Vibrio parahaemolyticus is a human pathogen, and it is a major cause of severe gastroenteritis in coastal areas. OmpU is one of the major outer membrane porins of V. parahaemolyticus.
KEYWORDS: OmpU, TLR, Vibrio parahaemolyticus, innate immunity, proinflammatory responses
ABSTRACT
Vibrio parahaemolyticus is a human pathogen, and it is a major cause of severe gastroenteritis in coastal areas. OmpU is one of the major outer membrane porins of V. parahaemolyticus. Host-immunomodulatory effects of V. parahaemolyticus OmpU (VpOmpU) have not been elucidated yet. In this study, in an effort towards characterizing the effect of VpOmpU on innate immune responses of the host, we observed that VpOmpU is recognized by the Toll-like receptor 1/2 (TLR1/2) heterodimer in THP-1 monocytes but by both TLR1/2 and TLR2/6 heterodimers in RAW 264.7 macrophages. To the best of our knowledge, this is the first report of a natural pathogen-associated molecular pattern (PAMP) recognized by both TLR1/2 and TLR2/6 heterodimers; so far, mainly the synthetic ligand Pam2CSK4 has been known to be recognized by both the TLR1/2 and TLR2/6 heterodimers. We also have shown that VpOmpU can activate monocytes and macrophages, leading to the generation of proinflammatory responses as indicated by tumor necrosis factor alpha (TNF-α), interleukin-6 (IL-6), and NO production in macrophages and TNF-α and IL-6 production in monocytes. VpOmpU-mediated proinflammatory responses involve MyD88-IRAK-1 leading to the activation of mitogen-activated protein (MAP) kinases (p38 and Jun N-terminal protein kinase [JNK]) and transcription factors NF-κB and AP-1. Further, we have shown that for the activation of macrophages leading to the proinflammatory responses, the TLR2/6 heterodimer is preferred over the TLR1/2 heterodimer. We have also shown that MAP kinase activation is TLR2 mediated.
INTRODUCTION
Vibrio parahaemolyticus is one of the human pathogens belonging to the genus Vibrio. It is a Gram-negative, motile, marine bacterium which has been a major cause of gastroenteritis worldwide. In addition to severe gastroenteritis, it can also cause septicemia and eventually death in cases of wound infection. V. parahaemolyticus infection occurs upon consumption of raw or undercooked seafood; hence, it is more prevalent in coastal areas.
The virulence property of V. parahaemolyticus has been attributed mainly to thermostable direct hemolysin (TDH) (1, 2) and TDH-related hemolysin (TRH) (3), but the observation that TDH and TRH deletion strains retain some virulence indicates the need for more research exploring other antigens of the bacterium for their contribution to its pathogenesis (4, 5).
One such potential antigen is Vibrio parahaemolyticus OmpU (VpOmpU), which is a major outer membrane protein of V. parahaemolyticus. OmpU is porin in nature, and it is present in all of the Vibrio species. In addition to its porin function, in various species of Vibrio, OmpU plays crucial roles in modulation of host cellular responses and bacterial pathogenesis. For example, the OmpU proteins of V. cholerae and V. splendidus help the bacteria in bile and antimicrobial peptide resistance (6, 7). In V. vulnificus, OmpU is reported to act as an adhesin (8), and in V. alginolyticus, OmpU has been reported to help the bacteria in acquiring antibiotic resistance (9). In V. splendidus, OmpU has been reported to help in adhesion and invasion (10). V. cholerae OmpU has been reported to cause programmed cell death (11). Further, the unique amino acid sequence of V. cholerae OmpU and its highly conserved nature distinguish epidemic strains from less pathogenic strains (12). This suggests that OmpU plays a critical role in V. cholerae pathogenesis.
OmpU proteins from various Vibrio spp. were also tested for their ability to modulate host immune responses and as potential vaccine candidates. OmpU has been developed as a vaccine candidate against V. alginolyticus, a fish pathogen (13), and against V. splendidus, an oyster pathogen (14). Antibodies against OmpU of V. cholerae in patient samples have also been reported (15). Though V. cholerae OmpU is one of the favorite candidates for development of vaccines against the disease cholera, reports from our lab challenge the candidacy, as they suggested that although V. cholerae OmpU can activate the innate immune response (16), it can also translocate to host cell mitochondria, thus inducing host cell death (11).
So far, no vaccine against V. parahaemolyticus has been reported. A report in 2007 indicated the production of antibody against V. parahaemolyticus OmpU (VpOmpU) when it was injected into yellow croaker fish (17). This suggests that VpOmpU could also be explored for its vaccine potential. For this, a detailed immunological characterization of VpOmpU to better understand its role in host immunomodulation and pathogenesis is required.
In this study, we have purified VpOmpU from wild-type and recombinant sources and further examined how VpOmpU modulates the innate immune response in vitro. We found that it generates a proinflammatory response in monocytes and macrophages via a Toll-like receptor (TLR)-mediated signaling pathway. Our study also shows the involvement of mitogen-activated protein (MAP) kinases and both NF-κB and AP-1 transcription factors in induction of the VpOmpU-mediated proinflammatory response.
RESULTS
Identification and analysis of putative OmpU from Vibrio parahaemolyticus.
Analysis of the V. parahaemolyticus genome revealed the presence of a gene sequence encoding a putative OmpU protein (18) composed of 330 amino acid residues with a predicted molecular mass of 35.6 kDa. This putative OmpU of V. parahaemolyticus (VpOmpU) showed 60 to 90% sequence similarity with the OmpU proteins of the related Vibrio species (Fig. 1A). Homology-based structural modeling based on the crystal structure of V. cholerae OmpU (19) showed a typical porin-like β-barrel architecture of VpOmpU (Fig. 1B).
FIG 1.
Amino acid sequence alignment and homology-based structural model of VpOmpU. (A) Amino acid sequence alignment of VpOmpU and OmpU porins from related Vibrio species. The region corresponding to the signal sequence is marked. (B) Homology-based structural model of VpOmpU based on the crystal structure of VcOmpU (PDB entry 6EHB).
Based on this analysis, this putative OmpU, which is probably a porin, was selected for purification from the outer membrane of V. parahaemolyticus. An attempt was also made to express and purify the recombinant form of the protein by employing the heterologous expression system in Escherichia coli. Wild-type and recombinant forms of VpOmpU were also subjected to functional characterization.
Purification of VpOmpU from the outer membrane of Vibrio parahaemolyticus.
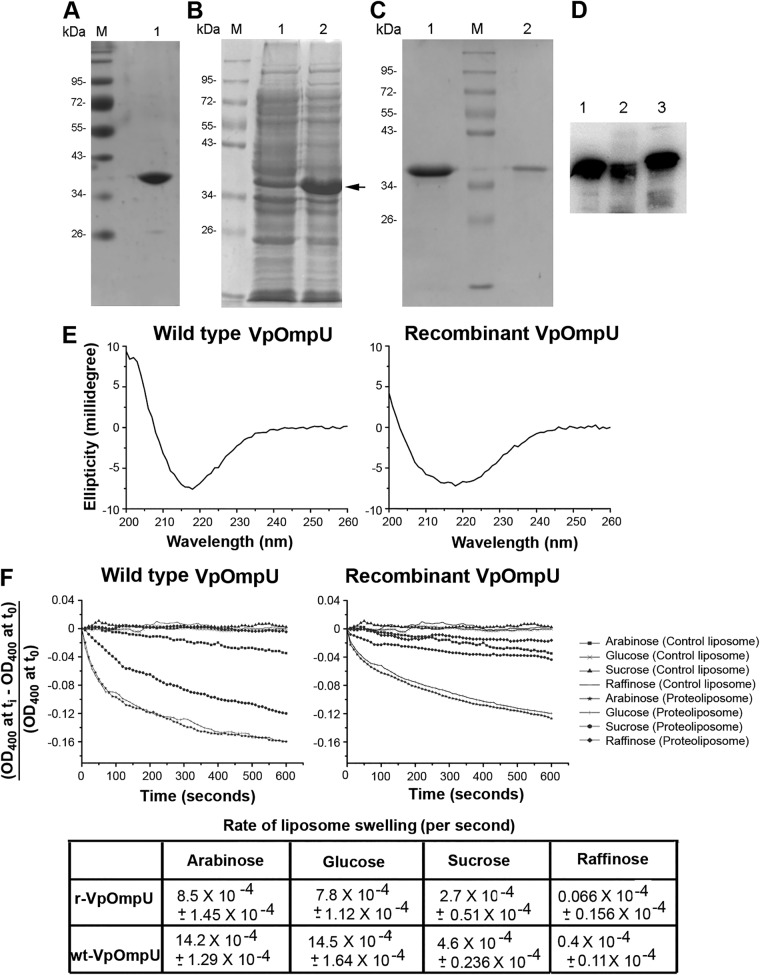
Wild-type VpOmpU (wt-VpOmpU) was purified from the outer membrane fraction of V. parahaemolyticus. Bacterial cells were grown in brain heart infusion (BHI) broth, the cells were lysed, and bacterial membrane fractions were harvested upon fractionation with 1% N-Sarkosyl. Subsequently, the outer membrane proteins were extracted via solubilization with 4% Triton X-100. VpOmpU was purified from the solubilized membrane fraction by anion-exchange chromatography on DEAE-cellulose in the presence of 0.1% Triton X-100, followed by size exclusion chromatography on Sephacryl S-200 in the presence of 0.5% lauryldimethylamine N-oxide (LDAO). Homogeneity of the purified wild-type VpOmpU preparation was analyzed by SDS-PAGE and Coomassie blue staining (Fig. 2A).
FIG 2.
Characterization of OmpU from Vibrio parahaemolyticus. (A) Lane 1, SDS-PAGE/Coomassie blue staining analysis of the purified form of wt-OmpU extracted from the V. parahaemolyticus outer membrane fraction. Lane M, molecular weight markers. (B) Overexpression of recombinant VpOmpU in E. coli. Protein overexpression was induced with 1 mM IPTG, cells were lysed, and the soluble fraction of the cell lysate (lane 1) and the insoluble inclusion body fraction (lane 2) were analyzed by SDS-PAGE and Coomassie blue staining. The band corresponding to the recombinant VpOmpU is marked. The majority of the VpOmpU protein was found to be associated with the insoluble inclusion body fraction. Lane M, molecular weight markers. (C) Purification of refolded recombinant VpOmpU as analyzed by SDS-PAGE and Coomassie blue staining. The recombinant form of VpOmpU was refolded by rapid dilution in a buffer containing 0.5% LDAO (lane 1). Refolded VpOmpU was further purified by size exclusion chromatography on Sephacryl S-200 (lane 2). Lane M, molecular weight markers. (D) Immunoblot analysis of recombinant and wild-type VpOmpU using anti-VpOmpU antiserum. Lane 1, purified form of refolded recombinant His-tagged VpOmpU; lane 2, purified form of refolded recombinant VpOmpU after removal of the N-terminal His tag; lane 3, wt-OmpU extracted from the V. parahaemolyticus outer membrane fraction. (E) Far-UV CD spectra of wild-type and recombinant VpOmpU show negative ellipticity minima at around 218 nm for both proteins. (F) The refolded recombinant form and the wild-type VpOmpU protein show nearly identical trends in the liposome-swelling response. Control liposome, liposome preparation lacking VpOmpU; proteoliposome, liposomes containing VpOmpU. The initial rates of the liposome-swelling response (calculated from the slope of the linear fit of the data over the initial period of 50 s) are shown in the table (r-VpOmpU, recombinant VpOmpU; wt-VpOmpU, wild-type VpOmpU).
Overexpression, purification, and refolding of r-VpOmpU.
To obtain a large quantity of the VpOmpU protein and also to reduce lipopolysaccharide (LPS) contamination, we cloned and expressed VpOmpU in Escherichia coli. The nucleotide sequence encoding VpOmpU without the N-terminal signal sequence was cloned into the bacterial expression vector pET14b and was transformed into the E. coli Origami B cells. The pET14b vector allowed expression of the recombinant VpOmpU (r-VpOmpU) with an N-terminal 6×His tag that would allow affinity purification of the recombinant protein using Ni affinity chromatography. The N-terminal signal sequence (Fig. 1A) was omitted from the cloned construct to avoid possible incorporation of the recombinant protein into the bacterial membrane.
