted from SYTO9 bound to nucleic acids, and dead bacteria with membrane damage were visualized by red or orange fluorescence generated by the binding of internalized propidium iodide (PI) to DNA. When the GPA was applied, green dots, representing the total bacteria, were observed on the host cell surface or inside the membrane (Fig. 2A). The red or orange dots, representing dead S. aureus cells with membrane damage, were also found both on the host cell membrane and within host cells (Fig. 2A). However, when the EPA was applied, there were no red dots but only green ones found inside living host cells (Fig. 2B). Red dots were found inside a few host cells that seemed to have lost cell integrity, possibly due to cell death. To validate the localization of dead S. aureus bacteria by a quantitative analysis of two-dimensional images, we measured the fluorescence intensity of the red dots as ∼210 ± 20 arbitrary units (AU) and ∼120 ± 20 AU for dead S. aureus cells present extracellularly and putative S. aureus cells inside the host cells, respectively. This analysis supports that dead S. aureus cells were found inside the host cells in the GPA (Fig. S3) but not inside the host cells in the EPA (Fig. S4).
FIG 2.
Visualization of intracellular killing of S. aureus during the GPA and EPA. The Live/Dead BacLight bacterial viability kit (catalog no. L7007) was used for the qualitative assessment of dead bacteria with SYTO9 (green fluorescence) and propidium iodide (PI) (red fluorescence), which stain all cells and membrane-damaged cells, respectively, and thus, all bacteria are shown in green, while only membrane-damaged bacterial cells are indicated by red dots. (A and B) Confocal images of RAW264.7 cells infected with S. aureus after GPA-mediated (A) and EPA-mediated (B) eradication of extracellular S. aureus showing that red dots are not found in lysostaphin-treated cells (B) but are found in gentamicin-treated cells (A), indicating the presence of intracellular killing of S. aureus during the GPA. (C and D) Central z-stack images of the host-pathogen complex (RAW264.7 cells and S. aureus) with gentamicin killing for 2 h (C) and the without-gentamicin-killing control (D) showing that red cells are found only under conditions of gentamicin treatment, while green cells are found under both conditions. These images indicate that gentamicin causes intracellular killing of S. aureus.
To further evaluate and confirm the localization of dead S. aureus cells, phagocytic RAW264.7 cells infected with S. aureus and subsequently treated with gentamicin were subjected to z-stack live-cell imaging by confocal microscopy (Fig. 2C and Fig. S5A). The RAW264.7-S. aureus (host-pathogen) complex after infection but without gentamicin treatment was used as a control (Fig. 2D). The z-stack imaging and analysis of the center stack showed a significant number of dead S. aureus bacteria inside RAW264.7 cells (Fig. 2C and Fig. S5A) as red dots, compared to the number detected when the same assay was conducted using cells without gentamicin treatment (Fig. 2D). To further evaluate whether the GPA method is suitable for monitoring intracellular bacteria, we repeated the same experiments with nonphagocytic human embryonic kidney cells, HEK293 cells (Fig. S5B and C). We were able to confirm the presence of dead S. aureus bacteria inside HEK293 host cells under GPA conditions (Fig. S5C and S5C″), compared to the HEK293-S. aureus (host-pathogen) complex control without gentamicin treatment (Fig. S5B and S5B′). These results conclusively demonstrate that intracellular S. aureus could be killed during the GPA.
Assessment of gentamicin internalization in host cells.
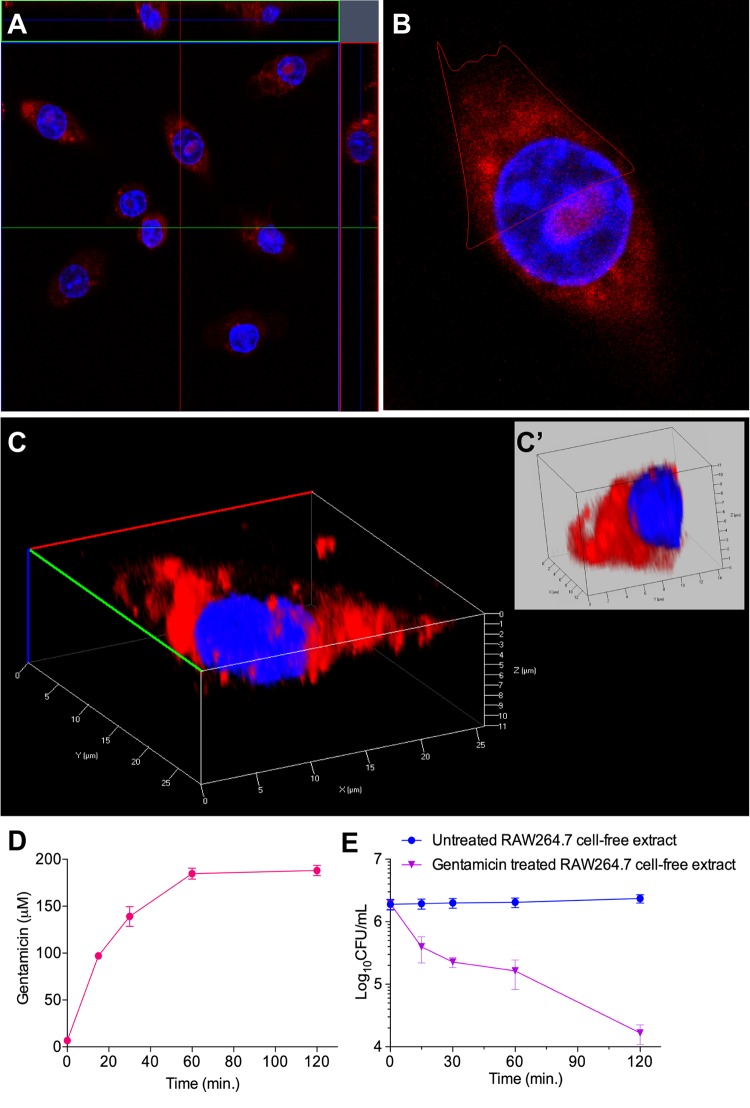
The presence of a significant number of dead bacteria inside the host cells during the GPA (Fig. 1D, Fig. 2, and Fig. S3 and S5) suggests the possibility of intracellular S. aureus being killed by the internalized gentamicin. Consistent with our results, the possibility of gentamicin being internalized into host cells during the GPA was questioned previously (13, 17–20). Therefore, we hypothesized that the effect of internalized gentamicin on intracellular bacteria might be the main cause for the discrepancy between the EPA and the GPA. To test this possibility, we first examined the entry of gentamicin into host cells by confocal microscopy after treatment of RAW264.7 and HEK293 cells with gentamicin labeled with a monoisomer of Texas Red-X succinimidyl ester (TR) (Fig. S6). The z-stack confocal images revealed that gentamicin-TR (GTTR) was internalized into both RAW264.7 (Fig. 3A to C) and HEK293 (Fig. S7A to S7D) cells. Furthermore, three-dimensional (3D) image analysis of single cells also clearly showed localization of gentamicin-TR in the host cell cytoplasm near the nucleus for both RAW264.7 (Fig. 3C and C′) and HEK293 (Fig. S7C and S7D) cells.
FIG 3.