Upon overexpression of the recombinant protein in the E. coli Origami B cells via IPTG (isopropyl-β-d-thiogalactopyranoside) induction, the majority of the VpOmpU was found to be present in the form of insoluble inclusion bodies (Fig. 2B). The recombinant form of His-tagged VpOmpU was solubilized from the insoluble inclusion bodies using 8 M urea and was purified by Ni-nitrilotriacetic acid (NTA) agarose affinity chromatography under denaturing conditions in 8 M urea.
The denatured form of purified recombinant VpOmpU in 8 M urea was refolded by dilution in a buffer containing 0.5% LDAO and 10% glycerol. Upon refolding, the majority of VpOmpU remained in the soluble form and was separated from the insoluble aggregated fractions of the protein via centrifugation (Fig. 2C). Refolded VpOmpU was further purified by size exclusion chromatography on Sephacryl S-200 (Fig. 2C). Homogeneity of the purified form of the recombinant His-tagged VpOmpU preparation was analyzed by SDS-PAGE and Coomassie blue staining (Fig. 2C). Further, the His tag was removed from recombinant VpOmpU by thrombin cleavage. The size of the recombinant VpOmpU without the His tag was compared to that of wild-type VpOmpU by Western blotting using antiserum generated against the recombinant VpOmpU (Fig. 2D).
The far-UV CD spectrum of VpOmpU reveals its β-sheet-rich structure.
Consistent with their β-barrel structural assembly, Gram-negative bacterial porins display far-UV circular dichroism (CD) profiles showing a typical signature of β-sheet-rich architecture. We examined the structural characteristics of the VpOmpU protein by monitoring the far-UV CD profiles of the recombinant and wild-type forms of the protein.
Wild-type VpOmpU displayed a symmetric far-UV CD spectrum, with a sharp negative ellipticity minimum at 218 nm, suggesting a β-sheet-rich structural organization of the protein (Fig. 2E). The far-UV CD profile of the refolded recombinant protein also showed an ellipticity minimum at 218 nm, suggesting a β-sheet-rich structure of recombinant VpOmpU. The β-sheet-rich structures of the wt-OmpU and the r-VpOmpU were similar to those observed with typical bacterial outer membrane porin proteins.
Liposome-swelling assay demonstrates a porin-like channel-forming property of VpOmpU.
Porins are typically characterized by their ability to form channels in the membrane lipid bilayer, allowing free diffusion of small molecules up to a definite size limit as per the size constraint imposed by the corresponding pore diameter. Therefore, we wanted to explore whether the VpOmpU protein could show any such porin-like channel-forming ability in the membrane lipid bilayer of synthetic lipid vesicles or liposomes. The channel-forming property of VpOmpU was assayed using the conventional liposome-swelling assay. In this assay, liposomes incorporating the VpOmpU protein were subjected to treatment in the presence of sugars of various molecular sizes. Formation of VpOmpU channels in the liposome membranes would allow free diffusion of water into the lipid vesicles, resulting in swelling of the liposomes. The VpOmpU channel, however, would not allow the passage of sugars/saccharides having molecular sizes larger than the pore diameter. Such saccharides would act as the osmoprotectants and thus would presumably suppress the liposome-swelling response. Based on this notion, both the wild-type and refolded recombinant forms of the VpOmpU proteins, incorporated in the liposome membranes, were examined using the liposome-swelling assay (Fig. 2F). It was observed in both cases that in the presence of monosaccharides, such as arabinose and glucose, there was a steady increase in the liposome-swelling response (Fig. 2F). In the presence of the disaccharide sucrose, the liposome-swelling responses of wild-type and recombinant VpOmpU were prominently suppressed, while the trisaccharide raffinose severely compromised the liposome swelling on proteoliposomes made up of the wild-type and the recombinant proteins (Fig. 2F). These results clearly suggested that VpOmpU indeed displayed a porin-like channel-forming property with a definite pore size, thus allowing free diffusion of solutes within the specific size limit. The VpOmpU channel allowed free diffusion of the monosaccharides, while the permeability efficiency decreased progressively in the case of the disaccharides and even further in the case of the trisaccharides of larger molecular size. It is also important to note here that the refolded form of the recombinant VpOmpU showed a trend in channel-forming ability (i.e., monosaccharide > disaccharide > trisaccharide) similar to that observed with the wild-type protein, thus showing functional integrity of the recombinant protein generated in our study (Fig. 2F). Based on these observations, it could be concluded that the recombinant VpOmpU was structurally and functionally similar to the native wild-type OmpU of V. parahaemolyticus.
VpOmpU induces proinflammatory responses in monocytes and macrophages.
As a part of the functional characterization, we explored the host-immunomodulatory role of VpOmpU. Generally, bacterial ligands known as pathogen-associated molecular patterns (PAMPs) are recognized by pattern recognition receptors (PRRs) of the innate immune cells such as macrophages and monocytes, leading to their activation, which results in the production of various proinflammatory molecules, such as nitric oxide (NO) (20, 21) and tumor necrosis factor alpha (TNF-α) and interleukin-6 (IL-6) (22).
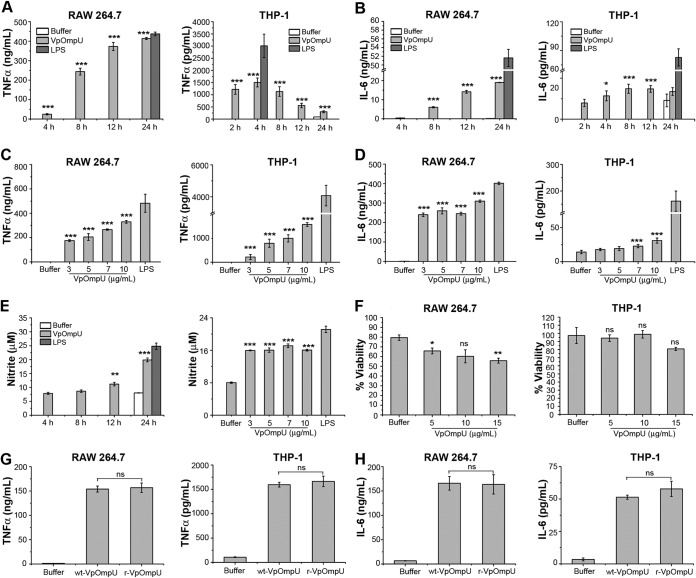
Therefore, to probe whether VpOmpU acts as a PAMP, RAW 264.7 cells (a murine macrophage cell line) and THP-1 cells (a human monocytic cell line) were treated with 10 μg/ml of recombinant VpOmpU. At different time points, supernatants were analyzed for TNF-α and IL-6 by enzyme-linked immunosorbent assay (ELISA). A time dependency analysis of TNF-α and IL-6 in response to 10 μg/ml of VpOmpU showed a maximum production of both cytokines at 24 h in RAW 264.7 cells (Fig. 3A and B). However, in THP-1 cells, maximum TNF-α production was observed at 4 h (Fig. 3A) and maximum IL-6 production was observed at 8 h (Fig. 3B). A similar time dependency analysis of NO production in RAW 264.7 cells treated with 10 μg/ml of VpOmpU showed a maximum NO production at 24 h (Fig. 3E). THP-1 cells did not produce NO in response to VpOmpU as observed by a time dependency analysis of VpOmpU-treated cells (see Fig. S1 in the supplemental material).
FIG 3.
VpOmpU induces proinflammatory responses in THP-1 monocytes and RAW 264.7 macrophages. (A and B) Maximum production of TNF-α (A) and IL-6 (B) was observed at 24 h in RAW 264.7 cells and at 4 h and 8 h, respectively, in THP-1 cells in response to VpOmpU. RAW 264.7 and THP-1 cells were treated with 10 μg/ml of recombinant VpOmpU for different times, and supernatants were analyzed for TNF-α (A) and IL-6 (B) production by ELISA. (C and D) A dose-dependent increase in TNF-α (C) and IL-6 (D) production is seen in both RAW 264.7 and THP-1 cells in response to different doses of VpOmpU. Cells were incubated with different doses of recombinant VpOmpU, and following incubation, supernatants were collected and analyzed for TNF-α and IL-6 production. RAW 264.7 cells and THP-1 cells were incubated with VpOmpU for 24 h and 4 h, respectively, for TNF-α production (C) and for 24 h and 8 h, respectively, for IL-6 production (D). (E) Nitric oxide (NO) production was observed in VpOmpU-treated RAW 264.7 cells. RAW 264.7 cells were treated with 10 μg/ml of recombinant VpOmpU and incubated for different times. Supernatants were analyzed for NO (in terms of nitrite), and significant production of NO was observed at 24 h. Similar extents of NO production were observed when RAW 264.7 cells were treated with different doses of recombinant VpOmpU and incubated for 24 h. For panels A to E, LPS (1 μg/ml) was used as a positive control for all of the experiments. (F) Cell viability in response to different doses of VpOmpU was minimally affected in both RAW 264.7 and THP-1 cells. For panels A to F, results are expressed as mean ± SEM from three or four independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus buffer-treated cells]). (G and H) Comparable production of TNF-α and IL-6 was observed in both RAW 264.7 and THP-1 cells in response to similar doses of purified wild-type VpOmpU (wt-VpOmpU) and recombinant VpOmpU (r-VpOmpU). RAW 264.7 and THP-1 cells were treated with 5 μg/ml of wt-VpOmpU or r-VpOmpU and incubated for 24 h and 4 h, respectively, for TNF-α (G) and for 24 h and 8 h, respectively, for IL-6 (H). Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus wt-VpOmpU-treated cells]).
Further, RAW 264.7 cells and THP-1 cells were treated with different concentrations of recombinant VpOmpU. Supernatants were analyzed for TNF-α and IL-6 by ELISA at the time point where maximum cytokine production had been observed. A dose-dependent increase in TNF-α (Fig. 3C) and IL-6 (Fig. 3D) was observed in both cell types. A similar dose dependency analysis of NO production was done with the supernatant of RAW 264.7 cells at 24 h posttreatment. RAW 264.7 cells produced comparable amounts of NO in response to different concentrations of VpOmpU ranging from 3 μg/ml to 10 μg/ml (Fig. 3E).
We also performed a 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay to examine the cell mortality rate in response to different doses of recombinant VpOmpU following 24 h of incubation. The cell mortality in response to VpOmpU was comparable at doses up to 10 μg/ml and was in the range of 30 to 40% for RAW 264.7 cells and about 5 to 15% for THP-1 cells (Fig. 3F).
These results show that VpOmpU can activate macrophages and monocytes to induce proinflammatory responses.
Further, we wanted to compare the proinflammatory responses generated by the cells upon treatment with wild-type and recombinant VpOmpU. For this, we treated both RAW 264.7 and THP-1 cells with an equal amount (5 μg/ml) of each of the proteins and analyzed the supernatants for TNF-α and IL-6 at the time point where the maximum amount of the respective cytokine was observed in the time dependency analysis (Fig. 3A and B). We observed that comparable amounts of TNF-α (Fig. 3G) and IL-6 (Fig. 3H) were produced in response to wild-type and recombinant VpOmpU in both RAW 264.7 and THP-1 cells.
Based on these results, the recombinant form of VpOmpU was used in all subsequent experiments.
VpOmpU elicits the proinflammatory responses via a TLR signaling pathway.
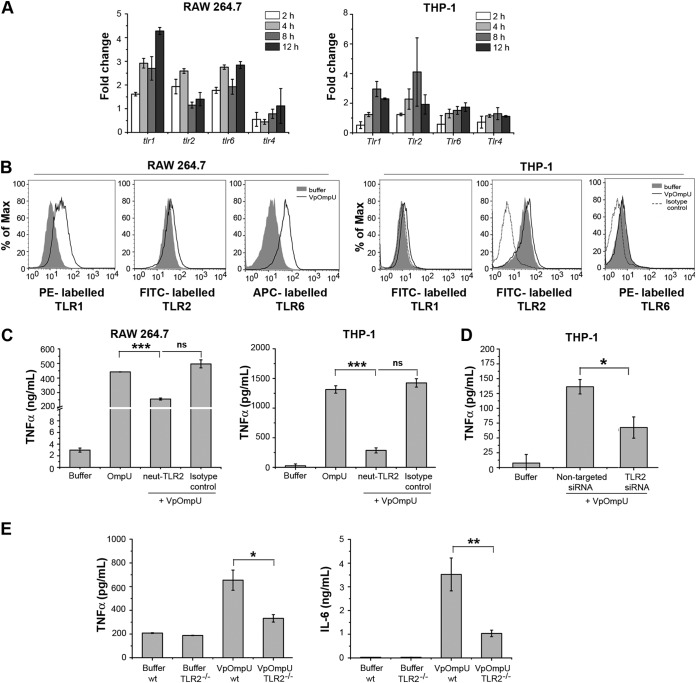
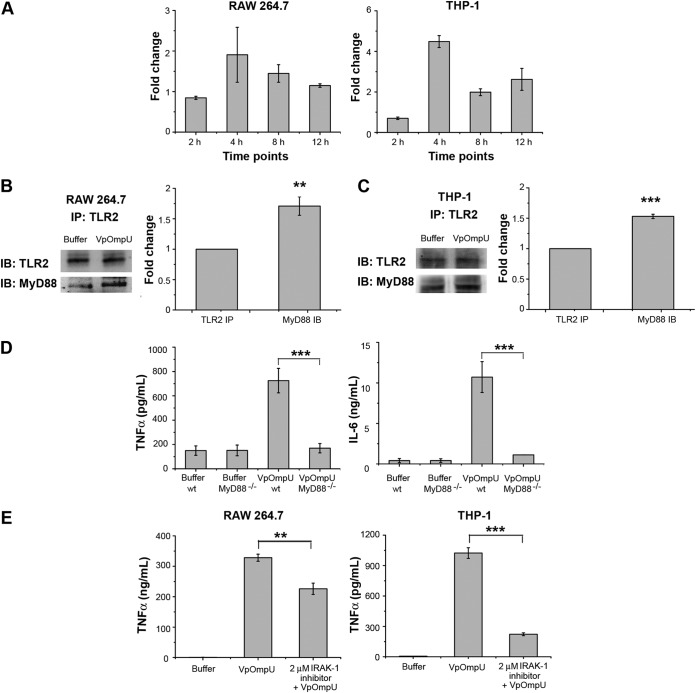
The observation that VpOmpU induces a proinflammatory response in monocytes and macrophages reflected the possibility that VpOmpU may be recognized as a PAMP by PRRs on the host cells. One of the major classes of PRRs that recognize bacterial PAMPs is the Toll-like receptors (TLRs) (22). Since V. parahaemolyticus is a noninvasive bacterium, VpOmpU will be most likely recognized as a PAMP by the surface TLRs present on the cells. Therefore, a time course analysis of gene expression of surface TLRs was done in VpOmpU-treated RAW 264.7 and THP-1 cells. An increase in gene expression of TLR2, TLR1, and TLR6 was observed in RAW 264.7 cells, whereas an upregulation of TLR2 and TLR1 was observed in THP-1 cells (Fig. 4A). In accordance with this, we observed an increase in the surface expression of TLR1, TLR2, and TLR6 in RAW 264.7 cells, while in THP-1 cells, an increase in surface expression of TLR1 and TLR2 was observed (Fig. 4B).
FIG 4.
The VpOmpU-mediated proinflammatory response is TLR2 dependent. (A) Upregulation of gene expression of TLR1, TLR2, and TLR6 at different time points in response to VpOmpU in RAW 264.7 cells and of TLR1 and TLR2 in THP-1 cells. Following treatment with VpOmpU, cells were harvested to isolate total RNA. cDNA was generated and subjected to RT-PCR. The CT values obtained were normalized to the respective CT values of the housekeeping genes. Fold change was calculated above buffer-treated cells, and results are expressed as mean ± SD from three independent experiments. (B) Increase in surface expression of TLR1, TLR2, and TLR6 in RAW 264.7 cells and of TLR1 and TLR2 in THP-1 cells in response to VpOmpU compared to the buffer-treated cells. Cells were treated with VpOmpU and analyzed for surface expression of TLRs by flow cytometry. (C) Decrease in TNF-α production upon TLR2 neutralization in VpOmpU-treated RAW 264.7 and THP-1 cells compared to the isotype-pretreated VpOmpU-treated cells and cells treated with only VpOmpU. Cells were pretreated with neutralizing antibody and the isotype for 1 h, followed by VpOmpU treatment for 24 h in RAW 264.7 cells and for 4 h in THP-1 cells. Supernatants were analyzed for TNF-α by ELISA. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus cells treated with VpOmpU only]). (D) Decrease in TNF-α production in response to VpOmpU upon knockdown of TLR2 by siRNA in VpOmpU-treated THP-1 cells compared to the nontargeted siRNA-treated VpOmpU-treated cells. Following siRNA knockdown, cells were treated with VpOmpU for 4 h, and supernatant was analyzed for TNF-α by ELISA. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus nontargeted siRNA-transfected VpOmpU-treated cells]). (E) Decrease in TNF-α and IL-6 production by VpOmpU-treated BMDMs differentiated from TLR2−/− mice compared to the wild-type control. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus VpOmpU-treated cells from wild-type mice]).
The above data indicate that TLR1 and TLR2 could be involved in recognition of VpOmpU in THP-1 cells, whereas in RAW 264.7 cells, TLR1, TLR2, and TLR6 could be involved in recognition of VpOmpU.
To confirm the involvement of TLR2, we used a neutralizing antibody. Upon neutralizing TLR2, we observed a significant decrease in the proinflammatory cytokine production in both THP-1 and RAW 264.7 cells, compared to that in isotype-treated cells, in response to VpOmpU (Fig. 4C). This result confirms that TLR2 is probably involved in recognition of VpOmpU in both monocytes and macrophages. To further confirm the involvement of TLR2 in monocytes, we performed small interfering RNA (siRNA)-mediated knockdown of TLR2 in THP-1 cells (see Fig. S2A in the supplemental material) and observed a decrease in cytokine production upon treatment of TLR2-knocked-down cells with VpOmpU (Fig. 4D). To confirm the involvement of TLR2 in macrophages, we used bone marrow-derived macrophages (BMDMs) from TLR2−/− mice and wild-type mice. We treated the BMDMs with VpOmpU and compared the cytokine production following 24 h of incubation. We observed significantly less production of TNF-α and IL-6 in TLR2−/− BMDMs than in wild-type BMDMs (Fig. 4E). These data confirmed that TLR2 is involved in recognition of VpOmpU in both monocytes and macrophages.
In response to a PAMP, TLRs are known to dimerize and activate the downstream signaling, and TLR2 is known to make heterodimers with TLR1 or TLR6. The gene expression profile (Fig. 4A) and surface expression profile (Fig. 4B) suggested that in RAW 264.7 cells, VpOmpU could be recognized by both TLR1/2 and TLR2/6 heterodimers, while in THP-1 cells, VpOmpU could be recognized by the TLR1/2 heterodimer only.
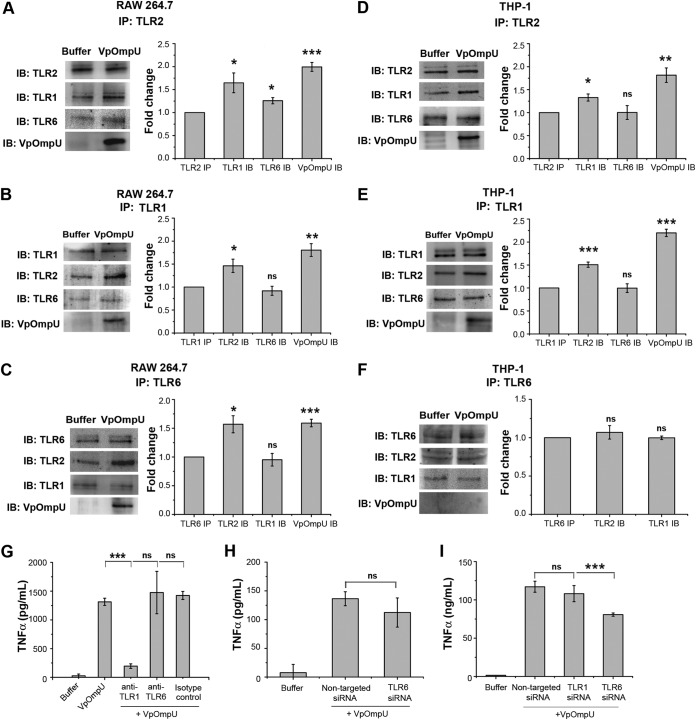
Therefore, to determine the heterodimerizing partner of TLR2, we pulled down the VpOmpU-treated cell lysates with anti-TLR2 antibody, and we observed that both TLR1 and TLR6 coimmunoprecipitated with TLR2 in RAW 264.7 cells (Fig. 5A). To further confirm this result, we immunoprecipitated VpOmpU-treated RAW 264.7 cell lysates with anti-TLR1 and anti-TLR6 antibodies, and we observed coimmunoprecipitation (co-IP) of TLR2 along with both TLR1 (Fig. 5B) and TLR6 (Fig. 5C). VpOmpU also coimmunoprecipitated with TLR2, TLR1, and TLR6 (Fig. 5A to C). These data indicated that TLR2 probably heterodimerizes with both TLR1 and TLR6 in response to VpOmpU in RAW 264.7 macrophages. In the case of THP-1 cells we observed that TLR1, but not TLR6, coimmunoprecipitated with TLR2 in response to VpOmpU treatment (Fig. 5D). To further confirm this result, we did reverse co-IP of VpOmpU-treated THP-1 cell lysates with anti-TLR1 and anti-TLR6 antibodies, and we observed coimmunoprecipitation of TLR2 along with TLR1 (Fig. 5E) but not with TLR6 (Fig. 5F). Consistent with this, VpOmpU coimmunoprecipitated only with TLR2 and TLR1 and not with TLR6 (Fig. 5D to F). These data indicated that in THP-1 cells, TLR2 probably heterodimerizes with TLR1 but not TLR6 in response to VpOmpU.
FIG 5.
Recognition of VpOmpU by TLR1/2 in THP-1 cells and by TLR1/2 and TLR2/6 in RAW 264.7 cells. (A to C) In RAW 264.7 cells, VpOmpU associates with TLR2, TLR1, and TLR6 as evidenced in coimmunoprecipitation (co-IP) and reverse co-IP studies using anti-TLR2, anti-TLR1, and anti-TLR6 antibodies. (A) TLR1 and TLR6 were coimmunoprecipitated with TLR2 in VpOmpU-treated RAW 264.7 cells as observed by immunoblotting (IB) and densitometric analysis of the bands. (B) TLR2 was coimmunoprecipitated with TLR1 in VpOmpU-treated RAW 264.7 cells as observed by immunoblotting and densitometric analysis of the bands. (C) TLR2 was coimmunoprecipitated with TLR6 in VpOmpU-treated RAW 264.7 cells as observed by immunoblotting blotting and densitometric analysis of the bands. (D to F) VpOmpU associates with TLR1 and TLR2 in THP-1 cells as observed by co-IP and reverse co-IP using anti-TLR2, anti-TLR1, and anti-TLR6 antibodies. (D) TLR1 was coimmunoprecipitated with TLR2 in VpOmpU-treated THP-1 cells as observed by immunoblotting and densitometric analysis of the bands. (E) TLR2 was coimmunoprecipitated with TLR1 in VpOmpU-treated THP-1 cells as observed by immunoblotting and densitometric analysis of the bands. (F) TLR2 did not coimmunoprecipitate with TLR6 in VpOmpU-treated THP-1 cells as observed by immunoblotting and densitometric analysis. For panels A to F, cell lysates were prepared after 15 min of VpOmpU treatment and immunoprecipitated with anti-TLR1, anti-TLR2, or anti-TLR6 antibody, and the IP lysates were further analyzed for TLRs coimmunoprecipitated together to identify the receptor complex. For the densitometric analysis, the band intensities of TLR1, TLR2, TLR6, or VpOmpU in the VpOmpU-treated samples above the respective band intensities in the buffer-treated samples were estimated. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus band intensities in the buffer-treated cells]). (G) Decrease in TNF-α production with anti-TLR1, but not anti-TLR6, neutralizing antibody in VpOmpU-treated THP-1 cells compared to isotype-pretreated VpOmpU-treated cells and cells treated only with VpOmpU. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus cells treated with VpOmpU only]). (H) No decrease in TNF-α production in THP-1 cells with knockdown of TLR6 by siRNA, compared to the nontargeted siRNA-transfected control, in response to VpOmpU. (I) Decrease in TNF-α production with siRNA-mediated knockdown of TLR6, but not TLR1, compared to the nontargeted siRNA-transfected control in RAW 264.7 cells in response to VpOmpU. For panels H and I, results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus VpOmpU-treated cells transfected with nontargeted siRNA]).