Qualitative and quantitative assessment of internalization of gentamicin. (A) Central z-stack image of RAW264.7 cells indicating the internalization of gentamicin conjugated with Texas Red (GTTR). RAW264.7 cells seeded for 24 h were stained with 20 μg/ml Hoechst 33258 (blue) and 50 μg/ml GTTR (red) at 37°C for 1 h. (B) Enlarged single image of the image in panel A. (C) Three-dimensional image of the cell in panel B. (C′) Dissected section of the single cell marked in panel B. (D) Time-dependent internalization of gentamicin in host cells. RAW264.7 cells were treated with gentamicin at a concentration of 840 μM for various time periods (0, 15, 30, 60, and 120 min) in six-well microtiter plates. The host cells were then harvested and intensively washed to avoid gentamicin contamination. The concentration of intracellular gentamicin was calculated at each time point and plotted. (E) Antibacterial activity of gentamicin-treated host cells. RAW264.7 cells were treated with gentamicin (840 μM) for various time periods. The host cells were harvested and intensively washed to avoid gentamicin contamination. The host cell lysates were applied to S. aureus cells to investigate the antibacterial activity of internalized gentamicin. RAW264.7 cells treated with water at an equivalent volume were used as a control for each time point. CFU count is decreased up to 100-fold in gentamicin-treated host cell lysate compared to its corresponding control.
The concentration of gentamicin inside RAW264.7 cells without S. aureus infection was measured by an enzyme-linked immunosorbent assay (ELISA) using an antigentamicin antibody after treating cells with 400 μg/ml (840 μM) gentamicin in the growth medium at various time points (Fig. 3D). RAW264.7 cells without gentamicin treatment were used as a control. To eliminate the chance of measuring any residual extracellular gentamicin, the gentamicin-treated host cells were washed four times with PBS. Through this approach, we confirmed that the concentration of intracellular gentamicin increased in proportion to the incubation time and reached a maximum of 89.6 μg/ml (188 μM), which corresponds to 22.4% of the extracellular gentamicin, at 60 min (Fig. 3D). Although the concentration of intracellular gentamicin in RAW264.7 cells appeared lower than the extracellular concentration, this concentration of gentamicin could nevertheless potentially affect the survival of internalized pathogens. To corroborate this result, we evaluated the killing effect of 89.6 μg/ml gentamicin on S. aureus by incubating gentamicin with log-phase-grown S. aureus cells (OD600 of 0.01, equivalent to 1.0 × 107 cells) for 2 h under shaking culture conditions. The viable cell count of S. aureus was significantly decreased by ∼3 log10 CFU in Dulbecco’s modified Eagle’s medium (DMEM) devoid of fetal bovine serum (FBS) (Fig. S8). Next, to eliminate the possibility of the inactivation of the internalized gentamicin, we tested the antibiotic activity of the internalized gentamicin. First, we pretreated RAW264.7 cells with 400 μg/ml of gentamicin and without gentamicin (control; treated with an equivalent volume of water) for various periods of time (0, 15, 30, 60, and 120 min) in six-well plates and washed the cells with PBS four times to remove any residual extracellular gentamicin. The cell lysates of control and gentamicin-treated RAW264.7 cells were exposed to S. aureus for 2 h. We observed that the CFU count decreased by up to 100-fold (2 log10 CFU) when the gentamicin-treated cell lysates were incubated with S. aureus, compared to the control lysates (Fig. 3E). These results together demonstrated that internalized gentamicin has the potency to kill intracellular S. aureus, and thus, the CFU count of internalized bacteria can be affected by gentamicin treatment during the GPA.
Assessment of permeation of lysostaphin into the host cell.
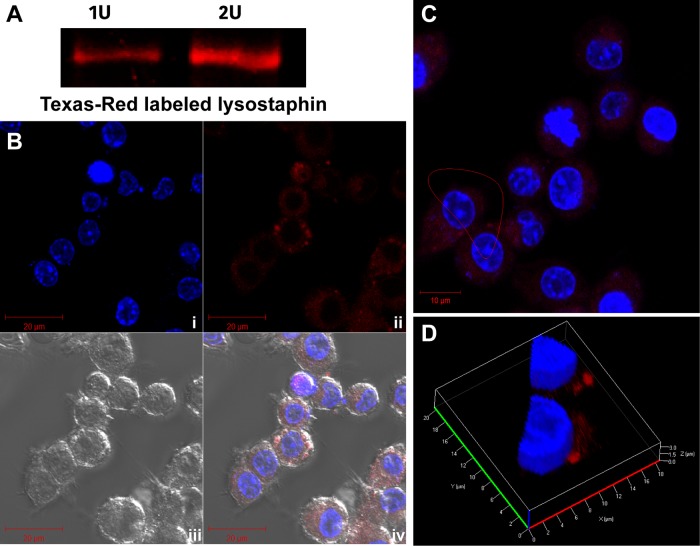
To visualize lysostaphin permeation, lysostaphin was conjugated with TR. The fluorescence of the lysostaphin-TR conjugate was confirmed by SDS-PAGE (Fig. 4A). In addition, lysostaphin was also labeled with fluorescein isothiocyanate (FITC) for testing lysostaphin permeation (Fig. S9). FITC-labeled lysostaphin has an advantage for assessing the internalization of lysostaphin in live versus dead host cells, which could not be achieved using lysostaphin-TR, owing to the overlap of the red fluorescence signal of PI and Texas Red.
FIG 4.
Assessment of lysostaphin internalization into the host cell. (A) Red fluorescence signals (excitation/emission wavelength of 588/615 nm) of 1 U (5 μl; 8.8 nM) and 2 U (10 μl; 17.6 nM) lysostaphin-TR on SDS-PAGE gels. (B) RAW264.7 cells imaged by confocal microscopy shown in split channels, with (i) host cell nucleus stained with 20 μg/ml Hoechst 33258 (blue), (ii) host cells stained with 2 U of lysostaphin-TR (red), (iii) bright-field images, and (iv) merged image of bright-field, blue, and red signals. (C and D) Image in panel B enlarged in a z-stack (C) and analyzed in a 3D dissected section (D), confirming that the red signal from lysostaphin-TR was located on the cell surface and not inside the host cells.
Lysostaphin-TR (2 U) was added to RAW264.7 cells for 30 min, along with Hoechst 33258 (2 μl of 10 mg/ml) for nucleus staining. After 30 min of incubation, excess lysostaphin-TR and Hoechst 33258 were removed, and cells were washed using PBS. The washed cells were fixed with 4% paraformaldehyde, followed by PBS washing three times (2 ml each). The confocal images of lysostaphin-TR-treated RAW264.7 cells in z-stack mode proved that lysostaphin could not be detected inside the host cells but rather were mostly localized on the host cell surface (Fig. 4B and C); the conceivable reason behind this may be that protein macromolecules cannot passively enter the live cell membrane (39). The enlarged three-dimensional dissected images also showed that the lysostaphin-TR signal was not found in the cytoplasm but was found only on the host cell surface (Fig. 4D). Next, we performed an experiment to detect the internalization of lysostaphin-FITC. After briefly exposing FITC-lysostaphin to host cells (10 min), the host cells were imaged either without fixation (live-cell imaging) (Fig. S9A) or with immediate fixation after incubation using 4% paraformaldehyde (Fig. S9B). The fixation of host cells was necessary to cease the progression of the endosomal network toward acidification, which may cause the quenching of pH-sensitive FITC fluorescence. In both live- and fixed-cell images, the fluorescence signal from FITC-labeled lysostaphin was observed only in the extracellular milieu and not inside cells (Fig. S9A and S9B). It is noteworthy that FITC fluorescence was detected inside cells that were also stained by PI. Considering the fact that PI can stain only dead cells, FITC-labeled lysostaphin was detected only in the cytoplasm of dead host cells (Fig. S9A). To further support our observation, FITC-labeled lysostaphin was applied to RAW264.7 cells for 30 min at 37°C with 5% CO2, and each cell fraction was collected for SDS-PAGE analysis, followed by highly sensitive fluorescence imaging. With this approach, it was consistently confirmed that FITC-lysostaphin was observed only in the extracellular fraction and not in the intracellular fraction (Fig. S9C).