Therefore, the coimmunoprecipitation data suggested the involvement of both the TLR1/2 and TLR2/6 heterodimers in recognizing VpOmpU in RAW 264.7 cells but involvement of only the TLR1/2 heterodimer in THP-1 cells.
To confirm the above results, we used neutralizing antibodies against TLR1 and TLR6 in THP-1 cells followed by treatment with VpOmpU. We observed reduced production of TNF-α in cells pretreated with TLR1- but not with TLR6-neutralizing antibodies, confirming the involvement of the TLR1/2 heterodimer and not the TLR2/6 heterodimer in VpOmpU-mediated proinflammatory responses in THP-1 cells (Fig. 5G). Also, no decrease in TNF-α was observed after siRNA knockdown of TLR6 in THP-1 cells (Fig. 5H and S2B). Altogether, these results confirm the involvement of the TLR1/2 dimer in the VpOmpU-mediated proinflammatory response in THP-1 cells.
To confirm the TLR heterodimers involved in recognition of VpOmpU in macrophages, we knocked down TLR1 and TLR6 using siRNA in RAW 264.7 cells (Fig. S2C). Upon TLR6 knockdown, a decrease in TNF-α production was observed (Fig. 5I). However, when TLR1 was knocked down, there was no significant decrease in TNF-α production (Fig. 5I).
Although both TLR1/2 and TLR2/6 heterodimers are indicated by coimmunoprecipitation (Fig. 5A to C), the siRNA knockdown (Fig. 5I) implies that TLR6 is the major player in the recognition of VpOmpU in RAW 264.7 cells.
MyD88 and IRAK-1 are involved in TLR-mediated signaling by VpOmpU.
The TLR activation signal can be mediated through MyD88-dependent or -independent pathways (23). If MyD88 is recruited to the receptor complex as a result of TLR activation, it most certainly recruits IRAK-1 to the receptor complex (23). Gene expression analysis showed an upregulation of MyD88 in VpOmpU-treated THP-1 and RAW 264.7 cells, indicating its involvement in VpOmpU-mediated TLR-signaling (Fig. 6A). Further, we observed that MyD88 coimmunoprecipitated along with TLR2 when lysates of VpOmpU-treated RAW 264.7 (Fig. 6B) and THP-1 cells (Fig. 6C) were pulled down using anti-TLR2 antibody. These data suggested that VpOmpU-mediated TLR signaling involves MyD88. Further, we have confirmed the involvement of MyD88 by using MyD88-deficient (MyD88−/−) transgenic mice, where the TNF-α and IL-6 production in response to VpOmpU by BMDMs from the MyD88−/− mice was significantly lower than that with the wild-type mice (Fig. 6D).
FIG 6.
MyD88 and IRAK-1 are involved in VpOmpU-mediated proinflammatory responses. (A) Increase in gene expression of MyD88 in VpOmpU-treated RAW 264.7 and THP-1 cells at different time points. Fold change is calculated as value above that for buffer-treated cells, and results are expressed as mean ± SD from three independent experiments. (B) MyD88 coimmunoprecipitated with TLR2 upon VpOmpU treatment in RAW 264.7 cells as evidenced by immunoblotting and densitometric analysis of the bands. (C) MyD88 coimmunoprecipitated with TLR2 upon VpOmpU treatment in THP-1 cells as evidenced by immunoblotting and densitometric analysis of bands. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus band intensities in the buffer-treated cells]). (D) Decrease in TNF-α and IL-6 production in VpOmpU-treated BMDMs differentiated from MyD88−/− mice compared to the wild-type control. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus VpOmpU-treated BMDMs from wild-type mice]). (E) Decrease in TNF-α production with use of IRAK-1 inhibitor in response to VpOmpU treatment in RAW 264.7 and THP-1 cells. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus cells treated with VpOmpU only]).
To examine whether IRAK-1 is involved in VpOmpU-mediated signaling, we used a pharmacological inhibitor of IRAK-1. We observed a considerable decrease in the proinflammatory cytokine production in the VpOmpU-treated cells with the use of the IRAK-1 inhibitor in both RAW 264.7 and THP-1 cells (Fig. 6E).
These observations confirmed that in macrophages and monocytes, the proinflammatory responses generated on recognition of VpOmpU by the TLRs involve MyD88 and IRAK-1.
The NF-κB transcription factor is involved in TLR-mediated signaling of VpOmpU.
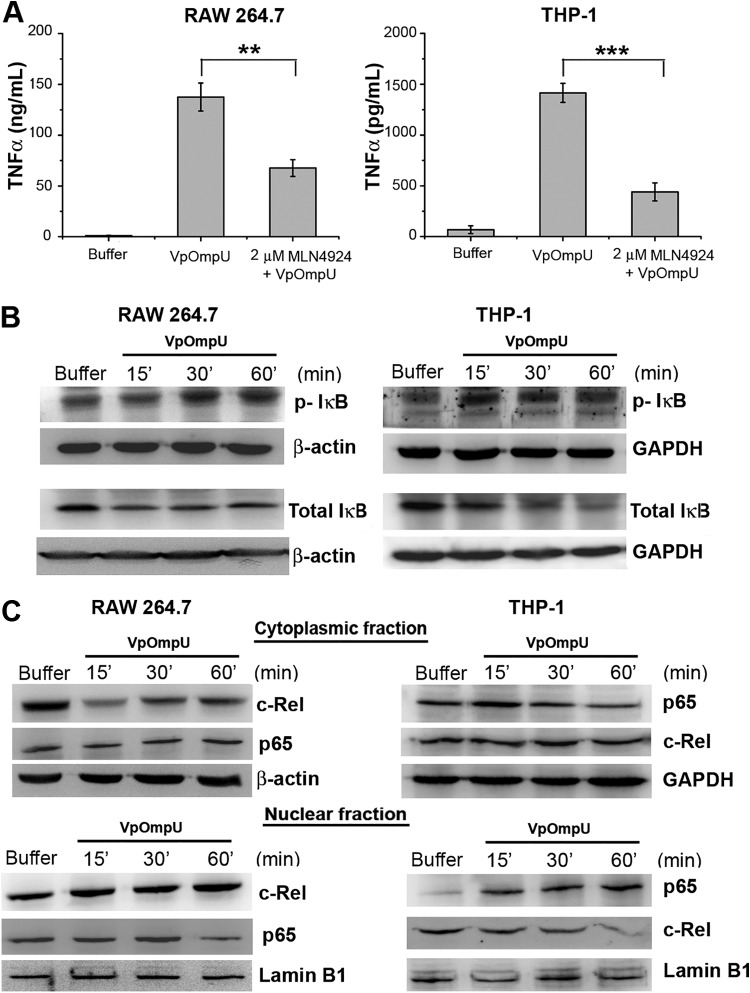
In general, TLR activation ultimately leads to the activation of transcription factor NF-κB (22). Upon pretreatment of cells with an NF-κB inhibitor (MLN4924) followed by VpOmpU stimulation, a decrease in the production of TNF-α was observed in both RAW 264.7 cells and THP-1 cells compared to VpOmpU-treated cells (Fig. 7A). These data indicated that NF-κB might be involved in VpOmpU-mediated proinflammatory responses.
FIG 7.
The NF-κB transcription factor is involved in VpOmpU-mediated proinflammatory responses. (A) Decrease in production of TNF-α in response to VpOmpU with the use of an inhibitor of NF-κB (MLN4924) in both RAW 264.7 and THP-1 cells. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus cells treated with VpOmpU only]). (B) Increase in phosphorylated IκB and decrease in total IκB in response to VpOmpU with increase in time in both RAW 264.7 and THP-1 cells. Cells were treated with VpOmpU and incubated for different times. Whole-cell lysates were prepared and analyzed by Western blotting for phosphorylated and total IκB. β-Actin and GAPDH were used as loading controls for the whole-cell lysates. (C) Translocation of NF-κB subunits c-Rel in RAW 264.7 and p65 in THP-1 from the cytoplasm to the nucleus in response to VpOmpU. Lamin B1 was used as a loading control for nuclear lysates, and β-actin and GAPDH were used as loading controls for the cytoplasmic lysates.
In an unstimulated cell, NF-κB remains bound to its inhibitor IκB and is present in the cytoplasm. Activation of the IRAK-1/4 complex results in the activation of signaling cascades that lead to the phosphorylation and degradation of IκB, rendering NF-κB free to translocate to the nucleus and transcribe the genes for proinflammatory cytokines. There are five members in the NF-κB subfamily. They either heterodimerize or homodimerize, and depending on the subunits and their nature of dimerization, the cell exerts pro- or anti-inflammatory responses (24). In the majority of cases, p65 and/or c-Rel is the member of the NF-κB family which dimerizes with p50 to transcribe proinflammatory genes. Therefore, phosphorylation and degradation of IκB are crucial for activation of NF-κB (24). In VpOmpU-treated RAW 264.7 and THP-1 cells, we observed phosphorylation and degradation of IκB, indicating the probable activation of NF-κB (Fig. 7B). Further, we also observed nuclear translocation of c-Rel in RAW 264.7 cells and of p65 in THP-1 cells (Fig. 7C).
Altogether, these experiments confirm that NF-κB is involved in VpOmpU-mediated proinflammatory responses.
The AP-1 transcription factor is also involved in VpOmpU-mediated signaling.
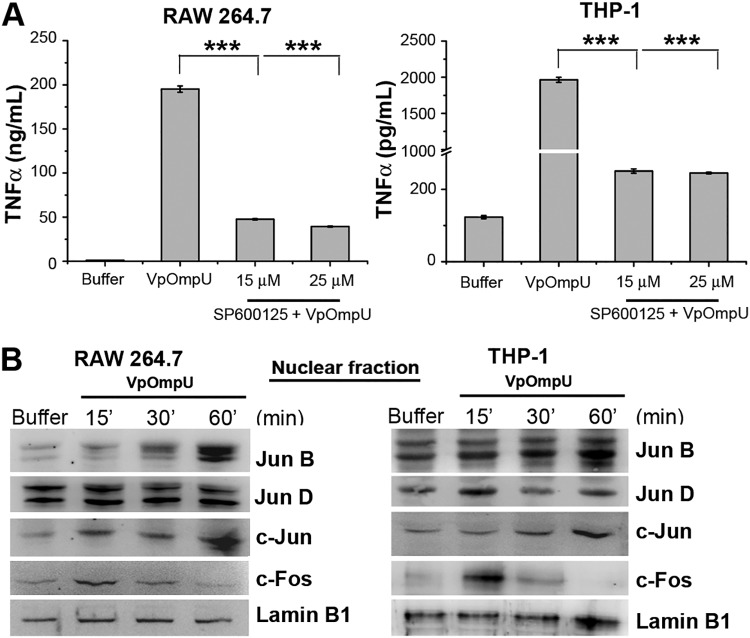
Apart from NF-κB, another transcription factor, AP-1, is also known to be involved in proinflammatory responses (23). To probe whether AP-1 has a role in VpOmpU-mediated proinflammatory response, we first used a chemical inhibitor against AP-1 (SP600125). We observed a decrease in production of TNF-α in VpOmpU-treated RAW 264.7 and THP-1 cells in the presence of the inhibitor (Fig. 8A), indicating a role of AP-1 transcription in the VpOmpU-mediated proinflammatory response.
FIG 8.
The AP-1 transcription factor is involved in VpOmpU-mediated proinflammatory responses. (A) Decrease in the production of TNF-α in response to VpOmpU in the presence of an AP-1 inhibitor (SP600125) in both RAW 264.7 and THP-1 cells. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus cells treated with VpOmpU only]). (B) Increase in AP-1 family members in the nuclear lysates of VpOmpU-treated cells, namely, JunB, c-Jun, and c-Fos in RAW 264.7 cells and JunB, c-Jun, c-Fos, and JunD in THP-1 cells. Lamin B1 was used as a loading control for nuclear lysates.
The majority of the subunits of the AP-1 transcription factor are in one of two families, Fos and Jun. The members of the Fos and Jun families generally heterodimerize and form AP-1 to function as a transcription factor (25). Nuclear fractions of VpOmpU-treated RAW 264.7 and THP-1 cells were analyzed for the involvement of AP-1 subunits in VpOmpU-mediated proinflammatory processes. By observing the AP-1 subunit levels in nuclear lysates, we came to the conclusions that in VpOmpU-treated RAW 264.7 cells, AP-1 subunits JunB, c-Jun, and c-Fos could be involved and that in THP-1 cells, AP-1 subunits JunB, JunD, c-Jun, and c-Fos could be involved (Fig. 8B).
TLR2 activation by VpOmpU leads to the MAP kinase activation.
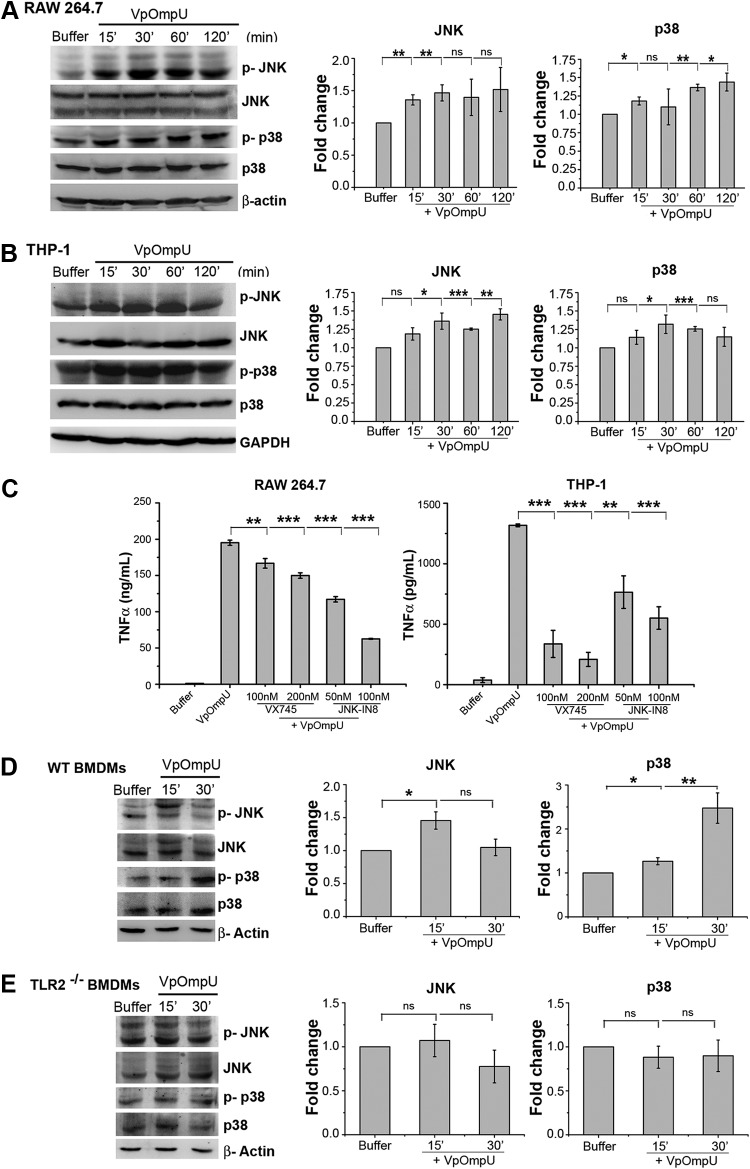
Generally, MAP kinase activation leads to the activation of AP-1 (23). To probe whether MAP kinases are involved in VpOmpU-mediated proinflammatory responses in RAW 264.7 and THP-1 cells, we checked the phosphorylation levels of both JNK and p38. We observed phosphorylation (indicative of activation status) of both p38 and JNK in RAW 264.7 (Fig. 9A) and THP-1 (Fig. 9B) cells. Further, we observed a decrease in cytokine production in VpOmpU-treated cells pretreated with chemical inhibitors against JNK and p38 (Fig. 9C). To determine whether activation of MAP kinases was TLR2 mediated, we investigated the phosphorylation levels of p38 and JNK in BMDMs from TLR2−/− mice. We found that in the absence of TLR2, MAP kinases (p38 and JNK) were not activated in response to VpOmpU (Fig. 9D and E).
FIG 9.
Proinflammatory response by VpOmpU involves TLR-mediated p38 and JNK MAP kinase activation. (A and B) Increased phosphorylation of p38 and JNK in RAW 264.7 (A) and THP-1 (B) cells in response to VpOmpU as evidenced by immunoblotting and densitometric analysis of the bands. Whole-cell lysates were analyzed for the phosphorylation of p38 and JNK at different times following VpOmpU treatment. Total p38 and JNK in the cells were also determined. Densitometric analysis of the immunoblots confirmed increased phosphorylation of p38 and JNK in RAW 264.7 and THP-1 cells in response to VpOmpU. For the densitometric analysis, the band intensities of p-p38 or p-JNK in the samples were calculated as those above the band intensities of p38 or JNK, respectively, and fold changes upon VpOmpU treatment were estimated with respect to the buffer-treated cells. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus band intensities in the buffer-treated cells]). (C) Decrease in TNF-α in response to VpOmpU upon pretreatment with inhibitors of the MAP kinases p38 (VX745) and JNK (JNK-IN8) in RAW 264.7 and THP-1 cells. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus cells treated with VpOmpU only]). (D and E) Increased phosphorylation of p38 and JNK in VpOmpU-treated BMDMs from wild-type (WT) mice but no change in phosphorylation status of p38 and JNK in VpOmpU-treated BMDMs from TLR2−/− mice. Densitometric analysis of the immunoblots shows increased phosphorylation of JNK at 15 min and of p38 at both 15 min and 30 min in the wild-type BMDMs and no change in the phosphorylation status of JNK and p38 in the TLR2−/− BMDMs in response to VpOmpU. For the densitometric analysis, the band intensities of p-p38 or p-JNK in the samples were calculated as those above the band intensities of p38 or JNK, respectively, and fold changes upon VpOmpU treatment were estimated with respect to the buffer-treated cells. Results are expressed as mean ± SEM from three independent experiments (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, P > 0.05 [versus band intensities in buffer-treated cells]). β-Actin and GAPDH were used as the loading controls in the immunoblots of the whole-cell lysates.
DISCUSSION
OmpU is a major outer membrane protein which is present across the Vibrio species. In most of the Vibrio species it plays a role in pathogenesis and modulation of host immune function. In V. alginolyticus and V. harveyi, OmpU has been considered as a vaccine candidate (13, 26). OmpU of V. parahaemolyticus (VpOmpU) has also been reported to be immunogenic in yellow croaker fish (17), though this has not been well elucidated.
Many porins, such as PorB of Neisseria meningitidis (27) and OmpS1 and OmpS2 of Salmonella enterica serovar Typhi (28), have been found to induce proinflammatory responses. In this study, we have characterized the role of VpOmpU in modulating the host’s innate immune responses. We have observed that VpOmpU activates macrophages and monocytes as indicated by production of proinflammatory mediators, such as TNF-α and IL-6, in both macrophages and monocytes and of nitric oxide (NO) in macrophages (Fig. 3). Further, to identify the pattern recognition receptor (PRR) which recognizes VpOmpU, we started with the cue that since VpOmpU is present on the outer membrane of V. parahaemolyticus, which is a noninvasive bacterium, it must be recognized by the surface PRRs of the cell. Of the surface PRRs, TLRs are mostly involved in the recognition of bacterial ligands (22). TLR1, TLR2, TLR4, TLR5, and TLR6 are the common surface TLRs (29). Generally, upon activation, TLRs form homo- or heterodimers; e.g., TLR2 forms heterodimers with either TLR1 or TLR6, whereas TLR4 forms homodimers (30). Various other outer membrane proteins, such as PorB of Neisseria meningitidis (31) and OspA of Borrelia spirochetes (32), have been shown to be recognized by TLR1/2 heterodimers. OmpA, an outer membrane protein of Shigella flexneri (33), and OmpS2 of S. Typhi (28) also are known to be recognized by TLR2/6 heterodimers. We observed that VpOmpU mediates its proinflammatory response through TLR2 in both monocytes and macrophages (Fig. 4). Further, to find the binding partner of TLR2, we did coimmunoprecipitation of VpOmpU-treated cell lysates with anti-TLR2 antibody and observed that only TLR1 is present along with TLR2 in VpOmpU-treated THP-1 monocytes, whereas both TLR1 and TLR6 are present along with TLR2 in macrophages. Reverse coimmunoprecipitation with anti-TLR1 and anti-TLR6 antibodies confirmed the result (Fig. 5). This result is particularly intriguing since, to the best of our knowledge, this is the first report of a natural PAMP, i.e., VpOmpU, being recognized by both TLR1/2 and TLR2/6 heterodimers. As mentioned above, while TLR1/2 and TLR2/6 heterodimers have been known to recognize discrete ligands, a previous report indicated the recognition of a synthetic lipoprotein (Pam2CSK4, which is a diacylated lipoprotein) by both the TLR1/2 and TLR2/6 heterodimers (34, 35). Buwitt-Beckmann et al. (34) also showed that recognition of a ligand by TLR6 depends not only on the acylation of the ligand but also on the peptide sequence and the overall structure of the protein ligand. However, Farhat et al. (35) showed that this difference in ligand recognition did not alter the downstream signaling elicited by the dimers. They also speculated that this difference in ligand recognition was necessary to increase the array of ligands recognized by the cells.
Bacterial porins of S. enterica serovar Typhimurium (36), PorB of N. meningitidis (27), OmpA of Shigella flexneri (33), P2 porin of Haemophilus influenzae (37), and OmpU of V. cholerae (38) have previously been reported to induce NF-κB- or AP-1-mediated proinflammatory responses. We have shown in the present study that VpOmpU-mediated activation of TLR signaling leads to activation of the transcription factors NF-κB and AP-1 in both THP-1 monocytes and RAW 264.7 macrophages (Fig. 7 and 8). AP-1 is activated by upstream MAP kinases (Fig. 9). Furthermore, in this study we have also shown that the activation of MAP kinases (p38 and JNK) is TLR2 dependent (Fig. 9). These data suggest that a difference in the recognition of VpOmpU by the TLR1/2 or TLR2/6 heterodimer does not affect the downstream signaling elicited by it.
VpOmpU shares about 70% sequence identity with V. cholerae OmpU (VcOmpU) (Fig. 1). Like VcOmpU, VpOmpU is also porin in nature (Fig. 2). Extensive study from our laboratory and others confirmed that VcOmpU is recognized only by TLR1/2 in both monocytes and macrophages (38), whereas our present study shows that VpOmpU can be recognized by both TLR1/2 and TLR2/6 in macrophages, with TLR2/6 being favorable for induction of proinflammatory responses. Further, in the case of VpOmpU, the activation of both MAP kinases (p38 and JNK) is mainly TLR dependent, but in the case of VcOmpU, only p38 MAP kinase activation is dependent on TLR-mediated signaling in macrophages (unpublished data).
Altogether, our study shows that VpOmpU elicits proinflammatory responses via TLR1/2 in monocytes and via TLR2/6 in macrophages. Recognition of VpOmpU by the TLR heterodimer leads to MyD88-IRAK-1-dependent activation of NF-κB and AP-1 transcription factors for such proinflammatory response generation.
MATERIALS AND METHODS
Bacterial strains and chemicals.
The V. parahaemolyticus strain was obtained from the Microbial Type Culture Collection (MTCC) and Gene Bank facility (MTCC code 451) of the Institute of Microbial Technology, Chandigarh, India. All of the DNA-modifying enzymes were obtained from New England Biolabs (USA). Luria-Bertani (LB) broth and antibiotics were from Himedia, Mumbai, India. Brain heart infusion (BHI) medium was from Fluka, USA. Plasmid and DNA isolation kits were obtained from Qiagen. Ni-NTA agarose was obtained from Qiagen.
PCR amplification and DNA manipulation.