Although we could not detect lysostaphin-TR and lysostaphin-FITC conjugates under our experimental conditions, we cannot completely rule out the possibility of the internalization of lysostaphin inside host cells. Alternatively, it is also the case that internalized lysostaphin that is present at concentrations lower than the detection limit or that has degraded may not have any practical impact on the intracellular CFU. For instance, a fraction of lysostaphin internalized into host cells through pinocytosis, or any uncharacterized internalization mechanism, is highly likely to be degraded/modified by host cells. To validate this possibility, we tested the killing efficiency of host cell-internalized lysostaphin by treating S. aureus with cell lysates, which contain supposedly an intracellular fraction of lysostaphin. For the preparation of such cell lysates, RAW264.7 cells were treated with buffer (control) or 2 U of lysostaphin for 30 min at 37°C in a CO2 incubator. DMEM was then aspirated, and host cell or microtiter plate surface-adhered lysostaphin was washed with PBS or PBS with 100 μM 1,10-phenanthroline, followed by two additional gentle washing cycles with PBS. Control and lysostaphin-treated RAW264.7 cells were lysed in 1.0 ml of lysis buffer. Log-phase-grown S. aureus cells (OD600 of 0.01) were exposed to 1 ml of host cell lysates in PBS for 2 h at 37°C in a 5% CO2 incubator, followed by CFU enumeration. There were no significant changes observed in CFU counts between the control and lysostaphin-treated host cell extracts, further suggesting that lysostaphin was not internalized into the host cells or had been inactive in the host cells (Fig. S10). Several lines of evidence (Fig. 1D and Fig. 2 and 4) suggest that lysostaphin-mediated killing of intracellular bacteria during the EPA is negligible (nearly zero, if any), in comparison to gentamicin-mediated killing during the GPA; this is due to gentamicin being quite stable and, thus, maintaining bactericidal properties inside the host cells (Fig. 3E).
The present results collectively support the hypothesis that accurate counting of internalized bacteria can be confounded in the GPA due to the killing of intracellular bacteria by internalized gentamicin. In contrast, the EPA could be useful for the accurate measurement of internalized bacteria, since lysostaphin activity inside host cells is nearly zero. Furthermore, the EPA has an advantage over the GPA in terms of counting internalized cells on a minute time scale, since the lysostaphin bactericidal activity is exceptionally high and the killing kinetics are significantly faster than those of gentamicin.
Applicability and advantage of the EPA.
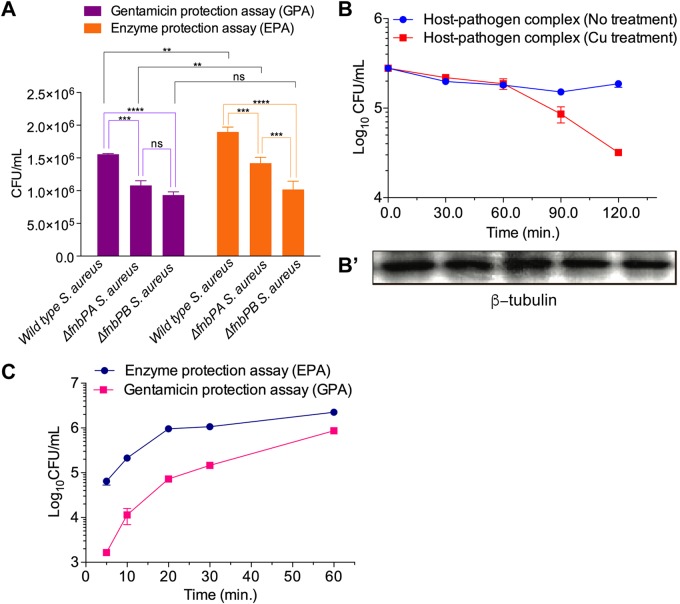
Next, we tested the applicability of the EPA to the study of host-pathogen interactions. First, we investigated the phenotypic changes of mutant strains of S. aureus (fnbPAΩEmr and fnbPBΩEmr), which lack fibronectin binding protein A (FnbPA) and FnbPB, respectively; this has been so because the internalization of bacteria into host cells (40) via bridging fibronectin to α5β1 integrin has been well established in nonprofessional phagocytic cells (41–43). The internalization potentials of the wild-type (WT), fnbPAΩEmr, and fnbPBΩEmr strains were investigated in HEK293 and RAW264.7 by employing the EPA and GPA. The mutant strains showed reduced internalized CFU in HEK293 cells compared to WT S. aureus, suggesting that both fibronectin binding proteins A and B contribute to the internalization potential of S. aureus in HEK293 cells (Fig. 5A). Of the two proteins, FnbPB seems to have greater importance since the fnbPBΩEmr mutant displayed less internalization potential than the fnbPAΩEmr mutant (Fig. 5A). It is noteworthy that the FnbPB mutant showed marginally reduced internalization compared to the WT in RAW264.7 cells (Fig. S11), although it is known that internalization of bacteria into macrophages is not dependent on fibronectin bridging events between FnbP and integrins. This result suggests that FnbP might be involved in the internalization of bacteria into mouse macrophages through an unknown mechanism. For example, in the case of nonprofessional phagocytes, Hsp60 has been reported to interact with FnbPs directly without the bridging events of α5β1 integrin of host cells (44), which might also be relevant for the murine macrophage RAW264.7 cell line.
FIG 5.
Applications of the lysostaphin-mediated enzymatic protection assay. (A) Assessment of the internalizing potential of wild-type S. aureus and its isogenic mutants lacking functional FnbPA and FnbPA (fnbPAΩEmr and fnbPBΩEmr). Wild-type, fnbPAΩEmr, and fnbPBΩEmr S. aureus strains were added to nonphagocytic HEK293 cells for 30 min in a 5% CO2 environment at 37°C. After killing of the extracellular bacteria with gentamicin (300 μg/ml for 1 h and 100 μg/ml) and lysostaphin (2 U for 5 min), the internalized S. aureus cells were assessed by counting CFU. Both fnbP mutant strains showed differential reductions in internalization potential, demonstrating that the EPA can precisely discern a phenotypic change between mutant and WT bacteria. Internalization potentials of S. aureus strains in nonphagocytic HEK293 cells were compared by one-way ANOVA with Bonferroni’s multiple-comparison test. ns, nonsignificant; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001. (B) Measurement of the bactericidal activity of RAW264.7 cells by the EPA. After 30 min of infection of RAW264.7 cells with S. aureus, the extracellular bacteria were killed by lysostaphin. The infected RAW264.7 cells were treated with 100 μM Cu2+ for various time periods (0, 30, 60, 90, and 120 min) in Hanks’ balanced salt solution (HBSS). The number of intracellular S. aureus cells was monitored by measuring CFU. As a control, β-tubulin was monitored by Western blotting. The time-dependent reduction of intracellular CFU suggests that internalized bacteria were killed by a copper-mediated defense mechanism of the host. This experiment demonstrates that the EPA can be applied to monitor changes in intracellular bacteria. (C) Internalization kinetics of S. aureus entering RAW264.7 cells using the GPA and the EPA. RAW264.7 cells were infected with S. aureus for various time periods (5, 10, 20, 30, and 60 min). At each time point, the extracellular bacteria were killed by gentamicin (400 μg/ml for 1 h of incubation, followed by gentamicin removal by washing) or lysostaphin (2 U for 10 min of incubation, followed by EDTA quenching and removal by washing), and measurement of cell lysate CFU was performed. The internalization kinetics measured by applying the GPA were significantly lower than those measured via the EPA.