The full-length nucleotide sequence encoding the putative OmpU protein of V. parahaemolyticus (VpOmpU) was retrieved from the NCBI server (available online at https://www.ncbi.nlm.nih.gov/pubmed) (GenBank accession no. HM042874.1). The N-terminal signal sequence of the VpOmpU protein was determined using SignalP (available online at http://www.cbs.dtu.dk/services/SignalP/). The nucleotide sequence encoding VpOmpU, omitting the N-terminal signal sequence, was amplified by PCR using V. parahaemolyticus MTCC 451 genomic DNA as the template. The following primer sequences were designed based on the VpOmpU nucleotide sequence (GenBank accession no. HM042874.1), omitting the sequence for the N-terminal signal peptide: forward primer, 5′-TACATATGGCTGAACTTTACAACCAAG-3′; reverse primer, 5′-CGGGATCCTTAGAAGTCGTAACGTAGAC-3′. NdeI and BamHI restriction endonuclease sites (in italic) were incorporated in the forward and reverse primers, respectively, so as to allow cloning of the amplified nucleotide sequence into the target expression vector.
The amplified PCR product was first cloned into the TA cloning vector (pTZ57R/T; Fermentas Life Sciences) and transformed into E. coli TOP10 cells (Invitrogen). Transformed cells were screened for the positive plasmid harboring the cloned nucleotide sequence of the VpOmpU gene by PCR, using the gene-specific primers mentioned above. Cloned sequence encoding VpOmpU was excised from the pTZ57R/T vector by restriction digestion with NdeI and BamHI and recloned into the pET-14b bacterial expression vector (Novagen), and the recombinant pET-14b vector (pET-14b/VpOmpU) was transformed into the E. coli TOP10 cells. The transformants were selected on ampicillin (50 μg/ml)-supplemented LB agar plates. Bacterial colonies were screened for the recombinant pET-14b/VpOmpU plasmid by colony PCR using the gene-specific primers. Recombinant pET-14b/VpOmpU plasmid was purified, and the construct was verified by DNA sequencing of the cloned nucleotide segment for VpOmpU.
Overexpression and purification of VpOmpU.
Recombinant pET-14b/VpOmpU plasmid was transformed into E. coli Origami B cells. A single colony of E. coli Origami B cells harboring the pET-14b/VpOmpU plasmid was inoculated into 20 ml LB medium containing ampicillin (50 μg/ml), and the cells were grown at 37°C overnight. For large-scale protein expression, 1 liter of LB medium supplemented with 50 μg/ml ampicillin was inoculated with 20 ml of the overnight-grown seed culture and was grown at 37°C. Protein overexpression was induced by addition of 1 mM IPTG when the optical density at 600 nm (OD600) of the culture medium reached 0.4 to 0.6. After induction for 3 h at 37°C, the cells were harvested by centrifugation at 3,220 × g for 30 min. The cells were resuspended in 10 ml of 20 mM sodium phosphate buffer (pH 7.0) containing bacterial protease inhibitor cocktail (Sigma-Aldrich, USA). The cells were disrupted by sonication. Insoluble inclusion bodies were separated from the soluble fraction of the cell lysate by centrifugation at 18,500 × g for 30 min. The presence of the majority of the recombinant VpOmpU protein in the insoluble inclusion body fraction was observed by SDS-PAGE and Coomassie blue staining. The crude inclusion bodies were washed twice with phosphate-buffered saline (PBS) (20 mM sodium phosphate buffer [pH 7.0] containing 150 mM sodium chloride). Subsequently, the inclusion bodies were solubilized in PBS containing 8 M urea (10 ml of the urea-containing buffer was used to solubilize the inclusion bodies obtained from 1 liter of culture) by constant stirring at 25°C. Insoluble debris was separated by centrifugation at 18,500 × g for 30 min at 4°C. Recombinant VpOmpU protein was further purified from the crude urea-solubilized inclusion body fraction by passage through Ni-NTA agarose affinity chromatography resins (Qiagen) under denaturing conditions in the presence of 8 M urea. The urea-solubilized inclusion body fraction was adjusted with 10 mM imidazole and was applied to Ni-NTA agarose resin preequilibrated with PBS containing 8 M urea (4 ml Ni-NTA agarose resin/10 ml of solubilized inclusion body fraction). After washing with 10 volumes of PBS containing 8 M urea and 40 mM imidazole, the bound protein was eluted with PBS containing 8 M urea and 300 mM imidazole. The purity of the protein was examined by SDS-PAGE and Coomassie blue staining.
Refolding of the recombinant VpOmpU protein.
Purified recombinant VpOmpU protein was subjected to refolding by the rapid-dilution method. The purified protein in 8 M urea was diluted into refolding buffer (PBS containing 10% glycerol, 0.5% lauryldimethylamine N-oxide [LDAO] [Sigma-Aldrich, USA]) at 25°C with constant shaking for 10 min. The diluted protein was further incubated at 4°C overnight to allow optimal refolding of the protein. Thereafter, the refolding mixture was centrifuged at 18,500 × g for 30 min at 4°C to remove the insoluble aggregates formed during the refolding process. The soluble fraction was immediately subjected to size exclusion chromatography by passage through a Sephacryl S-200 column (1.6 by 60 cm; bed volume, 120 ml; flow rate, 1 ml/min) (GE Healthcare) equilibrated with 10 mM Tris-HCl buffer (pH 7.6) containing 10 mM NaCl and 0.5% LDAO. Eluted fractions containing the VpOmpU protein (eluted at around 40 ml of elution volume) were analyzed by SDS-PAGE and Coomassie blue staining, pooled, and concentrated by ultrafiltration using Millipore Ultra 10-kDa-cutoff filters. The protein concentration was estimated using the Bradford reagent (Sigma-Aldrich, USA), using bovine serum albumin (BSA) as the standard. The final soluble fraction of the purified form of the refolded recombinant VpOmpU protein was analyzed by SDS-PAGE and Coomassie blue staining. The N-terminal 6×His tag of the recombinant protein was removed by treatment with thrombin (1 unit of enzyme/300 μg of protein) for 2 h at 37°C. The reaction was stopped with 2 mM phenylmethylsulfonyl fluoride (PMSF).
Purification of wild-type VpOmpU from the V. parahaemolyticus outer membrane fraction.
V. parahaemolyticus was grown in 2 liters brain heart infusion (BHI) medium until the OD600 of the culture reached 1.0. The bacterial culture was subsequently centrifuged at 2,050 × g for 30 min. The bacterial cell pellet was resuspended in 20 mM Tris-HCl (pH 7.6), containing protease inhibitor cocktail (Sigma-Aldrich, USA), and the bacterial cells were lysed by sonication and subjected to centrifugation at 18,500 × g for 50 min. The supernatant was collected and subjected to ultracentrifugation at 320,000 × g for 20 min. The pellet thus obtained was resuspended in PBS and ultracentrifuged at 320,000 × g for 20 min. The pellet was further subjected to treatment with 1% N-Sarkosyl in PBS for 30 min at 37°C and was ultracentrifuged at 105,000 × g for 1 h. The pellet was washed twice with 20 mM Tris-HCl (pH 7.6) at 320,000 × g for 20 min and was then subjected to treatment with 4% Triton X-100 in 20 mM Tris-HCl (pH 8.0) for 20 min at 37°C. The supernatant thus obtained contained the outer membrane proteins. The extracted protein fractions were incubated with 10 mM dithiothreitol (DTT) for 10 min at room temperature and were further diluted 5-fold with 20 mM Tris-HCl buffer (pH 7.6) containing 0.1% Triton X-100. The protein sample was loaded onto a DEAE-cellulose column equilibrated with 20 mM Tris-HCl (pH 7.6) containing 0.1% Triton X-100. Bound proteins were eluted with a gradient of 50 to 250 mM NaCl in Tris-HCl (pH 7.6) buffer at a flow rate of 1 ml/min. The eluted fractions were subjected to SDS-PAGE and Coomassie blue staining analysis for detection of the eluted proteins. The eluted fractions were collected, selectively pooled, and subjected to size exclusion chromatography on a Sephacryl S-200 column (1.6 by 60 cm; bed volume, 120 ml; flow rate, 1 ml/min) (GE Healthcare) equilibrated with 10 mM Tris-HCl (pH 7.6) buffer containing 10 mM sodium chloride and 0.5% LDAO. The fractions that eluted at around 36 ml contained the purified form of VpOmpU protein as analyzed by SDS-PAGE and Coomassie blue staining.
Immunological relatedness between the wild-type and recombinant VpOmpU proteins.
Polyclonal antiserum was generated in rabbit using the purified refolded form of the recombinant His-tagged VpOmpU protein as the antigen. The antiserum was raised in a New Zealand White rabbit using the Polyclonal Antibody Production Service of Bangalore Genei, India. The antiserum recognized the wild-type VpOmpU extracted from the V. parahaemolyticus outer membrane fraction in the immunoblot assay, confirming the antigenic identity/immunological relatedness between the wild-type and recombinant forms of VpOmpU generated in our study.
Far-UV CD.
The far-UV circular dichroism (CD) spectra were collected on an Applied Photophysics Chirascan spectropolarimeter equipped with a Peltier-based temperature control system, using a 5-mm-path-length quartz cuvette. Far-UV CD spectra of the purified refolded recombinant VpOmpU and wild-type VpOmpU (extracted from the V. parahaemolyticus outer membrane fraction) were recorded between 200 and 260 nm. Protein concentrations were adjusted to 1.5 to 3.0 μM in 10 mM Tris-HCl buffer (pH 7.6) containing 10 mM NaCl and 0.5% LDAO. The resulting spectra were averaged (average of three spectra) and were corrected with the corresponding buffer spectra. All of the spectra were recorded at 25°C.
Liposome-swelling assay.
Functional channel-forming activity of VpOmpU (wild-type and refolded recombinant proteins) in the membrane lipid bilayer was determined by the conventional liposome-swelling assay following the method described by Nikaido and Rosenberg (39), with slight modifications. Liposomes were prepared with egg yolk phosphatidylcholine (Sigma-Aldrich, USA) and dicetylphosphate (Sigma-Aldrich, USA) following the method described earlier, with modifications. Phosphatidylcholine (4.7 mg dissolved in 500 μl chloroform) and dicetylphosphate (0.10 mg dissolved in 1 ml of a chloroform-methanol mixture [1:1, vol/vol]) were mixed together in a round-bottom flask, and the lipid film was allowed to generate by evaporation in a vacuum desiccator for 4 h. To this completely dried and solvent-free lipid film, 1 ml of the VpOmpU protein solution (in 5 mM Tris-HCl buffer [pH 7.6]) was added. The refolded recombinant VpOmpU protein was used at up to a 25 μg/ml final concentration, and the wild-type protein was used at up to a 30 μg/ml final concentration. Control liposomes were prepared by mixing the dried lipid film in 5 mM Tris-HCl buffer (pH 7.6) without the protein. Proteoliposomes and the control liposomes were subjected to ultracentrifugation at 350,000 × g for 15 min at 4°C and washed thrice with 5 mM Tris-HCl buffer (pH 7.6). The pellets were dried under reduced pressure in a vacuum desiccator overnight and then resuspended in 0.6 ml of same buffer containing 15% dextran 40,000 (Sigma-Aldrich, USA) by incubating for 2 h at 25°C. For the liposome-swelling assay, 20 μl of the proteoliposomes or the control liposomes was mixed rapidly in a cuvette with 580 μl of 30 mM sugar solution (arabinose, glucose, sucrose, and raffinose [Sigma-Aldrich, USA]) in 5 mM Tris-HCl buffer (pH 7.6), and the OD400 was continuously monitored for 10 min at intervals of 10 s. The assay was carried out at room temperature in triplicate.
Sequence alignment and protein structural model.
An amino acid sequence alignment was generated with CLUSTALW using the server available online (https://www.genome.jp/tools-bin/clustalw) and was visualized with ESPript (http://espript.ibcp.fr/ESPript/ESPript/).
The VpOmpU polypeptide sequence was subjected to a BLAST search in the NCBI server (http://blast.ncbi.nlm.nih.gov/Blast.cgi) against protein sequences having experimentally determined three-dimensional structures in the Protein Data Bank (PDB) (http://www.rcsb.org/pdb/home/home.do). The most suitable template was found to be OmpU of Vibrio cholerae (PDB entry 6EHB). Therefore, a homology-based structural model of VpOmpU was generated based on the structural coordinates of 6EHB using the SWISS-MODEL server (http://www.expasy.org/spdbv/). A membrane-inserted structural model of VpOmpU was constructed with the OPM server found online (http://opm.phar.umich.edu/server.php). Protein structural models were visualized with PyMOL (http://pymol.org).