Nonetheless, the difference in the internalization potentials between WT and fnbP knockout strains was found to be less prominent in the GPA than in the EPA (Fig. 5A and Fig. S11). Especially, this difference is less significant when CFU were compared in RAW264.7 cells, presumably due to the fact that more gentamicin is internalized in RAW264.7 cells than in nonphagocytic HEK293 cells (Fig. 5A and Fig. S11). Thus, precise measurement of internalized bacteria is possible with the EPA, even if a knockout strain has marginal phenotypic defects in invasion potential (Fig. 5A).
Next, we applied the EPA to examine the intracellular bactericidal capacity of host cells by counting the number of intracellular bacteria remaining under a host defense system in a time-dependent manner. For this purpose, we first prepared RAW264.7 cells with internalized S. aureus by applying the EPA and then exposed the S. aureus-infected RAW264.7 cells to cupric ion to trigger a host defense system, since it is known that copper plays a crucial role in host defense against pathogens (45). The CFU count was monitored over time up to 120 min in comparison to conditions with no treatment with cupric ion as a control (Fig. 5B). With this approach, we were able to trace the Cu-mediated killing of intracellular S. aureus in a time-dependent manner, wherein the log10 CFU value decreased from 5.5 to 4.5 over a period of 120 min. As a control, β-tubulin was monitored by Western blotting to evaluate the host cell physiology and the number of living host cells (Fig. 5B′). Using the GPA, it can be difficult to measure such small changes in CFU counts on a minute time scale due to the long duration of extracellular bacterial eradication and nonspecific killing of intracellular bacteria by gentamicin (Fig. 5B). It is conceivable that Cu-mediated bactericidal activity can be mimicked by the antimicrobial activity of antibiotics. Therefore, determination of the in vitro efficacy of new antibiotics using the GPA would presumably give misleading results due to the combined effect with internalized gentamicin. We thus confirmed that the EPA is useful for studying host-pathogen interactions and determining the in vitro antibacterial efficacy of newly discovered antimicrobial agents. Specifically, the EPA could be used to evaluate the infection potential of bacterial pathogens, quantitatively monitor small phenotypic changes of mutants, and measure time-dependent host defense activities triggered by copper (Fig. 5A and B).
The most important application of the EPA in comparison to the GPA is the measurement of internalization kinetics of bacteria. Extracellular bacteria can be killed on a minute time scale (Fig. 1B and C and Fig. S1B and S2), and the zinc metallopeptidase activity of lysostaphin can be completely abated immediately after metal ion chelation. When we tested the effects of two metal chelators, EDTA and 1,10-phenanthroline, on lysostaphin activity, we found that both chelators inhibited lysostaphin activity at their nonlethal concentrations, but higher quenching activity was attained by 1,10-phenanthroline (Fig. S12), which is expected since 1,10-phenanthroline is a zinc-specific metal chelator (46). These results suggest that it is possible to count the number of internalized bacteria at any time point, which is not possible with the GPA due to the slow killing kinetics of gentamicin (Fig. 1A and Fig. S1A) and the lack of an optimal method to cease gentamicin activity. We monitored the internalization kinetics of S. aureus entering RAW264.7 cells by measuring CFU after first infecting host cells with S. aureus for given time intervals and then treating cells with lysostaphin or gentamicin (Fig. 5C). Consistent with the results in Fig. 1D, the CFU values measured by the EPA at the initial time points were approximately higher, in the range of 1 log10 unit, than those obtained by applying the GPA. For example, while 955,256 bacterial cells were counted as internalized into 4 × 105 host cells for the initial 20 min when the EPA was applied, only 72,684 bacterial cells were found to enter host cells using the GPA under the same conditions. These results once more suggested that bacterial cell entry kinetics estimated by the GPA seem to deviate largely from the real values. Interestingly, the difference between numbers of intracellular bacteria and internalization kinetics obtained by the GPA and the EPA becomes smaller as time passes, suggesting that the GPA is not suitable for counting internalized bacteria or measuring their internalization kinetics in the early stages of infection (Fig. 5C).
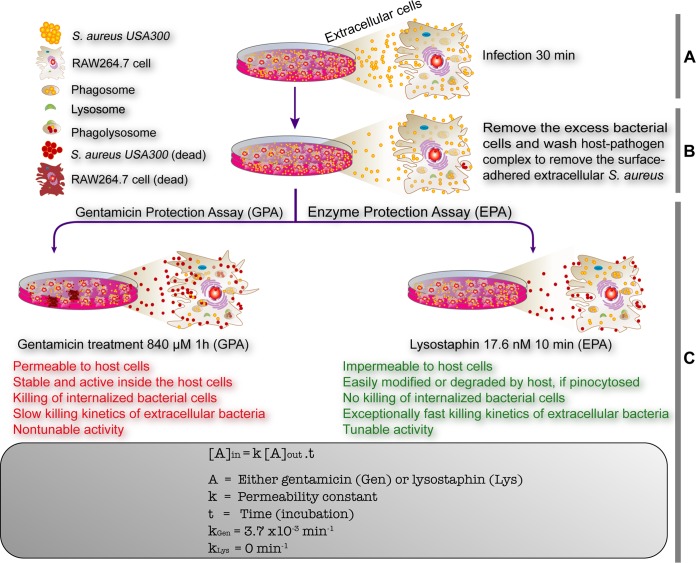
The difference between the entry kinetics obtained by the EPA and GPA originates from the death of internalized bacteria by gentamicin. Moreover, the slow killing kinetics of gentamicin also seem to contribute to the misleading results obtained by the GPA, since host cells are exposed to gentamicin for more than 1 h. Both internal and external gentamicin can affect the internalization of bacteria by altering various host cellular processes, such as protein trafficking (4) and translational termination (47), which are relevant to the defense and offense mechanisms of host cells against pathogens. To explore the effect of gentamicin on host-pathogen interactions, we monitored the gentamicin-mediated killing of S. aureus infecting RAW264.7 cells by time-lapse live-cell imaging for 1 h (see Video SV1 and Fig. S13 in the supplemental material) and compared the results to those obtained for lysostaphin-mediated killing (Video SV2 and Fig. S14). Various steps of the GPA and EPA are depicted and summarized in Fig. 6A to C. In this experiment, despite the removal and washing of excess cells (Fig. 6A), we observed entry of the remaining adhered S. aureus cells into RAW264.7 cells during the gentamicin killing step. However, such internalization was not observed in the EPA because of lysostaphin-mediated rapid lysis of S. aureus cells. In this experimental setting, we cannot exclude the possibility of phagocytosis-mediated death of bacteria in RAW264.7 cells, since green fluorescence represents both live and dead bacteria, whereas red fluorescence represents only dead bacteria with membrane damage. Nevertheless, it is clear that the GPA causes miscounting of internalized bacteria due to the entry of gentamicin and the internalization of bacteria during gentamicin killing.
FIG 6.