Cell lines and culture conditions.
The RAW 264.7 (a murine macrophage cell line) and THP-1 (a human monocytic cell line) cells (NCCS, Pune, India) used in this study were maintained in RPMI 1640 supplemented with 10% fetal bovine serum (FBS), 100 units/ml of penicillin, and 100 μg/ml of streptomycin (Invitrogen, Life Technologies, USA) at 37°C and 5% CO2. In all of the experiments with VpOmpU, cells were pretreated with 10 μg/ml of polymyxin B (Sigma-Aldrich, USA) for 30 min to prevent any LPS contamination from interfering with the responses.
Cell viability assay.
Cells (1 × 105) were plated in 100 μl medium in a 96-well plate and treated with different concentrations of VpOmpU for 24 h. The buffer (10 mM Tris-HCl with 0.5% LDAO) was used as a negative control. The amount of buffer added to the cells was same as that for the highest concentration of VpOmpU. The cell viability was quantified using the MTT assay (EZ Count kit; HiMedia, Mumbai, India) according to the manufacturer’s protocol.
Quantification of NO.
RAW 264.7 and THP-1 cells were plated at a density of 1 × 106/ml in a 6-well plate and treated with VpOmpU. At different time points, supernatant was collected for quantification of nitric oxide (NO) using Griess reagent (Sigma-Aldrich, USA). Equal volumes of Griess reagent and the supernatant were mixed and incubated in the dark for 15 min, and the optical density was measured at 540 nm. The NO was quantified using a standard curve prepared with different concentrations of sodium nitrite (Sigma-Aldrich, USA), and 1 μg/ml of E. coli lipopolysaccharide (LPS) (Sigma-Aldrich, USA) was used as a positive control in the experiments. Buffer (10 mM Tris-HCl with 0.5% LDAO) was used a negative control. For time dependency analysis, buffer was added in an equal amount as for VpOmpU, and for dose dependency studies, the amount of buffer added was equivalent to that for the highest dose of VpOmpU.
Quantification of TNF-α and IL-6.
Cells were plated at a density of 1 × 106/ml in a 6-well plate and treated with VpOmpU. At different time points, supernatant was collected. TNF-α and IL-6 were quantified by ELISA (BD Biosciences, USA) by the manufacturer’s protocol; 1 μg/ml of E. coli lipopolysaccharide (LPS) (Sigma-Aldrich, USA) was used as a positive control in the experiments. Buffer (10 mM Tris-HCl with 0.5% LDAO) was used as a negative control. For time dependency analysis, buffer was added in an equal amount as for VpOmpU, and for dose dependency studies, the amount of buffer added was equivalent to that for the highest dose of VpOmpU.
Gene expression analysis.
Cells (2 × 106) were plated in 2 ml medium and treated for different time with 5 μg/ml of VpOmpU. At all time points, buffer (equivalent to the amount of VpOmpU being added) was used as a control. Cells were then harvested, and RNA was isolated according to the manufacturer’s protocol using the Nuclepore RNA isolation kit (Genetix). The cDNA was then synthesized using the Verso-cDNA kit (Thermo-Fischer). PCRs were done using the Maxima SYBR green quantitative PCR (qPCR) master mix (Thermo Fischer) and the Eppendorf Realplex master cycler. The primer sequences used were from the primer bank and were synthesized by Integrated DNA Technologies. The fold change of gene expression in VpOmpU-treated cells was calculated as that above expression in the buffer-treated cells for each time point. The threshold cycle (CT) values of both the buffer-treated and VpOmpU-treated cells were normalized to the respective CT values of the housekeeping genes.
Analysis of surface expression of TLRs.
Cells (1 × 106) were plated in a 6-well plate and treated with 5 μg/ml of VpOmpU for different times. Buffer (10 mM Tris-HCl with 0.5% LDAO)-treated cells were used as a control. Cells were then harvested and washed twice with flow cytometry buffer (PBS supplemented with 1% FBS and 0.05% sodium azide). RAW 264.7 cells were incubated with mouse Fc block for 15 min at 4°C and in THP-1 cells, isotype controls were used. RAW 264.7 and THP-1 cells were then incubated with fluorescein isothiocyanate (FITC)-, phycoerythrin (PE)-, or allophycocyanin (APC)-tagged anti-TLR antibody or the isotype control for 1 h at 4°C. Cells were then washed twice with ice-cold flow cytometry buffer and analyzed on a FACSCalibur flow cytometer (BD Biosciences, USA). FITC-tagged anti-mouse/human TLR2 antibody, PE-tagged anti-mouse TLR1, FITC-tagged anti-human TLR1, and PE-tagged anti-human TLR6 antibodies and the isotype controls were from eBioscience, and APC-tagged anti-mouse TLR6 antibody was from R&D Technologies.
Neutralization of TLRs.
Cells were plated in a 96-well plate at a density of 1 × 106 cells/ml. Cells were pretreated for 1 h with 5 μg/ml anti-TLR neutralizing antibodies or the isotype controls and then treated with 5 μg/ml of VpOmpU. Supernatants were collected after 4 h for THP-1 cells and after 24 h for RAW 264.7 cells. TNF-α was then quantified using ELISA. Neutralizing antibodies against mouse/human TLR2 and their isotypes were from BioLegend, and neutralizing antibodies against human TLR1 and TLR6 and the isotypes were from InvivoGen.
Inhibitor studies.
Cells were plated at a density of 1 × 106 cells/ml. Cells were then pretreated for 1 h with different pharmacological inhibitors and treated with 5 μg/ml of VpOmpU for 24 h for RAW 264.7 cells and for 4 h for THP-1 cells. The supernatants were then collected and analyzed for TNF-α by ELISA. The inhibitors for IRAK-1, AP-1 (SP600125), JNK (JNK-IN8), and p38 (VX745) were from Sigma-Aldrich, USA, and the inhibitor for NF-κB (MLN4924) was from Boston Biochem, United Kingdom.
siRNA knockdown.
For THP-1 cells, 7 × 104 cells were plated in a 24-well plate and transfected using ONTARGETplus siRNA against human TLR2 or human TLR6 or the ONTARGETplus nontargeted siRNA pool (Dharmacon, GE), using DharmaFECT 1 according to the manufacturer’s protocol. Knockdown was observed using flow cytometry at 48 h (see Fig. S1A and B in the supplemental material). After 48 h of transfection, cells were treated with 5 μg/ml of VpOmpU for 4 h, after which the supernatants were collected and analyzed for TNF-α using ELISA. For RAW 264.7 cells, 2.5 × 105 cells were plated in a 24-well plate and transfected with ONTARGETplus siRNA against mouse TLR1 or mouse TLR6 or with the ONTARGETplus nontargeted siRNA pool (Dharmacon, GE) using FuGENE HD (Promega) at a ratio of 1:5 (RNA to FuGENE) for 24 h, after which knockdown was analyzed by semiquantitative PCR (Fig. S1C) and cells were treated with 10 μg/ml of VpOmpU for 12 h. Supernatants were then collected, and TNF-α was quantified using ELISA.
BMDMs from wild-type, TLR2−/−, and MyD88−/− mice.
The protocols for animal handling were approved by our Institutional Animal Ethics Committee (IISERM/SAFE/PRT/2017-2018/001). Six- to 8-week-old mice (C57BL6 wild type, TLR2−/−, or MyD88−/−) were euthanized, and their femur and tibia bones were extracted. The femurs and tibias were then cleaned of any muscle tissue and washed with ice-cold PBS. They were then dipped in 70% alcohol for 2 min and transferred to RPMI medium. The epiphyses of the bones were then cut using sterile scissors, and the bones were flushed with RPMI medium, releasing the bone marrow cells into the medium. These bone marrow cells were then differentiated to bone marrow-derived macrophages (BMDMs) using macrophage colony-stimulating factor (M-CSF). Briefly, cells were suspended in bone marrow differentiation medium (RPMI 1640 supplemented with 10% FBS, 100 units/ml of penicillin and streptomycin each, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids [NEAA], 1% β-mercaptoethanol, and 20 ng/ml of M-CSF) and plated in 24-well plates. They were then incubated at 37°C with 5% CO2. The medium was changed every 2 days, and fresh bone marrow differentiation medium was added. On day 7, the medium was changed to medium without M-CSF, and cells were treated with 5 μg/ml of VpOmpU or the equivalent amount of buffer (10 mM Tris-HCl with 0.5% LDAO) for 24 h. The supernatants were collected and analyzed for TNF-α and IL-6 using ELISA.
Whole-cell lysates and nuclear lysates.
Cells were harvested at 2,000 × g and washed twice with PBS. The pellet was then resuspended in whole-cell lysis buffer (50 mM Tris-Cl, 150 mM NaCl, 0.1% SDS, and 0.1% TritonX-100, pH 8) with mammalian protease inhibitor cocktail (Sigma-Aldrich, USA). This mixture was then subjected to sonication at 10 A for 15 s with 3 pulses of 5 s each. It was then centrifuged at 16,000 × g for 30 min, and the supernatant was collected as the whole-cell lysate.
For nuclear lysates, cells were harvested and washed twice with PBS. The pellet volume was measured, and the pellet was dissolved in 5 times the pellet volume in hypotonic buffer (10 mM HEPES [pH 7.9] with 1.5 mM MgCl2 and 10 mM KCl) and centrifuged at 1,850 × g for 5 min at 4°C. The pellet was then dissolved in hypotonic buffer with 0.5 M DTT and mammalian protease inhibitor. After incubation on ice for 15 min, the mixture was sonicated at 10 A for 15 s with 3 pulses of 5 s each and then centrifuged at 3,300 × g for 15 min at 4°C. The supernatant obtained contained the cytoplasmic fraction of the cell. The pellet was resuspended in 70 μl of low-salt buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.02 M KCl, 0.2 mM EDTA, and 25% glycerol) with 0.5 M DTT and mammalian protease inhibitor. To this, 30 μl of high-salt buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 0.8 M KCl, 0.2 mM EDTA, and 25% glycerol) was added dropwise. After 10 min of incubation on ice, the mixture was sonicated at 10 A for 15 s with 3 pulses of 5 s each and then incubated on ice for 30 min with periodic shaking and subjected to centrifugation at 24,000 × g for 30 min at 4°C. The supernatant obtained contained the nuclear fraction.
Immunoprecipitation and immunoblotting.
Whole-cell lysates were prepared after VpOmpU treatment, and 3 μg/ml of antibody was added. This mixture was then subjected to continuous low-speed shaking at 4°C for 3 h. To this, 20 μl of protein A/G beads (Santa Cruz) was added, and the mixture was subjected to low-speed shaking overnight at 4°C. The beads were then pelleted at 6,000 × g for 5 min at 4°C and washed thrice with whole-cell lysis buffer. Beads were then resuspended in SDS loading buffer and boiled for 10 min. This mixture was then subjected to immunoblotting. For immunoblotting, lysates were loaded on SDS-polyacrylamide gels and transferred onto polyvinylidene difluoride (PVDF) membranes using wet transfer. Antibodies against TLR1, TLR2, MyD88, IκB, GAPDH (glyceraldehyde-3-phosphate dehydrogenase), β-actin, p65, c-Rel, JunD, and lamin B1 were from Santa Cruz Biotechnology, and antibodies against TLR6, p-p38, p-JNK, p38, JNK, c-Jun, c-Fos, and JunB were from Cell Signaling Technologies.
The blots were developed using Clarity ECL substrate from Bio-Rad, and the images were acquired using an ImageQuant LAS 4000 instrument (GE Healthcare Life Sciences).
Densitometric analysis.
Densitometric analysis of the Western blots was done using the Image J software (https://imagej.nih.gov/ij/). For coimmunoprecipitation studies, the fold changes of the band intensities in the VpOmpU-treated cells above the band intensities in the buffer-treated cells were calculated. For analysis of the phosphorylation status of p38 and JNK, the band intensities of p-p38 or p-JNK above the band intensities of p38 or JNK, respectively, were calculated.
Statistical analysis.