Schematic diagrams showing the summary of drawbacks of the gentamicin protection assay and its comparison with the newly established enzyme protection assay. The GPA is one of the key techniques in infectious disease biology to assess the invasion/infection potential of bacterial pathogens and the efficacy of newly identified antibiotics. The major drawbacks of the GPA (text in red font), which kills internalized bacterial cells, are recognized. The EPA has been devised to alleviate the drawbacks of the GPA with added advantages of measuring the internalization/invasion kinetics due to its exceptionally fast and tunable enzymatic killing efficiency (text in green font). (A and B) Both the GPA and EPA share the first two steps: infection (A) and removal of excess bacterial cells (B). (C) The killing of extracellular and surface-bound bacterial pathogens is the key step wherein gentamicin or lysostaphin was used, based on the results that the killing agent cannot enter mammalian host cells. Gentamicin seeps into the cells (kGen = 3.7 × 10−3 min−1) and kills the internalized bacteria, resulting in the misleading infection potential of the bacterial pathogen and precise drug efficacy. In the EPA, lysostaphin efficiently kills host cell surface-bound bacterial cells and cannot enter the host cells (kLys = 0), which avoids any misleading calculation and interpretation of both the invasion potential and antibiotic drug efficacy.
In summary, we have shown that intracellular bacteria can be miscounted when the GPA or an antibiotic protection assay is applied due to the diffusion of the antibiotic into host cells. As an alternative approach, we introduced the EPA for precise enumeration of living intracellular bacteria. As an example, we used lysostaphin for counting S. aureus bacteria, chosen for its rapid killing of extracellular and surface-bound bacteria, instantaneous quenching of enzyme-mediated killing activity, and inability to permeate host cells. We demonstrate that the enzyme protection assay is more beneficial in the study of host-pathogen interactions than the gentamicin protection assay, particularly for measurement of internalization kinetics and in determination of antimicrobial efficacy of newly discovered antibiotics. By the same principle, this method can be expanded to count other species of intracellular bacteria by applying enzymes that have killing activity against those specific pathogens. For example, bacterium-specific endolysin, which acts as a hydrolase and is produced from bacteriophages, can be used for this purpose (12, 48). The gentamicin protection assay might still be useful in comparative studies of the invasion potential of various pathogens or the efficiency of different drugs against bacteria under in vitro conditions. However, we propose that the EPA provides a viable alternative to the GPA for the absolute and precise quantitative investigation of host-pathogen interactions.
MATERIALS AND METHODS
Growth and culture conditions of the pathogen and host cells.
Wild-type S. aureus strain USA300 FPR3757 (here referred to as S. aureus) and its insertional mutants of the fibronectin binding proteins FnbPA and FnbPB (fnbPAΩEmr and fnbPBΩEmr) were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA), BEI Resources (49, 50). S. aureus strains were grown in tryptic soy broth (TSB) under shaking culture conditions at 200 rpm or on tryptic soy agar (TSA) (1.5% agar) plates at 37°C. The fnbPAΩEmr and fnbPBΩEmr insertion mutants were grown in TSB supplemented with 10 μg/ml erythromycin (50). Bacterial growth was monitored by measuring the optical density at 600 nm (OD600). The bacterial cultures were allowed to grow to logarithmic phase and were washed with Dulbecco’s phosphate-buffered saline (PBS) (pH 7.4; Invitrogen) before infection. The mouse macrophage cell line (RAW264.7) and the human embryonic kidney cell line (HEK293) were obtained from the American Type Culture Collection (ATCC). RAW264.7 and HEK293 cells were grown in DMEM containing 10% fetal bovine serum (FBS). Mammalian cells were grown at 37°C in a humidified 5% CO2 incubator, by maintaining a cell density of 2 × 106 cells/ml.
Measurement of gentamicin and lysostaphin killing efficiencies.
Log-phase S. aureus cells were washed with PBS, and the cell number was adjusted to an OD600 of 0.01 (equivalent to ∼1.0 × 107 cells/ml). The actual number of bacterial cells taken was calculated by serial dilution, plating, and counting of CFU. The calculated number (OD600 of 0.01) of S. aureus cells was resuspended in 24-well plates filled with 1 ml DMEM. Various concentrations of gentamicin or lysostaphin were applied to S. aureus in triplicate for each time point, and cells were incubated at 37°C in a 5% CO2 incubator. Aliquots were taken at various time points and diluted in PBS. To obtain CFU values, 100 μl of each serially diluted sample was plated on a TSA plate and incubated at 37°C for 18 h.
Bacterial viability staining and confocal microscopy.
S. aureus cells at log phase (OD600 of 1.0, equivalent to 1 × 109 cells) were harvested and washed with 0.9% saline (0.9% sodium chloride). The harvested cells were stained using the Live/Dead BacLight bacterial viability kit (catalog no. L7007; Invitrogen, USA), in which SYTO9 (green fluorescence) and propidium iodide (PI) (red fluorescence) were used for staining total and dead cells, respectively. The stained bacterial cells were washed twice with saline before confocal microscopy.
Infection and enumeration of intracellular bacteria.
RAW264.7 and HEK293 cells were prepared in 6-well culture plates by seeding 1.0 × 105 cells into 2 ml medium and allowed to grow for 24 h. Two hours before bacterial infection, the medium was changed to 1 ml fresh medium. For the infection experiment, S. aureus cells at logarithmic phase were washed with ice-cold PBS and resuspended in RPMI 1640 medium. The multiplicity of infection (MOI) was adjusted to 10. Both host and pathogen cells were kept on ice for 15 min for synchronization. After synchronization, S. aureus cells were mixed with mammalian cells on ice. Infection was initiated by moving the host-pathogen mixture to a 37°C incubator with 5% CO2. After 30 min of infection, the host-pathogen mixture was washed three times with PBS to remove nonadherent host cells and excess extracellular S. aureus bacteria. The remaining extracellular bacteria and the bacteria associated with the host cell surface were eradicated by using 100 to 400 μg/ml gentamicin (210 to 840 μM) for various times or by using 8.8 and 17.6 nM (1 and 2 U) lysostaphin for various times at 37°C to determine the optimized concentrations and incubation time. Lysostaphin activity was quenched with 50 mM EDTA (pH 8.7) or 100 μM 1,10-phenanthroline. The host-pathogen mixture was washed three times at 4°C and centrifuged at 500 × g for 5 min to remove any remaining extracellular bacteria and antibacterial agents. Internalized bacteria in the host cells were enumerated by measuring the CFU in host cell lysates, prepared by treating harvested host cells with 1 ml of 0.02% Triton X-100 in water, followed by serial dilutions in PBS, plating, and incubation at 37°C for 18 h. The CFU of internalized mutants of S. aureus lacking a functional fibronectin binding protein, FnbPA or FnbPB (fnbPAΩEmr or fnbPBΩEmr), were counted in the same way as for wild-type S. aureus. For kinetic measurement of internalization, S. aureus cells were incubated with RAW264.7 cells for the indicated time periods. Before counting the intracellular bacteria, the extracellular bacteria were killed with 400 μg/ml gentamicin (840 μM) for 60 min or with 17.6 nM (2 U) lysostaphin for 10 min.
Conjugation of gentamicin with Texas Red-X succinimidyl ester.
Gentamicin was conjugated with a monoisomer of Texas Red-X succinimidyl ester (TR) (AAT Bioquest, USA), as described previously (51). Liquid chromatography (LC) and LC/mass spectrometry (MS) for separation and mass analysis of gentamicin and TR-labeled gentamicin (GTTR) were performed as described previously (51). Briefly, a gentamicin sulfate (Sigma-Aldrich, USA) solution (50 mg/ml or 70.6 mM) in 100 mM sodium carbonate buffer (pH 10.0) and a 3.9 mM stock of a monoisomer of TR in dimethylformamide (DMF) were prepared separately. Next, to conjugate gentamicin with TR, a 1.46-ml gentamicin solution (103.1 μmol) and a 0.24-ml TR solution (0.94 μmol) were mixed. The reaction mixture was continuously mixed using a rotator mixer at 4°C in a cold room for up to 8 days until the reaction was worked up. Separation (LC) and mass analysis of the gentamicin conjugate (GTTR) were performed as described previously (51). The solvent of the gentamicin-TR conjugate was evaporated completely by purging with nitrogen gas, and gentamicin-TR was dissolved in an equivalent volume of PBS. It is noteworthy that the separated gentamicin-TR conjugate hardly possessed any visible TR colors, compared to the initial reaction mixture of gentamicin and Texas Red. RAW264.7 and HEK293 host cells were seeded in a confocal disc 24 h prior to the confocal imaging experiment. The nuclei of the host cells were stained with Hoechst 33258 (2 μl of 10 mg/ml), 50 μg/ml of the GTTR conjugate was added, and the host cells were then incubated at 37°C for 1 h in a CO2 incubator. The excess GTTR and Hoechst 33258 were washed off with PBS three times at room temperature. The cells were fixed with 4% paraformaldehyde for 5 min, followed by three cycles of washing with 2 ml of PBS, and cells were then visualized under a confocal laser scanning fluorescence microscope using laser excitation for Hoechst 33258 (excitation wavelength [λexcitation] at 355 nm and λemission at 465 nm) and Texas Red (λexcitation at 591 nm and λemission at 615 nm).
Measurement of intracellular gentamicin in RAW264.7 cells.
RAW264.7 cells (∼5.3 × 106 cells) were incubated with gentamicin at 400 μg/ml (840 μM) and without gentamicin but with an equivalent volume of autoclaved water (control) for various times (0, 15, 30, 60, and 120 min). At each time point, the medium was removed by aspiration, and RAW264.7 cells were washed four times using PBS. Both control and gentamicin-treated RAW264.7 cells were harvested by scraping the adhered cells using a soft rubber policeman and mixing the cells with 1.0 ml of lysis solution (ice-chilled distilled water containing 130 mM trichloroacetic acid [TCA] and 0.04% Triton X-100). Cells were lysed by repetitive pipetting. The cell lysates were incubated on ice for 15 min and centrifuged at 10,000 × g for 10 min at 4°C. From the supernatant, a volume of 200 μl of the cell-free lysate was diluted with the sample dilution buffer in the gentamicin ELISA kit (product no. ABIN400596; Antibodies-Online, Germany) according to the manufacturer’s instructions. The amount of gentamicin in the solution was estimated by measuring the absorbance at 450 nm (A450). The RAW264.7 cell lysate treated with an equivalent volume of autoclaved water was kept as a control for background correction. A standard curve was prepared by plotting the absorbance against standard gentamicin at various concentrations. The cell volume of 5.3 × 106 RAW264.7 cells was estimated to be 50.8 μl by subtracting the volume of the lysis solution from the volume of cell lysates. Therefore, intracellular gentamicin concentration was calculated as follows: concentration (micrograms per milliliter) = [total amount of gentamicin in 200 μl of the cell lysate (micrograms) × 1,051/200]/51 (microliters).
Assessment of the bactericidal activity of internalized gentamicin.
RAW264.7 cells (1.0 × 106) were treated with gentamicin at 400 μg/ml (840 μM) in 1 ml overlaid DMEM for various time periods (0, 15, 30, 60, and 120 min). RAW264.7 cells treated with an amount of autoclaved water (control) equivalent to the gentamicin volume were kept as the control for each time point (0, 15, 30, 60, and 120 min). RAW264.7 cells were washed 4 times with PBS to remove any extracellular gentamicin and then harvested by scraping using a rubber policeman. The cells were lysed by 1 cycle of freezing and thawing, followed by treatment with cell lysis buffer (1 ml). The cell debris was removed by centrifugation at 16,800 × g for 10 min at 4°C. S. aureus cells were grown to log phase in a culture tube, and 10 μl of the log-phase-grown culture was aliquoted into a 24-well microtiter plate. The host cell lysates of RAW264.7 cells (1 ml) were applied to S. aureus cells in a microtiter plate. The bacterial cells were diluted and plated accordingly to count the CFU.
FITC labeling of lysostaphin to assess the possibility of passive internalization into host cells.
Lysostaphin (Sigma, USA) was labeled with fluorescein isothiocyanate (FITC) by incubating FITC with lysostaphin in carbonate buffer (pH 9.0) at a molar ratio of 2:1 for 20 min at 25°C using an orbital shaker (400 rpm). FITC-labeled lysostaphin was separated from unbound FITC using a PD-10 column (GE Healthcare, USA). The labeling of lysostaphin was confirmed using SDS-PAGE. FITC-labeled lysostaphin was used to validate the impermeability of the host cells to lysostaphin via confocal fluorescence microscopy.
Conjugation of Texas Red-X succinimidyl ester with lysostaphin.
The amino terminus of lysostaphin (25 U; 220 nM) was labeled with TR (AAT Bioquest, USA) to obtain lysostaphin-TR. Briefly, a native suspension buffer containing 25 U of lysostaphin was exchanged with 100 mM sodium carbonate buffer (pH 10) using a 3-kDa-cutoff Amicon Ultra-4 centrifugal filter (Millipore, USA) by centrifugation (16,880 × g) at 4°C. After exchanging the buffer, 220 nM lysostaphin was diluted in 100 mM sodium carbonate buffer to maintain the volume up to 700 μl. The concentration of Texas Red-X succinimidyl ester in the stock solution was 3.9 mM. To maintain the 1:200 ratio of lysostaphin to TR, 11.3 μl of TR was added, and the volume of the reaction mixture was maintained at 1 ml using 100 mM sodium carbonate buffer (pH 10). The reaction mixture was continuously mixed overnight using a rocker mixer at 4°C in a cold room. The unconjugated TR was removed from the lysostaphin-TR conjugate using a 3-kDa-cutoff Centricon microcentrifuge by centrifugation (16,880 × g) at 4°C. The labeled lysostaphin-TR conjugate was washed four times with 4 ml of 0.1 M phosphate buffer (pH 7.2) using a 5-ml Centricon tube (3-kDa cutoff) and centrifugation (3,220 × g) at 4°C. Finally, the labeled lysostaphin-TR was concentrated by centrifugation (16,880 × g) in a 3-kDa-cutoff Centricon microcentrifuge tube and resuspended to 125 μl (25 U; 1 U/5 μl) in 0.1 M phosphate buffer (pH 7.2). The conjugation of lysostaphin with TR was confirmed using SDS-PAGE.
Monitoring copper-induced intracellular killing of S. aureus.
The copper-induced intracellular bactericidal activity of host cells against S. aureus was monitored. After allowing the internalization of S. aureus into RAW264.7 cells by incubation at 37°C for 30 min, the culture was treated with 17.6 nM lysostaphin for 10 min to remove extracellular bacteria (this step was followed by EDTA quenching). Subsequently, cells were washed three times with fresh DMEM. The infected RAW264.7 cells were treated with 100 μM Cu2+ for various times in Hanks’ balanced salt solution (HBSS). The host cells were lysed, and the intracellular bacteria were counted using the above-mentioned CFU counting method. From this analysis, the Cu2+-mediated bactericidal activity of mouse macrophages (RAW264.7 cells) was confirmed by a reduction in the intracellular CFU. The β-tubulin level was measured alongside each CFU measurement by Western blotting as a control.
Real-time monitoring of S. aureus infection of RAW264.7 cells using time-lapse confocal microscopy during gentamicin and enzymatic protection assays.