Data were expressed as mean ± standard deviation (SD) or standard error of the mean (SEM). Statistical analysis was done using Student’s two-sided t test, and P values of less than 0.05 were considered significant. Significance is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001; and not significant (ns), P > 0.05.
Supplementary Material
ACKNOWLEDGMENTS
Aakanksha Gulati designed and performed experiments, analyzed the data, and wrote the paper. Ranjai Kumar designed and performed experiments and wrote the paper. Arunika Mukhopadhaya designed experiments, analyzed data, supervised the study, and wrote the paper.
We acknowledge the Indian Council of Medical Research (ICMR), Government of India, for a research fellowship to A.G. We acknowledge and thank the Indian Institute of Science Education and Research Mohali for financial support.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00809-18.
REFERENCES
- 1.Miyamoto Y, Obara Y, Nikkawa T, Yamai S, Kato T, Yamada Y, Ohashi M. 1980. Simplified purification and biophysicochemical characteristics of Kanagawa phenomenon-associated hemolysin of Vibrio parahaemolyticus. Infect Immun 28:567–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Honda T, Ni Y, Miwatani T, Adachi T, Kim J. 1992. The thermostable direct hemolysin of Vibrio parahaemolyticus is a pore-forming toxin. Can J Microbiol 38:1175–1180. doi: 10.1139/m92-192. [DOI] [PubMed] [Google Scholar]
- 3.Honda T, Ni YX, Miwatani T. 1988. Purification and characterization of a hemolysin produced by a clinical isolate of Kanagawa phenomenon-negative Vibrio parahaemolyticus and related to the thermostable direct hemolysin. Infect Immun 56:961–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ceccarelli D, Hasan NA, Huq A, Colwell RR. 2013. Distribution and dynamics of epidemic and pandemic Vibrio parahaemolyticus virulence factors. Front Cell Infect Microbiol 3:97. doi: 10.3389/fcimb.2013.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones JL, Ludeke CH, Bowers JC, Garrett N, Fischer M, Parsons MB, Bopp CA, DePaola A. 2012. Biochemical, serological, and virulence characterization of clinical and oyster Vibrio parahaemolyticus isolates. J Clin Microbiol 50:2343–2352. doi: 10.1128/JCM.00196-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mathur J, Waldor MK. 2004. The Vibrio cholerae ToxR-regulated porin OmpU confers resistance to antimicrobial peptides. Infect Immun 72:3577–3583. doi: 10.1128/IAI.72.6.3577-3583.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duperthuy M, Binesse J, Le Roux F, Romestand B, Caro A, Got P, Givaudan A, Mazel D, Bachere E, Destoumieux GD. 2010. The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ Microbiol 12:951–963. doi: 10.1111/j.1462-2920.2009.02138.x. [DOI] [PubMed] [Google Scholar]
- 8.Goo SY, Lee HJ, Kim WH, Han KL, Park DK, Lee HJ, Kim SM, Kim KS, Lee KH, Park SJ. 2006. Identification of OmpU of Vibrio vulnificus as a fibronectin-binding protein and its role in bacterial pathogenesis. Infect Immun 74:5586–5594. doi: 10.1128/IAI.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong XP, Zhang BW, Yang MJ, Ye MZ, Peng XX, Li H. 2010. Identification of vaccine candidates from differentially expressed outer membrane proteins of Vibrio alginolyticus in response to NaCl and iron limitation. Fish Shellfish Immunol 29:810–816. doi: 10.1016/j.fsi.2010.07.027. [DOI] [PubMed] [Google Scholar]
- 10.Duperthuy M, Schmitt P, Garzón E, Caro A, Rosa RD, Le Roux F, Lautrédou-Audouy N, Got P, Romestand B, de Lorgeril J, Kieffer-Jaquinod S, Bachère E, Destoumieux-Garzón D. 2011. Use of OmpU porins for attachment and invasion of Crassostrea gigas immune cells by the oyster pathogen Vibrio splendidus. Proc Natl Acad Sci U S A 108:2993–2998. doi: 10.1073/pnas.1015326108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta S, Prasad GV, Mukhopadhaya A. 2015. Vibrio cholerae porin OmpU induces caspase-independent programmed cell death upon translocation to the host cell mitochondria. J Biol Chem 290:31051–31068. doi: 10.1074/jbc.M115.670182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paauw A, Trip H, Niemcewicz M, Sellek R, Heng JM, Mars-Groenendijk RH, de Jong AL, Majchrzykiewicz-Koehorst JA, Olsen JS, Tsivtsivadze E. 2014. OmpU as a biomarker for rapid discrimination between toxigenic and epidemic Vibrio cholerae O1/O139 and non-epidemic Vibrio cholerae in a modified MALDI-TOF MS assay. BMC Microbiol 14:158. doi: 10.1186/1471-2180-14-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai SH, Lu YS, Wu ZH, Jian JC. 2013. Cloning, expression of Vibrio alginolyticus outer membrane protein-OmpU gene and its potential application as vaccine in crimson snapper, Lutjanus erythropterus Bloch. J Fish Dis 36:695–702. doi: 10.1111/jfd.12036. [DOI] [PubMed] [Google Scholar]
- 14.Jung CR, Park MJ, Heo MS. 2005. Immunization with major outer membrane protein of Vibrio vulnificus elicits protective antibodies in a murine model. J Microbiol 43:437–442. [PubMed] [Google Scholar]
- 15.Das M, Chopra AK, Cantu JM, Peterson JW. 1998. Antisera to selected outer membrane proteins of Vibrio cholerae protect against challenge with homologous and heterologous strains of V. cholerae. FEMS Immunol Med Microbiol 22:303–308. doi: 10.1111/j.1574-695X.1998.tb01219.x. [DOI] [PubMed] [Google Scholar]
- 16.Sakharwade SC, Sharma PK, Mukhopadhaya A. 2013. Vibrio cholerae porin OmpU induces pro-inflammatory responses, but down-regulates LPS-mediated effects in RAW 264.7, THP-1 and human PBMCs. PLoS One 8:e76583. doi: 10.1371/journal.pone.0076583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mao Z, Yu L, You Z, Wei Y, Liu Y. 2007. Cloning, expression and immunogenicty analysis of five outer membrane proteins of Vibrio parahaemolyticus zj2003. Fish Shellfish Immunol 23:567–575. doi: 10.1016/j.fsi.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 18.Makino K, Oshima K, Kurokawa K, Yokoyama K, Uda T, Tagomori K, Iijima Y, Najima M, Nakano M, Yamashita A, Kubota Y, Kimura S, Yasunaga T, Honda T, Shinagawa H, Hattori M, Iida T. 2003. Genome sequence of Vibrio parahaemolyticus: a pathogenic mechanism distinct from that of V cholerae. Lancet 361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 19.Pathania M, Acosta-Gutierrez S, Bhamidimarri SP, Basle A, Winterhalter M, Ceccarelli M, van den Berg B. 2018. Unusual constriction zones in the major porins OmpU and OmpT from Vibrio cholerae. Structure 26:708–721 e4. doi: 10.1016/j.str.2018.03.010. [DOI] [PubMed] [Google Scholar]
- 20.Bogdan C. 2001. Nitric oxide and the immune response. Nat Immunol 2:907–916. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 21.MacMicking J, Xie QW, Nathan C. 1997. Nitric oxide and macrophage function. Annu Rev Immunol 15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 22.Akira S, Uematsu S, Takeuchi O. 2006. Pathogen recognition and innate immunity. Cell 124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 23.Kawai T, Akira S. 2010. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol 11:373–384. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- 24.Oeckinghaus A, Ghosh S. 2009. The NF-kappaB family of transcription factors and its regulation. Cold Spring Harb Perspect Biol 1:a000034. doi: 10.1101/cshperspect.a000034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zenz R, Eferl R, Scheinecker C, Redlich K, Smolen J, Schonthaler HB, Kenner L, Tschachler E, Wagner EF. 2008. Activator protein 1 (Fos/Jun) functions in inflammatory bone and skin disease. Arthritis Res Ther 10:201. doi: 10.1186/ar2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Q, Chen J, Liu R, Jia J. 2011. Identification and evaluation of an outer membrane protein OmpU from a pathogenic Vibrio harveyi isolate as vaccine candidate in turbot (Scophthalmus maximus). Lett Appl Microbiol 53:22–29. doi: 10.1111/j.1472-765X.2011.03062.x. [DOI] [PubMed] [Google Scholar]
- 27.Massari P, Henneke P, Ho Y, Latz E, Golenbock DT, Wetzler LM. 2002. Immune stimulation by neisserial porins is Toll-like receptor 2 and MyD88 dependent. J Immunol 168:1533–1537. doi: 10.4049/jimmunol.168.4.1533. [DOI] [PubMed] [Google Scholar]
- 28.Moreno-Eutimio MA, Tenorio-Calvo A, Pastelin-Palacios R, Perez-Shibayama C, Gil-Cruz C, López-Santiago R, Baeza I, Fernández-Mora M, Bonifaz L, Isibasi A, Calva E, López-Macías C. 2013. Salmonella Typhi OmpS1 and OmpS2 porins are potent protective immunogens with adjuvant properties. Immunology 139:459–471. doi: 10.1111/imm.12093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawasaki T, Kawai T. 2014. Toll-like receptor signaling pathways. Front Immunol 5:461. doi: 10.3389/fimmu.2014.00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Botos I, Segal DM, Davies DR. 2011. The structural biology of Toll-like receptors. Structure 19:447–459. doi: 10.1016/j.str.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Massari P, Visintin A, Gunawardana J, Halmen KA, King CA, Golenbock DT, Wetzler LM. 2006. Meningococcal porin PorB binds to TLR2 and requires TLR1 for signaling. J Immunol 176:2373–2380. doi: 10.4049/jimmunol.176.4.2373. [DOI] [PubMed] [Google Scholar]
- 32.Oosting M, Ter Hofstede H, Sturm P, Adema GJ, Kullberg BJ, van der Meer JW, Netea MG, Joosten LA. 2011. TLR1/TLR2 heterodimers play an important role in the recognition of Borrelia spirochetes. PLoS One 6:e25998. doi: 10.1371/journal.pone.0025998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bhowmick R, Pore D, Chakrabarti MK. 2014. Outer membrane protein A (OmpA) of Shigella flexneri 2a induces TLR2-mediated activation of B cells: involvement of protein tyrosine kinase, ERK and NF-kappaB. PLoS One 9:e109107. doi: 10.1371/journal.pone.0109107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Buwitt-Beckmann U, Heine H, Wiesmuller KH, Jung G, Brock R, Akira S, Ulmer AJ. 2005. Toll-like receptor 6-independent signaling by diacylated lipopeptides. Eur J Immunol 35:282–289. doi: 10.1002/eji.200424955. [DOI] [PubMed] [Google Scholar]
- 35.Farhat K, Riekenberg S, Heine H, Debarry J, Lang R, Mages J, Buwitt-Beckmann U, Röschmann K, Jung G, Wiesmüller K-H, Ulmer AJ. 2008. Heterodimerization of TLR2 with TLR1 or TLR6 expands the ligand spectrum but does not lead to differential signaling. J Leukoc Biol 83:692–701. doi: 10.1189/jlb.0807586. [DOI] [PubMed] [Google Scholar]
- 36.Galdiero M, Vitiello M, Sanzari E, D'Isanto M, Tortora A, Longanella A, Galdiero S. 2002. Porins from Salmonella enterica serovar Typhimurium activate the transcription factors activating protein 1 and NF-kappaB through the Raf-1-mitogen-activated protein kinase cascade. Infect Immun 70:558–568. doi: 10.1128/IAI.70.2.558-568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galdiero S, Capasso D, Vitiello M, D'Isanto M, Pedone C, Galdiero M. 2003. Role of surface-exposed loops of Haemophilus influenzae protein P2 in the mitogen-activated protein kinase cascade. Infect Immun 71:2798–2809. doi: 10.1128/IAI.71.5.2798-2809.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan J, Sharma PK, Mukhopadhaya A. 2015. Vibrio cholerae porin OmpU mediates M1-polarization of macrophages/monocytes via TLR1/TLR2 activation. Immunobiology doi: 10.1016/j.imbio.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 39.Nikaido H, Rosenberg EY. 1983. Porin channels in Escherichia coli: studies with liposomes reconstituted from purified proteins. J Bacteriol 153:241–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.