S. aureus infection of RAW264.7 cells and gentamicin or lysostaphin killing were monitored by confocal microscopy. Log-phase bacteria were stained using the BacLight bacterial viability staining kit. For confocal live-cell imaging, RAW264.7 cells were cultured for 1 day in a confocal disc and placed on the stage-top incubator of the confocal microscope at 37°C in 5% CO2. The host cells were infected with S. aureus at an MOI of 200 by adding the S. aureus cells directly to the host cells in the confocal disc, placed on the stage-top incubator of the confocal microscope. Before the addition of S. aureus bacteria to RAW264.7 cells, the host cells were imaged for 1 min (one image per 2 s). After 30 min of infection, excess S. aureus cells were removed by washing twice with PBS, and 1 ml DMEM was then added to the RAW364.7 cells in the confocal disc. To observe gentamicin killing, the confocal disc was monitored for 60 min with an 1,800-image acquisition after the addition of gentamicin (400 μg/ml) and mixing with gentle pipetting (see Video SV1 and Fig. S13 in the supplemental material). Similarly, lysostaphin killing was observed for 10 min after the addition of 2 U of lysostaphin (Video SV2 and Fig. S14).
Statistical analysis.
All the data are means ± standard deviations. Statistical significance was determined using Student’s t test or one-way analysis of variance (ANOVA) with Bonferroni’s correction for multiple-comparison tests.
Supplementary Material
ACKNOWLEDGMENTS
The fnbPAΩEmr and fnbPBΩEmr mutants of S. aureus were provided by the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for distribution by BEI Resources, NIAID, NIH (Staphylococcus aureus subsp. aureus strain JE2).
This work was supported by grants from the National Research Foundation of Korea to J.-H.K. (2017R1A6A3A11033617), A.K.C. (2017R1D1A1B03035720), and K.K.K. (2017M3A9E4078553).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00119-19.
REFERENCES
- 1.Ribet D, Cossart P. 2015. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect 17:173–183. doi: 10.1016/j.micinf.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 2.Thammavongsa V, Kim HK, Missiakas D, Schneewind O. 2015. Staphylococcal manipulation of host immune responses. Nat Rev Microbiol 13:529–543. doi: 10.1038/nrmicro3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Kessel KP, Bestebroer J, van Strijp JA. 2014. Neutrophil-mediated phagocytosis of Staphylococcus aureus. Front Immunol 5:467. doi: 10.3389/fimmu.2014.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cossart P, Sansonetti PJ. 2004. Bacterial invasion: the paradigms of enteroinvasive pathogens. Science 304:242–248. doi: 10.1126/science.1090124. [DOI] [PubMed] [Google Scholar]
- 5.Monack DM, Mueller A, Falkow S. 2004. Persistent bacterial infections: the interface of the pathogen and the host immune system. Nat Rev Microbiol 2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- 6.Croxen MA, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 7.Proctor RA, von Eiff C, Kahl BC, Becker K, McNamara P, Herrmann M, Peters G. 2006. Small colony variants: a pathogenic form of bacteria that facilitates persistent and recurrent infections. Nat Rev Microbiol 4:295–305. doi: 10.1038/nrmicro1384. [DOI] [PubMed] [Google Scholar]
- 8.Sinha B, Herrmann M, Krause KH. 2000. Is Staphylococcus aureus an intracellular pathogen? Trends Microbiol 8:343–344. doi: 10.1016/S0966-842X(00)01813-8. [DOI] [PubMed] [Google Scholar]
- 9.Lowy FD. 2000. Is Staphylococcus aureus an intracellular pathogen? Trends Microbiol 8:341–343. doi: 10.1016/S0966-842X(00)01803-5. [DOI] [PubMed] [Google Scholar]
- 10.Shiratsuchi A, Osada Y, Nakanishi Y. 2013. Differences in the mode of phagocytosis of bacteria between macrophages and testicular Sertoli cells. Drug Discov Ther 7:73–77. doi: 10.5582/ddt.2013.v7.2.73. [DOI] [PubMed] [Google Scholar]
- 11.Segal AW, Dorling J, Coade S. 1980. Kinetics of fusion of the cytoplasmic granules with phagocytic vacuoles in human polymorphonuclear leukocytes. Biochemical and morphological studies. J Cell Biol 85:42–59. doi: 10.1083/jcb.85.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmelcher M, Donovan DM, Loessner MJ. 2012. Bacteriophage endolysins as novel antimicrobials. Future Microbiol 7:1147–1171. doi: 10.2217/fmb.12.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sokolovska A, Becker CE, Stuart LM. 2012. Measurement of phagocytosis, phagosome acidification, and intracellular killing of Staphylococcus aureus. Curr Protoc Immunol Chapter 14:Unit 14.30. doi: 10.1002/0471142735.im1430s99. [DOI] [PubMed] [Google Scholar]
- 14.Lecaroz C, Campanero MA, Gamazo C, Blanco-Prieto MJ. 2006. Determination of gentamicin in different matrices by a new sensitive high-performance liquid chromatography-mass spectrometric method. J Antimicrob Chemother 58:557–563. doi: 10.1093/jac/dkl258. [DOI] [PubMed] [Google Scholar]
- 15.Imbuluzqueta E, Lemaire S, Gamazo C, Elizondo E, Ventosa N, Veciana J, Van Bambeke F, Blanco-Prieto MJ. 2012. Cellular pharmacokinetics and intracellular activity against Listeria monocytogenes and Staphylococcus aureus of chemically modified and nanoencapsulated gentamicin. J Antimicrob Chemother 67:2158–2164. doi: 10.1093/jac/dks172. [DOI] [PubMed] [Google Scholar]
- 16.Elsinghorst EA. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol 236:405–420. doi: 10.1016/0076-6879(94)36030-8. [DOI] [PubMed] [Google Scholar]
- 17.Drevets DA, Canono BP, Leenen PJ, Campbell PA. 1994. Gentamicin kills intracellular Listeria monocytogenes. Infect Immun 62:2222–2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Menashe O, Kaganskaya E, Baasov T, Yaron S. 2008. Aminoglycosides affect intracellular Salmonella enterica serovars Typhimurium and Virchow. Antimicrob Agents Chemother 52:920–926. doi: 10.1128/AAC.00382-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.VanCleave TT, Pulsifer AR, Connor MG, Warawa JM, Lawrenz MB. 2017. Impact of gentamicin concentration and exposure time on intracellular Yersinia pestis. Front Cell Infect Microbiol 7:505. doi: 10.3389/fcimb.2017.00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flannagan RS, Heit B, Heinrichs DE. 2016. Intracellular replication of Staphylococcus aureus in mature phagolysosomes in macrophages precedes host cell death, and bacterial escape and dissemination. Cell Microbiol 18:514–535. doi: 10.1111/cmi.12527. [DOI] [PubMed] [Google Scholar]
- 21.Pratten MK, Lloyd JB. 1986. Pinocytosis and phagocytosis: the effect of size of a particulate substrate on its mode of capture by rat peritoneal macrophages cultured in vitro. Biochim Biophys Acta 881:307–313. doi: 10.1016/0304-4165(86)90020-6. [DOI] [PubMed] [Google Scholar]
- 22.Edwards RA, Maloy SR. 2001. Inside or outside: detecting the cellular location of bacterial pathogens. Biotechniques 30:304–306, 308–311. doi: 10.2144/01302st03. [DOI] [PubMed] [Google Scholar]
- 23.Wang MC, Chien HF, Tsai YL, Liu MC, Liaw SJ. 2014. The RNA chaperone Hfq is involved in stress tolerance and virulence in uropathogenic Proteus mirabilis. PLoS One 9:e85626. doi: 10.1371/journal.pone.0085626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L, Michel R, Cohen J, Decarlo A, Kozarov E. 2008. Intracellular survival and vascular cell-to-cell transmission of Porphyromonas gingivalis. BMC Microbiol 8:26. doi: 10.1186/1471-2180-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards AM, Massey RC. 2011. Invasion of human cells by a bacterial pathogen. J Vis Exp 2011:2693. doi: 10.3791/2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yokogawa K, Kawata S, Takemura T, Yoshimura Y. 1975. Purification and properties of lytic enzymes from Streptomyces globisporus 1829. Agric Biol Chem 39:1533–1543. doi: 10.1271/bbb1961.39.1533. [DOI] [Google Scholar]
- 27.Blattner S, Das S, Paprotka K, Eilers U, Krischke M, Kretschmer D, Remmele CW, Dittrich M, Muller T, Schuelein-Voelk C, Hertlein T, Mueller MJ, Huettel B, Reinhardt R, Ohlsen K, Rudel T, Fraunholz MJ. 2016. Staphylococcus aureus exploits a non-ribosomal cyclic dipeptide to modulate survival within epithelial cells and phagocytes. PLoS Pathog 12:e1005857. doi: 10.1371/journal.ppat.1005857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grosz M, Kolter J, Paprotka K, Winkler AC, Schafer D, Chatterjee SS, Geiger T, Wolz C, Ohlsen K, Otto M, Rudel T, Sinha B, Fraunholz M. 2014. Cytoplasmic replication of Staphylococcus aureus upon phagosomal escape triggered by phenol-soluble modulin alpha. Cell Microbiol 16:451–465. doi: 10.1111/cmi.12233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Munzenmayer L, Geiger T, Daiber E, Schulte B, Autenrieth SE, Fraunholz M, Wolz C. 2016. Influence of Sae-regulated and Agr-regulated factors on the escape of Staphylococcus aureus from human macrophages. Cell Microbiol 18:1172–1183. doi: 10.1111/cmi.12577. [DOI] [PubMed] [Google Scholar]
- 30.Sendi P, Proctor RA. 2009. Staphylococcus aureus as an intracellular pathogen: the role of small colony variants. Trends Microbiol 17:54–58. doi: 10.1016/j.tim.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 31.Garzoni C, Kelley WL. 2009. Staphylococcus aureus: new evidence for intracellular persistence. Trends Microbiol 17:59–65. doi: 10.1016/j.tim.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 32.Tong SY, Davis JS, Eichenberger E, Holland TL, Fowler VG Jr. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations, and management. Clin Microbiol Rev 28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiramatsu K, Katayama Y, Matsuo M, Sasaki T, Morimoto Y, Sekiguchi A, Baba T. 2014. Multi-drug-resistant Staphylococcus aureus and future chemotherapy. J Infect Chemother 20:593–601. doi: 10.1016/j.jiac.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 34.Bastos MD, Coutinho BG, Coelho ML. 2010. Lysostaphin: a staphylococcal bacteriolysin with potential clinical applications. Pharmaceuticals (Basel) 3:1139–1161. doi: 10.3390/ph3041139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cha HY, Moon DC, Choi CH, Oh JY, Jeong YS, Lee YC, Seol SY, Cho DT, Chang HH, Kim SW, Lee JC. 2005. Prevalence of the ST239 clone of methicillin-resistant Staphylococcus aureus and differences in antimicrobial susceptibilities of ST239 and ST5 clones identified in a Korean hospital. J Clin Microbiol 43:3610–3614. doi: 10.1128/JCM.43.8.3610-3614.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu F, Liu Y, Lv J, Qi X, Lu C, Ding Y, Li D, Liu H, Wang L. 2015. Antimicrobial susceptibility, virulence determinant carriage and molecular characteristics of Staphylococcus aureus isolates associated with skin and soft tissue infections. Braz J Infect Dis 19:614–622. doi: 10.1016/j.bjid.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hau SJ, Kellner S, Eberle KC, Waack U, Brockmeier SL, Haan JS, Davies PR, Frana T, Nicholson TL. 2018. Methicillin-resistant Staphylococcus aureus sequence type (ST) 5 isolates from health care and agricultural sources adhere equivalently to human keratinocytes. Appl Environ Microbiol 84:e02073-17. doi: 10.1128/AEM.02073-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grundling A, Missiakas DM, Schneewind O. 2006. Staphylococcus aureus mutants with increased lysostaphin resistance. J Bacteriol 188:6286–6297. doi: 10.1128/JB.00457-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Watson H. 2015. Biological membranes. Essays Biochem 59:43–69. doi: 10.1042/bse0590043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sinha B, Francois P, Que YA, Hussain M, Heilmann C, Moreillon P, Lew D, Krause KH, Peters G, Herrmann M. 2000. Heterologously expressed Staphylococcus aureus fibronectin-binding proteins are sufficient for invasion of host cells. Infect Immun 68:6871–6878. doi: 10.1128/IAI.68.12.6871-6878.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flock JI, Fröman G, Jönsson K, Guss B, Signäs C, Nilsson B, Raucci G, Höök M, Wadström T, Lindberg M. 1987. Cloning and expression of the gene for a fibronectin-binding protein from Staphylococcus aureus. EMBO J 6:2351–2357. doi: 10.1002/j.1460-2075.1987.tb02511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jonsson K, Signas C, Muller HP, Lindberg M. 1991. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem 202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 43.Josse J, Laurent F, Diot A. 2017. Staphylococcal adhesion and host cell invasion: fibronectin-binding and other mechanisms. Front Microbiol 8:2433. doi: 10.3389/fmicb.2017.02433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dziewanowska K, Carson AR, Patti JM, Deobald CF, Bayles KW, Bohach GA. 2000. Staphylococcal fibronectin binding protein interacts with heat shock protein 60 and integrins: role in internalization by epithelial cells. Infect Immun 68:6321–6328. doi: 10.1128/IAI.68.11.6321-6328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.White C, Lee J, Kambe T, Fritsche K, Petris MJ. 2009. A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J Biol Chem 284:33949–33956. doi: 10.1074/jbc.M109.070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Day TA, Chen GZ. 1998. The metalloprotease inhibitor 1,10-phenanthroline affects Schistosoma mansoni motor activity, egg laying and viability. Parasitology 116:319–325. doi: 10.1017/S0031182097002370. [DOI] [PubMed] [Google Scholar]
- 47.Murphy GJ, Mostoslavsky G, Kotton DN, Mulligan RC. 2006. Exogenous control of mammalian gene expression via modulation of translational termination. Nat Med 12:1093–1099. doi: 10.1038/nm1376. [DOI] [PubMed] [Google Scholar]
- 48.Yuan Y, Peng Q, Gao M. 2012. Characteristics of a broad lytic spectrum endolysin from phage BtCS33 of Bacillus thuringiensis. BMC Microbiol 12:297. doi: 10.1186/1471-2180-12-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. 2013. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. mBio 4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bae T, Banger AK, Wallace A, Glass EM, Aslund F, Schneewind O, Missiakas DM. 2004. Staphylococcus aureus virulence genes identified by bursa aurealis mutagenesis and nematode killing. Proc Natl Acad Sci U S A 101:12312–12317. doi: 10.1073/pnas.0404728101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Woiwode U, Sievers-Engler A, Lämmerhofer M. 2016. Preparation of fluorescent labeled gentamicin as biological tracer and its characterization by liquid chromatography and high resolution mass spectrometry. J Pharm Biomed Anal 121:307–315. doi: 10.1016/j.jpba.2015.12.053. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.